Laidlaw et al. show that Ephrin-B1 is a specific marker of mature germinal center (GC) B cells. Use of Ephrin-B1 allows for the identification of phenotypically distinct GC B cell subsets, including a population that may represent memory precursor B cells.
Abstract
Identification of germinal center (GC) B cells is typically reliant on the use of surface activation markers that exhibit a wide range of expression. Here, we identify Ephrin-B1, a ligand for Eph-related receptor tyrosine kinases, as a specific marker of mature GC B cells. The number of Ephrin-B1+ GC B cells increases during the course of an immune response with Ephrin-B1+ GC B cells displaying elevated levels of Bcl6, S1pr2, and Aicda relative to their Ephrin-B1– counterparts. We further identified a small proportion of recently dividing, somatically mutated Ephrin-B1+ GC B cells that have begun to down-regulate Bcl6 and S1pr2 and express markers associated with memory B cells, such as CD38 and EBI2. Transcriptional analysis indicates that these cells are developmentally related to memory B cells, and likely represent a population of GC memory precursor (PreMem) B cells. GC PreMem cells display enhanced survival relative to bulk GC B cells, localize near the edge of the GC, and are predominantly found within the light zone. These findings offer insight into the significant heterogeneity that exists within the GC B cell population and provide tools to further dissect signals regulating the differentiation of GC B cells.
Introduction
Germinal centers (GCs) are tightly confined clusters of cells within the follicle, in which GC B cells compete for signals necessary for their survival and continued maturation into memory B cells or plasma cells. GC B cells highly express the transcription factor Bcl6 and the G protein–coupled receptor sphingosine-1-phosphate receptor (S1PR2) that promotes their confinement within the GC (Green et al., 2011; Muppidi et al., 2014; Huang and Melnick, 2015). The GC is divided into a light zone (LZ), where GC B cells interact with antigen-bearing follicular DCs (FDCs) and follicular helper T cells, and a dark zone (DZ) in which GC B cells rapidly divide and undergo somatic hypermutation (SHM). Through regulated expression of the chemokine receptor CXCR4, GC B cells rapidly transit between these compartments, allowing for continued selection of high affinity GC B cells via competition for T cell help (Allen et al., 2007; Victora and Nussenzweig, 2012).
Memory B cells can arise from both GC-independent and -dependent pathways, with the majority of memory B cells against T cell–dependent antigens thought to originate within the GC (McHeyzer-Williams et al., 2011; Tarlinton and Good-Jacobson, 2013; Kurosaki et al., 2015). Memory B cells emerge early during the GC response and derive from lower affinity GC B cells that receive less T cell help and, accordingly, maintain higher expression of the transcription factor Bach2 (Shinnakasu et al., 2016; Weisel et al., 2016). Expression of Bach2 predisposes GC B cell to differentiate into memory B cells, whereas expression of Blimp1 promotes the development of plasma cells (Turner et al., 1994; Shinnakasu et al., 2016). Memory B cells are a heterogeneous population with distinctly functioning subsets arising within the GC at different times (Zuccarino-Catania et al., 2014; Adachi et al., 2015; Weisel et al., 2016). The exact signals regulating GC B cell differentiation into memory B cells are poorly understood.
GC B cells are typically defined through their low expression of IgD or CD38 and their positive staining for one or two surface markers. Most studies use the rat monoclonal antibody GL7, which recognizes α2,6-linked N-acetylneuraminic acid (Neu5Ac) on glycan chains, Fas (CD95), and/or peanut agglutinin (PNA), a lectin that binds exposed galactose-β(1–3)-N-acetylgalactosamine (Gal-β(1–3)-GalNac; László et al., 1993; Naito et al., 2007). However, these markers have a wide range of expression and are also expressed on activated B cells before entry into the GC, and thus do not faithfully mark only cells that have already entered the GC (Wang et al., 1996; Shinall et al., 2000; Naito et al., 2007). A marker with discrete expression that specifically labels B cells residing in the GC would be a useful tool for separating maturation occurring within the GC from events occurring before GC entry. This level of resolution is particularly important for identifying GC B cells in the process of differentiating into memory B cells and could inform efforts to more precisely decipher the signals regulating this process.
In this study, we identify Ephrin-B1, a ligand for Eph-related receptor tyrosine kinases, as a specific marker of B cells residing within the GC. Ephrin-B1 functions as a repulsive guidance cue in mice and humans and is highly expressed on GC B cells (Bush and Soriano, 2009), but we find that it is not essential for GC B cell development or positioning. Using Ephrin-B1, we found that GC B cells, defined as B220+ IgDloGL7+CD95+ cells, represent at least four cell subsets of different maturation states based on expression of markers such as Bcl6, CXCR4, S1pr2, EBI2, CD38, and CD73. Importantly, we were able to identify a recently divided, somatically mutated GC B cell population in the process of down-regulating Bcl6 and S1pr2 and up-regulating CD38 and Ebi2. Transcriptional analysis indicates that these cells are developmentally related to memory B cells and likely represent a population of GC memory precursor (PreMem) cells. Finally, we found that PreMem cells localize near the edge of the GC and are predominantly found within the LZ.
Results and discussion
Ephrin-B1 is a specific marker of mature GC B cells
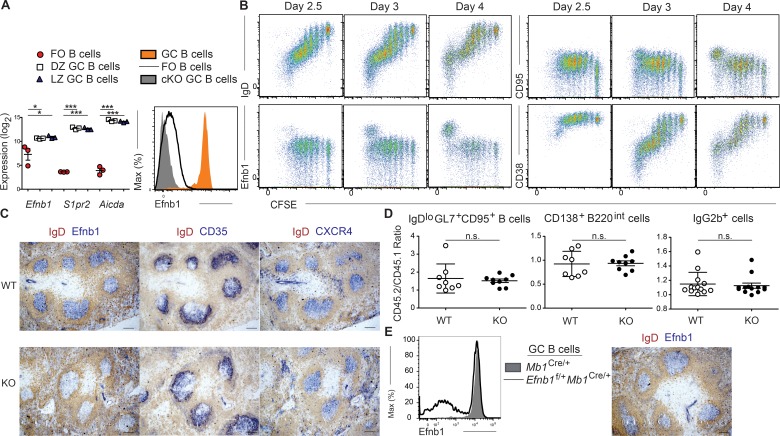
Microarray gene expression analysis of CXCR4hi (DZ) and CXCR4lo (LZ) GC B cells, gated as IgDloGL7+CD95+ cells, and IgD+CD23+GL7–CD95− follicular B cells, identified Efnb1 transcripts as being highly expressed in GC B cells relative to their follicular counterparts (Fig. 1 A). Ephrin-B1 protein was highly expressed on IgDloGL7+CD95+ cells after protein antigen or sheep RBC (SRBC) immunization, but was minimally expressed by other B cell subsets in the spleen or BM, including memory B cells (Fig. 1 A, Fig. S1 A, and not depicted). Ephrin-B1 began to become up-regulated after ∼7 cell divisions in B cells responding to a T cell–dependent antigen in vivo, with its expression preceded by loss of CD38 and IgD expression and occurring well after the start of CD95 up-regulation (Fig. 1 B). Ephrin-B1 has a critical role as a repulsive guidance cue during tissue development, and mutations in the gene result in a wide spectrum of developmental abnormalities constituting craniofrontonasal syndrome in humans and related defects in mice (Bush and Soriano, 2009). Ephrin-B1 is also important in bone formation and in thymocyte development (Xing et al., 2010; Luo et al., 2011; Cejalvo et al., 2013). To test whether Ephrin-B1 may have a functional role in GC B cell development we generated mice in which Efnb1 was specifically deleted in B cells (Efnb1f/fMb1Cre/+). GC B cells from conditional KO (cKO) mice did not express Ephrin-B1 (Fig. 1 A). After SRBC immunization, cKO GCs developed that were of normal size and location and that exhibited correct organization as determined by the polarized distribution of CD35+ FDCs and CXCR4hi GC B cells (Fig. 1 C). Ephrin-B1 staining of tissue sections confirmed that Ephrin-B1 was expressed by GC B cells while also being expressed by endothelial cells of blood vessels (Fig. 1 C). To test whether Ephrin-B1 influenced the competitiveness of B cells in the GC, adoptive cell transfer experiments were performed. We generated Efnb1f/fMb1Cre/+ Hy10 and control Mb1Cre/+ Hy10 mice, which have B cells that are specific for hen egg lysozyme (HEL). Equal numbers of cKO or control HEL-specific Hy10 cells were transferred, along with WT HEL-specific Hy10 cells and OVA-specific OT-II T cells, into congenically mismatched mice 1 d before immunization with duck egg lysozyme (DEL) conjugated to OVA. We detected no defect in the development of IgDloGL7+CD95+ B cells, plasmablasts, or class switching in cKO Hy10 cells (Fig. 1 D). Furthermore, female Efnb1f/+Mb1Cre/+ mice, which have mosaic expression of Efnb1 caused by the gene’s location on the X chromosome, displayed no apparent difference in development or positioning of Efnb1-deficient or -sufficient GC B cells after SRBC immunization (Fig. 1 E). We also did not detect defects in the GC B cell response in mediastinal LNs or spleen after influenza HKx31 infection (two experiments, cKO versus WT and mixed BM chimeras, with at least three mice per group), in the spleen after acute lymphocytic choriomeningitis virus strain Armstrong (LCMV Arm) infection (two mixed BM chimera experiments with three mice per group), in skin draining LNs after subcutaneous immunization with protein antigen in complete Freund’s adjuvant, or in the chronic GCs occurring in Peyer’s patches in response to commensal-derived antigens (three experiments, cKO versus WT and mixed BM chimeras, at least four mice per group; unpublished data). These observations suggest that Efnb1 is not essential for the development or organization of GCs.
