Rommel et al. reveal a novel RAG1/2-mediated insertion pathway, which has the potential to destabilize the lymphocyte genome and shares features with DNA insertions observed in human cancer.
Abstract
The RAG recombinase (RAG1/2) plays an essential role in adaptive immunity by mediating V(D)J recombination in developing lymphocytes. In contrast, aberrant RAG1/2 activity promotes lymphocyte malignancies by causing chromosomal translocations and DNA deletions at cancer genes. RAG1/2 can also induce genomic DNA insertions by transposition and trans-V(D)J recombination, but only few such putative events have been documented in vivo. We used next-generation sequencing techniques to examine chromosomal rearrangements in primary murine B cells and discovered that RAG1/2 causes aberrant insertions by releasing cleaved antibody gene fragments that subsequently reintegrate into DNA breaks induced on a heterologous chromosome. We confirmed that RAG1/2 also mobilizes genomic DNA into independent physiological breaks by identifying similar insertions in human lymphoma and leukemia. Our findings reveal a novel RAG1/2-mediated insertion pathway distinct from DNA transposition and trans-V(D)J recombination that destabilizes the genome and shares features with reported oncogenic DNA insertions.
Introduction
Antigen receptor diversity enables lymphocytes to initiate effective immune responses against a virtually limitless array of pathogens. The diversity in the primary antigen receptor repertoire is achieved by V(D)J recombination, a site-specific reaction catalyzed by a heterotetrameric protein complex encoded by the recombination-activating genes RAG1 and RAG2 (Kim et al., 2015; Ru et al., 2015). The RAG recombinase (RAG1/2) joins randomly selected variable, diversity, and joining (V, D, and J) gene segments to assemble a V(D)J exon that encodes the variable region of antibodies and T cell receptors (Schatz and Ji, 2011; Schatz and Swanson, 2011). RAG1/2 does so in part by recognizing and cleaving conserved recombination signal sequences (RSSs) that flank each V, D, and J gene segment. RAG1 is the principal DNA binding and cleavage component of the recombinase. RAG2 is an essential cofactor and consists of a core portion (RAG2core) minimally required for its activity and a C-terminal region important for efficiency, fidelity, and ordering of V(D)J rearrangements (Sekiguchi et al., 2001; Liang et al., 2002; Akamatsu et al., 2003; Talukder et al., 2004; Curry and Schlissel, 2008).
RSSs are comprised of a conserved palindromic heptamer (consensus: 5′-CACAGTG-3′) that is required for DNA cleavage, a degenerate spacer of 12 or 23 bp, and a less-conserved A-rich nonamer (consensus: 5′-ACAAAAACC-3′) that is important for RAG1/2 binding (Schatz and Ji, 2011; Schatz and Swanson, 2011). RSSs with 12- or 23-bp spacers are termed 12RSSs and 23RSSs, respectively. During V(D)J recombination, RAG1/2 first binds to a single 12- or 23RSS (signal complex) and then captures a “complementary” 23- or 12RSS (paired complex) according to the “12/23 rule.” Upon synapsis, the recombinase introduces DNA double-strand breaks between coding sequences and flanking RSSs by making a single-strand nick that is used to catalyze a transesterification that produces a hairpin-sealed coding end and a blunt-cut signal end. After cleavage, RAG1/2 remains associated with paired coding and signal ends in a post-cleavage complex, thereby scaffolding their repair by nonhomologous end joining (NHEJ). Coding ends are fused to produce V(D)J-coding exons, and ligation of signal ends generates noncoding signal joints. Depending on the orientation of paired RSSs, RAG1/2 catalyzes either inversional (head-to-tail RSSs) or deletional (convergent RSSs) recombination. During inversional recombination, signal joints remain in the genome, whereas they are excised as episomal signal joints during deletional recombination (Helmink and Sleckman, 2012).
In addition to its essential role in adaptive immunity, RAG1/2 has been implicated in the genesis of chromosome translocations and deletions associated with lymphoid malignancy (Roth, 2003; Lieber, 2016). Mice deficient for ataxia-telangiectasia mutated kinase (ATM) or both the tumor suppressor protein p53 and components of the NHEJ machinery develop RAG1/2-dependent chromosome translocations associated with pro–B cell lymphomas (Nussenzweig and Nussenzweig, 2010; Alt et al., 2013). In humans, RAG1/2 is implicated in the genesis of follicular lymphoma (FL), mantle cell lymphoma, and acute lymphoblastic leukemia (ALL), all of which carry genome aberrations in the proximity of RSSs in antigen receptor genes or nonphysiological cryptic RSSs (cRSSs) with conserved heptamer motifs (Küppers and Dalla-Favera, 2001; Nussenzweig and Nussenzweig, 2010; Alt et al., 2013). Predicted cRSSs are broadly distributed throughout the genome, and so are RAG1/2 binding sites, as assayed by chromatin immunoprecipitation (Lewis et al., 1997; Ji et al., 2010; Merelli et al., 2010; Teng et al., 2015). Consistent with the idea that RAG1/2 can induce DNA damage at cRSSs, it causes chromosomal deletions, and in the context of ATM deficiency also translocations, between engineered RSSs and genomic cRSSs in primary pro–B cells and pro–B cell lines (Hu et al., 2015). The reported off-target mechanism involves directional, linear tracking of RAG1/2 within chromosomal loop domains to locate RSS/cRSS pairs (Hu et al., 2015).
Biochemical experiments as well as episomal assays in cell lines and yeast suggest that RAG1/2 can mediate DNA transposition by inserting RSS-containing donor sequences into target DNA (Agrawal et al., 1998; Hiom et al., 1998; Lee et al., 2002; Neiditch et al., 2002; Clatworthy et al., 2003; Elkin et al., 2003; Tsai et al., 2003; Chatterji et al., 2006; Posey et al., 2006; Reddy et al., 2006). Experiments with reporter cell lines indicate that RAG1/2 can also catalyze trans-V(D)J recombination, during which episomal signal joints are reinserted at an endogenous RSS or cRSS (Reddy et al., 2006). Nevertheless, only a few RAG1/2-mediated genomic insertions have been documented in vivo (Messier et al., 2003; Curry et al., 2007; Vanura et al., 2007). Moreover, RAG1/2-mediated DNA insertions contributing to cancer display characteristics that are not compatible with either DNA transposition or trans-V(D)J recombination (Navarro et al., 2015). Hence, how RAG1/2 causes genomic DNA insertions is still largely unknown.
Here we use translocation capture sequencing (TC-Seq) and insertion capture sequencing (IC-Seq) to analyze chromosomal rearrangements in primary murine developing B cells. We identify aberrant RAG1/2-dependent DNA deletions at immunoglobulin (Ig) genes, whose products are reinserted at DNA breaks generated by the I-SceI endonuclease on a heterologous chromosome. The existence of similar insertions in human cancer indicates that RAG1/2 also mobilizes genomic DNA into independent physiological breaks. Thus, our findings reveal a novel pathway through which RAG1/2 causes DNA insertions independent of DNA transposition and trans-V(D)J recombination. Importantly, this pathway has the potential to destabilize the lymphocyte genome by causing aberrant signal-end, hybrid-end, and coding-end insertions at RAG1/2-independent DNA breaks and shares features with reported oncogenic DNA insertions.
Results
Chromosomal rearrangements in pro–B cells
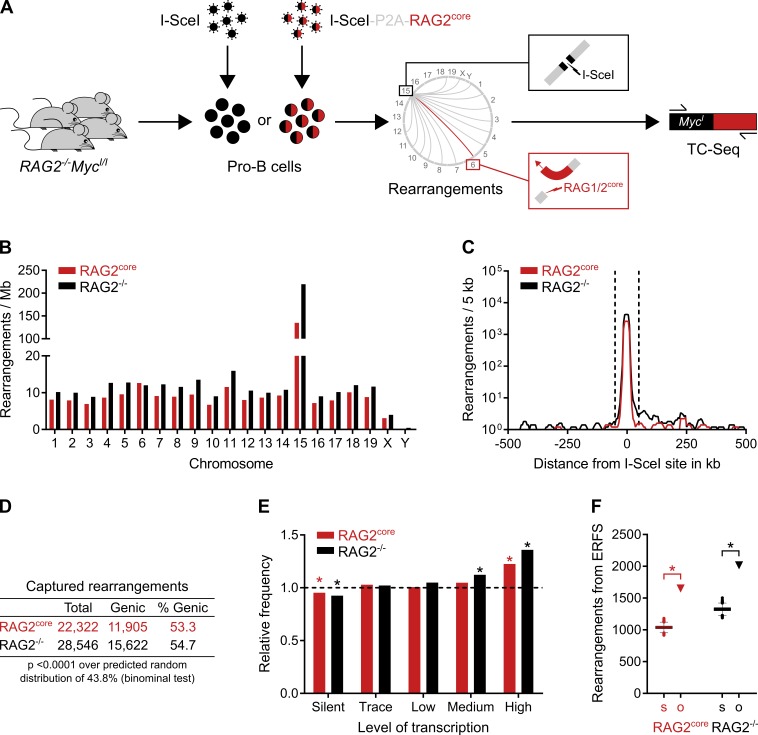
To examine RAG1/2-induced chromosomal rearrangements in pro–B cells, we adapted a previously described next-generation TC-Seq method (Klein et al., 2011; Oliveira et al., 2012). TC-Seq captures genome-wide chromosomal rearrangements to a unique DNA double-strand break created by the I-SceI endonuclease. DNA rearrangements between the I-SceI break and the genome are amplified by PCR, deep-sequenced, and analyzed computationally. We prepared TC-Seq libraries from cell cultures of primary murine pro–B cells deficient for RAG2 and harboring I-SceI sites at c-myc (RAG2−/−MycI/I) that were infected with retroviruses expressing either I-SceI alone (RAG2−/− TC-Seq libraries) or I-SceI together with murine RAG2core (RAG2core TC-Seq libraries; Fig. 1 A and Fig. S1 A; also see Materials and methods). RAG2core was used because it promotes aberrant V(D)J recombination and causes genomic instability at T cell receptor (TCR) loci in thymocytes (Sekiguchi et al., 2001; Talukder et al., 2004; Curry and Schlissel, 2008; Deriano et al., 2011). Moreover, mice expressing RAG2core and deficient for either p53 alone or in combination with XRCC4-like factor develop thymic or pro–B cell lymphomas, respectively, with translocations involving antigen receptor genes (Deriano et al., 2011; Mijušković et al., 2015; Lescale et al., 2016).
Figure 1.
Landscape of chromosomal rearrangements in primary pro–B cells by TC-Seq. (A) Detection of RAG1/2core-induced chromosomal rearrangements by TC-Seq. Primary RAG2−/−MycI/I pro–B cells were infected ex vivo with retroviruses that express either I-SceI alone (RAG2−/− TC-Seq libraries) or I-SceI together with murine RAG2core (RAG2core TC-Seq libraries) by using a “self-cleaving” P2A peptide. DNA breaks, such as those induced by RAG1/2core at Igκ on chromosome 6 (red lightning), rearrange to the I-SceI break at c-myc on chromosome 15 (black lightning) and are subsequently amplified by PCR, deep-sequenced, and analyzed computationally. RAG2core and RAG2−/− TC-Seq libraries were prepared in independent duplicates from infected pro–B cells of 180 mice. (B) Chromosomal distribution of rearrangements. Events were normalized per megabase to account for different chromosome sizes. (C) Profile of rearrangements around the I-SceI site in 5-kb intervals. Dashed lines indicate the ±50-kb region excluded from the analysis for D–F because of saturation. (D) Proportion of genic rearrangements. (E) Frequency of rearrangements derived from differentially transcribed genes compared with a random model (dashed line). Asterisks indicate values significantly different from random (P < 0.01, binominal test). (F) Observed number of rearrangements (o, triangle) originating from ERFSs compared with the random Monte Carlo simulation (s, Tukey boxplot). Asterisks indicate significant enrichment (P < 0.0001, binominal test). For D–F, events from the saturated I-SceI region, cryptic I-SceI sites, and other portions of the genome were excluded (see Materials and methods). Data analysis was performed with pooled RAG2core and RAG2−/− TC-Seq libraries (two independent experiments each). See also Fig. S1 (A and B).
