Schjerven et al. compare mouse and human models of pre–B ALL to define conserved target genes and pathways of the tumor suppressor Ikaros, revealing CTNND1 and the early hematopoietic cell-surface receptors SPN (CD43) and CD34 as novel Ikaros targets that each confer oncogenic growth advantage.
Abstract
Inactivation of the tumor suppressor gene encoding the transcriptional regulator Ikaros (IKZF1) is a hallmark of BCR-ABL1+ precursor B cell acute lymphoblastic leukemia (pre–B ALL). However, the mechanisms by which Ikaros functions as a tumor suppressor in pre–B ALL remain poorly understood. Here, we analyzed a mouse model of BCR-ABL1+ pre–B ALL together with a new model of inducible expression of wild-type Ikaros in IKZF1 mutant human BCR-ABL1+ pre–B ALL. We performed integrated genome-wide chromatin and expression analyses and identified Ikaros target genes in mouse and human BCR-ABL1+ pre–B ALL, revealing novel conserved gene pathways associated with Ikaros tumor suppressor function. Notably, genetic depletion of different Ikaros targets, including CTNND1 and the early hematopoietic cell surface marker CD34, resulted in reduced leukemic growth. Our results suggest that Ikaros mediates tumor suppressor function by enforcing proper developmental stage–specific expression of multiple genes through chromatin compaction at its target genes.
INTRODUCTION
Acute lymphoblastic leukemia (ALL) is the most common childhood malignancy and the leading cause of childhood cancer–related mortality (Smith et al., 2010; Hunger and Mullighan, 2015). The majority of childhood ALL cases are of the precursor B cell subtype of acute leukemia (pre–B ALL), which arise upon transformation of developing progenitor B cells in the BM. pre–B ALL is characterized by genomic lesions, including the Philadelphia chromosome, a translocation involving the breakpoint cluster region (BCR) and the ABL1 gene (Ph+ or BCR-ABL1+; Wong and Witte, 2004). In addition, loss-of-function mutations in hematopoietic transcription factors occur in many pre–B ALL cases (Somasundaram et al., 2015). More than 80% of BCR-ABL1+ pre–B ALL harbor deletions or mutations in the IKZF1 gene, encoding the zinc finger (ZnF) transcription factor Ikaros (Mullighan et al., 2008). The majority of Ikaros lesions in pre–B ALL involve an aberrant Rag-mediated deletion of the exons encoding the DNA-binding ZnFs, resulting in expression of a dominant-negative (DN) isoform called IK6 (Mullighan et al., 2008). Furthermore, deletion or mutation of the IKZF1 gene correlate with poor prognosis in other subgroups of pre–B ALL, providing evidence that Ikaros is an important tumor suppressor in pre–B ALL (Mullighan et al., 2009; van der Veer et al., 2013). Despite an established tumor suppressor role for Ikaros, it remains unclear how Ikaros functions as a tumor suppressor, and insight into the mechanism of action and its downstream target genes might aid in the development of targeted therapies for treatment of aggressive IKZF1-mutated pre–B ALL.
Ikaros is an important regulator of hematopoiesis and is required for B cell development, as demonstrated by the lack of B cells in Ikzf1null mice (Wang et al., 1996). Ikaros tumor suppressor activity in the lymphoid lineage has been demonstrated in mouse models, as Ikzf1 mutant mice develop spontaneous thymic lymphoma caused by activating Notch1 mutations in the developing precursor T cells (Winandy et al., 1995; Papathanasiou et al., 2003; Dumortier et al., 2006). Although the complete lack of B lymphoid lineage cells in the original germline Ikzf1null and Ikzf1DN mice precluded the use of these homozygote mutants as models for studies of B cell malignancies, a heterozygote hypomorphic Ikzf1L mutant was shown to collaborate with a BCR-ABL1-Tg mouse model, indicating that the loss of Ikaros in human BCR-ABL1+ pre–B ALL can be modeled in mice (Georgopoulos et al., 1994; Wang et al., 1996; Virely et al., 2010).
Recently, we reported the generation of two Ikzf1 mutant mice with targeted deletions of the exons encoding the DNA-binding ZnF1 or ZnF4 (Ikzf1ΔF1/ΔF1 and Ikzf1ΔF4/ΔF4, respectively), and found that whereas the exon encoding ZnF1 was expendable, the exon encoding ZnF4 was required for Ikaros tumor suppressor function (Schjerven et al., 2013). Furthermore, both mutant strains of mice retained B cell lymphopoiesis, but displayed different partial defects in B cell development associated with deregulation of distinct subsets of genes, suggesting that they can be useful in elucidating Ikaros function and target genes. Upon retroviral transduction of BCR-ABL1 in BM cells from these mice, Ikzf1ΔF4/ΔF4 mutant cells gave rise to a more aggressive growth phenotype than either WT or Ikzf1ΔF1/ΔF1 BCR-ABL1–transformed pre–B ALL cells. This demonstrated selective ZnF4-dependent loss of Ikaros tumor suppressor function and presents a new mouse model of BCR-ABL1+ pre–B ALL.
Herein, we use the Ikzf1ΔF4/ΔF4 mutant mouse strain to gain insight into the functional consequence of loss of Ikaros tumor suppression in BCR-ABL1+ pre–B ALL. Furthermore, we present a new model of inducible expression of WT Ikaros in IKZF1 mutant human patient–derived BCR-ABL1+ pre–B ALL. We have defined the Ikaros target genes in human BCR-ABL1+ pre–B ALL and compared this to the Ikzf1ΔF4/ΔF4 mouse model of BCR-ABL1+ pre–B ALL to uncover conserved functions of Ikaros. Our analysis reveals new target genes and pathways not previously associated with Ikaros function. Specifically, we found that repression of key developmentally restricted cell surface receptors, as well as the intracellular protein p120-catenin, are conserved functions of Ikaros that restrict leukemic growth. Overall, these results further our understanding of how Ikaros functions as a tumor suppressor and define downstream targets of Ikaros that promote leukemic growth.
RESULTS
Targeted deletion of the fourth Ikaros DNA-binding ZnF domain in a mouse model of BCR-ABL1+ pre–B ALL results in enhanced cell proliferation
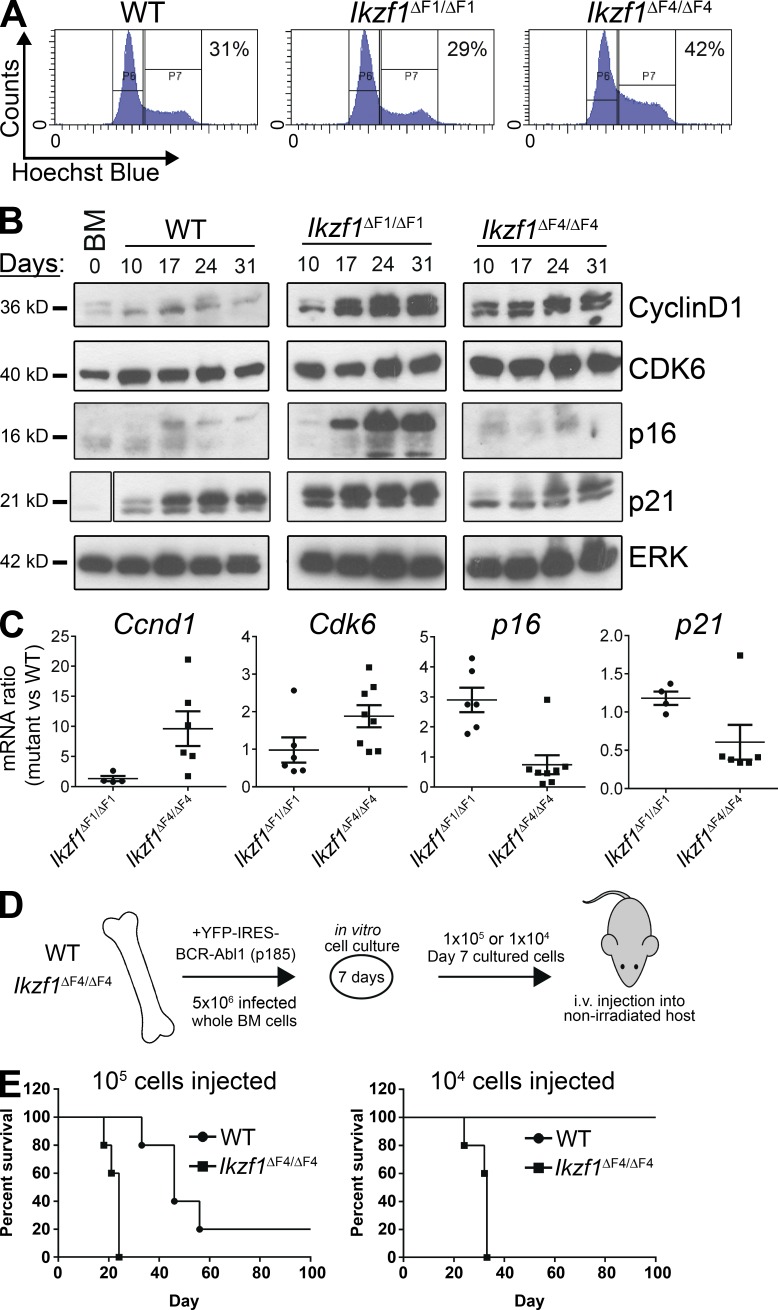
BCR-ABL1–transduced pre–B cells from Ikzf1ΔF4/ΔF4 mice lacking the exon encoding the fourth Ikaros ZnF domain (ZnF4) exhibited an increased growth rate relative to transduced pre–B cells from WT or Ikzf1ΔF1/ΔF1 mice lacking the exon encoding the first ZnF domain (ZnF1; Schjerven et al., 2013). To investigate the cellular process underlying the increased growth rate, we examined cell cycle status and apoptosis by flow cytometry analysis and found that BCR-ABL1–transduced Ikzf1ΔF4/ΔF4 pre–B ALL cultures had a consistently increased fraction of cells in S/G2-M as compared with WT and Ikzf1ΔF1/ΔF1 cultures (Fig. 1 A). However, we did not see any evidence of increased growth caused by reduced apoptosis in Ikzf1ΔF4/ΔF4 cultures. The increased fraction of cells engaged in active cell cycle corresponded to an increase in the protein and mRNA levels of the cell cycle regulators Cyclin D1 and Cdk6, and reduced levels of the negative cell cycle regulator p21 (Fig. 1, B and C). In contrast, the negative cell cycle regulator p16 was selectively induced in Ikzf1ΔF1/ΔF1 cultures (Fig. 1, B and C), corresponding to the observed reduced rate of growth and senescence of the Ikzf1ΔF1/ΔF1 cultures.
Figure 1.
Loss of Ikaros tumor suppression in Ikzf1ΔF4/ΔF4 mutant mice results in an increase in active cell cycle and aggressive leukemia in a nonirradiated in vivo model. (A–C) BM from WT, Ikzf1ΔF1/ΔF1, and Ikzf1ΔF4/ΔF4 mutant mice were transduced with BCR-ABL1-p185-IRES-YFP and grown in vitro on BM stroma–derived feeder layers. (A) Cell cycle flow cytometry analysis was performed by Hoechst incorporation, and one representative experiment (at day 14 of cell culture) is shown. (B) Cells were harvested on different days of in vitro cell culture and protein was extracted for Western blot analysis of Cyclin D1, Cdk6, p16, and p21, with ERK as loading control. Space indicates that intervening lanes have been spliced out. (C) RNA was isolated from the in vitro cultures, cDNA was prepared, and mRNA for Ccnd1, Cdk6, p16, and p21 was analyzed by quantitative real-time PCR. Ubc was used as a reference gene. (D) Schematic of the experimental set up for the in vivo transplantation assay. BM cells from WT and Ikzf1ΔF4/ΔF4 mutant mice were transduced and cultured in vitro as in A–C for 7 d before i.v. injection into nonirradiated immunocompetent WT c57BL/6 recipient mice. Animals were monitored and euthanized upon signs of leukemia development. (E) Kaplan-Meyer curve of cohorts receiving 105 (left) or 104 (right) BCR-ABL1–transformed cells. (A–C) are representative of three independent experiments and (E) represents one experiment with five recipient mice for each of the four cohorts. The result in right panel was repeated in a separate independent experiment displayed in Fig. 6 F.
Ikzf1ΔF4/ΔF4 BCR-ABL1+ cells give more aggressive leukemia in vivo
To examine the leukemic potential of the Ikzf1ΔF4/ΔF4-mutated model of BCR-ABL1+ pre–B ALL, we performed i.v. transplantation of BCR-ABL1+ pre–B ALL cells into nonirradiated immunocompetent WT-recipient mice (Williams et al., 2006). Whole BM from WT or Ikzf1ΔF4/ΔF4 mice were transduced with BCR-ABL1, and cultured in vitro for 7 d to allow for expansion of BCR-ABL1–transformed pre–B ALL cells before i.v. injection (Fig. 1 D). Transplantation of 105 BCR-ABL1–transformed cells was sufficient to give rise to leukemia with both WT and Ikzf1ΔF4/ΔF4 donor cells (Fig. 1 E, left), but mice that received Ikzf1ΔF4/ΔF4 BCR-ABL1+ cells developed a more aggressive leukemia and had a shorter lifespan than those that received WT BCR-ABL1+ cells. Furthermore, whereas 104 WT BCR-ABL1+ cells were not sufficient to induce leukemia, 104 Ikzf1ΔF4/ΔF4 BCR-ABL1+ cells were sufficient to induce aggressive leukemia at 100% penetrance (Fig. 1 E, right). This indicates that loss of Ikaros tumor suppression in the Ikzf1ΔF4/ΔF4 mutant leads to a higher frequency of cells that can initiate leukemia upon transplantation to a nonirradiated immunocompetent host.