Figure 1.
Ephrin-B1 is highly expressed on GC B cells. (A) Analysis of the expression of Efnb1, S1pr2, and Aicda transcripts in IgD+CD23+GL7–CD95− follicular B cells, CXCR4hi (DZ), and CXCR4lo (LZ) GC B cells, gated as IgDloGL7+CD95+ cells at day 8 after SRBC immunization (left). Expression was determined using an Affymetrix Mouse Genome 430 2.0 Array with data combined from three independent experiments in which each population was sorted from multiple pooled mice. Expression of Ephrin-B1 in Mb1Cre/+ follicular and GC B cells and in Efnb1f/fMb1Cre/+ (cKO) GC B cells at day 12 after SRBC immunization (right). Data are representative of three independent experiments with at least three mice per group. (B) Analysis of Ephrin-B1 up-regulation in antigen-specific B cells. CFSE-labeled lysozyme-specific (MD4) transgenic B cells and OT-II T cells were transferred to mice 1 d before immunization with DEL-OVA. Splenocytes were analyzed at days 2.5, 3, and 4 after immunization, and expression of IgD, CD95, Ephrin-B1, and CD38 was determined as compared with CFSE dilution in the transferred MD4 B cell population. Data are representative of at least two independent experiments at each time point. (C) Representative images of GCs in spleens from Efnb1f/fMb1Cre/+ (KO) and Mb1Cre/+ (WT) mice at day 12 after SRBC immunization. Data are representative of many imaged GCs from at least three mice of each type. All GC clusters in WT mice were Efnb1+. Bar, 50 µm. (D) Analysis of the GC response in mice in which CD45.2+ Efnb1f/fMb1Cre/+ or Mb1Cre/+ Hy10 cells were transferred, along with CD45.1+ wild-type Hy10 cells and OT-II T cells, 1 d before immunization with DEL-OVA. GC B cells were defined as IgDloGL7+CD95+ cells. Data are pooled from three independent experiments with two to three mice per group analyzed at day 7 after immunization. No statistical differences were found between groups. (E) Representative histogram of Ephrin-B1 expression in GC B cells from female Efnb1f/+Mb1Cre/+, and Mb1Cre/+ mice at day 12 after SRBC immunization (left). Representative Efnb1 staining in an Efnb1f/+Mb1Cre/+ mouse spleen (right). Data are representative of at least two mice of each type. Bar, 50 µm. Statistical analyses were performed using the unpaired two-tailed Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
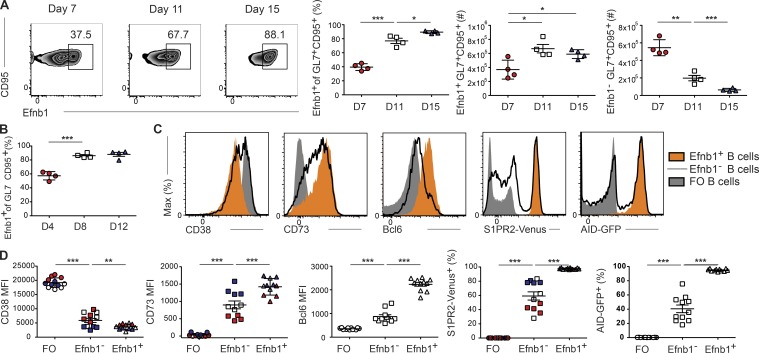
Investigating the kinetics of Ephrin-B1 expression after LCMV Arm infection revealed that there was a marked increase in the proportion and numbers of IgDloGL7+CD95+ B cells that were Efnb1+ during the course of the GC response (Fig. 2 A). Conversely, the number of Efnb1– GL7+CD95+IgDlo B cells decreased over time (Fig. 2 A). Similar findings were obtained when using PNA in place of GL7 (unpublished data) and when analyzing Ephrin-B1 expression kinetics after SRBC immunization (Fig. 2 B). To examine whether Eprhin-B1 may mark distinct GC B cell populations, we determined the expression of markers associated with GC B cell fate and function in these populations. Efnb1+ cells displayed reduced expression of CD38 and increased expression of CD73 relative to their Efnb1– counterparts (Fig. 2, C and D, red, white and blue symbols in D correspond to days 7, 11, and 15, respectively). CD73 expression increases within GC B cells over time, whereas CD38 expression decreases, suggesting that Ephrin-B1 may distinguish more temporally mature GC B cells (Conter et al., 2014). Consistent with this notion, Efnb1– cells expressed intermediate levels of Bcl6 and heterogeneous amounts of S1pr2 (as detected using S1pr2Venus/+ reporter mice) and activation-induced cytidine deaminase (AID; as revealed using AID-GFP reporter mice), indicating that they were not fully confined to the GC or uniformly undergoing SHM (Fig. 2, C and D; Crouch et al., 2007; Moriyama et al., 2014). In contrast, Efnb1+ cells expressed high levels of Bcl6, S1pr2, and Aicda (Fig. 2, C and D). Collectively, these data indicate that Ephrin-B1 is a specific marker of mature GC-resident B cells.
Figure 2.
