Stanley et al. show that loss of Nol3 in mice leads to a myeloproliferative phenotype resembling primary myelofibrosis, with activation of JAK–STAT signaling and significant cellular and molecular resemblance to human disease. These findings provide a novel role for Nol3 in hematopoiesis and myeloid malignancies.
Abstract
Despite the identification of several oncogenic driver mutations leading to constitutive JAK–STAT activation, the cellular and molecular biology of myeloproliferative neoplasms (MPN) remains incompletely understood. Recent discoveries have identified underlying disease-modifying molecular aberrations contributing to disease initiation and progression. Here, we report that deletion of Nol3 (Nucleolar protein 3) in mice leads to an MPN resembling primary myelofibrosis (PMF). Nol3−/− MPN mice harbor an expanded Thy1+LSK stem cell population exhibiting increased cell cycling and a myelomonocytic differentiation bias. Molecularly, this phenotype is mediated by Nol3−/−-induced JAK–STAT activation and downstream activation of cyclin-dependent kinase 6 (Cdk6) and Myc. Nol3−/− MPN Thy1+LSK cells share significant molecular similarities with primary CD34+ cells from PMF patients. NOL3 levels are decreased in CD34+ cells from PMF patients, and the NOL3 locus is deleted in a subset of patients with myeloid malignancies. Our results reveal a novel genetic PMF-like mouse model and identify a tumor suppressor role for NOL3 in the pathogenesis of myeloid malignancies.
Introduction
Myeloproliferative neoplasms (MPNs) are a group of related but heterogeneous clonal hematopoietic malignancies that can be classified by the presence of the Philadelphia chromosome (Ph). Ph− MPNs include essential thrombocythemia (ET), polycythemia vera (PV), and primary myelofibrosis (PMF). ET and PV are characterized by increased platelet counts and erythrocyte counts, respectively, whereas PMF is characterized by peripheral cytopenias, extramedullary hematopoiesis, and bone marrow fibrosis (Tefferi and Pardanani, 2015). Furthermore, of the Ph− MPNs, PMF has the most severe morbidity and greatest mortality, with the highest risk of leukemic transformation (Tefferi et al., 2014).
Ph− MPNs are clonal stem cell diseases united by the discovery of recurrent oncogenic driver mutations in JAK2, CALR, and MPL that lead to constitutive activation of the JAK–STAT signaling pathway (Baxter et al., 2005; James et al., 2005; Kralovics et al., 2005; Levine et al., 2005; Pikman et al., 2006; Klampfl et al., 2013; Nangalia et al., 2013). The advent of clinically approved JAK inhibitors such as ruxolitinib to treat Ph− MPNs has shown promising results in ameliorating symptoms; however, it does not affect allele burden or significantly alter the course of disease (Tefferi, 2012; Tefferi and Pardanani, 2015). Whereas oncogenic JAK2, CALR, and MPL all play key roles in disease initiation (James et al., 2005; Pikman et al., 2006; Elf et al., 2016), there are many unknown cooperating molecular and genetic aberrations that contribute to disease pathogenesis. Furthermore, the determination of clonal hierarchy and temporal order of mutational occurrence in Ph− MPNs has proven complex (Tefferi, 2010). Recently, several mutations in epigenetic regulators (IDH1/2, DNMT3A, andTET2), signaling molecules (SOCS, LNK, and CBL), splicing factors (SRSF2 and SF3B1), and even whole chromosomal rearrangements have been shown to precede driver mutation acquisition (Kralovics et al., 2006; Schaub et al., 2009, 2010; Wang et al., 2009; Abdel-Wahab et al., 2012; Tibes et al., 2012), highlighting the importance of these factors in contributing to Ph− MPN disease formation and pathogenesis. The identification and study of novel cellular and molecular mechanisms that contribute to Ph− MPN disease initiation is critical for understanding the molecular pathways that drive these diseases, and may also offer new opportunities for therapeutic intervention.
Nucleolar protein 3 (also known as apoptosis repressor with caspase recruitment domain [ARC]) is encoded by the Nol3 gene and is highly expressed in cardiac and skeletal myocytes, neurons, and β-cells of the pancreas (Geertman et al., 1996; Koseki et al., 1998; McKimpson et al., 2013). In these tissues, ARC is a potent inhibitor of cell death and has the unique ability to antagonize both the intrinsic and extrinsic apoptosis pathways (Nam et al., 2004). ARC has been shown to have increased expression in solid tumors and in blast cells of patients with acute myeloid leukemia (AML), and to mediate cellular responsiveness to pharmacologic apoptosis induction (Wang et al., 2005; Mercier et al., 2008; Carter et al., 2011; Medina-Ramirez et al., 2011; Mak et al., 2014a,b). Interestingly, a recent study also revealed that ARC may play a tumor suppressor role in renal cell carcinoma cells, suggesting dual roles for ARC in oncogenesis that may be cell type–dependent (Gobe et al., 2016). Despite its name, NOL3/ARC primarily resides in the cytoplasm in most cell types, but was reported to localize to the nucleus in some solid tumor cell lines (Mercier et al., 2005; Wang et al., 2005). ARC protein has been reported to suppress NF-κB pathway activation and to interact directly with p53 to disrupt its transcriptional activity in cancer cells (Foo et al., 2007; Kung et al., 2014). The role of ARC in normal and malignant hematopoiesis is largely unclear and has not yet been assessed using genetically engineered mouse models.
In this study, we investigated the functional and molecular consequences of loss of Nol3/ARC on the hematopoietic system. Unexpectedly, we found that Nol3−/− mice develop a progressive MPN with features resembling PMF, including thrombocytopenia, anemia, extramedullary hematopoiesis, bone marrow fibrosis, and an expanded stem cell compartment. Moreover, we show that increased JAK–STAT activation in the expanded stem cell compartment leads to enhanced cell cycling and a myelomonocytic differentiation bias that is dependent on CDK6 and Myc activation. Furthermore, we find that the Nol3−/− MPN phenotype shares significant molecular similarities with CD34+ cells from patients with PMF. Additionally, NOL3 levels are decreased in CD34+ cells of patients with PMF, and NOL3 is deleted in a subset of patients with myeloid malignancies. Our study provides a novel PMF-like mouse model with similarities to human PMF, implicates Nol3 as a negative modulator of JAK–STAT signaling, and reveals a tumor suppressor role for NOL3 in the pathogenesis of myeloid malignancies.
Results
Loss of Nol3 leads to peripheral cytopenias and extramedullary hematopoiesis
Loss of ARC protein expression in Nol3-null mice (Nol3−/−) was confirmed by Western blot analysis of heart tissue and bone marrow cells (Fig. S1, A and B). ARC is expressed in total bone marrow cells from Nol3+/+ mice and purification into mature lineage positive (Lin+) and immature (Lin−cKit+) cells markedly enriched for ARC expression in the immature bone marrow fraction (Fig. S1 B). Consistent with this, qPCR analysis of wild-type animals showed low or absent Nol3 transcript expression in sorted B cells, T cells, erythrocyte precursors, and mature myeloid blood cells; however, we detected elevated expression in the Lin−Sca-1+cKit+ (LSK) stem and progenitor cell compartment. Further purification of stem cell populations based on FLK2 and THY1 expression showed stable Nol3 expression in multipotent progenitor cells (MPPs; Lin−Sca-1+cKit+Flk2+Thy1−), short-term hematopoietic stem cells (ST-HSCs; Lin−Sca-1+cKit+Flk2+Thy1+), and long-term HSCs (LT-HSCs; Lin−Sca-1+cKit+Flk2−Thy1+; Fig. S1 C).
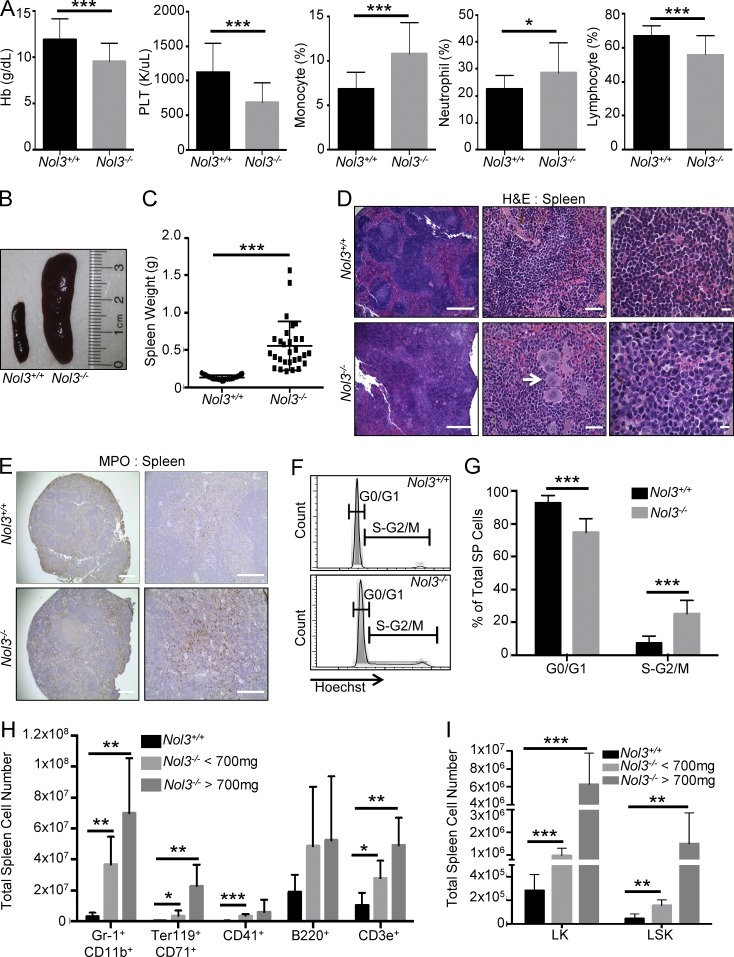
Peripheral blood analysis of 11–18-mo-old Nol3−/− mice showed anemia and thrombocytopenia, as well as significant increases in peripheral blood monocyte and neutrophil percentages with a decrease in lymphocyte percentage in comparison to age-matched Nol3+/+ mice (Fig. 1 A and Table S1). This phenotype was detectable in ∼20% of Nol3−/− analyzed mice. Strikingly, these mice displayed significant splenomegaly (>200 mg) and increased spleen cell number compared with Nol3+/+ mice (Fig. 1, B and C; and Fig. S1 D). Hematoxylin and eosin (H&E) staining of Nol3−/− spleens revealed severely disordered splenic architecture, including disruption of lymphoid follicles, an infiltration of larger cells, and megakaryocyte hyperplasia and clustering, which is a hallmark of MPN (Fig. 1 D). Additionally, Nol3−/− spleens showed increased staining for myeloperoxidase, indicative of active splenic myelopoiesis (Fig. 1 E). Interestingly, no significant difference in the percentage of apoptotic spleen cells in Nol3+/+ and Nol3−/− mice was observed (Fig. S1 E). Additionally, hematopoietic cell infiltration was present in the livers of Nol3−/− mice (Fig. S1 F). To determine if cells in Nol3−/− spleens were actively proliferating, we stained spleen cells with Hoechst 33342 and detected a significant increase in the percentage of cells in the S-G2/M phase of the cell cycle compared with Nol3+/+ (Fig. 1, F and G). We next used flow cytometry to identify the expanded cell types in Nol3−/− spleens and detected significant increases in the absolute number of almost all mature cell types analyzed, including granulocytes (Gr-1+CD11b+), erythrocyte precursors (Ter119+CD71+), megakaryocytes (CD41+), B cells (B220+), and T cells (CD3e+; Fig. 1 H). Elevation of numbers of myelo-erythroid cells correlated with overall spleen weight. We also observed significant increases in immature LK (Lin−Sca-1−cKit+) and LSK cells in the spleen (Fig. 1 I). Collectively, these findings are consistent with active extramedullary hematopoiesis in the spleens of Nol3−/− mice.
Figure 1.