In agreement with previous TC-Seq studies in other cell types, chromosomal rearrangements in pro–B cells were especially abundant near the I-SceI cleavage site on chromosome 15 (Fig. 1, B and C; Klein et al., 2011; Wang et al., 2014; Robbiani et al., 2015). Moreover, rearrangements were enriched at genic regions, highly transcribed genes, and early replication fragile sites (ERFSs), which define regions particularly susceptible to DNA damage during early replication (Fig. 1, D–F; Barlow et al., 2013).
DNA damage at physiological RSSs and cRSSs
To identify the DNA damage caused by RAG1/2core, we compared chromosomal rearrangements in RAG2core and RAG2−/− TC-Seq libraries. In brief, genomic hotspots of rearrangement were identified, and those unique to RAG1/2core were analyzed for the occurrence of breakpoint clusters (see Materials and methods). Overall, 33 RAG1/2core-dependent rearrangement breakpoint clusters were detected throughout the genome (Table S1).
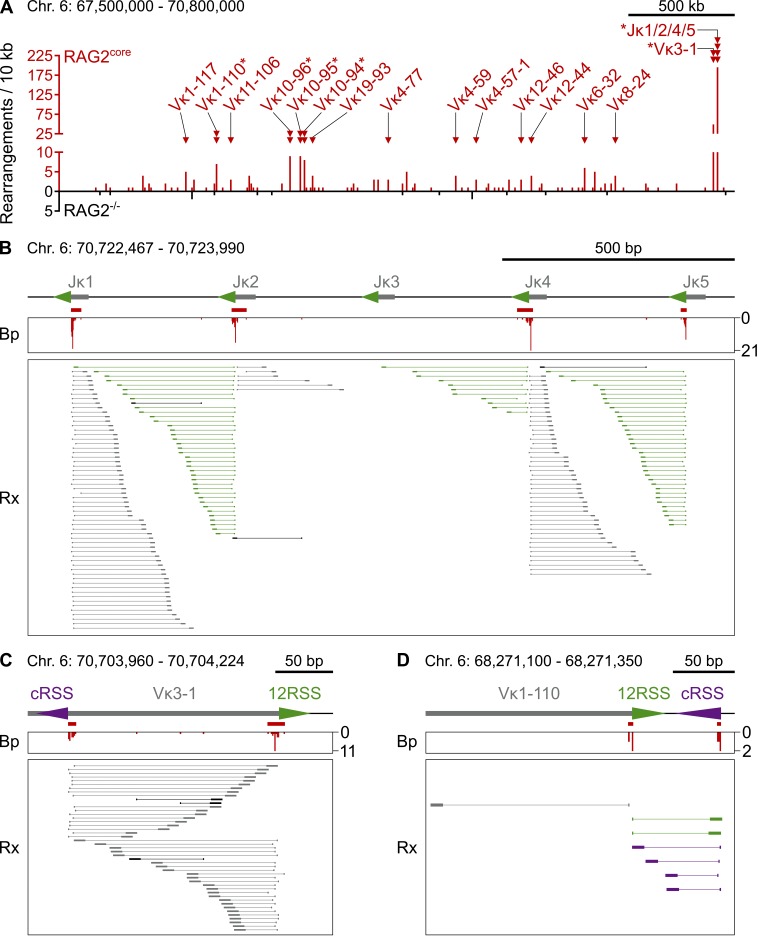
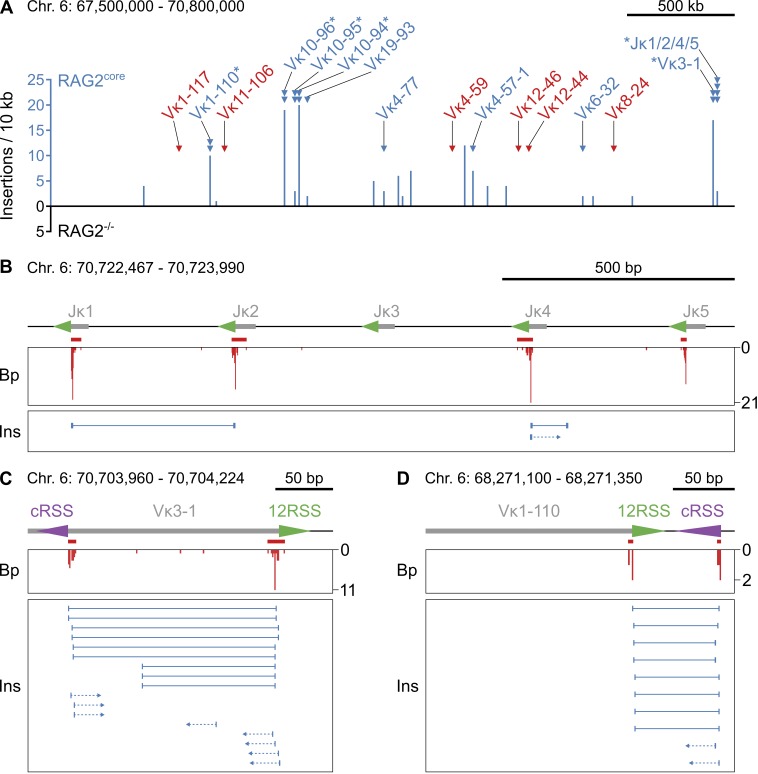
In agreement with previous studies, we observed limited recombination of the Igh locus by RAG1/2core and consequently detected only few disperse breakpoints at Vh, Dh, and Jh gene segments (Fig. S1 B and not depicted; Liang et al., 2002; Akamatsu et al., 2003). In contrast, 24 RAG1/2core-dependent breakpoint clusters were identified at Igκ (Fig. 2 A and Table S1). Each functional Jκ segment (Jκ1, Jκ2, Jκ4, and Jκ5) had a single cluster at its 23RSS cleavage site (Fig. 2 B). Surprisingly, DNA at these clusters recombined with the I-SceI break in a biased manner. Although in principle both DNA ends of a RAG1/2core-induced break would have an equal probability of joining to the cleaved I-SceI site, most rearrangements occurred with only one of the two ends for any RSS. For example, rearrangements between the I-SceI break and RAG1/2core breaks at Jκ1 exclusively involved the coding end (Fig. 2 B, rearrangements in gray), whereas those at the neighboring Jκ2 predominantly (86%) contained the signal end (Fig. 2 B, rearrangements in green). Moreover, rearrangements at Jκ1 did not extended beyond the 23RSS cleavage site of Jκ2, and vice versa. A similar phenomenon was observed for Jκ4/Jκ5 (Fig. 2 B).
Figure 2.
Rearrangements between I-SceI and RAG1/2core-induced DNA breaks at Igκ. (A) Overview of rearrangement breakpoints at the Igκ locus on chromosome 6. Histogram of the number of breakpoints in the presence or absence of RAG2core (red and black, respectively) in 10-kb intervals. RAG1/2core-dependent rearrangement breakpoint clusters are indicated by red triangles and labeled with the corresponding Jκ or Vκ gene segment. Asterisks mark breakpoint clusters with biased rearrangements (see B–D). Chromosome coordinates and scale bar are indicated on top. (B–D) Examples of RAG1/2core-dependent breakpoint clusters at Jκs and Vκs. On top is a diagram of the region, with gray boxes representing Ig segments, triangles indicating 12/23RSSs (green) or cRSSs (purple), and red bars indicating the breakpoint clusters. In the middle is a histogram showing the number and position of breakpoints (Bp, red). At the bottom, each horizontal line indicates a unique rearrangement (Rx), with its breakpoint represented by the vertical line and its sheared end (which determines the uniqueness of the event) shown by the box. Color coding indicates whether rearrangements contain RSSs/cRSSs (green/purple, signal ends) or not (gray, coding ends). Rearrangements in black are undefined. Chromosome coordinates and scale bar are indicated on top. Data analysis was performed with pooled RAG2core and RAG2−/− TC-Seq libraries (two independent experiments each). See also Fig. S2 (A–C) and Tables S1 and S2.
In addition to Jκs, breakpoint clusters were also found at 15 Vκ gene segments. Strikingly, although 10 of these had a single cluster at their physiological 12RSS cleavage sites, the other 5 (Vκ3-1, Vκ10-94, Vκ10-95, Vκ10-96, and Vκ1-110) revealed an additional cluster at a nearby cRSS (Fig. 2 A and Table S1). Overall, the heptamer sequences of these cRSSs were similar to the physiological consensus and to those identified in previous studies (Fig. S2 A; Hu et al., 2015). However, none of the cRSSs were detectable by computational tools because of their low RSS information content (RIC) scores (Table S2; Cowell et al., 2002; Merelli et al., 2010). Similar to the biased recombination pattern observed at Jκs, Vκ rearrangements at neighboring 12RSS/cRSS clusters were biased for coding or signal ends and limited in length by both cleavage sites (Fig. 2, C and D).
The remaining breakpoint clusters (nine) mapped to off-target regions outside of Ig loci (Table S1). Off targets were preferentially in transcribed genes (six) but not enriched in histone H3 lysine-4 trimethylation (H3K4me3), an active chromatin mark (unpublished data). Off-target clusters occurred near cRSS motifs that were similar to those identified at Vκ segments and also undetectable by computational tools (Fig. S2, A–C; and Table S2).
We conclude that RAG1/2core damages the B cell genome at physiological RSSs and cRSSs, and that some of the resulting DNA breaks at Jκs and Vκs recombine with the cleaved I-SceI site in a biased manner.
Aberrant deletions at Igκ
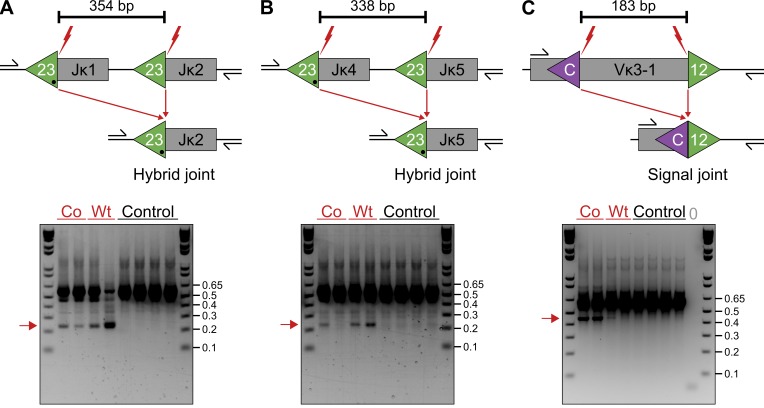
The peculiar rearrangement pattern observed at Jκ and some of the Vκ clusters suggested that RAG1/2core may mediate aberrant deletions by recombining neighboring RSSs and cRSSs at these sites. To examine this possibility, we searched for deletions by poison primer PCR (see Materials and methods; Edgley et al., 2002). Strikingly, aberrant deletions mediated by either RAG1/2core or endogenous wild-type RAG1/2 were readily detected at Jκs, where the RSSs at Jκ1 and Jκ4 were joined to the neighboring Jκ2 and Jκ5 exons, respectively (Fig. 3, A and B). The resulting deletion junctions (hybrid joints) represent aberrant joining events because physiological recombination of head-to-tail RSSs induces inversions and only involves 12/23RSS pairs (Helmink and Sleckman, 2012). However, some of these joints could also originate from two sequential inversions involving nearby Vκs, similar to those observed between D segments at TCRδ in thymocytes and between engineered RSS/cRSS pairs in ATM-deficient pro–B cell lines (Hu et al., 2015; Zhao et al., 2016). In addition to those at Jκs, deletions mediated by either RAG1/2core or RAG1/2 wild type were also identified at Vκ3-1, where joining of the 12RSS to the nearby cRSS described above generated aberrant signal joints (Fig. 3 C). We conclude that both RAG1/2core and RAG1/2 wild type cause aberrant genomic deletions at Jκ and Vκ segments.
