Abstract
Scythians were nomadic and semi-nomadic people that ruled the Eurasian steppe during much of the first millennium BCE. While having been extensively studied by archaeology, very little is known about their genetic identity. To fill this gap, we analyzed ancient mitochondrial DNA (mtDNA) from Scythians of the North Pontic Region (NPR) and successfully retrieved 19 whole mtDNA genomes. We have identified three potential mtDNA lineage ancestries of the NPR Scythians tracing back to hunter-gatherer and nomadic populations of east and west Eurasia as well as the Neolithic farming expansion into Europe. One third of all mt lineages in our dataset belonged to subdivisions of mt haplogroup U5. A comparison of NPR Scythian mtDNA linages with other contemporaneous Scythian groups, the Saka and the Pazyryks, reveals a common mtDNA package comprised of haplogroups H/H5, U5a, A, D/D4, and F1/F2. Of these, west Eurasian lineages show a downward cline in the west-east direction while east Eurasian haplogroups display the opposite trajectory. An overall similarity in mtDNA lineages of the NPR Scythians was found with the late Bronze Age Srubnaya population of the Northern Black Sea region which supports the archaeological hypothesis suggesting Srubnaya people as ancestors of the NPR Scythians.
The Eurasian Steppe is a vast grassland region that stretches from the Carpathian foothills to Outer Mongolia. For millennia, the steppe was home to human populations that had a significant and long-lasting impact on the cultural history of the Eurasian continent. One such group that emerged at the beginning of the Iron Age had developed into an ethno-cultural agglomerate commonly referred to as the Scythians. The Scythians are best known from ancient Persian, Greek and Assyrian literary sources mainly for their nomadic warrior lifestyle, but they are also known to have actively practiced farming, pastoralism and may have been among the earliest peoples to master the art of horseback riding1.
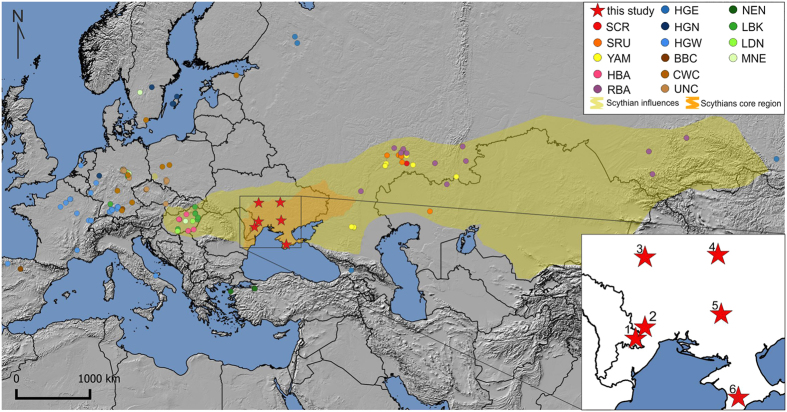
The territory occupied by contemporaneous nomadic and semi-nomadic Scythian groups with shared elements of material culture extended between the lower stretches of the Danube River in the west and the Yenisei River in the east2,3 (Fig. 1). Most archaeology and history researchers suggest that the core territory of the tribes designated as “Scythians” in historic literature of Antiquity covered the steppe and forest-steppe regions of the northern Black Sea (the North Pontic region, NPR) between the lower Danube and Don rivers4,5. Together with other contemporaneous groups they formed the ‘Scythian horizon’. These groups are collectively referred to by some researchers as Scytho-Siberians, who inhabited the steppe regions to the east of NPR Scythians and included Saka from Kazakhstan (7th–3rd century BCE)6,7 and Pazyryks (5th–3rd century BCE) from the Sayano-Altai region of Siberia2,8. The forest-steppe zone of the core Scythian territory in the NPR was settled by populations with agro-pastoral economy, while the nomadic and semi-nomadic Scythian tribes occupied the steppe regions adjacent to the northern Black Sea. Archaeological studies of the Scythian and pre-Scythian period sites in the forest-steppe zone of the NPR pointed towards autochthonous origins of local agro-pastoral Scythian populations. Those groups which first emerged between the middle 7th–3rd century BCE, may have been formed on the foundation of pre-existing groups belonging to such cultures as Srubnaya (Timber Grave) and Thracian Hallstatt9. The origins of nomadic Scythians in the steppes of Central Asia10 were supported by recent archaeological findings of elements of developed Scythian material culture in a series of kurgans in the western Sayan Mountains in southern Siberia dated to 9th–7th century BCE11.
Figure 1. Map of the Scythian core region and territories of their possible influences in 7th–3rd century BCE.
Locations of sites investigated are marked as red stars: Glinoe (1), Vapnyarka (2), Nesterivka (3), Svetlovodsk (4), Starosillya (5), Simferopol (6). Colored circles show locations of comparative ancient populations. Population abbreviations: SCR, Scythians from Russia; SRU, Srubnaya culture; YAM, Yamnaya culture; HBA, Bronze Age populations from Hungary; RBA, Bronze Age populations from Russia; HGE, Hunter-Gatherers Eastern; HGN, Hunter-Gatherers Northern; HGW, Hunter-Gatherer Western; BBC, Bell Beaker culture; CWC, Corded Ware culture; UNC, Unetice culture; NEN, Near Eastern Neolithic; LBK, Linear Pottery culture; LDN, Late Danubian cultures; MNE, Middle Neolithic cultures. The map was created using QGIS 2.12.243.
The relationships between the groups of the Scythian horizon are not fully understood. Some researchers regard the NPR Scythians and Scytho-Siberians as one society on account of similar animal motifs on the products of their material culture, while others consider them to be different populations each having a distinct origin and geographic specificity yet sharing cultural traditions12. Moreover, the ancestral relationships between the NPR Scythians and local predecessor populations in the Ponto-Caspian region are neither fully resolved.
Most of the current knowledge about the genetic relations among the populations belonging to the Scythian cultural horizon is based on studies of non-coding mitochondrial DNA (mtDNA) fragments from Scythian remains from the lower Don and southern Ural regions in Russia13 as well as Pazyryks from Altai and Inner Mongolia7,14,15,16,17,18. To date, only one complete mt genome of a Scythian individual from Russia has been published19. Mitochondrial lineages in the studied populations consist of an overlapping mix of haplogroups of east and west Eurasian descent, which does not clarify their ultimate origins. Here we aim to identify the maternal origin of the NPR Scythians and their genetic affinities to other contemporary Scythian groups through the analyses of complete mtDNA genomes. The investigated human remains were excavated from kurgans, crypts and ground burials in the main area of Scythian distribution, including territories in the lower Dniester, lower and middle Dnieper and Crimea.
