Abstract
Microphthalmia transcription factor (MITF) and STAT3 are two transcription factors that play a major role in the regulation of growth and function in mast cells and melanocytes. In the present study, we explored the MITF-PIAS3-STAT3 network of interactions, how these interactions regulate gene expression, and how cytokine-mediated phosphorylation of MITF and STAT3 is involved in the in vivo interplay between these three proteins. In NIH 3T3 cells stimulated via gp130 receptor, transfected MITF was found to be phosphorylated at S409. Such phosphorylation of MITF leads to PIAS3 dissociation from MITF and its association with STAT3. Activation of mouse melanoma and mast cells through gp130 or c-Kit receptors induced the mobilization of PIAS3 from MITF to STAT3. In mast cells derived from MITFdi/di mice, whose MITF lacks the Zip domain (PIAS3-binding domain), we found downregulation in mRNA levels of genes regulated by either MITF or STAT3. This regulatory mechanism is of considerable importance since it is likely to advance the deciphering of a role for MITF and STAT3 in mast cells and melanocytes.
Microphthalmia transcription factor (MITF) is a basic helix-loop-helix leucine zipper (bHLH-Zip) DNA-binding protein (17). Its gene resides at the mi locus in mice (19), and mutation of this gene results in deafness, bone loss, small eyes, and poorly pigmented eyes and skin (32). The primary cell types affected in MITF-deficient mice are mast cells, osteoclasts, and melanocytes (32). In humans, mutation in this gene causes Waardenburg syndrome type II (40). MITF regulates the expression of mouse mast cell protease 6 (mMCP) (35), mMCP5 (33), c-Kit (20), p75 nerve growth factor (33), granzyme B (9), and tryptophan hydroxylase (21). MITF regulates gene transcription by binding to E-box-type enhancers in the 5′-flanking regions of MITF-responsive genes (35). Like many other DNA-binding proteins, the transcription-enhancing activity of MITF is influenced in a complex manner by an array of different intracellular proteins. We previously identified two MITF-interacting proteins, PKCI/Hint (37) and PIAS3 (27), by using the yeast two-hybrid system with the bHLH-Zip domain as a bait. These two MITF-associated proteins were shown to be repressors of MITF transcriptional activity (27, 37). We have also shown by pull-down assay and by using cells derived from MITF truncated mice (di/di) that the MITF Zip domain plays an essential role in the interaction between MITF and PIAS3 (28).
Three serine sites for MITF phosphorylation have been reported (14, 42, 46). Phosphorylation of S73 (14) and S409 (46) occurs as a result of kit ligand stimulation and activation of mitogen-activated protein kinase and Rsk-1, respectively. These serine sites are of interest since, upon their phosphorylation, MITF transcriptional activity is upregulated, and this phosphorylation also serves as a signal for the degradation of MITF by ubiquitin-dependent proteolysis in melanocytes (46). In our previous study, we observed that phosphorylation of MITF at S73 and S409 plays a major role in its association with PIAS3 (28). This effect was profound with the phosphorylation of MITF on S409, which significantly reduced PIAS3-mediated inhibition of MITF transcriptional activity.
STAT3 is involved in signal transduction pathways that are activated by the interleukin-6 (IL-6) family of cytokines. It is tyrosine phosphorylated by janus kinase (JAK), translocates as a dimer into the nucleus, and activates specific genes (4). STAT3 signaling has been shown to prevent programmed cell death and enhance cell proliferation through regulating genes such as Bcl-XL, Mcl-l, c-Myc, and cyclin D1 (1, 3, 10, 39). Targeted disruption of the mouse gene encoding STAT3 leads to early embryonic lethality (41). It is interesting that the human mast cell line HMC-1.1 harbors a single V560G mutation in the kit receptor that causes constitutive phosphorylation and therefore the activation of the JAK/STAT pathway (29). Since PIAS3 was identified as an inhibitor of both activated STAT3 and MITF, its interplay between these two proteins is of great importance. In the present study, we report that the MITF-PIAS3-STAT3 network of interactions is mediated by the cytokines IL-6 and stem cell factor (SCF).
MATERIALS AND METHODS
Mice.
Mouse colonies were established from transgenic mice (MITFsp/sp and MITFsp/di) kindly provided by Lynne Lamoreux from the College of Veterinary Medicine, Texas A&M University. The MITF encoded by the mutant midi allele is lacking the Zip domain (13) due to a C-to-T transition in exon 8 at position 916, which introduces a premature stop codon between the bHLH and the leucine zipper domain.
Cell culture and treatments.
B16 melanoma cells and NIH 3T3 cells were cultured and maintained in a growth medium containing Dulbecco modified Eagle medium, 10% fetal calf serum, 2 mM l-glutamine, 2 mM nonessential amino acids, 100 μg of penicillin/ml, 100 μg of streptomycin/ml, and 50 μM β-mercaptoethanol.
Femoral bone marrow cells derived from mice were cultured in IL-3-containing medium for 3 weeks to generate bone marrow-derived mast cells (BMMC) as previously described (36). All of the cells were grown in a humidified incubator at 37°C with 5% CO2.
Cells were stimulated with a chimeric molecule of IL-6/IL-6 receptor (IL-6R) kindly provided by Michel Revel (Weizmann Institute, Rehovot, Israel).
Plasmid construction.
Mouse MITF (1,129 bp) was subcloned into the XbaI and HindIII sites of the pcDNA3.1 (−) vector (Invitrogen Life Technologies, Carlsbad, Calif.). This vector was used for the production of all MITF mutants by site-directed mutagenesis (Invitrogen), in which serine was replaced by alanine at position 73 or 409 (S73A or S409A, respectively). A double mutation at S73 and S409 produced the S73/409A mutant. The cDNA encoding the open reading frame of mouse PIAS3 was subcloned into the XbaI and HindIII sites of the pcDNA3.1(−) vector (Invitrogen).
Mouse MITF (1,129 bp) was inserted into the pGEX-4T-3 vector (Stratagene, La Jolla, Calif.). PGEX2-mouse STAT3 plasmid was kindly provided by Dov Tzipori (Weizmann Institute). The luciferase reporter plasmid, pSP72, containing the MITF binding region of the promoter and the first exon of the mMCP6 gene (−191 to +26), was generously provided by Y. Kitamura (Osaka, Japan). pEBB-MITF and its mutant (S73A, S409A, and S73/409A) plasmids were kindly provided by David Fisher (Boston, Mass.). Flag-tagged pcDNA-STAT3, as well as its mutants STAT3-Y705F, STAT3-C, and the M67 pTATA tk-Luc reporter gene, was kindly provided James E. Darnell (The Rockefeller University, New York, N.Y.). The green fluorescent protein (GFP)-PIAS3 construct was kindly provided by Lothar Vassen (Universitatsklinikum, Essen, Germany). The fidelity of all constructs was verified by direct sequencing.
Transient cotransfection and luciferase assay.
NIH 3T3 cells (5 × 105) were used in various luciferase assay experiments. Cells were cotransfected with liposomes (Promega, San Luis Obispo, Calif.) with 0.1 μg of luciferase reporter gene (MITF or STAT3), 0.1 μg of pcDNA-MITF (wild type or mutant), 0.1 μg of pcDNA-PIAS3, and 0.1 μg of pcDNA-STAT3 (wild type or mutant). Then, 0.1 μg of pcDNA alone was used as a nonspecific control. The cells were incubated in 24-well plates for 48 h, treated or not with IL-6/IL-6R, lysed, and assayed for luciferase activity. The luciferase activity was normalized to the total protein concentration. The normalized value was then divided by the luciferase activity obtained by cotransfection of the reporter with pcDNA alone. The ratio was expressed as the relative luciferase activity.