Nol3 deletion leads to peripheral cytopenias and extramedullary hematopoiesis. (A) Hemoglobin (Hb), platelet count (PLT), monocyte, neutrophil, and lymphocyte percentage in the peripheral blood of Nol3+/+ and Nol3−/− mice (Nol3+/+, n = 20; Nol3−/−, n = 25). (B) Representative images of spleens from Nol3+/+ and Nol3−/− mice. (C) Spleen weights of Nol3+/+ and Nol3−/− mice (Nol3+/+, n = 23; Nol3−/−, n = 29). (D) H&E staining of Nol3+/+ and Nol3−/− spleens. White arrow highlights megakaryocyte clustering in Nol3−/− spleens. Bars: (left) 400 µm; (middle) 80 µm; (right) 10 µm. (E) Representative images of myeloperoxidase staining of Nol3+/+ and Nol3−/− spleens. Bars: (left) 1,000 µm; (right) 200 µm. (F and G) Representative histogram plots and quantification of Hoechst-stained spleen cells for cell cycle analysis of Nol3+/+ and Nol3−/− spleens (Nol3+/+, n = 7; Nol3−/−, n = 5). (H) Total cell numbers of mature cells of different lineages in Nol3+/+ and Nol3−/− spleens (Nol3+/+, n = 6; Nol3−/− < 700 mg, spleen, n = 5; Nol3−/− > 700 mg, spleen, n = 3). (I) Total cell numbers of LK and LSK cells in Nol3+/+ and Nol3−/− spleens (Nol3+/+ n = 7, Nol3−/− < 700 mg spleen weight n = 4, Nol3−/− > 700 mg spleen weight n = 2). Bars represent mean values. Error bars represent ±SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Myeloproliferation and myelofibrosis in the bone marrow of Nol3−/− mice
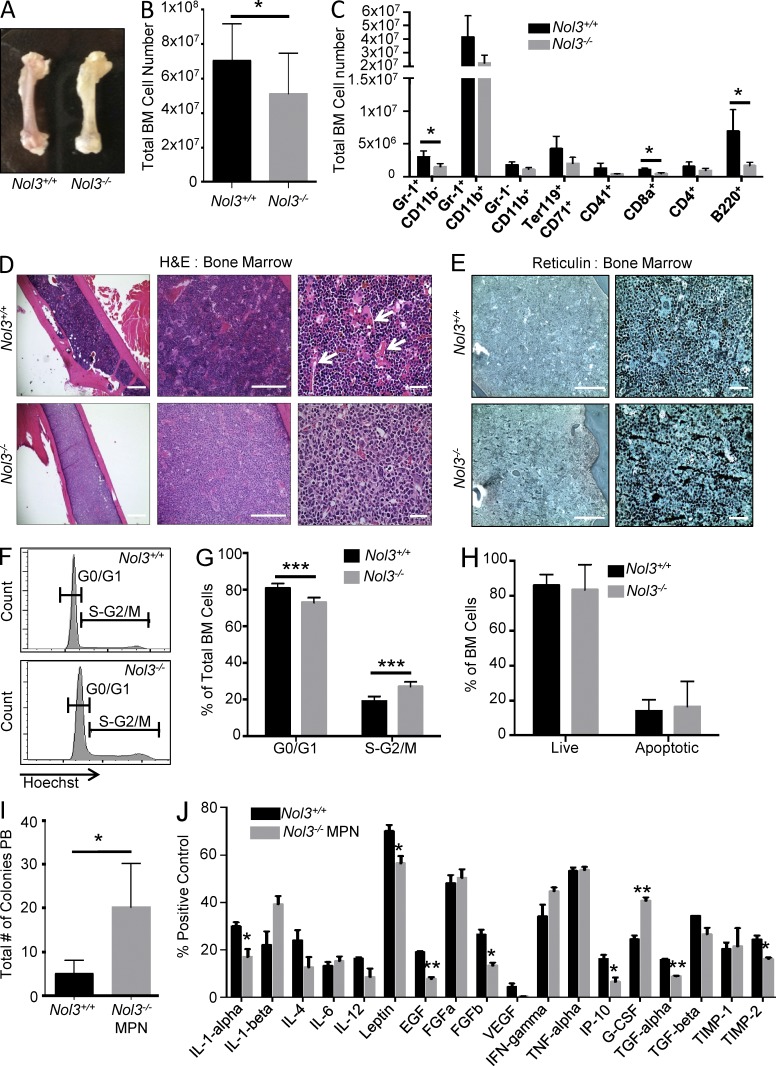
Investigation of femurs from Nol3−/− animals showed a macroscopically pale appearance, and cell counts revealed a reduction in total bone marrow, including both myeloid and lymphoid cell populations compared with Nol3+/+ mice (Fig. 2, A–C). H&E staining of femur sections showed a reduction in blood sinusoids and significant reticulin staining, a marker of bone marrow fibrosis that is frequently found in patients with PMF (Fig. 2, D and E). Cell cycle analysis of Nol3−/− bone marrow cells showed a significant increase in the percentage of cells in the S-G2/M phase of the cell cycle compared with Nol3+/+ cells (Fig. 2, F and G). As in the spleen, comparison of Nol3+/+ and Nol3−/− cells in the bone marrow showed no significant difference in the percentage of cells undergoing apoptosis, suggesting that ARC may not be essential to apoptosis inhibition in hematopoietic cells (Fig. 2 H; also see Fig. S1 E).
Figure 2.
Increased proliferation and myelofibrosis in Nol3−/− bone marrow. (A) Representative femurs from Nol3+/+ and Nol3−/− mice. (B) Total number of cells in Nol3+/+ and Nol3−/− bone marrow (Nol3+/+, n = 17; Nol3−/−, n = 10). (C) Absolute number of mature cell types in Nol3+/+ and Nol3−/− bone marrow (Nol3+/+, n = 4; Nol3−/−, n = 5). (D) H&E staining of Nol3+/+ and Nol3−/− bone marrow sections. Arrows highlight blood sinusoids present in Nol3+/+ femurs. Bars: (left) 1,000 µm; (middle) 200 µm; (right) 80 µm. (E) Reticulin staining of Nol3+/+ and Nol3−/− bone marrow sections. Bars: (left) 200 µm; (right) 80 µm. (F and G) Representative histogram plots and quantification of Hoechst stained bone marrow cells for cell cycle analysis of Nol3+/+ and Nol3−/− bone marrow (Nol3+/+, n = 7; Nol3−/−, n = 5). (H) Percentage of live and apoptotic cells in the bone marrow of Nol3+/+ and Nol3−/− mice measured by Annexin V and DAPI staining (n = 6). (I) Peripheral blood colony assay of Nol3+/+ and Nol3−/− MPN mice (Nol3+/+, n = 5; Nol3−/− MPN, n = 4). (J) Cytokine measurements from pooled serum samples of Nol3+/+ or Nol3−/− mice (n = 6). Bars represent mean values. Error bars represent ±SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The abnormal peripheral blood counts, extramedullary hematopoiesis with megakaryocyte hyperplasia and clustering, increased cell proliferation, and presence of bone marrow fibrosis in Nol3-/- mice are features consistent with an MPN phenotype that resembles PMF (now termed Nol3−/− MPN). Congruous with this PMF-like phenotype, we observed a significant increase in colony formation from peripheral blood cells of Nol3−/− MPN mice, indicative of increased circulating stem and progenitor cells, which is frequently observed in patients with PMF (Fig. 2 I; Andréasson et al., 2002). In addition, it has been shown that aberrant expression and activity of cytokines plays an integral role in the pathogenesis of MPNs and PMF. To identify cytokines involved in Nol3−/− MPN, we profiled the serum of Nol3+/+ and Nol3−/− MPN mice and found a significant increase in circulating granulocyte-colony stimulating factor (G-CSF), as well as elevated circulating IL-1-β (Fig. 2 J), both of which have been previously implicated in the pathogenesis of human PMF (Tefferi et al., 2011).
Nol3−/−-induced MPN is intrinsic to hematopoietic cells and transplantable to irradiated congenic recipients
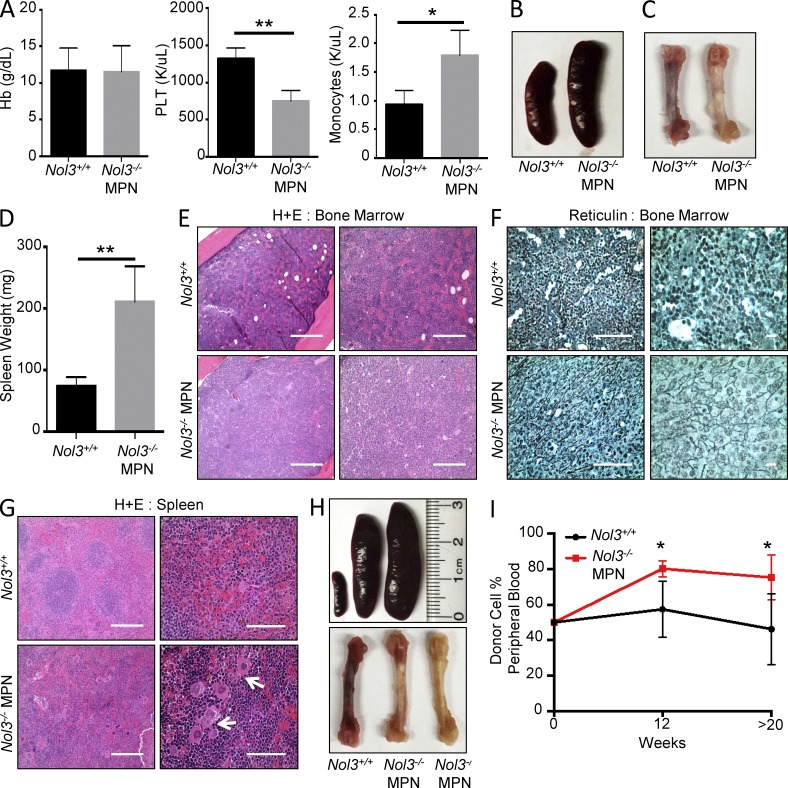
To determine whether the MPN observed in Nol3−/− mice was intrinsic to hematopoietic cells, we transplanted total bone marrow cells from Nol3+/+ and Nol3−/− MPN (CD45.2+) mice into lethally irradiated CD45.1+ wild-type recipients. At 6–8 mo after transplantation all recipient mice were sacrificed for detection of donor-derived disease. All recipient mice evaluated for MPN initiation had >93% donor cell chimerism in the peripheral blood at time of sacrifice (Fig. S1 G). Transplantation of bone marrow from 11 individual Nol3−/− MPN mice resulted in four recipient mice (36.4%) developing donor-derived disease, whereas no Nol3+/+ recipients developed MPN. The MPN phenotype in Nol3−/− recipient mice was indistinguishable from the MPN in donor mice, with pale femurs, splenomegaly, decreased platelet counts, and increased monocyte counts in the peripheral blood, in comparison to Nol3+/+ cell recipients (Fig. 3, A–D). In addition, H&E and reticulin staining showed reduced blood sinuses and increased reticulin staining in the bone marrow, as well as severely disorganized splenic architecture, including disruption of lymphoid follicles and clustering of megakaryocytes, further confirming the MPN phenotype in recipient mice (Fig. 3, E–G). Interestingly, congenic transplantation of total spleen cells from three individual Nol3−/− MPN mice also generated donor-derived MPN in two recipient mice (66.6%) at time of sacrifice, indicating that disease-initiating cells of Nol3−/− MPN are present and transplantable from either hematopoietic compartment (Fig. 3 H and Fig. S1 H).
Figure 3.
Nol3−/−-induced MPN is cell intrinsic and transplantable into congenic recipients. (A) Peripheral blood counts of mice transplanted with either Nol3+/+ or Nol3−/− MPN whole bone marrow cells. Hemoglobin (Hb), platelet (PLT), and monocyte counts are shown (n = 4). (B and C) Representative spleen and bone marrow images of recipient mice after transplantation with Nol3+/+ or Nol3−/− MPN total bone marrow cells. (D) Spleen weight of mice transplanted with either Nol3+/+ or Nol3−/− MPN cells (n = 4). (E) Representative H&E of bone marrow sections from congenic recipient mice after transplantation with Nol3+/+ or Nol3−/− MPN whole bone marrow cells. Bars: (left) 400 µm; (right) 200 µm. (F) Representative reticulin staining of bone marrow sections from congenic recipient mice after transplantation with Nol3+/+ or Nol3−/− MPN whole bone marrow cells. Bars: (left) 80 µm; (right) 10 µm. (G) Representative H&E staining of spleen sections from congenic recipient mice after transplantation with Nol3+/+ or Nol3−/− MPN cells. White arrows highlight megakaryocyte clustering in mice transplanted with Nol3-deficient cells. Bars: (left) 200 µm; (right) 80 µm. (H) Images of spleen and femoral bones of congenic recipient mice after transplantation with Nol3+/+ and Nol3−/− MPN total spleen cells. (I) Percentage of peripheral blood donor cell engraftment at 12 and >20 wk after competitive transplantation of either Nol3+/+ or Nol3−/− cells with wild-type competitor cells (Nol3+/+, n = 6; Nol3−/− MPN, n = 5). Bars represent mean values. Error bars represent ±SD; *, P < 0.05; **, P < 0.01.
To determine whether Nol3−/− MPN bone marrow cells have a competitive advantage, we performed competitive repopulation transplantation assays and found that Nol3−/− MPN outcompeted Nol3+/+ bone marrow cells in recipient mice at 12 and >20 wk after transplantation (Fig. 3 I). Consistent with this, mice that received Nol3−/− MPN marrow had significantly larger spleens and higher amounts of pale bone marrow compared with mice that received Nol3+/+ marrow, suggesting that Nol3−/− MPN bone marrow cells exhibit increased self-renewal capacity compared with Nol3+/+ cells in vivo (Fig. S1, I and J). Interestingly, competitive stem cell transplantation experiments revealed no significant difference in engraftment between sorted Nol3+/+ and Nol3−/− MPN stem cell populations at 16 wk after transplantation (Fig. S1 K), suggesting that the competitive advantage of Nol3−/− MPN bone marrow cells is caused by the expansion of immature cellular subsets rather than increased self-renewal of individual stem cells.