Loss of Ikaros tumor suppressor function corresponds to a less mature cell surface phenotype, but does not block successful IgH V(D)J recombination
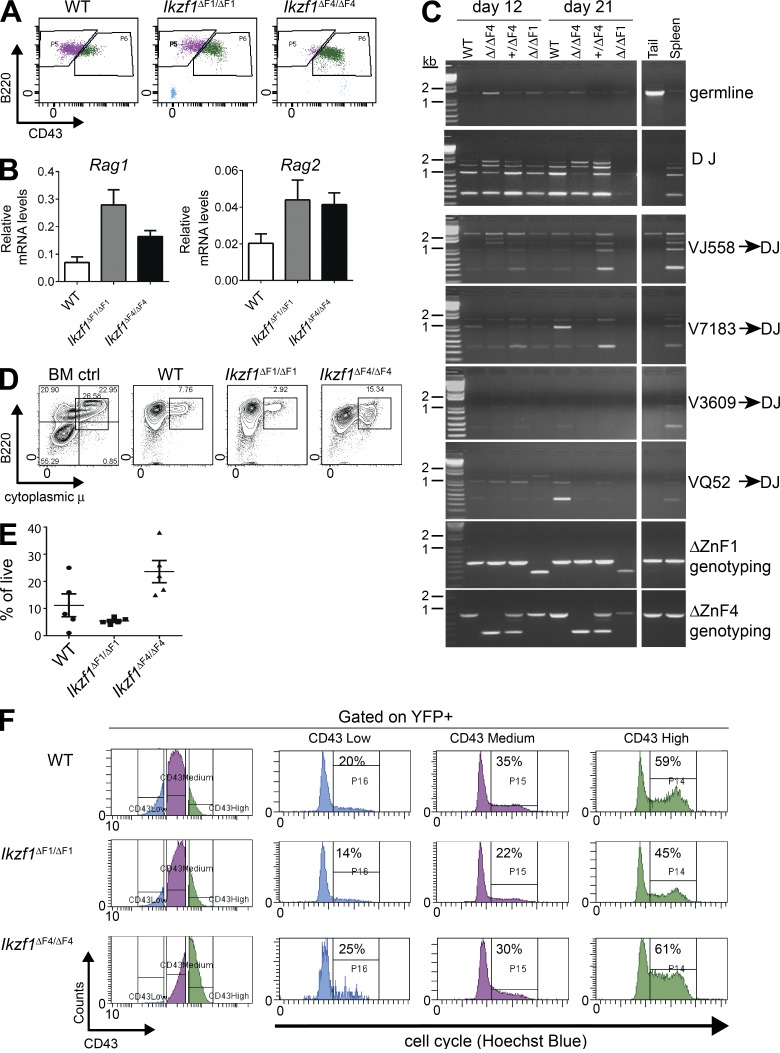
Previous phenotypic analysis had revealed an aberrant expression of c-Kit and lack of CD25 expression on BCR-ABL1–transformed Ikzf1ΔF4/ΔF4 cells (Schjerven et al., 2013). We found that this was also accompanied by a higher expression level of CD43 than on WT and Ikzf1ΔF1/ΔF1 cells (Fig. 2 A). Together, this suggests that the BCR-ABL1–transformed Ikzf1ΔF4/ΔF4 cells are at a less mature stage of B cell development than WT and Ikzf1ΔF1/ΔF1 (Hardy et al., 1991; Rolink et al., 1994). Because Ikaros has been reported to be required for Rag1 and Rag2 expression as well as IgH recombination (Reynaud et al., 2008), we tested the expression of Rag1 and Rag2 by RT-qPCR and found that Ikzf1ΔF1/ΔF1 and Ikzf1ΔF4/ΔF4 cells expressed Rag1 and Rag2 mRNA at higher levels than WT cells (Fig. 2 B). We also found that BCR-ABL1–transformed Ikzf1ΔF4/ΔF4 cells were able to undergo IgH V(D)J recombination to the same extent as BCR-ABL1–transformed WT and Ikzf1ΔF1/ΔF1 cells (Fig. 2 C), thus demonstrating that the cells are not defective in Rag-mediated recombination of IgH. Furthermore, we analyzed the levels of intracellular μ chain by flow cytometry analysis, and found that WT, as well as both mutants, were able to express cytoplasmic μ (Fig. 2, D and E). These results indicate that Ikzf1ΔF4/ΔF4 pre–B ALL cells can develop to the stage of successful IgH recombination and expression of cytoplasmic μ, but fail to down-regulate c-Kit and induce expression of CD25.
Figure 2.
Ikzf1ΔF4/ΔF4mutant BCR-ABL1+ cells display immature cell surface phenotype but undergo successful IgH V(D)J recombination, and increased cell cycle correlates with CD43 expression. (A) Flow cytometry analysis of CD43 on in vitro grown BCR-ABL1–transformed WT, Ikzf1ΔF1/ΔF1, and Ikzf1ΔF4/ΔF4 pre–B ALL cells. (B) Quantitative RT-PCR analysis of Rag1 and Rag2 mRNA expression on in vitro grown BCR-ABL1–transformed WT, Ikzf1ΔF1/ΔF1, and Ikzf1ΔF4/ΔF4 pre–B ALL cells. UBC was used as a reference gene. (C) IgH V(D)J recombination was analyzed by PCR (Schlissel et al., 1991) with genomic DNA from BCR-ABL1–transformed WT, Ikzf1ΔF1/ΔF1, and Ikzf1ΔF4/ΔF4 cells, and genomic DNA from WT tail and spleen was used as negative and positive controls, respectively. (D and E) Expression of cytoplasmic μ chain was analyzed by flow cytometry. Whole mouse BM was used as positive control. One representative is shown in (D) and results from multiple samples analyzed in independent experiments are summarized in E as fraction of cells expressing cytoplasmic μ (% of live; E). (F) Flow cytometry analysis of CD43 and cell cycle analysis on in vitro cultures of BCR-ABL1–transformed WT, Ikzf1ΔF1/ΔF1, and Ikzf1ΔF4/ΔF4 cells (gated on YFP+), displaying cell cycle activity of cells gated on low, medium, or high expression of CD43, respectively. All results are reproduced in two or more independent experiments.
Correlation between cell cycle profile and the developmentally regulated cell surface marker CD43
Given the less mature cell surface phenotype of the Ikzf1ΔF4/ΔF4 pre–B ALL cells, we hypothesized that BCR-ABL1–transformed Ikzf1ΔF4/ΔF4 cells are maintained at a highly proliferative stage of B cell development (C' or transitional pre–BI/large pre–BII; Hardy et al., 1991; Rolink et al., 1994; Mårtensson et al., 2010), with the failure of repressing key genes from the earlier progenitor stages (such as c-Kit). In contrast, WT and Ikzf1ΔF1/ΔF1 cells are able (at a certain rate) to mature beyond this proliferative stage, express CD25, and exit cell cycle, thus limiting the aggressiveness of growth. This model is in agreement with reports that Ikaros is required for the normal progenitor B cell differentiation at this point and pre–BCR-mediated cell cycle exit, as well as earlier reports that Ikaros is required to turn off the gene expression programs for self-renewal and multipotency (Ng et al., 2009; Trageser et al., 2009; Ferreirós-Vidal et al., 2013; Heizmann et al., 2013).
To test if proliferation is linked to developmental cell surface markers, we analyzed the cell cycle profile of various sub-populations in the BCR-ABL1–transformed in vitro cultures. We found that CD43 expression correlated well with cell cycle in all three genetic backgrounds (Fig. 2 F). CD43+High cells had a higher percentage of cells in S/G2-M phase, whereas cells with CD43+low expression had the lowest percentage of cells in S/G2-M phase (Fig. 2 F). Although CD43 could merely be a marker for these more highly proliferative cells, this observation is also consistent with the reported growth-promoting function of CD43 (Balikova et al., 2012). Collectively, the loss of tumor suppressor function in Ikzf1ΔF4/ΔF4 leukemia cells correlated with a failure to repress the expression of progenitor cell surface markers, resulting in a less mature cell surface phenotype as compared with WT and Ikzf1ΔF1/ΔF1 leukemia cells.
Expression profiling of Ikzf1ΔF4/ΔF4 BCR-ABL1–transformed cells reveals Ikaros target genes relevant for tumor suppressor function
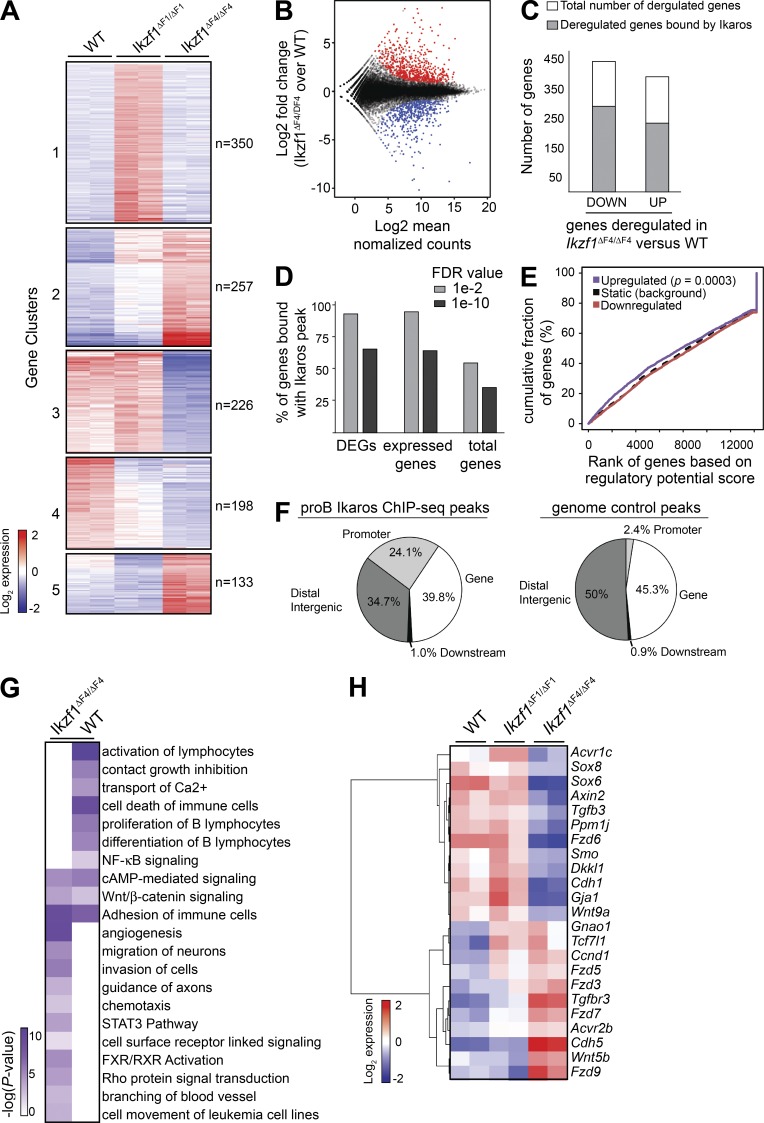
To investigate the changes in gene expression associated with the increased proliferative growth and loss of tumor suppression, we compared global expression profiles of replicate BCR-ABL1–transduced WT, Ikzf1ΔF1/ΔF1 and Ikzf1ΔF4/ΔF4 mutant cell cultures from independent experiments at 14 d after transduction. Clustering of differentially expressed genes demonstrated that the Ikzf1ΔF4/ΔF4 mutant pre–B ALL expression patterns were distinct from the WT and Ikzf1ΔF1/ΔF1 profiles, and also revealed a cluster of genes that were selectively up-regulated in Ikzf1ΔF1/ΔF1 cells compared with either WT or Ikzf1ΔF4/ΔF4 mutant cells (Fig. 3 A; cluster 1). As the Ikzf1ΔF1/ΔF1 cells retain tumor suppressor function, the cluster of differentially expressed genes specific for Ikzf1ΔF1/ΔF1 mutant cells are not expected to be tumor suppressor target genes, whereas clusters of genes deregulated in Ikzf1ΔF4/ΔF4 mutant cells represent candidate genes regulated by Ikaros tumor suppressor function. This selective ZnF-dependent gene regulation underscores the utility of ZnF mutant mice to define the subset of Ikaros-regulated genes associated with Ikaros tumor suppressor function.
Figure 3.
BCR-ABL1–transformed Ikzf1 mutant cells show distinct sets of deregulated genes and display altered expression of canonical signaling pathways. (A) RNA-Seq libraries were prepared from two biological replicates of day 14 in vitro cultures of BCR-ABL1–transformed WT, Ikzf1ΔF1/ΔF1 and Ikzf1ΔF4/ΔF4 cells from independent experiments. Heat map based on k-means clustering ofdifferentially expressed genes (fold change ≥ 2; P ≤ 0.005) across Ikzf1ΔF1/ΔF1 and Ikzf1ΔF4/ΔF4 compared with WT. (B) MA-plot showing the distribution of differentially expressed genes between Ikzf1ΔF4/ΔF4 and WT. Up-regulated and down-regulated genes in Ikzf1ΔF4/ΔF4 compared with WT samples are indicated in red and blue, respectively. (C) Ikaros ChIP-Seq data from mouse BM-derived progenitor B cells (Bossen et al., 2015) were analyzed and the number of differentially expressed genes with Ikaros binding annotated within 100 kb is displayed in gray, whereas genes without any annotated Ikaros peaks are displayed in white (open) columns. (D) MACS2 peak calling was performed on the Bossen dataset using default (FDR of 1 × 10−2) or a stringent cutoff (FDR of 1 × 10−10), resulting in 33,959 or 7,656 peaks, respectively. All genes within 100 kb of a peak were then annotated and the percentage of differentially expressed genes (DEGs) between Ikzf1ΔF4/ΔF4 versus WT cells was determined. The percentage of expressed genes bound by Ikaros was determined by dividing the number of genes bound by Ikaros by the number of genes in our mouse RNA-Seq data that have a mean regularized log normalized count > 5 across all Ikzf1ΔF4/ΔF4 and WT samples. The percentage of total genes bound by Ikaros was determined by dividing the total number of genes bound by Ikaros by the total 34,588 RefSeq genes. (E) Activating/repressive function prediction of Ikaros by ChIP BETA. The purple and the red lines represent the up-regulated and down-regulated genes in Ikzf1ΔF4/ΔF4 (versus WT) cells, respectively. The dashed line indicates the nondifferentially expressed genes as background. Genes are cumulated by the rank on the basis of the regulatory potential score from high to low. P values that represent the significance of the up-regulated (P = 0.000302) or down-regulated (P = 0.983) group distributions are compared with the nondifferentially expressed group by the Kolmogorov-Smirnov test. (F) Relative distribution of Ikaros enrichment peaks of the shared 1,704 pro–B Ikaros peaks (as defined in Fig. S1) or control genomic regions relative to RefSeq gene features. (G) IPA revealed several enriched deregulated pathways. Several representative pathways and biological processes are shown, with –log10 P values in heat map. (H) Heatmap of deregulated genes that fall into the Wnt gene pathway list.
To investigate the tumor suppressor function of Ikaros, we examined genes that were differentially expressed in Ikzf1ΔF4/ΔF4 mutant cells as compared with WT cells, and found that 365 and 414 genes were up- and down-regulated, respectively (P < 0.005; fold change ≥ 2; Fig. 3, B and C). To identify potential direct regulatory target genes of a transcription factor, a common analysis is to evaluate the binding sites within a given distance to gene transcription start sites (TSS; Wang et al., 2013). We found that 68 and 62% of the Ikzf1ΔF4/ΔF4 down- and up-regulated genes, respectively, were bound by Ikaros in mouse BM progenitor B cells within 100 kb of the gene TSS (Bossen et al., 2015; Fig. 3 C). We noted that Ikaros was not enriched at deregulated genes compared with all expressed genes, and this was the case whether analyzing the aforementioned dataset in isolation, or when using a high-confidence dataset of Ikaros peaks obtained by analysis of datasets from three different laboratories (Fig. 3 D and Fig. S1; Ferreirós-Vidal et al., 2013; Schwickert et al., 2014; Bossen et al., 2015). However, further analysis by integrating differential gene expression between the Ikzf1ΔF4/ΔF4 and WT pre–B ALL cells with Ikaros ChIP-Seq data using BETA (Wang et al., 2013), predicted a significant repressive function for WT Ikaros for genes that were up-regulated in Ikzf1ΔF4/ΔF4 BCR-ABL1+ cells compared with WT BCR-ABL1+ cells (P = 0.0003; Fig. 3 E). This supports the transcriptional repressor function of Ikaros, and although it still remains difficult to distinguish direct from indirect Ikaros targets, we nevertheless note that several of the Ikaros-dependent genes defined in this study contain Ikaros binding sites in proximity of the gene loci, including at gene promoters (Fig. 3 F).