Primers.
Bacteriophage T7 transcription regulatory element and ribosome-binding site were fused with the PIAS3 primers. pcDNA3.1-mouse PIAS3 was used as a template. Primers for the various domains (D1 to D5) were as follows: D1 (sense, 5′-CTAATACGACTCACTATAGGGAAGGAGATATACATATGATGGTGATGAGTTTC; antisense, 5′-TTAGGTGTAATCACACTTGGCTCCTGGCAG), D2 (sense, 5′-CTAATACGACTCACTATAGGGAAGGAGATATACATATGATACAAGTGCAGCTCAGATTC; antisense, 5′-TTACTTCCCTAGCGGGCACATGAGTGACAC), D3 (sense, 5′-CTAATACGACTCACTATAGGGAAGGAGATATACATATGCGCCTGACTGTCCCG; antisense,5′-TTAAGAGGTCAGGGCTCCTTTGCTTCCAGG), D4 (sense, 5′-CTAATACGACTCACTATAGGGAAGGAGATATACATATGGGTCACCAGCCATCTTCGGTG; antisense, 5′-TCAGTCCAAGGAAATGACGTCTGACCG), and D5 (sense, 5′-CTAATACGACTCACTATAGGGAAGGAGATATACATATGGTCACCATG; antisense, 5′-TTACCTGGACGTGAGAATCTGCTGCAGCTGCTG).
In vitro GST pull-down assay.
Glutathione S-transferase (GST) fusion protein MITF or GST fusion protein STAT3 was expressed in protease-deficient Escherichia coli strain B12 and purified on glutathione-Sepharose beads (Amersham Biosciences) essentially as described before (27). Pull-down assays (8) were performed with GST-MITF or GST-STAT3 fusion protein (1 to 5 μg) bound to Sepharose beads and preincubated for 1 h at 4°C in 1 ml of binding buffer (phosphate-buffered saline [PBS], 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol, 0.1% NP-40). Then, 1 to 10 μl of 35S-labeled full-length PIAS3 or its fragments were synthesized by using the TNT-coupled rabbit reticulocyte lysate system (Promega) and then added to each preincubation mix, and the binding reaction was carried out overnight at 4°C. Beads were washed four times in 1 ml of PBS-290 mM NaCl and boiled for 7 min in sample buffer, and aliquots were examined by electrophoresis. The integrity and quantity of the GST fusions were confirmed by Blue stain reagent (Pierce Biotechnology, Inc., Rockford, Ill.), and autoradiography detected the amount of retained full-length PIAS3 and its fragments.
Coimmunoprecipitation.
BMMC, B16 cells, and NIH 3T3 cells were used in various coimmunoprecipitation experiments. NIH 3T3 cells were cotransfected with PIAS3, MITF (wild type or mutants), and STAT3 (wild type or mutants). Cells were activated with IL-6/IL-6R or SCF for 15 or 30 min. Cells (5 × 106 to 10 × 106) were lysed by the addition of 300 μl of cold lysis buffer (0.01 M Tris-HCl [pH 7.4], 1% deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 0.15 M NaCl, 0.25 μM phenylmethylsulfonyl fluoride) and 15 μl of protease inhibitor (Sigma-Aldrich Corp., St. Louis, Mo.). Cells were then homogenized, and their supernatants were collected after a 15-min centrifugation in a microcentrifuge at 4°C. Recovered lysates were incubated with either MITF monoclonal antibody C5 (43) or with anti-mouse PIAS3 antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.) prebound to 15 mg of protein A/G agarose (Pierce) and incubated with agitation overnight at 4°C. Recovered immune complexes were washed three times with lysis buffer.
Gel electrophoresis and Western blots.
Proteins were resolved by SDS-10% polyacrylamide gel electrophoresis (PAGE) under reducing conditions and transferred to 0.45-μm-pore-size nitrocellulose membranes. Blots were probed with anti-phosphoserine, anti-PIAS3, anti-MITF, anti-RSK, or anti-STAT3 antibody (Santa Cruz). Visualization of reactive proteins was done by enhanced chemiluminescence (27).
Indirect fluorescence immunocytochemistry.
NIH 3T3 or B16 melanoma cells were cotransfected with GFP-PIAS3, Flag-tagged STAT3 (wild type or mutants), and MITF. Cells were grown on glass coverslips in six-well plates. After extensive washing with PBS, the cells were fixed with 1.5 ml of 3.7% formaldehyde in PBS for 10 min. The fixed cells were then washed with PBS and permeabilized with 1.5 ml of Triton X-100 diluted 1:2 with PBS containing 7.5 mg of bovine serum albumin. After 45 min of blocking with normal donkey serum, the cells were stained with mouse anti-MITF and rabbit anti-Flag antibodies, followed by the addition of rhodamine-conjugated goat anti-mouse immunoglobulin G (Jackson Immunoresearch) and Cy5-conjugated goat anti-rabbit immunoglobulin G. Fluorescence analysis was performed by using the Zeiss LSM 410 confocal laser-scanning system connected to a Zeiss Axiovert 135M microscope. The green fluorescence of GFP-labeled PIAS3 was excited with argon laser (488-nm excitation line with 515-nm long-pass barrier filter). Rhodamine-conjugated goat anti-mouse was excited with a helium-neon laser (543-nm excitation line with a 570-nm long-pass barrier filter). Cy5-conjugated goat anti-rabbit antibody was simultaneously excited with a helium-neon laser (633-nm excitation line).
Statistical image analysis.
The software we used was written in MATLAB, mainly by using the image-processing tool. Each color image was split into its three components: red, green, and blue. The software enabled us to select an identical area of interest in all three components for statistical comparisons. We compared the color distribution of the three components, selecting the nucleus versus the entire cell. In each we measured the color by calculating the integral or the area below the histogram plot of the gray level intensities of the pixels in the selected areas.
Real-time quantitative PCR.
MITF- and STAT3-responsive genes were measured by using real-time quantitative PCR. Total RNA was extracted from BMMC and B16 melanoma cells, and mRNA levels of various genes were quantified by measuring SYBR-Green incorporation (SYBR-Green PCR Master Mix; Applied Biosystems, Foster City, Calif.). SYBR-Green incorporation into double-stranded DNA permits the direct detection of PCR product after each amplification cycle (ABI Prism 7000 Sequence Detection System; Applied Biosystems). The genes whose mRNA levels were quantified by real-time PCR were as follows: β-actin; STAT3 target genes Bcl-XL, VEGF, and c-Myc; and MITF target genes Bcl-2, c-Kit, and tryptophan hydroxylase.
RESULTS
Characterization of the domain(s) in PIAS3 that directly associates with MITF and STAT3.
We previously showed a direct association between MITF and PIAS3 by using an in vitro pull-down assay (27) and found that the Zip domain of MITF is responsible for the direct interaction between these two proteins (28). In the present study, the same system was used in order to identify the regions in PIAS3 that are responsible for its interaction with MITF and STAT3.
PCR fragments coding for the various domains of PIAS3 (D1, amino acids 1 to 143; D2, amino acids 144 to 286; D3, amino acids 287 to 443; and D4, amino acids 443 to 585) were used for this purpose (Fig. 1A). MITF and STAT3 were expressed in bacteria as GST fusion proteins, immobilized on glutathione-Sepharose beads, and assayed for their ability to retain the in vitro-translated PIAS3 domains labeled with [35S]methionine.