Ephrin-B1 marks mature GC B cells. (A) Representative FACS plots of Ephrin-B1 expression (left) and percentages and numbers of Efnb1+ and Efnb1– (right) splenic IgDloGL7+CD95+ B cells at days 7, 11, and 15 after LCMV Arm infection. (B) Percentage of Efnb1+ splenic IgDloGL7+CD95+ B cells at days 4, 8, and 12 after SRBC immunization. (C and D) Histograms (C) and plots of mean fluorescence intensity (MFI; D) of CD38, CD73, Bcl6, S1pr2 (as defined using S1pr2Venus/+ mice), and Aicda (as defined using AID-GFP mice) expression in follicular (IgD+GL7–) and Efnb1+ and Efnb1– IgDloGL7+CD95+ B cells. Data are from day 7 (red), 11 (white), and 15 (dark blue) after LCMV Arm infection. Data for the CD38, CD73, and S1pr2 plots are from one experiment with three to four mice per time point at days 7, 11, and 15 and are representative of two independent experiments at days 7 and 15, and four experiments at day 11. Data for the Bcl6 and Aicda plots are pooled from three independent experiments with 3–4 mice per group at day 11 after infection. Statistical analyses were performed using the unpaired two-tailed Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Transitional GC B cells can be identified using Ephrin-B1 and S1pr2
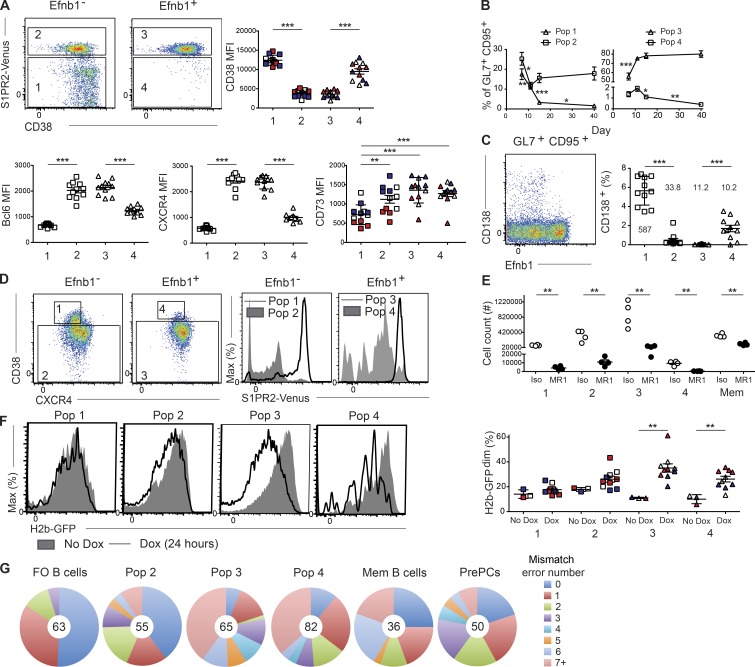
In the course of these studies, we observed that whereas the great majority of Efnb1+ cells were S1pr2hi, a small fraction of the cells (1–2%) had low S1pr2 expression (Fig. 2, C and D). Low S1pr2 expression in both Efnb1– and Efnb1+ IgDloGL7+CD95+ cells positively correlated with high CD38 expression, suggesting that these cells might represent transitional populations in the process of entering or exiting the GC, as CD38 is highly expressed on both follicular and memory B cells (Fig. 3 A). Both Efnb1– and Efnb1+ S1pr2lo cells (populations 1 and 4, respectively) expressed low levels of Bcl6 and CXCR4, consistent with these cells being in a distinct state from the bulk GC B cell population (Fig. 3 A). Efnb1– and Efnb1+ S1pr2hi cells (populations 2 and 3, respectively) conversely expressed high levels of Bcl6 and CXCR4, and had a similar fraction of DZ and LZ B cells (Fig. 3 A; detailed further in Fig. 5 A). S1pr2lo Efnb1– (Pop 1) cells had low expression of CD73, suggesting that they may largely be precursor GC cells (Fig. 3 A). Importantly, S1pr2lo Efnb1+ (Pop 4) cells expressed similar levels of CD73 as their S1pr2hi (Pop 3) counterparts, raising the possibility that they may represent a cell population in the process of exiting the GC (Fig. 3 A). The percentage of IgDloGL7+CD95+ B cells comprising Pop 1 and Pop 4 peaked at day 7 and 11, respectively, before declining, as Pop 3 increasingly dominated the GC response (Fig. 3 B). Pop 2 remained a small but stable fraction of the IgDloGL7+CD95+ B cells, even at late GC time points (Fig. 3 B). Together, these results suggest a model in which newly activated IgDloGL7+CD95+ B cells up-regulate S1pr2 before Ephrin-B1 as they enter the GC response, with Efnb1+ cells losing S1pr2 expression as they differentiate from the bulk GC state.
Figure 3.
Ephrin-B1 and S1pr2 can distinguish transitional populations of GC B cells. (A) Representative plots (left) of S1pr2 (as defined using S1pr2Venus/+ mice) and CD38 expression in splenic Efnb1– and Efnb1+ IgDloGL7+CD95+ B cells. Expression of CD38 (right), Bcl6, CXCR4, and CD73 (bottom) was determined at day 7 (red), 11 (white), or 15 (dark blue) after LCMV Arm infection in Efnb1– S1pr2lo (Pop 1), Efnb1– S1pr2hi (Pop 2), Efnb1+ S1pr2hi (Pop 3), and Efnb1+ S1pr2lo (Pop 4) IgDloGL7+CD95+ B cells. Data for the Bcl6 and CXCR4 plots are pooled from three experiments with three to four mice per group at day 11 after infection. (B) Percentage of IgDloGL7+CD95+ B cells that comprise the populations defined in A at day 7, 11, 15, and 40 after LCMV Arm infection. Data for the CD38 and CD73 plots are from one experiment with three to four mice per time point at days 7, 11, and 15 and are representative of two independent experiments for days 7 and 15, one experiment with four mice at day 40, and four experiments at day 11. (C) Representative plot (left) of CD138 and Ephrin-B1 expression in splenic B220+ IgDloGL7+CD95+ B cells. Percentage of CD138+ cells within each population as defined in A (right). Number indicates the mean number of CD138+ cells in each population per million B cells. Data are pooled from three experiments with three to four mice per group at day 11 after infection. (D) Representative plots (left) of IgDloGL7+CD95+ GC B cells are divided into four subsets (labeled 1–4) with the following marker profile: Efnb1– CD38+CXCR4lo (Pop 1), Efnb1– CD38– (Pop 2), Efnb1+ CD38– (Pop 3), and Efnb1+ CD38+CXCR4lo (Pop 4; left). Representative plots (right) of S1pr2 (as defined using S1pr2Venus/+ mice) in the populations defined using CD38 and CXCR4. (E) Number of Efnb1– CD38+CXCR4lo (Pop 1), Efnb1– CD38– (Pop 2), Efnb1+ CD38– (Pop 3), and Efnb1+ CD38+CXCR4lo (Pop 4) IgDloGL7+CD95+ B cells and memory (IgDloGL7–CD95+CD73+) B cells at day 13 after LCMV Arm infection in mice treated with anti-CD40L (MR1) or an isotype control antibody from days 9–12. Data are from one experiment representative of two independent experiments with four mice per group. (F) Representative histograms (left) of H2b-GFP expression in the populations defined in D in untreated or doxycycline treated Tet-off H2b-GFP mice. Analysis (right) of the percentage of H2b-GFPdim at day 7 (red), day 11 (white), or day 15 (dark blue) after LCMV Arm infection. Mice were treated with doxycycline (dox) 24 h before analysis with one mouse per time point not treated to determine background GFP dilution. Data are from one experiment representative of two independent experiments with three to four mice per time point. (G) Analysis of mismatch error rate frequency in 700 bp of the JH558 intronic sequence in follicular (IgD+GL7–) B cells, Efnb1– S1pr2hi (Pop 2), Efnb1+ S1pr2hi (Pop 3), Efnb1+ S1pr2lo (Pop 4) IgDloGL7+CD95+ B cells, memory (IgDloGL7–CD95+CD73+) B cells, and pre plasma (IgDloGL7+CD95+CD138+) cells sorted at day 11 after LCMV infection. Number of sequences analyzed for each population is listed in the center of each circle. Cells were pooled from two independent experiments with four mice per experiment. Statistical analyses were performed using the unpaired two-tailed Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
As plasma cells and memory B cells can both derive from the GC, we investigated whether S1pr2lo Efnb1+ (Pop 4) cells included plasma cell precursors by examining expression of the plasma cell marker CD138. Although the vast majority of CD138-expressing cells were B220lo/int (not depicted), we identified a small population of B220+ IgDloGL7+CD95+ B cells expressing CD138, likely representing plasma cell precursors (prePCs; Fig. 3 C; Fooksman et al., 2010). PrePCs were largely Efnb1– and found in the S1pr2lo Efnb1– (Pop 1) population, with only ∼2% of S1pr2lo Efnb1+ (Pop 4) cells expressing CD138 (Fig. 3 C). Similar results were found using Blimp1GFP mice to identify prePCs (unpublished data; three experiments, four mice per experiment). This result suggested that, at this time point of LCMV Arm infection, the majority of prePCs differentiate from activated rather than GC-resident B cells, with GC-derived prePCs perhaps arising at later GC time points (Weisel et al., 2016). Alternatively, GC-derived prePCs could lose expression of Ephrin-B1 more rapidly than those of GL7 or CD95. These findings also indicated that by defining transitional GC populations based on high expression of CD38 and low expression of CXCR4 (Fig. 3 A), we could evaluate these populations without significant contamination from prePCs, which express low CD38 levels and maintain elevated CXCR4 expression (Bhattacharya et al., 2007). Transitional populations defined in this way strongly enriched for S1pr2lo cells, allowing these populations to be evaluated in nonreporter mice (Fig. 3 D and Fig. S1, B and C). All four populations identified using CD38 and CXCR4 displayed a similar dependence on CD40-mediated signals for their survival, as treatment of mice with αCD40L antibody (MR1) from day 9–12 after LCMV Arm infection resulted in roughly equivalent decreases in the number of these cells (Fig. 3 E). This treatment only resulted in about a twofold decrease in memory B cells (CD73+CD95+CD38+IgDloGL7– B cells; Fig. S1 B), likely caused by a significant fraction of these cells exiting the GC before treatment (Fig. 3 E; Anderson et al., 2007; Dogan et al., 2009).