Figure 3.
Aberrant deletions at Igκ mediated by RAG1/2core and RAG1/2 wild type. (A–C) Deletions between neighboring RSSs or RSS/cRSS at Jκs and Vκs, respectively. (top) Diagram of the locus before and after deletion by RAG1/2core or RAG1/2 wild type. The predicted size of the deletion is shown above. Gray boxes represent Ig segments, triangles indicate 12/23RSSs (green) or cRSSs (purple), red lightning points to RAG1/2 cleavage sites, and black arrows indicate the location of the internal primers used for the deletion PCR (see Materials and methods). (bottom) Ethidium bromide–stained agarose gel with deletion PCR amplicons from cultured RAG1/2core (Co), RAG1/2 wild type (Wt), or RAG2−/− (Control) bone marrow B cells and water control (0). Red arrows point to amplified deletion junctions. DNA ladder with fragment sizes in kilobases is shown alongside. Deletions were captured from 20,000 (A and B) or 40,000 (C) cells/well. Selected amplicons were extracted and confirmed by sequencing. The frequency of aberrant deletions was subsequently determined by dilutional PCR. For Co: 1 in 4,000 cells in A, 1 in 20,000 cells in B, and 1 in 600 cells in C; for Wt: 1 in 3,600 cells in A, 1 in 2,700 cells in B, and 1 in 39,300 cells in C. The frequency of deletions by RAG1/2core was lower in the repeat experiment (see Materials and methods). All results were verified by at least two independent experiments.
Excised Igκ fragments insert into I-SceI breaks
Based on the colocalization of biased rearrangements and aberrant deletions, we hypothesized that Jκ/Vκ fragments might be aberrantly excised by RAG1/2core and subsequently reintegrate at the I-SceI break (Fig. 4 A; see Discussion). To test this hypothesis, we searched TC-Seq libraries computationally for bona fide insertions, which would have been excluded from our initial bioinformatic analysis geared at identifying translocations. In brief, insertions at the I-SceI site are flanked by MycI sequence on both ends, whereas translocations contain MycI sequence only on one end (Fig. 4 A). Thus, all sequences with MycI on both ends were examined for intervening DNA originating from elsewhere in the genome (see Materials and methods).
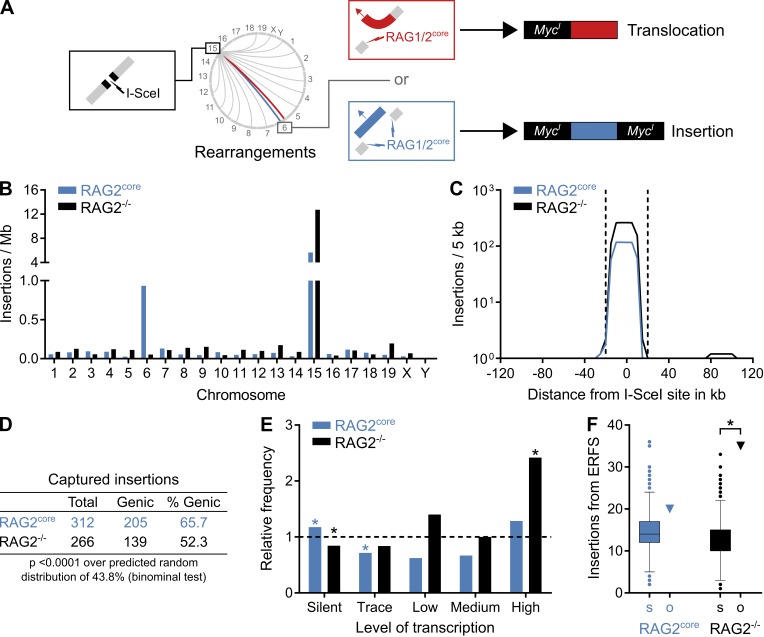
Figure 4.
Landscape of insertions in primary pro–B cells by TC-Seq. (A) Cartoon diagram comparing RAG1/2core-induced translocations and insertions. In a translocation (red), RAG1/2core introduces a single DNA break (red lightning) that recombines with the cleaved I-SceI site at MycI (black lightning) on chromosome 15. The resulting translocation contains MycI only on one side. In an insertion (blue), RAG1/2core causes tandem DNA breaks (blue lightning), thereby excising a DNA fragment that subsequently reintegrates into the cleaved I-SceI site. The resulting insertion is flanked by MycI on both sides. (B) Origin of insertions by chromosome. Events were normalized per megabase to account for different chromosome sizes. (C) Profile of insertions near the I-SceI site in 5-kb intervals. Dashed lines indicate the ±20-kb region excluded from the analysis for D–F because of saturation. (D) Proportion of insertions from genic regions. (E) Frequency of insertions derived from differentially transcribed genes compared with a random model (dashed line). Asterisks indicate values significantly different from random (P < 0.01, binominal test). (F) Observed number of insertions (o, triangle) originating from ERFSs compared with the random Monte Carlo simulation (s, Tukey boxplot). Asterisks indicate significant enrichment (P < 0.0001, binominal test). For D–F, events from the saturated I-SceI region, cryptic I-SceI sites, and other portions of the genome were excluded (see Materials and methods). Data analysis was performed with pooled RAG2core and RAG2−/− TC-Seq libraries (two independent experiments each).
We detected I-SceI insertions in both RAG2core and RAG2−/− TC-Seq libraries. Independent of RAG2core expression, inserted DNA fragments originated predominantly from a ±20-kb region around the I-SceI cleavage site on chromosome 15, similar to the chromosomal rearrangements described above (Fig. 1, B and C; and Fig. 4, B and C). Overall, inserted DNA fragments ranged from 36 to 354 bp in RAG2core and from 36 to 232 bp in RAG2−/− cells (36 bp being the minimum detection limit; see Materials and methods). Moreover, genic regions acted as preferred donors for insertions, particularly in RAG2core-expressing cells (Fig. 4 D). In contrast, insertions originating from highly transcribed regions and ERFSs were significantly enriched only in the absence of RAG2core, indicating that its expression alters the insertion landscape (Fig. 4, E and F). Thus, we found more insertions from chromosome 6 in RAG2core compared with RAG2−/− cells (140 vs. 8 events; Fig. 4 B), and with RAG2core nearly all of those (96%) originated from Igκ, whereas none derived from this locus in RAG2−/− cells (Fig. 5 A). Overall, Igκ insertions represented nearly half (43%) of all insertions in RAG2core cells and exclusively originated from regions flanked by RSSs and/or cRSSs (Fig. 5, B–D). Interestingly, donor regions included all of the Igκ gene segments displaying biased breakpoint clusters, suggesting that DNA insertions from these sites are responsible for the observed recombination pattern (see Figs. 2 and 5 and Discussion).
Figure 5.
Insertions of RAG1/2core-mobilized DNA into the I-SceI site. (A) Overview of insertions originating from the Igκ locus on chromosome 6. Histogram of the number of insertions derived from each site in the presence or absence of RAG2core (blue and black, respectively) in 10-kb intervals. RAG1/2core-dependent rearrangement breakpoint clusters at Jκs and Vκs (triangles, same as in Fig. 2 A) are color coded to indicate whether insertions from these sites are detected (blue) or not (red). Asterisks mark breakpoint clusters with biased rearrangements (see Fig. 2 [B–D]). No insertions from Igκ were detected in RAG2−/− cells. Chromosome coordinates and scale bar are indicated on top. (B–D) Examples of insertions derived from RAG1/2core-dependent breakpoint clusters at Jκs and Vκs. On top is a diagram of the region, with gray boxes representing Ig segments, triangles indicating 12/23RSSs (green) or cRSSs (purple), and red bars indicating the breakpoint clusters (same as in Fig. 2 [B–D]). In the middle is a histogram showing the number and position of breakpoints (Bp, red). At the bottom, each horizontal line indicates a unique insertion (Ins), with its breakpoints represented by the vertical lines at the ends. Arrows represent insertions for which only one of the two breakpoints could be identified. Chromosome coordinates and scale bar are indicated on top. Data analysis was performed with pooled RAG2core and RAG2−/− TC-Seq libraries (two independent experiments each). See also Table S3.
For 67% of Igκ insertions, we obtained sequence information on both junctions, providing insight into the original deletion events (Table S3). Overall, Igκ insertions originated from DNA excision between pairs of divergent, convergent, or head-to-tail RSSs, leading to insertions flanked by coding ends (coding-end insertions, 77), signal ends (signal-end insertions, 8), or both (hybrid-end insertions, 6), respectively (Fig. 5, B–D; and Table S3). Most deletions (87 out of 91) occurred between RSS/cRSS pairs, three resulted from excisions between two cRSSs, and one derived from a deletion between two 23RSSs. We conclude that RAG1/2core generates aberrant Ig fragments that are mobile and can be reinserted into I-SceI breaks on a heterologous chromosome.
Insertion of Igκ fragments excised by wild-type RAG1/2
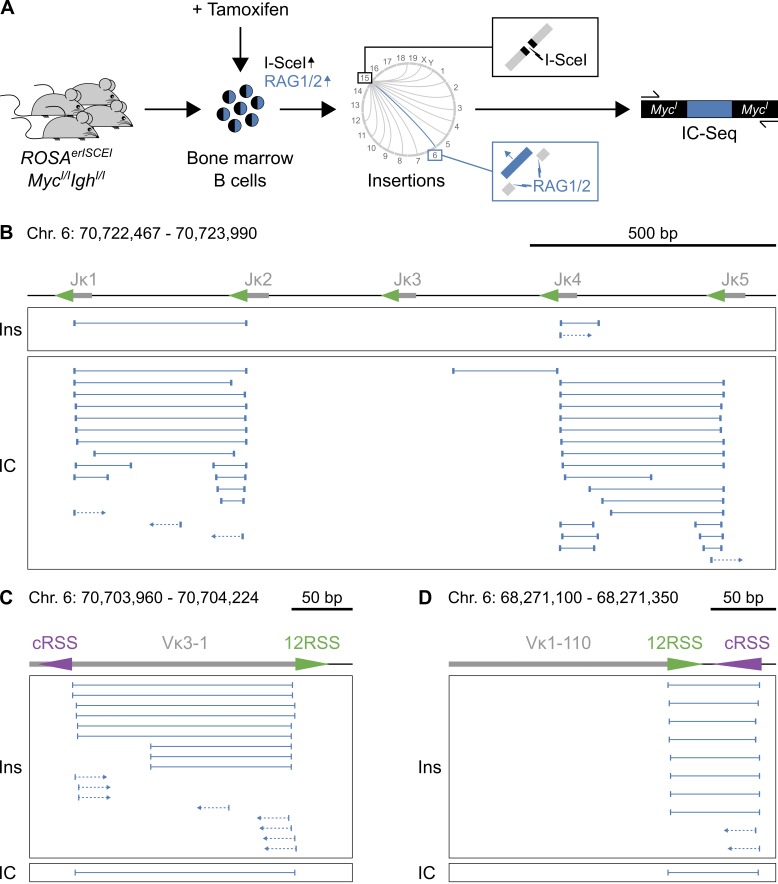
As demonstrated by our deletion PCR assays, RAG1/2 can produce aberrant Igκ deletions analogous to RAG1/2core. Thus, mobilization and insertion of Igκ DNA could in principle also occur in wild-type B cells. To test this possibility, we developed a next-generation IC-Seq method that qualitatively documents chromosomal insertions at an I-SceI site under physiological conditions. We prepared IC-Seq libraries from primary bone marrow B cells expressing a tamoxifen-inducible I-SceI transgene and bearing I-SceI cleavage sites (ROSAerISCEIMycI/IIghI/I and ROSAerISCEIMycI/IIghI/IAID−/−; see Materials and methods; Robbiani et al., 2015) that were treated ex vivo with tamoxifen to induce I-SceI breaks in the presence of wild-type RAG1/2. DNA insertions at the I-SceI site in c-myc were amplified by PCR, deep-sequenced, and analyzed computationally (Fig. 6 A; see Materials and methods).