Although there is strong evidence that MPNs are clonal stem cell diseases (Adamson et al., 1976; Jacobson et al., 1978; Fialkow et al., 1981), a possible involvement of bone marrow stromal cells in MPN disease pathogenesis has been suggested in several mouse models (Walkley et al., 2007a,b; Zimmer et al., 2011). To determine if nonhematopoietic stromal cell populations of Nol3−/− mice can induce the MPN phenotype, we performed reciprocal transplantation experiments. Congenic FACS-purified wild-type CD45.1+ cells were transplanted into 3-mo-old lethally irradiated Nol3+/+ (n = 6) or Nol3−/− (n = 13) CD45.2+ mice and sacrificed 11–12 mo after transplantation. There were no significant differences in spleen size or peripheral blood counts in Nol3 experimental mice with >65% donor cell engraftment, and 0/13 Nol3−/− recipient mice developed donor-derived disease (Figs. S1, L and M; and Table S1). Collectively, these results support a hematopoietic cell–intrinsic process in which the Nol3−/− MPN is induced by deletion of Nol3 in hematopoietic cells and can be recapitulated in transplantation experiments in congenic mice.
Hematopoietic stem and progenitor cells of Nol3−/− MPN mice show active proliferation and a myelomonocytic differentiation bias
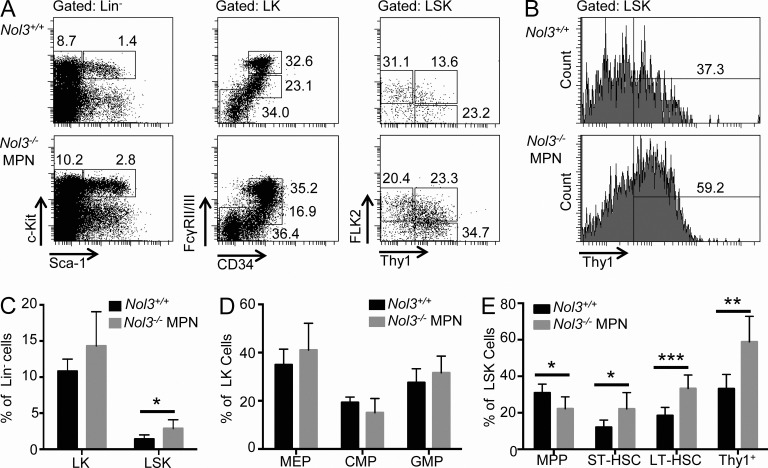
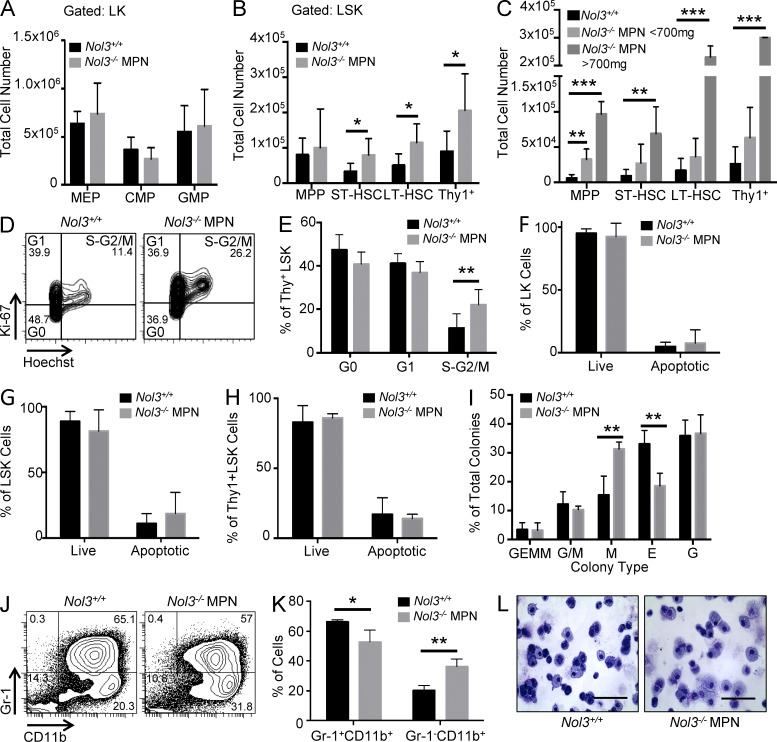
Investigation of hematopoietic stem and progenitor cell compartments by FACS revealed an increase in the percentage of LSK cells in Nol3−/− MPN mice compared with Nol3+/+ mice but no significant changes in phenotypically defined myeloid progenitor subpopulations (Fig. 4, A–D). Further analysis of the LSK population revealed a decrease in the percentage of MPPs but an increase in ST- and LT-HSCs (Fig. 4 E). Overall, there was a significant expansion in the percentage of Thy1+LSK cells in Nol3−/− MPN mice (Fig. 4, B and E). To verify an expanded stem cell population in the setting of an overall reduction in total bone marrow cellularity in Nol3−/− MPN (Fig. 2 B), we determined absolute numbers of stem and progenitor cells. We found a statistically significant increase in the absolute numbers of ST- and LT-HSC, as well as Thy1+LSK cells, with no change in the absolute numbers of progenitor cell populations (Fig. 5, A and B). Notably, absolute numbers of MPP, ST-HSC, LT-HSC, and Thy1+LSK cells were also elevated in spleens of Nol3−/− MPN mice (Fig. 5 C).
Figure 4.
Increased stem cell populations in Nol3−/− MPN mice. (A) Representative FACS plots of Nol3+/+ and Nol3−/− MPN bone marrow cells. (left) Lineage− bone marrow cells gated on c-Kit and Sca-1 to distinguish LK and LSK cells. (middle) LK cells co-stained with FcγRII/III and CD34 to distinguish common myeloid progenitor cells (CMPs; Lin−cKit+Sca-1− CD34+FcγRII/IIIlo), granulocyte/monocyte progenitor cells (GMPs; Lin−cKit+Sca-1−CD34+FcγRII/IIIhi), and megakaryocytic/erythroid progenitor cells (MEPs; Lin−cKit+Sca-1−CD34−FcγRII/IIIlo). (right) LSK cells co-stained with FLK2 and THY1 to distinguish MPP, ST-HSC, and LT-HSC. Percentages of cell populations are indicated. (B) Histogram plot of Thy1+LSK cells from Nol3+/+ and Nol3−/− MPN mice. (C) Percentage of LK and LSK cells in bone marrow of Nol3+/+ and Nol3−/− MPN mice (Nol3+/+, n = 7; Nol3−/− MPN, n = 6). (D) Percentage of MEP, CMP, and GMP cells in bone marrow of Nol3+/+ and Nol3−/− MPN mice (Nol3+/+, n = 7; Nol3−/− MPN, n = 6). (E) Percentage of MPP, ST-HSC, LT-HSC, and Thy1+LSK cells in bone marrow of Nol3+/+ and Nol3−/− MPN mice (Nol3+/+, n = 7, Nol3−/− MPN, n = 6). Bars represent mean values. Error bars represent ±SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 5.
Expanded Thy1+LSK cells exhibit increased cycling and myeloid differentiation bias in Nol3−/− MPN. (A) Absolute numbers of MEP, CMP, and GMP cells in bone marrow of Nol3+/+ and Nol3−/− MPN mice (Nol3+/+, n = 7; Nol3−/− MPN, n = 6). (B) Absolute numbers of MPP, ST-HSC, LT-HSC, and Thy1+LSK cells in bone marrow of Nol3+/+ and Nol3−/− MPN mice (Nol3+/+, n = 7; Nol3−/− MPN, n = 6). (C) Total cell number of MPP, ST-HSC, LT-HSC, and Thy1+LSK in spleens of Nol3+/+ and Nol3−/− MPN mice (Nol3+/+, n = 7; Nol3−/− MPN < 700 mg, spleen, n = 4; Nol3−/− MPN > 700 mg, spleen, n = 2). (D and E) Representative FACS plots and quantification of the cell cycle status of Nol3+/+ and Nol3−/− MPN Thy1+LSK bone marrow cells based on Ki-67 and Hoechst staining (Nol3+/+, n = 7; Nol3−/− MPN, n = 8). (F–H) Percentage of live and apoptotic cells in bone marrow LK, LSK, and Thy1+LSK cells, respectively, from Nol3+/+ and Nol3−/− MPN mice measured by Annexin V and DAPI staining (n = 3). (I) Colony-forming capacity of sorted Nol3+/+ and Nol3−/− MPN Thy1+LSK (Nol3+/+, n = 6; Nol3−/− MPN, n = 3). (J) Representative FACS plots of Gr-1 and CD11b staining of colonies derived from Nol3+/+ and Nol3−/− MPN cells. (K) Percentage of Gr-1+CD11+ and Gr-1−CD11b+ cells after colony formation assay of Nol3+/+ and Nol3−/− MPN cells (n = 3). (L) Wright-Giemsa staining of cytospun cells after colony assay of Nol3+/+ and Nol3−/− MPN Thy1+LSK cells. Bars, 80 µm. Bars represent mean values. Error bars represent ±SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To investigate the cell biological phenotype of the expanded Thy1+LSK cells, we performed cell cycle analysis using Hoechst 33342 and the proliferation marker Ki67. Nol3−/− MPN Thy1+LSK cells showed significantly increased cell cycling compared with Nol3+/+ Thy1+LSK cells (Fig. 5, D and E). Remarkably, analysis of the percentage of apoptotic cells showed no significant difference between Nol3+/+ and Nol3−/− MPN Thy1+LSK cells as well as LK or LSK cells (Fig. 5, F–H). Methylcellulose colony assays demonstrated a commitment bias toward myeloid colony formation, with a significant increase in the percentage of CFU-M (monocyte) over CFU-E (erythroid) colonies from Nol3−/− MPN Thy1+LSK cells compared with Nol3+/+ Thy1+LSK control cells (Fig. 5 I). FACS analysis of colony assay cells showed a significant increase in the percentage of Gr-1−CD11b+ monocytic cells derived from Nol3−/− MPN Thy1+LSK cells, which was also evident by morphological analysis after May-Grünwald Giemsa staining (Fig. 5, J–L).
JAK–STAT signaling is activated in stem and progenitor cells in Nol3−/− MPN, and Cdk6 and Myc are functionally relevant downstream targets
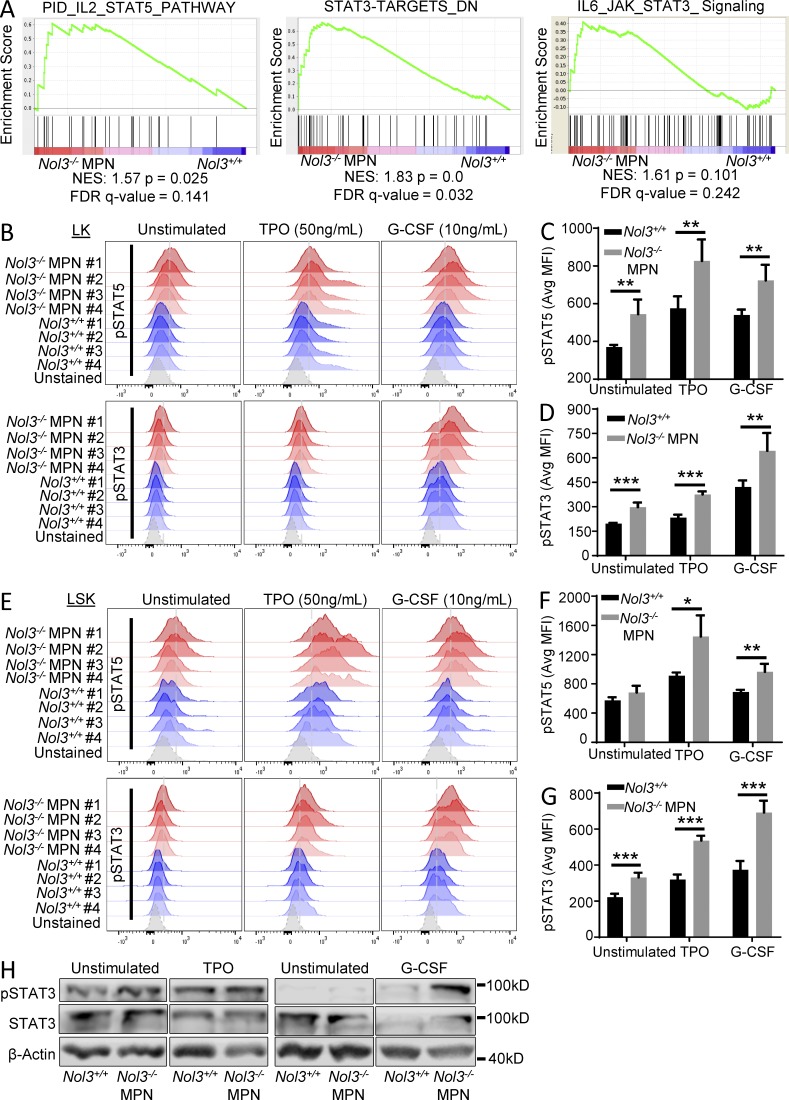
To obtain insight into the molecular consequences of Nol3 deletion in the expanded Thy1+LSK cell population, we performed gene expression profiling on FACS sorted Nol3+/+ and Nol3−/− MPN Thy1+LSK cells (complete data available in Gene Expression Omnibus [GEO] under accession no. GSE76121). Gene set enrichment analysis (GSEA) identified significant enrichment of STAT5 and STAT3 gene signaling pathways in Nol3−/− MPN compared with Nol3+/+ Thy1+LSK cells, suggesting JAK–STAT signaling involvement in Nol3−/− MPN (Fig. 6 A). We performed phospho-flow cytometry experiments of stem and progenitor cells and indeed found significant activation of pSTAT5 and pSTAT3 in Nol3−/− MPN bone marrow LK and LSK cells, demonstrating activation of JAK–STAT signaling in the Nol3−/− MPN model (Fig. 6, B–G). Western blot analysis confirmed increased activation of pSTAT3 in spleen cells from Nol3−/− MPN (Fig. 6 H). Notably, increased pSTAT5 and pSTAT3 activation in Nol3−/− MPN cells was also seen upon stimulation with thrombopoietin (TPO) and G-CSF (Fig. 6, B–H), cytokines that have been implicated in PMF and act on JAK–STAT signaling pathways (Oh et al., 2010; Tefferi et al., 2011).