Efnb1+ S1pr2lo cells represent a post-GC B cell population
We next evaluated the extent of cell division occurring in these populations 24 h before analysis. To do this, we used a tet-off-H2b-GFP mouse that allows proliferation to be assessed over multiple cell divisions (Foudi et al., 2009; Gitlin et al., 2014; Bannard et al., 2016). Treatment of transgenic mice with doxycycline results in the H2b-GFP gene being turned off and dividing cells diluting their expression of GFP as histone segregation occurs during cell division. Within the IgDloGL7+CD95+ compartment, Efnb1–CD38+CXCR4lo (Pop 1) cells displayed minimal GFP dilution further indicating that these cells are precursor GC cells that have not yet entered a rapidly dividing state (Fig. 3 F). Efnb1–CD38– (Pop 2) cells displayed some dilution of GFP, indicative of a small amount of division, but did not have a sizeable population that had fully lost GFP expression (Fig. 3 F). Efnb1+CD38– (Pop 3) cells, in contrast, displayed robust loss of GFP, consistent with this population representing mature GC B cells (Fig. 3 F). Efnb1+CD38+CXCR4lo (Pop 4) cells demonstrated a similar dilution of GFP (Fig. 3 F). These data indicate that Pop 4 is derived from recently dividing GC B cells and could represent a population in the process of differentiating away from the GC B cell state.
We further tested the proximity of these populations to the GC state by examining the extent of SHM. SHM was assessed by sequencing of the IgH JH4-intronic enhancer downstream of the rearranged VJ558DJH4 element in DNA from the populations (Park et al., 2009; Bannard et al., 2013). The mutation frequency in this intronic region provides a measurement of AID activity (Jolly et al., 1997). As expected, follicular B cells were largely germline, with some sequences having a small number of mutations, likely caused by errors introduced during the nested PCR reactions (Fig. 3 G; Bannard et al., 2013). Memory B cells were split between cells with low and high levels of mutation consistent with some of these cells emerging from the GC during early time points after infection, before significant mutations have occurred (Fig. 3 G). PrePCs had predominantly low to intermediate numbers of mutations (Fig. 3 G). Efnb1– S1pr2hi (Pop 2) cells displayed low levels of mutations, suggesting that these cells have either only recently adopted a GC state or are intrinsically impaired in their ability to fully participate in the GC reaction and undergo rapid division and SHM. Both Efnb1+ S1pr2hi (Pop 3) and Efnb1+ S1pr2lo (Pop 4) cells had a high fraction of mutated cells (Fig. 3 G). The mutational frequency in Efnb1+ S1pr2lo (Pop 4) cells was slightly less than in that of the Efnb+ S1pr2hi (Pop 3) cells, perhaps indicative of AID activity being lost in this population as it transitions from the GC state. This slightly reduced mutational frequency could also be caused by GC B cells of lower affinity being more prone to differentiate into memory B cells (Shinnakasu et al., 2016).
Efnb1+ S1pr2lo cells are developmentally related to memory B cells
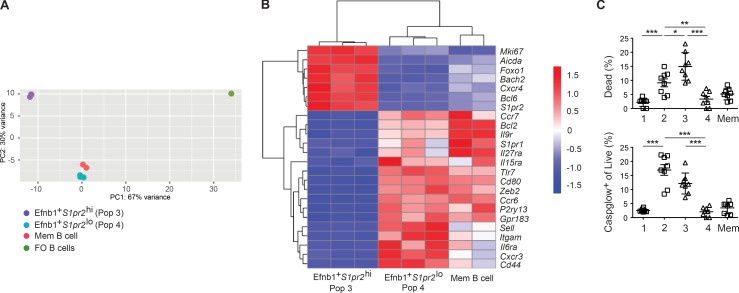
To further probe the developmental relationship between Efnb1+ S1pr2hi (Pop 3), Efnb1+ S1pr2lo (Pop 4), and memory B cells, we performed RNA-seq analysis at day 11 after LCMV Arm infection. Principal component analysis (PCA) indicated that Efnb1+ S1pr2hi (Pop 3) and Efnb1+ S1pr2lo (Pop 4) cells had vastly different transcriptional profiles, despite both expressing surface markers associated with GC B cells (Fig. 4 A). Instead, Efnb1+ S1pr2lo (Pop 4) cells clustered tightly with memory B cells and displayed striking similarities in their gene expression profile based on an unbiased analysis of genes with the greatest variance between groups (Fig. 4, A and B; and Fig. S2 A). Published RNA-seq data from splenic follicular B cells was included in the PCA as a reference (Shi et al., 2015). In total, there were 497 differentially expressed genes (DEGs) between Efnb1+ S1pr2hi (Pop 3) and Efnb1+ S1pr2lo (Pop 4) cells, and only 38 DEGs between Efnb1+ S1pr2lo (Pop 4) cells and memory B cells, with DEGs being defined as those with a Padj < 0.1, base count > 100, and log2foldchange > 1.5 (Fig. S2, B and C). Among the DEGs that were down-regulated in Efnb1+ S1pr2lo (Pop 4) cells and memory B cells were those associated with the GC B cell state, including Bcl6, Aicda, Bach2, Cxcr4, and Mki67 (Fig. 4 B). Although Bach2 has been reported to have higher expression on GC B cells predisposed to adopt a memory fate, its low expression on both Efnb1+ S1pr2lo (Pop 4) cells and memory B cells suggests that Bach2hi precursor cells are still in a GC state and that further development must occur within the GC before they can complete their differentiation into memory B cells (Shinnakasu et al., 2016). Up-regulated DEGs include many commonly associated with cell positioning (Ccr7, S1pr1, Ccr6, Gpr183, Cxcr3, and Sell), activation (Itgam, Cd44, and Tlr7), cytokine signaling (Il9r, Il27ra, Il15ra, and Il6ra), transcriptional regulation (Zeb2), and cell survival (Bcl2; Fig. 4 B). In several cases, these DEGs were validated by protein expression analysis (Fig. S2 D). Efnb1+CD38+CXCR4lo (Pop 4) cells displayed similar levels of death and apoptosis to memory B cells after in vitro culture, whereas Efnb1–CD38– (Pop 2) and Efnb1+CD38– (Pop 3) cells demonstrated poor survival (Fig. 4 C). Collectively, these data indicate that Efnb1+ S1pr2lo (Pop 4) cells are transcriptionally and functionally similar to memory B cells and likely represent a population of GC memory precursor (PreMem) B cells.