Figure 6.
Insertions at the I-SceI site induced by wild-type RAG1/2. (A) Detection of chromosomal insertions by IC-Seq. Primary ROSAerISCEIMycI/IIghI/I (and ROSAerISCEIMycI/IIghI/IAID−/−, see Materials and methods) bone marrow B cells were treated ex vivo with tamoxifen to induce I-SceI breaks at c-myc on chromosome 15 (black lightning). Mobilized DNA fragments, such as those excised by endogenous RAG1/2 from Igκ on chromosome 6 (blue lightning), reinsert into the cleaved I-SceI site and are subsequently amplified by PCR, deep-sequenced, and analyzed computationally. Two RAG1/2 wild-type IC-Seq libraries were independently prepared from tamoxifen-treated bone marrow B cells of 12 mice. (B–D) Qualitative comparison of insertions obtained by TC-Seq (RAG1/2core) and IC-Seq (RAG1/2 wild type). On top is a diagram of the region, with gray boxes representing Ig segments and triangles indicating 12/23RSSs (green) or cRSSs (purple). Below, insertions detected by TC-Seq (Ins, same as in Fig. 5 [B–D]) and IC-Seq (IC). Each horizontal line indicates a unique insertion, with its breakpoints represented by the vertical lines at the ends. Arrows represent insertions for which only one of the two breakpoints could be identified. Chromosome coordinates and scale bar are indicated on top. Data analysis was performed with pooled IC-Seq libraries (two independent experiments). See also Table S3.
Overall, we detected I-SceI insertions from seven different Igκ gene segments (Jκ1, Jκ2, Jκ4, Jκ5, Vκ1-110, Vκ3-1, and Vκ4-69), of which six were also involved in the aforementioned insertions mediated by RAG1/2core (Table S3). Moreover, similar to RAG1/2core, Igκ insertions in the presence of RAG1/2 originated exclusively from donor regions flanked by RSSs/and or cRSSs and were comprised of coding-, signal-, and hybrid-end insertions (Fig. 6, B–D; and Table S3). We conclude that DNA insertions from Igκ are not limited to RAG1/2core but also occur during physiological V(D)J recombination by wild-type RAG1/2.
Insertion of IG and TCR fragments at physiological DNA breaks
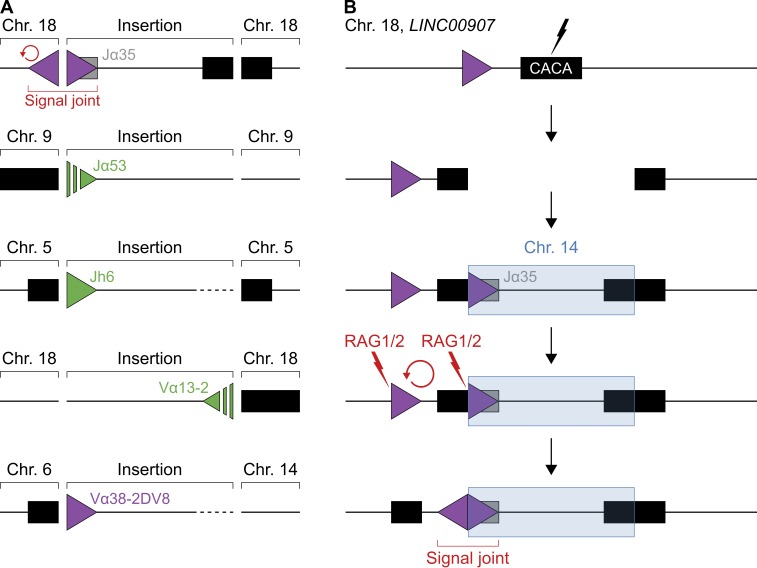
To determine whether RAG1/2 causes insertions at physiological DNA breaks in vivo, we searched published whole-genome sequences from ALL and FL patients for insertions deriving from IG and TCR loci (see Materials and methods). Overall, 5 out of 34 patients displayed genomic insertions of IG or TCR fragments at low frequency (Table S4). All insertions contained at least one RSS or cRSS motif and integrated near repetitive regions (Fig. 7 A and Fig. S3). Interestingly, DNA flanking one of the inserts was inverted to form a putative cRSS/cRSS signal joint, and in another case, a TCR fragment inserted at a translocation junction (Fig. 7, A and B). We conclude that RAG1/2 has the potential to destabilize the lymphocyte genome by mobilizing DNA that then reinserts at RAG1/2-independent, physiological DNA breaks in vivo.
Figure 7.
RAG1/2-induced insertions at physiological DNA breaks in vivo. (A) Diagram representing IG/TCR insertions detected in human cancer. From top to bottom: hypodiploid ALL (first), early T cell precursor ALL (second and third), and FL (fourth and fifth). Boxes indicate IG/TCR segments (gray) or repeat regions (black), and triangles represent 12/23RSSs (green) or cRSSs (purple). Inserted gene segments and RSSs/cRSSs are labeled with their corresponding IG/TCR segment of origin. Unresolved junctions are indicated by dashed lines/triangles. The insertion detected in hypodiploid ALL (top) is flanked by an upstream inversion (red arrow), forming a putative cRSS/cRSS signal joint. One of the insertions in FL (bottom) occurred at a translocation junction. Whole-genome sequences from 34 cancer patients were analyzed. (B) Diagram illustrating a putative RAG1/2-mediated DNA inversion caused by the insertion of a cRSS. Boxes represent IG segments (gray) or repeat regions (black), triangles indicate cRSSs (purple), and lightning indicates DNA cleavage induced by RAG1/2 (red) or unknown factors (black). From top to bottom: first, DNA is damaged at a simple CA-repeat region within LINC00907 on chromosome 18. Second, the locus is opened at the break. Third, RAG1/2 excises a DNA fragment containing a cRSS from the TCRα locus on chromosome 14, which subsequently reinserts into the break (blue). Fourth, RAG1/2 cleaves and inverts DNA between the cRSS in the insert and a cRSS near the insertion site (red arrow). Finally, the DNA inversion generates a putative cRSS/cRSS signal joint as observed in A (top). See also Fig. S3 and Table S4.
Discussion
RAG1/2 damages the pro–B cell genome at physiological RSSs and cRSSs
We used TC-Seq to examine chromosomal rearrangements in the pro–B cell genome and identified 33 RAG1/2core-dependent breakpoint clusters, of which 19 occurred at physiological RSS cleavage sites. Consistent with this finding, a previous study in ATM-deficient pro–B cell lines detected chromosomal rearrangements between I-SceI breaks at c-myc and RAG1/2-induced breaks at antigen receptor loci including Igκ (Zhang et al., 2012). Interestingly, off-target clusters at cRSSs were not detected in those experiments. In contrast, 14 of the 33 RAG1/2core-dependent breakpoint clusters identified herein were located near cRSS motifs at Vκs and off-target regions outside Ig loci. Off targets were not enriched in H3K4me3, an active chromatin mark that has been shown to colocalize with RAG1/2 binding and cleavage in developing B cells (Ji et al., 2010; Hu et al., 2015; Teng et al., 2015; unpublished data). Its absence might result from the fact that RAG2core lacks the C-terminal plant homeodomain, which normally mediates RAG1/2 binding to H3K4me3 (West et al., 2005; Liu et al., 2007; Matthews et al., 2007; Ramón-Maiques et al., 2007).
Our results demonstrate that RAG1/2core-mediated cleavage of cRSSs enables chromosomal rearrangements by producing cleaved ends that can recombine with RAG1/2-independent DNA breaks. Moreover, our data confirms that neighboring RSSs and cRSSs are substrates for aberrant genomic deletions, in agreement with previous studies using engineered RSSs (Mahowald et al., 2009; Hu et al., 2015). We speculate that the cRSSs at Vκs identified herein might also serve as beneficial substrates for secondary V-J rearrangements during V-gene replacement, similar to those described at Vhs (Rahman et al., 2006).
Aberrantly excised Igκ DNA reinserts at I-SceI breaks
We hypothesized that some of the observed rearrangements resembling translocations may actually represent insertions of deleted DNA into the I-SceI break. Because DNA is sonicated during preparation of TC-Seq libraries, a fraction of insertions would be randomly truncated and appear as translocations in the analysis (see comparison between translocation and insertion in Fig. 4 A). In agreement with this prediction, we identified bona fide insertions originating from all RAG1/2core breakpoint clusters with biased rearrangements. Furthermore, by using a novel next-generation sequencing method (IC-Seq), we confirmed Igκ insertions at I-SceI breaks in the presence of wild-type RAG1/2. Igκ insertions mediated by RAG1/2core and RAG1/2 wild type were similar in that they both originated from donor regions with RSSs/cRSSs and were comprised of all three insertion species (signal-end, coding-end, and hybrid-end).
Overall, insertions detected by both TC-Seq and IC-Seq were short (354 bp or shorter). The absence of larger insertions is likely caused by technical limitations. During TC-Seq, which was originally designed to detect chromosomal translocations, the size of insertions is mainly limited by the sonication of genomic DNA (see Materials and methods). We therefore expect long insertions to be truncated and appear as “translocations” in the computational analysis. In this regard, some of the apparent translocations at Vκs and Jκs could in principle result from the insertion of large physiological excision fragments (tens to hundreds of kilobases). Moreover, because even short insertions can be truncated, it is possible that TC-Seq considerably underestimates the actual frequency of insertions. During IC-Seq, which omits DNA sonication, the major factor limiting the detection of large insertions is PCR amplification. DNA templates with large insertions are likely outcompeted by those with small or no insertions. Finally, both TC-Seq and IC-Seq use high-throughput sequencing (see Materials and methods), which is inefficient for DNA fragments >1.5 kb.
Igκ insertions at I-SceI breaks are not mediated by DNA transposition or trans-V(D)J recombination
We observed three distinct insertion species from Igκ: those flanked by RSS/cRSS pairs (signal-end insertions), those lacking RSSs altogether (coding-end insertions), and those bearing only one RSS or cRSS (hybrid-end insertions).
Signal-end insertions derive from DNA deletion between convergent RSSs, which are normally joined to form episomal signal joints. There is some in vivo evidence that RAG1/2 can induce genomic insertions by recleaving and subsequently reintegrating episomal signal joints through either trans-V(D)J recombination or DNA transposition (Messier et al., 2003; Curry et al., 2007; Vanura et al., 2007). However, the observed signal-end insertions are not compatible with these two pathways because they occur at RAG1/2-independent DNA breaks generated by I-SceI. In contrast, during trans-V(D)J recombination, it is RAG1/2 that cleaves the RSS/cRSS at the insertion site, and in DNA transposition, RAG1/2 is responsible for catalyzing the nucleophilic attack required for insertion. Thus, the RAG1/2-induced signal-end insertions observed in our study are mediated by a pathway distinct from these previously described mechanisms.