Figure 6.
Nol3-deficient stem and progenitor cells show increased JAK–STAT activation. (A) GSEA of pathways enriched in Nol3−/− MPN Thy1+LSK cells compared with Nol3+/+ Thy1+LSK cells. (B) Phospho-flow analysis of pSTAT5 and pSTAT3 in LK cells of Nol3+/+ and Nol3−/− MPN mice, under unstimulated, TPO-stimulated (50 ng/ml), and G-CSF–stimulated (10 ng/ml) conditions (n = 4). (C and D) Quantification of phospho-flow analysis of pSTAT5 and pSTAT3, respectively, in LK cells from Nol3+/+ and Nol3−/− MPN mice (MFI, mean fluorescence intensity; n = 4). (E) Phospho-flow analysis of pSTAT5 and pSTAT3 in LSK cells of Nol3+/+ and Nol3−/− MPN mice, under unstimulated, TPO-stimulated (50 ng/ml), and G-CSF–stimulated (10 ng/ml) conditions (n = 4). (F and G) Quantification of phospho-flow analysis of pSTAT5 and pSTAT3, respectively, in LSK cells from Nol3+/+ and Nol3−/− MPN mice (MFI, mean fluorescence intensity; n = 4). (H) Western blot analysis of total spleen cells from Nol3+/+ and Nol3−/− MPN mice, under unstimulated, TPO-stimulated (50 ng/ml), G-CSF–stimulated (10 ng/ml) conditions (TPO, n = 3; G-CSF, n = 2). Bars represent mean values. Error bars represent ±SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
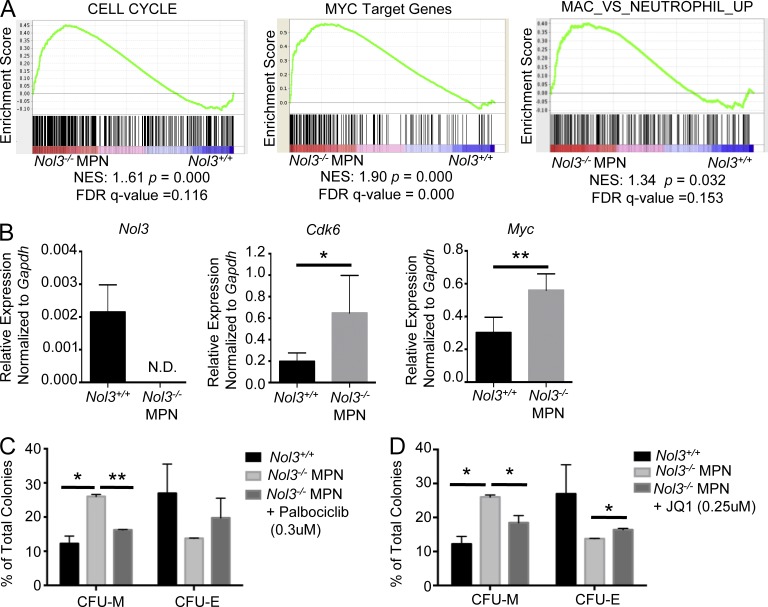
Further investigation of differentially expressed genes by GSEA uncovered signatures known to be downstream of JAK–STAT signaling that reflect the cellular phenotypes observed in Nol3−/− MPN Thy1+LSK cells, including cell cycle and Myc target signatures, as well as genes up-regulated in monocytic differentiation (Fig. 7 A). STAT5 and STAT3 have previously been shown to activate Myc and cell cycle regulators, including Cdk6 (Fukada et al., 1998; Kiuchi et al., 1999; Lin et al., 2012; Pinz et al., 2016). We validated Cdk6 and Myc up-regulation in Nol3−/− MPN Thy1+LSK cells by qRT-PCR (Fig. 7 B). To test whether the increase in Cdk6 and Myc expression in Nol3−/− MPN Thy1+LSK is functionally relevant, we treated Nol3−/− MPN LSK cells with two different small molecule inhibitors targeted toward these pathways and assessed colony formation capacity. Palbociclib is a CDK4/6 inhibitor that causes sustained cell cycle arrest (Fry et al., 2004) and JQ1 is a bromodomain/BRD4 inhibitor that causes repression of Myc transcription (Delmore et al., 2011). Strikingly, inhibition of CDK4/6 or Myc resulted in rescue of the myeloid bias observed in Nol3−/− MPN cells (Fig. 7, C and D). These data indicate that the Nol3−/−-induced phenotype in the immature stem cell population is, at least in part, mediated by activation of CDK4/CDK6 and Myc.
Figure 7.
Cdk6 and Myc are functionally relevant downstream targets in Nol3−/− MPN. (A) GSEA of pathways enriched in Nol3−/− MPN Thy1+LSK cells compared with Nol3+/+ Thy1+LSK cells. (B) Validation of Nol3 and dysregulated genes of interest from individual Nol3+/+ (n = 6) and individual Nol3−/− MPN Thy1+LSK (n = 4) cells by qPCR. (C) Colony formation assay of sorted Thy1+LSK or LSK cells isolated from Nol3+/+ and Nol3−/− MPN bone marrow or spleen and treated with DMSO or the CDK4/6 inhibitor Palbociclib Isethionate (n = 2). (D) Colony formation assay of sorted Thy1+LSK or LSK cells isolated from Nol3+/+ and Nol3−/− MPN bone marrow or spleen and treated with DMSO or JQ1 (n = 2). Bars represent mean values. Error bars represent ±SD; *, P < 0.05; **, P < 0.01.
NOL3 is functionally relevant in human PMF cells and frequently down-regulated in CD34+ cells of PMF patients, and deletions of the NOL3 locus are found in patients with myeloid malignancies
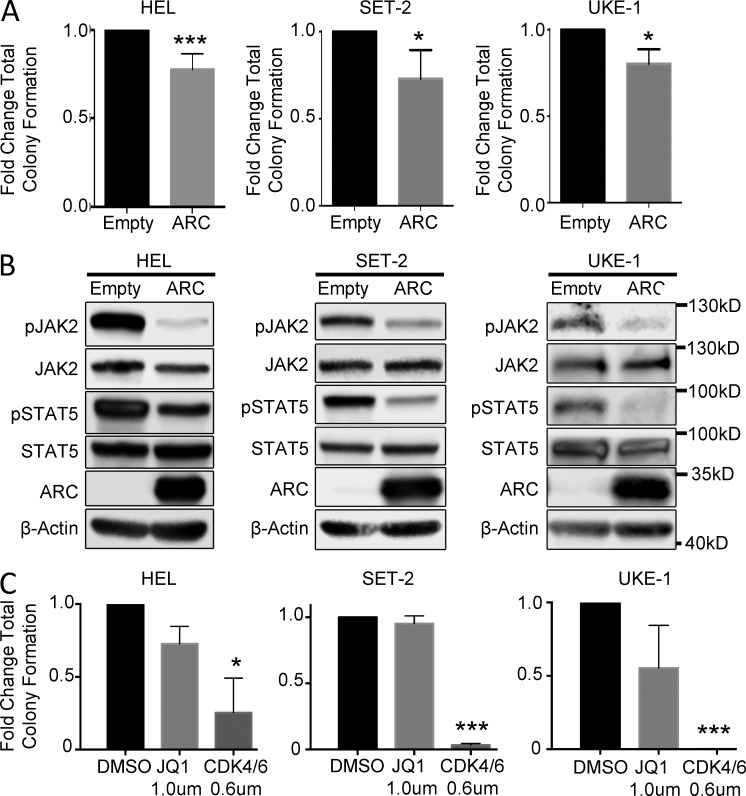
To interrogate the functional significance of NOL3/ARC in human MPN cells, we ectopically re-expressed ARC protein in HEL cells (which carry a deletion of the NOL3 locus), as well as SET-2 and UKE-1 cell lines. Ectopic expression of ARC protein led to a significant reduction in colony formation and inhibition of the JAK–STAT signaling pathway in all three cell lines compared with lentiviral vector control cells (Fig. 8, A and B). Additionally, colony formation capacity of all three cell lines was significantly inhibited with CDK4/6 inhibitor treatment (Fig. 8 C). These results implicate ARC as a negative modulator of JAK–STAT signaling and as functionally relevant in human MPN.
Figure 8.
Ectopic expression of ARC in human MPN cell lines leads to decreased colony formation and negative regulation of JAK–STAT signaling. (A) Colony formation assays of HEL (n = 5), SET-2 (n = 3), and UKE-1 (n = 3) cells transduced with either empty vector control or human ARC-expressing lentivirus. (B) Western blot analysis of HEL, SET-2, and UKE-1 cells transduced with either empty or human ARC-expressing vector (n = 3). (C) Colony formation assay of HEL, SET-2, and UKE-1 cells treated with DMSO, JQ1, or the CDK4/6 inhibitor Palbociclib Isethionate (n = 2). Bars represent mean values. Error bars represent ±SD; *, P < 0.05; ***, P < 0.001.
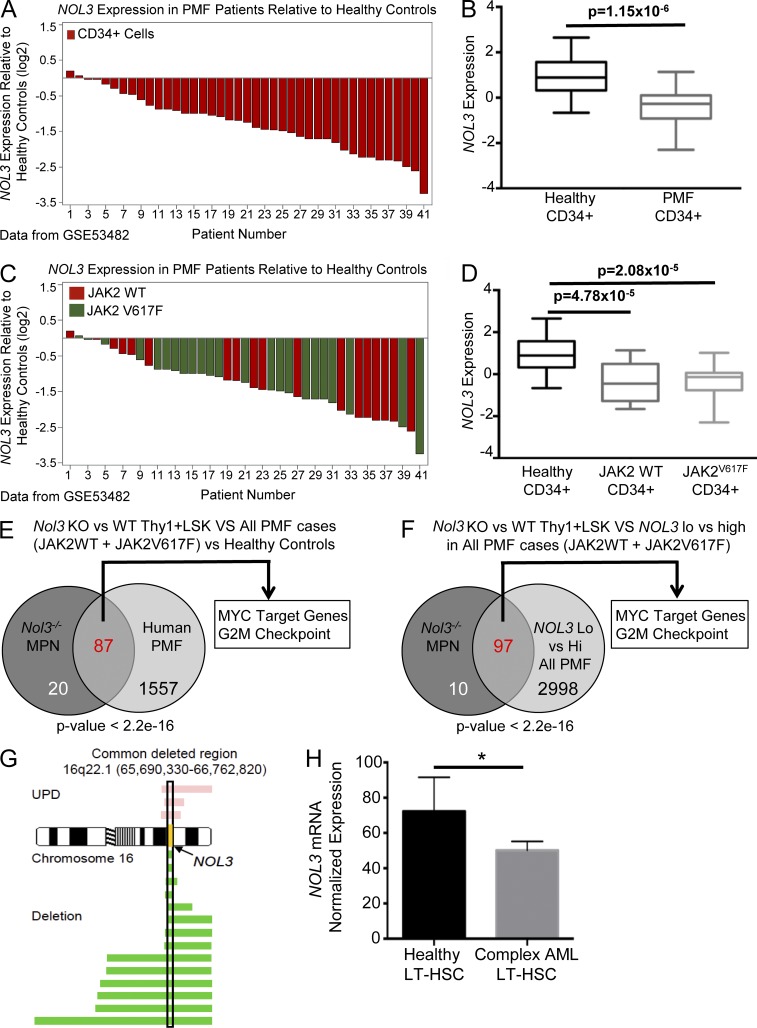
To interrogate whether the molecular alterations seen in the Nol3−/− MPN mouse model resemble human PMF, we analyzed gene expression data of isolated CD34+ cells from PMF patients (available in GEO under accession no. GSE53482; Norfo et al., 2014). Investigation of NOL3 mRNA expression in the context of human PMF (accession no. GSE53482) revealed a significant reduction in NOL3 expression in CD34+ cells from the majority of PMF patients compared with healthy control cells (Fig. 9, A and B). Interestingly, NOL3 expression was significantly decreased compared with healthy controls regardless of the absence (JAK2WT) or presence of JAK2V617F mutations and there was no significant difference between these two mutational groups (Fig. 9, C and D).