FIG. 1.
Domain of PIAS3 that directly binds to MITF and STAT3. (A) Schematic representation of the full-length mouse PIAS3, including the SAP domain, the zinc finger motifs, and the five domains of PIAS3 investigated in the present study. (B) Fragments D1, D2, D3, D4, and full-length PIAS3 (FL) labeled with [35S]methionine in vitro and added to GST-MITF or GST-STAT3 that was immobilized on glutathione-Sepharose beads. Retained 35S-labeled full-length PIAS3 or its fragments were determined by SDS-PAGE and autoradiography. Input and GST alone are shown as control. The results from one representative experiment of three experiments performed are shown. (C) Fragment D5 (common region of D1 and D2) and full-length PIAS3 (FL) labeled with [35S]methionine in vitro added to GST-MITF or GST-STAT3 that was immobilized on glutathione-Sepharose beads. Retained 35S-labeled full-length PIAS3 or D5 were determined by SDS-PAGE and autoradiography. The results from one representative experiment of three experiments performed are shown. (D) Full-length PIAS3 (FL) labeled with [35S]methionine in vitro added to GST-MITF or GST-STAT3 that were immobilized on glutathione-Sepharose beads. Increasing concentrations of in vitro-translated unlabeled D5 or D4 was added to the mixtures. Retained 35S-labeled full-length PIAS3 were determined by SDS-PAGE and autoradiography. The results from one representative experiment of three experiments performed are shown.
The results clearly show that domains D1 and D2 of PIAS3 are responsible for the direct interaction with both MITF and STAT3 (Fig. 1B). We went on to determine smaller fragments in these motifs that have the ability to associate with MITF and STAT3. Domain D5 (amino acids 82 to 156) was found to be the smallest fragment that can interact with both MITF and STAT3 (Fig. 1C). All PIAS3 domains tested were associated only with GST-MITF or GST-STAT3 but not with the control GST. The specificity of the binding was determined by using in vitro-translated D5 as a potential competitor with the full-length PIAS3 for binding to MITF and STAT3. Both D5 and full-length PIAS3 were in vitro translated, whereas only full-length PIAS3 was labeled with [35S]methionine (Fig. 1D). As a control we used the unlabeled D4 domain. The results clearly show that both MITF and STAT3 interact specifically with the same fragment (D5) of PIAS3.
Effect of IL-6/IL-6R on the association between MITF and PIAS3.
It was previously reported that the chimeric protein of IL-6 and its receptor (i.e., IL-6R) could mimic the effect of IL-6 alone by binding to the transmembrane receptor gp130 of the B16 melanoma cell line (24) and thus activate the intracellular signaling cascade, the JAK/STAT pathway and the mitogen-activated protein kinase pathway (15). Therefore, the effect of IL-6/IL-6R on MITF phosphorylation was determined in a system in which NIH 3T3 cells were transfected with MITF or its mutants: S73A, S409A, and S73/409A. These mutants could not be phosphorylated at the site of mutation. Cells were stimulated with IL-6/IL-6R for 15 min and lysed, and MITF was immunoprecipitated and immunoblotted with anti-phosphoserine antibody. As shown in Fig. 2A, an increase in serine phosphorylation of MITF was observed in cells transfected with either wild-type MITF or the S73A-MITF or S409A-MITF mutants, whereas cells transfected with the double mutant S73/409A-MITF did not show any increase in phosphorylation. Therefore, we concluded that S409 on MITF is phosphorylated upon stimulation of NIH 3T3 cells with IL-6/IL-6R.
FIG. 2.
Effect of IL-6/IL-6R on the association between MITF and PIAS3. (A) NIH 3T3 cells were transfected with MITF or its mutants S73A, S409A, and S73/409A-MITF and stimulated with 100 ng of IL-6/IL-6R/ml for 15 min at 37°C. The cells were then lysed and MITF immunoprecipitated (IP) with anti-MITF antibody, transferred to membrane, and immunoblotted with anti-phosphoserine antibody (pSer; top panel) or reprobed with anti-MITF antibody (lower panel). (B) NIH 3T3 cells were cotransfected with PIAS3, MITF, or its mutant S409A-MITF and then stimulated with IL-6/IL-6R. The cells were lysed, and PIAS3 was immunoprecipitated (IP) with anti-PIAS3 antibody, separated by SDS-PAGE, transferred to membrane, and immunoblotted with anti-MITF (top panel) or reprobed with anti-PIAS3 (indicated beneath the MITF blots). Cell lysates were blotted with anti-MITF as control. One representative of three experiments is shown. (C) NIH 3T3 cells were cotransfected with MITF or its mutant S409A-MITF and with luciferase reporter under control of the mMCP6 promoter. Cells were then treated with IL-6/IL-6R for 6 h, with the control being untreated cells. The luciferase activity of lysed cells was measured and normalized against the protein concentration. The mean ± the standard error of the mean (SEM) of four experiments is shown. (D) Percent inhibition of the MITF transcriptional activity by PIAS3. A luciferase reporter plasmid containing a promoter region of the mMCP6 was cotransfected to NIH 3T3 cells with either MITF or with the S409A-MITF mutant and PIAS3. Cells were then treated with IL-6/IL-6R for 6 h. The relative MITF transcriptional activity was determined as in Fig. 2C. The levels of transfected MITF expression are shown as a control. The mean ± the SEM of four experiments is shown.
In our previous study we showed that phosphorylation of MITF at S409 can lead to dissociation between MITF and its transcriptional inhibitor PIAS3 (28). In order to determine whether IL-6/IL-6R treatment, which leads to the phosphorylation of MITF at S409, causes dissociation between these two proteins, we transfected NIH 3T3 cells with MITF (wild type or S409A), together with PIAS3, and stimulated the cells with IL-6/IL-6R. The cells were then lysed, immunoprecipitated with anti-PIAS3 antibody, and immunoblotted with anti-MITF antibody. A significant decrease in the association between PIAS3 and MITF was observed only in IL-6/IL-6R-activated cells that were transfected with wild-type MITF (Fig. 2B). No dissociation between MITF and PIAS3 was observed in activated cells that contained S409A-MITF mutant. Stripping of the membrane and reprobing with anti-PIAS3 antibody revealed a similar amount of PIAS3 bound to the beads. Also, the same amount of MITF was detected in each of the transfected cells, as revealed by probing the lysates with anti-MITF antibody as a control. Therefore, we concluded that activation of cells with IL-6/IL-6R resulted in the dissociation of PIAS3 from MITF via phosphorylation of MITF at S409.
To quantitatively assess the role of IL-6/IL-6R treatment on the transcriptional activity of MITF, NIH 3T3 cells were cotransfected with a luciferase reporter plasmid containing the mouse mMCP6 promoter and with either MITF or its S409A-MITF mutant. The transcriptional activity of MITF was determined as the relative luciferase activity in cells with or without IL-6/IL-6R activation. No significant change in activity was detected regardless of whether the cells were transfected with MITF or its mutant (Fig. 2C).
The role of IL-6/IL-6R, if any, on PIAS3-mediated inhibition of MITF transcriptional activity was then determined from the relative luciferase activity (Fig. 2D). Treatment of cells with IL-6/IL-6R for 6 h caused a significant decrease in the ability of PIAS3 to inhibit MITF transcriptional activity compared to untreated cells. This decrease was much less in IL-6/IL-6R-activated cells that were transfected with S409A mutant. Similar protein levels of transfected MITF are shown as control.
