Abstract
Like eukaryotes, bacteria express one or more serine/threonine kinases (STKs) that initiate diverse signaling networks. The STK from Streptococcus suis is encoded by a single-copy stk gene, which is crucial in stress response and virulence. To further understand the regulatory mechanism of STK in S. suis, a stk deletion strain (Δstk) and its complementary strain (CΔstk) were constructed to systematically decode STK characteristics by applying whole transcriptome RNA sequencing (RNA-Seq) and phosphoproteomic analysis. Numerous genes were differentially expressed in Δstk compared with the wild-type parental strain SC-19, including 320 up-regulated and 219 down-regulated genes. Particularly, 32 virulence-associated genes (VAGs) were significantly down-regulated in Δstk. Seven metabolic pathways relevant to bacterial central metabolism and translation are significantly repressed in Δstk. Phosphoproteomic analysis further identified 12 phosphoproteins that exhibit differential phosphorylation in Δstk. These proteins are associated with cell growth and division, glycolysis, and translation. Consistently, phenotypic assays confirmed that the Δstk strain displayed deficient growth and attenuated pathogenicity. Thus, STK is a central regulator that plays an important role in cell growth and division, as well as S. suis metabolism.
Keywords: Streptococcus suis, eukaryote-like serine/threonine kinase, phosphorylation, RNA-Seq, phosphoproteome, growth, virulence, metabolism
Introduction
Upon sensing external stimuli, protein kinases, together with their cognate phosphatases, play a central role in signal transduction to quickly respond and adapt to constantly changing environments in both prokaryotes and eukaryotes. Reversible phosphorylation occurs on specific amino acid residues, most commonly serine (Ser), threonine (Thr), tyrosine (Tyr), histidine (His), and aspartate (Asp) (Pereira et al., 2011). Earlier classifications of prokaryote, kinases have been assumed to target only residues His and Asp, which are involved in two-component systems (TCS; Stock et al., 1990; Hoch, 2000). Increasing attention has been paid to the Ser/Thr kinases and their partner phosphatases. Some bacterial Ser/Thr kinases, which show conservation in their catalytic domains compared with eukaryotic Ser/Thr kinases, are designated as “eukaryote-like Ser/Thr kinases (eSTK)” (Pereira et al., 2011). Pkn1 of Myxococcus xanthus is the first characterized eSTK in bacteria (Muñoz-Dorado et al., 1991); subsequently a second eSTK, Pkn2, has been identified in M. xanthus (Udo et al., 1995). Furthermore, numerous bacterial eSTKs have been identified based on genome sequence databases (Galperin et al., 2010). Multiple eSTKs exist in most bacteria; therefore, comprehensively characterizing their essentiality and identifying their specific substrates are difficult. For example, 11 eSTKs exist in Mycobacterium tuberculosis that has functional redundancy and/or substrate promiscuity (Boitel et al., 2003; Sajid et al., 2015).
The eSTKs have been widely studied for their roles in diverse biological processes, including development (Zhang, 1993; Nádvorník et al., 1999; Inouye and Nariya, 2008), cell competence (Hussain et al., 2006), cell division, and cell wall synthesis (Deol et al., 2005; Fernandez et al., 2006; Ruggiero et al., 2012), central and secondary metabolism (Lee et al., 2002; Sawai et al., 2004), biofilm formation (Hussain et al., 2006; Liu et al., 2011), stress response (Neu et al., 2002; Mata-Cabana et al., 2012), and virulence (Madec et al., 2002; Rajagopal et al., 2003; Echenique et al., 2004). Gene expression profiles have proven their global regulatory roles in cellular processes (Sasková et al., 2007; Donat et al., 2009). Moreover, both phosphoproteomic analyses and kinase assays have identified eSTK substrates in Streptococcus pyogenes (Jin and Pancholi, 2006), Streptococcus pneumoniae (Nováková et al., 2005, 2010), Streptococcus agalactiae (Silvestroni et al., 2009), Staphylococcus aureus (Lomas-Lopez et al., 2007; Truong-Bolduc et al., 2008), Listeria monocytogenes (Archambaud et al., 2005), and M. tuberculosis (Arora et al., 2010). Most identified substrates are involved in cell growth/division and central metabolism of bacteria. Various microorganisms have been studied, but the profound effects of eSTKs and posttranslational modification on their targets remain poorly understood.
Streptococcus suis is a zoonotic Gram-positive pathogen that causes lethal infections in pigs and humans (Lun et al., 2007). Two large outbreaks of human S. suis infection have been reported in 1998 and 2005 in China, resulting in 229 infections and 52 deaths (Lun et al., 2007). Among the 33 S. suis serotypes, serotype 2 (SS2) is the most virulent and prevalent serotype isolated from diseased pigs (Smith et al., 1999). Moreover, SS2 is the prominent agent that caused adult human meningitis in Vietnam and Hong Kong (Wertheim et al., 2009). Numerous virulence-associated factors of S. suis have been identified over the past decade, such as capsular polysaccharide, muramidase-released protein, suilysin, extracellular factor, fibrinonectin- and fibrinogen-binding proteins, enolase, arginine deiminase system, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), inosine 5-monophosphate dehydrogenase (IMPDH), secreted nuclease A (SsnA), subtilisin-like protease A (Fittipaldi et al., 2012), H binding protein (Fhb; Pian et al., 2012), and so on. Compared with other Gram-positive bacteria, only a single-copy stk is present in the S. suis genome (Zhu et al., 2014). The STK of S. suis is involved in stress response and virulence. The disruption of stk in S. suis enables increased chain-length, reduced tolerance to high temperature, low acidic pH, oxidative stress, and decreased virulence (Zhu et al., 2014).
To further understand the regulatory mechanism of STK in S. suis, we constructed a stk-deletion mutant (Δstk) and investigated its biological characterizations using “-omics” approaches. By comparing the transcriptomic profiles, we identified differentially expressed genes (DEGs) between the Δstk strain and the wild-type parental strain, SC-19. Using phosphoproteome analyses, phosphorylation level of protein-coding sequences were systematically estimated. The analyses of both transcriptomic and phosphoproteomic provide functional context that STK can regulate cell growth and division, as well as metabolism of S. suis.
Materials and methods
Bacterial strains, plasmids, and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. The virulent S. suis strain SC-19 was isolated from a diseased pig during the 2005 outbreak in Sichuan, China (Li et al., 2009). Since the genome of SC-19 has not been sequenced, the genome sequence of the strain S. suis 05ZYH33 (GenBank accession number CP000407) was used as reference for gene clone, transcriptomic, and phosphoproteomic analysis. S. suis 05ZYH33 was isolated from an infected human during the same outbreak in Sichuan (Lun et al., 2007). Both of these two isolates are serotype 2. Bacteria were grown in TODD-Hewitt broth (THB; OXOID, England) medium or plated on THB Agar (THA; OXOID) with 5% (v/v) sheep blood at 37°C. Erythromycin (90 μg/ml) was added to screen the mutant strain and erythromycin (90 μg/ml) and spectinomycin (100 μg/ml) were added to select for a complementary strain.
Table 1.
Bacterial strains and plasmids used in this study.
| Stains and plasmids | Revelant characteristics and genotype* | Sources or references |
|---|---|---|
| S. suis STRAINS | ||
| SC-19 | S. suis serotype 2, the wild-type (Strepr) | Li et al., 2009 |
| Δstk | SC-19 stk::erm (Strepr Ermr) | This study |
| CΔstk | SC-19 stk::erm/stk+ (Strepr Ermr Spcr) | This study |
| E. coli | ||
| DH5α | Cloning host for recombinant vector | Trans |
| BL21 (DE3) | Expressing host for fusion protein | Trans |
| PLASMIDS | ||
| pET-28a | Expression vector (Kanr) | Novagen |
| pSTK | pET-28a containing stk, cloned from SC-19 genome | This study |
| pAT18 | A plasmid carrying an erythromycin resistance rRNA methylase (erm) gene expression cassette | Trieu-Cuot et al., 1991 |
| pSET4s | Temperature-sensitive E. coli-S. suis shuttle vector (Spcr) | Takamatsu et al., 2001b |
| pSET4s-S | Derived from pSET4s for deleting stk in SC-19 | This study |
| pSET2 | E. coli-S. suis shuttle vector (Spcr) | Takamatsu et al., 2001a |
| pSET2-CM | Derived from pSET2 for functional complementation of stk (Spcr) | This study |
Strepr, streptomycin resistant; Ermr, erythromycin resistant; Spcr, spectinomycin resistant; Kanr, kanamycin resistant.
Escherichia coli (E. coli) strain DH5α (Trans, China) was used as host strain for cloning and E. coli strain BL21 (DE3) (Trans) was used to express His-tag fusion proteins. E. coli strains were cultured in Luria-Bertani (LB) Broth (Difco, France) or plated on LB Agar at 37°C. If necessary, kanamycin (25 μg/ml) was added.
Construction of mutant and complementary strain
The stk deletion strain was obtained using an existing method (Takamatsu et al., 2001b; Zhu et al., 2014). Primers used in this study were designed according to the genome sequence of S. suis 05ZYH33 and are listed in Table S1. Primers SU-F/SU-R and SD-F/SD-R were used to amplify the upstream and downstream regions of stk. Moreover, the fragments were cloned into pSET4s, respectively. Finally, the erythromycin-resistance expression cassette (ermR) was amplified from pAT18 with primers Erm-F/Erm-R and cloned to achieve the stk-knockout vector pSET4s-S. The pSET4s-S was then electroporated into SC-19. Through homologous recombination, the stk gene was replaced the ermR via double-crossover incident.
The promoter sequence of IMPDH was selected to drive the expression of stk gene in SC-19 (Takamatsu et al., 2001a; Zhu et al., 2014). The promoter and the coding sequence of stk were cloned into pSET2 to obtain the recombinant plasmid pSET2-S. The pSET2-S was transformed into the mutant strain Δstk to acquire the complementary strain CΔstk.
To further confirm the mutant strain Δstk and the complementary strain CΔstk, we performed Western blot analysis. The overnight cultures of SC-19, Δstk and CΔstk, cultured in THB were collected by centrifugation and resuspended in bacterial lysis buffer (50 mM Tris/HCl, pH 8.5, 100 mM NaCl, 2 mM ethylene diamine tetraacetic acid, 100 μg/ml lysozyme, 1 mM phenylmethanesulfonyl fluoride, 0.5% Triton X-100). Bacterial cells were lysed with a French pressure cell press. Quantified bacterial lysate was separated on SDS-PAGE and then electrotransferred to PVDF membrane (Invitrogen, USA) (Tan et al., 2015). Mouse anti-STK serum was produced as described previously (Li et al., 2011) by using recombinant STK protein. The PVDF membranes (Invitrogen, USA) were probed with primary antibodies against STK (1:1000) or GidA as control (1:1000; stored at −80°C in our laboratory). After wash, the membranes were incubated with goat anti-mouse IgG (H+L)-HPR (1:5000; Southern Biotech, USA). Antibody-tagged protein bands were detected by using Western ECL Substrate Kit (Bio-Rad, USA).
RNA extraction and RT-PCR
To confirm the mutant strain Δstk and the complementary strain CΔstk, we also performed RT-PCR (Tan et al., 2015). Briefly, RNA was isolated using SV Total RNA Isolation System (Promega, USA) according to the manufacturer's instructions. Then, cDNA was synthesized using HiScript Q Select RT SuperMix (Vazyme, China) according to the manufacturer's instructions.