Figure 4.
Efnb1+ S1pr2lo cells are transcriptionally and functionally similar to memory B cells. (A) Principal component analysis of RNA-seq data from splenic Efnb1+ S1pr2hi (Pop 3), Efnb1+ S1pr2lo (Pop 4), and memory (IgDloGL7–CD95+CD73+) B cells at day 11 after LCMV Arm infection. Published RNA-seq data from splenic follicular (FO) B cells (small, B220+CD21+CD23+ cells) was equivalently analyzed and included as a reference (Shi et al., 2015). (B) Heat map of select DEGs among mRNA isolated from the aforementioned populations, presented as expression (log2) normalized by row. Genes with a Padj < 0.1 and log2fold change > 1.5 between the Efnb1+ S1pr2hi (Pop 3) and Efnb1+ S1pr2lo (Pop 4) groups, and that had a base mean count across all three groups >100, were considered DEGs. Data are from three independent experiments with four mice per experiment pooled for each sample. (C) Splenocytes from day 11 after LCMV Arm–infected mice were cultured for 5 h at 37°C. Percentage of dead cells (top) and caspase expression in live cells (bottom) in Efnb1− CD38+CXCR4lo (Pop 1), Efnb1– CD38– (Pop 2), Efnb1+ CD38– (Pop 3), and Efnb1+ CD38+CXCR4lo (Pop 4) IgDloGL7+CD95+ B cells and memory (IgDloGL7–CD95+CD73+) B cells. Data are pooled from two independent experiments with four mice per experiment. Statistical analyses were performed using the unpaired two-tailed Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Ingenuity Pathway Analysis (IPA; Ingenuity Systems) was also used to explore upstream regulators of the gene signature. IPA found that the Efnb1+ S1pr2hi (Pop 3) to Efnb1+ S1pr2lo (Pop 4) transition was driven primarily by a loss in Bcl6 activity and an increase in genes associated with inflammatory exposure, possibly resulting from the loss of Bcl6-mediated repression of inflammatory signaling (Fig. S3 A; Dent et al., 1997; Cui et al., 2011). The Efnb1+ S1pr2lo (Pop 4) to memory B cell transition was marked by a loss in genes associated with cell cycle progression and an increase in those associated with a more quiescent phenotype, consistent with the notion that Efnb1+ S1pr2lo cells are still in the process of completing their differentiation into long-lived memory B cells (Fig. S3 B).
Efnb1+ S1pr2lo cells localize near the edge of the GC LZ
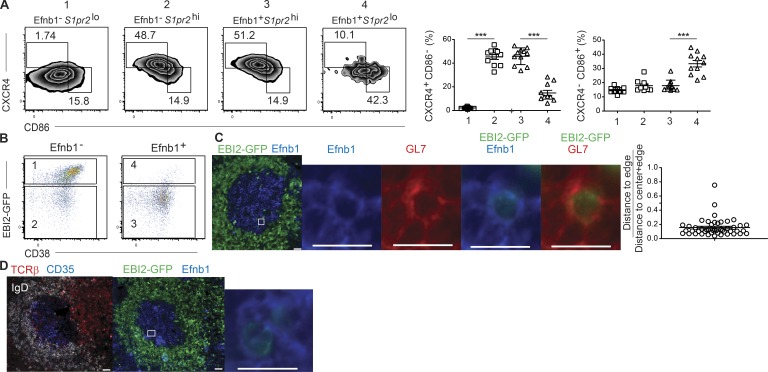
Finally, we sought to determine where PreMem B cells localize within the GC. Phenotypic analysis indicated that Efnb1+ S1pr2lo (Pop 4) cells had an increased propensity to adopt a LZ GC B cell phenotype and therefore might preferentially localize to this zone (Fig. 5 A). To positively identify these transitioning cells, we used EBI2GFP/+ mice, as the RNA-seq data indicated that Gpr183 (the gene encoding EBI2) was highly expressed within Efnb1+ S1pr2lo (Pop 4) cells. Indeed, use of EBI2-GFP along with Ephrin-B1 allowed for clear detection of four populations of GL7+CD95+IgDlo B cells with equivalent expression of CD38, CD73, Bcl6, and CXCR4 to those identified using S1pr2 and Ephrin-B1 (Fig. 5 B and not depicted). Identification of transitional cells using CD38 and CXCR4 also enriched for cells expressing EBI2-GFP (Fig. S1, B and C). Analysis of splenic sections from EBI2GFP/+ mice after LCMV Arm infection identified a small population of Efnb1+GL7+IgDlo cells within the GC that displayed intracellular expression of GFP, and these cells localized near the edge of the GC (Fig. 5 C). They were found within the LZ in 22 of 25 cases where they were identified, and GC polarization could be determined on serial sections (Fig. 5 D). These data are consistent with the notion that memory precursor cells predominantly arise in the LZ and exit the GC from this compartment.
Figure 5.
GC memory precursor cells localize near the edge of the LZ. (A) Representative plots (left) and summary graphs (right) of the percentages of splenic DZ (CXCR4+CD86−) and LZ (CXCR4–CD86+) phenotype cells in in Efnb1– S1pr2lo (Pop 1), Efnb1– S1pr2hi (Pop 2), Efnb1+ S1pr2hi (Pop 3), and Efnb1+ S1pr2lo (Pop 4) IgDloGL7+CD95+ B cells at day 11 after LCMV infection. Data are pooled from three independent experiments with three to four mice per group at day 11 after infection. Statistical analyses were performed using the unpaired two-tailed Student’s t test (***, P < 0.001). (B) Representative plots of EBI2 (as defined using EBI2GFP/+ mice) and CD38 expression in Efnb1– and Efnb1+ IgDloGL7+CD95+ B cells at day 11 after LCMV infection. Plots are representative of four independent experiments with three to four mice per experiment at days 11 or 15 after infection. (C) Representative images of Ephrin-B1, GL7, and EBI2 (as defined using EBI2GFP/+ mice). Distance of Efnb1+GL7+IgDlo EBI2+ GC B cells from the edge of the GC was determined using Imaris software to determine the GC center, and then determining distance from the center to the cell and from the cell to the GC edge. Distance was quantified in such a manner that the mean distance from the GC center to GC B cells divided by the total distance from the GC center to the edge was ∼0.375. (D) Representative images of GC polarization as determined by the distribution of CD35+ FDCs and positioning of the T cell zone, and Ephrin-B1 and EBI2 (as defined using EBI2GFP/+ mice) in serial sections. Analysis of CD35 staining in serial sections of 21 GCs where Efnb1+ EBI2+ cells were identified indicated that 22 of 25 identified cells were LZ resident. Data are representative of six independent EBI2GFP/+ mice at either day 11 or day 15 after LCMV infection. Bars: (left) 20 µm; (right) 10 µm (inset).
Concluding remarks
In summary, we find that Ephrin-B1 is a specific marker of mature GC-resident B cells and that it facilitates the identification of transitional populations of GC B cells. Despite the high expression, Ephrin-B1 was not required for B cell participation in the GC response. However, because Eph receptors are expressed on various stromal and hematopoietic cells, including T cells (Alfaro et al., 2007; Maddigan et al., 2011), it remains possible that Ephrin-B1 will influence GC B cell selection or differentiation events under some conditions. We demonstrate that Efnb1+ S1pr2lo cells are somatically mutated, derived from recently dividing GC B cells, display enhanced survival, and are transcriptionally similar to memory B cells and thus likely represent a population of GC memory precursor B cells. However, we do not exclude the possibility that the PreMem B cell population is heterogeneous in its origins and properties. Future studies with lineage reporter mice and single cell RNA sequencing will help further define the pathway(s) leading to memory cell commitment in the GC. Based on the striking overlap in gene signature between the PreMem B cells and memory B cells, we speculate that memory B cell differentiation occurs to a large extent within the GC. As these cells lose GC confinement signals and become responsive to migration cues found outside the GC, they likely will exit the GC and migrate to niches, where they can complete their development into long-lived memory B cells. Several cell surface receptors, including Il9r, Il6ra, Il15ra, and Il27ra, were up-regulated on PreMem B cells and could be involved in promoting memory cell differentiation. Our work suggests that the GL7, PNA, CD95, and IgD marker combination is insufficient to precisely identify GC B cells. Incorporation of Ephrin-B1 in the marker combination highly enriches for GC B cells that display elevated expression of S1pr2, Bcl6, and Aicda, and which are undergoing rapid cell division. Further addition of markers such as CD38 and CXCR4 allow for segregation of GC precursor and PreMem B cells, the latter being identified as Efnb1+CD38hiCXCR4lo cells. The use of Ephrin-B1 together with these established markers should prove beneficial in studies aimed at gaining a more precise understanding of memory B cell differentiation.