Figure 9.
Nol3-deficient Thy1+LSK cells have a molecular phenotype similar to human PMF, and NOL3 is deleted and down-regulated in patients with myeloid malignancies. (A) Waterfall plot of NOL3 expression in CD34+ cells from 41 patients with PMF relative to the mean expression of NOL3 in CD34+ cells from 16 healthy controls (GEO accession no. GSE53482). (B) Box plot summary of NOL3 expression in CD34+ cells from patients with PMF in comparison to CD34+ cells from healthy controls (GSE53482). (C) Waterfall plot of relative expression of NOL3 in CD34+ cells from 41 patients with PMF (same data as in A) with indicated JAK2V617F mutation status relative to the mean expression of NOL3 in CD34+ cells from 16 healthy controls (GSE53482). (D) Box plot summary of NOL3 expression in CD34+ cells from patients with PMF with or without JAK2V617F mutations in comparison to CD34+ cells from healthy donors (GSE53482). (E) Comparative GSEA analysis depicting significantly positively enriched gene sets (FDR q-value < 0.01) in Nol3−/− MPN (vs. Nol3+/+) Thy1+LSK cells and CD34+ cells from PMF patients (vs. healthy controls). P < 2.2 × 10−16; odds ratio, 28.19. (F) Comparative GSEA analysis depicting significantly positively enriched gene sets (FDR q-value < 0.01) in Nol3−/− MPN (vs. Nol3+/+) Thy1+LSK cells and CD34+ cells from PMF patients dichotomized by high and low NOL3 expression. P < 2.2 × 10−16; odds ratio, 27.99. (G) Commonly deleted region (hg18) on chromosome 16 in human patients with myeloid malignancies. Yellow, NOL3 locus; pink bars, length of region with UPD; green bars, length of chromosomal deletion. (H) NOL3 mRNA normalized expression in sorted LT-HSCs from healthy control subjects (n = 4) and from patients with complex karyotype AML (n = 5). Bars represent mean values. Error bars represent ±SD; *, P < 0.05.
Comparative GSEA analysis of differentially expressed gene sets in Nol3−/− MPN Thy1+LSK cells (relative to age-matched Nol3+/+ controls) identified overlapping gene sets that are also enriched in human CD34+ cells from patients with PMF (in comparison to healthy controls). We found significant positive enrichment of 87 shared gene sets between murine Nol3−/− MPN and human PMF cells (Fig. 9 E; FDR < 0.01; P < 2.2 × 10−16; odds ratio, 28.19), including enrichment of gene sets involved in cell cycle regulation and MYC target gene expression. Furthermore, dichotomization of PMF patients into higher and lower NOL3 expression showed a significant enrichment of 97 gene sets in NOL3-low expressing patients compared with Nol3−/− MPN mice (Fig. 9 F; FDR < 0.01; P < 2.2 × 10−16; odds ratio, 27.99). Collectively, these findings show that the expanded Thy1+LSK cells in Nol3−/− MPN share significant molecular features with CD34+ cells from patients with PMF.
To investigate whether deletion or loss of heterozygosity of NOL3 occurs in human myeloid malignancies, we analyzed single nucleotide polymorphism (SNP) array data of patients with MPN, AML, chronic myelogenous leukemia, and myelodysplastic syndrome (MDS). We identified 17 patients (1.57% of all patients analyzed) with deletion or uniparental disomy (UPD) of NOL3 (Fig. 9 G; for patient characteristics see Table S2). 7 of the 17 patients with NOL3 deletion were diagnosed with AML with myelodysplasia-related changes (AML-MRC), an AML that occurs in patients with a previous history of MDS or MDS/MPN. For 7 out of the 17 patients with NOL3 deletion or UPD, diagnostic fibrosis data were available. Strikingly, five out of these seven patients were positive for bone marrow fibrosis (not depicted).
We also determined if NOL3 expression is decreased in more stringently defined human hematopoietic stem cells from patients with myeloid malignancies. Microarray data from our laboratory revealed an ∼30% reduction in NOL3 mRNA expression in highly purified human LT-HSCs (Lin−CD34+CD38−CD90+) isolated from patients with AML with complex karyotype (three or more cytogenetic abnormalities) compared with age-matched healthy control cells (Fig. 9 H; Barreyro et al., 2012).
Collectively, these results support a role for NOL3, including at the HSC level in human MPN and other myeloid malignancies.
Discussion
The discovery of oncogenic driver mutations that constitutively activate JAK–STAT signaling has been vital to understanding the pathophysiology of Ph− MPNs. Targeting of the JAK–STAT pathway has shown promising results in alleviating symptoms; however, it has not significantly altered the course of disease, suggesting that additional underlying factors play key roles in Ph− MPN pathophysiology.
Our results show that Nol3 is primarily expressed in hematopoietic stem and progenitor cells and that loss of Nol3 in mice confers phenotypic and molecular similarities to human PMF. Nol3−/− MPN mice develop an expanded Thy1+LSK stem cell population that exhibits functional alterations in cell cycling, as well as a bias toward myelomonocytic differentiation. Cytokines play a major role in the pathogenesis of PMF, and our results show elevated levels of circulating G-CSF in Nol3−/− MPN mice. Furthermore, Nol3−/− MPN stem cells show increased activation of JAK–STAT signaling in response to stimulation by either TPO or G-CSF, cytokines elevated and implicated in MPN (Panteli et al., 2005). Activation of both STAT5 and STAT3 is present in Nol3−/− MPN mice and subsequently leads to elevation of downstream effectors Cdk6 and Myc (Fukada et al., 1998; Kiuchi et al., 1999; Lin et al., 2012; Pinz et al., 2016), which we found to be functionally relevant targets in Nol3−/− MPN stem cells.
ARC has primarily been described in the context of apoptosis inhibition of the extrinsic and intrinsic apoptosis pathways in many tissues. Our study has uncovered a novel tumor suppressor-like function for ARC within the hematopoietic system. In support of new functions of ARC, several recent papers have shown that ARC can negatively regulate NF-κB signalingand that it may also serve a tumor suppressor role in renal cell carcinoma cells (Kung et al., 2014; Gobe et al., 2016). Our study further suggests dual roles for Nol3/ARC in oncogenesis and hematopoiesis that may be cell context– and cell type–dependent. ARC has been reported to localize to both the cytoplasm and the nucleus, depending on cell type. ARC is primarily cytoplasmic in most cell types that have been studied; however, it has been reported to localize to the nucleus in some solid tumor cell lines in one study (Mercier et al., 2005; Wang et al., 2005). Interestingly, Western blot fractionation experiments of four AML cell lines show that ARC protein expression is primarily localized to the cytoplasm (Fig. S1 N), supporting a cytoplasmic function for ARC in normal and malignant hematopoiesis.
Given that loss of Nol3/ARC leads to increased JAK–STAT activation in mouse hematopoiesis, and that ectopic expression of ARC in MPN cell lines leads to repression of JAK–STAT signaling, our data provide evidence for ARC as a negative modulator of JAK–STAT signaling in the context of the hematopoietic system. ARC could, for instance, be behaving as a scaffolding protein downstream of hematopoietic cytokine receptors that has the ability to inhibit signaling, similar to other scaffolding proteins involved in MPN, such as LNK (Tong and Lodish, 2004; Oh et al., 2010). Further studies are warranted to investigate these interesting possibilities.
Gene expression analysis showed that NOL3 is decreased in CD34+ cells in the majority of PMF patients, and SNP array data showed NOL3 deletion occurs, albeit not at a high frequency, in patients with myeloid malignancies. Importantly, 0/72 healthy controls assayed by SNP array had UPD/deletion of NOL3, suggesting that NOL3 loss specifically occurs in patients with myeloid malignancies. Mutational data were available in four of the 17 patients with NOL3 deletion (Table S2), and additional investigation of TCGA AML patient data showed five patients with either deep or shallow deletions of NOL3. 3/9 NOL3 deletion patients from these two datasets in which mutational data were available showed mutations in TP53. All other leukemia-related mutations did not occur more than once. Interestingly, JAK2V617F and TP53 mutations coexist in the SET-2 cell line, as well as the HEL cell line, which additionally harbors a deletion containing the NOL3 locus (Zhao et al., 2012). The association of NOL3 deletion and TP53 mutational status may be relevant, as ARC has been shown to bind and inhibit p53 function in the context of malignancy (Foo et al., 2007). Interestingly, there was no obvious correlation with NOL3 expression and deletion; however, the samples used in the SNP array were unfractionated bulk cells, and our data suggest that NOL3 is primarily expressed at the stem and progenitor cell level; further studies using precisely fractionated immature cell types will be required.
In summary, our studies provide a novel mouse model in which genetic deletion of Nol3 leads to development of a transplantable MPN that resembles PMF and shares key phenotypic, cell biological, and molecular characteristics with human disease. Nol3−/−-induced disease is characterized by an expanded immature Thy1+LSK cell population that is actively proliferating and skewed toward myelomonocytic differentiation. Molecularly, this phenotype is mediated by increased activation of STAT5 and STAT3, leading to functionally relevant up-regulation of Cdk6 and Myc. Furthermore, we found NOL3 levels to be decreased in CD34+ cells of the majority of patients with PMF, and identified deletions of the NOL3 gene locus in a subset of patients with different myeloid malignancies. Our study provides a new PMF-like mouse model and reveals a novel tumor suppressor role of NOL3 in the pathogenesis of MPN.
Materials and methods
Mice
Nol3−/− mice (deletion encompassing the entire open reading frame of ARC) were kindly provided by R.N. Kitsis (Albert Einstein College of Medicine, Bronx, NY) (Zaiman et al., 2011). Nol3+/+ and Nol3−/− mice were age matched for all experimental studies. C57BL/6 SJL-Ptprca Pepcb/BoyJ wild-type CD45.1+ mice were purchased from The Jackson Laboratory. All mice were housed in a special pathogen–free (SPF) barrier facility. All animal experiments were performed in compliance with institutional guidelines and approved by the Animal Institute Committee of the Albert Einstein College of Medicine (#20130102).
Cell counts
Total bone marrow cell counts were determined from tibiae, pelvic bones, and femurs for each mouse after erythrocyte lysis using ACK buffer, pH 7.4 (0.15 M NH4Cl, 10 mM KHCO3, and 1.0 mM EDTA). Spleen cell counts were determined from entire spleen tissue after erythrocyte lysis.
Complete blood counts
Peripheral blood was obtained from the facial vein of living mice or heart of euthanized mice and analyzed using the Forcyte Hematology Analyzer (Oxford Science Inc.) according to the manufacturer’s instructions.
Cell lines
The AML cell lines HEL 92.1.7, HL-60, MV4-11, and THP-1 were purchased from the American Type Culture Collection and grown in IMDM or RPMI medium supplemented with 10% FBS and 1% penicillin/streptomycin. The MOLM-14 cell line was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ) and grown in RPMI supplemented with 10% FBS and 1% penicillin/streptomycin. The SET-2 and UKE-1 cell lines were gifts from R. Levine (Memorial Sloan-Kettering Cancer Center, New York, NY). SET-2 cells were grown in RPMI medium supplemented with 10% FBS and 1% penicillin/streptomycin. UKE-1 cells were grown in IMDM supplemented with 10% FBS, 10% Horse Serum, 1 µM hydrocortisone, and 1% penicillin/streptomycin.