To confirm whether the upstream and downstream genes of stk in the mutant were affected or not, the genes were amplified using the primers SSU05_0427-F/SSU05_0427-R (for upstream gene) and SSU05_0429-F/SSU05_0429-R (for downstream gene; Table S1) using the SC-19 cDNA as template.
Protein expression and purification
The coding sequence of stk was amplified using primers stk-F/stk-R from the SC-19 genome. The PCR product was restricted with EcoRI and XhoI and then inserted into the digested pET-28a vector to generate the recombinant plasmid pET-stk, which was then transformed into E. coli BL21 (DE3) cells. Expression was induced by 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) (Sigma, USA) at 18°C for 12 h. His-tagged STK was purified using Ni-NTA columns (GE Healthcare, Sweden) according to the manufacturer's recommendation. The purified protein was identified by Western blot analysis using the His-tag rabbit-polyclonal antibody (Sigma, USA) and STK-mouse polyclonal antibody (made in our laboratory), respectively.
Protein kinase assay
The kinase assay was performed as described previously (Boitel et al., 2003; Fernandez et al., 2006). The reaction was conducted in 50 μl of kinase buffer (50 mM Hepes, 1 mM DTT, 0.01% Brij35, pH 7.0) containing 2 mM MnCl2, 100 μM ATP and 1 μCi of [γ-32P]-ATP (PerkinElmer, UK). The reaction was initiated by adding the kinase at 30°C for 10 min and stopped by adding SDS-PAGE sample buffer plus EDTA (25 mM final). A total of 20 μl of the reaction was subjected to electrophoresis. In each case, the reaction products were separated on a 12% SDS-polyacrylamide gel and the radiolabelled proteins were visualized using auto-radiography. To obtain relative quantification of the radiolabelled ATP incorporation, the radioactive samples were analyzed using a PhosphorImager apparatus (Fujifilm, Japan). For the substrate phosphorylation, myelin basic protein (MBP; Sigma, USA) was used as a positive control to test the kinase activity. The enzyme/substrate ratio was 1:10 with 0.4 μM kinase.
Microscopy image
The strains SC-19, Δstk and CΔstk at the mid-log phase were collected and then resuspended twice using ddH2O. Each sample (20 μl) was fixed on glass slides (Shitai, China) through flaming. A Gram staining kit (Jiancheng, China) was utilized according to the manufacturer's instruction. The stained samples were observed under light microscope.
Scanning electron microscope (SEM) observation was performed following previous methods (Wang et al., 2011; Fleurie et al., 2012). The bacteria were grown in THB broth at 37°C and harvested at OD600 of 0.6. Bacterial suspensions were spotted onto glass coverslips (0.17 mm in thickness and 20 mm in diameter; WHB, China) and washed with phosphate-buffered saline. The cells were fixed with 2.5% glutaraldehyde overnight at 4°C. All samples were dehydrated with a series of gradient ethanol and air dried. The dried samples were covered with a 10 nm gold/platinum layer (JSM-6390LV, JEOL, Japan) and observed via SEM (JFC-1600, JEOL, Japan).
All SEM images were analyzed with Image J. The sizes (in pixels) of bacterial chains were defined as their two-dimensional area based on the “Auto Threshold” function, which defined the cell chain outline. Shapes that were not chains and chains on the border of images were manually excluded from subsequent analyses (Dalia and Weiser, 2011). For chain length analysis, data are the result of at least 100 chains analyzed per sample from at least 100 fields.
Detection of growth characterizations
Growth rates of the SC-19, Δstk and CΔstk were detected through the measurement of the density changes represented by OD600 nm values and CFU counts of the cultures. Different strains were grown overnight in THB medium and then the initial OD600 nm of all subcultures were adjusted to 0.014. The diluted cells were incubated at 180 rpm/m at 37°C in the same medium and OD600 nm values were read every hour until the growth process entered stationary phase. Meanwhile, 100 μl of the bacterial culture was diluted and then vortexed to break the chains. Bacteria numbers were recorded every hour by viable count. All the growth data were analyzed with Origin 8.0. The growth rate at each time point was calculated based on the function of “Non-linear Curve Fit” and “Mathematics.” The average growth rate (AGR) was calculated from data in mid-log phase based on the function of “Linear Curve Fit” (OriginLab, Northampton, MA, USA).
Virulence assay in mice
All mice used in our study were purchased from the Wuhan Institute of Biological Products (Wuhan, China). The study was performed strictly in accordance with the Ethics Committee of Huazhong Agricultural University according to Hubei Province Laboratory Animal Management Regulations-2005. All efforts were made to minimize suffering.
To probe the possible role of the stk gene in S. suis virulence, 24 female specific-pathogen-free (SPF) Kunming mice (4–6 weeks old) were divided into three groups. Groups 1 and Group 2 were intraperitoneally injected with 1 × 109 colony-forming unit (CFU) mid-log-phase cells of either SC-19 or Δstk. Saline was applied in Group 3 as negative control. Clinical signs and survival time were recorded. The mice were observed for 7 days to obtain steady survival curves.
To better evaluate the pathogenicity of Δstk, we performed a determination of viable bacteria in organs assay as described previously (Gao et al., 2016). Six-week-old female SPF Kunming mice (6 mice per group) were intraperitoneally inoculated with 5 × 108 CFU of a 1:1 mixture of mid-log phase SC-19 and Δstk. Saline was applied as negative control in six mice. At 6, 18, and 52 h post-infection (hpi), blood samples, brains, and lungs were obtained from each group. The samples were homogenized after weighing, and serial dilutions were plated onto THA. To count the colonies, we used streptomycin (20 μg/ml) for SC-19, whereas 20 μg/ml streptomycin and 90 μg/ml erythromycin were used for Δstk.
RNA-Seq analysis
To investigate gene expression profiles between SC-19 and Δstk, RNA-Seq was performed in the BGI, Shenzhen. Three biological replicates were mixed as one sample before RNA-Seq. RNA isolation and purification were the same as aforementioned. RNA-Seq based transcriptomic profiles were conducted as previous cited (Wilhelm and Landry, 2009). After the quality inspection using an Agilent 2100 Bioanalyzer and ABI StepOnePlus Real-Time PCR System, the library was sequenced using Illumina HiSeq 2000. Quality control of raw reads was performed and clean reads were mapped onto the complete reference S. suis 05ZYH33 genome.
The gene expression level was calculated by using RPKM (reads per kb per million reads) method. The number of reads that uniquely aligned to a unique gene was normalized to RPKM (Mortazavi et al., 2008). The RPKM method eliminates the effect of different gene lengths and sequencing levels on the calculation of gene expression. Therefore, the gene expression can be directly calculated by comparing the different gene expression level among samples (Chen et al., 2014). To identify DEGs between two samples, a statistical analysis of the frequency of each unique-match read in each library was performed by referring to “the significance of digital gene expression profiles” (Audic and Claverie, 1997; Chen et al., 2014). The probability of gene A expressed equally between two samples can be calculated with the following formula, wherein the total clean tag number of the sample 1 is N1, and total clean tag number of sample 2 is N2; gene A holds x tags in sample 1 and y tags in sample 2; p-value corresponds to differential gene expression test (Audic and Claverie, 1997; Chen et al., 2014).
or
False Discovery Rate (FDR) was used in the multiple hypothesis testing to correct for p-value (Chen et al., 2014). Following the formula below, assuming W DEGs had been selected, M genes of those were really differential expressed, whereas H genes indicated no difference which were false positive. If we decide that the error ratio “Q = H/W” must stay below a cutoff (e.g., 5%), we should preset the FDR to a number no larger than 0.05. FDR-values were calculated according to the previous algorithm (Li J. et al., 2015)
To detecte DEGs, we used FDR ≤ 0.001 and log transformed fold change (the ratio of RPKM values) > 2.0. Moreover, enrichment analysis based on KEGG pathway database (http://www.genome.jp/kegg/) was done using the R package GAGE v2.22 (Luo et al., 2009). Pathway-level differential expression was visually checked using Pathview (Luo and Brouwer, 2013). Functional classification of gene products was predicted using BLASTP v2.5.0+ (Altschul et al., 1997) by the clusters within the orthologous group (COG) database v2014 (Galperin et al., 2015). The q-value cutoff for BLAST searching was set to 0.1. Putative virulence-associated genes (VAGs) of S. suis were screened by searching all protein sequences against Virulence Factor Database (VFDB; Chen et al., 2005) using BLASTP. The E-value cutoff for BLAST searching was set to 1e-20. The RNA-Seq data have been submitted to the NCBI Gene Expression Omnibus (GEO) database under the accession number GSE87759.
Quantitative RT-PCR (qRT-PCR)
A subgroup (n = 10) of DEGs was selected to cross-validate the RNA-Seq data with SYBR green detection. The primers (Table S1) were designed according to the genomic sequence of S. suis 05ZYH33. RNA extraction was carried out as described above. qRT-PCR was performed on an ABI 7300 HT Sequence Detection System using the ABI Power SYBE Green PCR Master Mix. The 16S rRNA served as an internal reference gene. The relative expression level was measured using the 2−ΔΔct method. Data were reported as mean relative expression levels (± standard deviation) between SC-19 and Δstk. Five depressed genes in Δstk (SSU05_1776, SSU05_0272, SSU05_1815, SSU05_0309, and SSU05_2154) and five stimulative genes in Δstk (SSU05_0792, SSU05_0358, SSU05_1011, SSU05_0906, and SSU05_1532) were chosen to do qRT-PCR for RNA-Seq confirmation. The assays were replicated thrice.
Subsequently, we performed qRT-PCR to compare the transtriptional levels of the above genes between SC-19 and CΔstk related to Δstk. The relative expression level was measured using the 2−ΔΔct method. Data were reported as mean relative expression levels (± standard deviation) between SC-19 and Δstk, as well as between CΔstk and Δstk.
Protein extraction, digestion, labeling with iTRAQ reagents, and phosphopeptide enrichment
SC-19 and Δstk cells at mid-log phase were cultured in THB as described above. Three independent biological replicates of bacterial pellets were then treated with SDT buffer (4% SDS, 100 mM Tris-HCl, 1 mM DTT, pH 7.6) and heated for 15 min at 100°C. The cell suspensions were sonicated for 5 min (10 s sonication with 15 s interval, 80 W) on ice and protein concentrations in the supernatants were determined through Bradford protein assay (Gao et al., 2016). Each sample (20 μg) was then prefractionated using 12% SDS-PAGE. Subsequently, the gel was processed using 200 μl UA buffer (8 M Urea, 150 mM Tris-HCl, pH 8.0) and subjected to in-gel tryspin digestion at 37°C for 16 h (Hu et al., 2014). The resulting tryptic peptides were labeled according to the protocol of iTRAQ Reagent-8 plex Multiplex Kit (Applied Biosystems, USA). The labeled peptides were mixed, concentrated using a vacuum concentrator and resuspended in 500 μl 1 × DHB buffer (3% DHB, 80% CAN, 0.1% TFA). Afterward, TiO2 beads were added and agitated for 40 min. The centrifugation was performed, resulting in the first beads. The supernatant from the first centrifugation was mixed with additional TiO2 beads, resulting in the second beads that were collected as before. Both bead groups were combined and washed thrice with 50 μl of washing buffer I (30% ACN, 3% TFA) and then washed thrice with 50 μl washing buffer II (80% ACN, 3% TFA) to remove the non-absorbed material. Finally, the phosphopeptides were eluted with 50 μl of elution buffer (40% ACN, 15% NH4OH), followed by lyophilization and MS analysis.