Deregulated signaling pathways in Ikzf1ΔF4/ΔF4 leukemic cells
Consistent with recent findings from both progenitor T cells (thymocytes) and progenitor B cells in Ikzf1 mutant mouse models (Schjerven et al., 2013; Joshi et al., 2014; Schwickert et al., 2014; Churchman et al., 2015), adhesion was one of the most significantly enriched pathways in Ikzf1ΔF4/ΔF4 compared with WT pre–B ALL cells (Fig. 3 G). In agreement with our phenotypic analysis of Ikzf1ΔF4/ΔF4 mutant cells demonstrating the failure to down-regulate the cell surface markers c-Kit and CD43, the deregulated genes in Ikzf1ΔF4/ΔF4 mutant cells were enriched for genetic pathways involving cell surface receptor linked signaling (Fig. 3 G). In addition, genes associated with cell movement, migration, invasion of cells, and chemotaxis were also enriched, suggesting an increased potential of Ikzf1-mutated leukemia to leave the blood stream and invade tissues (Fig. 3 G). In contrast, genes involved in differentiation of B lymphocytes and contact growth inhibition were enriched in WT cells (i.e., down-regulated in Ikzf1ΔF4/ΔF4 cells), in agreement with the more immature cell surface phenotype and aggressive leukemic properties of the Ikzf1ΔF4/ΔF4 cells.
Comparative Ingenuity Pathway Analysis (IPA) of genes deregulated in Ikzf1ΔF4/ΔF4 and WT cells revealed alterations in the expression patterns of different cellular signaling pathways (Fig. 3 G). The signaling pathways that were enriched in the Ikzf1ΔF4/ΔF4 expression profiles and not in the WT profiles included the Rho, STAT3, and FXR–RXR pathways. Signaling pathways decreased in Ikzf1ΔF4/ΔF4 compared with WT included the NF-κB and Ca2+ signaling pathways. Adhesion, cAMP and Wnt–β-catenin pathways were enriched in both BCR-ABL1–transduced WT and Ikzf1ΔF4/ΔF4 expression sets. The Wnt–β-catenin pathway functions in normal hematopoiesis and has been linked to leukemia development (Staal and Clevers, 2005; Misaghian et al., 2009; Luis et al., 2012; McCubrey et al., 2014). The Rho signaling pathway is part of the noncanonical Wnt pathway and has been reported to be involved in BCR-ABL1–mediated leukemogenesis (Thomas et al., 2008). When we further examined the differential gene expression patterns of the Wnt–β-catenin pathway, we found that several members were indeed differentially expressed between WT, Ikzf1ΔF1/ΔF1, and Ikzf1ΔF4/ΔF4 datasets (Fig. 3 H).
Inducible restoration of Ikaros tumor suppressor function in human pre–B ALL cells
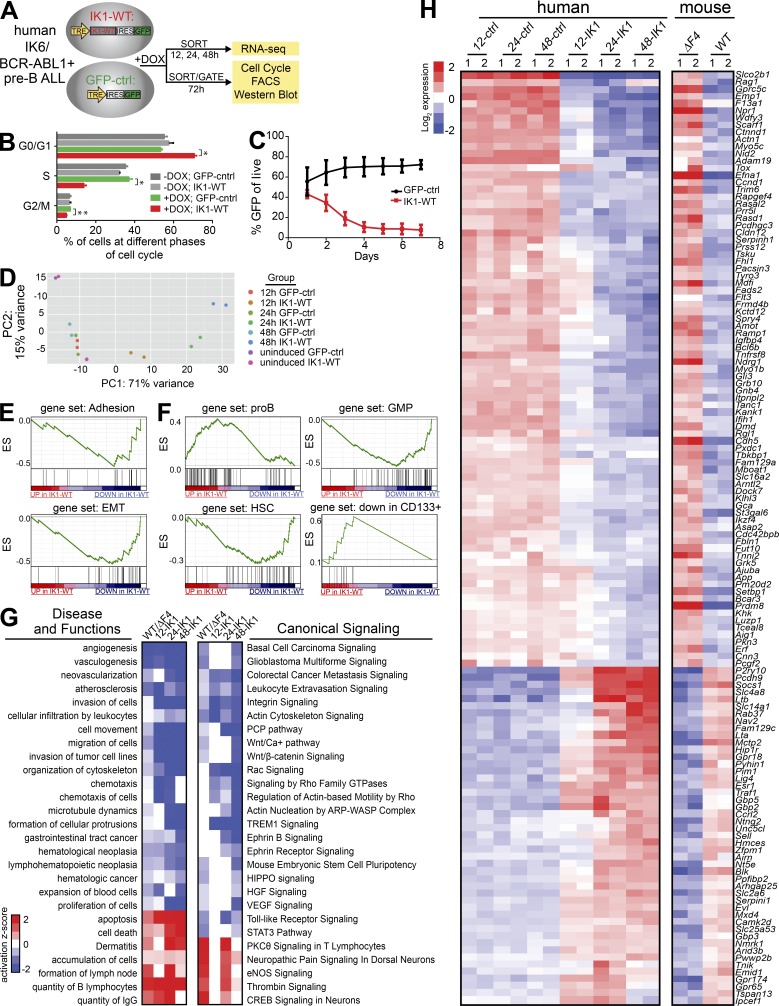
To relate our findings of Ikaros tumor suppressor function to human pre–B ALL cells, we established inducible Ikaros expression in human patient-derived BCR-ABL1+ pre–B ALL cells that naturally express the DN-IK6 isoform. Doxycycline-inducible (DOX) expression was monitored by (IRES-) GFP, and induced cells were gated or sorted for analysis based on GFP expression (Fig. 4 A). Upon expression of WT Ikaros, the IKZF1 mutant pre–B ALL cells exited cell cycle with a significantly reduced fraction in active S-phase and increased fraction in G0/G1 (Fig. 4 B; P < 0.005), resulting in reduced growth rate and out-competition by GFP-negative cells (Fig. 4 C). We reproducibly observed Ikaros-induced reduced growth rate and out-competition by GFP-negative cells when performing the same experiment in another IKZF1-mutated patient-derived BCR-ABL1+ pre–B ALL (unpublished data), indicating that the tumor suppressor role of Ikaros is restored in this system.
Figure 4.
Profiling the Ikaros target genes upon inducible expression of WT Ikaros in patient-derived IKZF1-mutated BCR-ABL1+ pre–B ALL reveals conserved Ikaros target genes and genetic pathways in BCR-ABL1+ pre–B ALL. (A) Schematic representation of experimental setup. Patient-derived BCR-ABL1+ pre–B ALL expressing DN-IK6 were stably transduced with TET-inducible expression of either WT Ikaros (IK1-WT-IRES-GFP) or GFP control. Cells were induced with DOX, and harvested at different time points for analysis. GFP+ cells were sorted for RNA-Seq analysis and protein analysis by Western blotting, and gated for flow cytometry analysis. (B) The bar graph representation of results from cell cycle analysis by flow cytometry from two independent experiments demonstrate that induction of WT Ikaros (3 d DOX treatment) leads to cell-cycle exit (*, P < 0.005; **, P < 0.006; multiple Student’s t test). The same result was obtained in a third independent experiment at day 2 after DOX. (C) Growth curve analysis after GFP+ cells over time demonstrate growth disadvantage of cells expressing WT Ikaros in liquid culture (n = 4). (D) The experimental variation between two independent RNA-Seq experiments with IK1-WT or GFP-control for each time point was compared by Principle Component Analysis (PCA) using the DESeq2 regularized log transformation for each individual dataset (Love et al., 2014). (E and F) GSEA preranked analysis of WT Ikaros-responsive genes are (E) negatively correlated with adhesion and EMT genesets, (F) enriched with B cell development gene signatures, and negatively correlated with HSC gene sets. (G) IPA comparison of Ikaros-regulated genes in BCR-ABL1+ pre–B ALL in the mouse and human system revealed common deregulated disease and functions pathways and canonical signaling pathways. The heat map is based on IPA activation z-scores. (H) Directly conserved Ikaros-regulated genes (P < 0.01; fold change ≥ 2).
Expression profiling after restoration of Ikaros tumor suppression in human BCR-ABL1+ pre–B ALL reveals Ikaros-regulated target genes and pathways
To elucidate the Ikaros target genes associated with restored Ikaros tumor suppressor function in human pre–B ALL, we analyzed the gene expression changes that occurred after induction of WT Ikaros in human BCR-ABL1+ pre–B ALL. We compared global expression profiles of induced WT Ikaros expressing cells to control cells at 12, 24, and 48 h post-induction in duplicate (Fig. 4 A). Principal component analysis showed high concordance between the replicates from independent experiments and highlighted the kinetics of the differential expression response following induction of WT Ikaros (Fig. 4 D). Gene set enrichment analysis (GSEA) demonstrated that adhesion was one of the most significantly enriched biological processes regulated by Ikaros in pre–B ALL (Fig. 4 E), consistent with findings from the mouse model of pre–B ALL (Fig. 3 G) as well as prior studies of mouse Ikzf1-mutated developing lymphoid B and T cells (Schjerven et al., 2013; Joshi et al., 2014; Schwickert et al., 2014; Churchman et al., 2015). Furthermore, there was an enrichment of Ikaros-repressed genes associated with the epithelial to mesenchymal transition (EMT) pathway, suggesting that Ikaros is important for repression of invasion-promoting gene programs (Fig. 4 E).
Based on the observation that Ikzf1ΔF4/ΔF4-mutated BCR-ABL1+ pre–B ALL cells failed to down-regulate progenitor cell surface markers (Fig. 2; Schjerven et al., 2013), we performed GSEA analysis using gene sets that correspond to hematopoiesis and associated signatures (Laurenti et al., 2013). Induction of WT Ikarosaltered the expression ofhuman hematopoietic progenitor, early myeloid and progenitor B cell population gene sets. As early as 12 h after induction, we found that the pro–B gene set was enriched, whereas the granulocyte-macrophage progenitor (GMP) gene set was down-regulated. After 48 h of Ikaros induction, these two gene sets were further enriched (Fig. 4 F), and the hematopoietic stem cell (HSC) and myeloid progenitor gene sets were down-regulated, whereas genes reported to be down in CD133+ stem cells were up-regulated by Ikaros (Fig. 4 F). This indicates that IKZF1-mutated human pre–B ALL cells display lineage infidelity/plasticity, with failure to down-regulate genes from the earlier progenitor programs, as well as promiscuous expression of early myeloid lineage progenitor gene signatures.
Conserved Ikaros-regulated pathways and target genes
To determine the Ikaros-regulated pathways conserved between the mouse and human models of Ikaros-mutated pre–B ALL, we performed comparative IPA analysis between the Ikzf1ΔF4/ΔF4 mutant pre–B ALL and the human IKZF1-mutated pre–B ALL expression profiles (Fig. 4 G). In line with a tumor suppressor function for Ikaros, proliferation of cells, cancer, and neoplasia pathways were predicted to be repressed (a negative activation Z-score) in cells with WT Ikaros compared with cells with mutated Ikaros in both mouse and human models (Fig. 4 G, left). In addition, cell migration, invasion, and organization of cytoskeleton were among the conserved pathways that were repressed by WT Ikaros, indicating a role for Ikaros in control of extravasation/invasion-promoting properties of cells. Analysis of canonical signaling pathways in both human and mouse pre–B ALL models, revealed that repression of carcinoma, metastasis signaling and leukocyte extravasation signaling pathways are repressed by Ikaros (Fig. 4 G, right). In addition, the Wnt pathways (including PCP, calcium, and β-catenin), as well as pathways that are known to be connected with Wnt signaling (integrin, actin cytoskeletal, and Rac/Rho signaling), were repressed by Ikaros in both mouse and human models (Fig. 4 G; right).
Next, we compared the deregulated genes from the mouse model to the human pre–B ALL cells to determine the degree of direct conservation at the level of individual genes. We found a set of 83 genes up-regulated in the mouse Ikzf1ΔF4/ΔF4 mutant pre–B ALL cells and down-regulated upon expression of WT Ikaros in the human pre–B ALL cells, and a set of 47 genes that were down-regulated in the mouse Ikzf1ΔF4/ΔF4 mutant pre–B ALL cells and up-regulated upon expression of WT Ikaros in the human pre–B ALL cells (Fig. 4 H; P < 0.01; fold change ≥ 2). Together, this reveals conserved Ikaros-regulated target genes and pathways connected to the observed Ikaros tumor suppressor function in mouse and human BCR-ABL1+ pre–B ALL.
Genome-wide analysis of Ikaros binding in human pre–B ALL cells
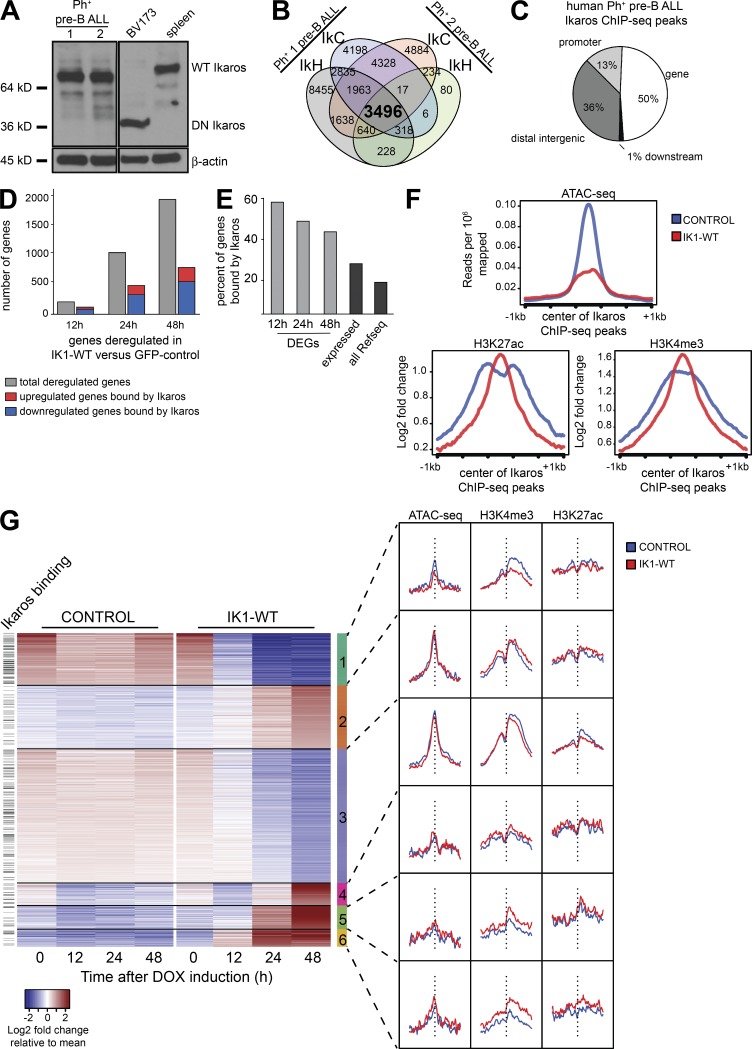
To further our understanding of Ikaros gene targets in human pre–B ALL cells, we performed Ikaros ChIP-Seq analysis in two different human patient–derived BCR-ABL1+ pre–B ALL samples that naturally express WT Ikaros (Fig. 5 A). We used two different Ikaros antibodies that recognize N-terminal (IkH) and C-terminal (IkC) epitopes, respectively, and performed duplicate independent experiments for one antibody (IkH). We then compared the significantly enriched binding sites across these six datasets (Fig. 5 B and Fig. S2). The result was a high-confidence set of 3,496 Ikaros binding sites in human pre–B ALL common to all six experimental datasets (Fig. 5 B). Although location analysis revealed a relative enrichment of Ikaros binding at gene promoters (13% compared with a random distribution of 2.4%; Fig. 5 C), the majority of Ikaros binding sites were located in intragenic or distal regions. Similar to what was observed in mouse progenitor B cells, a high proportion of the WT Ikaros-responsive genes were also bound by Ikaros (56% of differentially expressed genes at 12 h; Fig. 5, D and E). Notably, Ikaros binding was enriched at genes responsive to WT Ikaros expression, relative to all expressed genes (Fig. 5, D and E).
