One of the brain manifestations of tuberous sclerosis complex (TSC) is reduced myelination, but the underlying mechanism remains unclear. Ercan et al. demonstrate that mutant neurons overexpress a protein, connective tissue growth factor (CTGF), which impairs oligodendrocyte maturation and myelination.
Abstract
Disruption of myelination during development has been implicated in a range of neurodevelopmental disorders including tuberous sclerosis complex (TSC). TSC patients with autism display impairments in white matter integrity. Similarly, mice lacking neuronal Tsc1 have a hypomyelination phenotype. However, the mechanisms that underlie these phenotypes remain unknown. In this study, we demonstrate that neuronal TSC1/2 orchestrates a program of oligodendrocyte maturation through the regulated secretion of connective tissue growth factor (CTGF). We characterize oligodendrocyte maturation both in vitro and in vivo. We find that neuron-specific Tsc1 deletion results in an increase in CTGF secretion that non–cell autonomously stunts oligodendrocyte development and decreases the total number of oligodendrocytes. Genetic deletion of CTGF from neurons, in turn, mitigates the TSC-dependent hypomyelination phenotype. These results show that the mechanistic target of rapamycin (mTOR) pathway in neurons regulates CTGF production and secretion, revealing a paracrine mechanism by which neuronal signaling regulates oligodendrocyte maturation and myelination in TSC. This study highlights the role of mTOR-dependent signaling between neuronal and nonneuronal cells in the regulation of myelin and identifies an additional therapeutic avenue for this disease.
Introduction
Tuberous sclerosis complex (TSC) is an autosomal-dominant, multisystem disorder caused by loss of either TSC1 or TSC2 function (Tsai and Sahin, 2011). TSC affects 1/6,000 newborns worldwide and involves multiple organs including the brain, skin, eyes, kidneys, heart, and lungs (Crino et al., 2006). TSC patients have a high prevalence of epilepsy (∼90%), intellectual disability and autism (∼50%), sleep disruption, attention-deficit hyperactivity disorder, and anxiety (Han and Sahin, 2011). Neuropathological findings in TSC include cortical tubers, subependymal nodules, and subependymal giant cell astrocytomas (DiMario, 2004).
TSC1 and TSC2 proteins bind to each other to form a complex that regulates protein synthesis and cell size (Kwiatkowski and Manning, 2005). This is consistent with the fact that one of the hallmarks of the disease is the presence of giant cells in the brain, particularly within cortical tubers. A key function of the TSC1/2 complex is to inhibit Rheb, the Ras homologue enriched in brain GTPase, which targets the serine-threonine kinase mechanistic target of rapamycin (mTOR), a master regulator of protein synthesis. mTOR kinase functions in two distinct complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), which are defined by distinct binding partners (Huang and Manning, 2009; Lipton and Sahin, 2014). mTORC1 controls protein homeostasis and is constituted by mTOR, raptor, PRAS40, and mLT8. mTORC1 activity is responsive to the inhibitor rapamycin. mTORC2 controls cellular shape by modulating actin function and contains mTOR, rictor, mLST8, and mSIN1 (mammalian stress-activated protein kinase–interacting protein 1). Inhibition of this complex requires prolonged rapamycin treatment (Sarbassov et al., 2006; Lipton and Sahin, 2014). Without the functional TSC1/2 complex, mTORC1 is hyperactive, resulting in disinhibited protein synthesis and subsequent cell growth (Ruvinsky and Meyuhas, 2006; Wullschleger et al., 2006).
The pathophysiological basis of seizures, behavioral disorders, and autism remains largely unknown. Neuronal abnormalities have been considered the dominant basis of TSC neuropathology. However, accumulating evidence also implicates nonneuronal cells, for instance astrocytes, in the control of neuronal function (Uhlmann et al., 2002). In contrast, white matter (WM) abnormalities in TSC have received surprisingly little attention. Recent neuroimaging studies have demonstrated abnormal WM microstructures in patients with TSC that have autism compared with TSC patients without autism (Lewis et al., 2013; Peters et al., 2013), suggesting WM pathology. This is corroborated by a mouse model of TSC in which neuronal loss of Tsc1 renders a strong hypomyelination phenotype (Meikle et al., 2007).
Previous studies have used oligodendrocyte-specific knockout mouse models of Raptor or Rictor to investigate the role of mTORC1 and mTORC2 in myelination (Bercury et al., 2014; Lebrun-Julien et al., 2014). Based on these studies, mTORC1 in oligodendrocytes demonstrates a more prominent role than mTORC2 in promoting initiation of myelination, myelin thickness by controlling lipogenesis, and translation of myelin proteins (Lebrun-Julien et al., 2014). In addition to abatement of mTOR pathway activity, the effect of hyperactivation of mTORC1 in oligodendrocytes was assessed in oligodendrocyte-specific Tsc1 and Tsc2 knockout mouse models. Surprisingly, both inactivation and hyperactivation of mTORC1 through loss of raptor and loss of Tsc1/2, respectively, resulted in a hypomyelination phenotype (Lebrun-Julien et al., 2014; Carson et al., 2015). These observations highlight that a delicate balance of mTOR activity in oligodendrocytes is required for proper myelination.
Complementary to these findings, our work using a mouse model that lacks Tsc1 only in neurons also showed a hypomyelination phenotype (Meikle et al., 2008). Therefore, we hypothesized that in addition to the oligodendroglial mTOR activity, the non–cell autonomous effects of mTOR could also regulate myelination. Here, we show that, both in vitro and in vivo, the loss of functional TSC1/2 in neurons results in a block in oligodendrocyte development and myelination, respectively. We find that this process is mediated by neuronal connective tissue growth factor (CTGF; also known as CCN2). CTGF is highly expressed and secreted from the neurons lacking TSC1/2 and blocks the development of oligodendrocytes. Furthermore, we elucidate the molecular mechanism regulating the expression of CTGF in Tsc-deficient neurons by demonstrating decreases in the levels of serum response factor (SRF), which is the transcriptional repressor of Ctgf (Stritt et al., 2009). Finally, we show that myelination is improved by genetic ablation of Ctgf in neurons that also lack Tsc1. Electron microscopy (EM) data suggest that this rescue is caused by the rescue of number of myelinated axons, rather than changes in thickness of myelin. Collectively, our data show that loss of TSC1/2, and thus mTOR hyperactivity, in neurons results in a block of oligodendrocyte development and hypomyelination, uncovering a non–cell autonomous mechanism of mTOR in myelination.
Results
Loss of Tsc1 in neurons alone results in hypomyelination in vivo
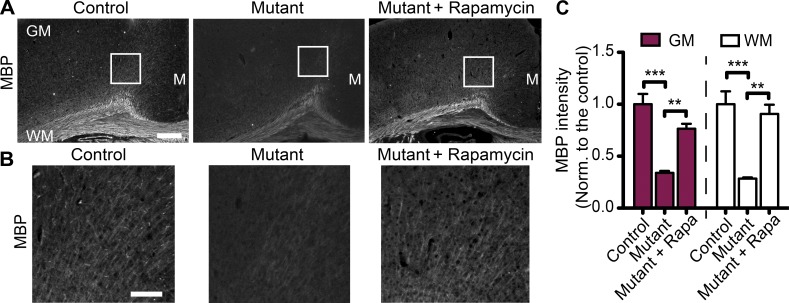
We have previously demonstrated hypomyelination in the Tsc1c−SynICre+ mouse brain, which lacks both alleles of Tsc1 in neurons and one allele of Tsc1 in all other cells, including astrocytes and oligodendrocytes (Meikle et al., 2007). In these mice, myelin basic protein (MBP) expression was reduced throughout the brain from postnatal day 7 (P7) to P21, suggesting a failure of myelination, as opposed to a demyelinating process (Meikle et al., 2007). In the current study, we compared the myelin-specific marker MBP staining in an alternative conditional knockout model of Tsc1 that lacks Tsc1 in all neurons but still expresses wild-type levels of Tsc1 in all other cell types, Tsc1ccSynICre+(mutant). Compared with the control Tsc1wwSynICre+ mice at P21, mutant mice showed lower MBP levels. This difference was reversed in both gray matter (GM; cortex) and WM (corpus callosum) by treating mutants with the mTOR inhibitor rapamycin, suggesting that mTOR hyperactivity in neurons suppresses myelination (Fig. 1, A–C). Moreover, mutant mice displayed fewer CC1+ mature oligodendrocytes in both the GM and WM; this difference was reversible by treatment of mutants with rapamycin (Fig. S1, A and B). In addition to the cortex and corpus callosum, we assessed the degree of myelination by MBP staining in other regions of the brain, such as striatum and internal capsule, and found that hypomyelination in mutant brains occurs in those regions as well (Fig. S1, C and D). Finally, we checked whether the heterozygous mutants show a similar hypomyelination phenotype. Compared with the controls, neither the MBP intensity nor the number of Olig2+ cells was different in the heterozygous mutants (Fig. S1, E–H). These results suggest that the loss of both alleles of Tsc1 in neurons results in hypomyelination and a reduction in the number of mature oligodendrocytes in an mTOR-dependent manner.
Figure 1.
Loss of neuronal Tsc1 results in hypomyelination. (A) Representative images of MBP staining of brain sections from control (Tsc1wwSynICre+; n = 3), mutant (Tsc1ccSynICre+; n = 4), and rapamycin-treated mutant (n = 4) mice at P21. GM, WM, and midline (M) regions are depicted. (B) Enlarged images corresponding to the white squares in A are shown. Bars: (A) 500 µm; (B) 150 µm. (C) Quantification of MBP staining of corresponding mice normalized (Norm.) to the control brains in arbitrary units. Rapa, rapamycin. n = 5 independent experiments. The error bars represent SEM. **, P < 0.01; ***, P < 0.001; Bonferroni’s test.
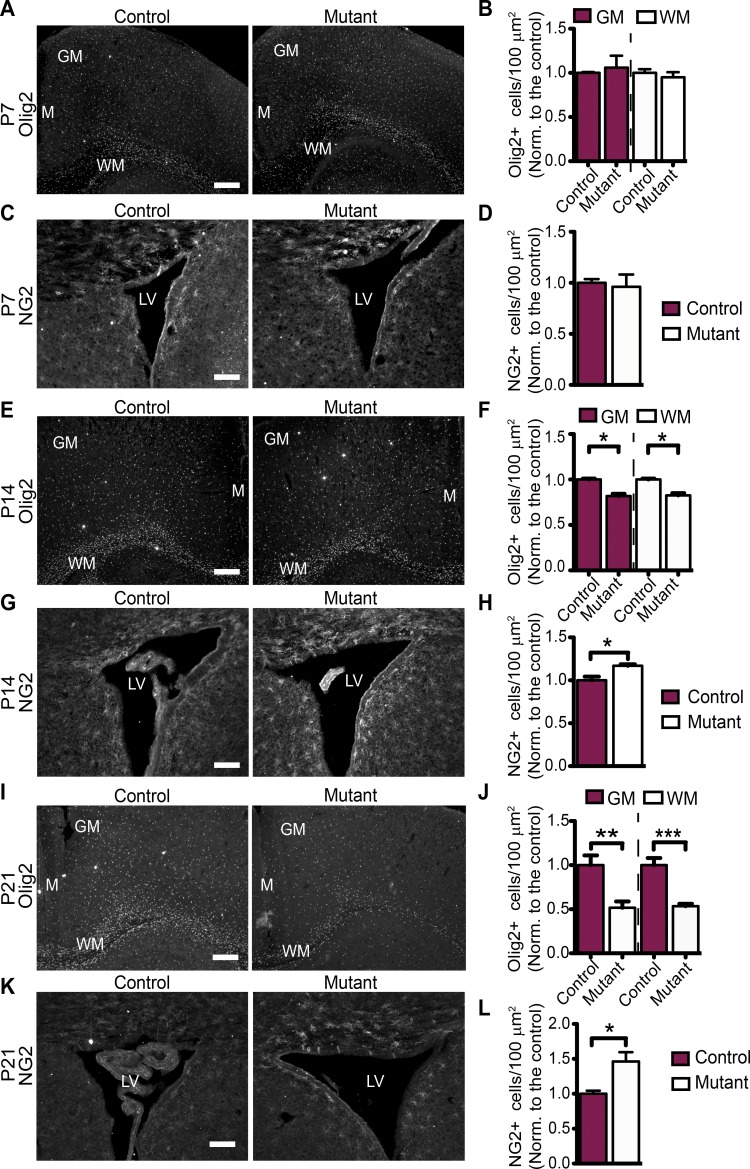
To further investigate the hypomyelination phenotype, we evaluated the number and distribution of oligodendrocytes in the mutant and control mice throughout development. We analyzed the number of total (Olig2+), immature (NG2+), and mature (CC1+) oligodendrocyte numbers in P7, P14, and P21 control and mutant brains. We observed no differences in Olig2+ and NG2+ oligodendrocyte numbers between control and the mutants at P7 (Fig. 2, A–D). However, at P14, the number of Olig2+ oligodendrocytes was decreased, whereas the number of NG2+ oligodendrocytes was increased in the mutants compared with controls (Fig. 2, E–H). In addition, at P14, the number of CC1+ oligodendrocytes was decreased in the mutants both in the WM and GM, compared with controls (Fig. S1, I and J). The differences in relative numbers of Olig2+, NG2+, and CC1+ oligodendrocytes were even greater by P21 (Fig. 2, I–L; Fig. S1, A and B; and Fig. S1 K for summary table). Together, these data suggest that loss of Tsc1 in neurons results in a decrease in the number of oligodendrocytes, and these existing oligodendrocytes can develop to mature (CC1+) oligodendrocytes. However, even the mature (CC1+) oligodendrocytes in Tsc1 mutants do not myelinate as efficiently as the controls, as assessed by MBP (Fig. 1).