In summary, the data suggest that IL-6/IL-6R treatment induced signal transduction that caused phosphorylation of MITF at S409 and its dissociation from PIAS3.
Effect of mutation of MITF at S409 on PIAS3-STAT3 association.
NIH 3T3 cells were transfected with STAT3, PIAS3, and with or without MITF (wild type or S409A). The lysates were immunoprecipitated with anti-PIAS3 and then immunoblotted with anti-STAT3. In cells that were not transfected with MITF, as in wild type-MITF-transfected cells, an increase in the association between STAT3 and PIAS3 was observed after IL-6/IL-6R treatment (Fig. 3A). This was in contrast to cells that were transfected with the S409A-MITF mutant, in which no such increase, was observed. Stripping of the membrane and reprobing with anti-PIAS3 revealed a similar amount of PIAS3 bound to the beads. The amounts of STAT3 and MITF were detected by probing the lysates with their appropriate antibodies.
FIG. 3.
Effect of mutation of MITF at S409 on PIAS3-STAT3 association. (A) NIH 3T3 cells were cotransfected with STAT, PIAS3, MITF, or its mutant S409A-MITF and then stimulated with IL-6/IL-6R for 15 min. The cells were lysed, and PIAS3 was immunoprecipitated (IP) with anti-PIAS3 antibody, separated by SDS-PAGE, transferred to membrane, and immunoblotted with anti-STAT3 (top panel) or reprobed with anti-PIAS3 (indicated beneath STAT3 blots). Cell lysates were blotted with anti-MITF and anti-STAT3 antibodies as control. The results of one representative of three experiments performed are shown. (B) NIH 3T3 cells were cotransfected with STAT3, STAT3 reporter gene (M67), PIAS3 and with different doses (0.05 μg, 0.1 μg) of MITF (wild type [WT] or S409A). Cells were triggered with IL-6/IL-6R for 6 h. Luciferase activity of lysed cells was measured and normalized against the protein concentration. The mean ± the SEM of three experiments is shown.
To quantitatively assess whether MITF interferes with inhibition of STAT3 transcriptional activity, NIH 3T3 cells were transfected with STAT3 and STAT3 reporter gene (M67), with or without PIAS3, and with different doses of MITF (wild type or S409A). Cells were treated with IL-6/IL-6R for 6 h, and the luciferase activity was measured and normalized against protein concentration. As shown in Fig. 3B, the STAT3 luciferase activity induced by IL-6/IL-6R was substantially reduced in cells transfected with PIAS3 compared to controls. However, presence of wild-type MITF and S409A-MITF decreased the inhibition effect of PIAS3 on STAT3 transcriptional activity in a dose-response manner, with the effect being greater in the mutated MITF.
Constitutively expressed activated STAT3 interferes with the association of MITF with PIAS3.
STAT3-C is a constitutively activated form of STAT3 that was constructed by the substitution of two cysteine residues for A661 and N663, which are within the C terminus loop of the SH2 domain (3). These mutations enable STAT3 to dimerize without phosphorylation at Y705. We hypothesized that this constitutively activated STAT3 would interfere with the association between MITF and PIAS3. Thus, STAT3-C Flag-tagged, MITF, and PIAS3-GFP were transfected to NIH 3T3 cells, and the STAT3-Y705F-Flag-tagged construct was used as a control. This construct cannot be phosphorylated at tyrosine 705 and can hardly dimerize (25). The cells were lysed; lysates were immunoprecipitated with anti-GFP and immunoblotted with anti-Flag antibody. As shown in Fig. 4A, there was a significant increase in PIAS3-GFP protein bound to the STAT3-C mutant compared to that bound to the Y705F-STAT3 mutant. Furthermore, a decrease in the association of PIAS3 with MITF was observed in cells that were transfected with STAT3-C mutant but not in cells that were transfected with the Y705F-STAT3 mutant. Stripping of the membrane and reprobing with anti-GFP revealed a similar amount of GFP-PIAS3 bound to the beads. Western blots of the lysates that were probed with anti-MITF or anti-Flag showed the same amounts of the transfected proteins as control.
FIG. 4.
Constitutively expressed activated STAT3 interferes with the association between MITF and PIAS3. (A) NIH 3T3 cells were cotransfected with GFP-PIAS3, MITF, and STAT3 Flag-tagged mutants (STAT3-C and Y705F-STAT3). The cells were lysed, and PIAS3 was immunoprecipitated (IP) with anti-GFP antibody, separated by SDS-PAGE, transferred to membrane, and immunoblotted with either anti-Flag antibody (top panel) or reprobed with anti-GFP antibody. Cell lysates were blotted with anti-MITF and anti-flag antibodies as a control for transfection. The results from one representative experiment of four experiments performed are shown. (B) NIH 3T3 cells were cotransfected with the luciferase reporter under the control of the mMCP6 promoter MITF, PIAS3, and STAT3 mutants (STAT3-C and Y705F-STAT3). The luciferase activity of lysed cells was measured and normalized against the protein concentration. The mean ± the SEM of four experiments is shown.
In order to find out whether STAT3-C interferes with the MITF transcriptional inhibitory effect induced by PIAS3, NIH 3T3 cells were cotransfected with MITF luciferase reporter plasmid containing the mouse mMCP6 promoter, STAT3-C, or its control, with or without PIAS3. No significant differences were observed in the relative luciferase activity of MITF in cells that were transfected with STAT3-C compared to cells transfected with Y705F-STAT3 (Fig. 4B). However, when PIAS3 was transfected to the cells, a twofold induction in the relative luciferase activity of MITF was observed in cells transfected with STAT3-C compared to those transfected with Y705F-STAT3 mutant. The minimal transcriptional activity of STAT3 constructs on mMCP6 promoter is also shown (Fig. 4B).
Colocalization of PIAS3, STAT3, and MITF in NIH 3T3 transfected cells.
The in vivo interactions between PIAS3, MITF, and STAT3 were determined in NIH 3T3 cells stimulated with IL-6/IL-6R by using confocal laser scanning. The cells were cotransfected with GFP-PIAS3, MITF, and Flag-tagged STAT3. Resting and activated cells were fixed and stained with mouse anti-MITF and rabbit anti-Flag, which were detected by staining with rhodamine-conjugated goat anti-mouse and Cy5-conjugated goat anti-rabbit antibodies, respectively. As shown in Fig. 5A, MITF and PIAS3 in resting cells were colocalized in the nucleus, whereas most of the Flag-tagged STAT3 was detected in the cytoplasm. IL-6/IL-6R treatment induced all three proteins to colocalize in the nucleus (Fig. 5A).
FIG. 5.
Intracellular localization of PIAS3, STAT3, and MITF. (A) NIH 3T3 cells were cotransfected with GFP-PIAS3, MITF, and Flag-tagged STAT3. Cells were incubated with IL-6/IL-6R for 15 min. The immunostaining was performed with anti-MITF and anti-Flag antibodies by using rhodamine (green)- and Cy5 (red)-labeled secondary antibodies, respectively. The cells were analyzed by laser-scanning confocal microscopy. The nuclear colocalization is shown in the right panels (white dots for triple colocalization and cyan for MITF and PIAS3 colocalization). (B) NIH 3T3 cells were cotransfected with GFP-PIAS3, MITF, and STAT3 Flag-tagged mutants STAT3-C and Y705F-STAT3. Cells were immunostained and labeled as in panel A. (C) Analysis of intracellular localization of STAT3 from 20 color images. The software we used was written in MATLAB by using mainly the image-processing tool.