Lentiviral vectors and transduction
For reexpression studies, we introduced a DNA coding sequence that codes for human ARC protein into the EcoRI site of a pCAD-IRES-GFP lentiviral construct (Steidl et al., 2007). For production of lentiviral particles, lentiviral expression constructs were transfected together with packaging vectors into 293T producer cells using Fugene HD transfection reagent (Roche), supernatants were harvested after 48 and 72 h. The HEL, SET-2, and UKE-1 cell lines were transduced with human ARC coding IRES-GFP containing lentivirus (MOI 10). For expression of ARC in cell lines, spinfection of the lentiviral supernatant, incubated with 10 µg/ml polybrene for 1 h at 37°C and 1,000 RCF was performed. Cells were then put in culture and media was added 3 h after spinfection. After 48 h in culture at 37°C, the cells were washed with PBS and transduction efficiency was determined by flow cytometry. GFP+ cells were sorted using a FACS Aria II sorter (BD) and used for experiments.
Histology
Femurs, spleens, and livers were fixed for >24 h in 10% neutral buffered formalin at room temperature and subject to paraffin embedding according to standard protocols. Sections were cut using a microtome and were stained for H&E for histological analysis, reticulin stain for detection of reticulin fibers, and myeloperoxidase for detection of active myelopoiesis. Sections were imaged using EVOS Cell Imaging Station (Life Technologies) with 4×, 10×, 20×, and 50× objective lenses.
Western blotting
Lysates from whole heart tissue, whole bone marrow (after erythrocyte lysis), FACS-purified Lin+, Lin−c-kit+ bone marrow cells, and cell lines were prepared using modified RIPA buffer with protease and phosphatase inhibitors. Cells were lysed for 10 minat 4°C and sonicated at 4°C for 10 min. 8 µg of heart protein lysates, 150 µg of total bone marrow protein lysates, and 125 µg of sorted cell protein lysates were resolved on 10% polyacrylamide gels for SDS-PAGE. For fractionation, cells were first lysed with a modified cytoplasmic RIPA buffer, and then cytoplasmic fractions were isolated. Remaining nuclear pellets were lysed with a complete RIPA buffer and then sonicated for 10 min. For phospho-Western analysis, HEL, SET-2, and UKE-1 cells were lysed in a modified RIPA buffer with protease and phosphatase inhibitors, and 50 µg of lysate was used. From total spleen, 2 × 106 cells were stimulated with 50 ng/ml TPO, 10 ng/ml G-CSF, or PBS (unstimulated), and then lysed in modified RIPA buffer with protease and phosphatase inhibitors and subsequently resolved on 10% polyacrylamide gels. Immunoblotting was performed with a rabbit polyclonal antibody against ARC (1:200; Cayman Chemical) or Lamin-B1 (1:1,000; Abcam), and a monoclonal rabbit antibody against β-actin (1:1,000; Sigma-Aldrich). For phospho-western, membranes were incubated with polyclonal rabbit antibodies against JAK2 (1:1,000; Cell Signaling Technology), phospho-JAK2 (Tyr1007/1008; 1:1,000; Cell Signaling Technology), STAT5 (1:1,000; Cell Signaling Technology), phospho-STAT5 (Tyr694; 1:1,000; Cell Signaling Technology), STAT3 (1:1,000; Cell Signaling Technology), and phospho-STAT3 (Tyr705; 1:1,000; Millipore). Primary antibodies were incubated overnight at 4°C in blocking solution (PBS containing 0.1% [vol/vol] Tween-20 with 5% [wt/vol] nonfat dry milk powder) or 2% BSA in PBS. Secondary antibody anti–rabbit IgG horseradish peroxidase (HRP)–conjugated (Santa Cruz Biotechnology, Inc.) at either 1:5,000 or 1:10,000 dilution was used for detection of primary antibodies. Membranes were washed in blocking solution or PBS 0.1% Tween-20 (last wash) and primary antibody signal was detected using the Pierce ECL Western Blotting Substrate or SuperSignal West Femto Maximum Sensitivity Substrate kit (Thermo Fisher Scientific).
Flow cytometry
All antibodies used were purchased from eBioscience unless otherwise stated. Total bone marrow cells were isolated from tibiae, femurs, and pelvic bones by gentle crushing in PBS, followed by erythrocyte lysis with ACK buffer.
For analysis of mature spleen, bone marrow and peripheral blood cell populations: Cells were stained with antibodies Gr-1 (RB6-8C5), CD11b (M1/70), TER-119 (TER-119), CD71 (R17217), CD41 (eVioMWReg30), CD8A (53–6.7), CD4 (GK1.5), and B220 (RA3-6B2).
For analysis of hematopoietic stem and progenitor cells: bone marrow or spleen cells were stained with a cocktail for lineage markers CD3e (145-2C11), CD4 (GK1.5), CD8a (53–6.7), CD19 (eBio1D3), Ter119 (TER-119), B220 (RA3-6B2), Gr-1 (RB6-8C5), CD11b (M1/70), and lineage-negative cells were analyzed for cKit (2B8), SCA-1 (D7; BioLegend), CD34 (RAM34), CD16/CD32 (93), FLT-3/FLK2 (A2F10), and Thy-1.2 (53–2.1) expression and streptavidin. Cells were sorted and analyzed using either a FACSAria or 5-laser FACSAria II Special Order System flow cytometer (BD). Analysis of FACS data were performed using FACSDiva (BD) and FlowJo (Tree Star) software.
Phospho-flow cytometry
Protocol adapted from (Kalaitzidis and Neel, 2008). In brief, lineage-depleted bone marrow cells from 10–14-mo-old wild-type and Nol3−/− MPN mice were incubated in IMDM 2% FBS for 30 min before cytokine stimulation. Cells were then stimulated with 50 ng/ml TPO, 10 ng/ml G-CSF, or PBS (unstimulated) for 15 min. After stimulation, cells were fixed using equal volume of Phosflow Fix Buffer I (BD) for 10 min at 37°C. Cells were pelleted by centrifugation for 5 min at 300 g, and after removal of supernatant, permeabilized using ice-cold acetone dropwise and incubated for 10 min on ice. Cells were then washed twice with 2% FBS in PBS, and stored in 2% FBS in PBS at 4°C until flow cytometry analysis. Samples were analyzed at the same time using a BD LSR II flow cytometer and antibodies against phospho-STAT5 (BD) and phospho-STAT3 (BD).
Ki-67 and Hoechst staining
All antibodies used were purchased from eBioscience unless otherwise stated. Total bone marrow cells were stained with a cocktail for lineage markers CD3e (145-2C11), CD4 (GK1.5), CD8a (53–6.7), CD19 (eBio1D3), Ter119 (TER-119), B220 (RA3-6B2), Gr-1 (RB6-8C5), CD11b (M1/70), and lineage-negative cells were analyzed for c-Kit (2B8), SCA-1 (D7), FLT-3 (A2F10), and Thy-1.2 (53–2.1) expression. Cells were washed with PBS 2% FBS and fixed with Cytofix/Cytoperm (BD) for 20 min at 4°C. Cells were then washed twice with PermWash buffer (BD), and resuspended in 100 µl of PermWash buffer with FITC conjugated Anti–Ki-67 antibody (BD) and incubated overnight at 4°C. Before analysis bone marrow cells were incubated with Hoechst 33342 (1:400 dilution) for 10 min on ice.
Annexin V staining
Bone marrow and spleen cells were isolated as previously described. Staining was performed according to the manufacturer’s instructions with the Annexin-V FLUOS Staining kit (Roche). 49,6-diamidino-2-phenylindole (DAPI; 1:1,000) was used to identify live cells. Viable cells were defined as Annexin-V−/DAPI−.
Transplantation experiments
Total bone marrow or spleen single-cell suspensions were harvested and erythrocytes were lysed using ACK buffer. Live cells were counted via Trypan blue exclusion and were resuspended in HBSS (Thermo Fisher Scientific). For all transplantation experiments unless otherwise specified, recipient mice were lethally irradiated (950–1,050 rads) using a Shepherd 6810 137Cs irradiator and received retroorbital injections for cell transplantation 4 h after irradiation. For all transplantation experiments engraftment of donor cells was monitored by FACS analysis of peripheral blood after transplantation using CD45.1 (A20) and CD45.2 (104) antibodies.
Transplantation of Nol3−/− MPN
Lethally irradiated SJL-Ptprca Pepcb/BoyJ wild-type CD45.1+ recipient mice were injected with Nol3+/+ or Nol3−/− MPN CD45.2+ 1–5 × 106 bone marrow cells or 6–10 × 106 spleen cells. For each individual Nol3+/+ or Nol3−/− MPN donor mouse 1–5 recipient mice were transplanted. All mice evaluated for MPN disease initiation in recipient mice had (89–99%) donor peripheral blood engraftment. Mice were sacrificed 6–8 mo after transplantation for bone marrow transplantations or 5–7 mo after transplantation for spleen transplantations for detection of donor-derived disease.
Competitive transplantation experiments
Lethally irradiated wild-type SJL-Ptprca Pepcb/BoyJ CD45.1+ recipient mice were co-injected intravenously with a mixture of 1–2 × 106 wild-type CD45.1+ or wild-type CD45.1/CD45.2+ whole bone marrow competitor cells and 1–2 × 106 of either Nol3+/+ or Nol3−/− MPN CD45.2+ whole bone marrow cells in a 1:1 ratio. Each individual recipient mouse was the recipient of bone marrow cells from one individual Nol3+/+ or Nol3−/− MPN mouse for each experiment, in addition to the competitor bone marrow, which was aliquoted from a pool that was common for the whole experiment. The experiment was performed twice with two independent Nol3+/+ or Nol3−/− MPN bone marrows. For LT-HSC competitive transplantation, 1,325 sorted Thy1+LSK cells from CD45.1+ wild-type mouse bone marrow was mixed in a 1:1 ratio with 1,325 sorted Thy1+LSK cells from CD45.2+ Nol3+/+ or Nol3−/− MPN bone marrow and transplanted via direct femoral injection into lethally irradiated SJL-Ptprca Pepcb/BoyJ CD45.1+ recipient mice, along with 100,000 CD45.1 supporting cells. Peripheral blood engraftment was analyzed every 4 wk for 16 wk.
Reciprocal transplantation experiments
Lethally irradiated 3-mo-old CD45.2+ Nol3+/+ or Nol3−/− mice were injected intravenously with 3–5 × 106 FACS purified wild-type CD45.1+ total bone marrow cells. Donor cell engraftment was detected via peripheral blood FACS analysis after transplantation. Mice were sacrificed 11–12 mo after transplantation for analysis of donor-derived MPN. 2/13 Nol3−/− recipient mice had <35% donor cell chimerism and were excluded for evaluation of donor-derived MPN including spleen size and complete blood count.
Colony assays
For stem and progenitor cell colony assays, cells were FACS purified and 400–1,200 cells/ml were plated in HSC007 methylcellulose (R&D Systems). Cytospins of single-cell suspensions from colony assays were stained using a modified Giemsa stain (Shandon Kwik-Diff Stains; Thermo Fisher Scientific) according to the manufacturer’s protocol. Cell morphology was evaluated using an EVOS FL Auto microscope (Life Technologies). For peripheral blood colony assays, 30 µl of peripheral blood freshly isolated from the heart of euthanized mice was added to 270 µl of IMDM 2% FBS and resuspended in HSC007 methylcellulose. For HEL, SET-2, and UKE-1 cell line colony assays, sorted GFP+ cells were plated in HSC002SF methylcellulose (R&D Systems). All scored colonies had >40 cells/colony. All colonies were scored after 7–10 d of culture at 37°C, 5% CO2.
Inhibitor treatment
FACS-sorted Nol3+/+ and Nol3−/− MPN Thy1+LSK or LSK cells isolated from bone marrow or spleen were plated in HSC007 in the presence of either DMSO, 300 nM CDK4/CDK6 inhibitor Palbociclib Isethionate (Sigma-Aldrich), or 250 nM Myc inhibitor JQ1 (Sigma-Aldrich). Inhibitors were prepared in DMSO and plated in a total volume of 2 µl/ml. For HEL, SET-2, and UKE-1 cell lines, cells were plated in HSC002SF in the presence of either DMSO, 1 µM JQ1, or 600 nM Palbociclib. All colonies were scored after 7–10 d of culture at 37°C, 5% CO2.
Isolation of serum and serum cytokine array
Serum was isolated from peripheral blood harvested from the heart of euthanized mice and was spun down at 450 g for 10 min. The supernatant was transferred and the spin was repeated. The supernatant was then stored at −80°C until use. 6 Nol3+/+ and 6 Nol3−/− MPN serum samples were pooled separately and diluted 1:10 and used as lysate according to the manufacturer’s instructions for Mouse Angiogenesis Antibody Array (Affymetrix, MA6320). Immunoblots were quantified using ImageJ software (National Institutes of Health).