Figure 2.
Decrease in total oligodendrocyte number and increase in OPCs in Tsc1 mutants (Tsc1ccSynICre+) start at P14. (A) Representative images of Olig2 staining of brain sections from control (Tsc1wwSynICre+; n = 3) and mutant (Tsc1ccSynICre+; n = 3) mice at P7. (B) Quantification of Olig2-positive (Olig2+) cell density in corresponding mice normalized (Norm.) to the control brains. n = 3 independent experiments. (C) NG2 staining of brain sections from control (n = 3) and mutant (n = 3) mice around the lateral ventricle (LV) at P7. (D) The graph shows the NG2-positive (NG2+) oligodendrocyte density normalized to the control brains. n = 3 independent experiments. (E) Representative images of Olig2 staining of brain sections from control (n = 3) and mutant (n = 3) mice at P14. (F) The graph shows the Olig2 positive (Olig2+) cell density normalized to the control brains. n = 3 independent experiments. (G) NG2 staining of brain sections from control (n = 3) and mutant (n = 3) mice around the lateral ventricle at P14. (H) NG2-positive (NG2+) oligodendrocyte numbers are quantified around the lateral ventricle. n = 3 independent experiments. (I) Olig2 staining of brain sections from control (n = 4) and mutant (n = 5) mice at P21. (J) The graph shows the Olig2-positive (Olig2+) cell density normalized to the control brains at P21. n = 5 independent experiments. (K) NG2 staining of brain sections from control (n = 3) and mutant (n = 3) mice around the lateral ventricle at P21. (L) NG2-positive (NG2+) oligodendrocyte numbers are quantified around the lateral ventricle. n = 3 independent experiments. The graphs show the cell density normalized to the control brains. The error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test. (A, E, and I) GM, WM, and midline (M) regions are depicted. Bars: (A, E, and I) 500 µm; (C, G, and K) 250 µm.
Loss of Tsc2 in neurons results in an arrest in oligodendrocyte development in vitro
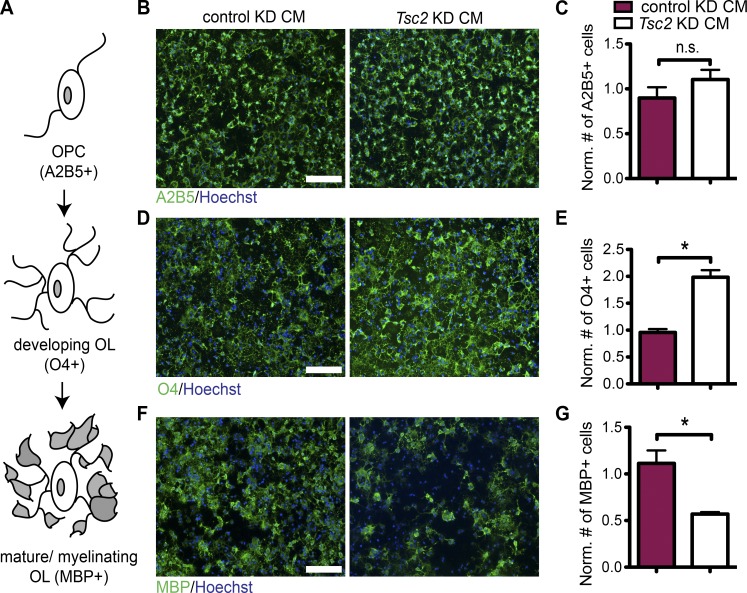
Because oligodendrocytes express Tsc1 normally in neuron-specific Tsc1 deletion mutants, we hypothesized that a secreted signal generated by Tsc1-deficient neurons could be responsible for the observed dysregulation of oligodendrocyte development and myelination. To test this hypothesis, we treated cultured oligodendrocyte precursor cells (OPCs) with conditioned medium (CM) from cortical neurons transduced with a viral construct expressing a Tsc2 shRNA (knockdown [KD]) and compared them with control shRNA–transduced cortical neurons. We assessed oligodendrocyte differentiation using stage-specific markers (Fig. 3 A). Of note, we had previously shown that the Tsc1 and Tsc2 loss in neurons render very similar phenotypes (Choi et al., 2008). Although treatment with Tsc2 KD CM did not change the number of OPCs (A2B5+ and NG2+ cells; Fig. 3, B and C; and Fig. S2, A and B), Tsc2 KD CM treatment increased the number of developing oligodendrocytes (O4+ and GalC+ cells; Fig. 3, D and E; and Fig. S2, C and D). Conversely, the number of mature oligodendrocytes stained with MBP antibody was markedly reduced when treated with Tsc2 KD CM (Fig. 3, F and G). Tsc2 KD CM treatment did not induce apoptosis, as measured by cleaved caspase-3 staining (Fig. S2, E and F). Together, these data suggest that a factor secreted by Tsc-deficient neurons can suppress oligodendrocyte differentiation in culture by blocking the transition from developing to myelinating oligodendrocytes.
Figure 3.
CM collected from Tsc2 KD neurons blocks the maturation of wild-type oligodendrocytes. (A) Schematic cartoon of oligodendrocyte differentiation, showing OPC and developing and mature/myelinating oligodendrocyte (OL) stages. The stage-specific markers are displayed in parentheses. OPCs, which are treated by either control KD CM or Tsc2 KD CM are stained for different stage-specific markers as shown. (B) The cells are stained for A2B5 (green) and Hoechst (blue). (C) The graph shows the quantification of A2B5+ cells. (D) The cells are stained for O4 (green) and Hoechst (blue). (E) The graph shows the quantification of O4+ cells. (F) The cells are stained for MBP (green) and Hoechst (blue). (G) The graph shows the quantification of MBP+ cells. Quantifications of cells normalized (Norm.) to Hoechst+ cells (total cell number) and to the cells treated with CM from noninfected neurons are shown. The error bars represent SEM. *, P < 0.05; Student’s t test. n = 3 independent experiments were performed for all. Bars, 100 µm.
Ctgf is up-regulated in Tsc-deficient neurons in vivo and in vitro
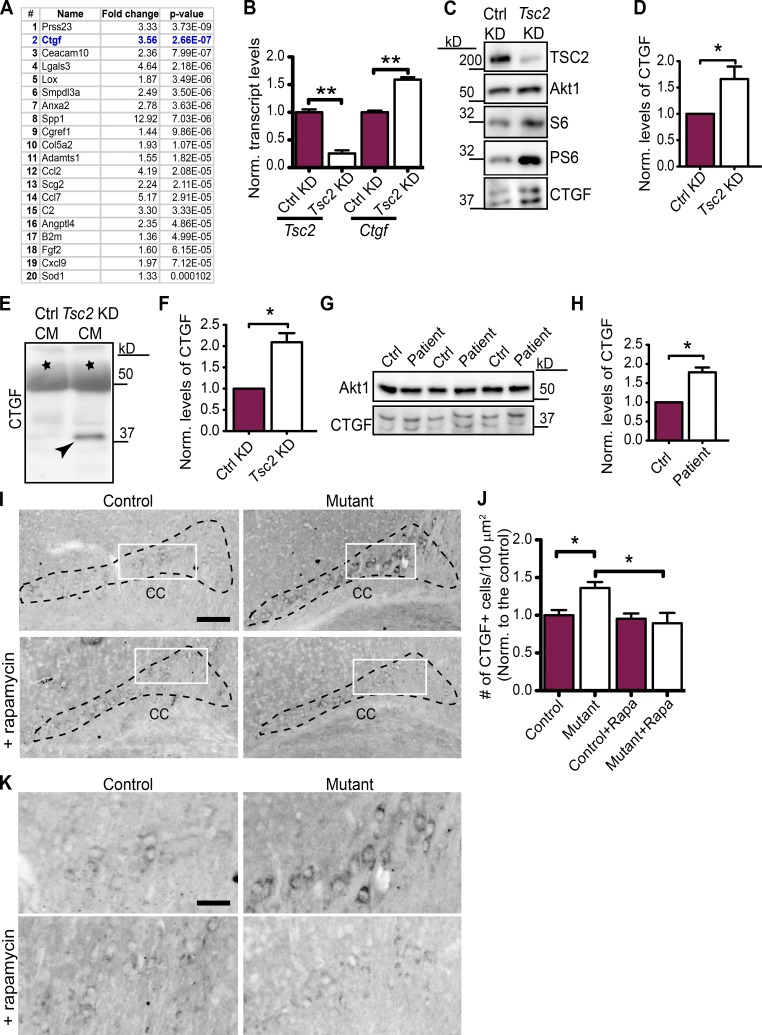
To identify the neuronal signals that regulate myelination, we performed gene expression studies of primary rat cortical neurons treated with a control KD or Tsc2 KD using Illumina microarrays. We identified several modules of coexpressed genes (Fig. S2 G and Table S1) and focused on the module that had the best correlation with the difference between control KD and Tsc2 KD. This group of genes was consistently up-regulated because of Tsc2 loss (Fig. S2 H). Because our prior results implicated a secreted factor, we focused our attention on genes that were up-regulated in Tsc2 KD neurons and encoded for secreted proteins (Fig. 4 A). From this analysis, Ctgf emerged as a promising candidate that had been previously implicated in myelination (Stritt et al., 2009; Lamond and Barnett, 2013). Interestingly, other genes within this list that affect oligodendrocyte development, such as Spp1 (osteopontin) and Lgals3 (galectin-3), were previously shown to improve rather than block myelination (Selvaraju et al., 2004; Zhao et al., 2009; Pasquini et al., 2011) and, thus, were unlikely to mediate this myelin deficiency phenotype.
Figure 4.
CTGF is up-regulated in Tsc-deficient neurons. (A) Gene expression profiling from Tsc2 KD primary cortical neurons, displaying the top 20 mRNAs encoding secreted proteins that are up-regulated in order of significance. (B) qRT-PCR of Tsc2 and Ctgf in control (Ctrl) KD and Tsc2 KD primary cortical neurons. n = 3 independent experiments. (C) Immunoblotting of control and Tsc2 KD cortical neurons showing Tsc2, S6, PS6 (activation of mTOR pathway), Akt1 (loading control), and CTGF levels. (D) Quantification of the CTGF protein levels normalized to Akt1. n = 4 independent experiments. (E) Immunoblotting of CTGF (depicted with an arrowhead) secreted in the control and Tsc2 KD CM from cortical neurons. (F) Quantification of the secreted CTGF protein levels normalized to a background band (indicated by black stars in E). n = 3 independent experiments. (G) Immunoblotting of iPSC-derived human neuron lysates from control and TSC patients for Akt1 (loading control) and CTGF. (H) Quantification of the CTGF protein levels normalized to Akt1. n = 3 independent experiments, three different cell line sets from two patient and control pairs. (I) CTGF staining of brain sections from control (Tsc1wwSynICre+; n = 5) and mutant (Tsc1ccSynICre+; n = 6) and rapamycin-treated control (n = 3) and mutant (n = 4) mice at P21. The corpus callosum (CC) is depicted. Bar, 500 µm. (J) Densities of CTGF+ cells are quantified (areas are depicted with black dashed lines in I). n = 5 independent experiments. Rapa, rapamycin. (K) Enlarged images from the white rectangles in I. Bar, 150 µm. The error bars represent SEM. *, P < 0.05; **, P < 0.01; Student’s t test (A, B, D, F, and H) or Bonferroni’s test (J). Norm., normalized.