We further investigated the interactions between PIAS3, MITF, and the STAT3 Flag-tagged mutants, STAT3-C and Y705F-STAT3. As shown in Fig. 5B, PIAS3 and MITF colocalize in the nucleus, whereas Y705F-STAT3 was detected predominantly in the cytoplasm. Cells that were transfected with STAT3-C showed colocalization between PIAS3, MITF, and STAT3-C in the nucleus.
In order to quantitatively assess the localization of the different proteins and the degree of their colocalization in NIH 3T3 cells, we used a MATLAB program by using mainly the image-processing tool. Twenty color images were split into their three components: red, green, and blue. We then selected the identical area of interest in all three components for statistical comparisons and determined the localization of the STAT3 protein. As shown in Fig. 5C, STAT3 is predominantly localized in the nucleus after the IL-6/IL-6R treatment, as observed for STAT3-C, whereas the Y705F-STAT3 mutant shows less localization in the nucleus.
Interactions between PIAS3, STAT3, and MITF in mast cells.
The phosphorylation of endogenous MITF at Ser409 upon BMMC stimulation by either IL-6/IL-6R or SCF was studied. Cells deprived from IL-3 for 4 h were activated either by SCF or IL-6/IL-6R for 30 min. MITF was immunoprecipitated with anti-MITF and immunoblotted with anti-pSer and anti-RSK. MITF was previously shown to be phosphorylated at Ser409 by RSK (46). As shown in Fig. 6A, an increase in serine phosphorylation of MITF was observed in cells activated with either SCF or IL-6/IL-6R. Moreover, an increase in the association of MITF with RSK was observed after cells were activated with either SCF or IL-6/IL-6R.
FIG. 6.
Interactions between PIAS3, STAT3, and MITF in mast cells. (A) BMMC were deprived of IL-3 for 4 h and then exposed to either SCF or IL-6/IL-6R. Cells were lysed, and MITF was immunoprecipitated with anti-MITF antibody, immunoblotted with anti-pSER (top blot) or anti-RSK (lower blot) antibody, and reprobed with anti-MITF antibody. (B) BMMC were starved for 4 h and then treated with either SCF or IL-6/IL-6R. Cells were lysed, and PIAS3 was immunoprecipitated with anti-PIAS3 antibody, immunoblotted with anti-STAT3 (top blot) or anti-MITF (lower blot) antibody, and reprobed with anti-PIAS3 antibody (indicated beneath the MITF blots). NC, isotype-matched control antibody. Cell lysates were blotted with anti-MITF and anti-STAT3 antibodies as a control. The results of one representative experiment of three experiments performed are shown. (C) BMMC derived from either wild-type or MITFdi/di mice were deprived of IL-3 for 4 h and then exposed to SCF. Cells were lysed, and PIAS3 was immunoprecipitated with anti-PIAS3 antibody, immunoblotted with anti-STAT3 (top blot) or anti-MITF (lower blot) antibody, and reprobed with anti-PIAS3 antibody (indicated beneath the MITF blots). Cell lysates were blotted with anti-MITF and anti-STAT3 antibodies as controls. (D and E) Quantitative reverse transcription-PCR analysis of MITF target genes (c-Kit and TPH) and STAT3 target genes (c-Myc and VEGF) from BMMC activated for 0.5 or 4 h with SCF derived from either wild-type or MITFdi/di mice. RNA levels were normalized to actin and were determined in triplicate.
Stripping of the membranes and reprobing with anti-MITF revealed a similar amount of MITF bound to the beads. The same amount of RSK was detected in each of the lysates, as revealed by probing the lysates with anti-RSK as a control (not shown).
In order to investigate the interactions between STAT3, PIAS3, and MITF in mast cells, the cells were lysed, and PIAS3 was immunoprecipitated with anti-PIAS3 and immunoblotted with anti-MITF and anti-STAT3 antibodies. As shown in Fig. 6B, an increase in the association of PIAS3 and STAT3 was observed after activation of mast cells with either SCF or IL-6/IL-6R. A decrease in the association between PIAS3 and MITF was observed after activation of the cells with the same cytokines. Stripping of the membranes and reprobing with anti-PIAS3 antibody revealed a similar amount of PIAS3 bound to the beads. The same amount of MITF or STAT3 was detected in each of the lysates, as revealed by probing the lysates with anti-MITF and anti-STAT3 as a control.
We have recently reported that the zip domain in MITF is responsible for its binding with PIAS3 by using BMMC derived from either wild-type mice or MITFdi/di mice (28). The association between PIAS3 and STAT3 was then determined in BMMC derived from these MITF-Zip domains in truncated mice. PIAS3 was immunoprecipitated from SCF-activated cells by using anti-PIAS3 and immunoblotted with anti-MITF and anti-STAT3. As expected, a decrease in the association between PIAS3 and MITF was observed in SCF-activated cells derived from the wild-type mice. However, in BMMC derived from MITFdi/di mice almost no association between PIAS3 and MITF was observed, whereas an increase in the association of PIAS3 and STAT3 was observed.
Real-time PCR was performed in order to investigate the role of MITF-PIAS3-STAT3 interactions on the transcriptional regulation of genes that are known to be regulated by these two transcription factors (1, 3, 10, 20, 21, 39). SCF-activated BMMC were derived from either wild-type mice or MITFdi/di mice. The real-time PCR analysis clearly shows that the expression of MITF target genes (c-kit and tryptophan hydroxylase) and STAT3 target genes (c-Myc and VEGF) were significantly elevated upon SCF stimulation (Fig. 6D and E). No such elevation was observed in MITF target genes, as well as in STAT3 target genes in SCF-activated BMMC derived from MITFdi/di mice. This suggests that PIAS3 that cannot bind MITF in these cells is accessible for binding and therefore inhibition of STAT3.
Interactions between PIAS3, STAT3, and MITF in melanoma cells.
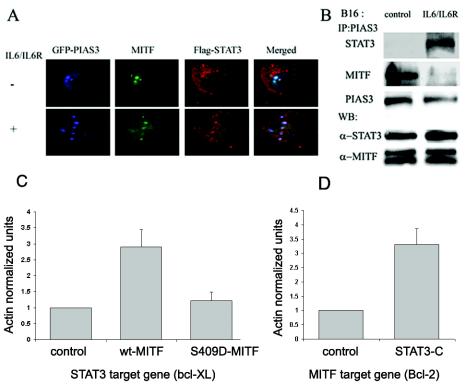
Interactions in vivo between PIAS3, MITF, and STAT3 were determined in B16 melanoma cells stimulated with IL-6/IL-6R by using confocal laser scanning. The cells were cotransfected with GFP-PIAS3, MITF, and Flag-tagged STAT3. Resting and activated cells were fixed and stained with mouse anti-MITF and rabbit anti-Flag antibodies, which were detected by staining with rhodamine-conjugated goat anti-mouse and Cy5-conjugated goat anti-rabbit antibodies, respectively. As shown in Fig. 7A, MITF and PIAS3 in resting cells were colocalized in the nucleus, whereas most of the Flag-tagged STAT3 was detected in the cytoplasm. IL-6/IL-6R treatment induced STAT3 to colocalize with PIAS3 and MITF in the nucleus.