Figure 5.
Genome-wide analysis of Ikaros binding and Ikaros-induced changes in chromatin in human BCR-ABL1+ pre–B ALL. (A) Western blot analysis of extracts from two in vitro growing patient-derived xenograft-expanded Ph+ pre–B ALL cells that express WT Ikaros and that were used for Ikaros ChIP-Seq. Also shown are the controls from BV173 cells and mouse spleen cells that express the DN IK6 isoform and full-length WT Ikaros, respectively. (B) Analysis of Ikaros ChIP-Seq datasets from two patient-derived BCR-ABL1+ pre–B ALL samples (Fig. S2). (C) Genome-wide distribution of human Ph+ pre–B ALL ChIP-Seq peaks in relation to RefSeq genes. (D) Total number of deregulated genes and the proportion with annotated Ikaros ChIP-Seq binding at each time point. (E) The percentage of differentially expressed genes (DEG) with an Ikarospeak within 100 kb in each comparison (12, 24, or 48 h IK1-WT versus GFP (control); FDR < 0.005; fold change > 2) was determined (light gray). The percentage of expressed genes bound by Ikaros was determined by defining all genes with an Ikaros peak having a mean of the regularized log value > 1 across all RNA-Seq samples. The percentage of all genes bound by Ikaros was determined by dividing the unique Ikaros bound genes by the total 50,801 RefSeq genes in the ChIP BETA database (Wang et al., 2013). (F) Analysis of chromatin accessibility by ATAC-Seq and histone modification ChIP-Seq in WT Ikaros-induced and control human pre–B ALL cells relative to Ikaros-enriched regions (G) Heat map based on k-means clustering of normalized log transformed read counts of differentially expressed genes in GFP-sorted cells after IK1-WT induction compared with control after a period of 48 h after induction. The signal from IK1-WT and control samples for ATAC-Seq and ChIP-Seq are shown for each cluster on the right. The vertical dashed line represents the center of the TSS of each gene, and the genes with an Ikarospeak within 10 kb of the TSS are shown on the left.
Genome-wide analysis of Ikaros-induced chromatin changes in human pre–B ALL cells
To evaluate the mechanisms by which Ikaros regulates gene expression, we analyzed chromatin accessibility by ATAC-Seq (Buenrostro et al., 2015) after 24 h of WT Ikaros induction, as well as the genome-wide enrichment of histone modifications H3K4me3 and K3K27ac by ChIP-Seq. Induction of WT Ikaros resulted in a reduction in ATAC-Seq signal at Ikaros-enriched ChIP-Seq regions (Fig. 5 F). This suggests that Ikaros regulates target gene expression through localized alterations in chromatin accessibility, resulting in chromatin compaction upon Ikaros binding, and supports a role for Ikaros in gene repression. Although a significant subset of differentially expressed genes were up-regulated upon Ikaros induction, analysis of active chromatin marks surrounding Ikaros-binding sites displayed only modest changes upon induction of WT Ikaros (Fig. 5 F). To further investigate the regulation of Ikaros-responsive genes, we performed clustering analysis of differentially expressed genes after induction of WT Ikaros and evaluated the corresponding changes of chromatin structure surrounding the TSS of differentially regulated genes. This analysis uncovered a cluster of down-regulated genes that displayed decreases in chromatin accessibility and H3K4me3 levels (Fig. 5 G; cluster 1, on top). Additionally, two clusters of genes that were up-regulated upon Ikaros induction displayed enhanced H3K4me3 signal centered at the gene TSS (Fig. 5 G; clusters 5 and 6, bottom). In contrast, other clusters of genes had significant changes in gene expression but displayed little or no changes in chromatin accessibility or in the enrichment of active histone marks, suggesting that Ikaros regulates gene expression also through alternative molecular mechanisms (Fig. 5 G; clusters 2–4, middle).
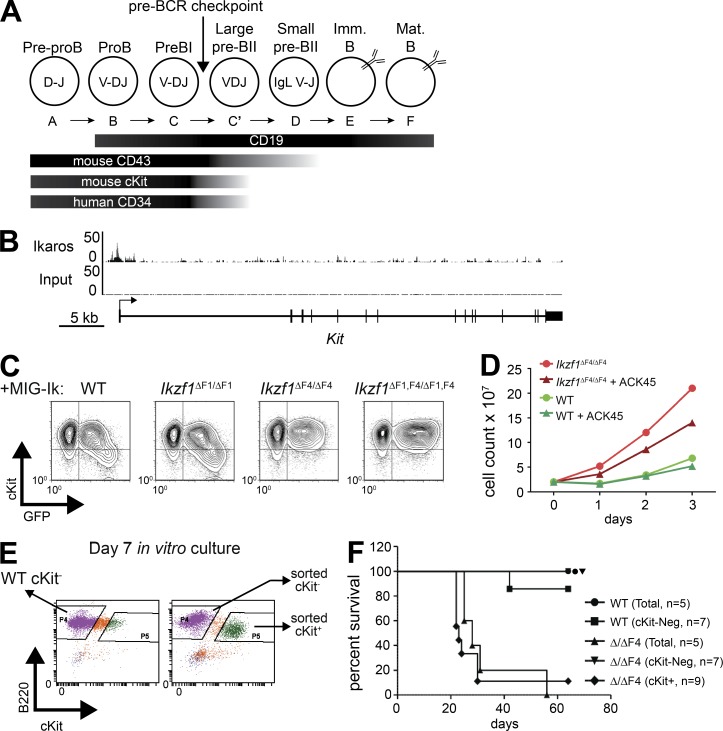
Developmental down-regulation of c-Kit on pro-B cells is dependent on Ikaros ZnF4
Both Ikaros mutant pre–B ALL models exhibited aberrant expression of early hematopoietic programs, including genes involved in developmental and adhesion-mediated receptor signaling. Among these was the gene encoding c-Kit (Kit), which is expressed on HSC and early B cell progenitors, but is normally down-regulated as cells progress through B cell development (Fig. 6 A). c-Kit was aberrantly expressed in Ikzf1ΔF4/ΔF4-mutated mouse pre–B ALL (Schjerven et al., 2013), and WT Ikaros binds to the Kit promoter in mouse progenitor B cells (Fig. 6 B; data from Bossen et al., 2015). To test if ZnF4 of Ikaros is required for the down-regulation of c-Kit in progenitor B cells, EBF1-rescued Ikzf1null pro–B cells (Reynaud et al., 2008) were transduced with retroviral vectors expressing either full-length Ikaros (Ik1, WT) or Ikaros missing the exon encoding ZnF1, ZnF4, or both. As a positive control, c-Kit was down-regulated in cells upon re-expression of full-length Ikaros, as indicated by GFP+High cells (Fig. 6 C, left). We found that the Ikaros isoform lacking ZnF1 was also sufficient for down-regulation of c-Kit. In contrast, Ikaros lacking ZnF4 or Ikaros lacking both ZnF1 and ZnF4, were unable to down-regulate c-Kit expression (Fig. 6 C, two rightmost panels). This indicates that Ikaros ZnF4 is required for the developmental down-regulation of c-Kit in progenitor B cells, and aberrant c-Kit expression correlates with the observed loss of tumor suppressor function in Ikzf1ΔF4/ΔF4 mutant BCR-ABL1+ pre–B ALL.
Figure 6.
Kit is a ZnF4-dependent Ikaros target gene that confers growth advantage and correlates with aggressive leukemia. (A) Schematic diagram of relevant cell surface markers expressed on developing B cells. (B) ChIP-seq tracks of Ikaros and input from mouse pro-B cells (Bossen et al., 2015) demonstrates binding of Ikaros at the Kit promoter. (C) Retroviral overexpression of Ikaros isoforms containing different subsets of the DNA-binding zinc fingers (IRES-GFP) in Ikzf1null, EBF1-rescued noncommitted pro–B cells (Reynaud et al., 2008). GFP and c-Kit expression was analyzed by flow cytometry. (D) Mouse pre–B ALL cells from WT and Ikzf1ΔF4/ΔF4 mutant mice were created and cultured as described in Fig. 1 A. Cell growth with and without c-Kit blocking Ab (ACK45; 10 µg/ml) was measured by daily cell count. (C and D) One representative of two independent experiments. (E) Day 7 cultures of mouse pre–B ALL cells from WT and Ikzf1ΔF4/ΔF4 mutant mice were sorted as indicated based on c-Kit expression. (F) Sorted cells from E or bulk unsorted day 7 cultures were i.v. injected into nonirradiated, immunocompetent WT C57BL/6 mice. Mice were monitored and euthanized upon development of leukemia. Leukemia was confirmed by necropsy and flow cytometry analysis, and survival of indicated cohorts is represented by Kaplan-Meyer curve.
c-Kit confers partial growth advantage and marks the Ikzf1ΔF4/ΔF4 leukemic cells that are able to give in vivo leukemia
c-Kit is a potent growth factor receptor known to provide a proliferative signal and has been recognized as an oncogene in several cancers (Lennartsson and Rönnstrand, 2012). To test if the sustained expression of c-Kit on Ikzf1ΔF4/ΔF4 cells contributed to the increased growth rate and leukemogenesis, we first used a blocking antibody (Ab) against c-Kit (ACK45). We observed a partial, but reproducible reduction in growth rate of the c-Kit+ BCR-ABL1–transformed Ikzf1ΔF4/ΔF4 cells, and as expected, the c-Kit blocking Ab had no effect on the c-Kit− WT cells (Fig. 6 D). This indicates that the derepressed expression of c-Kit on the BCR-ABL1–transformed Ikzf1ΔF4/ΔF4 cells confers some growth advantage; however, it is not the only factor contributing to the increased growth rate in this in vitro model of Ikzf1ΔF4/ΔF4-mutated BCR-ABL1+ pre–B ALL.
We observed that the BCR-ABL1–transformed BM cells started as a mixed population of c-Kit− and c-Kit+ cells. Over time, the c-Kit+ cells disappeared in the WT cultures and the dominating cell population was c-Kit−. In contrast, the c-Kit+ cell population was predominant over time in the BCR-ABL1–transformed Ikzf1ΔF4/ΔF4 cell cultures. To test if expression of c-Kit marked the BCR-ABL1+ Ikzf1ΔF4/ΔF4 cells able to confer leukemia upon in vivo transplantation, we sorted c-Kit+ and c-Kit− BCR-ABL1–transformed Ikzf1ΔF4/ΔF4 cells at day 7 of in vitro culture, for injection into nonirradiated immunocompetent mice (Fig. 6, E and F). Bulk WT and Ikzf1ΔF4/ΔF4 cells were used as control, as well as sorted c-Kit− WT cells (there were not sufficient numbers of WT c-Kit+ cells to sort for in vivo injections). Although transplantation of 104 cells was sufficient for Ikzf1ΔF4/ΔF4 cells to lead to pre–B ALL in vivo, 104 WT cells were not able to initiate disease (Fig. 1 E). We therefore injected 104 c-Kit+ or c-Kit− Ikzf1ΔF4/ΔF4 BCR-ABL+ cells, as well as sorted c-Kit− WT and bulk unsorted WT and Ikzf1ΔF4/ΔF4 cells as controls. We found that 104 c-Kit+ Ikzf1ΔF4/ΔF4 cells were sufficient to induce disease, comparable to the total unsorted Ikzf1ΔF4/ΔF4 cells. However, the c-Kit− Ikzf1ΔF4/ΔF4 cells were not able to initiate disease (Fig. 6 F). This demonstrated that the c-Kit+ population contains the cells in the Ikzf1ΔF4/ΔF4 BCR-ABL1+ pre–B ALL cell culture that confer in vivo leukemia. Collectively, these results indicate that Ikaros tumor suppressor function is linked to the developmental stage-specific regulation of the c-Kit hematopoietic growth factor receptor.
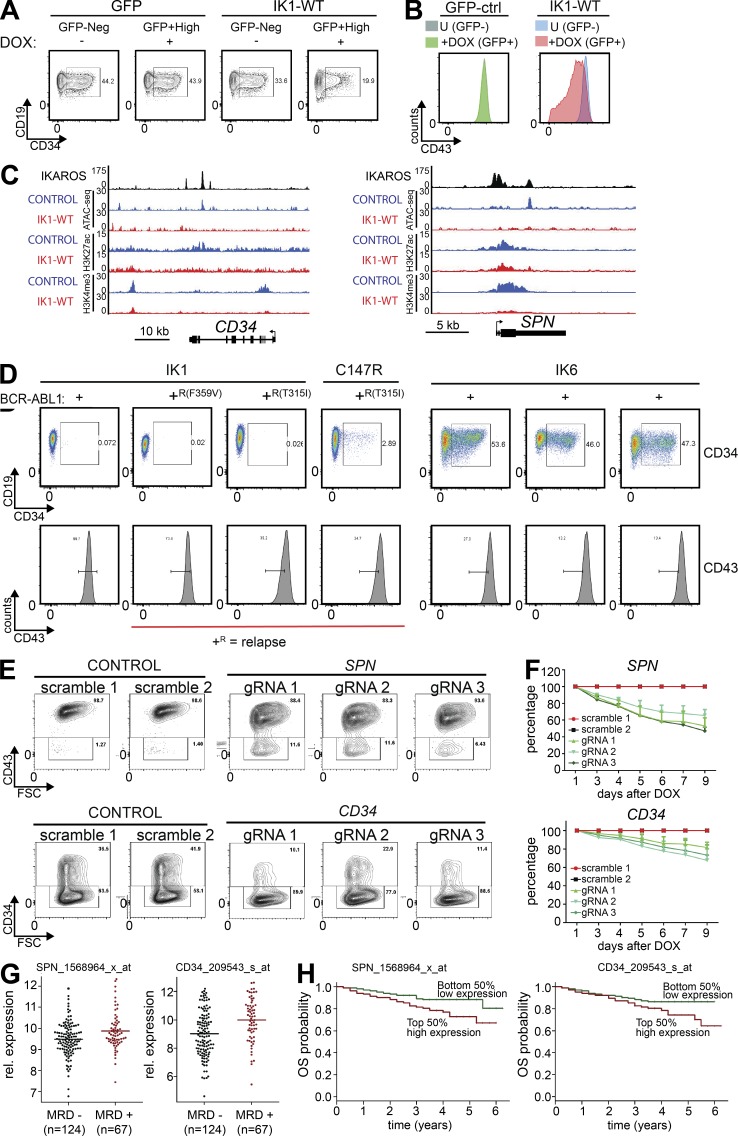
Ikaros regulates the developmental stage-specific expression of CD34 and CD43 in human pre–B ALL
Among the early hematopoietic progenitor cell surface receptors down-regulated during B cell development, we found CD34 and SPN (encoding CD43) to be deregulated in human IKZF1-mutated pre–B ALL cells. CD34 is commonly used for sorting or enrichment of human hematopoietic progenitor cells, and is proposed to be a common marker for progenitors of different lineages (Fig. 6 A; van Zelm et al., 2005; Sidney et al., 2014). In spite of its widespread use as a marker, the function of CD34 on hematopoietic cells is poorly understood. However, it is thought to be involved in adhesion to stromal cells, and similar to c-Kit, CD34 is reported to be associated with CRKL and is thus likely to transmit intracellular signaling (Felschow et al., 2001). We found that a high fraction of the IKZF1-mutated pre–B ALL cells expressed CD34, and both the fraction of CD34+ cells and the cell surface expression level of CD34 were reduced upon induction of WT Ikaros (Fig. 7 A). Similarly, expression of WT Ikaros in human IKZF1-mutated pre–B ALL cells decreased the levels of CD43 cell surface expression (Fig. 7 B). ChIP-Seq analysis revealed that Ikaros bound to both the CD34 and the SPN gene loci, and induction of Ikaros in IKZF1-mutated pre–B ALL resulted in a reduction in chromatin accessibility and H3K4me3 signal at these loci (Fig. 7 C). Together, this suggests that both CD34 and SPN are direct regulatory targets of Ikaros.