Coding-end insertions do not fit previously proposed RAG1/2 insertion mechanisms either because both trans-V(D)J recombination and DNA transposition require RSS-containing donor fragments (Agrawal et al., 1998; Hiom et al., 1998; Curry et al., 2007; Vanura et al., 2007). Coding-end insertions originate from DNA deletions between divergent RSSs, whose products are predicted to circularize into episomal coding joints. Because these cannot be recleaved by RAG1/2, coding-end insertions likely originate from noncircularized, linear deletion products.
Hybrid-end insertions derive from deletions between head-to-tail RSSs. In principle, such deletions produce episomal hybrid joints that contain a single RSS or cRSS. Although in vitro assays have shown that RAG1/2 can induce breaks at single RSSs, the extent to which this occurs in vivo is unclear (McBlane et al., 1995; Eastman and Schatz, 1997; Yu and Lieber, 2000; Rahman et al., 2006). Hence, similar to coding-end insertions, those with hybrid ends likely derive from linear deletion products.
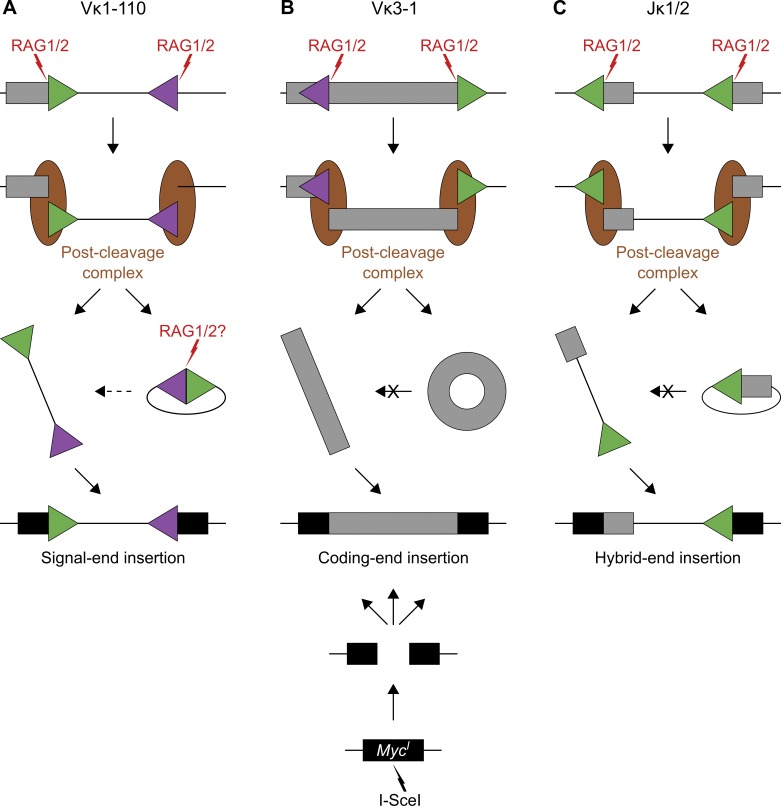
Although Igκ insertions originate from distinct types of RAG1/2 deletions, we propose a model in which they all share a common intermediate: excised linear DNA fragments that escaped from the post-cleavage complex before end joining (Fig. 8). This model agrees with biochemical experiments and studies with reporter cell lines showing that cleaved ends can prematurely escape the post-cleavage complex upon destabilization by RAG2core, nonconsensus RSS heptamers, or absence of the DNA damage response kinase ATM (Bredemeyer et al., 2006; Arnal et al., 2010; Deriano et al., 2011; Coussens et al., 2013). Our data support this model in two ways. First, the occurrence of coding- and hybrid-end insertions speaks against DNA circularization and points to the existence of stable, linear DNA deletion products. Second, because DNA integration is independent of RAG1/2, neither donor fragments nor insertion sites require RSSs/cRSSs for the insertion process. In agreement with our findings, previous studies in reporter cell lines detected a few insertions of RSS-flanked donor substrates that were not mediated by DNA transposition or by trans-V(D)J recombination (Chatterji et al., 2006; Reddy et al., 2006). Similarly, a study in primary T cells reported a few cases in which the insertion of a specific RSS-flanked TCRβ fragment occurred independently of both pathways (Curry et al., 2007). We conclude that RAG1/2 likely mobilizes linear deletion products, which are stable and have the capacity to reinsert back into the genome at independently generated DNA breaks on heterologous chromosomes. Thus, our findings reveal a novel RAG1/2-mediated insertion pathway distinct from DNA transposition and trans-V(D)J recombination.
Figure 8.
RAG1/2 mobilizes DNA from antibody gene segments into RAG1/2-independent DNA breaks. (A–C) Cartoon diagram to illustrate the pathways leading to insertion of RAG1/2-mobilized DNA into I-SceI breaks. Aberrant RAG1/2-mediated DNA excision at Vκ1-110 (A), Vκ3-1 (B), and Jκ1/2 (C) generates signal-end, coding-end, and hybrid-end insertions, respectively. Boxes represent Ig segments (gray) or MycI (black), triangles indicate RSSs (green) or cRSSs (purple), red lightning points to RAG1/2 cleavage sites, and brown ellipses represent the post-cleavage complex. From top to bottom: first, RAG1/2 induces DNA breaks at paired RSSs/cRSSs. Second, DNA is aberrantly excised, and cleaved ends remain bound to the post-cleavage complex to support their repair by the NHEJ machinery. Third, excised DNA is either circularized and released from the post-cleavage complex as episomal joint (right arrow) or it escapes before end joining as linear fragment (left arrow). For signal-end insertions (A), linear DNA fragments might also originate from recleavage of episomal signal joints by RAG1/2 (dashed arrow). For coding-end and hybrid-end insertions (B and C), recleavage of episomal joints is unlikely because of the absence of paired RSSs (crossed arrows). Finally, mobilized linear DNA fragments reinsert into the genome at the I-SceI break.
Insertions derived from non-Ig loci
Although RAG2core expression significantly alters the landscape of chromosomal insertions at I-SceI breaks, the majority of events originates from outside the Igκ locus in both RAG2core and RAG2−/− pro–B cells (57% and 100% of total, respectively). Those insertions possibly derive from regions prone to genomic instability caused by DNA transcription, replication, or other sources of DNA damage. Consistent with this possibility, chromosomal insertions in RAG2−/− cells preferentially originate from highly transcribed genes and ERFSs. Alternatively or in addition, non-Igκ insertions may represent “templated-sequence insertions” that derive from reverse-transcribed RNA (Onozawa et al., 2014). Finally, we cannot exclude that some insertions originate from RAG1/2-mediated deletions at off-target sites. In this context, it is intriguing that insertions of non-Ig DNA into antibody receptor genes were recently shown to contribute to antibody diversification (Tan et al., 2016).
RAG1/2 causes insertions at independent, physiological DNA breaks
As demonstrated by our computational analysis of human cancers, RAG1/2-induced DNA insertions are not limited to I-SceI breaks but also occur at physiological DNA breaks in vivo. The low number of IG/TCR insertions detected in our tumor analysis is likely caused by limitations of currently available datasets as well as general limitations of whole-genome sequencing techniques. Many of the publicly available tumor datasets either do not have a sufficient coverage or are not sequenced using long enough reads (e.g., 100 bp and longer) to allow for robust detection of insertion junctions. Moreover, the preparation of genomic libraries generally involves DNA fragmentation, which inevitably truncates existing insertions, thereby causing them to appear as translocations in the computational analysis.
Nevertheless, the detection of RAG1/2-induced insertions is particularly important because they pose a threat to genomic stability in at least two ways. First, they provide functional RSS and/or cRSS substrates for secondary rearrangements. In fact, introducing a RSS outside of Ig loci has been shown to cause aberrant RAG1/2-mediated deletions and inversions (Mahowald et al., 2009; Hu et al., 2015). Consistent with this, one of the tumor-associated insertions was accompanied by the formation of a putative cRSS/cRSS signal joint, which likely originated from a secondary RAG1/2-mediated DNA inversion between the cRSS in the insert and a nearby cRSS. These and other downstream recombinations (e.g., deletions and translocations) might also render RAG1/2-induced insertions especially difficult to detect. Second, although none of the insertions in the patients we analyzed are cancer drivers, the oncogenic insertion of an excised TCR fragment was recently described (Navarro et al., 2015). In the reported T-ALL patient, a DNA fragment flanked by two RSSs was excised from the TCRβ locus and reinserted upstream of the TAL1 oncogene, causing its activation. Notably, the TCRβ fragment inserted at a RAG1/2-independent DNA break, analogous to the insertions detected in our study. Furthermore, the oncogenic insertion of an IGH fragment was described in a patient with diffuse large B cell lymphoma (Chaganti et al., 1998). In the reported patient, a rearranged DJ fragment inserted into a translocation junction involving the BCL6 oncogene led to the expression of an aberrant BCL6-IGH fusion transcript. Similarly, we detected an inserted TCR fragment at a translocation junction in our cancer analysis. Thus, RAG1/2 has the capacity to destabilize the lymphocyte genome by producing cancer-associated DNA insertions.
Materials and methods
Mice
Mutant mice used in this study include RAG2−/−MycI/I (B6(Cg)-Rag2tm1.1Cgn/J (The Jackson Laboratory; Robbiani et al., 2008), ROSAerISCEIMycI/IIghI/I, and ROSAerISCEIMycI/IIghI/IAID−/− (Robbiani et al., 2015). All mice were in a C57BL/6 background or backcrossed to it for at least 10 generations. All experiments were performed in agreement with protocols approved by the Rockefeller University Institutional Animal Care and Use Committee.
Retroviruses
Murine RAG2 (RAG2full) and RAG2core sequences were amplified from mouse genomic DNA using primers p2/p6 and p3/p6, respectively (Table S5). I-SceI was amplified from pMX-I-SceI-EGFP using primers p4/p5 (Table S5; Robbiani et al., 2008). Overlap extension PCRs of the above products with primers p2/p4 and p3/p4 generated I-SceI-P2A-RAG2full and I-SceI-P2A-RAG2core, respectively (Table S5). Finally, both constructs were cloned into pMX-EGFP to generate pMX-I-SceI-P2A-RAG2full-EGFP and pMX-I-SceI-P2A-RAG2core-EGFP, respectively.
Cell culture and infection for TC-Seq
Pro–B cells were isolated from tibias, femurs, and humeri of RAG2−/−MycI/I mice at 4–10 wk of age by immunomagnetic enrichment with anti-B220 MicroBeads (Miltenyi Biotec). Cells were cultured at 2.0 × 106 cells/ml in the presence of IL-7 (5 ng/ml; Sigma-Aldrich) in complete RPMI (RPMI-1640 supplemented with l-glutamine [Gibco], sodium pyruvate [Gibco], antibiotic/antimycotic [Gibco], Hepes [Gibco], 55 µM β-mercaptoethanol [Gibco], and 10% fetal calf serum [HyClone]). IL-7 was replenished on day 2. On days 3 and 4, cell supernatants were replaced with retroviral supernatants resulting from cotransfection (Fugene-6; Roche) of BOSC23 cells with pCL-Eco and pMX-I-SceI-P2A-RAG2core-EGFP or pMX-I-SceI-EGFP plasmids 3 d before (Robbiani et al., 2008). Spinoculation was at 1,111 g for 1.5 h in the presence of 2.5 µg/ml polybrene, 5 ng/ml IL-7, and 20 mM Hepes. After 6–8 h at 37°C, on day 3, retroviral supernatants were replaced with original supernatants, whereas on day 4, cells were collected for IL-7 washout and replating in fresh complete RPMI. Cells were harvested after 2.5 d of IL-7 depletion, sorted for EGFP expression with a FACSAria instrument (BD), pelleted, and snap-frozen on dry ice. Samples infected with pMX-I-SceI-P2A-RAG2core-EGFP are referred to as RAG2core, and those infected with pMX-I-SceI-EGFP are referred to as RAG2−/−.