Materials and Methods
Materials
We extracted ancient DNA (aDNA) from 29 Iron Age Scythian individuals excavated in present-day Moldova (n = 21) and Ukraine (n = 8). Moldavian samples were obtained from the Archaeological Laboratory collection at the Taras Shevchenko University in Tiraspol. Ukrainian Scythians came from the Archeology Archives of the Institute of Archaeology, National Academy of Sciences of Ukraine in Kyiv and Odessa, and the Department of Culture and Tourism of the Cherkasy Regional State Administration. Detailed information about the ancient individuals can be found in Supplementary Information Text (Materials and Methods) and Supplementary Table S1.
From each Scythian individual from Moldova two teeth were collected and transferred to the Department of Human Evolutionary Biology at the Adam Mickiewicz University in Poznań (AMU) in Poland. Ukrainian samples consisted of both bone fragments (n = 5) and teeth (n = 3), which were analyzed at the Archaeological Research Laboratory (AFL), Department of Archaeology and Classical Studies, Stockholm University (SU) in Sweden and the Archeogenetics Laboratory at Grand Valley State University (GVSU) in Allendale, Michigan, USA.
DNA extraction, library construction and Illumina sequencing
All procedures leading to Next Generation Sequencing (NGS) were performed in the ancient DNA laboratories located at AMU and the AFL. Mitochondrial hypervariable region I (HVRI) and haplogroup-diagnostic coding regions of mtDNA of the Ukrainian samples were analyzed at GVSU using the methods described in the Supplementary Information Text (Materials and Methods). Osteological samples underwent decontamination procedures including NaOCl and UV treatments (Supplementary Information Text (Materials and Methods)), followed by DNA extraction as described by refs 20 and 21. Illumina-compatible blunt-end libraries were prepared following22 and screened on Illumina HiSeq 2500 High Output v4 (2 × 125 bp). For more details, see Supplementary Information Text (Materials and Methods).
Illumina sequencing was performed at the National Genomics Infrastructure (NGI) in Stockholm, Sweden. Sequence data were merged and mapped to human genome as previously published23. All primary pipeline computations were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under projects b2013240 and b201530724.
Mitochondrial DNA capture enrichment and Ion Torrent PGM sequencing
We used commercially biotinylated probes (MYbaits®) for human mtDNA provided by MYcroarray® (Ann Arbor, MI, USA; www.mycroarray.com) for mtDNA enrichment by hybridization capture on the Illumina sequencing libraries. The procedure was undertaken on 25 libraries, which did not yield sufficient mtDNA genome coverage (less than 5x), after initial Illumina screening (Supplementary Table S1). We performed two rounds of target enrichment following the manufacturer’s protocol version 2.3.1 (http://www.mycroarray.com/pdf/MYbaits-manual-v2.pdf). More details about mitochondrial genome capture can be found in Supplementary Information Text (Materials and Methods). Enriched and amplified indexed libraries were purified using MinElute spin columns (Qiagen) and quantified on 2200 TapeStation (Agilent Technologies, Inc.). Prior to the amplification by emulsion PCR, the indexed libraries were pooled in equimolar concentrations and adjusted to a final concentration of 20 pM25. Clonal template amplification on Ion Sphere Particles (ISPs) was performed using the Ion Torrent One Touch System II and the Ion One Touch 200 template kit v2 DL with regard to manufacturer’s instructions. Sequencing of the templated ISPs was conducted with the use of Ion PGM HI−Q Seq kit and Ion Torrent Personal Genome Machine (Ion PGM) system (Ion Torrent, Thermo Fisher Scientific, Inc.) at Molecular Biology Techniques Laboratory, Faculty of Biology, AMU.
Bioinformatic analysis
Customizable analytical pipeline was used to process Illumina sequencing data as described in ref. 23. Read pairs were merged, and adapters were trimmed according to ref. 22. Merged reads were mapped as single-end reads against the revised Cambridge Reference Sequence (rCRS)26,27 (GenBank: NC_012920) with the use of BWA software package version 0.7.828.
We processed sequence data from PGM Ion Torrent using a pipeline adjusted specifically to the Ion Torrent reads. Sequences were demultiplexed by barcodes using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Long (−M 110), short (−m 35), and low-quality sequences (−q 20) were removed using Cutadapt v.1.8.129. The filtered sequence reads were analyzed with FastQC v 0.11.330, followed by mapping against the rCRS using TMAP v3.4.131. For more details, see Supplementary Information Text (Materials and Methods).
FilterUniqueSAMCons.py was used to collapse clonal reads with identical start and end coordinates, for both PGM and Illumina sequence data as in ref. 22. Misincorporation patterns were determined with the use of mapDamage v2.0.532. Final sequences were visualized using Biomatters IGV software v2.3.6633. ANGSD v0.91034 was applied to build consensus sequence accepting only reads with mapping score of 30, a minimum base quality of 20, and a minimum coverage of 3.
Mitochondrial haplotypes were determined for each sample with the use of HAPLOFIND35, and the PhyloTree phylogenetic tree build 1736. The mutations reported as unexpected or missing were visually inspected in the original binary alignment map (BAM) files in IGV33.
The ratio of reads mapping to Y and X chromosomes (Ry) was calculated to determine molecular sex of individuals sequenced on the Illumina platform37. Individuals with the Ry ratio ≤ 0.016 were identified as females, while those with RY ≥ 0.077 were determined as males (Supplementary Information Text Table S7). The molecular sex calculation was restricted only to the DNA sequence reads with mapping qualities of at least 30.
Statistical analysis
Comparative ancient samples used in the statistical analyses were retrieved from the web depositories (NCBI Nucleotide, European Nucleotide Archive) and literature. Detailed information about comparative samples used in principal component analysis (PCA), pairwise genetic distances (FST) and median network is shown in Supplementary Tables S2–S4.
To calculate PCA we additionally included Asian origin mtDNA haplogroups (A, B, D, F, G, M) and their frequencies to cover suggested Asian influences into Scythian populations (Supplementary Table S2). PCA was computed using RapidMiner Studio 7 (RapidMiner Inc., Boston, MA, USA) and plotted using Matplotlib 1.5.1 Python package38.
We have performed top-down clustering analysis in the form of k-means algorithm to explore the unbiased population relations in the PCA based on haplogropup frequencies sample-set. K-means analysis was performed in RapidMiner Studio 7 (RapidMiner Inc., Boston, MA, USA).