Figure 7.
Ikaros regulates developmentally restricted signaling-competent cell surface receptors that confer leukemic growth advantage. (A and B) Inducible expression of WT Ikaros in human patient-derived BCR-ABL1+ pre–B ALL results in down-regulation of CD34 (A) and CD43 (B). (C) Ikaros binds to the CD34 and SPN (CD43) loci in human patient-derived BCR-ABL1+ pre–B ALL cells, and chromatin accessibility (ATAC-Seq) and histone modification ChIP-Seq (H3K4me3 and H3K27ac) signals are shown for control (GFP) and Ikaros-induced cells (Ik-WT). (D) Flow cytometry analysis of CD34 and CD43 in a panel of human patient-derived BCR-ABL1+ pre–B ALL cells. IKZF1 status and BCR-ABL1 relapse mutations are indicated on top and bottom. (E and F) Cas9/CRISPR-mediated depletion of CD34 and CD43 was confirmed by flow cytometry of cell surface expression of the respective proteins, as demonstrated by both reduced mean fluorescence intensity (MFI) and fraction of CD34+ (bottom) and CD43+ (top) cells, respectively (E), and resulted in reduced growth in liquid culture (P = 0.0032 for CD34; P = 0.0010 for CD43; unpaired Student’s t test; F). (G) Analysis of published clinical data (COG, P9906) showed that presence of MRD after 29 d of treatment correlated with higher expression of CD34 and SPN (encoding CD43; P = 7 × 10−5 and P < 0.007, respectively; two-tailed Wilcoxon test; H) OS of patients (COG, P9906) separated above or below median mRNA expression level of CD34 and SPN (encoding CD43; P < 0.07 and P < 0.02, respectively; Log-rank test). A, B, and D display one representative from three independent experiments performed. For E and F, data from three repeats from two independent experiments are displayed (F) with one representative flow cytometry plot (E).
Analysis of a panel of BCR-ABL1+ pre–B ALL cells showed that samples that express DN-IK6 displayed high levels of CD34, whereas samples that express full-length Ikaros (IK1) had low levels of CD34 (Fig. 7 D, top). The expression of CD43 was high in IKZF1-mutated (DN-IK6+) samples and also in the tyrosine kinase inhibitor (TKI) treatment-relapse samples (T315I or F359V-mutated BCR-ABL1; Fig. 7 D, bottom). These findings parallel the observed increased cell surface expression of CD43 and c-Kit on Ikzf1ΔF4/ΔF4 mutant mouse pre–B ALL cells (Figs. 2 F and 6 E). Together, this supports our findings that a conserved function of Ikaros is to regulate the proper developmental stage-specific expression of cell surface proteins.
Deregulated expression of CD43 and CD34 on IKZF1-mutated pre–B ALL confers leukemic growth advantage
To determine if CD34 or SPN gene expression has a functional effect on leukemic growth, we performed Cas9/CRISPR-mediated depletion of CD34 or SPN in human IKZF1-mutated BCR-ABL1+ pre–B ALL cells. We found that depletion of either of these cell surface receptors (Fig. 7 E) resulted in a growth disadvantage in a growth competition assay (Fig. 7 F). Furthermore, analysis of clinical expression data from the Children’s Oncology Group (COG) Clinical Trial P9906 (available from GEO under accession no. GSE11877; Harvey et al., 2010; Kang et al., 2010) showed that presence of minimal residual disease (MRD) after 29 d of treatment correlated with higher expression of CD34 or SPN (P = 7 × 10−5 and P < 0.007, respectively; Fig. 7 G). When comparing patient samples based on CD34 or SPN expression above or below the median expression level, high expression of either gene correlated with reduced probability of overall survival (OS; P < 0.07 and P < 0.02, respectively; Fig. 7 H). Overall, this indicates that the aberrant expression of CD34 and CD43 on IKZF1-mutated pre–B ALL represent biologically relevant targets of Ikaros tumor suppressor function.
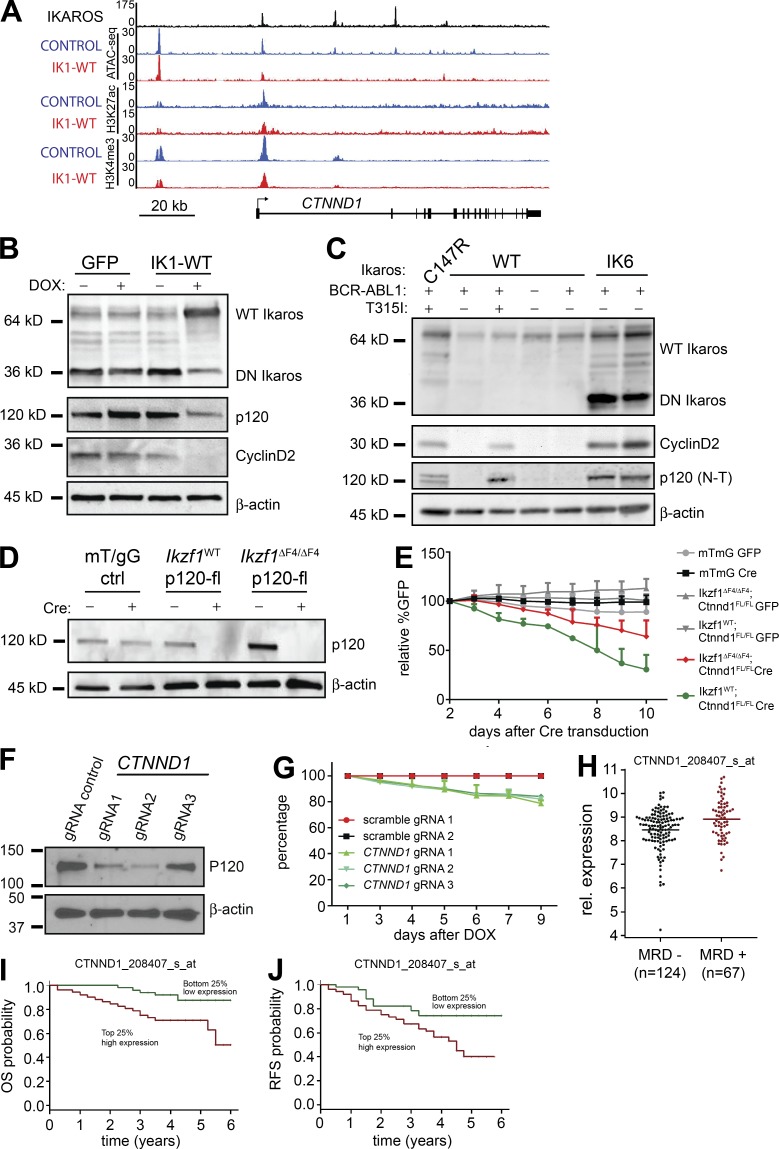
CTNND1 is a conserved Ikaros target gene that confers leukemic growth advantage
Restoration of WT Ikaros in IKZF1-mutated human pre–B ALL resulted in a growth arrest and outcompetition within 4 d (Fig. 4 C), and individual depletion of SPN and CD34 resulted in significant but only partial growth inhibition in these cells (Fig. 7 F). From the list of conserved Ikaros target genes, CTNND1 was among the most highly deregulated genes (Fig. 4 H). CTNND1 expression is reported to be high in human BCR-ABL1+ compared with other types of pre–B ALL (Juric et al., 2007), and higher in IKZF1-mutated than in Ikaros WT pre–B ALL (Mullighan et al., 2009). CTNND1 encodes p120-catenin (also known as p120 or p120-ctn), a multifunctional protein that regulates cadherin stability at the cell membrane, Rac–Rho–cdc42 pathway in the cytoplasm and Wnt/β-catenin target genes in the nucleus (Reynolds, 2007; Kourtidis et al., 2013; Schackmann et al., 2013; McCrea and Gottardi, 2016). We found that Ikaros bound to the CTNND1 promoter and intronic regions and that induction of WT Ikaros resulted in partial reduction of chromatin accessibility and H3K4me3 enrichment at this locus (Fig. 8 A). Furthermore, we found that p120-catenin protein levels were reduced upon induction of WT Ikaros expression (Fig. 8 B). Interestingly, CCND1 (encoding Cyclin D1), which is reported to be an indirect target of p120-catenin through relief of Kaiso-mediated repression (Park et al., 2005; Donaldson et al., 2012; Liu et al., 2014), was also among the list of conserved genes regulated by Ikaros (Fig. 4 H). CCND2 (encoding Cyclin D2) was more highly expressed than CCND1 in the human cells, and it was also repressed by WT Ikaros in the human pre–B ALL cells. Consistent with this, the protein level of Cyclin D2 was greatly reduced upon expression of WT Ikaros (Fig. 8 B). Screening a panel of patient-derived pre–B ALL samples, we found that expression of DN-IK6, as well as TKI-treatment relapse of BCR-ABL1+ pre–B ALL, correlated with both p120-catenin and Cyclin D2 expression (Fig. 8 C). Although a small sample size, this suggests that the selection for high p120-catenin expression in relapsed clones might provide a growth or survival advantage to leukemic cells.
Figure 8.
CTNND1 is a conserved Ikaros target gene that confers leukemic growth advantage. (A) Ikaros binding to the CTNND1 gene locus in human patient-derived BCR-ABL1+ pre–B ALL cells. ATAC-Seq and ChIP-Seq signals are shown for control (GFP) and Ikaros-induced cells (Ik-WT). (B) Expression of WT Ikaros in DN-IK6+ BCR-ABL1+ pre–B ALL resulted in reduced p120-catenin and Cyclin D2protein levels. β-Actin was used for loading control. (C) Expression of Ikaros, p120-catenin, and Cyclin D2 protein was evaluated in a panel of human patient-derived pre–B ALL samples. (D) Cre-mediated inactivation by deletion ofCtnnd1 exon 7 (Elia et al., 2006) was confirmed at the protein level in sorted GFP+ BCR-ABL1–transduced murine pre–B ALL cells. (E) Growth competition assay displayed growth disadvantage of BCR-ABL1+ pre–B ALL cells upon cre-mediated (GFP+) inactivation of Ctnnd1. mTmG cells that lose Red fluorescence (Tomato) upon cre-mediated deletion were used as controls (Muzumdar et al., 2007). Statistical analysis used 2-way ANOVA on mean values. Data in A–E are summaries or representative of two or more independent experiments. (F and G) Cas9/CRISPR-mediated depletion of p120-catenin (encoded by CTNND1) was confirmed by Western blotting for p120-catenin (F) and resulted in reduced growth in liquid culture (P = 0.0011; unpaired Student’s t test; G). Data from three repeats from two independent experiments are displayed (G) with one representative Western blot in (F). (H) Analysis of published clinical data (COG, P9906) showed that presence of MRD after 29 d of treatment correlated with higher expression of CTNND1 (P < 0.002, two-tailed Wilcoxon test; I and J) OS (I) and RSF (J) of patients (COG, P9906) separated on above the top or below the bottom first quartile of mRNA expression level of CTNND1 (P < 0.005 and P < 0.02, respectively; Log-rank test).
We then tested whether p120-catenin provides a growth advantage to cells by performing loss-of-function experiments through retroviral cre-mediated deletion of Ctnnd1 exon 7 (Elia et al., 2006) in mouse BCR-ABL1-transduced pre–B ALL cells. Upon cre-mediated inactivation of Ctnnd1 and loss of p120-catenin protein expression (Fig. 8 D), we observed decreased growth rates of p120-catenin depleted cells in GFP growth-competition assays (Fig. 8 E). Compared with the cre-expressing mTmG control cells, both WT Ikaros cells and Ikzf1ΔF4/ΔF4 cells displayed reduced growth upon deletion of p120-catenin (P < 0.0001 and P < 0.0008, respectively; Fig. 8 E). Of note, the effect was stronger in the Ikaros-WT cells than in the Ikzf1ΔF4/ΔF4 cells (P < 0.0001), indicating that the Ikzf1ΔF4/ΔF4 cells may harbor additional Ctnnd1-independent growth-promoting alterations. Moreover, Cas9/CRISPR-mediated depletion of p120-catenin in human IKZF1-mutated BCR-ABL1+ pre–B ALL resulted in reduced growth (Fig. 8, F and G). Finally, analysis of expression data from the COG Clinical Trial P9906 (available from GEO under accession no. GSE11877; Harvey et al., 2010; Kang et al., 2010) showed that presence of MRD after 29 d of treatment correlated with higher expression of CTNND1 (P < 0.002; Fig. 8 H). When comparing patient samples based on CTNND1 expression above the top or below the bottom first quartile, high CTNND1 expression correlated with reduced probability of OS (P < 0.005; Fig. 8 I) and relapse-free survival (RFS; P < 0.02; Fig. 8 J). Collectively, these results indicate that CTNND1 is a direct target of Ikaros that confers oncogenic growth properties. Collectively, our analysis of conserved Ikaros tumor suppressor function indicate that IKZF1 mutations lead to deregulation of multiple cellular functions that, together, confer increased leukemogenesis.
DISCUSSION
We have investigated the mechanisms of Ikaros tumor suppressor function using our mouse model of Ikzf1ΔF4/ΔF4-mutated BCR-ABL1+ pre–B ALL combined with a new model of inducible expression of WT Ikaros in human IKZF1-mutated BCR-ABL1+ pre–B ALL. We have defined the Ikaros target genes associated with tumor suppression in both mouse and human BCR-ABL1+ pre–B ALL, and have demonstrated conserved Ikaros target genes and a converging network of genetic pathways. In particular, Ikaros mutant BCR-ABL1+ pre–B ALL cells display lineage plasticity and fail to down-regulate developmentally restricted cell surface receptors, including c-Kit, CD34 and CD43, likely retaining the functions and signaling abilities of these receptors. Genetic loss-of-function experiments demonstrated that these cell surface receptors confer part of the observed growth advantage in Ikaros-mutated pre–B ALL. Although CD34 has been widely studied as a cell surface marker, it was not known whether this receptor has any functional activity on pre–B ALL. Here we present the novel finding that the aberrant expression of CD34 on IKZF1-mutated pre–B ALL not only represents a biomarker, but also functionally influences the aggressiveness of the disease. CD43 is reported to interact and cooperate with β-catenin and promote cell growth in nonhematopoietic cancer cells (Andersson et al., 2004; Balikova et al., 2012), and its expression has been shown to correlate with poor prognosis also in other cancers (Mitrovic et al., 2013; Ma et al., 2015). In addition to the identified hematopoietic cell surface receptors, the intracellular protein p120-catenin was found to be a conserved Ikaros target, and conferred a growth advantage to the leukemic cells. Although p120-catenin is extensively studied in other tissues, very little is known about p120-catenin in hematopoietic cells. However, microarray data from the Immgen consortium indicates that CTNND1 is expressed in early hematopoietic progenitor cells and is normally down-regulated as cells differentiate down the B cell lineage (Heng et al., 2008). Collectively, a conserved function of Ikaros is to restrict expression of developmental stage-specific genes that promote growth.