CTGF is a multimodular extracellular matrix protein having an N-terminal secretory signal, insulin-like growth factor–binding protein module, von Willebrand factor type C repeat, thrombospondin type I repeat, and a carboxy-terminal domain containing a cysteine knot. Although CTGF is known to be up-regulated in response to trauma, injury, and excitotoxicity in the central nervous system (Brigstock et al., 1997; Lau and Lam, 1999; Hertel et al., 2000; Moussad and Brigstock, 2000; Schwab et al., 2001; Leask and Abraham, 2003; Conrad et al., 2005), its function during brain development has not been previously explored.
To confirm the increase in Ctgf mRNA, we performed quantitative RT-PCR (qRT-PCR) with mRNA from Tsc2 KD neurons and found increased transcript levels compared with control KD neurons (Fig. 4 B). CTGF protein was also increased in Tsc2 KD neurons compared with the control neurons in cell lysates (Fig. 4, C and D) and CM (Fig. 4, E and F). Given the effect of Tsc deficiency in rat cortical neurons, we asked whether CTGF levels are also elevated in induced pluripotent stem cell (iPSC)–derived human neurons from TSC patients. We generated iPSC lines from fibroblasts collected from TSC patients and unaffected family members to use as controls and differentiated these cells into neurons (Fig. S2, I–N). Compared with unaffected controls, iPSC-derived neurons from fibroblasts of TSC patients showed elevated levels of CTGF protein (Fig. 4, G and H). Together, these data demonstrate that Ctgf mRNA and protein are up-regulated in Tsc-deficient neurons in vitro. Recently, in two epilepsy-associated pathologies, focal cortical dysplasia (FCD) and TSC, myelin deficiencies caused by decreased number of oligodendrocytes and insufficient oligodendrocyte maturation have been observed (Scholl et al., 2016). Then, we asked whether CTGF is expressed in human brains with epilepsy. For this analysis, we used epileptic tissue resected from patients with FCD, from TSC patients, and from patients with epilepsy but no FCD or TSC. The Western blot analysis in these brain samples showed that CTGF is expressed in all samples (Fig. S2 O).
CTGF expression has been used as a marker for subplate neurons in both humans and in rodents (Fig. S3 A; Heuer et al., 2003). Therefore, we used immunostaining to evaluate CTGF expression in this region in brain sections from mutant and control mice to evaluate the effect of neuronal Tsc1 ablation in vivo. We analyzed the expression of CTGF in P7, P14, and P21 mice. We found the number of cells in the subplate region expressing CTGF above a fixed threshold intensity was not changed between controls and mutants at P7 (Fig. S3, B and C). However, at P14, we observed a significant increase in CTGF expression, which was elevated at P21, in the mutant brains compared with controls (Fig. S3, D and E; and Fig. 4, I and J), and CTGF-positive cells expressed higher levels of protein in the mutants (enlarged images in Fig. 4 K). Increased CTGF expression was reversed by rapamycin treatment, indicating that this process is mTOR dependent in vivo (Fig. 4, I and J; and enlarged images in Fig. 4 K).
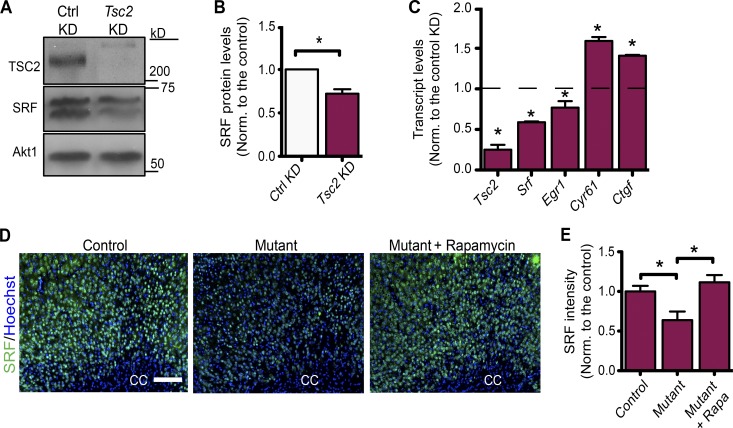
To investigate the molecular mechanism that regulates CTGF expression in Tsc-deficient neurons, we chose to evaluate the role of SRF. Previous studies show that the SRF regulates the expression CTGF in different cell types (Muehlich et al., 2007; Haak et al., 2014; Hinkel et al., 2014; Angelini et al., 2015). Neurons lacking SRF have higher expression of CTGF and thus block the maturation of oligodendrocytes (Stritt et al., 2009). Therefore, we analyzed the levels of SRF both in our mutant mice and in primary cortical neurons lacking Tsc2. Both SRF protein and transcript levels were decreased in Tsc2 KD neurons compared with the control neurons (Fig. 5, A–C). In addition, we examined the transcript levels of other targets of SRF that included Egr1 and Cyr61. SRF functions as a transcription activator of Egr1 (Kim et al., 2008), whereas it suppresses the transcription of Cyr61 (Stritt et al., 2009), which is also in the same CCN family of proteins with CTGF (Jun and Lau, 2011). Consistent with these functions, we found that, in Tsc2 KD cortical neurons, both Srf and Egr1 transcripts are decreased, whereas Cyr61 transcript is increased as Ctgf (Fig. 5 C). Moreover, when stained with SRF antibody, compared with the control mice, the mutant mice brain sections showed decreased intensity levels, suggesting that SRF expression is diminished also in vivo (Fig. 5, D and E; and Fig. S3 F). Together, our data indicate that the up-regulation of CTGF in Tsc-deficient neurons may be caused by the down-regulation of its suppressor, SRF.
Figure 5.
SRF is down-regulated in Tsc-deficient neurons. (A) Representative immunoblots of control (Ctrl) and Tsc2 KD cortical neurons probed with Tsc2, SRF, and Akt1 (loading control) antibodies. (B) Quantification of the SRF protein levels normalized (Norm.) to Akt1. n = 3 independent experiments. (C) qRT-PCR of Tsc2, Srf, Egr1, Cyr61, and Ctgf in Tsc2 KD primary cortical neurons. n = 3 independent experiments. The dashed line represents the transcript levels in the control KD, which is set to 1. (D) SRF staining of brain sections from control (Tsc1wwSynICre+; n = 3), Tsc1-mutant (Tsc1ccSynICre+; n = 3), and rapamycin-treated mutant (Tsc1ccSynICre+; n = 3) mice at P21. n = 3 independent experiments. The corpus callosum (CC) is depicted. Bar, 200 µm. (E) Quantifications of SRF intensity of the corresponding mice in arbitrary units. n = 3 for each. Rapa, rapamycin. The error bars represent SEM. *, P < 0.05; Student’s t test (B and C) or Bonferroni’s test (E).
CTGF negatively regulates oligodendrocyte development in vitro and myelination in vivo
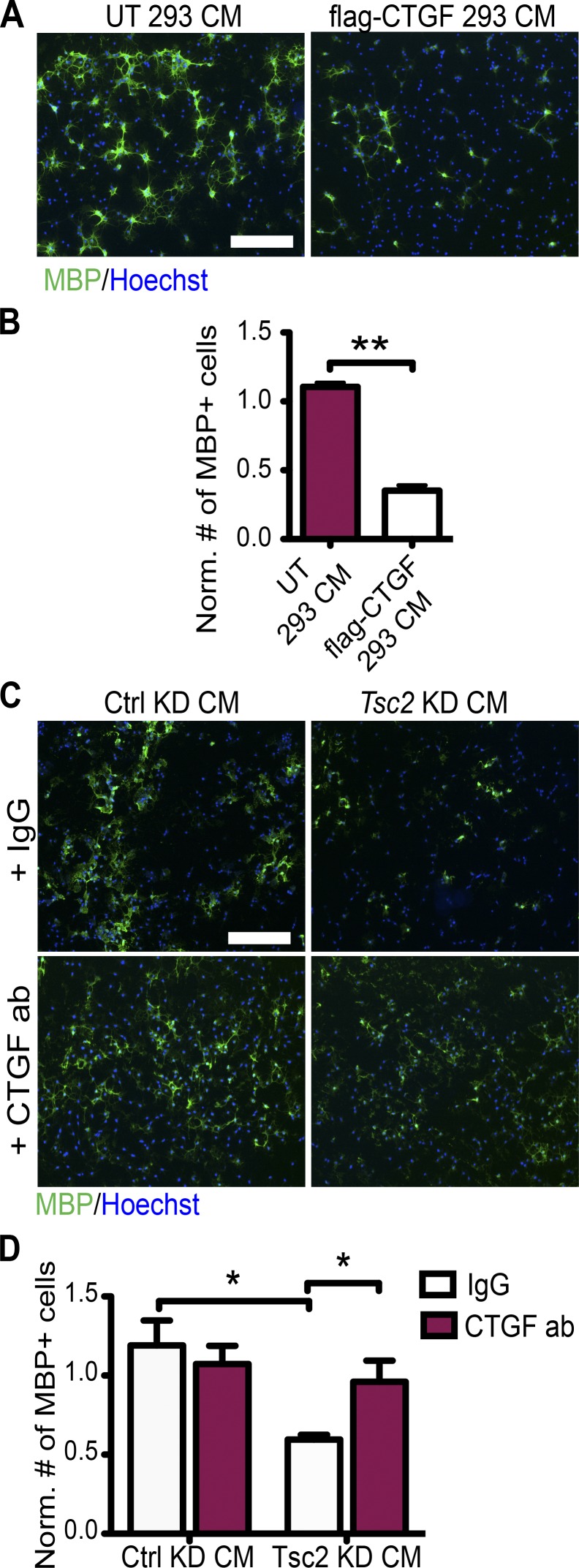
The role of CTGF on maturation of oligodendrocytes was evaluated by treating OPC cultures with CM from HEK293T cells expressing FLAG-tagged full-length CTGF and control CM. CTGF-containing CM induced a marked reduction of MBP-expressing oligodendrocytes (Fig. 6, A and B; and Fig. S3 G). Consistent with this finding, OPCs treated with purified recombinant full-length CTGF protein showed a dose-dependent reduction in the number of MBP-expressing oligodendrocytes (Fig. S3, H and I). CTGF treatment did not induce apoptosis, as measured by cleaved caspase-3 staining (Fig. S3, J and K). Then, we evaluated whether CTGF up-regulation in the Tsc2 KD CM produced a dysregulation of oligodendrocyte development. When we neutralized CTGF in Tsc2 KD CM by adding CTGF antibody, we observed no inhibitory effect of the CM on the number of MBP-positive mature oligodendrocytes (Fig. 6, C and D). Together these data suggest that up-regulation of CTGF and secretion from neurons is both necessary and sufficient to block oligodendrocyte maturation in vitro.
Figure 6.
CTGF is necessary and sufficient for oligodendrocyte maturation arrest in vitro. (A) OPCs, which are treated with either CM from untransfected HEK293T cells (UT 293T CM) or CM from HEK293T cells expressing flag-CTGF (flag-CTGF CM), are stained for MBP (green) and Hoechst (blue). (B) Quantifications of the cell numbers normalized (Norm.) to Hoechst+ cells. n = 3 independent experiments. (C) OPCs, which are treated by either control (Ctrl) KD CM or Tsc2 KD CM are treated with either IgG or CTGF antibody (ab) and stained for MBP (green) and Hoechst (blue). (D) Quantifications of the cell numbers in C normalized to Hoechst+ cells (total cell number) and MBP+ cells treated with CM from noninfected neurons. n = 5 independent experiments. Error bars represent SEM. *, P < 0.05; **, P < 0.01; Student’s t test. Bars, 100 µm.