Pairwise genetic distances (FST) were applied only to samples with complete ancient mt genomes (Supplementary Table S3). FST values and Nei’s average number of pairwise differences39 were computed in Arlequin 3.540. We tested the hierarchical partitioning of the genetic variance in different setups by analysis of molecular variance (AMOVA)41 as implemented in Arlequin. Populations used in AMOVA were the same as for the FST analysis and the list is provided in Supplementary Table S3. We have tested several configurations of population groupings to evaluate the best matching position of Scythian population from the NPR and its relation to other populations. Multidimensional scaling (MDS) of FST values was computed using Python scikit-learn 0.17 package42. The maps were created using QGIS 2.12.243.
The median network was calculated and computed for the haplogroup U5 using the Networks 4.614 software (fluxus-engineering.com) with the most common mutations weighted reversely to their frequency.
Results
We successfully retrieved complete mt genomes for 19 out of the 29 Scythian individuals (Table 1 and Supplementary Table S1). Shotgun screening of genomic DNA libraries generated mt genomes for four individuals with the coverage ranging from 6.5x to 70x. For the remaining samples, the mt capture enrichment followed by Ion Torrent sequencing, provided additional 15 complete mt genomes with the depth of coverage from 7x to 103.9x. Mitochondrial DNA genome data were deposited in GenBank under accession numbers KX977302-KX977320.
Table 1. Description of studied individuals.
| Sample | Archaeological site | Dating of the sites based on typochronology | Age at death | Molecular sex | mtDNA haplotype |
|---|---|---|---|---|---|
| SCY192 | Glinoe, Moldova | 4th-2nd BCE | 15–20 yrs | XX | H8c |
| SCY193 | Glinoe, Moldova | 4th-2nd BCE | n.a. | XY | U5a2a1 |
| SCY196 | Glinoe, Moldova | 4th-2nd BCE | 10–11 yrs | n.r. | W3a |
| SCY197 | Glinoe, Moldova | 4th-2nd BCE | n.a. | XY | U5a1a1 |
| SCY300 | Glinoe, Moldova | 4th-2nd BCE | 35–50 yrs | XX | H5b |
| SCY303 | Glinoe, Moldova | 4th-2nd BCE | 6–7 yrs | XX | U5a1a2b |
| SCY305 | Glinoe, Moldova | 4th-2nd BCE | 50 + yrs | n.r. | U5a2b |
| SCY308 | Glinoe, Moldova | 4th-2nd BCE | 20–35 yrs | n.r. | F1d |
| SCY311 | Glinoe, Moldova | 4th-2nd BCE | 35–50 yrs | XX | T2b |
| SCY332 | Glinoe, Moldova | 4th-2nd BCE | 30–35 yrs | XX | M10a1a1a |
| SCY334 | Glinoe, Moldova | 4th-2nd BCE | 20–35 yrs | n.r. | H5b |
| SCY001 | Svetlovodsk, Ukraine | 4th BCE | n.a. | n.r. | U5a1b |
| SCY002 | Svetlovodsk, Ukraine | 4th BCE | n.a. | n.r. | J1c2 |
| SCY006 | Starosillya, Ukraine | 7th BCE | n.a. | XX | D4j2 |
| SCY011 | Nesterivka, Ukraine | 4th BCE | n.a. | XX | A |
| SCY012 | Vapnyarka, Ukraine | 4th-3rd BCE | n.a. | n.r. | U5b2a1a2 |
| SCY009 | Starosillya, Ukraine | 7th BCE | n.a. | XY | J2b1a6 |
| SCY010 | Starosillya, Ukraine | 7th BCE | n.a. | XX | N1b1a |
| SCY005 | Simferopol, Ukraine | 3rd-2nd BCE | n.a. | XX | H |
n.a. - not available. n.r. - no result.
The analysis of damage patterns revealed the presence of damages typical for aDNA, i.e. C-T and G-A transitions accumulated at 5′ and 3′ ends, respectively (Supplementary Fig. S11).
Mitochondrial lineages in the NPR Scythians analyzed in this study appear to consist of a mixture of west and east Eurasian haplogroups. West Eurasian lineages were represented by subdivisions of haplogroup U5 (U5a2a1, U5a1a1, U5a1a2b, U5a2b, U5a1b, U5b2a1a2, six individuals total, 31.6%), H (H and H5b, three individuals total, 15.8%), J (J1c2 and J2b1a6, two individuals, 10.5%), as well as haplogroups N1b1a, W3a and T2b (one individual each, 5.3% each specimen). East Eurasian mt lineages were represented by haplogroups A, D4j2, F1b, M10a1a1a, and H8c (represented by a single individual), in total, comprising 26.3% of our sample set.
The results of the low-resolution mtDNA screening at GVSU were consistent with whole mt capture results from AMU (see Supplementary Table S1). Polymorphism patterns uncovered in the coding and HVRI regions in specimens SCY009 and SCY011 identified their lineages to belong to haplogroups J and A respectively. Specimens SCY006 and SCY010 were assigned to the M* and N* clades respectfully.
To trace genetic affinities between Scythians from present day Moldova and Ukraine (SCU) and other European and Asian ancient populations, their mt haplogroup frequencies were visualized in the space of principle components. The PCA plot of the first two components accounted for 43.4% of the total variance (Fig. 2). The SCU group was located in direct proximity to the central European Neolithic Corded Ware culture (CWC). It also grouped near a number of Bronze Age populations from eastern and central Europe (Srubnaya (SRU), Yamnaya (YAM) and Unetice (UNC)), as well as from central Asia (Bronze Age Kazakhstan (BAK)). Finally, k-means clustering (k value = 5), grouped SCU within a cluster further encompassing Scythians from Russia (SCR) and Tagar culture from southern Siberia (TAG). The Pazyryks from Mongolia (SCM) and Altai (SCA) were separated from SCU and grouped within the k-mean cluster consisting of Central and East Asian populations.
Figure 2. PC analysis based on mitochondrial DNA haplogroup frequencies.