LC-MC/MS analysis
Phosphopeptide solution (6 μl) was mixed with 0.1% (v/v) fluoroacetic acid (20 μl). Equal amounts of digested protein were loaded into a Thermo Scientific EASY column (2 cm*100 μm 5 μm-C18) and then washed with solvent A (99% H2O, and 0.1% formic acid). By applying solvent B (84% acetonitrile, 16% H2O, and 0.1% formic acid), the peptides were eluted from the trapping column over a Thermo scientific EASY column (75 μm* 100 mm 3 μm-C18) with a gradient (0–55% B for 220 min at 250 nL/min, 55–100% B for 8 min, and 100% B for 12 min) using Thermo scientific Easy nLC system. For MS analysis, peptides were analyzed in positive ion mode. MS/MS was carried out with a Q-Exactive mass spectrometer (Thermo Finnigan, USA) setting in a positive ion mode and data-dependent manner choosing the most abundant precursor ions using a full MS scan from m/z 350–1800, with a resolution of 70,000 at m/z 200. Target value determination was based on automatic gain control and dynamic exclusion duration was 30 s. MS/MS scan was acquired at a resolution of 17,500 at m/z 200. Normalized collision energy was 29 eV and the underfill ratio was set to 0.1%. Quantitation was achieved by comparing the peak areas and resultant peak ratios among the four MS/MS reporter ions from 114 to 117 Da.
Phosphoproteomic data analysis
The data files produced by LC-MS/MS were processed by Proteome Discoverer 1.4 and searched by Mascot 2.2 (Matrix Science, MA) against 26,496 S. suis protein-coding sequences deposited in the Uniprot database (downloaded on August 4, 2015). For peptides after phosphopeptide enrichment, the following options were used. Peptide mass tolerance: ± 20 ppm; MS/MS tolerance: 0.1 Da; enzyme: trypsin; max missed cleavages: 2; fixed modifications: carbamidomethyl (C), iTRAQ8plex (K) and iTRAQ8plex (N-term); variable modifications: oxidation (M), phosphorylation (Ser/Thr/Tyr). For peptides, proteins, and phosphosites identification, FDR was estimated and the threshold was set to 1%. The peptides were determined as true phosphorylation based on the following criteria: Phospho RS score > 50 and Phospho RS site probabilities > 75%. Proteome Discoverer 1.4 software was used to extract the peak intensity within 20 ppm of each expected iTRAQ reporter ion from each analyzed fragmentation spectrum. Only spectra in which all the expected iTRAQ reporter ions detected were used for quantification.
The intensity of the reporter ions was used for phosphopeptide quantification. We normalized the phosphopeptide ratios by dividing by the median ratio of all peptides identified. As for the quantitative analysis, the log2 fold-change values (SC-19/Δstk) were calculated for each phosphopeptide. Only phosphopeptides detected in at least two out of the three biological replicates were used for assessment of significant change. The t-test was employed to identify significant changes between the wild type strain and the mutant strain among the three biological replicates. The phosphopeptides that passed t-test with p < 0.05 were considered to be significantly regulated (Fan et al., 2014). We also included the cutoff for the log2 fold change values, in which the phosphorylation changes were considered highly significant if the log2 value ≥ 0.26 or ≤ −0.26. The data were then normalized and logarithmically transformed as FC. FC (Δstk/SC-19) ≥ 1.2 and ≤ 0.83 were used to represent up- or down-regulations. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the dataset identifier PXD005663.
Statistical analysis
The means of two groups were compared using Student's t-test (unpaired, two-tailed) in GraphPad Prism 5 (San Diego, USA), with p < 0.05 considered to be statistically significant. Unless otherwise specified, all the experiments were performed in triplicate at least thrice.
Results
Characterization of STK in S. suis
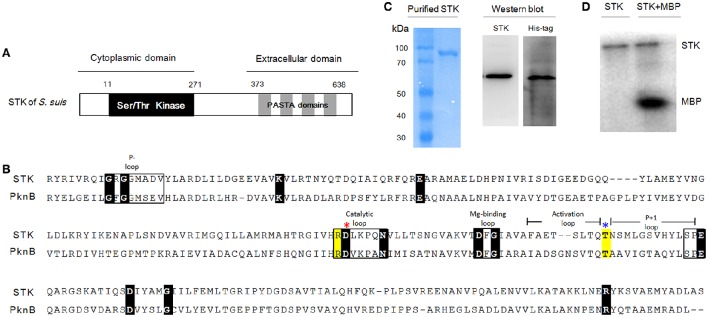
As previously described, the STK in S. suis consists of a cytoplasmic kinase domain (residues 11–267 aa) and an extracellular C-terminal region composed of four penicillin-binding proteins (residues 373–434 aa; 441–500 aa; 507–568 aa; 575–638 aa) and a Ser/Thr kinase-associated domain (PASTA; residues 348–629 aa; Figure 1A; Beilharz et al., 2012). Protein sequence alignments between STK and its homolog PknB (Rv0014c in M. tuberculosis H37Rv; NC_000962.3; Boitel et al., 2003) showed that Ser/Thr kinase domains share 43% similarity (Figure 1B). The STK expression was confirmed by Western blot analysis (Figure 1C). The ex vivo kinase assay demonstrated that STK possesses autophosphorylation and substrate phosphorylation activities (Figure 1D).
Figure 1.
Kinase domain of STK in S. suis is conserved compared with other genera. (A) The predicted secondary structure of the STK protein. (B) Multiple sequence alignments of S. suis STK and M. tuberculosis PknB. Sequences used here are separately collected from S. suis 05ZYH33 (CP000407) and M. tuberculosis H37Rv (NC_000962.3). Conserved motifs are shown in boxes and the invariant residues are marked with black bottom color. Other important residues are highlighted in yellow. Red and blue asterisks indicate the catalytic Asp and phosphorylated Thr residues, respectively. (C) SDS-PAGE and Western blot analysis of STK. (D) Kinase activity of STK in vitro. Left lane, STK was incubated with [γ-32P]-ATP; Right lane, STK and MBP were mixed and incubated with [γ-32P]-ATP.
Construction and characterization of Δstk and CΔstk
We constructed a deletion mutant of stk, as confirmed by RT-PCR (Figure S1A) and Western blot (Figure S1B). The RT-PCR showed that the stk upstream and downstream genes transcriptions were not affected by stk deletion. Western blot showed the STK protein did not expressed in Δstk, but presented in SC-19 and CΔstk.
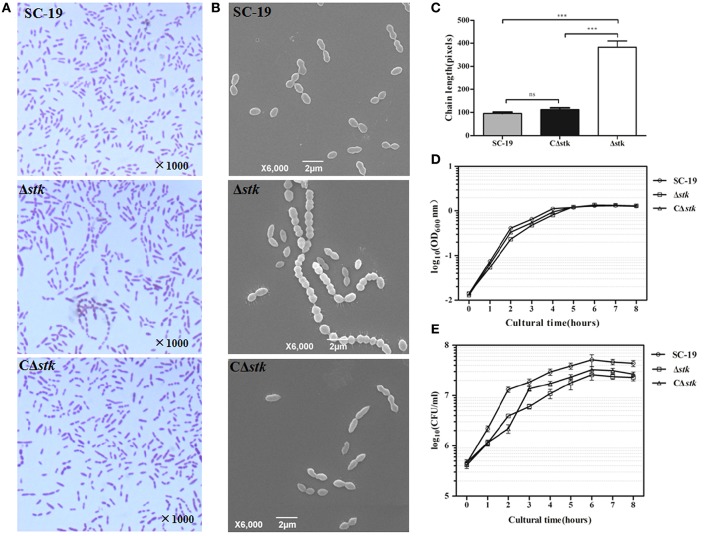
The effect of stk knockout on morphology of S. suis was investigated using Gram staining (Figure 2A) and SEM (Figure 2B). The results revealed that stk inactivation triggered significant chain elongation (Figure 2C).
Figure 2.
Micrographs and growth curves of SC-19, Δstk, and CΔstk. (A) Light microscope morphology of S. suis using Gram staining. (B) Scanning electron micrographs of the strains. (C) The average size of bacterial chains. Data are the result of at least 100 chains analyzed per sample ± Standard Error of Mean. The mutant strain Δstk showed significantly increased chain sizes compared with those of SC-19 (***p < 0.001) and CΔstk (***p < 0.001). (D) Growth curves of the strains. Bacterial cell density was measured spectrometrically at 600 nm. During the exponential phase (2–4 h), AGRs of the three strains displayed no significant difference (SC-19 was 0.51 % per min; AGR of Δstk and CΔstk was 0.49 % per min). (E) CFU count of the strains. The CFU counts showed that Δstk grew much slower than SC-19 and CΔstk during the exponential phase (2–4 h) (AGR of SC-19 was 1.47 × 105 CFU per min; AGR of Δstk was 6.58 × 104 CFU per min; AGR of CΔstk was 1.18 × 105 CFU per min). Data are displayed on a logarithmic scale of Y-axis both in (D) and (E). Results are expressed as log10 mean ± Standard Error of Mean OD or CFU/ml obtained from three independent experiments.
To characterize SC-19, Δstk and CΔstk, we measured growth by detecting OD600 and CFU counts. The AGRs of mid-log phase (2–4 h) displayed no significant difference among the three strains detected by the OD600 value (AGR of SC-19 was 0.51% per min; AGR of Δstk and CΔstk was 0.49% per min; Figure 2D). Whereas, the AGRs of mid-log phase (2–4 h) detected by CFU counts showed that Δstk grew much slower than SC-19 (AGR of SC-19 was 1.47 × 105 CFU per min and Δstk was 6.58 × 104 CFU per min; Figure 2E). Although the growth ability of CΔstk was not fully restored by introducing a complementary plasmid in the mutant, CΔstk grew much faster than Δstk during 2–4 h (AGR of CΔstk was 1.18 × 105 CFU per min).
We tried to use the endogenous promoter of stk gene to construct the complementary strain, but it failed (no expression; the endogenous promoter needs further characterization). Therefore, we used a known promoter in S. suis genome, the promoter of IMPDH (Zhu et al., 2014). Maybe this promoter is not a strong promoter to restore all the phenotypes. Similar facts have also been reported in other studies (Ju et al., 2012).
Δstk pathogenicity was attenuated in mice
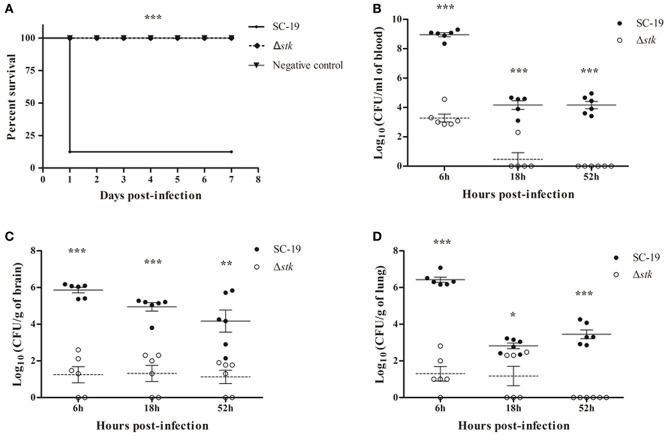
Experimental infection of mice was performed to estimate the difference of viability in vivo between Δstk and SC-19. All SC-19-infected mice displayed severe clinical symptoms, such as septicemia, and most (7/8) died during the first day of infection. By contrast, all Δstk-infected mice displayed slight clinical symptoms and had none mortality during the 7-day observation period (Figure 3A). Therefore, the pathogenicity of Δstk was markedly attenuated.