Materials and methods
Mice
Adult C57BL/6 CD45.1+ (stock number 564) mice at least 6 wk of age were purchased from the National Cancer Institute (NCI) or Charles River. Mb1Cre mice were provided by M. Reth (Max Planck Institute of Immunobiology and Epigenetics, Freiburg, Germany; Hobeika et al., 2006). Efnb1f/f mice were provided by A. Soriano (Mount Sinai School of Medicine, New York, NY; Davy et al., 2004). S1pr2Venus/+ were generated as previously described (Moriyama et al., 2014). Tet-off H2b-GFP mice were generated by crossing TetOp-H2b-GFP, ROSA:LNL:tTA, and Mb1Cre mice (Hobeika et al., 2006; Wang et al., 2008; Foudi et al., 2009). EBI2GFP/+ (containing a GFP reporter in place of the Gpr183 coding exon), HEL-specific Hy10, OVA-specific OTII TCR-transgenic, and MD4-Ig transgenic mice were from an internal colony and have been previously described (Pereira et al., 2009; Yi et al., 2012; Yi and Cyster, 2013). AID-GFP mice were purchased from Jackson ImmunoResearch Laboratories. Mice were housed in a specific pathogen–free environment in the Laboratory Animal Research Center at the University of California (San Francisco [UCSF], CA), and all animal procedures were approved by the UCSF Institutional Animal Care and Use Committee.
Adoptive transfer, immunization, infections, and treatments
For experiments involving transfer of Hy10 or OT-II cells, 105 cells of each population were adoptively transferred into mice 1 d before immunization. To visualize proliferation of antigen-specific B cells, 30–50 × 106 MD4 cells were CFSE-labeled according to the manufacturer’s instructions (Invitrogen) and transferred 1 d before immunization. Mice were immunized with 2 × 108 SRBC (Colorado serum) by i.p. injection or 50 µg of duck egg lysozyme conjugated to OVA (DEL-OVA) in Sigma or Ribi Adjuvant System (Sigma-Aldrich). Mice were infected with 2 × 105 plaque-forming units of LCMV Armstrong administered i.p. For anti-CD40L treatment, mice were treated with neutralizing antibody by i.p. injection of 250 µg anti–mouse CD40L (clone MR1) daily for 4 d before sacrifice. For tet-off experiments, mice were treated with 1.6 mg doxycycline (Sigma-Aldrich) administered i.p. in saline, and then maintained by including doxycycline (2 mg/ml) and sucrose (2%) in their drinking water until analysis.
Antibodies for flow cytometry and microscopy staining
Spleens were mashed through a 70-µm cell strainer, and RBCs were lysed with RBC lysing buffer. Lymphocytes were then washed and counted. The following antibodies were used for flow cytometry and microscopy staining: Phycoerythrin (PE) anti-CD86 (105008), phycoerythrin–indotricarbocyanine (PE-Cy7) anti-CD38 (102718), Brilliant Violet 605 (BV605) anti-CD45.1 (110738), allophycocyanin (APC) anti-GL7 (144606), Pacific Blue (PacBlue) anti-GL7 (144614), peridinin chlorophyll protein Cy5.5 (PerCpCy5.5) anti-CD73 (127214), PacBlue anti-IgD (405712), PerCpCy5.5 anti-IgD (405710), APC anti-CD80 (104718), PE anti-CD11b (101208), PE anti-CD44 (103008), PE anti-IgD (405705), PE anti-CD62L (104418), APC Cy7 anti-B220 (103224), FITC anti-GL7 (144604), PE anti-CD138 (142504; all from BioLegend); APC anti-CXCR4 (558644), PE anti-CD95 (554258), PE Cy7 anti-CD95 (557653), APC anti-Bcl6 (561525), APC anti-CCR6 (557976), Biotin anti-CD35 (553816), Biotin anti-CXCR4 (551968), APC anti-TCRβ (17–5961-82), BV605 Streptavidin (563260), APC Cy7 anti-CD19 (115530), PE Texas Red Streptavidin (551487; all from BD); PerCPCy5.5 anti-CD45.2 (65–0454-U100; Tonbo Biosciences); Biotin donkey anti–mouse polyclonal Ephrin-B1 (BAF473; R&D Systems); Alexa Fluor 488 Rabbit polyclonal anti-GFP (A-21311; Invitrogen/Life Technologies); Goat anti–mouse IgD (goat polyclonal GAM/IGD(FC)/7S), APC anti-CD23 (CL8910APC; Cedarlane Labs); aminomethylcoumarin-donkey anti–goat (705–156-147), Cy3 Streptavidin (016–160-084; Jackson ImmunoResearch Laboratories). Flow cytometry data were acquired on a LSRII with FACSDiva software (BD) and were analyzed with FlowJo software (Tree Star).
Immunohistochemistry (IHC) and immunofluorescence (IF) microscopy
For IHC, 7-µm cryosections were acetone fixed and stained as previously described (Allen et al., 2007) For IF microscopy, 7-µm cryosections were prepared as previously described (Reboldi et al., 2016). For staining of mouse Ephrin-B1 for IF images, a tyramide kit was used according to the manufacturer’s instructions (TSA Biotin System; Perkin Elmer). Images were captured with a Zeiss AxioObserver Z1 inverted microscope. Identification of the center of the GC and distance from the center to specific cells was performed using Imaris (Bitplane). If the GC is assumed to be a circle with GC B cells evenly distributed throughout, the mean distance from the edge of the GC to an individual cell divided by the total distance from the cell to the GC center, and the cell to the GC edge is predicted to be ∼0.375. This is a result of more cells being located in the outer part of the GC, which has greater area than the inner GC.
Analysis of somatic mutations in JH558 intron
20,000–40,000 cells for each population were FACS sorted, and DNA was isolated using a QIAamp DNA Micro kit (QIAGEN). DNA was eluted in 20 µl and used as a template for nested PCR (Park et al., 2009; Bannard et al., 2013). New primers and reagents were added directly to the first PCR product for the secondary reaction. Primers are specific for the JH558 family members, and only PCR products of ∼700 bp were excised and purified from 1.2% agarose gels using a QIAquick Gel Extraction kit (QIAGEN). The purified gel product was then cloned into the pCR4 Bunt-TOPO vector according to the manufacturer’s instructions (Invitrogen). Colonies were then submitted for preparation and sequencing (TACgen), and sequences aligned to the germline JH558 intronic sequence using standard nucleotide BLAST. Primers are as follows: Nested Forward 1, 5′-AGCCTGACATCTGAGGAC-3′; nested reverse 1, 5′-TCTGATCGGCCATCTTGACTC-3′; nested forward 2, 5′-CATCTGAGGACTCTGCGGTCT-3′; nested reverse 2, 5′-CTGTGTTCCTTTGAAAGCTGG-3′.