Real-time PCR
RNA for real-time quantitative PCR was extracted from isolated cell populations using the RNeasy Micro kit (QIAGEN) and RNA was reverse transcribed to cDNA using Superscript II (Invitrogen). For some samples RNA was extracted using Dynabeads mRNA DIRECT kit (Ambion) and converted to cDNA using RNA to cDNA EcoDRY Premix (Random Hexamers; Takara Bio Inc.). Amplification of target genes was measured using the Universal PCR Power SYBR Green mix (Applied Biosystems). cDNA was amplified in a final volume of 15 µl in 96-well or 384-well microtiter plates according to the manufacturer’s recommendation. Primers used for real-time PCR are as follows: Nol3 forward, 5′-AGTTCGAAGAAATGGGCAAC-3′; reverse, 5′-GCATCCAAGGCTTCGTACTC-3′; c-Myc forward, 5′-ACAGGACTCCCCAGGCTCCG-3′; reverse, 5′-CGTGGCTGTCTGCGGGGTTT-3′; Cdk6 forward, 5′-CCTTACCTCGGTGGTCGTC-3′; reverse, 5′-GAACTTCCACGAAAAAGAGGCT-3′; Gapdh forward, 5′-CCAGCCTCGTCCCGTAGAC-3′; reverse, 5′-GCCTTGACTGTGCCGTTGA-3′. All real-time PCR experiments were performed using a ViiA7 instrument (Life Technologies) with one cycle of 50°C (2 min) and 95°C (10 min), followed by 40 cycles of amplification, with each cycle comprising the steps: 95°C (15 s) and 60°C (for 1 min). Specific amplification of the target gene products was validated by melting curve analysis and Sanger sequencing. Target gene expression quantification was calculated using the Pfaffl model and normalized to Gapdh expression levels.
Analysis of microarray data
RNA for microarray was extracted from FACS purified Thy1+LSK cells using the RNeasy Micro kit (QIAGEN). RNA quantity and quality was assessed using a 2100 Bioanalyzer (Agilent). For global gene expression analysis, isolated RNA was amplified using the WT Ovation Pico RNA amplification system (Nugen). After labeling with the GeneChip WT terminal labeling kit (Affymetrix), labeled cRNA of each individual sample was hybridized to an Affymetrix Mouse Gene 2.0ST microarray (Affymetrix), stained and scanned by GeneChip Scanner 3000 7 G system (Affymetrix) according to standard protocols. Raw data were normalized using Expression Console Software (Affymetrix) and analyzed using Transcriptome Analysis Console Software (Affymetrix). Unannotated and sex-specific genes varying among samples were removed for analysis. Gene set enrichment analysis was performed using the gene set enrichment analysis tool (GSEA; Broad Institute).
GEO
Gene expression data generated from Thy1+LSK cells isolated from Nol3+/+ and Nol3−/− MPN mice are provided under accession no. GSE76121.
Datasets
For human PMF gene expression comparative analysis, we used the publicly available GEO dataset under accession no. GSE53482 (healthy controls, n = 16; PMF patients, n = 41) consisting of gene expression data of fractionated CD34+ cells isolated from peripheral blood of healthy controls and patients with PMF. Box plots of NOL3 expression data were generated using Qlucore Omics Explorer software (Qlucore). Waterfall plots for NOL3 expression in PMF patients were created using the SAS statistical analysis software (SAS Institute Inc.). NOL3 expression data in AML and healthy control LT-HSCs were analyzed using previously reported, publically available datasets (Barreyro et al., 2012; Schinke et al., 2015).
Comparative analysis with published datasets
For GSEA comparative analysis, we compared GSEA results of differentially expressed genes between murine Nol3−/− MPN Thy1+LSK cells (versus age-matched Nol3+/+) and human PMF CD34+ cells (GEO accession no. GSE53482) versus healthy controls. Data from PMF patients were then dichotomized into NOL3 high and low expression groups based on a threshold of NOL3 expression of 3.85. Significant positive enrichment was identified using FDR q-values < 0.01.
Statistics
For PMF and AML analyses, Fisher’s exact test of independence was used to assess correlations of overlapping GSEA pathways in human PMF and NOL3 expression in human de novo AML. Throughout this study, P values are by 2-tailed Student’s t test and error bars represent the mean ± SD unless otherwise indicated. Statistical analysis of group comparisons was performed using Student’s t test in Excel or GraphPad Prism. A value of P < 0.05 was used to determine whether a significant difference existed between two groups.
Online supplemental material
Fig. S1 shows ARC expression in different hematopoietic cell types, additional analyses of hematopoietic organs pre- and post-transplantation, and subcellular localization of ARC protein. Table S1 lists peripheral blood counts. Table S2 lists Characteristics of patients with myeloid malignancies with NOL3 deletion or UPD (hg18).
Acknowledgments
We thank J. Filipa Leite and J. Chan for their guidance and technical expertise. We also thank D. Sun and G. Simkin of the Stem Cell Isolation and Xenotransplantation Core Facility (funded by NYSTEM grant #C029154) of the Gottesman Institute for Stem Cell and Regenerative Medicine Research for expert technical support, W. Tran from the Einstein Genomics Core Facility, and P. Schultes from the Einstein Department of Cell Biology for technical assistance.
This work was supported by National Institutes of Health (NIH) grants to U. Steidl (R01CA166429) and K. Gritsman (R01CA196973), an MSTP training grant to R.F. Stanley and R.T. Piszczatowski. (T32GM007288), National Heart, Lung, and Blood Institute Fellowship to R.F. Stanley (F30HL122085), the NIH Cellular and Molecular Biology and Genetics Training Grant to R.F. Stanley, K. Mitchell, and W.M. McKimpson (T32GM007491). R.N. Kitsis was supported by the Dr. Gerald and Myra Dorros Chair in Cardiovascular Disease. U. Steidl is the Diane and Arthur B. Belfer Scholar in Cancer Research of the Albert Einstein College of Medicine, and is a Research Scholar of the Leukemia & Lymphoma Society.
The authors declare no competing financial interests.
Author contributions: R.F. Stanley designed and performed research, conducted experiments, analyzed data, made figures, and wrote the manuscript; R.T. Piszczatowski designed and performed research, conducted experiments, analyzed data, made figures, and wrote the manuscript; B. Bartholdy designed research, interpreted data, and performed statistical analyses; K. Mitchell conducted experiments and interpreted data; W.M. McKimpson provided reagents and interpreted data; S. Narayanagari conducted experiments; D. Walter conducted experiments and interpreted data; T.I. Todorova conducted experiments; C. Hirsch analyzed and interpreted data; C. McMahon interpreted data; K. Gritsman designed experiments and interpreted data; B. Will and R.N. Kitsis designed research and analyzed and interpreted data; H. Makishima, C. Hirsch, and J.P. Maciejewski provided clinical data; U. Stiedl designed research, analyzed data, and contributed to writing of the manuscript.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- AML-MRC
- AML with myelodysplasia-related changes
- ARC
- apoptosis repressor with caspase recruitment domain
- CMP
- common myeloid progenitor
- G-CSF
- granulocyte-colony stimulating factor
- GEO
- Gene Expression Omnibus
- GMP
- granulocyte/monocyte progenitor
- GSEA
- Gene Set Enrichment Analysis
- H&E
- hematoxylin and eosin
- LK
- Lin-Sca-1-c-kit+
- LSK
- Lin-Sca-1+c-kit+
- LT-HSC
- long-term hematopoietic stem cell
- MDS
- myelodysplastic syndrome
- MEP
- megakaryocyte/erythrocyte progenitor
- MPN
- myeloproliferative neoplasm
- MPP
- multipotent progenitor
- Ph
- Philadelphia chromosome
- PMF
- primary myelofibrosis
- SNP
- single nucleotide polymorphism
- ST-HSC
- short-term hematopoietic stem cell
- TPO
- thrombopoietin
- UPD
- uniparental disomy
References
- Abdel-Wahab O., Pardanani A., Bernard O.A., Finazzi G., Crispino J.D., Gisslinger H., Kralovics R., Odenike O., Bhalla K., Gupta V., et al. 2012. Unraveling the genetic underpinnings of myeloproliferative neoplasms and understanding their effect on disease course and response to therapy: proceedings from the 6th International Post-ASH Symposium. Am. J. Hematol. 87:562–568. 10.1002/ajh.23169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson J.W., Fialkow P.J., Murphy S., Prchal J.F., and Steinmann L.. 1976. Polycythemia vera: stem-cell and probable clonal origin of the disease. N. Engl. J. Med. 295:913–916. 10.1056/NEJM197610212951702 [DOI] [PubMed] [Google Scholar]
- Andréasson B., Swolin B., and Kutti J.. 2002. Patients with idiopathic myelofibrosis show increased CD34+ cell concentrations in peripheral blood compared to patients with polycythaemia vera and essential thrombocythaemia. Eur. J. Haematol. 68:189–193. 10.1034/j.1600-0609.2002.01610.x [DOI] [PubMed] [Google Scholar]
- Barreyro L., Will B., Bartholdy B., Zhou L., Todorova T.I., Stanley R.F., Ben-Neriah S., Montagna C., Parekh S., Pellagatti A., et al. 2012. Overexpression of IL-1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood. 120:1290–1298. 10.1182/blood-2012-01-404699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter E.J., Scott L.M., Campbell P.J., East C., Fourouclas N., Swanton S., Vassiliou G.S., Bench A.J., Boyd E.M., Curtin N., et al. Cancer Genome Project . 2005. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 365:1054–1061. 10.1016/S0140-6736(05)74230-6 [DOI] [PubMed] [Google Scholar]
- Carter B.Z., Qiu Y.H., Zhang N., Coombes K.R., Mak D.H., Thomas D.A., Ravandi F., Kantarjian H.M., Koller E., Andreeff M., and Kornblau S.M.. 2011. Expression of ARC (apoptosis repressor with caspase recruitment domain), an antiapoptotic protein, is strongly prognostic in AML. Blood. 117:780–787. 10.1182/blood-2010-04-280503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M., Kastritis E., Gilpatrick T., Paranal R.M., Qi J., et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 146:904–917. 10.1016/j.cell.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf S., Abdelfattah N.S., Chen E., Perales-Patón J., Rosen E.A., Ko A., Peisker F., Florescu N., Giannini S., Wolach O., et al. 2016. Mutant calreticulin requires both its mutant C-terminus and the thrombopoietin receptor for oncogenic transformation. Cancer Discov. 6:368–381. 10.1158/2159-8290.CD-15-1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fialkow P.J., Faguet G.B., Jacobson R.J., Vaidya K., and Murphy S.. 1981. Evidence that essential thrombocythemia is a clonal disorder with origin in a multipotent stem cell. Blood. 58:916–919. [PubMed] [Google Scholar]
- Foo R.S.-Y., Nam Y.-J., Ostreicher M.J., Metzl M.D., Whelan R.S., Peng C.-F., Ashton A.W., Fu W., Mani K., Chin S.-F., et al. 2007. Regulation of p53 tetramerization and nuclear export by ARC. Proc. Natl. Acad. Sci. USA. 104:20826–20831. 10.1073/pnas.0710017104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry D.W., Harvey P.J., Keller P.R., Elliott W.L., Meade M., Trachet E., Albassam M., Zheng X., Leopold W.R., Pryer N.K., and Toogood P.L.. 2004. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol. Cancer Ther. 3:1427–1438. [PubMed] [Google Scholar]
- Fukada T., Ohtani T., Yoshida Y., Shirogane T., Nishida K., Nakajima K., Hibi M., and Hirano T.. 1998. STAT3 orchestrates contradictory signals in cytokine-induced G1 to S cell-cycle transition. EMBO J. 17:6670–6677. 10.1093/emboj/17.22.6670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geertman R., McMahon A., and Sabban E.L.. 1996. Cloning and characterization of cDNAs for novel proteins with glutamic acid-proline dipeptide tandem repeats. Biochim. Biophys. Acta. 1306:147–152. 10.1016/0167-4781(96)00036-X [DOI] [PubMed] [Google Scholar]
- Gobe G.C., Ng K.L., Small D.M., Vesey D.A., Johnson D.W., Samaratunga H., Oliver K., Wood S., Barclay J.L., Rajandram R., et al. 2016. Decreased apoptosis repressor with caspase recruitment domain confers resistance to sunitinib in renal cell carcinoma through alternate angiogenesis pathways. Biochem. Biophys. Res. Commun. 473:47–53. 10.1016/j.bbrc.2016.03.048 [DOI] [PubMed] [Google Scholar]
- Jacobson R.J., Salo A., and Fialkow P.J.. 1978. Agnogenic myeloid metaplasia: a clonal proliferation of hematopoietic stem cells with secondary myelofibrosis. Blood. 51:189–194. [PubMed] [Google Scholar]