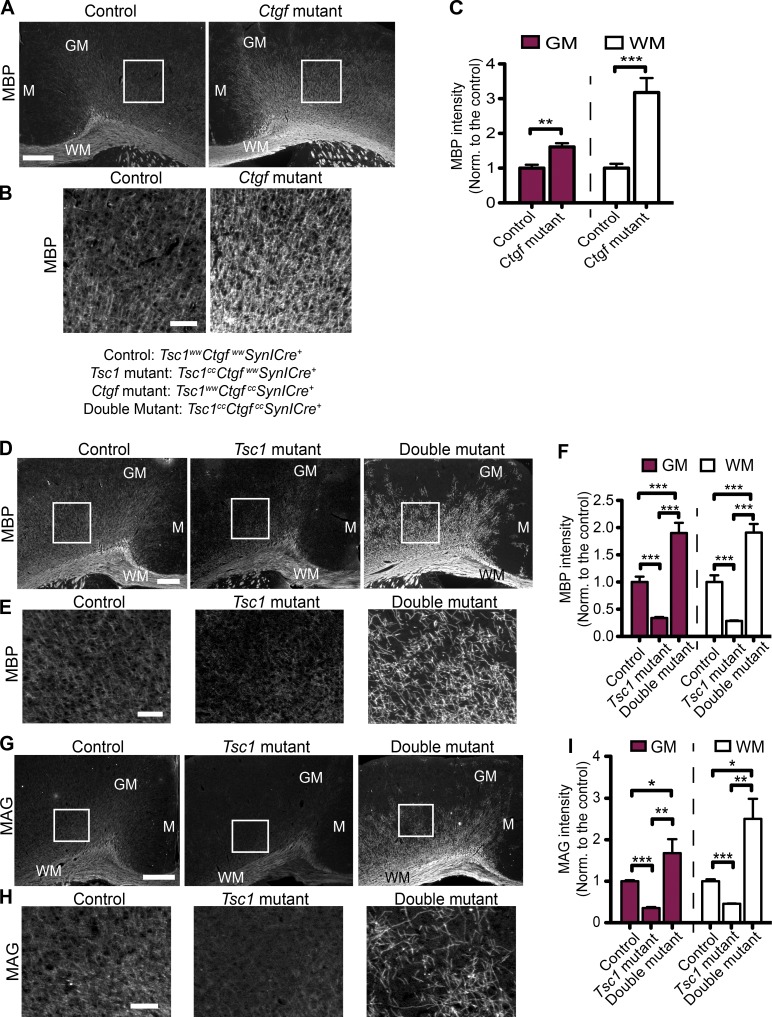
Next, we investigated the effect of neuronal CTGF on myelination in vivo. We crossed Tsc1;SynICre+ mice with Ctgfcc mice (Liu et al., 2011) to generate the following groups of mice: wild type for both genes (control: Tsc1wwCtgfwwSynICre+), mutant for Tsc1 and wild type for Ctgf (Tsc1 mutant: Tsc1ccCtgfwwSynICre+), mutant for both genes (double mutant: Tsc1ccCtgfccSynICre+), and wild type for Tsc1 and mutant for Ctgf (Ctgf mutant: Tsc1wwCtgfccSynICre+). We confirmed the loss of CTGF expression in neurons by staining Ctgf-mutant brain sections for CTGF (Fig. S4 A). To our knowledge, this is the first mouse model lacking CTGF only in neurons. The loss of Ctgf in neurons in a Tsc1 wild-type background enhanced myelination, as assessed by the increased MBP intensity and number of Olig2+ oligodendrocytes at P21 (Fig. 7, A–C; and Fig. S4 F). The increase in MBP intensity was significantly higher in the Ctgf mutants compared with controls at P30 but not in adult mice (at P60; Fig. S4, B–E). Interestingly, compared with the controls, the number of Olig2+ cells was not different in the Ctgf mutants at P30 and P60 (Fig. S4, F and G for summary table). Therefore, we analyzed the expression of CTGF in control mice at P21, P30, and P60 and found that the number of cells expressing CTGF above a fixed threshold intensity decreases at P60 (Fig. S4, H and I). This suggests that CTGF affects the survival and/or development of oligodendrocytes in an age-dependent manner. In younger animals, Ctgf-deficient mice have enhanced myelination, but this abnormality is resolved by older ages as CTGF expression is reduced in control mice with development.
Figure 7.
Loss of neuronal CTGF improves myelination. (A) Representative images of MBP staining of the brain sections from the control (Tsc1wwCtgfwwSynICre+; n = 3) and Ctgf-mutant (Tsc1wwCtgfccSynICre+; n = 3) mice at P21. GM and the WM and midline (M) regions are depicted. (B) Enlarged images from the depicted white squares in A. (C) Quantifications of MBP intensity in arbitrary units. n = 3 independent experiments. (D) MBP staining of brain sections from control (n = 5), Tsc1-mutant (Tsc1ccCtgfwwSynICre+; n = 3), and double-mutant (Tsc1ccCtgfccSynICre+; n = 4) mice at P21. GM and the WM and midline regions are depicted. (E) Enlarged images corresponding to the areas depicted by the white squares in D. (F) MBP intensity quantifications of corresponding mice for GM and WM in arbitrary units. n = 5 independent experiments. (G) MAG staining of brain sections from control (n = 3), Tsc1-mutant (n = 3), and double-mutant (n = 3) mice at P21. GM and the WM and midline regions are depicted. (H) Enlarged images corresponding to the areas depicted by white squares in G. (I) MAG intensity quantifications of corresponding mice for GM and WM in arbitrary units. n = 3 independent experiments. The error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test (C) or Bonferroni’s test (F and I). Norm., normalized. Bars: (A, D, and G) 500 µm; (B, E, and H) 150 µm.
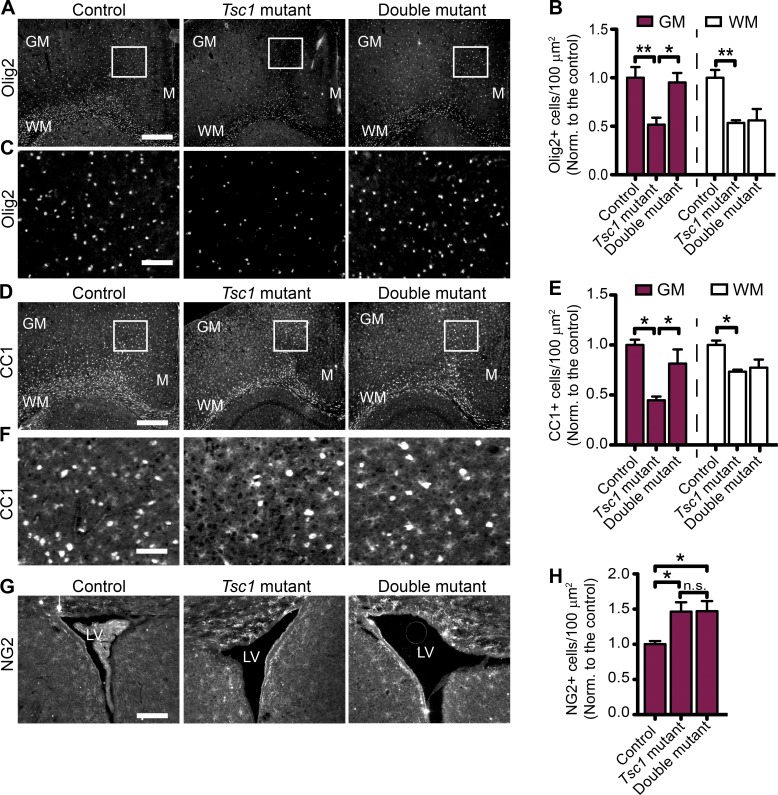
Next, we stained brain sections of control, Tsc1-mutant, and double-mutant mice with the MBP and myelin-associated glycoprotein (MAG) antibodies and found that loss of Ctgf in neurons improved the hypomyelination phenotype of Tsc1 deficiency. Moreover, the intensity of MBP and MAG stainings in both GM and WM of the double mutants was significantly higher than controls (Fig. 7, D–I). Then, we analyzed the number of CC1+, Olig2+, and NG2+ cells in these different genotypes. The numbers of total oligodendrocytes (Olig2+) and mature oligodendrocytes (CC1+) were also increased in the GM of the double mutants compared with Tsc1 mutants. However, in the WM, there was no difference in Olig2+ or CC1+ cells between the Tsc1 and double mutants (Fig. 8, A–F). Furthermore, we did not observe any difference in NG2+ oligodendrocyte numbers between the Tsc1-mutant and double-mutant mice (Fig. 8, G and H).
Figure 8.
Effect of CTGF on the number of total, mature, and precursor oligodendrocytes in vivo. (A) Olig2 staining of brain sections from control (Tsc1wwCtgfwwSynICre+; n = 3), Tsc1-mutant (Tsc1ccCtgfwwSynICre+; n = 3), and double-mutant (Tsc1ccCtgfccSynICre+; n = 4) mice at P21. GM and the WM and midline (M) regions are depicted. (B) Olig2+ oligodendrocyte numbers are quantified for the corresponding areas, and the graph shows the cell density normalized (Norm.) to the control brains. n = 5 independent experiments. (C) Enlarged images corresponding to the white squares in A. (D) CC1 staining of brain sections from control (n = 4), Tsc1-mutant (n = 4), and double-mutant (n = 5) mice at P21. GM and the WM and midline regions are depicted. (E) CC1+ oligodendrocyte numbers are quantified for the GM and WM, and the graph shows the cell density normalized to the control brains. n = 3 independent experiments. (F) Enlarged images corresponding to the white squares in D. (G) NG2 staining of brain sections from control (n = 3), Tsc1-mutant (n = 3), and double-mutant (n = 5) mice at P21. LV, lateral ventricle. (H) NG2-positive (NG2+) oligodendrocyte numbers are quantified around the lateral ventricle, and the graph shows the cell density normalized to the control mice. n = 3 independent experiments. The error bars represent SEM. *, P < 0.05; **, P < 0.01; Bonferroni’s test. Bars: (A and D) 500 µm; (C and F) 150 µm; (G) 250 µm.
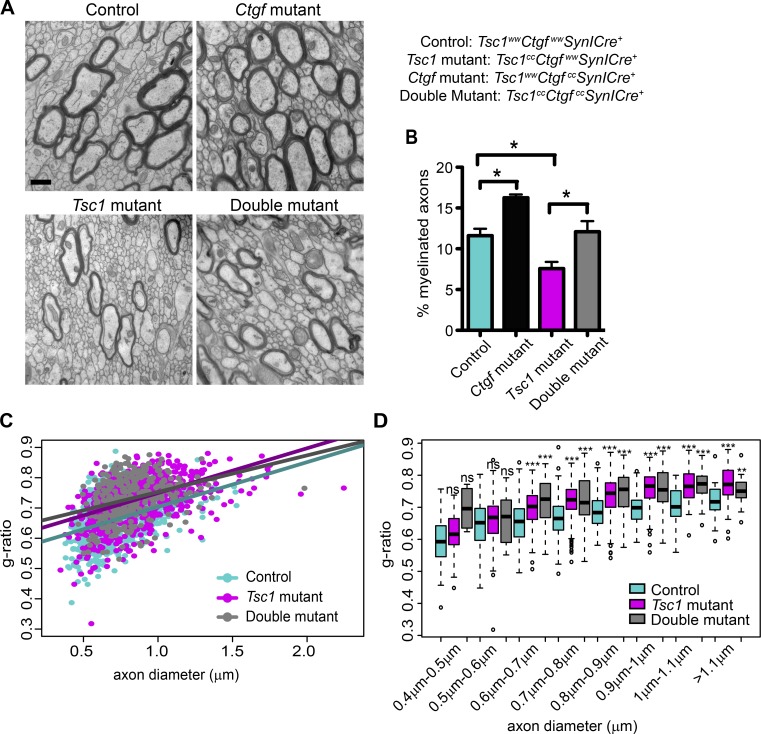
Given that the loss of Ctgf enhances myelination as assessed by improved MBP and MAG signals in both Ctgf and double mutants, we investigated the effect of CTGF on myelination further by using EM. We performed EM of the corpus callosum at the midline from control, Ctgf-mutant, Tsc1-mutant, and double-mutant mice at P21 (Fig. 9 A). We counted the number of myelinated axons and found that, compared with the controls, the percentage of myelinated axons was reduced in the Tsc1 mutants, whereas it was increased in Ctgf mutants. Loss of Ctgf in neurons rescued the decrease in the percentage of myelinated axons in Tsc1 deficiency (Fig. 9 B). In addition to the number of myelinated axons, we also analyzed the thickness of myelin by assessing the g-ratio, which is defined as the ratio of the inner and outer diameters of the myelin sheath. A high g-ratio corresponds to a thinner myelin sheath (Rushton, 1951). Compared with the controls, both the axon diameter of the myelinated axons and the g-ratio were increased in Tsc1 mutants. However, loss of Ctgf did not rescue this phenotype in the double mutants (Fig. 9, C and D; and Fig. S5, A and B). Interestingly, between the axon diameters of 0.7 and 0.8 µm, Ctgf mutants also showed a significant increase in the g-ratio (Fig. S5, C and D). These results indicate that loss of Ctgf results in an increase in the percentage of myelinated axons, but it does not enhance the myelin thickness.