RNA-seq library preparation and data analysis
Total RNA was purified from FACS sorted cells using the RNeasy Mini kit (QIAGEN). RNA quality was assessed with an Agilent 2100 Bioanalyzer (RNA integrity number > 9 for all samples). Barcoded sequencing libraries were generated with 100 ng of RNA with an Ovation RNA-seq System V2 (Nugen), KAPA Hyper Prep kit for Illumina (KAPA Biosystems), and NEXTflex DNA barcodes (Bioo Scientific). Single-end sequencing was performed on an HiSeq 2500 (Illumina; UCSF Center for Advanced Technology), and sequences reported as FASTQ files, which were aligned to the mm10 genome with STAR (Spliced Transcript Alignment to a Reference). Mappable reads were counted with HTseq and imported into RStudio software for analysis of differential expression with DESeq2 software. For the generation of the heat map, select genes with a difference in expression (log2) of 1.5-fold, p-adjusted value < 0.1, and mean count across all groups >100 were chosen for visualization with values scaled by row.
Pathway analysis
The pathway analysis was performed using the Ingenuity Pathway Analysis software (QIAGEN). The Upstream analysis was performed to identify upstream regulators across the Efnb1+S1pr2hi (Pop3) and Efnb1+S1pr2lo (Pop 4) groups as well as the Efnb1+S1pr2lo (Pop 4) and memory B cell groups. Upstream regulator predictions were made by the z-score algorithm. All upstream regulators shown have P < 10−9.
Accession nos.
The RNA-seq data reported in this paper are available at the Gene Expression Omnibus under accession no.GSE89897.
Statistical analysis
Results represent the mean ± SEM unless indicated otherwise. Statistical significance was determined by the unpaired Student’s t test. Statistical analyses were performed using Prism GraphPad software v5.0. (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Online supplemental material
Fig. S1 shows an example flow cytometric gating scheme to identify GC B cell subsets and memory B cells. Fig. S2 shows a heat map and volcano plots derived from the RNA-seq analysis, along with protein validation of select DEGs. Fig. S3 shows an IPA upstream regulator analysis of the RNA-seq data.
Acknowledgments
We thank Phil Soriano for Efnb1f/f mice, M. Matloubian for help in providing LCMV Arm virus, and Y. Xu and J. An for expert technical assistance.
This work was supported by grants from the National Institutes of Health (R01AI045073 [J.G. Cyster], R01AI040098 [J.G. Cyster], and T32AI07019 [B.J. Laidlaw]). B.J. Laidlaw is a recipient of a postdoctoral fellowship from the Damon Runyon Cancer Institute. J.G. Cyster is an investigator of the Howard Hughes Medical Institute.
The authors declare no competing financial interests.
Author contributions: B.J. Laidlaw and J.G. Cyster conceived and designed the experiment. B.J. Laidlaw, T.H. Schmidt, J.A. Green and C.D.C. Allen performed the experiments. B.J. Laidlaw and J.G. Cyster analyzed the data. T. Okada provided tools for the experiments. B.J. Laidlaw and J.G. Cyster wrote the manuscript.
Footnotes
Abbreviations used:
- AID
- activation-induced cytidine deaminase
- cKO
- conditional KO
- DEG
- differentially expressed gene
- DZ
- dark zone
- FDC
- follicular DC
- GC
- germinal center
- HEL
- hen egg lysozyme
- LCMV
- lymphocytic choriomeningitis virus
- LZ
- light zone
- PNA
- peanut agglutinin
- PreMem
- memory precursor
- SHM
- somatic hypermutation
- SRBC
- sheep RBC
References
- Adachi Y., Onodera T., Yamada Y., Daio R., Tsuiji M., Inoue T., Kobayashi K., Kurosaki T., Ato M., and Takahashi Y.. 2015. Distinct germinal center selection at local sites shapes memory B cell response to viral escape. J. Exp. Med. 212:1709–1723. 10.1084/jem.20142284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro D., García-Ceca J.J., Cejalvo T., Jiménez E., Jenkinson E.J., Anderson G., Muñoz J.J., and Zapata A.. 2007. EphrinB1-EphB signaling regulates thymocyte-epithelium interactions involved in functional T cell development. Eur. J. Immunol. 37:2596–2605. 10.1002/eji.200737097 [DOI] [PubMed] [Google Scholar]
- Allen C.D.C., Okada T., Tang H.L., and Cyster J.G.. 2007. Imaging of germinal center selection events during affinity maturation. Science. 315:528–531. 10.1126/science.1136736 [DOI] [PubMed] [Google Scholar]
- Anderson S.M., Tomayko M.M., Ahuja A., Haberman A.M., and Shlomchik M.J.. 2007. New markers for murine memory B cells that define mutated and unmutated subsets. J. Exp. Med. 204:2103–2114. 10.1084/jem.20062571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannard O., Horton R.M., Allen C.D.C., An J., Nagasawa T., and Cyster J.G.. 2013. Germinal center centroblasts transition to a centrocyte phenotype according to a timed program and depend on the dark zone for effective selection. Immunity. 39:912–924. 10.1016/j.immuni.2013.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannard O., McGowan S.J., Ersching J., Ishido S., Victora G.D., Shin J.-S., and Cyster J.G.. 2016. Ubiquitin-mediated fluctuations in MHC class II facilitate efficient germinal center B cell responses. J. Exp. Med. 213:993–1009. 10.1084/jem.20151682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D., Cheah M.T., Franco C.B., Hosen N., Pin C.L., Sha W.C., and Weissman I.L.. 2007. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J. Immunol. 179:6808–6819. 10.4049/jimmunol.179.10.6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush J.O., and Soriano P.. 2009. Ephrin-B1 regulates axon guidance by reverse signaling through a PDZ-dependent mechanism. Genes Dev. 23:1586–1599. 10.1101/gad.1807209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejalvo T., Munoz J.J., Tobajas E., Fanlo L., Alfaro D., García-Ceca J., and Zapata A.. 2013. Ephrin-B-dependent thymic epithelial cell-thymocyte interactions are necessary for correct T cell differentiation and thymus histology organization: relevance for thymic cortex development. J. Immunol. 190:2670–2681. 10.4049/jimmunol.1201931 [DOI] [PubMed] [Google Scholar]
- Conter L.J., Song E., Shlomchik M.J., and Tomayko M.M.. 2014. CD73 expression is dynamically regulated in the germinal center and bone marrow plasma cells are diminished in its absence. PLoS One. 9:e92009 10.1371/journal.pone.0092009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch E.E., Li Z., Takizawa M., Fichtner-Feigl S., Gourzi P., Montaño C., Feigenbaum L., Wilson P., Janz S., Papavasiliou F.N., and Casellas R.. 2007. Regulation of AID expression in the immune response. J. Exp. Med. 204:1145–1156. 10.1084/jem.20061952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W., Liu Y., Weinstein J.S., Craft J., and Kaech S.M.. 2011. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 35:792–805. 10.1016/j.immuni.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy A., Aubin J., and Soriano P.. 2004. Ephrin-B1 forward and reverse signaling are required during mouse development. Genes Dev. 18:572–583. 10.1101/gad.1171704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent A.L., Shaffer A.L., Yu X., Allman D., and Staudt L.M.. 1997. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 276:589–592. 10.1126/science.276.5312.589 [DOI] [PubMed] [Google Scholar]
- Dogan I., Bertocci B., Vilmont V., Delbos F., Mégret J., Storck S., Reynaud C.-A., and Weill J.-C.. 2009. Multiple layers of B cell memory with different effector functions. Nat. Immunol. 10:1292–1299. 10.1038/ni.1814 [DOI] [PubMed] [Google Scholar]