Cell culture for IC-Seq
Bone marrow B cells were isolated from tibias, femurs, and humeri of ROSAerISCEIMycI/IIghI/I and ROSAerISCEIMycI/IIghI/IAID−/− mice at 6–8.5 mo of age by immunomagnetic enrichment with anti-B220 MicroBeads (Miltenyi Biotec). Cells were pooled and cultured at 2.0 × 106 cells/ml in the presence of IL-7 (5 or 10 ng/ml; Sigma-Aldrich) and tamoxifen (1 µM; Sigma-Aldrich) in complete RPMI. On day 1, cells were collected for IL-7 washout and replated in fresh complete RPMI with 1 µM tamoxifen. On day 2, cultures were harvested, and cell pellets were snap-frozen on dry ice.
TC-Seq library preparation
TC-Seq libraries of RAG2core and RAG2−/− pro–B cells were prepared in duplicates from each of 50 million sorted cells, as previously described (Klein et al., 2011; Robbiani et al., 2015) with the exception that sonication of genomic DNA was performed with Covaris S220 (power 105, duty factor 5%, cycles 200, time 35 s, water level 12, temperature 7°C), yielding a core of DNA fragments between 500 and 850 bp. Each library was sequenced twice using Illumina MiSeq (300 cycles, paired-end).
IC-Seq library preparation
IC-Seq libraries of bone marrow B cells were prepared in duplicates from 40 million and 60 million cultured cells. Genomic DNAs were extracted with phenol-chloroform after Proteinase K digestion, washed twice with 70% ethanol, and resuspended in TE buffer (Invitrogen). For the first PCR, 1 µg of DNA was amplified in each reaction with Phusion polymerase (New England Biolabs, Inc.) and the MycI flanking primers p247/p251 with the following conditions: 98°C for 2 min; 35× (98°C for 10 s, 72°C for 1:30 min); and 72°C for 5 min (Table S5). Pooled PCR reactions were column purified (MACHEREY-NAGEL) and high molecular weight products (1,500–5,000 bp) were isolated by agarose gel electrophoresis. Extracted DNA was digested with I-SceI (New England Biolabs, Inc.) and column purified (MACHEREY-NAGEL). In the second PCR, 25 ng DNA were amplified in each reaction with Phusion polymerase (New England Biolabs, Inc.) and primers p274a/p275a, p274b/p275b, p274c/p275c, and p274d/p275d with the following conditions: 98°C for 2 min; 3× (98°C for 10 s, 65°C for 30 s, 72°C for 1 min); 32× (98°C for 10 s, 72°C for 1:15 min); and 72°C for 5 min (Table S5). PCR products were pooled, and high molecular weight amplicons (280–3,000 bp) were isolated by agarose gel electrophoresis. Extracted DNA was digested with I-SceI (New England Biolabs, Inc.) and column purified (MACHEREY-NAGEL). To add index adapters for sequencing, the PCR was similar to the second PCR but with primers pNextflex common/pNextflex index5 or pNextflex common/pNextflex index6 with the following conditions: 98°C for 2 min; 3× (98°C for 10 s, 67°C for 30 s, 72°C for 1 min); 32× (98°C for 10 s, 72°C for 1:15 min); and 72°C for 5 min (Table S5). PCR products were pooled, and high molecular weight amplicons (350–2,000 bp) were isolated by agarose gel electrophoresis. Extracted DNA was digested with I-SceI (New England Biolabs, Inc.) and column purified (MACHEREY-NAGEL), and high molecular weight products (300–2,000 bp) were isolated once more by agarose gel electrophoresis. Extracted DNA was sequenced twice using Illumina NextSeq (150 cycles, paired-end).
TC-Seq analysis
Two independent libraries were sequenced twice, and the data were pooled for analysis using a novel pipeline to identify rearrangement and insertion breakpoints. First, sequencing reads were trimmed for high quality with seqtk (error rate threshold of 0.01; Broad Institute), and those with primer sequences from the first PCR or <5 bp of MycI after the nested primer sequence were discarded. Second, reads were mapped against MycI with its repetitive regions masked using SMALT (v0.7.6; parameters: -c 11 -x -O; Sanger Institute). Paired reads that both aligned to MycI at their 5′ end were analyzed in “insertion mode”; otherwise, they were processed in “rearrangement mode.”
In rearrangement mode (Figs. 1 and 2), bases aligning to MycI were clipped from either the beginning or the end of the reads, and the remaining sequences were mapped to the mouse genome (mm10) with SMALT (parameters: -O -r -1). Only alignments with at least 36 bp and a Phred score of 20 were accepted. Reads with the same sheared ends, which derive from sonication during library preparation, were merged into one event, and single reads were preserved. Rearrangements that did not yield breakpoints were discarded. Finally, reads that crossed the I-SceI site by >3 bp were excluded.
In insertion mode (Figs. 4 and 5), bases aligning to MycI were clipped from both ends of the reads, and the remaining sequences were mapped to the mouse genome (mm10) with SMALT (parameters: -O -r -1). Only alignments with at least 36 bp and a Phred score of 20 were accepted. Pairs with incorrect genomic orientation (+/+ and −/−) were excluded. The alignment of insertions yielded either both genomic breakpoints (double junctions) or only one (single junctions). Because of saturation at MycI, events were merged if they possessed all of the following features: identical shears, genomic breakpoints within 5 bp, and same orientation. Events based on single reads were preserved. Finally, reads that crossed the I-SceI site by >3 bp were excluded.
IC-Seq analysis
Data from two independent libraries were pooled and analyzed similar to the “insertion mode” in TC-Seq, with minor modifications (Fig. 6). Only genomic alignments with at least 25 bp and a Phred score of 20 were accepted. Insertions were merged if they possessed genomic breakpoints within 5 bp of each other and occurred in the same orientation. Finally, reads that crossed the I-SceI site by more than 3 bp were excluded.
Analysis of rearrangements (TC-Seq) and insertions (IC-Seq)
To characterize chromosomal rearrangements and insertions derived from distal regions (Fig. 1, D–F; and Fig. 4, D–F), the following portions of the genome were excluded: 50 or 20 kb surrounding the I-SceI site at MycI (rearrangements or insertions, respectively), 2 kb surrounding cryptic I-SceI sites (consensus [TCA][AT]GGGATA[AC]CAGG[GCT][TC][ATC][AG][TAC]), RAG2 (likely representing retroviral integrations), 3 Mb at each centromere, and chromosome M (mitochondrial DNA).
To determine the enrichment at genic regions (Figs. 1 D and 4 D), the portion of DNA from −2 kb of the most upstream transcription start site to the end of the last exon was considered as genic. For transcription analysis (Figs. 1 E and 4 E), RNA-seq data (Revilla-i-Domingo et al., 2012) were mapped with STAR aligner (v2.4.2a; default parameters; Dobin et al., 2013) using the mouse genome (mm10) and removing multiple alignments. Transcripts were quantified and annotated using cufflinks (v2.2.1; cuffdiff parameters: –upper-quartile-norm –dispersion-method per-condition; Trapnell et al., 2013) and Ensembl annotation (release 80). Transcription groups were defined using the mclust R package: silent (0 FPKM), trace (0.000000522291–2.8443 FPKM), low (2.84555–11.9418 FPKM), medium (11.9476–47.115 FPKM), and high (47.1191–74.211 FPKM). To detect enrichment within ERFSs (Figs. 1 F and 4 F), previously reported sites (Barlow et al., 2013) were lifted over from mouse genome mm9 to mm10 (UCSC LiftOver tool).
Detection of rearrangement breakpoint clusters (TC-Seq)
RAG1/2core-dependent breakpoint clusters were detected by a three-step process. First, RAG2core and RAG2−/− TC-Seq libraries were screened for local enrichment of rearrangement breakpoints to identify breakpoint hotspots (at least three breakpoints and a combined p-value of <10−8; Klein et al., 2011). To prevent potential sonication artifacts, hotspots were excluded if their sheared ends were either within less than 18 bp of each other or overlapped with simple repeat regions. Second, breakpoint hotspots were defined as RAG1/2core dependent if they did not display any RAG2−/− breakpoints or sheared ends within ±1-kb distance. Third, breakpoint clusters containing three or more events within up to 25 bp distance of each other were identified within each RAG1/2core hotspot. Off-target clusters were manually filtered based on the location of recurrent breakpoints near CA motifs that were shared by at least three clusters (CACA, CACC, CACT, and CAGA). Simple CA-repeat regions were excluded. Putative cRSS sequences were manually detected and analyzed using Geneious (Kearse et al., 2012) and RSSsite (http://www.itb.cnr.it/rss; Merelli et al., 2010). Sequences of physiological RSSs were obtained from IMGT (http://www.imgt.org/) and published RSS datasets (Cowell et al., 2002). Annotation of V(D)J segments was based on Ensembl (release 80). Rearrangements crossing the I-SceI site were still allowed during the detection of breakpoint hotspots and clusters, but afterward manually removed from all sites in the final data.
Analysis of insertions in human tumors
We designed a novel pipeline to search whole-genome sequences for insertions derived from IG/TCR loci. First, IG/TCR baits were generated that correspond to regions spanning 150 bp upstream and downstream from each physiological RSS cleavage site of human V and J segments (Ensembl, release 84). D segments were excluded, and repeat regions were masked. Second, whole-genome sequences from published human cancer datasets (Table S4; Wang et al., 2011; Holmfeldt et al., 2013; Okosun et al., 2014) were mapped with bwa mem (v0.7.12-r1039; default parameters) using the IG/TCR baits as references. Third, paired reads aligning to the baits were mapped against the human genome (hg38) using bwa mem (v0.7.12-r1039; default parameters). Only alignments with a Phred score of at least 20 were accepted. Finally, reads containing junctions (chimeric alignments) were filtered to yield insertions that were then manually verified using Geneious (Kearse et al., 2012). The analysis of publicly available human cancer datasets was classified as exempt activity by the Rockefeller University Institutional Review Board.