Figure 9.
Loss of CTGF does not improve myelin thickness but increases the number of myelinated axons. (A) Representative EM images of control (Tsc1wwCtgfwwSynICre+; n = 5), Ctgf-mutant (Tsc1wwCtgfccSynICre+; n = 3), Tsc1-mutant (Tsc1ccCtgfwwSynICre+; n = 5), and double-mutant (Tsc1ccCtgfccSynICre+; n = 3) corpus callosum at P21. Bar, 500 µm. (B) Percentage of myelinated axons for control (n = 5), Ctgf mutant (n = 3), Tsc1 mutant (n = 5), and double mutant (n = 3) are calculated. (C) G-ratios as a function of axon diameter for control (n = 5), Tsc1-mutant (n = 5), and double-mutant (n = 3) mice. (D) The box plot represents the comparison of g-ratios versus axon diameter for control (n = 5), Tsc1 mutant (n = 5), and double mutant (n = 3). Within each axon diameter cohort, Tsc1-mutant and double-mutant axons are compared with control axons. The difference between the Tsc1 mutants and double mutants is not significant. The error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Bonferroni’s test (B) or Welch two-sample t test (D).
Discussion
Several studies have investigated the effect of mTOR activity on oligodendrocyte development and have collectively suggested that a delicate balance of mTOR activity in oligodendrocytes is required to achieve normal myelination (Bercury et al., 2014; Lebrun-Julien et al., 2014; Wahl et al., 2014). Our findings indicate that neuronal mTOR activity is also an important regulator of myelination through paracrine secretion of CTGF. Although CTGF has been implicated in myelination previously (Stritt et al., 2009), our results link this molecular mechanism for the first time to a human disease, TSC. Furthermore, we demonstrate that a neuron-specific knockout of Ctgf can improve myelination in vivo.
Goebbels et al. (2010) previously reported that the loss of Pten in oligodendrocytes, which results in hyperactivity of mTOR in oligodendrocytes, triggers myelination, and treatment of Pten adult mice with rapamycin (P43–P95) reduces the degree of myelination in the mutants. Presumably, the effect of rapamycin is directly on the Pten-deficient oligodendrocytes in this system. In contrast, we performed the rapamycin treatment between P7 and P21 and observed enhanced myelination in neuronal Tsc1 knockout mice. In our experiments, the time of rapamycin treatment (P7 to P21) and spatiotemporal expression of CTGF in subplate neurons (Heuer et al., 2003) coincide with the development of OPCs into mature oligodendrocytes. Rapamycin treatment during this period leads to enhanced myelination. This may be explained by the role of an axonal myelination signal (CTGF) that regulates the mTOR activity in oligodendrocytes in addition to the effect of rapamycin on oligodendrocytes. CTGF has been shown to bind to and antagonize insulin-like growth factor and stimulate oligodendrocyte development and, thus, can indirectly block the activation of the mTOR pathway in oligodendrocytes (Kim et al., 1997; Stritt et al., 2009). Rapamycin treatment may mitigate the expression and thus secretion of CTGF from neurons. The extracellular CTGF can no longer interfere with IGF1 binding to its receptor on oligodendrocytes. This, in turn, activates the Akt–mTOR pathway in oligodendrocytes, which enhances myelination. Together, these studies highlight the complexity of regulation of myelination by different signaling pathways (Pten vs. TSC1/2) and different cell types (neurons vs. oligodendrocytes) and at different times of development.
Our results show that the double knockout of Tsc1 and Ctgf in neurons improved the hypomyelination phenotype observed in the Tsc1-mutant mice in both the GM and WM as assessed by MBP and MAG immunostaining. However, in the WM, the total number of oligodendrocytes (Olig2+) was similar in both Tsc1- and double-mutant mice, suggesting that Ctgf loss does not rescue the number of oligodendrocytes. Moreover, in the WM and lateral ventricle of double mutants, we did not observe a rescue in the number of CC1+ and NG2+ cells, respectively. Collectively, these results indicate that neuronal Tsc1 loss results in a reduction in CC1+ cells and an increase in NG2+ cells. Therefore, it is likely that the impact of neuronal Tsc1 loss in vivo is on both the oligodendrocyte cell number and the ability of these cells to differentiate into mature oligodendrocytes (CC1+). However, in the WM, double knockout of Ctgf with Tsc1 rescues neither the total (Olig2+) nor the mature (CC1+) oligodendrocyte number. Nonetheless, we find that Ctgf loss does enhance myelination as assessed by MBP and MAG immunostaining and EM. As revealed by EM, the number of myelinated axons increases by the loss of Ctgf, whereas the myelin thickness decreases. It is possible that the oligodendrocytes now generate more internodes per cell and that each oligodendrocyte is able to wrap more axons. In the WM, the loss of Ctgf affects the number of oligodendrocyte processes but not their ability to differentiate. The rescue of MBP and MAG intensity in the double mutants is therefore not caused by the formation of a thicker myelin sheath; rather, it may be caused by the increased number of processes and, thus, myelinated axons. This remains to be investigated further at the cellular level.
We observed that the loss of Ctgf affects the number of oligodendrocytes in WM and GM somewhat differently. Compared with the GM, the intensity of MBP in WM increases more profoundly. In addition, the number of oligodendrocytes (Olig2+ and CC1+) is rescued in the GM but not in the WM. These region-specific differences in oligodendrocyte numbers and MBP intensity might be the result of the region-specific expression of CTGF. As CTGF is predominantly expressed in the subplate neurons, which reside in the cortical WM, the WM oligodendrocytes might be affected more profoundly than the GM oligodendrocytes. Although the subplate neurons reside in the deeper layer of cortex, they can project into layer 4 (Allendoerfer and Shatz, 1994). This might explain why we observe hypomyelination not only in WM but also in the GM.
In our in vitro oligodendrocyte maturation assay, we observed that a block in oligodendrocyte differentiation by addition of Tsc2 KD CM and depletion of CTGF from the medium improves the differentiation. Moreover, we found that the in-vitro effect of CTGF is dose dependent. In vivo, we also observed an effect of CTGF on oligodendrocyte number, which we did not observe in vitro. This difference may be because of the growth factor–rich conditions we used to grow oligodendrocytes in culture. Abundance of growth factors in culture may mask an effect on oligodendrocyte survival or proliferation. In addition, for the in vitro dose-response experiments, we used a recombinant full-length human CTGF; however, in-vivo proteolytic processing of CTGF might yield to different modules (Steffen et al., 1998; Yang et al., 1998; Hashimoto et al., 2002). Ultimately, these different forms might affect the oligodendrocyte development and/or process formation in distinct ways.
Furthermore, there might be additional inhibitory factors in vivo and/or compensatory factors in vitro that may play roles in oligodendrocyte development. For instance, from our microarray data, we found that the mRNA level of contactin-1 (cntn1), an axon–glia interaction protein, is decreased (unpublished data). Although such additional factors and their effects remain to be discovered, our data suggest that the neuronal CTGF is a strong determinant of myelination in vivo. Future studies of the downstream effects of CTGF on oligodendrocytes and identification of the protein modules within CTGF that are responsible for regulation of oligodendrocyte development will be a major goal for the discovery of new therapeutic targets. Our study provides the first description of a possible molecular mechanism that could underlie the aberrant WM microstructure in TSC patients. The non–cell autonomous effect of CTGF should also be investigated in other diseases associated with myelination deficits such as periventricular leukomalacia and cerebral palsy, as well in demyelinating diseases such as multiple sclerosis.
Materials and methods
Mouse models
All experimental procedures were performed in compliance with animal protocols approved by the Animal Research Committee at Boston Children’s Hospital. All mice were in mixed background, derived from C57BL/6, CBA, 129S4/SvJae, and BL/6 strains. We use c and w to denote the conditional (floxed) and wild-type alleles of Tsc1, respectively; the formal name of the c allele is Tsc1tm1Djk (Meikle et al., 2007). In the SynICre mouse model, Cre expression is controlled by the rat synapsin I promoter, which has been shown to drive transgene expression specifically in neuronal cells (Hoesche et al., 1993). Cre function can be detected as early as E12.5, when β-gal activity is restricted to brain, spinal cord, and dorsal root ganglia. Histological analysis showed that Cre activity is mainly distributed in differentiated neurons, outside the ventricular regions of the brain and spinal cord (Zhu et al., 2001). To generate Tsc1ccSynICre+ mice, first Tsc1cwSynCre+ females were crossed with Tsc1ccSynICre− male mice to eliminate the effect of SynCre expression in other cell types. To generate Tsc1ccCtgfccSynICre+ mice, Tsc1ccSynICre+ females were crossed with Ctgfcc mice (Liu et al., 2011). The progenies were crossed with one another to give litters consisting of control (Tsc1wwCtgfwwSynICre+), Tsc1-mutant (Tsc1ccCtgfwwSynICre+), Ctgf-mutant (Tsc1wwCtgfccSynICre+), or double-mutant (Tsc1ccCtgfccSynICre+) mice. Littermates were used as controls for all experiments. Rapamycin treatment was performed by injecting 6 mg/kg intraperitoneally every other day beginning at P7 until sacrifice (P21). Both female and male mice were used in the experiments. Mice were maintained on a 12-h light/dark cycle with free access to food and water according to the Animal Research Committee at Boston Children’s Hospital. This treatment timing and dosing was chosen based on pharmacokinetics and pharmacodynamics of rapamycin in the brain and has been shown to be effective in reversing the hypomyelination phenotype described previously (Meikle et al., 2008). Of note, this treatment differs in dosage, frequency, duration, and age of treatment from another study where the investigators started treating wild-type and Plp-Akt-DD mice at 6–8 wk of age (P21) and observed that young mice showed reduction in myelin accumulation (Narayanan et al., 2009).
Immunohistochemistry (IHC)
The slide-mounted sections were rinsed with PBS for 5 min followed by incubation for 1 h in 5% bovine serum albumin and 0.5% Triton X-100 diluted in PBS for blocking and permeabilization step before overnight primary antibody incubation in the same solution. The next day, sections were washed with 1× PBS three times and were incubated for 2 h in the same blocking solution containing secondary antibody (1:750 dilution; Molecular Probes) and Hoechst 33342 solution (1:50,000) for DNA staining. Then, the sections were washed three times for 10 min each with 1× PBS before being cover slipped and imaged using a microscope (80i; Nikon) equipped with epifluorescence and 4× or 10× objectives. The exposure times for the staining groups (such as MBP and CTGF) were kept the same, and the analysis was performed using ImageJ (National Institutes of Health). For CTGF calculations, the cells expressing CTGF above a certain threshold were counted by using the triangle threshold method. For MBP and MAG intensity calculation defaults and for SRF intensity calculations, Otsu Threshold methods were used.
Immunocytochemistry
The cells were fixed using 4% paraformaldehyde for 10 min and permeabilized for 10 min using PBS with Triton (0.075 M PBS, pH 7.3, and 0.1% Triton). Then, the coverslips were washed twice with 1× PBS and incubated for 1–2 h in blocking solution (5% normal goat serum [NGS] in 1× PBS). Next, the coverslips were incubated with primary antibodies in 2.5% NGS in PBS at 4°C overnight. The next day, coverslips were washed three times with 1× PBS and were incubated for 1 h at room temperature with fluorescence-conjugated secondary antibody solution (1:750 dilution in 2.5% NGS in PBS) and Hoechst 33342 solution. For O4 and GalC staining of the cells, no permeabilization was performed. Imaging was performed with a microscope (80i; Nikon) with a 10× objective. The total number of cells was calculated according to the Hoechst staining. The total number of oligodendrocyte marker–positive cells (such as O4+) was calculated from five different regions of a coverslip. The number of oligodendrocyte marker–positive cells was divided by the total number of Hoechst+ cells to obtain the ratio of marker-positive cells on one coverslip. At least three coverslips from three independent experiments were analyzed by using the ImageJ Particle Analysis tool.