FIG. 7.
Interactions between PIAS3, STAT3, and MITF in B16 melanoma cells. (A) B16 melanoma cells were cotransfected with GFP-PIAS3, MITF, and Flag-tagged STAT3. Cells were incubated with IL-6/IL-6R for 30 min. The immunostaining was performed with anti-MITF and anti-Flag antibodies by using rhodamine (green)- and Cy5 (red)-labeled secondary antibodies, respectively. The cells were analyzed by laser-scanning confocal microscopy. The nuclear colocalization is shown in the right panels (white dots for triple colocalization and cyan for MITF and PIAS3 colocalization). (B) B16 melanoma cells were treated with IL-6/IL-6R. Cells were lysed, and PIAS3 was immunoprecipitated with anti-PIAS3 antibody, immunoblotted with anti-STAT3 (top blot) or anti-MITF (lower blot) antibody, and reprobed with anti-PIAS3 (indicated beneath the MITF blots). Cell lysates were blotted with anti-MITF and anti-STAT3 antibodies as a control. The results of one representative experiment of three experiments performed are shown. (C and D) Quantitative reverse transcription-PCR analysis of MITF target gene (Bcl2) and STAT3 target gene (Bcl-XL) from B16 melanoma cells transfected with either MITF, S409D-MITF, or STAT3-C. RNA levels were normalized to actin and were determined in triplicate.
In order to investigate the interactions between endogenous STAT3, PIAS3, and MITF in B16 melanoma cells, cells were activated with IL-6/IL-6R for 30 min and lysed, and PIAS3 was immunoprecipitated with anti-PIAS3 and immunoblotted with anti-MITF or anti-STAT3 antibody. As shown in Fig. 7B, an increase in the association of PIAS3 and STAT3 was observed after activation of melanoma cells with IL-6/IL-6R. A decrease in the association between PIAS3 and MITF was observed after activation of the cells with the same cytokine. Stripping of the membranes and reprobing with anti-PIAS3 revealed a similar amount of PIAS3 bound to the beads. The same amount of MITF or STAT3 was detected in each of the lysates as revealed by probing the lysates with anti-MITF and anti-STAT3 as a control.
Real-time PCR was then carried out in melanoma cells in order to explore the role played by MITF-PIAS3-STAT3 interactions on MITF and STAT3 transcriptional activity. The accumulation of mRNAs of Bcl-XL (STAT3 target gene [2]) and Bcl2 (MITF target gene [31]) in cells transfected with STAT3-C or MITF was tested. Constructs were GFP labeled, and fluorescence analysis of the transfected cells revealed 75% efficiency in the transfection. The real-time PCR analysis clearly shows that the expression of Bcl2 was upregulated in STAT3-C-transfected cells and that Bcl-XL was upregulated in MITF-transfected cells (Fig. 7C and D). As a control we used the S409D-MITF mutant. Such an amino acid substitution of serine by aspartate was used to mimic the charge brought by the phosphate group (18, 45). This mutated MITF was previously shown to cause a significant decrease in the association between PIAS3 and MITF, although it has normal transcriptional activity and function (28). In S409D-MITF-transfected cells, almost no increase in Bcl-XL mRNA levels was observed. This suggests that MITF enhances STAT3-mediated transcriptional activation via its association with PIAS3.
DISCUSSION
We have presented here experimental evidence regarding the functional cross talk between MITF, PIAS3, and STAT3. Our results suggest that STAT3 acts in an indirect manner as a regulatory factor of MITF transcriptional activity. We showed that, upon IL-6/IL-6R or SCF activation, PIAS3 is mobilized from MITF to the activated STAT3 due to the phosphorylation of MITF at S409.
Such interplay of PIAS3 with other proteins has previously been reported. PIAS3 was found to interact with another zinc finger protein, Gfi-1 (38). This association enhances STAT3-mediated transcriptional activation in T cells. Furthermore, PIAS3 can serve as an enhancer by its association with androgen receptor, which activates its transcriptional activity in the human prostate (11).
The PIAS family of proteins contains several conserved domains: the SAP domain, which is required for repression of STAT1 by PIASy (30); the Miz-Zn finger/RING domain, which is essential for SUMO ligase activity (22); and the PINIT motif, which is required for nuclear retention of PIAS3 (6). In the present study, we showed that PIAS3 directly and specifically interacts with MITF and STAT3 via the same motif of 70 amino acids, which is located at the N terminus of the newly defined “PINIT” domain.
Previously, we showed that phosphorylation of MITF at S409 caused the dissociation of PIAS3 from MITF (28). Phosphorylation-dephosphorylation plays a major role in the assembly and disassembly of multiprotein complexes. For example, phosphorylation of MITF at S73 triggers the recruitment of the coactivator P300 (44). In activated melanocytes, an alanine substitution at either S73 or S409 produced transcriptionally inactive MITF, whereas mitogen-activated protein kinase and Rsk-1, which phosphorylate MITF at S73 and S409, respectively, promote its activation, followed by its degradation (14, 46).
Here we have shown that activation of NIH 3T3 cells by IL-6/IL-6R leads to the phosphorylation of MITF at the same S409 and thus to the dissociation of PIAS3 from MITF. Furthermore, when NIH 3T3 cells were transfected with the MITF-S409A mutant, there was no increase in STAT3-PIAS3 interactions upon IL-6/IL-6R activation. These results emphasize the biological importance of MITF phosphorylation at S409 in the cross talk between MITF, PIAS3, and STAT3 in cells activated by variety of stimuli.
Since NIH 3T3 cells, B16 melanocytes, and BMMC contain low amounts of IL-6R but do express the gp130 receptor (3, 12), we used the IL-6/IL-6R chimera as a ligand for our experiments. This chimera has been shown to associate with the membrane surface-expressed gp130 receptor in melanocytes and leads to the activation of intracellular signaling cascade, including the JAK/STAT pathway and the mitogen-activated protein kinase pathway (16). Kamaraju's group have demonstrated that the activation of melanoma cells with the IL-6/IL-6R chimera caused a loss of expression of MITF mRNA (24). This might be considered a cellular mechanism of modulating the posttranslational regulation of MITF expression caused by IL-6/IL-6R stimulation.
Constitutively activated STAT3 has been found in diverse cancer cell lines and tumor tissues (2, 3, 10). Various features of constitutively activated STAT3 may contribute to the increase in MITF transcriptional activity found in a variety of melanomas (5) since activated STAT3 might induce the recruitment of PIAS3 into the STAT system and thus indirectly increase the transcriptional activity of MITF. Vice versa, the observation that MITF enhances STAT3-mediated transcriptional activation could offer an explanation of the indirect oncogenic potential of MITF via its association with PIAS3.
Using confocal laser-scanning microscope, we observed that in resting cells MITF and PIAS3 were colocalized in the nucleus, whereas most of Flag-tagged STAT3 was detected in the cytoplasm. IL-6/IL-6R treatment induced all three proteins to colocalize in the nucleus in dotted complexes. Such triple colocalization does not necessarily mean direct interaction, although these results suggest that the cross talk between these three proteins occurs in close proximity. In this context, it is important to mention that no direct interaction between MITF and STAT3 was observed by coimmunoprecipitation or pull-down assay (27). Therefore, our results suggest that MITF-STAT3 colocalization is due to their interaction with PIAS3.