- Fooksman D.R., Schwickert T.A., Victora G.D., Dustin M.L., Nussenzweig M.C., and Skokos D.. 2010. Development and migration of plasma cells in the mouse lymph node. Immunity. 33:118–127. 10.1016/j.immuni.2010.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foudi A., Hochedlinger K., Van Buren D., Schindler J.W., Jaenisch R., Carey V., and Hock H.. 2009. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 27:84–90. 10.1038/nbt.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin A.D., Shulman Z., and Nussenzweig M.C.. 2014. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 509:637–640. 10.1038/nature13300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J.A., Suzuki K., Cho B., Willison L.D., Palmer D., Allen C.D.C., Schmidt T.H., Xu Y., Proia R.L., Coughlin S.R., and Cyster J.G.. 2011. The sphingosine 1-phosphate receptor S1P2 maintains the homeostasis of germinal center B cells and promotes niche confinement. Nat. Immunol. 12:672–680. 10.1038/ni.2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobeika E., Thiemann S., Storch B., Jumaa H., Nielsen P.J., Pelanda R., and Reth M.. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. USA. 103:13789–13794. 10.1073/pnas.0605944103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., and Melnick A.. 2015. Mechanisms of action of BCL6 during germinal center B cell development. Sci. China Life Sci. 58:1226–1232. 10.1007/s11427-015-4919-z [DOI] [PubMed] [Google Scholar]
- Jolly C.J., Klix N., and Neuberger M.S.. 1997. Rapid methods for the analysis of immunoglobulin gene hypermutation: application to transgenic and gene targeted mice. Nucleic Acids Res. 25:1913–1919. 10.1093/nar/25.10.1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T., Kometani K., and Ise W.. 2015. Memory B cells. Nat. Rev. Immunol. 15:149–159. 10.1038/nri3802 [DOI] [PubMed] [Google Scholar]
- László G., Hathcock K.S., Dickler H.B., and Hodes R.J.. 1993. Characterization of a novel cell-surface molecule expressed on subpopulations of activated T and B cells. J. Immunol. 150:5252–5262. [PubMed] [Google Scholar]
- Luo H., Charpentier T., Wang X., Qi S., Han B., Wu T., Terra R., Lamarre A., and Wu J.. 2011. Efnb1 and Efnb2 proteins regulate thymocyte development, peripheral T cell differentiation, and antiviral immune responses and are essential for interleukin-6 (IL-6) signaling. J. Biol. Chem. 286:41135–41152. 10.1074/jbc.M111.302596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddigan A., Truitt L., Arsenault R., Freywald T., Allonby O., Dean J., Narendran A., Xiang J., Weng A., Napper S., and Freywald A.. 2011. EphB receptors trigger Akt activation and suppress Fas receptor-induced apoptosis in malignant T lymphocytes. J. Immunol. 187:5983–5994. 10.4049/jimmunol.1003482 [DOI] [PubMed] [Google Scholar]
- McHeyzer-Williams M., Okitsu S., Wang N., and McHeyzer-Williams L.. 2011. Molecular programming of B cell memory. Nat. Rev. Immunol. 12:24–34. 10.1038/nri3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama S., Takahashi N., Green J.A., Hori S., Kubo M., Cyster J.G., and Okada T.. 2014. Sphingosine-1-phosphate receptor 2 is critical for follicular helper T cell retention in germinal centers. J. Exp. Med. 211:1297–1305. 10.1084/jem.20131666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muppidi J.R., Schmitz R., Green J.A., Xiao W., Larsen A.B., Braun S.E., An J., Xu Y., Rosenwald A., Ott G., et al. 2014. Loss of signalling via Gα13 in germinal centre B-cell-derived lymphoma. Nature. 516:254–258. 10.1038/nature13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y., Takematsu H., Koyama S., Miyake S., Yamamoto H., Fujinawa R., Sugai M., Okuno Y., Tsujimoto G., Yamaji T., et al. 2007. Germinal center marker GL7 probes activation-dependent repression of N-glycolylneuraminic acid, a sialic acid species involved in the negative modulation of B-cell activation. Mol. Cell. Biol. 27:3008–3022. 10.1128/MCB.02047-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-R., Zan H., Pal Z., Zhang J., Al-Qahtani A., Pone E.J., Xu Z., Mai T., and Casali P.. 2009. HoxC4 binds to the promoter of the cytidine deaminase AID gene to induce AID expression, class-switch DNA recombination and somatic hypermutation. Nat. Immunol. 10:540–550. 10.1038/ni.1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J.P., Kelly L.M., Xu Y., and Cyster J.G.. 2009. EBI2 mediates B cell segregation between the outer and centre follicle. Nature. 460:1122–1126. 10.1038/nature08226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboldi A., Arnon T.I., Rodda L.B., Atakilit A., Sheppard D., and Cyster J.G.. 2016. IgA production requires B cell interaction with subepithelial dendritic cells in Peyer’s patches. Science. 352:aaf4822 10.1126/science.aaf4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Liao Y., Willis S.N., Taubenheim N., Inouye M., Tarlinton D.M., Smyth G.K., Hodgkin P.D., Nutt S.L., and Corcoran L.M.. 2015. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat. Immunol. 16:663–673. 10.1038/ni.3154 [DOI] [PubMed] [Google Scholar]
- Shinall S.M., Gonzalez-Fernandez M., Noelle R.J., and Waldschmidt T.J.. 2000. Identification of murine germinal center B cell subsets defined by the expression of surface isotypes and differentiation antigens. J. Immunol. 164:5729–5738. 10.4049/jimmunol.164.11.5729 [DOI] [PubMed] [Google Scholar]
- Shinnakasu R., Inoue T., Kometani K., Moriyama S., Adachi Y., Nakayama M., Takahashi Y., Fukuyama H., Okada T., and Kurosaki T.. 2016. Regulated selection of germinal-center cells into the memory B cell compartment. Nat. Immunol. 17:861–869. 10.1038/ni.3460 [DOI] [PubMed] [Google Scholar]
- Tarlinton D., and Good-Jacobson K.. 2013. Diversity among memory B cells: origin, consequences, and utility. Science. 341:1205–1211. 10.1126/science.1241146 [DOI] [PubMed] [Google Scholar]
- Turner C.A. Jr., Mack D.H., and Davis M.M.. 1994. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 77:297–306. 10.1016/0092-8674(94)90321-2 [DOI] [PubMed] [Google Scholar]
- Victora G.D., and Nussenzweig M.C.. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457. 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Wang J., Taniuchi I., Maekawa Y., Howard M., Cooper M.D., and Watanabe T.. 1996. Expression and function of Fas antigen on activated murine B cells. Eur. J. Immunol. 26:92–96. 10.1002/eji.1830260114 [DOI] [PubMed] [Google Scholar]
- Wang L., Sharma K., Deng H.-X., Siddique T., Grisotti G., Liu E., and Roos R.P.. 2008. Restricted expression of mutant SOD1 in spinal motor neurons and interneurons induces motor neuron pathology. Neurobiol. Dis. 29:400–408. 10.1016/j.nbd.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Weisel F.J., Zuccarino-Catania G.V., Chikina M., and Shlomchik M.J.. 2016. A Temporal Switch in the Germinal Center Determines Differential Output of Memory B and Plasma Cells. Immunity. 44:116–130. 10.1016/j.immuni.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W., Kim J., Wergedal J., Chen S.-T., and Mohan S.. 2010. Ephrin B1 regulates bone marrow stromal cell differentiation and bone formation by influencing TAZ transactivation via complex formation with NHERF1. Mol. Cell. Biol. 30:711–721. 10.1128/MCB.00610-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi T., and Cyster J.G.. 2013. EBI2-mediated bridging channel positioning supports splenic dendritic cell homeostasis and particulate antigen capture. eLife. 2:e00757 10.7554/eLife.00757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi T., Wang X., Kelly L.M., An J., Xu Y., Sailer A.W., Gustafsson J.-A., Russell D.W., and Cyster J.G.. 2012. Oxysterol gradient generation by lymphoid stromal cells guides activated B cell movement during humoral responses. Immunity. 37:535–548. 10.1016/j.immuni.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccarino-Catania G.V., Sadanand S., Weisel F.J., Tomayko M.M., Meng H., Kleinstein S.H., Good-Jacobson K.L., and Shlomchik M.J.. 2014. CD80 and PD-L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nat. Immunol. 15:631–637. 10.1038/ni.2914 [DOI] [PMC free article] [PubMed] [Google Scholar]