Figure 3.
Mouse infection models. (A) Survival curves for mice in infection experiment. Eight mice in each group were separately injected intraperitoneally with 1 × 109 CFU/mouse of SC-19 and Δstk. Eight mice were inoculated with saline and served as negative control. Significant difference in survival between different groups was analyzed by Log Rank test (p < 0.001). (B) Bacteria loads in blood, (C) brain, and (D) lungs. Mice were inoculated intraperitoneally with 5 × 108 CFU of a 1:1 mixture of mid-log phase SC-19 and Δstk. The survival strains were enumerated by plating serial dilutions of the samples on selective plates. Data are the result of CFU/ml or CFU/g in different organs analyzed per sample ± Standard Error of Mean. Solid lines, the mean data of SC-19; dotted lines, the mean data of Δstk. Statistical significance was determined using the two-tailed t-test (ns, p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001).
To further evaluate the pathogenicity of Δstk, we determined the differences of viable bacteria in organs using intraperitoneal inoculation. Bacteria were recovered from blood (Figure 3B), brains (Figure 3C), and lungs (Figure 3D) at different time points post-infection. The bacterial loads in blood, brains, and lungs were lower in Δstk than SC-19, from 6 to 52 hpi. The mutant strain was almost cleared at the 52 hpi (Figures 3B–D). These results indicated that the survival ability of Δstk in the host was weakened compared with the parental strain SC-19.
DEGs of Δstk
To get a glimpse of transcriptomic dysregulation of Δstk, gene expression profiles were determined using RNA-Seq for Δstk and SC-19. A total of 2,116 genes were expressed with read counts greater than five in at least one sample. Among these, 539 were differentially expressed (25% of expressed genes) in Δstk compared with SC-19, with 320 up-regulated and 219 down-regulated (p < 0.05 and log transformed fold change > 2.0; Table S2). These genes were involved in diverse physiological activities, including carbohydrate metabolism, amino acid metabolism, nucleotide metabolism, and translation. As an internal control, stk transcript was not detected for Δstk but presented in SC-19.
The effectiveness of the RNA-Seq data was cross-validated using qRT-PCR. Ten genes with various expression levels, as determined by RNA-Seq analysis were chosen for qRT-PCR analysis (Table 2). The correlation between the two methods was high (R2 = 0.903; Figure S2A).
Table 2.
Validation of RNA-Seq results by real-time quantitative RT-PCR (qRT-PCR).
| Gene locus_tag | Function | Fold change of RNA-Seq | Fold change of qRT-PCR |
|---|---|---|---|
| SSU05_0272 | Translation initiation factor 2 | −3.47 | −4.62 ± 1.00 |
| SSU05_0309 | Cation transport ATPase | −2.64 | −3.48 ± 1.16 |
| SSU05_0358 | deoxyguanosinetriphosphate triphosphohydrolase-related protein | 7.10 | 3.23 ± 1.23 |
| SSU05_0792 | Carbamoylphosphate synthase large subunit | 17.58 | 14.17 ± 0.95 |
| SSU05_0906 | NisK | 4.13 | 2.98 ± 1.01 |
| SSU05_1011 | dihydroorotate dehydrogenase, electron transfer subunit | 11.74 | 52.16 ± 1.31 |
| SSU05_1532 | lipoprotein involved thiamine biosynthesis | 2.49 | 5.76 ± 1.24 |
| SSU05_1776 | Permease | −7.31 | −8.11 ± 1.16 |
| SSU05_1815 | Ribonucleases G and E | −7.40 | −12.21 ± 1.01 |
| SSU05_2154 | Succinate dehydrogenase/fumarate reductase, flavoprotein subunit | −5.46 | −3.14 ± 1.30 |
The transcription levels of 10 selected genes were compared between SC-19 and CΔstk related to Δstk (Figure S2B). The differential expression of these target genes of SC-19 and CΔstk showed identical trend (five up-regulated and five down-regulated), although the fold changes were displayed significant different between SC-19 and CΔstk (p < 0.05). This indicates that the STK complementation works but not completely restored.
Functional perturbation in Δstk vs. wild type SC-19
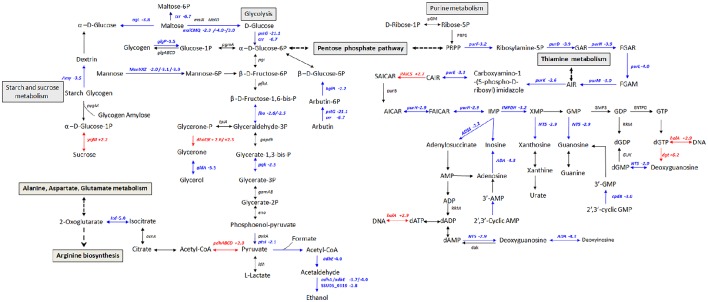
To get an overview of major perturbed functions in Δstk compared with the wild type SC-19, a KEGG pathway enrichment analysis was conducted. Seven metabolic pathways were significantly repressed by stk-deletion (q < 0.1; Table 3). Among these, five pathways, namely, carbohydrate metabolism (ssu00010 and ssu00500), amino acid metabolism (ssu00290), nucleotide metabolism (ssu00230), PTS (ssu02060), as well as the other two pathways (ssu03010 and ssu00970) are crucial for the central metabolism of bacterial cells and are vital in translation. Most genes in the glycolysis were significantly down-regulated, including ptsG (SSU05_0397), crr (SSU05_0398), pgk (SSU05_0157), adhA (SSU05_0279), and adhE (SSU05_0280) (Figure 4). Notably, expression levels of rpsN (SSU05_1535), rpsR (SSU05_1832), rpsP (SSU05_0796), rpsU (SSU05_1433), and glyS (SSU05_1764) translation-associated genes in the pathways of ribosome and aminoacyl-tRNA biosynthesis were decreased in Δstk. A total of 78% (28 out of 36) involved in various PTSs were down-regulated, including manX/Y/Z (SSU05_1778–1780)—encoding mannose transporter, ptsG/crr/bglF (SSU05_0397, SSU05_0398, and SSU05_1490) encoding maltose/glucose transporter, celC (SSU05_0709) encoding cellobiose transporter, and ulaA/ulaB (SSU05_2062, and SSU05_2063) encoding ascorbate transporter. Down-regulation of these pathways and related genes were potentially consistent with the decreased bacterial cell growth in the mutant strain.
Table 3.
Perturbated pathways significantly repressed in the Δstk mutant strain.
| Pathway | q.val | Set.size | Pathway ID |
|---|---|---|---|
| CARBOHYDRATE METABOLISM | |||
| Glycolysis/Gluconeogenesis | 0.086 | 27 | ssu00010 |
| Starch and sucrose metabolism | 0.010 | 38 | ssu00500 |
| TRANSLATION | |||
| Ribosome | 0.002 | 53 | ssu03010 |
| Aminoacyl-tRNA biosynthesis | 0.010 | 25 | ssu00970 |
| MEMBRANE TRANSPORT | |||
| Phosphotransferase system (PTS) | 0.010 | 35 | ssu02060 |
| AMINO ACID METABOLISM | |||
| Valine, leucine, and isoleucine biosynthesis | 0.063 | 11 | ssu00290 |
| NUCLEOTIDE METABOLISM | |||
| Purine metabolism | 0.086 | 54 | ssu00230 |
Figure 4.
Schematic representation of S. suis metabolic pathways differentially regulated in carbohydrate and purine metabolism. Differentially expressed genes encoding proteins involved in glycolysis, starch, sucrose, and purine metabolism. Red color, up-regulated genes; blue color, down-regulated genes.
Virulence-associated genes
The stk deletion significantly reduced virulence of S. suis. Among 180 VAGs predicted by VFDB and known VAGs of S. suis (Fittipaldi et al., 2012), 52 genes were DEGs in Δstk, including 32 down-regulated genes and 20 up-regulated genes (Table 4). These DEGs were mainly assigned into three classes: (i) 10 genes involved in adherence and immune evasion, including 9 down-regulated genes, such as adsA (SSU05_1000), clpC (SSU05_0389), and clpE (SSU05_0390); (ii) 17 genes involved in metal ion uptake, including 11 down-regulated genes, such as hitC (SSU05_1029), fbpC (SSU05_0551), and mgtB (ssu05_1418); (iii) 6 genes involved in ABC-type multidrug transport systems, including 5 up-regulated genes.
Table 4.
Virulence associated genes identified by RNA-Seq.