Statistical analysis
Cell counting was performed by ImageJ particle analysis, and statistics were performed in Excel (Microsoft) or Prism5 (GraphPad Software). The data were expressed as means ± SEM of at least three independent experiments for IHC experiments. For IHC of brains, at least three sections 200 µm apart from each other were imaged, and the cell number was counted for the corresponding areas (WM or GM) and divided by the area to calculate the cell density (cells/100 µm2). The mean cell densities for all regions from all different sections were calculated. At least three different animals per group were used in the imaging and calculations. For IHC experiments, the number of marker-positive oligodendrocytes (MBP+, O4+, etc.) was first normalized to the total number of oligodendrocytes (Hoechst+) and then normalized to the number of marker-positive oligodendrocytes, treated with a noninfected CM, which was set to 1. Statistical analysis was performed by using unpaired two-tailed Student’s t tests for comparisons between two groups. Analyses involving data from three or more groups were performed using one-way or two-way ANOVA with Bonferroni’s posthoc test. P < 0.05 was considered significant. The statistical analysis for EM was performed in R. For analysis of nonrandom distributed samples, permutation testing was applied. The labels of the two groups switched, and the mean difference was computed 10,000 times and then compared with the observed difference. Then, p-values were calculated with a Welch two-sample t test. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those generally used in the field. All experiments were performed at least three times, and data replication was observed in repeated experiments.
Cortical neuronal cultures
Cortical neuronal cultures were prepared as previously described (Sahin et al., 2005). In brief, the cortices from embryonic day 18 rats (CD1; Charles River) were isolated under the microscope and collected in Hank’s balanced salt solution containing 10 mM MgCl2, 1 mM kynurenic acid, 10 mM Hepes, and penicillin/streptomycin. The cortices were dissociated in 30 µ/ml papain at 37°C for 5 min, mechanically triturated, and plated in neurobasal medium (NB; BRL 21103-049; Gibco) containing B27 supplement (BRL 17504-044; Gibco), 2 mM l-glutamine (BRL 25030-081; Gibco), and penicillin/streptomycin (BRL 15140-122; Gibco). For CM collection, the cells were plated at 107 cells/10 cm plate coated with poly-d-lysine (P7280; Sigma-Aldrich).
Lentivirus production and infection
After 2 d of recovery and media changes, the neurons were infected with lentivirus against either Tsc2 or Gl3 (nontargeting control shRNAi construct against the luciferase gene; Flavell et al., 2006). The virus was made as previously published (Mostoslavsky et al., 2005; Di Nardo et al., 2009). In brief, the four packaging vectors (gift from R.C. Mulligan, Harvard Medical School, Boston, MA) were cotransfected into HEK293T cells with either the Tsc2-shRNAi or GL3-shRNAi plasmids using Lipofectamine 2000 according to the manufacturer’s instructions. The resulting viral particles were collected 48 and 72 h after the transfection and filtered through the 0.45-µm membrane, and the aliquots were stored at −80°C until usage.
Rat cortical neurons were infected at 2 d in vitro in the presence of 0.6 µg/ml polybrene. 6 h after infection, the virus was washed out with NB, and the medium was replaced with NB/B27. After infection, the neurons were kept in culture for 10 more days to achieve >90% KD efficiency of the Tsc2 gene. The Tsc2 targeting sequence was 5′-GGTGAAGAGAGCCGTATCACA-3′.
CM collection and cell treatment
7 d after the lentivirus infection, neuronal media was replaced with 5 ml of fresh DMEM media (BRL 11960-44; Gibco) per 10-cm plate. 4 d later, the media was collected, placed into prehydrated Vivaspin concentrator tubes (VS2092; Sartorius), and immediately centrifuged for 45 min at 3,400 rpm at 4°C. This allows for 10× concentration of media. For CM treatment experiments, the concentrated media was mixed 1:1 with normal OPC media (refer to the next section) and placed on the cells immediately after mitogenic dedifferentiation of the cells to OPCs. The media was replaced every other day for either 3 d (A2B5 and NG2 stage) or 5 d (O4, GalC, and MBP stage). The preparation of the HEK293T CM was performed as follows: HEK293T cells were transfected with flag-CTGF, the next day the media was replaced with 5 ml of fresh DMEM media, and 3 d later the media was collected and concentrated. To inhibit CTGF function, 2 µg neutralizing antibody to CTGF (L-20; sc14939; Santa Cruz Biotechnology, Inc.) per well was added and replaced each time when the media was refreshed. To treat OPC with the recombinant full-length CTGF (87314.10; Biomol), 2 µg/ml and other doses were prepared in OPC media, and the media containing recombinant full-length CTGF was refreshed every other day for 5 d.
Oligodendrocyte cultures
OPC purification cultures were prepared from mixed cortical glial cultures using established methods (McCarthy and de Vellis, 1980). Cortices of P1 Sprague-Dawley rat pups were mechanically dissociated using 22-gauge and 25-gauge needles. The dissociated cortices were filtered through the 70-µm cell strainer and spun down before being plated on poly-d-lysine–coated T75 flasks in 10% FBS and 25 µg/ml gentamicin in DMEM (BRL 11960-44; GIBCO). The cultures were placed in a humidified incubator at 5% CO2 for 7–10 d, changing the media every 3 d.
The OPCs were purified using the differential shake method. In brief, after 7–10 d, the T75 flasks of mixed glial cultures were shaken at 200 rpm for 1 h to remove the microglia, and the fresh media was added before the flasks were shaken overnight at 37°C in a humidified incubator. The next day, the resulting supernatant was filtered through a 40-µm mesh filter, spun down, reconstituted in 8 ml of media, and seeded on an uncoated 10-cm tissue culture dish for 1 h to allow any remaining fibroblasts to stick. The floating OPCs were collected after the hour and filtered through a 20-µm nylon filter. Then, OPCs were seeded onto poly-d-lysine– and mouse laminin–coated glass coverslips in chemically defined media (OPC media) consisting of 1 mM sodium pyruvate, 25 µg/ml gentamycin, 1× insulin-transferrin-selenium (Invitrogen), 1% vol/vol l-glutamine, and 0.5% FBS in DMEM. The OPCs were seeded at a density of 15,000/coverslip and allowed to recover and amplify in the presence of 10 ng/ml each of platelet-derived growth factor (100-13A; PeproTech) and basic fibroblast growth factor (FGF; bFGF; 233FB; R&D Systems) for 3 d before differentiation in platelet-derived growth factor– and FGF-free CM for 3–5 d.
Gene expression profiling
For the microarray experiments, 24 h after plating, the neurons were infected with the Tsc2 KD or GL3 KD (control) virus for 6 h. Fresh media was provided after 3 d until 11 d after infection. On the 11th day after infection, cells were collected by gentle scraping. The scraped cells were spun down in the cold room for 15 min until they were prepared for microarray. RNA was isolated using TRIzol reagent (Invitrogen) per the manufacturer’s instructions. In brief, the cells were lysed by direct application of TRIzol and 5-min room-temperature incubation. RNA was separated from the total lysate by the addition of chloroform. RNA was precipitated using isopropyl alcohol and 10-min room-temperature incubation. Precipitated RNA was washed with 75% ethanol and centrifuged to remove the ethanol. The final RNA pellet was dissolved in RNase-free water, and the concentration was measured using a spectrophotometer (ND-1000; software V3.1.0; NanoDrop).
Genome-wide expression profiles were obtained using the microarrays (RatRef12; Illumina). cDNA labeling and array hybridization and scanning were performed following the manufacturer’s protocol at University of California, Los Angeles, microarray core facility. Microarray data were imported into R, and Pearson correlations between all samples using all genes were calculated to identify outlier samples. After the removal of one sample because of lower correlations with all other samples, the data were normalized using the quantile normalization algorithm.
Weighted gene coexpression network analysis was performed as described previously (Zhang and Horvath, 2005; Oldham et al., 2008). In brief, the top 20% of genes with the highest variance across all samples were selected, representing 4,388 genes. Pearson correlations between all these genes were calculated, and this correlation matrix was scaled using an exponent (β = 8) to best approximate scale-free topology. Then, this scaled matrix was used to calculate the topological overlap matrix (Ravasz et al., 2002), which was used as an input into the hierarchical clustering algorithm. From these clusters, modules were identified using a dynamic tree-cutting algorithm (Langfelder et al., 2008), and we identified 17 modules of coexpressed genes (M1-17). Module eigengenes (MEs) were calculated using singular-value decomposition. The kME of a gene within a module was calculated using the Pearson correlation between the expressions of that gene with the ME of the module. Then, we evaluated correspondence between the modules and treatment by using both Student’s t test and Pearson correlation. The functions used here can be found in the weighted gene coexpression network analysis R package. Functional annotation was performed by obtaining all gene ontology terms associated with mouse genes and filtering for genes associated with the term extracellular region (GO: GO:0005576). We found that the M_1 module was enriched in genes associated with the extracellular region by gene ontology (n = 62; P = 3.9e-12, hypergeometric probability).
Western blotting
Blocking of the Western blots was performed in 3% milk in Tris-buffered saline–Tween 20. Incubation with primary antibody was performed in 3% milk or 3% BSA (for phospho-antibodies and CTGF antibody) overnight at 4°C. The blots were incubated in the secondary antibody solution HRP-conjugated secondary antibodies in 5% milk at 1:1,000 dilutions at room temperature for 2 h. Densitometry was performed to permit quantitative analysis of chemiluminescent bands using ImageJ software.
Antibodies
The following antibodies were used: NG2 (rabbit; AB5320; EMD Millipore), O4 (gift from P. Rosenberg, Boston Children’s Hospital, Boston, MA), A2B5 (mouse; MAB312R; EMD Millipore), GalC (mouse; MAB342; EMD Millipore), CTGF (for IHC: goat, L-20 sc14939, Santa Cruz Biotechnology, Inc.; and for WB: rabbit, ab6992, Abcam), S6 (mouse; 54D2; Cell Signaling Technology), pS6 (rabbit; S240/244; D69F8; Cell Signaling Technology), cleaved caspase-3 (rabbit; 9664S; Cell Signaling Technology), AKT1 (goat; C-20 sc-1618; Santa Cruz Biotechnology, Inc.), TSC2 (rabbit; D93F12; Cell Signaling Technology), MBP (mouse; SMI-99P-100; Covance), MAG (mouse; MAB1567; EMD Millipore), Olig2 (rabbit; AB9610; EMD Millipore), SRF (rabbit; D71A9; Cell Signaling Technology), anti-Oct4 (sc5279; Santa Cruz Biotechnology, Inc.), anti-Nanog (sc33759; Santa Cruz Biotechnology, Inc.), anti-SSEA4 (411000; Invitrogen), anti–TRA1-60 (414000; Invitrogen), anti-MAP2 (ab32454; Abcam), and NeuN (mouse; MAB377; EMD Millipore). Secondary antibodies for IHC were: Alexa Fluor 594 goat anti–rabbit IgG (A11037), Alexa Fluor 594 goat anti–mouse IgG (A11005), Alexa Fluor 594 rabbit anti–goat IgG (A11080), Alexa Fluor 488 goat anti–mouse IgG (A11001), Alexa Fluor 488 goat anti–rabbit IgG (A11008), and Alexa Fluor 568 donkey anti–rabbit. Secondary antibodies for Western blotting were: mouse IgG (H + L) antibody peroxidase conjugated (610-103-121; Rockland), rabbit IgG (H + L) antibody peroxidase conjugated (610-103-122; Rockland), and goat IgG (H + L) antibody peroxidase conjugated (605-4313; Rockland).
qRT-PCR
RNA extraction was performed by using an RNeasy Mini kit (QIAGEN) according to the manufacturer’s protocol. For reverse transcription, 1 µg poly(A) mRNA and a High-Capacity cDNA Reverse Transcription kit (4368814; Applied Biosystems) were used as per the manufacturer’s instructions. For quantitative PCR, we used the SYBR green kit (4309155; Applied Biosystems) in triplicate for each sample and normalized them against GAPDH as an endogenous control for each sample. Analysis was performed using 7300 System SDS software on a 7300 Real Time PCR system (Applied Biosystems). The data were expressed as means ± SEM of at least three independent experiments. Statistical analysis was performed by unpaired two-tailed Student’s t test and considered significant at P < 0.05. The sequences of the primers are available upon request.