An increasing body of work from mouse models supports the model that Ikaros is required for proper regulation of various developmentally regulated genes in primary lymphocytes (Schwickert et al., 2014; Arenzana et al., 2015), including the silencing of genes involved in stem-ness and self-renewal (Ng et al., 2009). Its many and critical roles in normal early B cell development are extensively studied (Wang et al., 1996; Reynaud et al., 2008; Ferreirós-Vidal et al., 2013; Heizmann et al., 2013; Schjerven et al., 2013; Joshi et al., 2014; Schwickert et al., 2014), but the mechanism underlying the tumor suppressor role in pre–B ALL remains incompletely understood. In the present study, we find that Ikaros exerts its tumor suppressor function in part by promoting differentiation of the pro-B cells past the highly proliferative fraction C' or large pre–BII cell stage, and importantly, by turning off the progenitor cell gene expression program. Our results are in support of a recent report highlighting the stem-cell like properties of Ikzf1-mutated mouse pre–B ALL (Churchman et al., 2015). In addition, our results uncovered novel Ikaros target genes not previously associated with Ikaros function. We have directly investigated the tumor suppressor role of Ikaros in human patient-derived BCR-ABL1+ pre–B ALL and we describe new Ikaros targets that confer growth advantage in functional assays and that correlate with poor outcome in clinical data.
Like many Ikaros-regulated genes defined in this study, the CD34, SPN, and CTNND1 genes were bound by Ikaros in human pre–B ALL cells. Although our Ikaros ChIP-Seq analysis in human pre–B ALL resulted in thousands of high-confidence Ikaros-binding sites, it is difficult to determine whether individual binding events represent functional or opportunistic interactions at accessible chromatin. To address this, we performed genome-wide analysis of chromatin structure after WT Ikaros induction to evaluate the functional consequence of Ikaros binding, and the mechanism by which Ikaros regulates genes. We found that Ikaros expression was associated with loss of chromatin accessibility at Ikaros-binding sites, supporting a direct repressive role at its down-regulated target genes. Surprisingly, although several genes were activated upon expression of WT Ikaros, our current analysis only revealed a modest effect on the active chromatin marks H3K4me3 and H3K27ac. Although Ikaros is known to interact with chromatin-modifying complexes, the mechanism by which Ikaros regulates gene expression is not fully understood, and several reports indicate that Ikaros might have an atypical mode of action (Kim et al., 1999; Sridharan and Smale, 2007; Zhang et al., 2011; Arenzana et al., 2015). Thus, it is likely that induction of WT Ikaros results in gene regulation also through other mechanisms, such as the modulation of other transcription factors and possibly higher-order chromatin structure. Nevertheless, our current analysis does not rely on defining the direct targets, but seeks to determine the gene expression changes leading to the observed biological phenotype of loss of Ikaros tumor suppressor function. As such, we present genome-wide data that can also be used for future analysis of Ikaros function in pre–B ALL and we provide examples of Ikaros-responsive genes with Ikaros-induced changes in chromatin as candidate direct Ikaros target genes associated with pathways of Ikaros tumor suppressor function.
We combined two different approaches, in two different species, each with a single variable parameter of Ikaros-perturbation. The mouse model used a small deletion of Ikaros DNA-binding zinc finger 4 that selectively leads to loss of tumor suppressor function, while retaining some of Ikaros’ other functions, and thus helps filter the list of target genes relevant for tumor suppression. In the human patient–derived BCR-ABL1+ pre–B ALL cells with endogenous expression of DN-IK6, we inducibly expressed WT Ikaros, resulting in cell cycle exit, and analyzed the resulting changes in chromatin and gene expression. Together, these approaches gave a robust and comprehensive analysis indicating conserved Ikaros-regulated pathways and genes in BCR-ABL1+ pre–B ALL.
Deregulated Ikaros target genes show several points of convergence on a cellular network coupling cell surface protein expression with intracellular Wnt and Rho signaling, and catenin-regulated gene regulation in the nucleus. Recently, it was proposed that members of the catenin family also mediate a network of gene regulation in the nucleus, similar to its well-studied network of interactions in regards to cadherin interactions at the cell membrane (McCrea and Gottardi, 2016). These deregulated pathways can in turn be linked to several aspects of oncogenesis, such as proliferation, loss of contact inhibition and adhesion-mediated drug resistance. CD43 has been linked to loss of contact inhibition of growth (Camacho-Concha et al., 2013) and we observed increased expression levels of CD43 in both mouse and human IKZF1-mutated pre–B ALL. The observed increased expression of p120-catenin could also contribute to increased growth and loss of contact inhibition through regulation of β-catenin signaling (Zhu et al., 2012). Indeed, the Wnt–β-catenin pathway has been implicated in drug resistance and suggested as a new target for therapy in pre–B ALL (Yang et al., 2013; Dandekar et al., 2014; Gang et al., 2014).
An interesting question that remains unanswered is the biological reason for the high occurrence of IKZF1 mutations in BCR-ABL1+ (Ph+) and Ph-like pre–B ALL. BCR-ABL1 interacts with and phosphorylates the adaptor protein CRKL, which also binds c-Kit and CD34, two cell surface receptors expressed on HSC that we here found to be specifically deregulated in Ikaros mutant BCR-ABL1+ pre–B ALL. Abl tyrosine kinases are reported to be involved in the cadherin-associated Rho signaling pathway, and BCR-ABL1 is found to interact with and activate Rho family GTPases (Harnois et al., 2003; Zandy and Pendergast, 2008), and to confer mobility, and potentially increased invasiveness, in leukemia cells in a Rho signaling–dependent process (Daubon et al., 2012). In BCR-ABL1+ chronic myelogenous leukemia (CML), cadherin-dependent adhesion to BM stroma is reported to provide protection against imatinib treatment through stromal-derived noncanonical Wnt–β-catenin signaling (Zhang et al., 2013; Chen et al., 2014). Furthermore, CTNND1, a key Ikaros target gene identified in this study, was found to be more highly expressed in human BCR-ABL1+ compared with other pre–B ALL (Juric et al., 2007). p120-catenin regulates cadherin stability at the cell membrane, as well as the Rac–Rho–cdc42 pathway in the cytoplasm and Wnt–β-catenin target genes in the nucleus (Kourtidis et al., 2013; Schackmann et al., 2013; McCrea and Gottardi, 2016). In addition to regulating CTNND1, Ikaros was found to regulate several genes involved in these integrated pathways. Collectively, Ikaros regulates several interacting partners of BCR-ABL1 that have been linked to growth advantage, protection against drug through stromal adherence, and enhanced invasiveness. This might explain the strong association of loss of Ikaros function in BCR-ABL1+ pre–B ALL, and the correlation of IKZF1-mutations with poor prognosis. Hence, understanding the network of Ikaros-regulated target genes and pathways may provide new insight to guide the development of new targeted therapies.
Although therapies have improved, relapse as well as secondary malignancies caused by initial cancer treatment still constitute major clinical problems. For these reasons, additional targeted therapies with reduced side-effects are needed. Collectively, our results presented herein suggest that Ikaros mediates tumor suppression, at least in part, by enforcing proper developmental stage-specific expression of multiple genes that each contribute to the aggressiveness of growth. This provides a rationale for exploring these new Ikaros target genes and pathways for development of targeted therapies for IKZF1-mutated pre–B ALL.
MATERIALS AND METHODS
Mice
The Ctnnd1Floxed (p120ctnflox), Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo (herein referred to as mTmG), Ikzf1ΔF1/ΔF1, and Ikzf1ΔF4/ΔF4 mice were described earlier (Elia et al., 2006; Muzumdar et al., 2007; Schjerven et al., 2013). The Ikzf1ΔF1/ΔF1 and Ikzf1ΔF4/ΔF4 mice were backcrossed onto C57BL/6 (n ≥ 12). The Ctnnd1Floxed mice were on a mixed background, but backcrossed to C57BL/6 through in-cross with Ikzf1ΔF4/ΔF4 mice at least three generations, and littermate controls were used for all WT-Ikzf1 or Ikzf1ΔF4/ΔF4 Ctnnd1Floxed mice. Two independent experiments were performed with two or more biological replicates for each experimental sample. mTmG control cells were obtained from a separate litter. WT C57BL/6 mice were obtained from The Jackson Laboratory. Animals were housed by the Division of Laboratory Animal Medicine (DLAM) at UCLA or by Laboratory Animal Resource Center (LARC) at UCSF. All experiments were approved by the UCLA and UCSF Animal Research Committees (ARC) and were performed according to UCLA and UCSF Institutional Animal Care and Use Committee (IACUC) guidelines.
Retroviral transduction, mouse cell culture, and BM transplantation
Retroviral supernatants for expression of BCR-ABL1 were produced with the plasmid pMSCV-YFP-IRES-p185 (Figs. 1–3 and 6, D–F) or pMSCV-p210-IRES-NEO (Fig. 8, D and E), and pMSCV-Ikaros-IRES-GFP for expression of Ikaros isoforms (Fig. 6 C). BM cells were harvested from the indicated mice and either transduced directly and plated on top of preestablished feeder cells from WT C57BL/6 BM stroma at 5 × 106 transduced cells/6-well (Figs. 1–3 and 6, D–F), or cultured for 4–5 d in vitro at 4 × 106/ml with IL-7 (10 ng/ml) for enrichment of progenitor B cells before transduction with BCR-ABL-p210-NEO (Fig. 8, D and E). Upon establishment of IL-7–independent BCR-ABL1+ pre–B ALL cells, the cells were subsequently transduced with retroviral MIG-CRE or empty vector MIG control for in vitro inactivation of Ctnnd1 by deletion of the floxed exon 7 in the Ctnnd1 gene (Fig. 8, D and E; Elia et al., 2006). For in vivo experiments, 105 or 104 of day 7 in vitro expanded BCR-ABL1+ pre–B ALL cells were injected i.v. into nonirradiated WT C57BL/6 recipient mice (Figs. 1 E and 6 F). Mice were monitored for development of B-ALL, and euthanized upon endpoint as evaluated by paralysis of hind legs or a moribund condition. EBF-rescued Ikzf1null pro-B cells were cultured on OP9 stromal cell support with recombinant IL-7, as previously described (Fig. 6 C; Reynaud et al., 2008). For in vitro c-Kit inhibition assay (Fig. 6 D), BCR-ABL1-p185-IRES-YFP+ cells were seeded at 5 × 105 cells/ml, counted and split every day, and c-Kit blocking Ab (ACK45) was added daily at 10 µg/ml.
Cloning of human Ikaros expression vectors, lentiviral transduction, and in vitro cell culture of human patient–derived BCR-ABL1+ pre–B ALL cells
Patient-derived, xenograft-expanded human BCR-ABL1+ pre–B ALL cells were cultured in vitro with or without irradiated OP9 cells as stromal support. Human full-length WT Ikaros (IK1) and the leukemia-derived DN IK6 isoform created by Rag-mediated intragenic deletion (Mullighan et al., 2008), were cloned from above patient-derived pre–B ALL cells and verified by Sanger sequencing. The two Ikaros isoforms and GFP were cloned into the TET-inducible expression vector pLVX-TRE3G-IRES (Takara Bio Inc.) with In-Fusion cloning reagents (Takara Bio Inc.). Lentiviral supernatants for expression of pLVX-EF1a-Tet3G (Takara Bio Inc.) and aforementioned cloned Ikaros and GFP-control pLVX-TRE3G-IRES were produced with the packaging vector pCDNLBH and EM140 envelope vector. Expression was induced with doxycycline, a tetracycline (TET) analogue. For Cas9/CRISPR-mediated depletion experiments, scrambled or gene-targeting gRNA sequences (Table S5) were purchased from Transomic technologies (cloned into the pCLIP-grNA-hCMV-RFP vector) and transduced into parental cells that had been stably transduced and selected for DOX-inducible expression of Cas9-IRES-ZsGreen.
Flow cytometry and cell sorting
Antibodies used for flow cytometry were from BioLegend, BD, or eBioscience, except for the antibody used to detect cytoplasmic immunoglobulin-µ, which was from Southern Biotech (Table S1). Cell cycle analysis was performed with Hoechst dye incorporation (Figs. 1 A and 2 F) or with BrdU (BD; BioLegend) and 7AAD incorporation (BioLegend; Fig. 4 B). GFP, ZsGreen, and RFP expression was monitored by flow cytometry and GFP+high cells were sorted for mRNA, protein analysis, ATAC-Seq and ChIP-Seq. A FACSCalibur, LSR I, LSR Fortessa, or FACSAria II SOP (BD) were used for flow cytometry or sorting, and data were analyzed with FlowJo and FACSDiva software.
Western blot, gDNA, IgH V(D)J recombination, and genotyping PCR
Cells were harvested and washed in PBS, and whole-cell lysate was prepared for Western blot analysis. Antibodies used are listed in Table S2. Cell pellets were snap frozen for subsequent gDNA extraction by genomic DNA isolation kit (QIAGEN and Macherey-Nagel) and PCR for analysis of IgH V(D)J recombination was performed as previously described (Schlissel et al., 1991) with primers listed in Table S3. For genotyping of mice, genotyping PCR was performed on either genomic DNA isolated with genomic DNA isolation kit (QIAGEN) or crude tissue lysis, or performed by direct use of Phusion Blood Direct PCR Master Mix (Thermo Fisher Scientific). Primers for genotyping PCR are listed in Table S3.
RNA isolation, qRT-PCR, and library preparation for RNA-Seq
RNA was isolated using TRI Reagent (Molecular Research Center, Inc.), followed by purification with an RNeasy kit (QIAGEN), or direct isolation by RNA-isolation kit (QIAGEN and Macherey-Nagel), with on-column RNase-free DNase I treatment. Isolated RNA was either reverse transcribed to cDNA (for qRT-PCR) or processed for RNA-Seq. Primers used for quantitative real-time RT-PCR of mouse mRNA are listed in Table S4. Ubc was used as reference gene, with Gapdh and β-Actin as additional control primers. For mouse RNA-Seq experiments, the Tru-Seq RNA Sample Prep kit (Illumina) was used to prepare sequencing libraries according to the manufacturer's protocol. For the human RNA-Seq experiments, rRNA was depleted using 1 µg of total RNA with the RiboErase kit (KAPA Biosystems). Strand-specific Illumina cDNA libraries were prepared using the KAPA Stranded RNA-Seq library preparation kit with nine cycles of PCR (KAPA Biosystems). Libraries with unique indexes were pooled and sequencing was performed on the Illumina HiSeq2000 instrument to generate 50-bp (mouse) or 100-bp single-end sequencing reads (human).