STAT3 participates in signal transduction pathways activated by the IL-6 family of cytokines in the hematopoietic system. STAT3 signaling has been shown to prevent programmed cell death and enhance cell proliferation through regulating genes involved in cell growth and apoptosis, including Bcl-XL, Mcl-l, c-Myc, and cyclin D1 (1, 3, 10, 39). IL-6 was found to be important for human mast cell survival (47), and an active (phosphorylated) form of STAT3 is presented in activated or transformed human mast cells (7, 29). The ability of activated STAT3 to enhance MITF activity by recruiting PIAS3 might play an important role in mast cells, since MITF has an essential role in mast cell function and development (23, 26, 34). An in-depth study is essential to reveal in detail the biological pathways in which STAT3 is involved in activated mast cells.
In conclusion, we suggest that the activation of MITF by phosphorylation at S409 leads to the release of PIAS3, which then potentially can bind to STAT3. Such activation by IL-6/IL-6R or SCF leads to fine-tuning the regulation of these transcription factors by shuttling of PIAS3 from MITF to STAT3, as shown in our model (Fig. 8).
FIG. 8.
Diagram of the cross talk between MITF, PIAS3, and STAT3 in activated cells. Activation of MITF by phosphorylation at S409 leads to the mobilization of PIAS3 to the nucleus translocated STAT3.
Acknowledgments
This study was supported by the U.S. Binational Science Foundation, the Israeli Academy of Science Foundation, and the Israel Cancer Association.
REFERENCES
- 1.Bowman, T., M. A. Broome, D. Sinibaldi, W. Wharton, W. J. Pledger, J. M. Sedivy, R. Irby, T. Yeatman, S. A. Courtneidge, and R. Jove. 2001. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc. Natl. Acad. Sci. USA 98:7319-7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowman, T., R. Garcia, J. Turkson, and R. Jove. 2000. STATs in oncogenesis. Oncogene 19:2474-2488. [DOI] [PubMed] [Google Scholar]
- 3.Bromberg, J. F., M. H. Wrzeszczynska, G. Devgan, Y. Zhao, R. G. Pestell, C. Albanese, and J. E. Darnell, Jr. 1999. Stat3 as an oncogene. Cell 98:295-303. [DOI] [PubMed] [Google Scholar]
- 4.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 5.Dorvault, C. C., K. N. Weilbaecher, H. Yee, D. E. Fisher, L. A. Chiriboga, Y. Xu, and D. C. Chhieng. 2001. Microphthalmia transcription factor: a sensitive and specific marker for malignant melanoma in cytologic specimens. Cancer 93:337-343. [DOI] [PubMed] [Google Scholar]
- 6.Duval, D., G. Duval, C. Kedinger, O. Poch, and H. Boeuf. 2003. The “PINIT” motif, of a newly identified conserved domain of the PIAS protein family, is essential for nuclear retention of PIAS3L. FEBS Lett. 554:111-118. [DOI] [PubMed] [Google Scholar]
- 7.Fumo, G., C. Akin, D. D. Metcalfe, and L. Neckers. 2004. 17-Allylamino-17-demethoxygeldanamycin (17-AAG) is effective in down-regulating mutated, constitutively activated KIT protein in human mast cells. Blood 103:1078-1084. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein, R. E., G. Jimenez, O. Cook, D. Gur, and Z. Paroush. 1999. Huckebein repressor activity in Drosophila terminal patterning is mediated by Groucho. Development 126:3747-3755. [DOI] [PubMed] [Google Scholar]
- 9.Gommerman, J. L., and S. A. Berger. 1998. Protection from apoptosis by steel factor but not interleukin-3 is reversed through blockade of calcium influx. Blood 91:1891-1900. [PubMed] [Google Scholar]
- 10.Grandis, J. R., S. D. Drenning, Q. Zeng, S. C. Watkins, M. F. Melhem, S. Endo, D. E. Johnson, L. Huang, Y. He, and J. D. Kim. 2000. Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc. Natl. Acad. Sci. USA 97:4227-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross, M., B. Liu, J. Tan, F. S. French, M. Carey, and K. Shuai. 2001. Distinct effects of PIAS proteins on androgen-mediated gene activation in prostate cancer cells. Oncogene 20:3880-3887. [DOI] [PubMed] [Google Scholar]
- 12.Gyotoku, E., E. Morita, Y. Kameyoshi, T. Hiragun, S. Yamamoto, and M. Hide. 2001. The IL-6 family cytokines, interleukin-6, interleukin-11, oncostatin M, and leukemia inhibitory factor, enhance mast cell growth through fibroblast-dependent pathway in mice. Arch. Dermatol. Res. 293:508-514. [DOI] [PubMed] [Google Scholar]
- 13.Hallsson, J. H., J. Favor, C. Hodgkinson, T. Glaser, M. L. Lamoreux, R. Magnusdottir, G. J. Gunnarsson, H. O. Sweet, N. G. Copeland, N. A. Jenkins, and E. Steingrimsson. 2000. Genomic, transcriptional and mutational analysis of the mouse microphthalmia locus. Genetics 155:291-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemesath, T. J., E. R. Price, C. Takemoto, T. Badalian, and D. E. Fisher. 1998. MAP kinase links the transcription factor microphthalmia to c-Kit signaling in melanocytes. Nature 391:298-301. [DOI] [PubMed] [Google Scholar]
- 15.Hirano, T. 1998. Interleukin 6 and its receptor: ten years later. Int. Rev. Immun. 16:249-284. [DOI] [PubMed] [Google Scholar]
- 16.Hirano, T., K. Ishihara, and M. Hibi. 2000. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene 19:2548-2556. [DOI] [PubMed] [Google Scholar]
- 17.Hodgkinson, C. A., K. J. Moore, A. Nakayama, E. Steingrimsson, N. G. Copeland, N. A. Jenkins, and H. Arnheiter. 1993. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 74:395-404. [DOI] [PubMed] [Google Scholar]
- 18.Hoeffler, W. K., A. D. Levinson, and E. A. Bauer. 1994. Activation of c-Jun transcription factor by substitution of a charged residue in its N-terminal domain. Nucleic Acids Res. 22:1305-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes, M. J., L. J.B., J. M. Krakowsky, and K. P. Anderson. 1993. A helix-loop-helix transcription factor-like gene is located at the mi locus. J. Biol. Chem. 268:20687-20690. [PubMed] [Google Scholar]
- 20.Isozaki, K., T. Tsujimura, S. Nomura, E. Morii, U. Koshimizu, Y. Nishimune, and Y. Kitamura. 1994. Cell type-specific deficiency of c-kit gene expression in mutant mice of mi/mi genotype. Am. J. Pathol. 145:827-836. [PMC free article] [PubMed] [Google Scholar]
- 21.Ito, A., E. Morii, D. K. Kim, T. R. Kataoka, T. Jippo, K. Maeyama, H. Nojima, and Y. Kitamura. 1999. Inhibitory effect of the transcription factor encoded by the mi mutant allele in cultured mast cells of mice. Blood 93:1189-1196. [PubMed] [Google Scholar]
- 22.Jackson, P. K. 2001. A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev. 15:3053-3058. [DOI] [PubMed] [Google Scholar]