| Gene name | Locus | Functions | Putative or confirmed function in virulence | Fold change | Up/Down | p-value |
|---|---|---|---|---|---|---|
| hitC | SSU05_1029 | Iron ABC transporter, ATP-binding protein | HitABC | 7.23 | down | 0 |
| hitC | SSU05_1543 | Iron ABC transporter, ATP-binding protein | HitABC | 3.37 | down | 0 |
| hitC | SSU05_0496 | Iron ABC transporter, ATP-binding protein | HitABC | 2.56 | down | 9.77E-77 |
| hitC | SSU05_1882 | Iron ABC transporter, ATP-binding protein | HitABC | 2.66 | down | 8.68E-26 |
| fbpC | SSU05_1253 | Iron ABC transporter, ATP-binding protein | FbpABC | 2.60 | down | 2.89E-77 |
| fbpC | SSU05_0551 | Iron ABC transporter, ATP-binding protein | FbpABC | 3.54 | down | 0 |
| fbpC | SSU05_2067 | Iron ABC transporter, ATP-binding protein | FbpABC | 2.28 | down | 0 |
| fbpC | SSU05_1544 | Iron ABC transporter, ATP-binding protein | FbpABC | 2.16 | down | 4.17E-28 |
| feoB | SSU05_1409 | Ferrous iron transporter | FeoAB | 2.79 | down | 0 |
| mgtB | SSU05_1418 | Mg2+ transport protein | MgtBC | 2.65 | down | 0 |
| mgtB | SSU05_1419 | Mg2+ transport protein | MgtBC | 2.12 | down | 1.08E-93 |
| pilB | SSU05_2100 | Major subunit PilB | PI-2a | 3.42 | down | 1.5E-173 |
| pilB | SSU05_2101 | Major subunit PilB | PI-2a | 3.78 | down | 1.91E-81 |
| sspA | SSU05_0811 | C5a peptidase precursor | C5a peptidase | 2.43 | down | 9.31E-35 |
| sspA | SSU05_0812 | C5a peptidase precursor | C5a peptidase | 2.53 | down | 2.23E-201 |
| permease | SSU05_0798 | Lipid transporter ATP-binding | LOS | 2.08 | down | 2.52E-62 |
| cylG | SSU05_1225 | 3-ketoacyl-ACP-reductase | Beta-hemolysin/cytolysin | 2.01 | down | 0.000471076 |
| cwhA | SSU05_0924 | P60 extracellular protein, Invasion associated protein | p60 | 2.61 | down | 0.0114929 |
| adsA | SSU05_1000 | Adenosine synthase A | AdsA | 2.85 | down | 0 |
| cyaB | SSU05_0800 | Cyclolysin secretion ATP-binding protein | Cya | 3.37 | down | 2.68E-138 |
| bexA | SSU05_0045 | ATP-dependent polysaccharide export protein | Capsule | 3.00 | down | 4.70E-08 |
| comGA | SSU05_0126 | General secretion pathway protein E | xcp secretion system | 2.82 | down | 8.08E-05 |
| clpC | SSU05_0389 | Endopeptidase Clp ATP-binding chain C | ClpC | 5.68 | down | 9.79E-56 |
| clpE | SSU05_0390 | ATP-dependent protease | ClpE | 11.12 | down | 2.26E-95 |
| adhE | SSU05_0280 | acetaldehyde dehydrogenase | Lap | 4.02 | down | 0 |
| purD | SSU05_0033 | Phosphoribosylamine-glycine ligase | Metabolism | 3.87 | down | 0 |
| purA | SSU05_1966 | Adenylosuccinate synthase | Metabolism | 2.15 | down | 0 |
| manN | SSU05_1779 | Mannose-specific PTS | Metabolism | 3.89 | down | 0 |
| fhb | SSU05_0272 | H binding protein | Antiphagocytic | 3.45 | down | 0 |
| ssnA | SSU05_1968 | DNA nuclease | Degradation of host DNA | 3.53 | down | 0 |
| impdh | SSU05_2183 | Inosine 5′-monophosphate dehydrogenase | Metabolism | 3.23 | down | 0 |
| – | SSU05_0550 | Glutamine ABC transporter | unknown | 2.40 | down | 4.77E-178 |
| fbpC | SSU05_0894 | Iron ABC transporter, ATP-binding protein | FbpABC | 4.40 | up | 8.28E-14 |
| fbpC | SSU05_0891 | Iron ABC transporter, ATP-binding protein | FbpABC | 2.14 | up | 0 |
| fbpC | SSU05_0946 | Iron ABC transporter, ATP-binding protein | FbpABC | 3.23 | up | 0 |
| hitC | SSU05_0741 | Iron ABC transporter, ATP-binding protein | HitABC | 3.80 | up | 0 |
| CiaR | SSU05_1095 | Two component system response transcriptional positive regulator | PhoP | 3.43 | up | 3.39E-12 |
| sboF | SSU05_0894 | Two component system response transcriptional positive regulator | PhoP | 4.40 | up | 8.28E-14 |
| fepC | SSU05_0650 | Ferrienterobactin ABC transporter ATPase | Enterobactin | 2.26 | up | 9.90E-13 |
| fepC | SSU05_1669 | Ferrienterobactin ABC transporter ATPase | Enterobactin | 2.14 | up | 0 |
| metQ | SSU05_1771 | Immunogenic lipoprotein A | IlpA | 2.80 | up | 0 |
| cps2J | SSU05_0573 | Glycosyl transferase | Capsule | 2.44 | up | 1.07E-13 |
| sodB | SSU05_1539 | Superoxide dismutase | SodB | 2.02 | up | 0 |
| gtrB | SSU05_1004 | Glycosyltransferase | LPS | 2.78 | up | 2.54E-12 |
| permease | SSU05_0947 | Lipid transporter ATP-binding | LOS | 5.33 | up | 2.44E-15 |
| permease | SSU05_1405 | Lipid transporter ATP-binding | LOS | 3.97 | up | 0 |
| permease | SSU05_1406 | Lipid transporter ATP-binding | LOS | 2.75 | up | 0 |
| permease | SSU05_0911 | Lipid transporter ATP-binding | LOS | 2.61 | up | 0 |
| permease | SSU05_0294 | Lipid transporter ATP-binding | LOS | 2.26 | up | 5.45E-13 |
| LuxS | SSU05_0420 | S-ribosyl homocysteinase | unknown | 2.04 | up | 1.48E-11 |
| GtfA | SSU05_1555 | Glycosidase | unknown | 2.94 | up | 0 |
| – | SSU05_0053 | Transcriptional regulator | unknown | 2.17 | up | 0 |
Analysis of phosphoproteomics
To investigate the Ser/Thr protein kinase activity and the substrates of STK in Δstk and SC-19, we conducted phosphoproteomic analysis. In total, 32 phosphoproteins were detected. Of these 12 were differentially expressed phosphoproteins (DEPPs), including 9 down-regulated phosphoproteins (FC < 0.83) and 3 up-regulated phosphoproteins (FC > 1.2; Table 5; Table S4). As an internal control, auto-phosphorylation of STK was not detected in Δstk. Among 12 DEPPs detected, all of the cell division associated proteins, including FtsA, GpsB, DivIVA, and MapZ were down-regulated. Another protein DnaK, which is a classical molecular chaperone, was down-regulated, as well as the predicted RNA-binding protein Jag. Moreover, down-regulated phosphorylation was detected in the predicted periplasmic solute-binding protein (SSU05_1717), an uncharacterized protein (SSU05_0066), and a hypothetical protein (SSU05_0636). Phosphorylation levels of three proteins, namely translation elongation factor (EF-Tu) involved in protein synthesis, fructose-bisphosphate aldolase (FBA) and GAPDH involved in glycolysis were up-regulated. Moreover, all of these three proteins were also involved in bacteria evasion of host defense, adhesion and invasion.
Table 5.
Differentially expressed phosphoproteins identified by phosphoproteomics.
| Protein name | Locus | Uniprot ID. | Functions | Ratio (Δstk/SC-19) | Phospho RS Site | p-value |
|---|---|---|---|---|---|---|
| FtsA | SSU05_0480 | A0A075SC62 | Actin-like ATPase involved in cell division | 0.212 | Ser310 | 0.043 |
| GpsB | SSU05_0417 | A4VZM2 | Cell division initiation protein | 0.215 | Ser73 | 0.005 |
| DivIVA | SSU05_0487 | B9WXH0 | Cell division initiation protein | 0.209 | Thr199 | 0.001 |
| MapZ | SSU05_0419 | B9WTU4 | Mid-cell-anchored protein Z | 0.057 | Thr66 | 0.000 |
| – | SSU05_1717 | A0A075SSZ7 | Predicted periplasmic solute-binding protein | 0.151 | Thr122/Thr197 | 0.005 |
| Jag | SSU05_2013 | B9WXK1 | Predicted RNA-binding protein | 0.054 | Thr116/Thr158 | 0.045 |
| DnaK | SSU05_0300 | U5UDB7 | Molecular chaperone | 0.531 | Thr591 | 0.029 |
| EF-Tu | SSU05_0530 | A4VZZ3 | Translation elongation factor EF-Tu | 1.442 | Ser58 | 0.017 |
| FBA | SSU05_0338 | A0A0F7FJU8 | Fructose-bisphosphate aldolase | 1.235 | Ser289 | 0.012 |
| GAPDH | SSU05_0155 | A2T9S8 | Glyceraldehyde-3-phosphate dehydrogenase | 1.696 | Thr244 | 0.008 |
| – | SSU05_0066 | A4VYN7 | Uncharacterized protein conserved in bacteria | 0.037 | Thr7 | 0.002 |
| – | SSU05_0636 | B9WVR3 | hypothetical protein | 0.089 | Thr72 | 0.006 |
Discussion
STK provides critical signaling that alters gene expression patterns in S. pneumoniae (Sasková et al., 2007), S. mutans (Banu et al., 2010), and S. pyogenes (Bugrysheva et al., 2011). In S. suis, STK is relevant in stress response and virulence. Down-regulating some VAGs in Δstk mutant strain have been experimentally validated using qRT-PCR (Zhu et al., 2014). In this study, through “-omics” approaches, we found that S. suis STK can regulate expression of genes involved in bacterial central metabolism and virulence. Moreover, STK directly or indirectly affects phosphorylation of 12 proteins that are involved in cell division, glycolysis, and translation. These findings can explain the elongation of bacterial chain length, attenuated growth and virulence of stk deleted strain.
Regulation by STK on cell growth and division
Inactivating S. suis STK affects the bacterial phenotypes, including cell chain length and growth kinetics (Figure 2). Consistently, the results have been reported in a previous study (Zhu et al., 2014). Several reports have also stated that stk-deficient could affect cell shapes and cell sizes in other Gram-positive bacteria, such as S. pneumoniae (Echenique et al., 2004; Fleurie et al., 2012), S. aureus (Donat et al., 2009), S. mutans (Banu et al., 2010), and M. tuberculosis (Kang et al., 2005).
In our study, we have used both OD600 value and CFU counts to measure the growth rate of SC-19, Δstk, and CΔstk. Through these two methods, we found during the mid-log phase, the growth rate of the three strains detected by OD600 value displayed no significant difference (Figure 2D), whereas the CFU counts showed the growth rate of Δstk was much slower than SC-19 and CΔstk during the mid-log phase (Figure 2E). Similar phenomenon has been also appeared in our previous study (Gao et al., 2016). The longer chains of the mutant strain may change the light-scattering properties of S. suis (Zheng et al., 2011) and other bacteria (Sha et al., 2004). Hence, growth detection through OD value was not adequate, and the CFU determination can accurately reflect the growth ability.
Phosphorylation of factors involved in cell division could function as an internal clock that regulates the sequence and timing of the cell cycle events (Grangeasse and Lesterlin, 2015). According to the phosphoproteomic analysis in this study, phosphoproteins associated with cell growth and division, such as DivIVA and MapZ, were down-regulated in Δstk (Table 5). This provides an explanation for the weakened growth ability and elongated chain length of Δstk. DivIVA has been identified as a STK-substrate in S. pneumoniae (Nováková et al., 2010). Moreover, S. pneumoniae with the non-phosphoablative form of DivIVA possess an elongated shape with a polar bulge and aberrant spatial organization of nascent peptidoglycan (Fleurie et al., 2012). These findings suggest that phosphoablative form of DivIVA is crucial for maintaining cell shape and division. MapZ is a newly discovered cell division protein essential in proper septum placement and likely functions as a marker of the cell division site (Holecková et al., 2014). S. pneumoniae with a non-phosphoablative form of MapZ exhibited cell shape and viability defects (Fleurie et al., 2014). Similar functions of DivIVA and MapZ may exist in S. suis. Hence, STK of S. suis may influence the cell growth and division of S. suis by regulating phosphorylation of DivIVA and MapZ. The other two down-regulated phosphoproteins in Δstk, FtsA, and GpsB involved in cell division, may also contribute to the STK regulated cell division process. In B. subtilis, GpsB is phosphorylated by STK and its phosphorylation regulates STK activity through a negative feedback loop (Pompeo et al., 2015). In E. coli, FtsA is crucial in recruiting of FtsZ filaments to the membrane and negative regulated FtsZ organization (Loose and Mitchison, 2014).
Regulation by STK on virulence
The deletion of stk in S. suis also resulted in the alteration of bacterial pathogenecity. This attenuation may result from the impaired growth of Δstk, and also can be due to direct effect on the expression of virulence genes. As previously reported, stk-deletion strain displayed reduced ability to adhere to and invade in epithelial cells and increased sensitivity to phagocytosis (Zhu et al., 2014). In this study, analysis of gene expression profiles indicated that several genes associated with adherence and immune evasion were repressed in Δstk (Table 4), such as adsA (−2.85-fold), clpC (−5.68-fold) and clpE (−11.12-fold). The gene adsA encodes an adenosine synthase AdsA. The adsA mutant of B. anthracis was easier to be cleared compared with the wild-type strain, implicating its role in immune evasion in the host (Thammavongsa et al., 2009). The gene clpC encodes a heat shock protein ClpC with ATPase activity. In L. monocytogenes, ClpC is a general stress response protein required for in vivo survival by promoting early bacterial escape from the phagosome of macrophages (Rouquette et al., 1998). Moreover, ClpC is required for adhesion and invasion. A clpC-deficient mutant of S. pneumoniae displayed decreased expression of CbpA (a structural adhesion) and pneumolysin (Nair et al., 2000). clpE acts synergistically with clpC in virulence and the clpE mutant of L. monocytogenes exhibited a significant reduction in virulence (Nair et al., 1999). Therefore, our results indicated that these down-regulated VAGs potentially contributed to decreased adherence to epithelial cells, increased immune evasion and increased sensitivity to phagocytosis of Δstk.