Human brain lysates
The institution review board protocol for all experiments was approved by Boston Children’s Hospital. Human brain protein extracts were lysed in 20 mM Tris-Cl, pH 7.5, 140 mM NaCl, 10 mM of NaF, 1 mM Na3VO4, 1 mM EDTA containing 1% Triton X-100, and a protease inhibitor mixture (Sigma-Aldrich) by 20 strokes in a tight-fitting homogenizer (Dounce). The lysates were cleared by a 30-min spin at 4°C and 14,000 rpm. Supernatants were mixed with SDS sample buffer, heated to 95°C for 5 min, and stored frozen. Protein concentration was measured by Bradford reagent, and before being subjected to discontinuous gel electrophoresis, equal amounts of all protein lysates were verified by Coomassie gel staining. For immunoblotting, equal amounts of protein lysates were subjected to SDS-PAGE.
iPSC generation
The institutional review board protocol for all experiments was approved by Boston Children’s Hospital. Human iPSCs were generated using episomal plasmids (pCXLE-hL-Myc-LIN28, pCXLE-hOct3/4-shp53, and pCXLE-hSox2-Klf4; Addgene) to introduce the OSKM factors (Oct4, Sox2, Klf4, and l-Myc) into TSC2 patient–derived fibroblasts (SAH0047-01; from a 4-yr-old male patient) and control fibroblasts (SAH0047-02; from patient’s mother), respectively. Embryonic stem–like colonies formed after 3 wk of transfection, and the observed embryonic stem–like colonies were handpicked and transferred onto mouse feeder cells to generate iPSC lines. The iPSCs were maintained on feeder cells (Globalstem) in human embryonic stem media (DMEM; Invitrogen), supplemented with 2 mM l-glutamine (Invitrogen), 1 mM β-mercaptoethanol, 1× nonessential amino acids (Invitrogen), 20% knockout serum replacement (Invitrogen), and 10 ng/ml bFGF (Invitrogen). Established iPSC cells were maintained in STEMPRO human embryonic stem cell culture medium (Invitrogen) supplemented with 10 ng/ml bFGF using Geltrex lactose dehydrogenase elevating virus–free human embryonic stem cell–qualified Reduced Growth Factor Basement Membrane Matrix (Invitrogen) and passaged using dispase (Invitrogen). All the iPSC lines showed pluripotency (qRT-PCR and immunostaining; Fig. S2, I–N). Established iPSC lines (TSC2 patient and control) were differentiated into neurons following the dual SMAD inhibition method. Karyotyping was done at the CytoGenomics Core Laboratory in Brigham and Women’s Hospital and showed normal karyotyping (Ebrahimi-Fakhari et al., 2016).
Neuronal differentiation
Human iPSCs were dissociated into single cells with Accutase and seeded on a Geltrex-coated 6-well plate. Cells were fed everyday with N2B27 media (Invitrogen) with 10 µM SB-431542 (Stemgent), 100 nM LDN-193189 (Stemgent), 1 µM retinoic acid (Sigma-Aldrich), and 1 µM smoothened agonist (Santa Cruz Biotechnology, Inc.) for 6 d. Cells were fed everyday with N2B27 media with 5 µM DAPT (N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester; Cayman Chemical), 4 µM SU-5402 (Abcam), 1 µM retinoic acid, and 1 µM smoothened agonist for 8 d. At day 14, cells were dissociated with Accutase and replated on a Geltrex-coated plate. Cells were grown with N2B27 media with 10 ng/ml brain-derived neurotrophic factor (PeproTech), 10 ng/ml glial-derived neurotrophic factor (PeproTech), and 10 ng/ml ciliary neurotrophic factor (PeproTech) for 7 d. The sequences of the primers are available upon request.
EM
Animals were perfused transcardially with 1× PBS for 4 min and then with 2% paraformaldehyde, 3% glutaraldehyde, and 0.03% picric acid in 0.15 M cacodylate buffer, pH 7.4, for 15 min. After dissection, brains were postfixed in the same fixative overnight. Then, samples were washed with 0.15 M cacodylate buffer and stained en bloc in 2% osmium tetroxide and 1.5% ferrocyanide in cacodylate buffer for 5 h, washed with water, and then stained in 1% aqueous uranyl acetate for 2 h. Then, samples were dehydrated in graded alcohols and propylene oxide and embedded in TAAB 812 resin (Canemco-Marivac). Blocks were kept for 48 h at 60°C to complete polymerization. Both semi- and ultrathin sections were prepared with Diatome Histo and Diatome Ultra 45° diamond knives, respectively, on an Ultramicrotome (UC7; Leica Biosystems). The digital images were taken with a charged-coupled device camera (2k; AMT) mounted on a transmission electron microscope (G2 Spirit BioTWIN; Tecnai).
At least 30 images were taken for each brain sample at a magnification of 9,300×. The total number of unmyelinated and myelinated axons was counted regardless of their diameters. The images were analyzed by ImageJ. The g-ratio was calculated by inner area divided by outer area of the myelinated axon. The circularity was determined by ImageJ, and only the axons having circularity >0.6 were used in calculations. Calculations and graphs were done in Excel and R, respectively.
Online supplemental material
Fig. S1 shows reduced myelination in Tsc1-mutant but not in heterozygous brains. Fig. S2 demonstrates that Tsc2 KD CM increases the number of developing oligodendrocytes. Fig. S3 shows that the number of CTGF-positive cells increases at P14 in Tsc1 mutants. Fig. S4 shows the effect of CTGF on myelination in vivo. Fig. S5 indicates that the g-ratio and axon diameter increases in Tsc1 mutant and double mutant. Table S1 is included as an Excel file and shows a list of modules and coexpressed genes from the microarray of control KD and Tsc2 KD primary cortical neurons.
Acknowledgments
We are indebted to Drs. Paul Rosenberg, Robin Kleiman, and members of the M. Sahin laboratory for critical reading of the manuscript, Balca Mardin for the statistical analysis of EM results, and the Cellular Imaging Core and EM Facility of Boston Children’s Hospital F.M. Kirby Neurobiology Department and Harvard Medical School, respectively.
This work was supported by the National Institute of Child Health and Human Development (5P30HD018655), U.S. Department of Defense (W81XWH-13-1-0040), Nancy Lurie Marks Family Foundation, and the Boston Children’s Hospital Translational Research Program (to M. Sahin).
The authors declare no competing financial interests.
Author contributions: E. Ercan, J.M. Han, and M. Sahin designed the study. E. Ercan performed the majority of the experiments and analyses. A. Di Nardo performed the experiments for the revision. M.-J. Han performed the iPSC generation and neuronal differentiation experiments. L. Hoyo helped with the IHC experiments and image analysis. A. Saffari helped with the analysis of EM data. K.D. Winden and D.H. Geschwind performed and analyzed the gene expression profiling experiment. A. Leask provided the Ctgf knockout mice and expertise on CTGF. E. Ercan and M. Sahin wrote the initial draft, and all authors edited the manuscript.
Footnotes
Abbreviations used:
- bFGF
- basic FGF
- CM
- conditioned medium
- CTGF
- connective tissue growth factor
- EM
- electron microscopy
- FCD
- focal cortical dysplasia
- FGF
- fibroblast growth factor
- GM
- gray matter
- IHC
- immunohistochemistry
- iPSC
- induced pluripotent stem cell
- KD
- knockdown
- MAG
- myelin-associated glycoprotein
- MBP
- myelin basic protein
- ME
- module eigengene
- mTOR
- mechanistic target of rapamycin
- NB
- neurobasal medium
- NGS
- normal goat serum
- OPC
- oligodendrocyte precursor cell
- qRT-PCR
- quantitative RT-PCR
- SRF
- serum response factor
- TSC
- tuberous sclerosis complex
- WM
- white matter
References
- Allendoerfer K.L., and Shatz C.J.. 1994. The subplate, a transient neocortical structure: its role in the development of connections between thalamus and cortex. Annu. Rev. Neurosci. 17:185–218. 10.1146/annurev.ne.17.030194.001153 [DOI] [PubMed] [Google Scholar]
- Angelini A., Li Z., Mericskay M., and Decaux J.F.. 2015. Regulation of connective tissue growth factor and cardiac fibrosis by an SRF/MicroRNA-133a axis. PLoS One. 10:e0139858 10.1371/journal.pone.0139858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercury K.K., Dai J., Sachs H.H., Ahrendsen J.T., Wood T.L., and Macklin W.B.. 2014. Conditional ablation of raptor or rictor has differential impact on oligodendrocyte differentiation and CNS myelination. J. Neurosci. 34:4466–4480. 10.1523/JNEUROSCI.4314-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigstock D.R., Steffen C.L., Kim G.Y., Vegunta R.K., Diehl J.R., and Harding P.A.. 1997. Purification and characterization of novel heparin-binding growth factors in uterine secretory fluids. Identification as heparin-regulated Mr 10,000 forms of connective tissue growth factor. J. Biol. Chem. 272:20275–20282. 10.1074/jbc.272.32.20275 [DOI] [PubMed] [Google Scholar]
- Carson R.P., Kelm N.D., West K.L., Does M.D., Fu C., Weaver G., McBrier E., Parker B., Grier M.D., and Ess K.C.. 2015. Hypomyelination following deletion of Tsc2 in oligodendrocyte precursors. Ann. Clin. Transl. Neurol. 2:1041–1054. 10.1002/acn3.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.J., Di Nardo A., Kramvis I., Meikle L., Kwiatkowski D.J., Sahin M., and He X.. 2008. Tuberous sclerosis complex proteins control axon formation. Genes Dev. 22:2485–2495. 10.1101/gad.1685008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad S., Schluesener H.J., Adibzahdeh M., and Schwab J.M.. 2005. Spinal cord injury induction of lesional expression of profibrotic and angiogenic connective tissue growth factor confined to reactive astrocytes, invading fibroblasts and endothelial cells. J. Neurosurg. Spine. 2:319–326. 10.3171/spi.2005.2.3.0319 [DOI] [PubMed] [Google Scholar]
- Crino P.B., Nathanson K.L., and Henske E.P.. 2006. The tuberous sclerosis complex. N. Engl. J. Med. 355:1345–1356. 10.1056/NEJMra055323 [DOI] [PubMed] [Google Scholar]
- DiMario F.J., Jr 2004. Brain abnormalities in tuberous sclerosis complex. J. Child Neurol. 19:650–657. [DOI] [PubMed] [Google Scholar]
- Di Nardo A., Kramvis I., Cho N., Sadowski A., Meikle L., Kwiatkowski D.J., and Sahin M.. 2009. Tuberous sclerosis complex activity is required to control neuronal stress responses in an mTOR-dependent manner. J. Neurosci. 29:5926–5937. 10.1523/JNEUROSCI.0778-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D., Saffari A., Wahlster L., Di Nardo A., Turner D., Lewis T.L. Jr., Conrad C., Rothberg J.M., Lipton J.O., Kölker S., et al. 2016. Impaired mitochondrial dynamics and mitophagy in neuronal models of tuberous sclerosis complex. Cell Reports. 17:1053–1070. 10.1016/j.celrep.2016.09.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell S.W., Cowan C.W., Kim T.K., Greer P.L., Lin Y., Paradis S., Griffith E.C., Hu L.S., Chen C., and Greenberg M.E.. 2006. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 311:1008–1012. 10.1126/science.1122511 [DOI] [PubMed] [Google Scholar]
- Goebbels S., Oltrogge J.H., Kemper R., Heilmann I., Bormuth I., Wolfer S., Wichert S.P., Möbius W., Liu X., Lappe-Siefke C., et al. 2010. Elevated phosphatidylinositol 3,4,5-trisphosphate in glia triggers cell-autonomous membrane wrapping and myelination. J. Neurosci. 30:8953–8964. 10.1523/JNEUROSCI.0219-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak A.J., Tsou P.S., Amin M.A., Ruth J.H., Campbell P., Fox D.A., Khanna D., Larsen S.D., and Neubig R.R.. 2014. Targeting the myofibroblast genetic switch: inhibitors of myocardin-related transcription factor/serum response factor-regulated gene transcription prevent fibrosis in a murine model of skin injury. J. Pharmacol. Exp. Ther. 349:480–486. 10.1124/jpet.114.213520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.M., and Sahin M.. 2011. TSC1/TSC2 signaling in the CNS. FEBS Lett. 585:973–980. 10.1016/j.febslet.2011.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto G., Inoki I., Fujii Y., Aoki T., Ikeda E., and Okada Y.. 2002. Matrix metalloproteinases cleave connective tissue growth factor and reactivate angiogenic activity of vascular endothelial growth factor 165. J. Biol. Chem. 277:36288–36295. 10.1074/jbc.M201674200 [DOI] [PubMed] [Google Scholar]