RNA sequencing analysis
Quality scores across sequenced reads were assessed using FASTQC v0.9.2. All samples were high quality. The average score (mean and median) at each base across reads in each sample was Q > 30. For alignment and transcript assembly, the sequencing reads were mapped to the mouse or human reference genomes (mm10 or hg38, respectively) using TopHat2 (Kim et al., 2013) with the vM7 and v23 releases of the comprehensive Gencode GTF transcript annotation files for either reference genome, respectively. The BAM files were sorted and converted to SAM files using SAMTOOLS (Li et al., 2009) and reads were counted using HTSEQ-count against the corresponding Gencode GTF file (Anders et al., 2015). Differential expression analysis was performed using the DESeq2 package (Love et al., 2014). Genes with a false discovery rate of <0.5% (i.e., FDR < 0.005) and a fold change > 2 were considered significantly differentially expressed (Figs. 3 A and B; and 4 D). For the mouse to human conservation analysis, P < 0.01 and fold change > 2 was used to filter the DESeq2 differentially expressed genes before comparison (Fig. 4 H). Heat maps were generated using the lists of differentially expressed genes unique to each comparison and the amount by which each of those genes deviate in each sample from the gene’s average across all samples. Clustering was performed using k-means method and Euclidian distance metrics using Cluster3 (de Hoon et al., 2004) and visualized using Java Treeview (Saldanha, 2004) or with custom R scripts.
Gene ontology, gene set enrichment and IPA
Genes differentially expressed (FDR < 0.005) between WT and mutant (mouse) or between IK1-WT and GFP-control (human) were included in pathway and functional analysis using IPA (Ingenuity Systems). The functional and canonical pathway analysis was used to identify the significant biological functions and pathways. Functions and pathways with P < 0.05 (Fischer’s exact test) were considered to be statistically significant. The activation z-score was used to predict activation or inhibition of pathways based on published findings accessible through the Ingenuity knowledge base. Canonical pathways or disease and function categories with z-score >1 or <−1 were considered to be significantly activated or inhibited, respectively. The IPA comparative analysis function was performed to relate mouse and human pathways. Gene ontology analysis was performed using DAVID (Huang et al., 2009). Pre-ranked GSEA was performed by ranking the differentially expressed genes (FDR < 0.005; fold change > 2) according to decreasing fold change (IK1-WT compared with GFP-control; Subramanian et al., 2005). The gene sets used for this analysis were the C2 Curated Molecular Signatures Database (MSigDB) genesets and the Dick laboratory human hematopoietic stem and progenitor cells genesets (Laurenti et al., 2013).
Patient outcome, gene expression microarray data, and survival analysis
Patient gene expression microarray and outcome data were obtained from the GEO database under accession no. GSE11877 (Harvey et al., 2010; Kang et al., 2010) of the COG Clinical Trial P9906. The end points of the clinical data include MRD after 29 d of treatment (for 207 COG samples), OS, and RFS. Kaplan-Meier survival analysis was used to estimate OS and RFS. Log-rank test was used to compare survival differences between patient groups. Patients with ALL in the clinical trial were segregated into two groups according to whether they had above or below the median expression level of a gene or above the top or below the bottom first quartile. For comparisons of MRD+ vs. MRD−, two-tailed Wilcoxon test was used to calculate P values. R package “survival” version 2.35–8 was used for the survival analysis (R Development Core Team, 2009).
Chromatin immunoprecipitation and ChIP-Seq analysis
Patient-derived, xenograft-expanded human BCR-ABL1+ pre–B ALL cells were expanded in vitro with or without irradiated OP9 cells. Ikaros and histone-modification specific ChIP was performed and ChIP-Seq libraries prepared (O’Geen et al., 2010) as previously described (Schjerven et al., 2013). In brief, cells were fixed in 1% formaldehyde for 10 min at room temperature, quenched with freshly prepared glycine, washed on ice with ice-cold PBS, and snap-frozen in liquid nitrogen as fixed cell pellets. Ikaros ChIP was performed on two different patient-derived pre–B ALL samples, and with antibodies against Ikaros N-terminal (clone H-100, Santa Cruz, cat # sc-13039-X; named IkH in figures) and C-terminal (Smale laboratory in-house Ab; Hahm et al., 1998; named IkC in figures). For the histone modification ChIP-Seq samples, WT Ikaros or control GFP cells were induced for >24 h and sorted for GFP signal before cross-linking. Two independent culture and sorting runs were used to generate independent replicates. The antibodies used were H3K4me3 and H3K27ac (Abcam ab8580 and ab4729). Libraries were sequenced at both the University of Southern California Epigenome Data Production Facility and the University of Vermont Advanced Genome Technologies Core on the Illumina HiSeq2000 instrument to generate 50-bp single-end sequencing reads. Mouse progenitor B cells Ikaros ChIP-Seq and corresponding input datasets were obtained from NCBI Gene Expression Omnibus accession GSE66978 (Bossen et al., 2015). Sequencing reads were mapped using Bowtie2 using the mm10 or hg19 reference genomes for mouse or human datasets, respectively (Langmead and Salzberg, 2012). Resulting SAM alignment files were used for peak calling using MACS2 against control input samples (Zhang et al., 2008). Pooled replicates were used to generate profile plots using ngs.plot (Shen et al., 2014). Target genes were annotated using significantly enriched peak datasets using ChIP BETA minus after lifting over ChIP-seq peaks from hg19 to hg38 genome coordinates (Wang et al., 2013). ChIP-Seq data were visualized using the Integrated Genome Browser (Nicol et al., 2009). The binding and expression target analysis (BETA) software package BETA tool was used to predict activating and repressive Ikaros ChIP-Seq targets (Wang et al., 2013), whereby significantly enriched ChIP-Seq peaks are associated to a differential expression dataset considering all genes having a peak within 100 kb of the gene TSS. For Ikaros we used pre–B Ikaros ChIP-Seq data (Bossen et al., 2015) and differential expression data comparing WT to Ikzf1ΔF4/ΔF4 pre–B ALL datasets.
ATAC-Seq
ATAC-Seq was performed as previously described (Buenrostro et al., 2015) on 50,000 WT Ikaros or GFP control FACS sorted cells. Libraries were generated using custom Nextera barcoded primers (Buenrostro et al., 2015) and were amplified for a total of 10 cycles. Libraries were purified with AMPure beads (Agencourt) and library quality was assessed using the Agilent Bioanalyzer High-Sensitivity DNA kit. Libraries were quantitated using Qubit (Life Technologies). ATAC-Seq libraries were sequenced on the Illumina HiSeq 2000 with 50 bp single-end reads. Illumina adapters were trimmed by Trimmomatic (Bolger et al., 2014), and the 50 bp reads were shortened by 10 bp with fastx_trimmer (http://hannonlab.cshl.edu/fastx_toolkit/). All reads for each sample were combined and aligned to hg19 with Bowtie2 (Langmead and Salzberg, 2012) using default settings.
The mouse and human RNA-Seq and human ATAC-Seq and histone ChIP-Seq and Ikaros ChIP-Seq datasets are deposited in the GEO database under accession nos. GSE39160, GSE90670, and GSE58825, respectively.
Online supplemental material
Table S1 and S2 list the antibodies used for flow cytometry and Western blotting, respectively. Table S3 shows the sequences of primers used for analysis of V(D)J recombination and mouse genotyping, whereas Table S4 shows the primers used for qRT-PCR of mouse RNA. Table S5 gives the sequence of the guide RNA for Cas9/CRISPR experiments. Fig. S1 shows the analysis of genes bound by Ikaros in mouse pro-B cells based on an overlap analysis of published Ikaros ChIP-Seq datasets from three independent laboratories. Fig. S2 shows the overlap analysis of the herein presented Ikaros ChIP-Seq experiments in human Ph+ pre–B ALL cells.
ACKNOWLEDGMENTS
We thank Division of Laboratory Animal Medicine (DLAM) at UCLA and Laboratory Animal Resource Center (LARC) at UCSF for ongoing care of mice, and Cesar Garcia, Juan Lorenzano, Carrie Lin and Biniam Tesfai for technical assistance with the mouse colonies. We thank Harinder Singh and Louis F. Reichardt for kind gift of cells and mice, respectively. This work benefitted from publicly available microarray RNA expression data deposited by the Immgen consortium. Human RNA-Seq, ChIP-Seq and ATAC-Seq libraries were sequenced at the University of Southern California Epigenome Data Production Facility and the University of Vermont Advanced Genome Technologies Core supported by the University of Vermont Cancer Center, Lake Champlain Cancer Research Organization, and the University of Vermont College of Medicine.
This work was supported by National Institutes of Health grants R21CA209229 (to H.S. and S.F.), RO1DK043726 (to S.T.S.), R01CA137060, R01CA139032, R01CA157644, R01CA169458 and R01CA172558 grants (to M.M), and by funding from an American Cancer Society IRG grant at the University of Vermont (to S.F. and J.L.H.). O.N.W. is supported in part by the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA. M.M. is an HHMI Faculty Scholar and supported by an NCI Outstanding Investigator Award through R35CA197628, and H.S. is an Arthritis National Research Foundation (ANRF) Scholar, a Hellman Fellow, and a Member of the UCSF Helen Diller Family Comprehensive Cancer Center.
The authors declare no conflicting financial interests.
Footnotes
Abbreviations used:
- ALL
- acute lymphoblastic leukemia
- COG
- Children’s Oncology Group
- DN
- dominant-negative
- EMT
- epithelial to mesenchymal transition
- GMP
- granulocyte-macrophage progenitor
- GSEA
- gene set enrichment analysis
- HSC
- hematopoietic stem cell
- IPA
- Ingenuity Pathway Analysis
- MRD
- minimal residual disease
- OS
- overall survival
- Ph
- Philadelphia chromosome
- pre-B ALL
- progenitor B-cell ALL
- RFS
- relapse-free survival
- TKI
- tyrosine kinase inhibitor
- TSS
- transcription start sites
- ZnF
- zinc finger
References
- Anders S., Pyl P.T., and Huber W.. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson C.X., Fernandez-Rodriguez J., Laos S., Sikut R., Sikut A., Baeckström D., and Hansson G.C.. 2004. CD43 has a functional NLS, interacts with beta-catenin, and affects gene expression. Biochem. Biophys. Res. Commun. 316:12–17. 10.1016/j.bbrc.2004.02.011 [DOI] [PubMed] [Google Scholar]
- Arenzana T.L., Schjerven H., and Smale S.T.. 2015. Regulation of gene expression dynamics during developmental transitions by the Ikaros transcription factor. Genes Dev. 29:1801–1816. 10.1101/gad.266999.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balikova A., Jääger K., Viil J., Maimets T., and Kadaja-Saarepuu L.. 2012. Leukocyte marker CD43 promotes cell growth in co-operation with β-catenin in non-hematopoietic cancer cells. Int. J. Oncol. 41:299–309. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., and Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossen C., Murre C.S., Chang A.N., Mansson R., Rodewald H.R., and Murre C.. 2015. The chromatin remodeler Brg1 activates enhancer repertoires to establish B cell identity and modulate cell growth. Nat. Immunol. 16:775–784. 10.1038/ni.3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Wu B., Chang H.Y., and Greenleaf W.J.. 2015. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol. 109:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho-Concha N., Olivos-Ortiz A., Nuñez-Rivera A., Pedroza-Saavedra A., Gutierrez-Xicotencatl L., Rosenstein Y., and Pedraza-Alva G.. 2013. CD43 promotes cells transformation by preventing merlin-mediated contact inhibition of growth. PLoS One. 8:e80806 10.1371/journal.pone.0080806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Zhang H.X., Wang M., Song X.G., Cao J., Wang L., Qiao J.L., Lu X.Y., Han Z.X., Zhu P., et al. 2014. Stromal cells attenuate the cytotoxicity of imatinib on Philadelphia chromosome-positive leukemia cells by up-regulating the VE-cadherin/β-catenin signal. Leuk. Res. 38:1460–1468. 10.1016/j.leukres.2014.09.012 [DOI] [PubMed] [Google Scholar]
- Churchman M.L., Low J., Qu C., Paietta E.M., Kasper L.H., Chang Y., Payne-Turner D., Althoff M.J., Song G., Chen S.C., et al. 2015. Efficacy of Retinoids in IKZF1-Mutated BCR-ABL1 Acute Lymphoblastic Leukemia. Cancer Cell. 28:343–356. 10.1016/j.ccell.2015.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar S., Romanos-Sirakis E., Pais F., Bhatla T., Jones C., Bourgeois W., Hunger S.P., Raetz E.A., Hermiston M.L., Dasgupta R., et al. 2014. Wnt inhibition leads to improved chemosensitivity in paediatric acute lymphoblastic leukaemia. Br. J. Haematol. 167:87–99. 10.1111/bjh.13011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubon T., Rochelle T., Bourmeyster N., and Génot E.. 2012. Invadopodia and rolling-type motility are specific features of highly invasive p190(bcr-abl) leukemic cells. Eur. J. Cell Biol. 91:978–987. 10.1016/j.ejcb.2012.04.006 [DOI] [PubMed] [Google Scholar]
- de Hoon M.J., Imoto S., Nolan J., and Miyano S.. 2004. Open source clustering software. Bioinformatics. 20:1453–1454. 10.1093/bioinformatics/bth078 [DOI] [PubMed] [Google Scholar]
- Donaldson N.S., Pierre C.C., Anstey M.I., Robinson S.C., Weerawardane S.M., and Daniel J.M.. 2012. Kaiso represses the cell cycle gene cyclin D1 via sequence-specific and methyl-CpG-dependent mechanisms. PLoS One. 7:e50398 10.1371/journal.pone.0050398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumortier A., Jeannet R., Kirstetter P., Kleinmann E., Sellars M., dos Santos N.R., Thibault C., Barths J., Ghysdael J., Punt J.A., et al. 2006. Notch activation is an early and critical event during T-Cell leukemogenesis in Ikaros-deficient mice. Mol. Cell. Biol. 26:209–220. 10.1128/MCB.26.1.209-220.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia L.P., Yamamoto M., Zang K., and Reichardt L.F.. 2006. p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron. 51:43–56. 10.1016/j.neuron.2006.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felschow D.M., McVeigh M.L., Hoehn G.T., Civin C.I., and Fackler M.J.. 2001. The adapter protein CrkL associates with CD34. Blood. 97:3768–3775. 10.1182/blood.V97.12.3768 [DOI] [PubMed] [Google Scholar]
- Ferreirós-Vidal I., Carroll T., Taylor B., Terry A., Liang Z., Bruno L., Dharmalingam G., Khadayate S., Cobb B.S., Smale S.T., et al. 2013. Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood. 121:1769–1782. 10.1182/blood-2012-08-450114 [DOI] [PubMed] [Google Scholar]
- Gang E.J., Hsieh Y.T., Pham J., Zhao Y., Nguyen C., Huantes S., Park E., Naing K., Klemm L., Swaminathan S., et al. 2014. Small-molecule inhibition of CBP/catenin interactions eliminates drug-resistant clones in acute lymphoblastic leukemia. Oncogene. 33:2169–2178. 10.1038/onc.2013.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos K., Bigby M., Wang J.H., Molnar A., Wu P., Winandy S., and Sharpe A.. 1994. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 79:143–156. 10.1016/0092-8674(94)90407-3 [DOI] [PubMed] [Google Scholar]