The Δstk mutants displayed defects in their ability to adapt to various environmental conditions, such as high temperature, high osmotic, oxidative, and low acidic pH stress (Zhu et al., 2014). These may due to the down-regulation of 11 genes encoding iron ABC transporters (HitC/FbpC, MgtB) that mediate the uptake of nutrients and several metabolism-related enzymes (i.e., ManN, PurA, and IMPDH; Table 4). S. suis requires nutrients, including trace metals, whose availability within the infected host is relatively low. The hitC/fbpC operon encodes an iron transport system that is responsible for utilizing iron bound to transferrin or iron chelates (Sanders et al., 1994). Mg2+ transporters encoded by mgtB are unique transport systems with unusual mechanisms for mediating Mg2+ movement through the membrane (Moncrief and Maguire, 1999). manN (−3.89-fold), whose product is the component of the mannose-specific PTS, can regulate hemolysin gene expression (Wilson et al., 2007). Down-regulation of these VAGs may hinder the acquisition of nutrients of S. suis, hence decrease the adaptation of S. suis to various conditions.
Other VAGs of S. suis, such as fhb (−3.45-fold) and ssnA (−3.53-fold), have been identified previously. The gene fhb encodes a factor H binding protein, which is a major antiphagocytic factor in preventing C3b deposition and blocking activation of the alternative pathway of the complement system (Pian et al., 2012). The cell wall-anchored DNase encoded by ssnA plays a role in disruption of neutrophil extracellular traps (De Buhr et al., 2014). Down-regulation of fhb and ssnA may be associated with the attenuated virulence.
According to the phosphoproteomic analysis, the phosphorylation levels of three proteins associated with virulence were up-regulated in the Δstk (Table 4). They are EF-Tu, GAPDH, and FBA. Interestingly, all of them are the bacterial outer surface proteins that contribute to evasion of host defense, adhesion and invasion (Tan et al., 2008; Li Q. et al., 2015). Ser/Thr phosphorylation of EF-Tu has been reported in E. coli, Thermus thermophilus, Streptomyces coelicolor, and B. subtilis (Pereira et al., 2011). EF-Tu phosphorylation could accelerate its release from the translation site (Absalon et al., 2009). In B. subtilis, phosphorylation impairs the essential GTPase activity of EF-Tu by preventing its release from the ribosome, leading to the overall protein synthesis inhibition (Pereira et al., 2015). However, the role of EF-Tu phosphorylation in pathogen virulence has not been reported. GAPDH is the surface expression protein with the ability to bind albumin (Brassard et al., 2004), which serves as a receptor for the plasmin receptor and contributes to host cytoskeletal protein binding and signal transduction between bacteria and host tissues (Gase et al., 1996; Jobin et al., 2004). FBA catalyzes the reversible reaction of fructose 1,6-bisphosphate transferring into glyceraldehydes-3-phosphate and dihydroxyacetone (Jado et al., 1999). The effects of GAPDH and FBA phosphorylation on virulence remain unknown. The biological significance of phosphorylation modulation by STK on these virulence associated proteins deserves further investigations.
Regulation by STK on metabolism
In our study, evaluating of the transcriptional status of gene pathways revealed that seven pathways are significantly repressed in Δstk (Table 3). These repressions could be an important reason of the impaired growth of Δstk.
Glycolysis is the most critical phase in glucose metabolism to produce energy in the form of ATP during bacterial cellular respiration (Kresge et al., 2005; Mulukutla et al., 2014). Our study found more than half of the genes (17 out of 27) involved in glycolysis were repressed in Δstk, such as crr (−6.67-fold), bglA (−4.61-fold), and pstG (−20.63-fold). The gene crr, which encodes a glucose-specific phosphotransferase enzyme component for glucose transport into the cytoplasm, is involved in glycolysis, starch and sucrose metabolism (Han et al., 2007). bglA encodes a 6-phospho-β-glucosidase to convert host derived-sugars into usable monosaccharaides that contribute to energy generation for bacterial survival (Terra et al., 2015). pstG codes for a glucose transporter that enables catalysis of glucose to transform into glucose-6p (Buhr et al., 1994). Down-regulating the enzymes mentioned above in Δstk may hinder glucose utilization, nutrient storage, and ATP synthesis.
More than two thirds of the genes (34 out of 54) involved in the purine metabolism pathway were repressed in Δstk (Table S3). Previous studies have emphasized the importance of nucleotide biosynthesis in bacteria. Auxotroph mutants of nucleotide biosynthesis in Salmonella (McFarland and Stocker, 1987), S. aureus (Mei et al., 1997), and S. pneumoniae (Polissi et al., 1998) have shown impaired growth.
Our results showed that translation-associated pathways (ssu00970 and ssu03010) are the most affected pathways in Δstk, which has not been reported in other bacteria (Table 3). In Δstk, 88% of the genes involved in ribosomes (ssu03010) were decreased (46 out of 52), and in aminoacyl-tRNA biosynthesis (ssu00970), 84% of the genes were decreased (21 out of 25). The ribosome plays a major role in determining the overall gene expression profile of the cell (Starosta et al., 2014). Meanwhile, glyS (−2.51-fold) encoding a glycyl-tRNA synthetase has been suggested to be involved in DNA stabilization under extreme environmental conditions (Kumar et al., 2011). It has been previously shown that alterations of genes involved in translation can provoke slower growth of E. coli (Ruusala et al., 1984; Starosta et al., 2014).
Phosphoproteome analysis in this study has revealed that the phosphorylations of two glycolytic enzymes (FBA, GAPDH) and a translation asscociated protein EF-Tu were up-regulated (Table 4). These proteins are also involved in S. suis virulence, which have been discussed above. The phosphorylation of a chaperone DnaK was down-regulated in Δstk. DnaK is induced in E. coli upon heat shock and promotes ATP-dependent refolding or degradation of damaged proteins (Sherman and Goldberg, 1993). Previously, in vivo phosphorylation has been shown to enhance DnaK binding to polypeptide substrates (Panagiotidis et al., 1994). Modulations of phosphorylation levels on these proteins may also lead to impaired metabolism of Δstk.
Conclusion
In our study, genome-wide transcriptional analyses identified 32 down-regulated VAGs in Δstk compared with the wild type strain. Additionally, seven pathways were significantly repressed in Δstk, particularly the translation-associated pathways. Phosphoproteomic analysis found that post-translational modifications of 12 proteins were significantly affected in Δstk. These DEPPs are involved in various biological processes, including cell growth and division, glycolysis, and translation. Consistently, phenotypic assays confirmed that the Δstk strain displayed deficient growth and attenuated pathogenicity. Therefore, STK-mediated signal transduction is important in cell growth and division, and metabolism of S. suis.
Author contributions
RZ conceived and designed this project and experiments. CZ, WS, MT, MD, and WL performed the experiments. CZ, TG, LL analyzed the data and contributed to the development of the figures and tables. CZ, RZ, and ZX wrote the manuscript. All authors reviewed the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Key Research and Development Plans, the National Basic Research Program of China (973 Program grant No. 2012CB518802), and Hubei Province Natural Science Foundation for Innovative Research Groups (2016CFA015). We are grateful to Dr. Yosuke Murakami for his pSET plasmids. We would like to thank Shanghai Applied Protein Technology Co. Ltd. for providing technical support.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2017.00066/full#supplementary-material
References
- Absalon C., Obuchowski M., Madec E., Delattre D., Holland I. B., Séror S. J. (2009). CpgA, EF-Tu and the stressosome protein YezB are substrates of the Ser/Thr kinase/phosphatase couple, PrkC/PrpC, in Bacillus subtilis. Microbiology 155, 932–943. 10.1099/mic.0.022475-0 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambaud C., Gouin E., Pizarro-Cerda J., Cossart P., Dussurget O. (2005). Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence of Listeria monocytogenes. Mol. Microbiol. 56, 383–396. 10.1111/j.1365-2958.2005.04551.x [DOI] [PubMed] [Google Scholar]
- Arora G., Sajid A., Gupta M., Bhaduri A., Kumar P., Basu-Modak S., et al. (2010). Understanding the role of PknJ in Mycobacterium tuberculosis: biochemical characterization and identification of novel substrate pyruvate kinase A. PLoS ONE 5:e10772. 10.1371/journal.pone.0010772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S., Claverie J. M. (1997). The significance of digital gene expression profiles. Genome Res. 7, 986–995. [DOI] [PubMed] [Google Scholar]
- Banu L. D., Conrads G., Rehrauer H., Hussain H., Allan E., Van Der Ploeg J. R. (2010). The Streptococcus mutans serine/threonine kinase, PknB, regulates competence development, bacteriocin production, and cell wall metabolism. Infect. Immun. 78, 2209–2220. 10.1128/IAI.01167-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz K., Nováková L., Fadda D., Branny P., Massidda O., Veening J. W. (2012). Control of cell division in Streptococcus pneumoniae by the conserved Ser/Thr protein kinase StkP. Proc. Natl. Acad. Sci. U.S.A. 109, E905–913. 10.1073/pnas.1119172109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitel B., Ortiz-Lombardía M., Durán R., Pompeo F., Cole S. T., Cervenansky C., et al. (2003). PknB kinase activity is regulated by phosphorylation in two Thr residues and dephosphorylation by PstP, the cognate phospho-Ser/Thr phosphatase, in Mycobacterium tuberculosis. Mol. Microbiol. 49, 1493–1508. 10.1046/j.1365-2958.2003.03657.x [DOI] [PubMed] [Google Scholar]
- Brassard J., Gottschalk M., Quessy S. (2004). Cloning and purification of the Streptococcus suis serotype 2 glyceraldehyde-3-phosphate dehydrogenase and its involvement as an adhesin. Vet. Microbiol. 102, 87–94. 10.1016/j.vetmic.2004.05.008 [DOI] [PubMed] [Google Scholar]
- Bugrysheva J., Froehlich B. J., Freiberg J. A., Scott J. R. (2011). Serine/threonine protein kinase Stk is required for virulence, stress response, and penicillin tolerance in Streptococcus pyogenes. Infect. Immun. 79, 4201–4209. 10.1128/IAI.05360-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr A., Flükiger K., Erni B. (1994). The glucose transporter of Escherichia coli. Overexpression, purification, and characterization of functional domains. J. Biol. Chem. 269, 23437–23443. [PubMed] [Google Scholar]
- Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328. 10.1093/nar/gki008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Long H., Gao P., Deng G., Pan Z., Liang J., et al. (2014). Transcriptome assembly and analysis of Tibetan Hulless Barley (Hordeum vulgare L. var. nudum) developing grains, with emphasis on quality properties. PLoS ONE 9:e98144. 10.1371/journal.pone.0098144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalia A. B., Weiser J. N. (2011). Minimization of bacterial size allows for complement evasion and is overcome by the agglutinating effect of antibody. Cell Host Microbe 10, 486–496. 10.1016/j.chom.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buhr N., Neumann A., Jerjomiceva N., Von Köckritz-Blickwede M., Baums C. G. (2014). Streptococcus suis DNase SsnA contributes to degradation of neutrophil extracellular traps (NETs) and evasion of NET-mediated antimicrobial activity. Microbiology 160, 385–395. 10.1099/mic.0.072199-0 [DOI] [PubMed] [Google Scholar]
- Deol P., Vohra R., Saini A. K., Singh A., Chandra H., Chopra P., et al. (2005). Role of Mycobacterium tuberculosis Ser/Thr kinase PknF: implications in glucose transport and cell division. J. Bacteriol. 187, 3415–3420. 10.1128/JB.187.10.3415-3420.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donat S., Streker K., Schirmeister T., Rakette S., Stehle T., Liebeke M., et al. (2009). Transcriptome and functional analysis of the eukaryotic-type serine/threonine kinase PknB in Staphylococcus aureus. J. Bacteriol. 191, 4056–4069. 10.1128/JB.00117-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echenique J., Kadioglu A., Romao S., Andrew P. W., Trombe M. C. (2004). Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect. Immun. 72, 2434–2437. 10.1128/IAI.72.4.2434-2437.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S., Meng Y., Song M., Pang C., Wei H., Liu J., et al. (2014). Quantitative phosphoproteomics analysis of nitric oxide-responsive phosphoproteins in cotton leaf. PLoS ONE 9:e94261. 10.1371/journal.pone.0094261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez P., Saint-Joanis B., Barilone N., Jackson M., Gicquel B., Cole S. T., et al. (2006). The Ser/Thr protein kinase PknB is essential for sustaining mycobacterial growth. J. Bacteriol. 188, 7778–7784. 10.1128/JB.00963-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fittipaldi N., Segura M., Grenier D., Gottschalk M. (2012). Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 7, 259–279. 10.2217/fmb.11.149 [DOI] [PubMed] [Google Scholar]
- Fleurie A., Cluzel C., Guiral S., Freton C., Galisson F., Zanella-Cleon I., et al. (2012). Mutational dissection of the S/T-kinase StkP reveals crucial roles in cell division of Streptococcus pneumoniae. Mol. Microbiol. 83, 746–758. 10.1111/j.1365-2958.2011.07962.x [DOI] [PubMed] [Google Scholar]
- Fleurie A., Lesterlin C., Manuse S., Zhao C., Cluzel C., Lavergne J. P., et al. (2014). MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature 516, 259–262. 10.1038/nature13966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Higdon R., Kolker E. (2010). Interplay of heritage and habitat in the distribution of bacterial signal transduction systems. Mol. Biosyst. 6, 721–728. 10.1039/b908047c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Makarova K. S., Wolf Y. I., Koonin E. V. (2015). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269. 10.1093/nar/gku1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T., Tan M., Liu W., Zhang C., Zhang T., Zheng L., et al. (2016). GidA, a tRNA modification enzyme, contributes to the growth, and virulence of Streptococcus suis serotype 2. Front. Cell. Infect. Microbiol. 6:44. 10.3389/fcimb.2016.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gase K., Gase A., Schirmer H., Malke H. (1996). Cloning, sequencing and functional overexpression of the Streptococcus equisimilis H46A gapC gene encoding a glyceraldehyde-3-phosphate dehydrogenase that also functions as a plasmin(ogen)-binding protein. Purification and biochemical characterization of the protein. Eur. J. Biochem. 239, 42–51. 10.1111/j.1432-1033.1996.0042u.x [DOI] [PubMed] [Google Scholar]
- Grangeasse C., Lesterlin C. (2015). Mapping mid-cell: mapZ shows the way. Cell Cycle 14, 937–938. 10.1080/15384101.2015.1010978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K. Y., Seo H. S., Song J. A., Ahn K. Y., Park J. S., Lee J. (2007). Transport proteins PotD and Crr of Escherichia coli, novel fusion partners for heterologous protein expression. Biochim. Biophys. Acta 1774, 1536–1543. 10.1016/j.bbapap.2007.09.012 [DOI] [PubMed] [Google Scholar]
- Hoch J. A. (2000). Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3, 165–170. 10.1016/S1369-5274(00)00070-9 [DOI] [PubMed] [Google Scholar]
- Holecková N., Doubravová L., Massidda O., Molle V., Buriánková K., Benada O., et al. (2014). LocZ is a new cell division protein involved in proper septum placement in Streptococcus pneumoniae. MBio 6, e01700–e01714. 10.1128/mBio.01700-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Liu Y., Yu K., Liu X. (2014). Decreasing the amount of trypsin in in-gel digestion leads to diminished chemical noise and improved protein identifications. J. Proteomics 109, 16–25. 10.1016/j.jprot.2014.06.017 [DOI] [PubMed] [Google Scholar]
- Hussain H., Branny P., Allan E. (2006). A eukaryotic-type serine/threonine protein kinase is required for biofilm formation, genetic competence, and acid resistance in Streptococcus mutans. J. Bacteriol. 188, 1628–1632. 10.1128/JB.188.4.1628-1632.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nariya H. (2008). Dual regulation with Ser/Thr kinase cascade and a His/Asp TCS in Myxococcus xanthus. Adv. Exp. Med. Biol. 631, 111–121. 10.1007/978-0-387-78885-2_7 [DOI] [PubMed] [Google Scholar]
- Jado I., Fenoll A., Cepeda T., Casal J., Pérez A. (1999). Cloning, sequencing, and chromosomal location of a putative class-II aldolase gene from Streptococcus pneumoniae. Curr. Microbiol. 39, 31–36. 10.1007/PL00006823 [DOI] [PubMed] [Google Scholar]
- Jin H., Pancholi V. (2006). Identification and biochemical characterization of a eukaryotic-type serine/threonine kinase and its cognate phosphatase in Streptococcus pyogenes: their biological functions and substrate identification. J. Mol. Biol. 357, 1351–1372. 10.1016/j.jmb.2006.01.020 [DOI] [PubMed] [Google Scholar]
- Jobin M. C., Brassard J., Quessy S., Gottschalk M., Grenier D. (2004). Acquisition of host plasmin activity by the Swine pathogen Streptococcus suis serotype 2. Infect. Immun. 72, 606–610. 10.1128/IAI.72.1.606-610.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C. X., Gu H. W., Lu C. P. (2012). Characterization and functional analysis of atl, a novel gene encoding autolysin in Streptococcus suis. J. Bacteriol. 194, 1464–1473. 10.1128/JB.06231-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C. M., Abbott D. W., Park S. T., Dascher C. C., Cantley L. C., Husson R. N. (2005). The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19, 1692–1704. 10.1101/gad.1311105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresge N., Simoni R. D., Hill R. L. (2005). Otto Fritz Meyerhof and the elucidation of the glycolytic pathway. J. Biol. Chem. 280:e3. [PubMed] [Google Scholar]
- Kumar M., Khan F. G., Sharma S., Kumar R., Faujdar J., Sharma R., et al. (2011). Identification of Mycobacterium tuberculosis genes preferentially expressed during human infection. Microb. Pathog. 50, 31–38. 10.1016/j.micpath.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Lee P. C., Umeyama T., Horinouchi S. (2002). afsS is a target of AfsR, a transcriptional factor with ATPase activity that globally controls secondary metabolism in Streptomyces coelicolor A3(2). Mol. Microbiol. 43, 1413–1430. 10.1046/j.1365-2958.2002.02840.x [DOI] [PubMed] [Google Scholar]
- Li J., Shi Y., Toga A. W. (2015). Controlling false discovery rate in signal space for transformation-invariant thresholding of statistical maps. Inf. Process. Med. Imaging 24, 125–136. 10.1007/978-3-319-19992-4_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Liu H., Du D., Yu Y., Ma C., Jiao F., et al. (2015). Identification of novel laminin- and fibronectin-binding proteins by far-western blot: capturing the adhesins of Streptococcus suis type 2. Front. Cell. Infect. Microbiol. 5:82. 10.3389/fcimb.2015.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Hu X., Liu L., Chen H., Zhou R. (2011). Induction of protective immune response against Streptococcus suis serotype 2 infection by the surface antigen HP0245. FEMS Microbiol. Lett. 316, 115–122. 10.1111/j.1574-6968.2010.02200.x [DOI] [PubMed] [Google Scholar]
- Li W., Liu L., Chen H., Zhou R. (2009). Identification of Streptococcus suis genes preferentially expressed under iron starvation by selective capture of transcribed sequences. FEMS Microbiol. Lett. 292, 123–133. 10.1111/j.1574-6968.2008.01476.x [DOI] [PubMed] [Google Scholar]
- Liu Q., Fan J., Niu C., Wang D., Wang J., Wang X., et al. (2011). The eukaryotic-type serine/threonine protein kinase Stk is required for biofilm formation and virulence in Staphylococcus epidermidis. PLoS ONE 6:e25380. 10.1371/journal.pone.0025380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomas-Lopez R., Paracuellos P., Riberty M., Cozzone A. J., Duclos B. (2007). Several enzymes of the central metabolism are phosphorylated in Staphylococcus aureus. FEMS Microbiol. Lett. 272, 35–42. 10.1111/j.1574-6968.2007.00742.x [DOI] [PubMed] [Google Scholar]
- Loose M., Mitchison T. J. (2014). The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 16, 38–46. 10.1038/ncb2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun Z. R., Wang Q. P., Chen X. G., Li A. X., Zhu X. Q. (2007). Streptococcus suis: an emerging zoonotic pathogen. Lancet Infect. Dis. 7, 201–209. 10.1016/S1473-3099(07)70001-4 [DOI] [PubMed] [Google Scholar]
- Luo W., Brouwer C. (2013). Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831. 10.1093/bioinformatics/btt285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Friedman M. S., Shedden K., Hankenson K. D., Woolf P. J. (2009). GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10:161. 10.1186/1471-2105-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madec E., Laszkiewicz A., Iwanicki A., Obuchowski M., Seror S. (2002). Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol. Microbiol. 46, 571–586. 10.1046/j.1365-2958.2002.03178.x [DOI] [PubMed] [Google Scholar]
- Mata-Cabana A., Garcia-Dominguez M., Florencio F. J., Lindahl M. (2012). Thiol-based redox modulation of a cyanobacterial eukaryotic-type serine/threonine kinase required for oxidative stress tolerance. Antioxid. Redox Signal. 17, 521–533. 10.1089/ars.2011.4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland W. C., Stocker B. A. (1987). Effect of different purine auxotrophic mutations on mouse-virulence of a Vi-positive strain of Salmonella dublin and of two strains of Salmonella typhimurium. Microb. Pathog. 3, 129–141. 10.1016/0882-4010(87)90071-4 [DOI] [PubMed] [Google Scholar]
- Mei J. M., Nourbakhsh F., Ford C. W., Holden D. W. (1997). Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26, 399–407. 10.1046/j.1365-2958.1997.5911966.x [DOI] [PubMed] [Google Scholar]