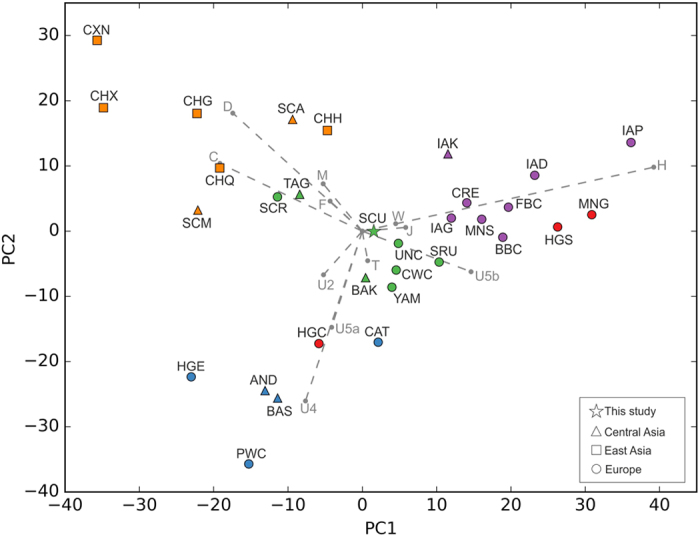
The two dimensions account for 43.4% of the total variance. Haplogroup contributions are represented by loading vectors marked on the plot as grey arrows. Populations are grouped into five clusters according to k-means. First cluster (in green): BAK, Bronze Age Kazakhstan; CWC, Corded Ware culture; SCR, Scythians from Russia; SCU, Scythians from Moldova and Ukraine, present study; SRU, Srubnaya culture; TAG, Tagar culture; UNC, Unetice culture; YAM, Yamnaya culture. Second cluster (in red): HGC, Hunter-Gatherers Central Europe; HGS, Hunter-Gatherers Southern Europe; MNG, Middle Neolithic Germany. Third cluster (in blue): AND, Andronovo culture; BAS, Bronze Age Siberia; CAT, Catacomb culture; HE, Hunter-Gatherers East Europe; PWC, Pitted Ware culture. Fourth cluster (in purple): BBC, Bell Beaker culture; CRE, Crete Minoans; FBC, Funnel Beaker culture; IAD, Iron Age Denmark; IAG, Iron Age Germany; IAK, Iron Age Kazakhstan; IAP, Iron Age Poland; MNS, Middle Neolithic Southern Europe. Fifth cluster (in orange): CHH, Western Hun, China; CHG, Gavaerg China; CHQ, Quin to Western Jin, China; CHX, Xiaohe, Xinjiang, China; CXN, Xiongnu, Mongolia; SCA, Scytho-Siberians from Altai; SCM, Pazyryk culture from Mongolia. Detailed descriptions and references of comparative populations are listed in Supplementary Table S2.
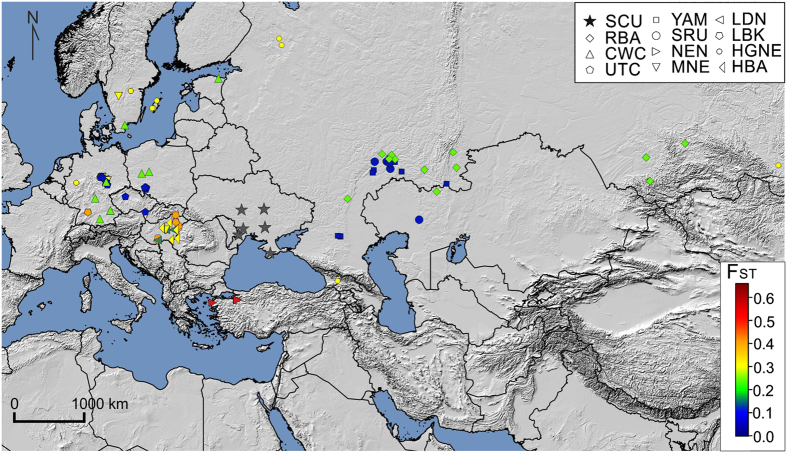
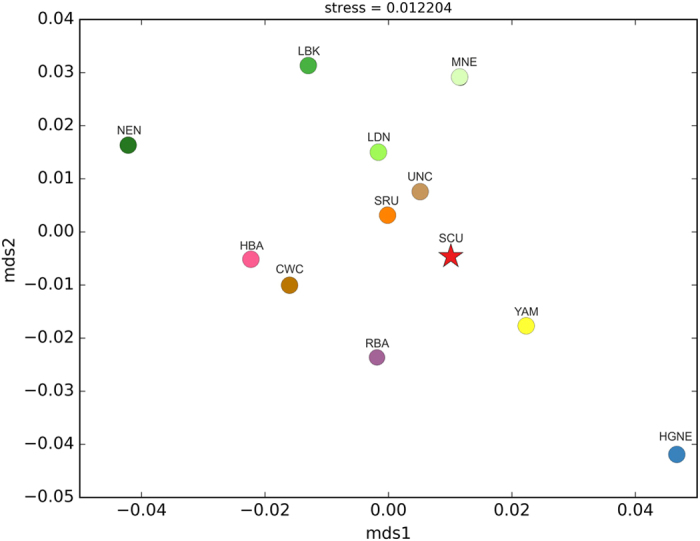
Slatkin’s linearized pairwise FST values calculated on complete mt genomes and visualized using MDS (Fig. 3) and heatmap (Fig. 4) indicate that the studied Scythian samples are closest to the Srubnaya (SRU) (FST = 0.00; p < 0.05) and Yamnaya (YAM) (FST = 0.006; p < 0.05) populations (see Supplementary Table S5), followed by Unetice (UNC) (FST = 0.008; p < 0.05) and Corded Ware Culture (CWC) (FST = 0.023; p < 0.05). The European Neolithic Linear Ware culture (LBK) (FST = 0.043; p > 0.05) and Near Eastern Neolithic populations (NEN) (FST = 0.062; p > 0.05) appeared most distant to the individuals in our dataset. Correspondingly, in the resulted FST based MDS plot (stress value of 0.012), our Scythian group positioned proximately to Srubnaya (SRU), Yamnaya (YAM) and Unetice (UNC) populations and distantly to Near Eastern and European Neolithic (NEN, LBK) and Near Eastern hunter-gatherer (HGNE) populations (Fig. 3).
Figure 3. MDS plot based on FST calculated from complete mitochondrial genomes.
Population abbreviations: CWC, Corded Ware culture; HBA, Bronze Age populations from the geographic area of modern Hungary including Maros, Vatya, Baden, Kyjatice cultures; HGNE, combined Hunter-Gatherers from the North and East; LBK, Linear Pottery culture; LDN, Late Danubian cultures; MNE, Middle Neolithic cultures; NEN, Near Eastern Neolithic; RBA, Bronze Age populations from the geographic area of present-day Russia including Afanasievo, Andronovo, Poltavka, Potapovka and Sintastha individuals; SRU, Srubnaya culture; UNC, Unetice culture; YAM, Yamnaya culture; SCU, Scythians from Moldova and Ukraine from present study combined with one individual from Rostov-on-Don59. Detailed information about each individual from particular comparative population is provided in Supplementary Table S3.