- Hahm K., Cobb B.S., McCarty A.S., Brown K.E., Klug C.A., Lee R., Akashi K., Weissman I.L., Fisher A.G., and Smale S.T.. 1998. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev. 12:782–796. 10.1101/gad.12.6.782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R.R., Carmack C.E., Shinton S.A., Kemp J.D., and Hayakawa K.. 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 173:1213–1225. 10.1084/jem.173.5.1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnois T., Constantin B., Rioux A., Grenioux E., Kitzis A., and Bourmeyster N.. 2003. Differential interaction and activation of Rho family GTPases by p210bcr-abl and p190bcr-abl. Oncogene. 22:6445–6454. 10.1038/sj.onc.1206626 [DOI] [PubMed] [Google Scholar]
- Harvey R.C., Mullighan C.G., Wang X., Dobbin K.K., Davidson G.S., Bedrick E.J., Chen I.M., Atlas S.R., Kang H., Ar K., et al. 2010. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 116:4874–4884. 10.1182/blood-2009-08-239681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heizmann B., Kastner P., and Chan S.. 2013. Ikaros is absolutely required for pre-B cell differentiation by attenuating IL-7 signals. J. Exp. Med. 210:2823–2832. 10.1084/jem.20131735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng T.S., and Painter M.W.. Immunological Genome Project Consortium . 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9:1091–1094. 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., and Lempicki R.A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Hunger S.P., and Mullighan C.G.. 2015. Acute Lymphoblastic Leukemia in Children. N. Engl. J. Med. 373:1541–1552. 10.1056/NEJMra1400972 [DOI] [PubMed] [Google Scholar]
- Joshi I., Yoshida T., Jena N., Qi X., Zhang J., Van Etten R.A., and Georgopoulos K.. 2014. Loss of Ikaros DNA-binding function confers integrin-dependent survival on pre-B cells and progression to acute lymphoblastic leukemia. Nat. Immunol. 15:294–304. 10.1038/ni.2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric D., Lacayo N.J., Ramsey M.C., Racevskis J., Wiernik P.H., Rowe J.M., Goldstone A.H., O’Dwyer P.J., Paietta E., and Sikic B.I.. 2007. Differential gene expression patterns and interaction networks in BCR-ABL-positive and -negative adult acute lymphoblastic leukemias. J. Clin. Oncol. 25:1341–1349. 10.1200/JCO.2006.09.3534 [DOI] [PubMed] [Google Scholar]
- Kang H., Chen I.M., Wilson C.S., Bedrick E.J., Harvey R.C., Atlas S.R., Devidas M., Mullighan C.G., Wang X., Murphy M., et al. 2010. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood. 115:1394–1405. 10.1182/blood-2009-05-218560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Sif S., Jones B., Jackson A., Koipally J., Heller E., Winandy S., Viel A., Sawyer A., Ikeda T., et al. 1999. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 10:345–355. 10.1016/S1074-7613(00)80034-5 [DOI] [PubMed] [Google Scholar]
- Kourtidis A., Ngok S.P., and Anastasiadis P.Z.. 2013. p120 catenin: an essential regulator of cadherin stability, adhesion-induced signaling, and cancer progression. Prog. Mol. Biol. Transl. Sci. 116:409–432. 10.1016/B978-0-12-394311-8.00018-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E., Doulatov S., Zandi S., Plumb I., Chen J., April C., Fan J.B., and Dick J.E.. 2013. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat. Immunol. 14:756–763. 10.1038/ni.2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennartsson J., and Rönnstrand L.. 2012. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol. Rev. 92:1619–1649. 10.1152/physrev.00046.2011 [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Caffrey T.C., Steele M.M., Mohr A., Singh P.K., Radhakrishnan P., Kelly D.L., Wen Y., and Hollingsworth M.A.. 2014. MUC1 regulates cyclin D1 gene expression through p120 catenin and β-catenin. Oncogenesis. 3:e107 10.1038/oncsis.2014.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis T.C., Ichii M., Brugman M.H., Kincade P., and Staal F.J.. 2012. Wnt signaling strength regulates normal hematopoiesis and its deregulation is involved in leukemia development. Leukemia. 26:414–421. 10.1038/leu.2011.387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.B., Zheng Y., Yuan H.P., Jiang J., and Wang Y.P.. 2015. CD43 expression in diffuse large B-cell lymphoma, not otherwise specified: CD43 is a marker of adverse prognosis. Hum. Pathol. 46:593–599. 10.1016/j.humpath.2015.01.002 [DOI] [PubMed] [Google Scholar]
- Mårtensson I.L., Almqvist N., Grimsholm O., and Bernardi A.I.. 2010. The pre-B cell receptor checkpoint. FEBS Lett. 584:2572–2579. 10.1016/j.febslet.2010.04.057 [DOI] [PubMed] [Google Scholar]
- McCrea P.D., and Gottardi C.J.. 2016. Beyond β-catenin: prospects for a larger catenin network in the nucleus. Nat. Rev. Mol. Cell Biol. 17:55–64. 10.1038/nrm.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubrey J.A., Steelman L.S., Bertrand F.E., Davis N.M., Abrams S.L., Montalto G., D’Assoro A.B., Libra M., Nicoletti F., Maestro R., et al. 2014. Multifaceted roles of GSK-3 and Wnt/β-catenin in hematopoiesis and leukemogenesis: opportunities for therapeutic intervention. Leukemia. 28:15–33. 10.1038/leu.2013.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misaghian N., Ligresti G., Steelman L.S., Bertrand F.E., Bäsecke J., Libra M., Nicoletti F., Stivala F., Milella M., Tafuri A., et al. 2009. Targeting the leukemic stem cell: the Holy Grail of leukemia therapy. Leukemia. 23:25–42. 10.1038/leu.2008.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovic Z., Iqbal J., Fu K., Smith L.M., Bast M., Greiner T.C., Aoun P., Armitage J.O., Vose J.M., Weisenburger D.D., and Chan W.C.. 2013. CD43 expression is associated with inferior survival in the non-germinal centre B-cell subgroup of diffuse large B-cell lymphoma. Br. J. Haematol. 162:87–92. 10.1111/bjh.12356 [DOI] [PubMed] [Google Scholar]
- Mullighan C.G., Miller C.B., Radtke I., Phillips L.A., Dalton J., Ma J., White D., Hughes T.P., Le Beau M.M., Pui C.H., et al. 2008. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 453:110–114. 10.1038/nature06866 [DOI] [PubMed] [Google Scholar]
- Mullighan C.G., Su X., Zhang J., Radtke I., Phillips L.A., Miller C.B., Ma J., Liu W., Cheng C., Schulman B.A., et al. Children’s Oncology Group . 2009. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N. Engl. J. Med. 360:470–480. 10.1056/NEJMoa0808253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., and Luo L.. 2007. A global double-fluorescent Cre reporter mouse. Genesis. 45:593–605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Ng S.Y., Yoshida T., Zhang J., and Georgopoulos K.. 2009. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 30:493–507. 10.1016/j.immuni.2009.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol J.W., Helt G.A., Blanchard S.G. Jr., Raja A., and Loraine A.E.. 2009. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 25:2730–2731. 10.1093/bioinformatics/btp472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Geen H., Frietze S., and Farnham P.J.. 2010. Using ChIP-seq technology to identify targets of zinc finger transcription factors. Methods Mol. Biol. 649:437–455. 10.1007/978-1-60761-753-2_27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papathanasiou P., Perkins A.C., Cobb B.S., Ferrini R., Sridharan R., Hoyne G.F., Nelms K.A., Smale S.T., and Goodnow C.C.. 2003. Widespread failure of hematolymphoid differentiation caused by a recessive niche-filling allele of the Ikaros transcription factor. Immunity. 19:131–144. 10.1016/S1074-7613(03)00168-7 [DOI] [PubMed] [Google Scholar]
- Park J.I., Kim S.W., Lyons J.P., Ji H., Nguyen T.T., Cho K., Barton M.C., Deroo T., Vleminckx K., Moon R.T., and McCrea P.D.. 2005. Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev. Cell. 8:843–854. 10.1016/j.devcel.2005.04.010 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2009) R: a language and environment for statistical computing. R Foundation for Statistical Computing: Vienna. URL http://www.R-project.org.
- Reynaud D., Demarco I.A., Reddy K.L., Schjerven H., Bertolino E., Chen Z., Smale S.T., Winandy S., and Singh H.. 2008. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat. Immunol. 9:927–936. 10.1038/ni.1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A.B. 2007. p120-catenin: Past and present. Biochim. Biophys. Acta. 1773:2–7. 10.1016/j.bbamcr.2006.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolink A., Grawunder U., Winkler T.H., Karasuyama H., and Melchers F.. 1994. IL-2 receptor α chain (CD25, TAC) expression defines a crucial stage in pre-B cell development. Int. Immunol. 6:1257–1264. 10.1093/intimm/6.8.1257 [DOI] [PubMed] [Google Scholar]
- Saldanha A.J. 2004. Java Treeview--extensible visualization of microarray data. Bioinformatics. 20:3246–3248. 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- Schackmann R.C., Tenhagen M., van de Ven R.A., and Derksen P.W.. 2013. p120-catenin in cancer - mechanisms, models and opportunities for intervention. J. Cell Sci. 126:3515–3525. 10.1242/jcs.134411 [DOI] [PubMed] [Google Scholar]
- Schjerven H., McLaughlin J., Arenzana T.L., Frietze S., Cheng D., Wadsworth S.E., Lawson G.W., Bensinger S.J., Farnham P.J., Witte O.N., and Smale S.T.. 2013. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat. Immunol. 14:1073–1083. 10.1038/ni.2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel M.S., Corcoran L.M., and Baltimore D.. 1991. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J. Exp. Med. 173:711–720. 10.1084/jem.173.3.711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickert T.A., Tagoh H., Gültekin S., Dakic A., Axelsson E., Minnich M., Ebert A., Werner B., Roth M., Cimmino L., et al. 2014. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat. Immunol. 15:283–293. 10.1038/ni.2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Shao N., Liu X., and Nestler E.. 2014. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 15:284 10.1186/1471-2164-15-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney L.E., Branch M.J., Dunphy S.E., Dua H.S., and Hopkinson A.. 2014. Concise review: evidence for CD34 as a common marker for diverse progenitors. Stem Cells. 32:1380–1389. 10.1002/stem.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.A., Seibel N.L., Altekruse S.F., Ries L.A., Melbert D.L., O’Leary M., Smith F.O., and Reaman G.H.. 2010. Outcomes for children and adolescents with cancer: challenges for the twenty-first century. J. Clin. Oncol. 28:2625–2634. 10.1200/JCO.2009.27.0421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasundaram R., Prasad M.A., Ungerbäck J., and Sigvardsson M.. 2015. Transcription factor networks in B-cell differentiation link development to acute lymphoid leukemia. Blood. 126:144–152. 10.1182/blood-2014-12-575688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R., and Smale S.T.. 2007. Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J. Biol. Chem. 282:30227–30238. 10.1074/jbc.M702541200 [DOI] [PubMed] [Google Scholar]
- Staal F.J., and Clevers H.C.. 2005. WNT signalling and haematopoiesis: a WNT-WNT situation. Nat. Rev. Immunol. 5:21–30. 10.1038/nri1529 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas E.K., Cancelas J.A., Zheng Y., and Williams D.A.. 2008. Rac GTPases as key regulators of p210-BCR-ABL-dependent leukemogenesis. Leukemia. 22:898–904. 10.1038/leu.2008.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trageser D., Iacobucci I., Nahar R., Duy C., von Levetzow G., Klemm L., Park E., Schuh W., Gruber T., Herzog S., et al. 2009. Pre-B cell receptor-mediated cell cycle arrest in Philadelphia chromosome-positive acute lymphoblastic leukemia requires IKAROS function. J. Exp. Med. 206:1739–1753. 10.1084/jem.20090004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veer A., Waanders E., Pieters R., Willemse M.E., Van Reijmersdal S.V., Russell L.J., Harrison C.J., Evans W.E., van der Velden V.H., Hoogerbrugge P.M., et al. 2013. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood. 122:2622–2629. 10.1182/blood-2012-10-462358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zelm M.C., van der Burg M., de Ridder D., Barendregt B.H., de Haas E.F., Reinders M.J., Lankester A.C., Révész T., Staal F.J., and van Dongen J.J.. 2005. Ig gene rearrangement steps are initiated in early human precursor B cell subsets and correlate with specific transcription factor expression. J. Immunol. 175:5912–5922. 10.4049/jimmunol.175.9.5912 [DOI] [PubMed] [Google Scholar]
- Virely C., Moulin S., Cobaleda C., Lasgi C., Alberdi A., Soulier J., Sigaux F., Chan S., Kastner P., and Ghysdael J.. 2010. Haploinsufficiency of the IKZF1 (IKAROS) tumor suppressor gene cooperates with BCR-ABL in a transgenic model of acute lymphoblastic leukemia. Leukemia. 24:1200–1204. 10.1038/leu.2010.63 [DOI] [PubMed] [Google Scholar]
- Wang J.H., Nichogiannopoulou A., Wu L., Sun L., Sharpe A.H., Bigby M., and Georgopoulos K.. 1996. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 5:537–549. 10.1016/S1074-7613(00)80269-1 [DOI] [PubMed] [Google Scholar]
- Wang S., Sun H., Ma J., Zang C., Wang C., Wang J., Tang Q., Meyer C.A., Zhang Y., and Liu X.S.. 2013. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc. 8:2502–2515. 10.1038/nprot.2013.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.T., Roussel M.F., and Sherr C.J.. 2006. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA. 103:6688–6693. 10.1073/pnas.0602030103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winandy S., Wu P., and Georgopoulos K.. 1995. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 83:289–299. 10.1016/0092-8674(95)90170-1 [DOI] [PubMed] [Google Scholar]
- Wong S., and Witte O.N.. 2004. The BCR-ABL story: bench to bedside and back. Annu. Rev. Immunol. 22:247–306. 10.1146/annurev.immunol.22.012703.104753 [DOI] [PubMed] [Google Scholar]
- Yang Y., Mallampati S., Sun B., Zhang J., Kim S.B., Lee J.S., Gong Y., Cai Z., and Sun X.. 2013. Wnt pathway contributes to the protection by bone marrow stromal cells of acute lymphoblastic leukemia cells and is a potential therapeutic target. Cancer Lett. 333:9–17. 10.1016/j.canlet.2012.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandy N.L., and Pendergast A.M.. 2008. Abl tyrosine kinases modulate cadherin-dependent adhesion upstream and downstream of Rho family GTPases. Cell Cycle. 7:444–448. 10.4161/cc.7.4.5452 [DOI] [PubMed] [Google Scholar]
- Zhang B., Li M., McDonald T., Holyoake T.L., Moon R.T., Campana D., Shultz L., and Bhatia R.. 2013. Microenvironmental protection of CML stem and progenitor cells from tyrosine kinase inhibitors through N-cadherin and Wnt-β-catenin signaling. Blood. 121:1824–1838. 10.1182/blood-2012-02-412890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Jackson A.F., Naito T., Dose M., Seavitt J., Liu F., Heller E.J., Kashiwagi M., Yoshida T., Gounari F., et al. 2011. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat. Immunol. 13:86–94. 10.1038/ni.2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.T., Chen H.C., Chen S.Y., and Tseng S.C.. 2012. Nuclear p120 catenin unlocks mitotic block of contact-inhibited human corneal endothelial monolayers without disrupting adherent junctions. J. Cell Sci. 125:3636–3648. 10.1242/jcs.103267 [DOI] [PMC free article] [PubMed] [Google Scholar]