Deletion PCR assays
Genomic DNAs of TC-Seq (RAG2core and RAG2−/−) and IC-Seq (RAG1/2 wild type) cultures were used for deletion PCR assays. Duplicates for RAG2core and RAG2−/− originated from cell cultures with modified conditions: control was infected with pMX-EGFP; cells were transferred onto irradiated S17 stroma cells after IL-7 washout on day 4 and depleted for 1.5 d. To detect small and rare deletion events, nested PCRs with a “poison” primer were performed (Edgley et al., 2002). For PCRI, 100 ng (Jκ1/2, Jκ4/5) or 200 ng (Vκ3-1) genomic DNA was amplified in 20-µl reactions with HotStarTaq polymerase (QIAGEN). For PCRII, 1 µl of PCRI was used as template. For deletions at Jκ1/2, primers p195/p256/p258 (PCRI) and p196/p257 (PCRII) were used with the following conditions: PCRI, 95°C for 15 min; 30× (95°C for 45 s, 63°C for 45 s, and 72°C for 25 s); and 72°C for 5 min; PCRII, 95°C for 15 min; 30× (95°C for 45 s, 63°C for 45 s, and 72°C for 10 s); and 72°C for 5 min (Table S5). For deletions at Jκ4/5, primers p199/p205/p255 (PCRI) and p200/p206 (PCRII) were used with the same cycling conditions as for Jκ1/2 (Table S5). For deletions at Vκ3-1, primers p243/p244/p245 (PCRI) and p207/p210 (PCRII) were used with the following conditions: PCRI, 95°C for 15 min; 30× (95°C for 45 s, 63°C for 45 s, and 72°C for 50 s); and 72°C for 5 min; PCRII, 95°C for 15 min; 30× (95°C for 45 s, 63°C for 45 s, and 72°C for 20 s); and 72°C for 5 min (Table S5). PCRII products were separated on 2% agarose gels stained with ethidium bromide. Fragments shorter than the expected size from the germline locus (Jκ1/2: <592 bp, Jκ4/5: <575 bp, and Vκ3-1: <635 bp) were extracted (MACHEREY-NAGEL) and sequenced (Genewiz). Deletion products were confirmed using Geneious (Kearse et al., 2012).
V(D)J PCR assays
Genomic DNAs of TC-Seq (RAG2core and RAG2−/−) cultures were used for V(D)J PCR assays. For RAG2full, cells were cultured as for TC-Seq but infected with pMX-I-SceI-P2A-RAG2full-EGFP. Duplicates originated from cell cultures with modified conditions: control was infected with pMX-EGFP; all cells were transferred onto irradiated S17 stroma cells after IL-7 washout on day 4 and depleted for 1.5 d. Semiquantitative V(D)J PCRs were performed as previously described (Schlissel et al., 1991; Dudley et al., 2003) with modifications: 100, 50, or 25 ng of template DNA was amplified in 20-µl reactions with HotStarTaq polymerase (QIAGEN). For V(D)J PCRs, primers p58/p96 (Dh-Jh PCR), p96/p98 (VhQ52-DJh PCR), and p305/p306 (Vκ-Jκ PCR) were used with the following conditions: 95°C for 15 min; 32× (95°C for 45 s, 62°C for 45 s, and 72°C for 2 min); and 72°C for 5 min (Table S5). For control PCRs (MycI) primers p113/p114 were used with the following conditions: 95°C for 15 min; 30× (95°C for 45 s, 58°C for 45 s, and 72°C for 20 s); and 72°C for 5 min (Table S5). PCR products were separated on 1.5% agarose gels stained with ethidium bromide.
Accession numbers
The TC-Seq and IC-Seq sequencing data generated in this study can be accessed from the SRA database (SRP077983).
Online supplemental material
Fig. S1 describes the RAG2-expressing retroviruses used in this study. Fig. S2 characterizes RAG1/2core-dependent rearrangement breakpoint clusters at cRSSs. Fig. S3 shows the sequences of inserted IG/TCR fragments identified in human cancers. Table S1 lists features of RAG1/2core-dependent breakpoint clusters detected by TC-Seq. Table S2 summarizes the RIC score analysis of identified cRSSs. Table S3 displays features of Igκ insertions detected by TC-Seq and IC-Seq. Table S4 summarizes the insertion analysis of human cancers. Table S5 lists primers used in this study. Tables S1–S5 are included as Excel files.
Acknowledgments
We thank all members of the Nussenzweig laboratory for discussions; Mila Jankovic for comments on the manuscript; Klara Velinzon, Yelena Shatalina, and Neena Thomas for FACS sorting; and David Bosque, Thomas Eisenreich, and Susan Hinklein for maintenance of the mouse colonies. We also thank Connie Zhao of the Rockefeller Genomics Resource Center for help with high-throughput sequencing and Patricia Cortes for V(D)J-PCR protocols and reagents. Finally, P.C. Rommel would like to thank his wife Sara Cuesta González for her endless patience and loving support.
This work was funded by National Institutes of Health grant AI112602 to D.F. Robbiani and in part by National Institutes of Health grants AI037526 and AI072529 to M.C. Nussenzweig. P.C. Rommel was supported by a fellowship of the German Academic Exchange Service (DAAD), and M.C. Nussenzweig is a Howard Hughes Medical Institute Investigator. The cancer datasets used in this study (dbGaP: phs000341.v2.p1 and phs000340.v3.p1; EBI: EGAS00001000399) were generated with the financial support of the National Cancer Institute, the St. Baldrick’s Foundation, Partners for Cures, the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital as part of the St. Jude/Washington University Pediatric Cancer Genome Project, Cancer Research UK, Bloodwise (now Leukemia and Lymphoma Research), and the Hungarian Scientific Research Fund (OTKA).
The authors declare no competing financial interests.
Author contributions: P.C. Rommel designed and performed experiments as well as data analysis, wrote the manuscript, and prepared the figures. T.Y. Oliveira designed and performed data analysis. M.C. Nussenzweig and D.F. Robbiani designed experiments and wrote the manuscript.
Footnotes
Abbreviations used:
- ALL
- acute lymphoblastic leukemia
- ATM
- ataxia-telangiectasia mutated kinase
- cRSS
- cryptic RSS
- ERFS
- early replication fragile site
- FL
- follicular lymphoma
- H3K4me3
- histone H3 lysine-4 trimethylation
- IC-Seq
- insertion capture sequencing
- NHEJ
- nonhomologous end joining
- RIC
- RSS information content
- RSS
- recombination signal sequence
- TC-Seq
- translocation capture sequencing
References
- Agrawal A., Eastman Q.M., and Schatz D.G.. 1998. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 394:744–751. 10.1038/29457 [DOI] [PubMed] [Google Scholar]
- Akamatsu Y., Monroe R., Dudley D.D., Elkin S.K., Gärtner F., Talukder S.R., Takahama Y., Alt F.W., Bassing C.H., and Oettinger M.A.. 2003. Deletion of the RAG2 C terminus leads to impaired lymphoid development in mice. Proc. Natl. Acad. Sci. USA. 100:1209–1214. 10.1073/pnas.0237043100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt F.W., Zhang Y., Meng F.-L., Guo C., and Schwer B.. 2013. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 152:417–429. 10.1016/j.cell.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnal S.M., Holub A.J., Salus S.S., and Roth D.B.. 2010. Non-consensus heptamer sequences destabilize the RAG post-cleavage complex, making ends available to alternative DNA repair pathways. Nucleic Acids Res. 38:2944–2954. 10.1093/nar/gkp1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow J.H., Faryabi R.B., Callén E., Wong N., Malhowski A., Chen H.T., Gutierrez-Cruz G., Sun H.-W., McKinnon P., Wright G., et al. 2013. Identification of early replicating fragile sites that contribute to genome instability. Cell. 152:620–632. 10.1016/j.cell.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer A.L., Sharma G.G., Huang C.-Y., Helmink B.A., Walker L.M., Khor K.C., Nuskey B., Sullivan K.E., Pandita T.K., Bassing C.H., and Sleckman B.P.. 2006. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 442:466–470. 10.1038/nature04866 [DOI] [PubMed] [Google Scholar]
- Chaganti S.R., Rao P.H., Chen W., Dyomin V., Jhanwar S.C., Parsa N.Z., Dalla-Favera R., and Chaganti R.S.K.. 1998. Deregulation of BCL6 in non-Hodgkin lymphoma by insertion of IGH sequences in complex translocations involving band 3q27. Genes Chromosomes Cancer. 23:328–336. [DOI] [PubMed] [Google Scholar]
- Chatterji M., Tsai C.L., and Schatz D.G.. 2006. Mobilization of RAG-generated signal ends by transposition and insertion in vivo. Mol. Cell. Biol. 26:1558–1568. 10.1128/MCB.26.4.1558-1568.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatworthy A.E., Valencia M.A., Haber J.E., and Oettinger M.A.. 2003. V(D)J recombination and RAG-mediated transposition in yeast. Mol. Cell. 12:489–499. 10.1016/S1097-2765(03)00305-8 [DOI] [PubMed] [Google Scholar]
- Coussens M.A., Wendland R.L., Deriano L., Lindsay C.R., Arnal S.M., and Roth D.B.. 2013. RAG2’s acidic hinge restricts repair-pathway choice and promotes genomic stability. Cell Reports. 4:870–878. 10.1016/j.celrep.2013.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell L.G., Davila M., Kepler T.B., and Kelsoe G.. 2002. Identification and utilization of arbitrary correlations in models of recombination signal sequences. Genome Biol. 3:research0072.1 10.1186/gb-2002-3-12-research0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry J.D., and Schlissel M.S.. 2008. RAG2's non-core domain contributes to the ordered regulation of V(D)J recombination. Nucleic Acids Res. 36:5750–5762. 10.1093/nar/gkn553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry J.D., Schulz D., Guidos C.J., Danska J.S., Nutter L., Nussenzweig A., and Schlissel M.S.. 2007. Chromosomal reinsertion of broken RSS ends during T cell development. J. Exp. Med. 204:2293–2303. 10.1084/jem.20070583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L., Chaumeil J., Coussens M., Multani A., Chou Y., Alekseyenko A.V., Chang S., Skok J.A., and Roth D.B.. 2011. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature. 471:119–123. 10.1038/nature09755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley D.D., Sekiguchi J., Zhu C., Sadofsky M.J., Whitlow S., DeVido J., Monroe R.J., Bassing C.H., and Alt F.W.. 2003. Impaired V(D)J recombination and lymphocyte development in core RAG1-expressing mice. J. Exp. Med. 198:1439–1450. 10.1084/jem.20030627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman Q.M., and Schatz D.G.. 1997. Nicking is asynchronous and stimulated by synapsis in 12/23 rule-regulated V(D)J cleavage. Nucleic Acids Res. 25:4370–4378. 10.1093/nar/25.21.4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley M., D’Souza A., Moulder G., McKay S., Shen B., Gilchrist E., Moerman D., and Barstead R.. 2002. Improved detection of small deletions in complex pools of DNA. Nucleic Acids Res. 30:e52 10.1093/nar/gnf051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkin S.K., Matthews A.G., and Oettinger M.A.. 2003. The C-terminal portion of RAG2 protects against transposition in vitro. EMBO J. 22:1931–1938. 10.1093/emboj/cdg184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmink B.A., and Sleckman B.P.. 2012. The response to and repair of RAG-mediated DNA double-strand breaks. Annu. Rev. Immunol. 30:175–202. 10.1146/annurev-immunol-030409-101320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiom K., Melek M., and Gellert M.. 1998. DNA transposition by the RAG1 and RAG2 proteins: A possible source of oncogenic translocations. Cell. 94:463–470. 10.1016/S0092-8674(00)81587-1 [DOI] [PubMed] [Google Scholar]