Figure 4. Heatmap of FST and geographic distribution.
Colors in gradient reflect FST values. Population abbreviations are as in Fig. 3. The map was created using QGIS 2.12.243.
Analysis of molecular variance (AMOVA) summarized and tested the distribution of genetic variability within and between subpopulations. We tested several other combinations of grouping Scythians with two or more populations. The results suggested the combination with the highest intragroup and lowest intergroup genetic variability to be the Scythians-Srubnaya sample-set (4.93% of variability among groups, −0.73% among populations within groups). Second best result was observed in combination of Scythians with Unetice sample-set (3.05% among groups, 1.14% among populations). The AMOVA results are summarized in Supplementary Table S6.
Considering the high number of Scythian individuals identified as belonging to haplogroup U5 (31.6%), we conducted median network analysis of published ancient haplotypes belonging to U5a and U5b subdivisions and ranging from the Mesolithic (excluding hunter-gatherers predating last glacial maximum) to the Iron Age (Fig. 5). Haplotypes used in the median network analysis are described in detail in Supplementary Table S4. The Scythians in the U5a cluster were located centrally in the median network, near the U5a ancestral node. The U5a Scythian samples (SCU) grouped together with Mesolithic hunter-gatherers from Sweden (haplotypes 3, 4 and 5), Germany (8), Russia (10) and France (45 and 46). Non-Mesolithic samples in those clusters were less numerous and included Srubnaya (SRU) (57) and Yamnaya (YAM) (13 and 14) from Samara region and one representative of Karasuk culture from the Altai territory (central-south Siberia) (24). A single U5b Scythian individual (31) was distant from the East European populations and was placed basally to a broader cluster composed of west hunter-gatherers (HGW) from France (41) and Germany (47) as well as chronologically younger samples from the Middle Neolithic (MNE) and Unetice (UNC) (both from Germany) (15 and 53, respectively).
Figure 5. Median network of U5 haplotypes.
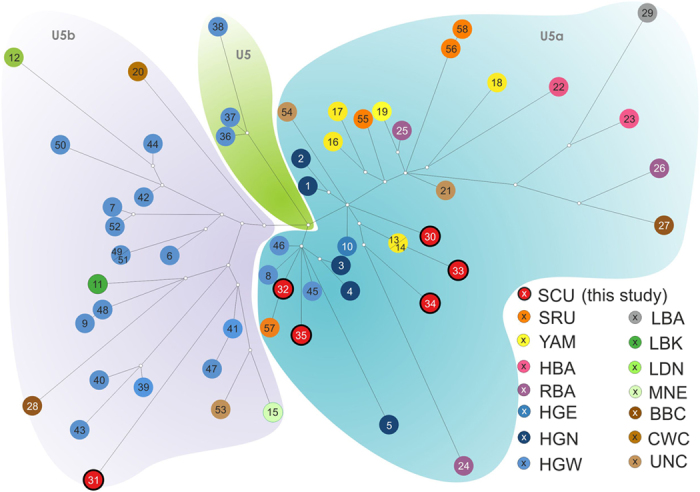
Colors of nodes represent archaeological cultures where a particular haplotype was found. Numbers in nodes indicate particular haplotype (1–58) summarized in Supplementary Table S4. Population abbreviations: CWC, Corded Ware culture; BBC, Bell Beaker culture; HBA, Bronze Age populations from the geographic area of modern Hungary including Maros and Vatya individuals; HGE, Hunter-Gatherers from the East; HGN, Hunter-Gatherers from the North; HGW, Hunter-Gatherers from the West; LBK, Linear Pottery culture; LDN, Late Danubian cultures including a Rössen individual; MNE, Middle Neolithic cultures; NEN, Near Eastern Neolithic; RBA, Bronze Age populations from the geographic area of present-day Russia including Afanasievo and Karasuk individuals; SRU, Srubnaya culture; UNC, Unetice culture; YAM, Yamnaya culture; SCU, Scythians from Moldova and Ukraine from present study.
Discussion
On the basis of published data concerning the phylogeography of mt lineages distribution in ancient populations of Europe and Asia, the 19 complete mt genomes of the NPR Iron Age Scythians produced in this study fall into three main groups of different ancestry. The first group of mt lineages is represented by U5 haplotypes that are considered to be a European Hunter-Gatherer genetic component44,45. The second group comprises haplotypes belonging to H, J, T, W and N1b, ultimately connected to the genetic package of the early Neolithic farmers44,46,47, and the third group includes A, D, M10 and F mt lineages considered to be of East Eurasian origin48,49,50,51.
Representatives of the U5 haplogroup account for one third of the mt haplotypes identified in analyzed Scythians. These ultimately relate to West Eurasian hunter-gatherers, whose descendants extended throughout the European subcontinent and into East Eurasia from the Mesolithic to the Bronze Age. We have combined the known diversity of the prehistoric U5 lineage from different sources19,45,46,47,52,53,54,55,56 into a median network, which allowed for identification of candidate source populations contributing to the U5a diversity in NPR Scythians. Although the network approach is still limited by the small number of available samples, our results indicate that the source populations with the most closely related haplotypes are the Bronze Age Srubnaya (SRU) (1900-1200 BCE) and the earlier Yamnaya (YAM) (3300-2700 BCE) from the Ponto-Caspian region and Bronze Age populations from the Altai Mountains, such as Karasuk (1500-800 BCE). Diverse U5a-carrying populations of the steppe such as Yamnaya, Srubnaya and Scythians shared the nomadic lifestyle, with the economic foundations transforming from wild game exploitation in the Mesolithic to pastoralic animal husbandry in the Bronze Age. Furthermore, both Yamnaya and Srubnaya were part of the Kurgan culture phenomenon57 with the Scythian cultural horizon being the most representative for the kurgan building tradition.
The second group of identified mt genomes in the NPR Scythians is comprised of lineages that suggest associations of the NPR Scythians with the Neolithic European farming groups. Although the mt lineage composition of analyzed Scythians significantly differs from that seen in NEN and LBK groups (FST = 0.06, FST = 0.04, respectively, p > 0.05), particular lineages such as J1c2, T2b, H/H5, H, W and N1b1a ultimately go back to the earliest European farmers. Lineages of J1c, H5 and T2b belong to the Neolithic farming package of mtDNA haplogroups which have been found in most Neolithic and Bronze Age European populations19,44,47,55,58. Noteworthy, individuals belonging to the N1b1a have been found in the Neolithic Anatolia19,59, but the lineage has not appeared in any other Eurasian Neolithic, Bronze or Iron Age populations to date. Therefore, the presence of N1b1a in Scythians could be attributed to population migrations along the southern boundaries of the Ponto-Caspian region60.