- 23.Jippo, T., E. Morii, A. Ito, and Y. Kitamura. 2003. Effect of anatomical distribution of mast cells on their defense function against bacterial infections: demonstration using partially mast cell-deficient tg/tg mice. J. Exp. Med. 197:1417-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamaraju, A. K., C. Bertolotto, J. Chebath, and M. Revel. 2002. Pax3 down-regulation and shut-off of melanogenesis in melanoma B16/F10.9 by interleukin-6 receptor signaling. J. Biol. Chem. 277:15132-15141. [DOI] [PubMed] [Google Scholar]
- 25.Kaptein, A., V. Paillard, and M. Saunders. 1996. Dominant negative stat3 mutant inhibits interleukin-6-induced Jak-STAT signal transduction. J. Biol. Chem. 271:5961-5964. [DOI] [PubMed] [Google Scholar]
- 26.Kitamura, Y., E. Morii, T. Jippo, and A. Ito. 2002. Effect of MITF on mast cell differentiation. Mol. Immunol. 38:1173-1176. [DOI] [PubMed] [Google Scholar]
- 27.Levy, C., H. Nechushtan, and E. Razin. 2002. A new role for the STAT3 inhibitor, PIAS3: a repressor of microphthalmia transcription factor. J. Biol. Chem. 277:1962-1966. [DOI] [PubMed] [Google Scholar]
- 28.Levy, C., A. Sonnenblick, and E. Razin. 2003. The role played by MITF phosphorylation and its Zip domain in its transcriptional inhibition by PIAS3. Mol. Cell. Biol. 23:9073-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linnekin, D. 1999. Early signaling pathways activated by c-Kit in hematopoietic cells. Int. J. Biochem. Cell Biol. 31:1053-1074. [DOI] [PubMed] [Google Scholar]
- 30.Liu, B., M. Gross, J. ten Hoeve, and K. Shuai. 2001. A transcriptional corepressor of Stat1 with an essential LXXLL signature motif. Proc. Natl. Acad. Sci. USA 98:3203-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGill, G. G., M. Horstmann, H. R. Widlund, J. Du, G. Motyckova, E. K. Nishimura, Y. L. Lin, S. Ramaswamy, W. Avery, H. F. Ding, S. A. Jordan, I. J. Jackson, S. J. Korsmeyer, T. R. Golub, and D. E. Fisher. 2002. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell 109:707-718. [DOI] [PubMed] [Google Scholar]
- 32.Moore, K. J. 1995. Insight into the microphthalmia gene. Trends Genet. 11:442-450. [DOI] [PubMed] [Google Scholar]
- 33.Morii, E., T. Jippo, K. Hashimoto, D.-K. Kim, Y.-M. Lee, H. Ogihara, K. Tsujino, H.-M. Kim, and Y. Kitamura. 1997. Abnormal expression of mouse mast cell protease 5 gene in cultured mast cells derived from mutant mi/mi mice. Blood 90:3057-3066. [PubMed] [Google Scholar]
- 34.Morii, E., H. Ogihara, K. Oboki, T. R. Kataoka, K. Maeyama, D. E. Fisher, M. L. Lamoreux, and Y. Kitamura. 2001. Effect of a large deletion of the basic domain of mi transcription factor on differentiation of mast cells. Blood 98:2577-2579. [DOI] [PubMed] [Google Scholar]
- 35.Morii, E., T. Tsujimura, T. Jippo, K. Hashimoto, K. Takebayashi, K. Tsujino, S. Nomura, M. Yamamoto, and Y. Kitamura. 1996. Regulation of mouse mast cell protease 6 gene expression by transcription factor encoded by the mi locus. Blood 88:2488-2494. [PubMed] [Google Scholar]
- 36.Razin, E., C. Cardon-Cardo, and R. A. Good. 1981. Growth of a pure population of mouse mast cells in vitro using conditioned medium derived from Con A-stimulated splenocytes. Proc. Natl. Acad. Sci. USA 78:2559-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Razin, E., Z. C. Zhang, H. Nechushtan, S. Frenkel, Y. N. Lee, R. Arudchandran, and J. Rivera. 1999. Suppression of microphthalmia transcriptional activity by its association with protein kinase C-interacting protein 1 in mast cells. J. Biol. Chem. 274:34272-34276. [DOI] [PubMed] [Google Scholar]
- 38.Rodel, B., K. Tavassoli, H. Karsunky, T. Schmidt, M. Bachmann, F. Schaper, P. Heinrich, K. Shuai, H. P. Elsasser, and T. Moroy. 2000. The zinc finger protein Gfi-1 can enhance STAT3 signaling by interacting with the STAT3 inhibitor PIAS3. EMBO J. 19:5845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinibaldi, D., W. Wharton, J. Turkson, T. Bowman, W. J. Pledger, and R. Jove. 2000. Induction of p21WAF1/CIP1 and cyclin D1 expression by the Src oncoprotein in mouse fibroblasts: role of activated STAT3 signaling. Oncogene 19:5419-5427. [DOI] [PubMed] [Google Scholar]
- 40.Tachibana, M., K. Takeda, Y. Nobukuni, K. Urabe, J. E. Long, K. A. Meyers, S. A. Aaronson, and T. Miki. 1996. Ectopic expression of MITF, a gene for Waardenburg syndrome type 2, converts fibroblasts to cells with melanocyte characteristics. Nat. Genet. 14:50-54. [DOI] [PubMed] [Google Scholar]
- 41.Takeda, K., K. Noguchi, W. Shi, T. Tanaka, M. Matsumoto, N. Yoshida, T. Kishimoto, and S. Akira. 1997. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 94:3801-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeda, K., C. Takemoto, I. Kobayashi, A. Watanabe, Y. Nobukuni, D. E. Fisher, and M. Tachibana. 2000. Ser298 of MITF, a mutation site in Waardenburg syndrome type 2, is a phosphorylation site with functional significance. Hum. Mol. Genet. 9:125-132. [DOI] [PubMed] [Google Scholar]
- 43.Weilbaecher, K. N., C. L. Hershey, C. M. Takemoto, M. A. Horstmann, T. J. Hemesath, A. H. Tashjian, and D. E. Fisher. 1998. Age-resolving osteopetrosis: a rat model implicating microphthalmia and the related transcription factor TFE3. J. Exp. Med. 187:775-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weilbaecher, K. N., G. Motyckova, W. E. Huber, C. M. Takemoto, T. J. Hemesath, Y. Xu, C. L. Hershey, N. R. Dowland, A. G. Wells, and D. E. Fisher. 2001. Linkage of M-CSF signaling to Mitf, TFE3, and the osteoclast defect in Mitfmi/mi mice. Mol. Cell 8:749-758. [DOI] [PubMed] [Google Scholar]
- 45.Wittekind, M., J. Reizer, J. Deutscher, M. H. Saier, and R. E. Klevit. 1989. Common structural changes accompany the functional inactivation of HPr by seryl phosphorylation or by serine to aspartate substitution. Biochemistry 28:9908-9912. [DOI] [PubMed] [Google Scholar]
- 46.Wu, M., T. J. Hemesath, C. M. Takemoto, M. A. Horstmann, A. G. Wells, E. R. Price, D. Z. Fisher, and D. E. Fisher. 2000. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 14:301-312. [PMC free article] [PubMed] [Google Scholar]
- 47.Yanagida, M., H. Fukamachi, K. Ohgami, T. Kuwaki, H. Ishii, H. Uzumaki, K. Amano, T. Tokiwa, H. Mitsui, H. Saito, et al. 1995. Effects of T-helper 2-type cytokines, interleukin-3 (IL-3), IL-4, IL-5, and IL-6 on the survival of cultured human mast cells. Blood 86:3705-3714. [PubMed] [Google Scholar]