- Hertel M., Tretter Y., Alzheimer C., and Werner S.. 2000. Connective tissue growth factor: a novel player in tissue reorganization after brain injury? Eur. J. Neurosci. 12:376–380. 10.1046/j.1460-9568.2000.00930.x [DOI] [PubMed] [Google Scholar]
- Heuer H., Christ S., Friedrichsen S., Brauer D., Winckler M., Bauer K., and Raivich G.. 2003. Connective tissue growth factor: a novel marker of layer VII neurons in the rat cerebral cortex. Neuroscience. 119:43–52. 10.1016/S0306-4522(03)00100-3 [DOI] [PubMed] [Google Scholar]
- Hinkel R., Trenkwalder T., Petersen B., Husada W., Gesenhues F., Lee S., Hannappel E., Bock-Marquette I., Theisen D., Leitner L., et al. 2014. MRTF-A controls vessel growth and maturation by increasing the expression of CCN1 and CCN2. Nat. Commun. 5:3970 10.1038/ncomms4970 [DOI] [PubMed] [Google Scholar]
- Hoesche C., Sauerwald A., Veh R.W., Krippl B., and Kilimann M.W.. 1993. The 5′-flanking region of the rat synapsin I gene directs neuron-specific and developmentally regulated reporter gene expression in transgenic mice. J. Biol. Chem. 268:26494–26502. [PubMed] [Google Scholar]
- Huang J., and Manning B.D.. 2009. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem. Soc. Trans. 37:217–222. 10.1042/BST0370217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J.I., and Lau L.F.. 2011. Taking aim at the extracellular matrix: CCN proteins as emerging therapeutic targets. Nat. Rev. Drug Discov. 10:945–963. 10.1038/nrd3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.S., Nagalla S.R., Oh Y., Wilson E., Roberts C.T. Jr., and Rosenfeld R.G.. 1997. Identification of a family of low-affinity insulin-like growth factor binding proteins (IGFBPs): characterization of connective tissue growth factor as a member of the IGFBP superfamily. Proc. Natl. Acad. Sci. USA. 94:12981–12986. 10.1073/pnas.94.24.12981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.J., Kang J.H., Chang S.Y., Jang H.J., Ryu G.R., Ko S.H., Jeong I.K., Kim M.S., and Jo Y.H.. 2008. Exendin-4 induction of Egr-1 expression in INS-1 β-cells: interaction of SRF, not YY1, with SRE site of rat Egr-1 promoter. J. Cell. Biochem. 104:2261–2271. 10.1002/jcb.21783 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski D.J., and Manning B.D.. 2005. Tuberous sclerosis: a GAP at the crossroads of multiple signaling pathways. Hum. Mol. Genet. 14:R251–R258. 10.1093/hmg/ddi260 [DOI] [PubMed] [Google Scholar]
- Lamond R., and Barnett S.C.. 2013. Schwann cells but not olfactory ensheathing cells inhibit CNS myelination via the secretion of connective tissue growth factor. J. Neurosci. 33:18686–18697. 10.1523/JNEUROSCI.3233-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Zhang B., and Horvath S.. 2008. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 24:719–720. 10.1093/bioinformatics/btm563 [DOI] [PubMed] [Google Scholar]
- Lau L.F., and Lam S.C.. 1999. The CCN family of angiogenic regulators: the integrin connection. Exp. Cell Res. 248:44–57. 10.1006/excr.1999.4456 [DOI] [PubMed] [Google Scholar]
- Leask A., and Abraham D.J.. 2003. The role of connective tissue growth factor, a multifunctional matricellular protein, in fibroblast biology. Biochem. Cell Biol. 81:355–363. 10.1139/o03-069 [DOI] [PubMed] [Google Scholar]
- Lebrun-Julien F., Bachmann L., Norrmén C., Trötzmüller M., Köfeler H., Rüegg M.A., Hall M.N., and Suter U.. 2014. Balanced mTORC1 activity in oligodendrocytes is required for accurate CNS myelination. J. Neurosci. 34:8432–8448. 10.1523/JNEUROSCI.1105-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis W.W., Sahin M., Scherrer B., Peters J.M., Suarez R.O., Vogel-Farley V.K., Jeste S.S., Gregas M.C., Prabhu S.P., Nelson C.A. III, and Warfield S.K.. 2013. Impaired language pathways in tuberous sclerosis complex patients with autism spectrum disorders. Cereb. Cortex. 23:1526–1532. 10.1093/cercor/bhs135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton J.O., and Sahin M.. 2014. The neurology of mTOR. Neuron. 84:275–291. 10.1016/j.neuron.2014.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Shi-wen X., Abraham D.J., and Leask A.. 2011. CCN2 is required for bleomycin-induced skin fibrosis in mice. Arthritis Rheum. 63:239–246. 10.1002/art.30074 [DOI] [PubMed] [Google Scholar]
- McCarthy K.D., and de Vellis J.. 1980. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 85:890–902. 10.1083/jcb.85.3.890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikle L., Talos D.M., Onda H., Pollizzi K., Rotenberg A., Sahin M., Jensen F.E., and Kwiatkowski D.J.. 2007. A mouse model of tuberous sclerosis: neuronal loss of Tsc1 causes dysplastic and ectopic neurons, reduced myelination, seizure activity, and limited survival. J. Neurosci. 27:5546–5558. 10.1523/JNEUROSCI.5540-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikle L., Pollizzi K., Egnor A., Kramvis I., Lane H., Sahin M., and Kwiatkowski D.J.. 2008. Response of a neuronal model of tuberous sclerosis to mammalian target of rapamycin (mTOR) inhibitors: effects on mTORC1 and Akt signaling lead to improved survival and function. J. Neurosci. 28:5422–5432. 10.1523/JNEUROSCI.0955-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky G., Kotton D.N., Fabian A.J., Gray J.T., Lee J.S., and Mulligan R.C.. 2005. Efficiency of transduction of highly purified murine hematopoietic stem cells by lentiviral and oncoretroviral vectors under conditions of minimal in vitro manipulation. Mol. Ther. 11:932–940. 10.1016/j.ymthe.2005.01.005 [DOI] [PubMed] [Google Scholar]
- Moussad E.E., and Brigstock D.R.. 2000. Connective tissue growth factor: what’s in a name? Mol. Genet. Metab. 71:276–292. 10.1006/mgme.2000.3059 [DOI] [PubMed] [Google Scholar]
- Muehlich S., Cicha I., Garlichs C.D., Krueger B., Posern G., and Goppelt-Struebe M.. 2007. Actin-dependent regulation of connective tissue growth factor. Am. J. Physiol. Cell Physiol. 292:C1732–C1738. 10.1152/ajpcell.00552.2006 [DOI] [PubMed] [Google Scholar]
- Narayanan S.P., Flores A.I., Wang F., and Macklin W.B.. 2009. Akt signals through the mammalian target of rapamycin pathway to regulate CNS myelination. J. Neurosci. 29:6860–6870. 10.1523/JNEUROSCI.0232-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham M.C., Konopka G., Iwamoto K., Langfelder P., Kato T., Horvath S., and Geschwind D.H.. 2008. Functional organization of the transcriptome in human brain. Nat. Neurosci. 11:1271–1282. 10.1038/nn.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquini L.A., Millet V., Hoyos H.C., Giannoni J.P., Croci D.O., Marder M., Liu F.T., Rabinovich G.A., and Pasquini J.M.. 2011. Galectin-3 drives oligodendrocyte differentiation to control myelin integrity and function. Cell Death Differ. 18:1746–1756. 10.1038/cdd.2011.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.M., Taquet M., Prohl A.K., Scherrer B., van Eeghen A.M., Prabhu S.P., Sahin M., and Warfield S.K.. 2013. Diffusion tensor imaging and related techniques in tuberous sclerosis complex: review and future directions. Future Neurol. 8:583–597. 10.2217/fnl.13.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasz E., Somera A.L., Mongru D.A., Oltvai Z.N., and Barabási A.L.. 2002. Hierarchical organization of modularity in metabolic networks. Science. 297:1551–1555. 10.1126/science.1073374 [DOI] [PubMed] [Google Scholar]
- Rushton W.A. 1951. A theory of the effects of fibre size in medullated nerve. J. Physiol. 115:101–122. 10.1113/jphysiol.1951.sp004655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I., and Meyuhas O.. 2006. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem. Sci. 31:342–348. 10.1016/j.tibs.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Sahin M., Greer P.L., Lin M.Z., Poucher H., Eberhart J., Schmidt S., Wright T.M., Shamah S.M., O’connell S., Cowan C.W., et al. 2005. Eph-dependent tyrosine phosphorylation of ephexin1 modulates growth cone collapse. Neuron. 46:191–204. 10.1016/j.neuron.2005.01.030 [DOI] [PubMed] [Google Scholar]
- Sarbassov D.D., Ali S.M., Sengupta S., Sheen J.H., Hsu P.P., Bagley A.F., Markhard A.L., and Sabatini D.M.. 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 22:159–168. 10.1016/j.molcel.2006.03.029 [DOI] [PubMed] [Google Scholar]
- Scholl T., Mühlebner A., Ricken G., Gruber V., Fabing A., Samueli S., Gröppel G., Dorfer C., Czech T., Hainfellner J.A., et al. 2016. Impaired oligodendroglial turnover is associated with myelin pathology in focal cortical dysplasia and tuberous sclerosis complex. Brain Pathol. 10.1111/bpa.12452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab J.M., Beschorner R., Nguyen T.D., Meyermann R., and Schluesener H.J.. 2001. Differential cellular accumulation of connective tissue growth factor defines a subset of reactive astrocytes, invading fibroblasts, and endothelial cells following central nervous system injury in rats and humans. J. Neurotrauma. 18:377–388. 10.1089/089771501750170930 [DOI] [PubMed] [Google Scholar]
- Selvaraju R., Bernasconi L., Losberger C., Graber P., Kadi L., Avellana-Adalid V., Picard-Riera N., Baron Van Evercooren A., Cirillo R., Kosco-Vilbois M., et al. 2004. Osteopontin is upregulated during in vivo demyelination and remyelination and enhances myelin formation in vitro. Mol. Cell. Neurosci. 25:707–721. 10.1016/j.mcn.2003.12.014 [DOI] [PubMed] [Google Scholar]
- Steffen C.L., Ball-Mirth D.K., Harding P.A., Bhattacharyya N., Pillai S., and Brigstock D.R.. 1998. Characterization of cell-associated and soluble forms of connective tissue growth factor (CTGF) produced by fibroblast cells in vitro. Growth Factors. 15:199–213. 10.3109/08977199809002117 [DOI] [PubMed] [Google Scholar]
- Stritt C., Stern S., Harting K., Manke T., Sinske D., Schwarz H., Vingron M., Nordheim A., and Knöll B.. 2009. Paracrine control of oligodendrocyte differentiation by SRF-directed neuronal gene expression. Nat. Neurosci. 12:418–427. 10.1038/nn.2280 [DOI] [PubMed] [Google Scholar]
- Tsai P., and Sahin M.. 2011. Mechanisms of neurocognitive dysfunction and therapeutic considerations in tuberous sclerosis complex. Curr. Opin. Neurol. 24:106–113. 10.1097/WCO.0b013e32834451c4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann E.J., Wong M., Baldwin R.L., Bajenaru M.L., Onda H., Kwiatkowski D.J., Yamada K., and Gutmann D.H.. 2002. Astrocyte-specific TSC1 conditional knockout mice exhibit abnormal neuronal organization and seizures. Ann. Neurol. 52:285–296. 10.1002/ana.10283 [DOI] [PubMed] [Google Scholar]
- Wahl S.E., McLane L.E., Bercury K.K., Macklin W.B., and Wood T.L.. 2014. Mammalian target of rapamycin promotes oligodendrocyte differentiation, initiation and extent of CNS myelination. J. Neurosci. 34:4453–4465. 10.1523/JNEUROSCI.4311-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., and Hall M.N.. 2006. TOR signaling in growth and metabolism. Cell. 124:471–484. 10.1016/j.cell.2006.01.016 [DOI] [PubMed] [Google Scholar]
- Yang D.H., Kim H.S., Wilson E.M., Rosenfeld R.G., and Oh Y.. 1998. Identification of glycosylated 38-kDa connective tissue growth factor (IGFBP-related protein 2) and proteolytic fragments in human biological fluids, and up-regulation of IGFBP-rP2 expression by TGF-β in Hs578T human breast cancer cells. J. Clin. Endocrinol. Metab. 83:2593–2596. [DOI] [PubMed] [Google Scholar]
- Zhang B., and Horvath S.. 2005. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4:e17. [DOI] [PubMed] [Google Scholar]
- Zhao C., Fancy S.P.J., Franklin R.J.M., and ffrench-Constant C.. 2009. Up-regulation of oligodendrocyte precursor cell αV integrin and its extracellular ligands during central nervous system remyelination. J. Neurosci. Res. 87:3447–3455. 10.1002/jnr.22231 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Romero M.I., Ghosh P., Ye Z., Charnay P., Rushing E.J., Marth J.D., and Parada L.F.. 2001. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 15:859–876. 10.1101/gad.862101 [DOI] [PMC free article] [PubMed] [Google Scholar]