Mitochondrial lineages T2b, W3a and J2b1a6 have been identified in representatives of the Bronze Age groups such as Srubnaya (T2b, J2b1a, H5)19, Sintashta (J2b1a, J1c)55, Yamnaya (W3a and T2b)47,61 and Mezhovskaya (J2b1a)55 along with East European Eneolithic Trypillian culture (T2b)62, suggesting genetic continuity of these lineages from at least the Bronze Age or even Neolithic times in NPR region. This is further supported by published data19 which showed that almost one fifth of the genetic makeup of the Late Bronze Age Srubnaya people of the Ponto-Caspian region is of the Early European Farmer or Anatolian Neolithic ancestry possibly resulting from the admixture of populations related to Early European Farmers and Yamnaya. Thus, if the NPR Scythians are the descendants of populations related to Srubnaya, the origin of the identified farming lineages would likely be within the steppe/forest-steppe region between the NPR and southern Ural. According to previous genomic studies47,55 the CWC people are likely to have arisen from Yamnaya background. Thus, genetic affinities of the NPR Scythians to the Yamnaya people might also explain their close genetic similarity to the CWC reflected in PCA and FST results. Close genetic relations of the NPR Scythians and Srubnaya are supported by the FST analyses revealing no significant differences between these two populations (Figs 3 and 4, Supplementary Table S5). AMOVA further corroborated these results showing that the combination of Scythians and Srubnaya results in the lowest genetic variability among populations in a group and the highest variability between groups (Supplementary Table S6). Our results support the archaeological interpretations concerning the origin of the Scythians. One of the versions of this theory suggests that the Srubnaya people have migrated in several waves from the Volga-Ural region to the NPR during the second half of the second millennium BCE where their descendants gave rise to the Scythians around the 7th century BCE63.
The third group of lineages identified in the NPR Scythians is derived from East Eurasian ancestry. Since, to our knowledge, there is no evidence of agricultural subsistence in East Eurasian Scythians, these lineages should be considered to be genetic components associated with nomadic populations. Mitochondrial haplogroups such as A, D and F have already been found in samples from the Mesolithic, Neolithic, Bronze and Iron Age southern Siberia and the Altai7,14,15,16,18,55,64,65,66,67. Notably, haplogroup M10 found among Scythians from Glinoe, is present not only in far East Asia but also in modern populations of the Altai68. Among ancient populations, the M10 lineages have been found in Chinese specimens from southern Xinjiang48 (8th -1st century BCE) and Xiongnu69 (3rd century BCE). Furthermore, haplogroup M have been identified in Pleistocene individuals from western Europe but it is thought that these lineages disappeared due to bottleneck effect in the Last Glacial Maximum56. Haplogroup H8c, identified in the NPR Scythians likely belongs to East Eurasian lineages as well. Sequence analysis of 830 modern Eurasian mt genomes suggested a distinct phylogeographc history for H8, with a clustering of Near Eastern and Central Asian haplotypes of H8 and a pronounced presence of carriers of H8c in the Altai region70. It was further hypothesized that the distribution of this lineage could have been facilitated by nomadic migrations along the NPR coast70. Although our PCA analysis showed Altai Pazyryk (SCA) to be distant from the NPR Scythians (SCU) (Fig. 2), it must be emphasized that the presence of haplogroups A and H8c in the analyzed population connects NPR Scythians to the Altai and identifies this region as a possible source of these East Eurasian mt lineages. The only Scythian mt genome from southern Urals published thus far also belonged to an East Eurasian lineage, G2a19. The Y chromosomal lineage (R1a1a1b2a2a) reconstructed for this individual was supposedly characteristic for members of the Srubnaya culture19 which additionally supports our conclusions concerning close genetic links between Scythians and people related to the late Bronze Age Srubnaya.
Previous analyses of mtDNA HVRI sequence data from Scythians inhabiting Rostov-on-Don region in eastern NPR also resulted in the identification of East Eurasian lineages, such as D13. Moreover, mt lineage D4j2 identified in the individual SCY006 was shown to have been dominant in the Pazyryk culture from Altai and Inner Mongolia (4th -2nd century BCE)7,14,16,17,18. The presence of East Eurasian mt lineages supports those archaeological theories that acknowledge the influence of an East Eurasian element in the formation of the Scythian horizon71,72.
Scythians from Rostov-on-Don as well as Pazyryks from Altai and Inner Mongolia were carriers of mixed east and west Eurasian lineages, with the dominant presence of the latter at 62.5% and 53.3%, respectively7,13,14,15,16,17,18. Mitochondrial haplogroup analyses of the NPR Scythians from this study and those from Rostov-on-Don and Pazyryks from Altai and Inner Mongolia, reveal that, for the most part, the same lineages are found in all three groups and are often singularly represented in each group. Noteworthy, comparing the frequencies of east and west Eurasian haplogroups in all three groups of the Scythian horizon, an east-west mtDNA lineage cline is visible, for east Eurasian lineages going west-east is from 26.3% (in present study) through 37.5% (in Scythians from Rostov-on-Don) to 46.7% (in Pazyryks) with the opposite trend for west Eurasian lineages. Otherwise, mt lineage composition is comparable in all three groups of the Scythian horizon which supports their shared maternal genetic roots founded on the common east and west Eurasian substrate with an input from neighboring populations. The genetic influx of East Eurasian haplotypes might be the result of establishing relationships between migrants with European ancestry and women of east Eurasian origin as was previously proposed by66 in case of Iron Age south Siberian populations. However, more detailed studies of autosomal DNA are needed to clearly resolve this issue.
Conclusions
Sequence data from whole mt genomes indicate three potential mtDNA lineage ancestries of the NPR Scythians. The first component traces back to west Eurasian hunter-gatherers and is represented by the lineages belonging to subdivisions of haplogroup U5. The second component is composed of mt lineages connected with Neolithic farming expansion into Europe (H, J, T, W and N1b). The last ancestral mt lineage component is comprised of east Eurasian haplotypes belonging to D, A, F1, H8 and M10 which point to association of the NPR Scythians with east Eurasian populations, in particular from the Altai region. A comparison of NPR Scythian mtDNA lineages with other ancient groups suggests close genetic affinities with representatives of the Bronze Age Srubnaya population, which is in agreement with the archaeological hypothesis suggesting Srubnaya people as the ancestors of the NPR Scythians. However, to provide additional genetic support for this hypothesis data from nuclear genomes are needed.