- James C., Ugo V., Le Couédic J.-P., Staerk J., Delhommeau F., Lacout C., Garçon L., Raslova H., Berger R., Bennaceur-Griscelli A., et al. 2005. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 434:1144–1148. 10.1038/nature03546 [DOI] [PubMed] [Google Scholar]
- Kalaitzidis D., and Neel B.G.. 2008. Flow-cytometric phosphoprotein analysis reveals agonist and temporal differences in responses of murine hematopoietic stem/progenitor cells. PLoS One. 3:e3776 10.1371/journal.pone.0003776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuchi N., Nakajima K., Ichiba M., Fukada T., Narimatsu M., Mizuno K., Hibi M., and Hirano T.. 1999. STAT3 is required for the gp130-mediated full activation of the c-myc gene. J. Exp. Med. 189:63–73. 10.1084/jem.189.1.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klampfl T., Gisslinger H., Harutyunyan A.S., Nivarthi H., Rumi E., Milosevic J.D., Them N.C., Berg T., Gisslinger B., Pietra D., et al. 2013. Somatic mutations of calreticulin in myeloproliferative neoplasms. N. Engl. J. Med. 369:2379–2390. 10.1056/NEJMoa1311347 [DOI] [PubMed] [Google Scholar]
- Koseki T., Inohara N., Chen S., and Núñez G.. 1998. ARC, an inhibitor of apoptosis expressed in skeletal muscle and heart that interacts selectively with caspases. Proc. Natl. Acad. Sci. USA. 95:5156–5160. 10.1073/pnas.95.9.5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralovics R., Passamonti F., Buser A.S., Teo S.-S., Tiedt R., Passweg J.R., Tichelli A., Cazzola M., and Skoda R.C.. 2005. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 352:1779–1790. 10.1056/NEJMoa051113 [DOI] [PubMed] [Google Scholar]
- Kralovics R., Teo S.-S., Li S., Theocharides A., Buser A.S., Tichelli A., and Skoda R.C.. 2006. Acquisition of the V617F mutation of JAK2 is a late genetic event in a subset of patients with myeloproliferative disorders. Blood. 108:1377–1380. 10.1182/blood-2005-11-009605 [DOI] [PubMed] [Google Scholar]
- Kung G., Dai P., Deng L., and Kitsis R.N.. 2014. A novel role for the apoptosis inhibitor ARC in suppressing TNFα-induced regulated necrosis. Cell Death Differ. 21:634–644. 10.1038/cdd.2013.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine R.L., Wadleigh M., Cools J., Ebert B.L., Wernig G., Huntly B.J., Boggon T.J., Wlodarska I., Clark J.J., Moore S., et al. 2005. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 7:387–397. 10.1016/j.ccr.2005.03.023 [DOI] [PubMed] [Google Scholar]
- Lin J.-X., Li P., Liu D., Jin H.T., He J., Ata Ur Rasheed M., Rochman Y., Wang L., Cui K., Liu C., et al. 2012. Critical Role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function. Immunity. 36:586–599. 10.1016/j.immuni.2012.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak P.Y., Mak D.H., Mu H., Shi Y., Ruvolo P., Ruvolo V., Jacamo R., Burks J.K., Wei W., Huang X., et al. 2014a Apoptosis repressor with caspase recruitment domain is regulated by MAPK/PI3K and confers drug resistance and survival advantage to AML. Apoptosis. 19:698–707. 10.1007/s10495-013-0954-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak P.Y., Mak D.H., Ruvolo V., Jacamo R., Kornblau S.M., Kantarjian H., Andreeff M., and Carter B.Z.. 2014b Apoptosis repressor with caspase recruitment domain modulates second mitochondrial-derived activator of caspases mimetic-induced cell death through BIRC2/MAP3K14 signalling in acute myeloid leukaemia. Br. J. Haematol. 167:376–384. 10.1111/bjh.13054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKimpson W.M., Weinberger J., Czerski L., Zheng M., Crow M.T., Pessin J.E., Chua S.C. Jr., and Kitsis R.N.. 2013. The apoptosis inhibitor ARC alleviates the ER stress response to promote β-cell survival. Diabetes. 62:183–193. 10.2337/db12-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Ramirez C.M., Goswami S., Smirnova T., Bamira D., Benson B., Ferrick N., Segall J., Pollard J.W., and Kitsis R.N.. 2011. Apoptosis inhibitor ARC promotes breast tumorigenesis, metastasis, and chemoresistance. Cancer Res. 71:7705–7715. 10.1158/0008-5472.CAN-11-2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier I, Vuolo M, Madan R, Xue X, Levalley A, Ashton A, Jasmin J, Czaja M, Lin E, Armstrong R, et al. 2005. ARC, an apoptosis suppressor limited to terminally differentiated cells, is induced in human breast cancer and confers chemo- and radiation- resistance. Cell Death and Differentiation. 682–686. [DOI] [PubMed] [Google Scholar]
- Mercier I., Vuolo M., Jasmin J.-F., Medina C.M., Williams M., Mariadason J.M., Qian H., Xue X., Pestell R.G., Lisanti M.P., and Kitsis R.N.. 2008. ARC (apoptosis repressor with caspase recruitment domain) is a novel marker of human colon cancer. Cell Cycle. 7:1640–1647. 10.4161/cc.7.11.5979 [DOI] [PubMed] [Google Scholar]
- Nam Y.-J., Mani K., Ashton A.W., Peng C.-F., Krishnamurthy B., Hayakawa Y., Lee P., Korsmeyer S.J., and Kitsis R.N.. 2004. Inhibition of both the extrinsic and intrinsic death pathways through nonhomotypic death-fold interactions. Mol. Cell. 15:901–912. 10.1016/j.molcel.2004.08.020 [DOI] [PubMed] [Google Scholar]
- Nangalia J., Massie C.E., Baxter E.J., Nice F.L., Gundem G., Wedge D.C., Avezov E., Li J., Kollmann K., Kent D.G., et al. 2013. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N. Engl. J. Med. 369:2391–2405. 10.1056/NEJMoa1312542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norfo R., Zini R., Pennucci V., Bianchi E., Salati S., Guglielmelli P., Bogani C., Fanelli T., Mannarelli C., Rosti V., et al. Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative Investigators . 2014. miRNA-mRNA integrative analysis in primary myelofibrosis CD34+ cells: role of miR-155/JARID2 axis in abnormal megakaryopoiesis. Blood. 124:e21–e32. 10.1182/blood-2013-12-544197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.T., Simonds E.F., Jones C., Hale M.B., Goltsev Y., Gibbs K.D. Jr., Merker J.D., Zehnder J.L., Nolan G.P., and Gotlib J.. 2010. Novel mutations in the inhibitory adaptor protein LNK drive JAK-STAT signaling in patients with myeloproliferative neoplasms. Blood. 116:988–992. 10.1182/blood-2010-02-270108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panteli K.E., Hatzimichael E.C., Bouranta P.K., Katsaraki A., Seferiadis K., Stebbing J., and Bourantas K.L.. 2005. Serum interleukin (IL)-1, IL-2, sIL-2Ra, IL-6 and thrombopoietin levels in patients with chronic myeloproliferative diseases. Br. J. Haematol. 130:709–715. 10.1111/j.1365-2141.2005.05674.x [DOI] [PubMed] [Google Scholar]
- Pikman Y., Lee B.H., Mercher T., McDowell E., Ebert B.L., Gozo M., Cuker A., Wernig G., Moore S., Galinsky I., et al. 2006. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 3:e270 10.1371/journal.pmed.0030270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinz S., Unser S., and Rascle A.. 2016. Signal transducer and activator of transcription STAT5 is recruited to c-Myc super-enhancer. BMC Mol. Biol. 17:10 10.1186/s12867-016-0063-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub F.X., Jäger R., Looser R., Hao-Shen H., Hermouet S., Girodon F., Tichelli A., Gisslinger H., Kralovics R., and Skoda R.C.. 2009. Clonal analysis of deletions on chromosome 20q and JAK2-V617F in MPD suggests that del20q acts independently and is not one of the predisposing mutations for JAK2-V617F. Blood. 113:2022–2027. 10.1182/blood-2008-07-167056 [DOI] [PubMed] [Google Scholar]
- Schaub F.X., Looser R., Li S., Hao-Shen H., Lehmann T., Tichelli A., and Skoda R.C.. 2010. Clonal analysis of TET2 and JAK2 mutations suggests that TET2 can be a late event in the progression of myeloproliferative neoplasms. Blood. 115:2003–2007. 10.1182/blood-2009-09-245381 [DOI] [PubMed] [Google Scholar]
- Schinke C., Giricz O., Li W., Shastri A., Gordon S., Barreyro L., Bhagat T., Bhattacharyya S., Ramachandra N., Bartenstein M., et al. 2015. IL8-CXCR2 pathway inhibition as a therapeutic strategy against MDS and AML stem cells. Blood. 125:3144–3152. 10.1182/blood-2015-01-621631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidl U., Steidl C., Ebralidze A., Chapuy B., Han H.-J., Will B., Rosenbauer F., Becker A., Wagner K., Koschmieder S., et al. 2007. A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J. Clin. Invest. 117:2611–2620. 10.1172/JCI30525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A. 2010. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 24:1128–1138. 10.1038/leu.2010.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A. 2012. JAK inhibitors for myeloproliferative neoplasms: clarifying facts from myths. Blood. 119:2721–2730. 10.1182/blood-2011-11-395228 [DOI] [PubMed] [Google Scholar]
- Tefferi A., and Pardanani A.. 2015. Myeloproliferative Neoplasms: A Contemporary Review. JAMA Oncol. 1:97–105. 10.1001/jamaoncol.2015.89 [DOI] [PubMed] [Google Scholar]
- Tefferi A., Vaidya R., Caramazza D., Finke C., Lasho T., and Pardanani A.. 2011. Circulating interleukin (IL)-8, IL-2R, IL-12, and IL-15 levels are independently prognostic in primary myelofibrosis: a comprehensive cytokine profiling study. J. Clin. Oncol. 29:1356–1363. 10.1200/JCO.2010.32.9490 [DOI] [PubMed] [Google Scholar]
- Tefferi A., Guglielmelli P., Larson D.R., Finke C., Wassie E.A., Pieri L., Gangat N., Fjerza R., Belachew A.A., Lasho T.L., et al. 2014. Long-term survival and blast transformation in molecularly annotated essential thrombocythemia, polycythemia vera, and myelofibrosis. Blood. 124:2507–2513, quiz:2615. 10.1182/blood-2014-05-579136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibes R., Bogenberger J.M., Benson K.L., and Mesa R.A.. 2012. Current outlook on molecular pathogenesis and treatment of myeloproliferative neoplasms. Mol. Diagn. Ther. 16:269–283. 10.1007/s40291-012-0006-3 [DOI] [PubMed] [Google Scholar]
- Tong W., and Lodish H.F.. 2004. Lnk inhibits Tpo-mpl signaling and Tpo-mediated megakaryocytopoiesis. J. Exp. Med. 200:569–580. 10.1084/jem.20040762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley C.R., Olsen G.H., Dworkin S., Fabb S.A., Swann J., McArthur G.A., Westmoreland S.V., Chambon P., Scadden D.T., and Purton L.E.. 2007a A microenvironment-induced myeloproliferative syndrome caused by retinoic acid receptor γ deficiency. Cell. 129:1097–1110. 10.1016/j.cell.2007.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley C.R., Shea J.M., Sims N.A., Purton L.E., and Orkin S.H.. 2007b Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell. 129:1081–1095. 10.1016/j.cell.2007.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Qanungo S., Crow M.T., Watanabe M., and Nieminen A.-L.. 2005. Apoptosis repressor with caspase recruitment domain (ARC) is expressed in cancer cells and localizes to nuclei. FEBS Lett. 579:2411–2415. 10.1016/j.febslet.2005.03.040 [DOI] [PubMed] [Google Scholar]
- Wang X., LeBlanc A., Gruenstein S., Xu M., Mascarenhas J., Panzera B., Wisch N., Parker C., Goldberg J.D., Prchal J., et al. 2009. Clonal analyses define the relationships between chromosomal abnormalities and JAK2V617F in patients with Ph-negative myeloproliferative neoplasms. Exp. Hematol. 37:1194–1200. 10.1016/j.exphem.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Zaiman A.L., Damico R., Thoms-Chesley A., Files D.C., Kesari P., Johnston L., Swaim M., Mozammel S., Myers A.C., Halushka M., et al. 2011. A critical role for the protein apoptosis repressor with caspase recruitment domain in hypoxia-induced pulmonary hypertension. Circulation. 124:2533–2542. 10.1161/CIRCULATIONAHA.111.034512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Du Y., Ho W.T., Fu X., and Zhao Z.J.. 2012. JAK2V617F and p53 mutations coexist in erythroleukemia and megakaryoblastic leukemic cell lines. Exp. Hematol. Oncol. 1:15 10.1186/2162-3619-1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer S.N., Zhou Q., Zhou T., Cheng Z., Abboud-Werner S.L., Horn D., Lecocke M., White R., Krivtsov A.V., Armstrong S.A., et al. 2011. Crebbp haploinsufficiency in mice alters the bone marrow microenvironment, leading to loss of stem cells and excessive myelopoiesis. Blood. 118:69–79. 10.1182/blood-2010-09-307942 [DOI] [PMC free article] [PubMed] [Google Scholar]