- Moncrief M. B., Maguire M. E. (1999). Magnesium transport in prokaryotes. J. Biol. Inorg. Chem. 4, 523–527. 10.1007/s007750050374 [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Mulukutla B. C., Yongky A., Daoutidis P., Hu W. S. (2014). Bistability in glycolysis pathway as a physiological switch in energy metabolism. PLoS ONE 9:e98756. 10.1371/journal.pone.0098756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Dorado J., Inouye S., Inouye M. (1991). A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a gram-negative bacterium. Cell 67, 995–1006. 10.1016/0092-8674(91)90372-6 [DOI] [PubMed] [Google Scholar]
- Nádvorník R., Vomastek T., Janecek J., Techniková Z., Branny P. (1999). Pkg2, a novel transmembrane protein Ser/Thr kinase of Streptomyces granaticolor. J. Bacteriol. 181, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S., Frehel C., Nguyen L., Escuyer V., Berche P. (1999). ClpE, a novel member of the HSP100 family, is involved in cell division and virulence of Listeria monocytogenes. Mol. Microbiol. 31, 185–196. 10.1046/j.1365-2958.1999.01159.x [DOI] [PubMed] [Google Scholar]
- Nair S., Milohanic E., Berche P. (2000). ClpC ATPase is required for cell adhesion and invasion of Listeria monocytogenes. Infect. Immun. 68, 7061–7068. 10.1128/IAI.68.12.7061-7068.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu J. M., Macmillan S. V., Nodwell J. R., Wright G. D. (2002). StoPK-1, a serine/threonine protein kinase from the glycopeptide antibiotic producer Streptomyces toyocaensis NRRL 15009, affects oxidative stress response. Mol. Microbiol. 44, 417–430. 10.1046/j.1365-2958.2002.02879.x [DOI] [PubMed] [Google Scholar]
- Nováková L., Bezousková S., Pompach P., Spidlová P., Saskova L., Weiser J., et al. (2010). Identification of multiple substrates of the StkP Ser/Thr protein kinase in Streptococcus pneumoniae. J. Bacteriol. 192, 3629–3638. 10.1128/JB.01564-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková L., Sasková L., Pallová P., Janecek J., Novotná J., Ulrych A., et al. (2005). Characterization of a eukaryotic type serine/threonine protein kinase and protein phosphatase of Streptococcus pneumoniae and identification of kinase substrates. FEBS J. 272, 1243–1254. 10.1111/j.1742-4658.2005.04560.x [DOI] [PubMed] [Google Scholar]
- Panagiotidis C. A., Burkholder W. F., Gaitanaris G. A., Gragerov A., Gottesman M. E., Silverstein S. J. (1994). Inhibition of DnaK autophosphorylation by heat shock proteins and polypeptide substrates. J. Biol. Chem. 269, 16643–16647. [PubMed] [Google Scholar]
- Pereira S. F., Gonzalez R. L., Jr., Dworkin J. (2015). Protein synthesis during cellular quiescence is inhibited by phosphorylation of a translational elongation factor. Proc. Natl. Acad. Sci. U.S.A. 112, E3274–E3281. 10.1073/pnas.1505297112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira S. F., Goss L., Dworkin J. (2011). Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol. Mol. Biol. Rev. 75, 192–212. 10.1128/MMBR.00042-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pian Y., Gan S., Wang S., Guo J., Wang P., Zheng Y., et al. (2012). Fhb, a novel factor H-binding surface protein, contributes to the antiphagocytic ability and virulence of Streptococcus suis. Infect. Immun. 80, 2402–2413. 10.1128/IAI.06294-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polissi A., Pontiggia A., Feger G., Altieri M., Mottl H., Ferrari L., et al. (1998). Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66, 5620–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompeo F., Foulquier E., Serrano B., Grangeasse C., Galinier A. (2015). Phosphorylation of the cell division protein GpsB regulates PrkC kinase activity through a negative feedback loop in Bacillus subtilis. Mol. Microbiol. 97, 139–150. 10.1111/mmi.13015 [DOI] [PubMed] [Google Scholar]
- Rajagopal L., Clancy A., Rubens C. E. (2003). A eukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J. Biol. Chem. 278, 14429–14441. 10.1074/jbc.M212747200 [DOI] [PubMed] [Google Scholar]
- Rouquette C., De Chastellier C., Nair S., Berche P. (1998). The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol. Microbiol. 27, 1235–1245. 10.1046/j.1365-2958.1998.00775.x [DOI] [PubMed] [Google Scholar]
- Ruggiero A., De Simone P., Smaldone G., Squeglia F., Berisio R. (2012). Bacterial cell division regulation by Ser/Thr kinases: a structural perspective. Curr. Protein Pept. Sci. 13, 756–766. 10.2174/138920312804871201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruusala T., Andersson D., Ehrenberg M., Kurland C. G. (1984). Hyper-accurate ribosomes inhibit growth. EMBO J. 3, 2575–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajid A., Arora G., Singhal A., Kalia V. C., Singh Y. (2015). Protein phosphatases of pathogenic bacteria: role in physiology and virulence. Annu. Rev. Microbiol. 69, 527–547. 10.1146/annurev-micro-020415-111342 [DOI] [PubMed] [Google Scholar]
- Sanders J. D., Cope L. D., Hansen E. J. (1994). Identification of a locus involved in the utilization of iron by Haemophilus influenzae. Infect. Immun. 62, 4515–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasková L., Novaková L., Basler M., Branny P. (2007). Eukaryotic-type serine/threonine protein kinase StkP is a global regulator of gene expression in Streptococcus pneumoniae. J. Bacteriol. 189, 4168–4179. 10.1128/JB.01616-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai R., Suzuki A., Takano Y., Lee P. C., Horinouchi S. (2004). Phosphorylation of AfsR by multiple serine/threonine kinases in Streptomyces coelicolor A3(2). Gene 334, 53–61. 10.1016/j.gene.2004.02.046 [DOI] [PubMed] [Google Scholar]
- Sha J., Kozlova E. V., Fadl A. A., Olano J. P., Houston C. W., Peterson J. W., et al. (2004). Molecular characterization of a glucose-inhibited division gene, gidA, that regulates cytotoxic enterotoxin of Aeromonas hydrophila. Infect. Immun. 72, 1084–1095. 10.1128/IAI.72.2.1084-1095.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M. Y., Goldberg A. L. (1993). Heat shock of Escherichia coli increases binding of dnaK (the hsp70 homolog) to polypeptides by promoting its phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 90, 8648–8652. 10.1073/pnas.90.18.8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestroni A., Jewell K. A., Lin W. J., Connelly J. E., Ivancic M. M., Tao W. A., et al. (2009). Identification of serine/threonine kinase substrates in the human pathogen group B streptococcus. J. Proteome Res. 8, 2563–2574. 10.1021/pr900069n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. E., Damman M., van der Velde J., Wagenaar F., Wisselink H. J., Stockhofe-Zurwieden N., et al. (1999). Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 67, 1750–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starosta A. L., Lassak J., Jung K., Wilson D. N. (2014). The bacterial translation stress response. FEMS Microbiol. Rev. 38, 1172–1201. 10.1111/1574-6976.12083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Stock A. M., Mottonen J. M. (1990). Signal transduction in bacteria. Nature 344, 395–400. 10.1038/344395a0 [DOI] [PubMed] [Google Scholar]
- Takamatsu D., Osaki M., Sekizaki T. (2001a). Construction and characterization of Streptococcus suis-Escherichia coli shuttle cloning vectors. Plasmid 45, 101–113. 10.1006/plas.2000.1510 [DOI] [PubMed] [Google Scholar]
- Takamatsu D., Osaki M., Sekizaki T. (2001b). Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid 46, 140–148. 10.1006/plas.2001.1532 [DOI] [PubMed] [Google Scholar]
- Tan C., Liu M., Jin M., Liu J., Chen Y., Wu T., et al. (2008). The key virulence-associated genes of Streptococcus suis type 2 are upregulated and differentially expressed in vivo. FEMS Microbiol. Lett. 278, 108–114. 10.1111/j.1574-6968.2007.00980.x [DOI] [PubMed] [Google Scholar]
- Tan M. F., Gao T., Liu W. Q., Zhang C. Y., Yang X., Zhu J. W., et al. (2015). MsmK, an ATPase, contributes to utilization of multiple carbohydrates and host colonization of Streptococcus suis. PLoS ONE 10:e0130792. 10.1371/journal.pone.0130792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terra V. S., Zhi X., Kahya H. F., Andrew P. W., Yesilkaya H. (2015). Pneumococcal 6-phospho-beta-glucosidase (BglA3) is involved in virulence and nutrient metabolism. Infect. Immun. 84, 286–292. 10.1128/IAI.01108-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thammavongsa V., Kern J. W., Missiakas D. M., Schneewind O. (2009). Staphylococcus aureus synthesizes adenosine to escape host immune responses. J. Exp. Med. 206, 2417–2427. 10.1084/jem.20090097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Carlier C., Poyart-Salmeron C., Courvalin P. (1991). Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 102, 99–104. 10.1016/0378-1119(91)90546-N [DOI] [PubMed] [Google Scholar]
- Truong-Bolduc Q. C., Ding Y., Hooper D. C. (2008). Posttranslational modification influences the effects of MgrA on norA expression in Staphylococcus aureus. J. Bacteriol. 190, 7375–7381. 10.1128/JB.01068-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udo H., Munoz-Dorado J., Inouye M., Inouye S. (1995). Myxococcus xanthus, a gram-negative bacterium, contains a transmembrane protein serine/threonine kinase that blocks the secretion of beta-lactamase by phosphorylation. Genes Dev. 9, 972–983. 10.1101/gad.9.8.972 [DOI] [PubMed] [Google Scholar]
- Vizcaino J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., et al. (2016). 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 11033 10.1093/nar/gkw880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang W., Wu Z., Lu C. (2011). Reduced virulence is an important characteristic of biofilm infection of Streptococcus suis. FEMS Microbiol. Lett. 316, 36–43. 10.1111/j.1574-6968.2010.02189.x [DOI] [PubMed] [Google Scholar]
- Wertheim H. F., Nghia H. D., Taylor W., Schultsz C. (2009). Streptococcus suis: an emerging human pathogen. Clin. Infect. Dis. 48, 617–625. 10.1086/596763 [DOI] [PubMed] [Google Scholar]
- Wilhelm B. T., Landry J. R. (2009). RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods 48, 249–257. 10.1016/j.ymeth.2009.03.016 [DOI] [PubMed] [Google Scholar]
- Wilson T. L., Jeffers J., Rapp-Gabrielson V. J., Martin S., Klein L. K., Lowery D. E., et al. (2007). A novel signature-tagged mutagenesis system for Streptococcus suis serotype 2. Vet. Microbiol. 122, 135–145. 10.1016/j.vetmic.2006.12.025 [DOI] [PubMed] [Google Scholar]
- Zhang C. C. (1993). A gene encoding a protein related to eukaryotic protein kinases from the filamentous heterocystous cyanobacterium Anabaena PCC 7120. Proc. Natl. Acad. Sci. U.S.A. 90, 11840–11844. 10.1073/pnas.90.24.11840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng F., Ji H., Cao M., Wang C., Feng Y., Li M., et al. (2011). Contribution of the Rgg transcription regulator to metabolism and virulence of Streptococcus suis serotype 2. Infect. Immun. 79, 1319–1328. 10.1128/IAI.00193-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Zhou J., Ni Y., Yu Z., Mao A., Hu Y., et al. (2014). Contribution of eukaryotic-type serine/threonine kinase to stress response and virulence of Streptococcus suis. PLoS ONE 9:e91971. 10.1371/journal.pone.0091971 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.