Additional Information
How to cite this article: Juras, A. et al. Diverse origin of mitochondrial lineages in Iron Age Black Sea Scythians. Sci. Rep. 7, 43950; doi: 10.1038/srep43950 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The project was supported by the Swedish Research Council and Riksbankens Jubileumsfond. Computations were performed at the Swedish National Infrastructure for Computing (SNIC-UPPMAX). E.E. was supported by PRVOUK P15 and Mobility Fund of the Charles University in Prague, Czech Republic.
Footnotes
The authors declare no competing financial interests.
Author Contributions Conceived and designed the experiments: A.J., M.K., S.Ł., M.K.N., M.C., A.G.N., M.D. Performed the experiments: A.J., M.C., M.K., A.G.N. Analyzed the data: A.J., E.E., M.K. Contributed reagents/materials/analysis tools: A.J., M.K., A.G.N., S.Ł., M.K.N., V.S., S.I., M.D., A.G., J.P. Wrote the paper: A.J., A.G.N., M.K., S.Ł., E.E.
References
- Bokovenko N. A. The origins of horse riding and the development of ancient Central Asian nomadic riding harnesses. Kurgans Ritual Sites Settl. Eurasian Bronze Iron Age. BAR International Series 890, 304–310 (2000). [Google Scholar]
- Parzinger H. Die Reiternomaden der eurasischen Steppe während der Skythenzeit. In Im Zeich. Goldenen Greifen Köönigsgräber Skythen (eds Menghin W. & Parzinger H.) 30–48 (Prestel, 2007). [Google Scholar]
- Parzinger H. Die Skythen der ukrainischen Steppe und ihre Stellung in der Welt der eurasischen Reiterno- maden, In Im Gol- Dener Horiz. 4000 Jahre Ukr. (eds Leskovar J. & Zingerle M.-C.) 58–66 (Bibliothek der Provinz, 2010). [Google Scholar]
- Rawlinson G. The History of Herodotus. (London: John Murray, Albemarle Street, 1859). [Google Scholar]
- Skoryj S. A. Skify v Dneprovskoj Pravoberezhnoj stepi (problema vydeleniya iranskogo etnokul’turnogo elementa) (ed. Skoryj S. A.) (1A NANU, 2003). [Google Scholar]
- Pipan M., Baradello L., Forte E. & Finetti I. Ground penetrating radar study of iron age tombs in southeastern Kazakhstan. Archaeol. Prospect. 8, 141–155 (2001). [Google Scholar]
- Ricaut F.-X., Keyser-Tracqui C., Bourgeois J., Crubézy E. & Ludes B. Genetic analysis of a Scytho-Siberian skeleton and its implications for ancient Central Asian migrations. Hum. Biol. 76, 109–125 (2004). [DOI] [PubMed] [Google Scholar]
- Jordana X. et al. The warriors of the steppes: osteological evidence of warfare and violence from Pazyryk tumuli in the Mongolian Altai. J. Archaeol. Sci. 36, 1319–1327 (2009). [Google Scholar]
- Liberov P. D. & Gulyaev V. I. Problemy Skifskoj Archeologii (eds Liberov P. D. & Gulyaev V. I. (Nauka, 1971). [Google Scholar]
- Tierenzokin A. I. Skifskaya kul’tura (ed. Tierenzokin A. I.) (MIA, 1976). [Google Scholar]
- Gulyaev V. I. Skify: Rastsvet i padenie velikogo tsarstva (ed. Gulyaev V. I.) (Mocow: Aleteia, 2005). [Google Scholar]
- Watt J. Y. C. Introduction, In Nomadic Art of the Eastern Eurasian Steppes: The Eugene V. Thaw and Other New York Collections (eds Bunker E. C., Watt J. C. Y., Sun Z.) 3 (Metropolitan Museum of Art, New York, 2002). [Google Scholar]
- Der Sarkissian C. Mitochondrial DNA in ancient human populations of Europe (University of Adelaide, 2011).
- Ricaut F.-X., Keyser-Tracqui C., Cammaert L., Crubézy E. & Ludes B. Genetic analysis and ethnic affinities from two Scytho-Siberian skeletons. Am. J. Phys. Anthropol. 123, 351–360 (2004). [DOI] [PubMed] [Google Scholar]
- González-Ruiz M. et al. Tracing the origin of the east-west population admixture in the Altai region (Central Asia). PloS One 7, e48904 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikisheva T. A. et al. A paleogenetic study of the prehistoric populations of the Altai. Archaeol. Ethnol. Anthropol. Eurasia 32, 130–142 (2007). [Google Scholar]
- Pilipenko A. S., Romaschenko A. G., Molodin V. I., Parzinger H. & Kobzev V. F. Mitochondrial DNA studies of the Pazyryk people (4th to 3rd centuries BC) from northwestern Mongolia. Archaeol. Anthropol. Sci. 2, 231–236 (2010). [Google Scholar]
- Pilipenko A. S., Trapezov R. O., Zhuravlev A. A., Molodin V. I. & Romaschenko A. G. MtDNA Haplogroup A10 Lineages in Bronze Age Samples Suggest That Ancient Autochthonous Human Groups Contributed to the Specificity of the Indigenous West Siberian Population. PloS One 10, e0127182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I. et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D. Y., Eng B., Waye J. S., Dudar J. C. & Saunders S. R. Technical note: improved DNA extraction from ancient bones using silica-based spin columns. Am. J. Phys. Anthropol. 105, 539–543 (1998). [DOI] [PubMed] [Google Scholar]
- Malmström H. et al. More on contamination: the use of asymmetric molecular behavior to identify authentic ancient human DNA. Mol. Biol. Evol. 24, 998–1004 (2007). [DOI] [PubMed] [Google Scholar]
- Meyer M. & Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, pdb.prot5448 (2010). [DOI] [PubMed] [Google Scholar]
- Günther T. et al. Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc. Natl. Acad. Sci. USA 112, 11917–11922 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampa S., Dahlö M., Olason P. I., Hagberg J. & Spjuth O. Lessons learned from implementing a national infrastructure in Sweden for storage and analysis of next-generation sequencing data. GigaScience 2, 9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juras A. et al. Investigating kinship of Neolithic post-LBK human remains from Krusza Zamkowa, Poland using ancient DNA. Forensic Sci. Int. Genet. doi: 10.1016/j.fsigen.2016.10.008. [DOI] [PubMed] [Google Scholar]
- Anderson S. et al. Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 (1981). [DOI] [PubMed] [Google Scholar]
- Andrews R. M. et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23, 147 (1999). [DOI] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma. Oxf. Engl. 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011). [Google Scholar]
- Andrews S. A. Quality control tool for high throughput sequence data (2012).