- Holmfeldt L., Wei L., Diaz-Flores E., Walsh M., Zhang J., Ding L., Payne-Turner D., Churchman M., Andersson A., Chen S.-C., et al. 2013. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat. Genet. 45:242–252. 10.1038/ng.2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Zhang Y., Zhao L., Frock R.L., Du Z., Meyers R.M., Meng F.L., Schatz D.G., and Alt F.W.. 2015. Chromosomal loop domains direct the recombination of antigen receptor genes. Cell. 163:947–959. 10.1016/j.cell.2015.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Resch W., Corbett E., Yamane A., Casellas R., and Schatz D.G.. 2010. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 141:419–431. 10.1016/j.cell.2010.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. 2012. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.-S., Lapkouski M., Yang W., and Gellert M.. 2015. Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature. 518:507–511. 10.1038/nature14174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein I.A., Resch W., Jankovic M., Oliveira T., Yamane A., Nakahashi H., Di Virgilio M., Bothmer A., Nussenzweig A., Robbiani D.F., et al. 2011. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 147:95–106. 10.1016/j.cell.2011.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küppers R., and Dalla-Favera R.. 2001. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 20:5580–5594. 10.1038/sj.onc.1204640 [DOI] [PubMed] [Google Scholar]
- Lee G.S., Neiditch M.B., Sinden R.R., and Roth D.B.. 2002. Targeted transposition by the V(D)J recombinase. Mol. Cell. Biol. 22:2068–2077. 10.1128/MCB.22.7.2068-2077.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescale C., Abramowski V., Bedora-Faure M., Murigneux V., Vera G., Roth D.B., Revy P., de Villartay J.-P., and Deriano L.. 2016. RAG2 and XLF/Cernunnos interplay reveals a novel role for the RAG complex in DNA repair. Nat. Commun. 7:10529 10.1038/ncomms10529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.M., Agard E., Suh S., and Czyzyk L.. 1997. Cryptic signals and the fidelity of V(D)J joining. Mol. Cell. Biol. 17:3125–3136. 10.1128/MCB.17.6.3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H.-E., Hsu L.-Y., Cado D., Cowell L.G., Kelsoe G., and Schlissel M.S.. 2002. The “dispensable” portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity. 17:639–651. 10.1016/S1074-7613(02)00448-X [DOI] [PubMed] [Google Scholar]
- Lieber M.R. 2016. Mechanisms of human lymphoid chromosomal translocations. Nat. Rev. Cancer. 16:387–398. 10.1038/nrc.2016.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Subrahmanyam R., Chakraborty T., Sen R., and Desiderio S.. 2007. A plant homeodomain in Rag-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 27:561–571. 10.1016/j.immuni.2007.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald G.K., Baron J.M., Mahowald M.A., Kulkarni S., Bredemeyer A.L., Bassing C.H., and Sleckman B.P.. 2009. Aberrantly resolved RAG-mediated DNA breaks in Atm-deficient lymphocytes target chromosomal breakpoints in cis. Proc. Natl. Acad. Sci. USA. 106:18339–18344. 10.1073/pnas.0902545106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews A.G.W., Kuo A.J., Ramón-Maiques S., Han S., Champagne K.S., Ivanov D., Gallardo M., Carney D., Cheung P., Ciccone D.N., et al. 2007. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 450:1106–1110. 10.1038/nature06431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBlane J.F., van Gent D.C., Ramsden D.A., Romeo C., Cuomo C.A., Gellert M., and Oettinger M.A.. 1995. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 83:387–395. 10.1016/0092-8674(95)90116-7 [DOI] [PubMed] [Google Scholar]
- Merelli I., Guffanti A., Fabbri M., Cocito A., Furia L., Grazini U., Bonnal R.J., Milanesi L., and McBlane F.. 2010. RSSsite: A reference database and prediction tool for the identification of cryptic Recombination Signal Sequences in human and murine genomes. Nucleic Acids Res. 38:W262–W267. 10.1093/nar/gkq391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier T.L., O’Neill J.P., Hou S.-M., Nicklas J.A., and Finette B.A.. 2003. In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. EMBO J. 22:1381–1388. 10.1093/emboj/cdg137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijušković M., Chou Y.-F., Gigi V., Lindsay C.R., Shestova O., Lewis S.M., and Roth D.B.. 2015. Off-target V(D)J recombination drives lymphomagenesis and is escalated by loss of the Rag2 C terminus. Cell Reports. 12:1842–1852. 10.1016/j.celrep.2015.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro J.-M., Touzart A., Pradel L.C., Loosveld M., Koubi M., Fenouil R., Le Noir S., Maqbool M.A., Morgado E., Gregoire C., et al. 2015. Site- and allele-specific polycomb dysregulation in T-cell leukaemia. Nat. Commun. 6:6094 10.1038/ncomms7094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiditch M.B., Lee G.S., Huye L.E., Brandt V.L., and Roth D.B.. 2002. The V(D)J recombinase efficiently cleaves and transposes signal joints. Mol. Cell. 9:871–878. 10.1016/S1097-2765(02)00494-X [DOI] [PubMed] [Google Scholar]
- Nussenzweig A., and Nussenzweig M.C.. 2010. Origin of chromosomal translocations in lymphoid cancer. Cell. 141:27–38. 10.1016/j.cell.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okosun J., Bödör C., Wang J., Araf S., Yang C.-Y., Pan C., Boller S., Cittaro D., Bozek M., Iqbal S., et al. 2014. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 46:176–181. 10.1038/ng.2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira T.Y., Resch W., Jankovic M., Casellas R., Nussenzweig M.C., and Klein I.A.. 2012. Translocation capture sequencing: A method for high throughput mapping of chromosomal rearrangements. J. Immunol. Methods. 375:176–181. 10.1016/j.jim.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onozawa M., Zhang Z., Kim Y.J., Goldberg L., Varga T., Bergsagel P.L., Kuehl W.M., and Aplan P.D.. 2014. Repair of DNA double-strand breaks by templated nucleotide sequence insertions derived from distant regions of the genome. Proc. Natl. Acad. Sci. USA. 111:7729–7734. 10.1073/pnas.1321889111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey J.E., Pytlos M.J., Sinden R.R., and Roth D.B.. 2006. Target DNA structure plays a critical role in RAG transposition. PLoS Biol. 4:e350 10.1371/journal.pbio.0040350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman N.S., Godderz L.J., Stray S.J., Capra J.D., and Rodgers K.K.. 2006. DNA cleavage of a cryptic recombination signal sequence by RAG1 and RAG2. Implications for partial V(H) gene replacement. J. Biol. Chem. 281:12370–12380. 10.1074/jbc.M507906200 [DOI] [PubMed] [Google Scholar]
- Ramón-Maiques S., Kuo A.J., Carney D., Matthews A.G.W., Oettinger M.A., Gozani O., and Yang W.. 2007. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc. Natl. Acad. Sci. USA. 104:18993–18998. 10.1073/pnas.0709170104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy Y.V.R., Perkins E.J., and Ramsden D.A.. 2006. Genomic instability due to V(D)J recombination-associated transposition. Genes Dev. 20:1575–1582. 10.1101/gad.1432706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla-i-Domingo R., Bilic I., Vilagos B., Tagoh H., Ebert A., Tamir I.M., Smeenk L., Trupke J., Sommer A., Jaritz M., and Busslinger M.. 2012. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J. 31:3130–3146. 10.1038/emboj.2012.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Bothmer A., Callen E., Reina-San-Martin B., Dorsett Y., Difilippantonio S., Bolland D.J., Chen H.T., Corcoran A.E., Nussenzweig A., and Nussenzweig M.C.. 2008. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 135:1028–1038. 10.1016/j.cell.2008.09.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Deroubaix S., Feldhahn N., Oliveira T.Y., Callen E., Wang Q., Jankovic M., Silva I.T., Rommel P.C., Bosque D., et al. 2015. Plasmodium infection promotes genomic instability and AID-dependent B cell lymphoma. Cell. 162:727–737. 10.1016/j.cell.2015.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D.B. 2003. Restraining the V(D)J recombinase. Nat. Rev. Immunol. 3:656–666. 10.1038/nri1152 [DOI] [PubMed] [Google Scholar]
- Ru H., Chambers M.G., Fu T.-M., Tong A.B., Liao M., and Wu H.. 2015. Molecular mechanism of V(D)J recombination from synaptic RAG1-RAG2 complex structures. Cell. 163:1138–1152. (published erratum appears in Cell 2015. 163:1807) 10.1016/j.cell.2015.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D.G., and Ji Y.. 2011. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 11:251–263. 10.1038/nri2941 [DOI] [PubMed] [Google Scholar]
- Schatz D.G., and Swanson P.C.. 2011. V(D)J recombination: Mechanisms of initiation. Annu. Rev. Genet. 45:167–202. 10.1146/annurev-genet-110410-132552 [DOI] [PubMed] [Google Scholar]
- Schlissel M.S., Corcoran L.M., and Baltimore D.. 1991. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J. Exp. Med. 173:711–720. 10.1084/jem.173.3.711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi J.A., Whitlow S., and Alt F.W.. 2001. Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol. Cell. 8:1383–1390. 10.1016/S1097-2765(01)00423-3 [DOI] [PubMed] [Google Scholar]
- Talukder S.R., Dudley D.D., Alt F.W., Takahama Y., and Akamatsu Y.. 2004. Increased frequency of aberrant V(D)J recombination products in core RAG-expressing mice. Nucleic Acids Res. 32:4539–4549. 10.1093/nar/gkh778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Pieper K., Piccoli L., Abdi A., Foglierini M., Geiger R., Tully C.M., Jarrossay D., Ndungu F.M., Wambua J., et al. 2016. A LAIR1 insertion generates broadly reactive antibodies against malaria variant antigens. Nature. 529:105–109. 10.1038/nature16450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng G., Maman Y., Resch W., Kim M., Yamane A., Qian J., Kieffer-Kwon K.-R., Mandal M., Ji Y., Meffre E., et al. 2015. RAG represents a widespread threat to the lymphocyte genome. Cell. 162:751–765. 10.1016/j.cell.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., and Pachter L.. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31:46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.-L., Chatterji M., and Schatz D.G.. 2003. DNA mismatches and GC-rich motifs target transposition by the RAG1/RAG2 transposase. Nucleic Acids Res. 31:6180–6190. 10.1093/nar/gkg819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanura K., Montpellier B., Le T., Spicuglia S., Navarro J.-M., Cabaud O., Roulland S., Vachez E., Prinz I., Ferrier P., et al. 2007. In vivo reinsertion of excised episomes by the V(D)J recombinase: A potential threat to genomic stability. PLoS Biol. 5:e43 10.1371/journal.pbio.0050043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Mullighan C.G., Easton J., Roberts S., Heatley S.L., Ma J., Rusch M.C., Chen K., Harris C.C., Ding L., et al. 2011. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat. Methods. 8:652–654. 10.1038/nmeth.1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Oliveira T., Jankovic M., Silva I.T., Hakim O., Yao K., Gazumyan A., Mayer C.T., Pavri R., Casellas R., et al. 2014. Epigenetic targeting of activation-induced cytidine deaminase. Proc. Natl. Acad. Sci. USA. 111:18667–18672. 10.1073/pnas.1420575111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West K.L., Singha N.C., De Ioannes P., Lacomis L., Erdjument-Bromage H., Tempst P., and Cortes P.. 2005. A direct interaction between the RAG2 C terminus and the core histones is required for efficient V(D)J recombination. Immunity. 23:203–212. 10.1016/j.immuni.2005.07.004 [DOI] [PubMed] [Google Scholar]
- Yu K., and Lieber M.R.. 2000. The nicking step in V(D)J recombination is independent of synapsis: Implications for the immune repertoire. Mol. Cell. Biol. 20:7914–7921. 10.1128/MCB.20.21.7914-7921.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., McCord R.P., Ho Y.-J., Lajoie B.R., Hildebrand D.G., Simon A.C., Becker M.S., Alt F.W., and Dekker J.. 2012. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 148:908–921. 10.1016/j.cell.2012.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Frock R.L., Du Z., Hu J., Chen L., Krangel M.S., and Alt F.W.. 2016. Orientation-specific RAG activity in chromosomal loop domains contributes to Tcrd V(D)J recombination during T cell development. J. Exp. Med. 213:1921–1936. 10.1084/jem.20160670 [DOI] [PMC free article] [PubMed] [Google Scholar]