- Merriman B. & Rothberg J. M., Ion Torrent R&D Team. Progress in ion torrent semiconductor chip based sequencing. Electrophoresis 33, 3397–3417 (2012). [DOI] [PubMed] [Google Scholar]
- Jónsson H., Ginolhac A., Schubert M., Johnson P. L. F. & Orlando L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinforma. Oxf. Engl. 29, 1682–1684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen T. S., Albrechtsen A. & Nielsen R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vianello D. et al. HAPLOFIND: a new method for high-throughput mtDNA haplogroup assignment. Hum. Mutat. 34, 1189–1194 (2013). [DOI] [PubMed] [Google Scholar]
- van Oven M. & Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 30, E386–E394 (2009). [DOI] [PubMed] [Google Scholar]
- Skoglund P., Stora J., Götherström A. & Jakobsson M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci. 40, 4477–4482 (2013). [Google Scholar]
- Hunter J. D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 9, 90–95 (2007). [Google Scholar]
- Nei M. & Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76, 5269–73 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L. & Lischer H. E. L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010). [DOI] [PubMed] [Google Scholar]
- Excoffier L., Smouse P. E. & Quattro J. M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F. et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011). [Google Scholar]
- QGIS Development Team. QGIS Geographic Information System. Open Source Geospatial Foundation Project http://www.qgis.org (2015).
- Brandt G. et al. Ancient DNA reveals key stages in the formation of central European mitochondrial genetic diversity. Science 342, 257–261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamba C. et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 5, 5257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak W. et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. et al. Prehistorical East–west admixture of maternal lineages in a 2,500-year-old population in Xinjiang. Am J Phys Anthr. 142, (2010). [DOI] [PubMed] [Google Scholar]
- Ricaut F.-X. et al. Comparison between morphological and genetic data to estimate biological relationship: The case of the Egyin Gol necropolis (Mongolia). Am. J. Phys. Anthropol. 143, 355–364 (2010). [DOI] [PubMed] [Google Scholar]
- Keyser-Tracqui C., Crubézy E. & Ludes B. Nuclear and Mitochondrial DNA Analysis of a 2,000-Year-Old Necropolis in the Egyin Gol Valley of Mongolia. Am. J. Hum. Genet. 73, 247–260 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyser-Tracqui C., Crubézy E., Pamzsav H., Varga T. & Ludes B. Population origins in Mongolia: Genetic structure analysis of ancient and modern DNA. Am. J. Phys. Anthropol. 131, 272–281 (2006). [DOI] [PubMed] [Google Scholar]
- Fu Q. et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature 524, 216–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollongino R. et al. 2000 Years of Parallel Societies in Stone Age Central Europe. Science 342, 479–481 (2013). [DOI] [PubMed] [Google Scholar]
- Sánchez-Quinto F. et al. Genomic Affinities of Two 7,000-Year-Old Iberian Hunter-Gatherers. Curr. Biol. 22, 1494–1499 (2012). [DOI] [PubMed] [Google Scholar]
- Allentoft M. E. et al. Population genomics of Bronze Age Eurasia. Nature 522, 167–172 (2015). [DOI] [PubMed] [Google Scholar]
- Posth C. et al. Pleistocene Mitochondrial Genomes Suggest a Single Major Dispersal of Non-Africans and a Late Glacial Population Turnover in Europe. Curr. Biol. 26, 827–833 (2016). [DOI] [PubMed] [Google Scholar]
- Mallory J. P. & Adams D. Q. Encyclopedia of Indo-European Culture, Fitzroy Dearborn Publishers (eds Mallory J. P. & Adams D. Q.) (London – Chicago 1997). [Google Scholar]
- Malmström H. et al. Ancient mitochondrial DNA from the northern fringe of the Neolithic farming expansion in Europe sheds light on the dispersion process. Phil Trans R Soc B 370, 20130373 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kılınç G. M. et al. The Demographic Development of the First Farmers in Anatolia. Curr. Biol. doi: 10.1016/j.cub.2016.07.057 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dönmez S. In Knowledge Production From the Black Sea to the Euphrates. Studies Presented in Honour of Önder Bilgi. 129–146 (Bilgin Kültür Sanat Yayınları: Ankara, 2011). [Google Scholar]
- Wilde S. et al. Direct evidence for positive selection of skin, hair, and eye pigmentation in Europeans during the last 5,000 y. Proc Natl Acad Sci 111, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin A. G., Sokhatsky M. P., Kovaliukh M. M. & Videiko M. Y. Comprehensive site chronology and ancient mitochondrial DNA analysis from verteba cave—a trypillian culture site of eneolithic Ukraine. Interdiscip. Archaeol. 1, 9–18 (2010). [Google Scholar]
- Sinor D. The Cambridge history of early inner Asia (ed. Sinor D.) 99 and 111 (Cambridge University Press, 1990).
- Lalueza-Fox C. et al. Unravelling migrations in the steppe: mitochondrial DNA sequences from ancient Central Asians. Proc Biol Sci 271, 941–947 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooder K. P., Schurr T. G., Bamforth F. J., Bazaliiski V. I. & Savel’ev N. A. Population affinities of Neolithic Siberians: a snapshot from prehistoric Lake Baikal. Am. J. Phys. Anthropol. 129, 349–361 (2006). [DOI] [PubMed] [Google Scholar]
- Keyser C. et al. Ancient DNA provides new insights into the history of south Siberian Kurgan people. Hum. Genet. 126, 395–410 (2009). [DOI] [PubMed] [Google Scholar]
- Molodin V. I. et al. Human migrations in the southern region of the West Siberian Plain during the Bronze Age: Archaeological, palaeogenetic and anthropological data, In Population Dynamics in Prehistory and Early History (eds Kaiser E., Burger J. & Schier W.) 90–111 (2012). [Google Scholar]
- Derenko M. et al. Complete mitochondrial DNA analysis of eastern Eurasian haplogroups rarely found in populations of northern Asia and eastern Europe. PloS One 7, e32179 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. Genetic data suggests that the Jinggouzi people are associated with the Donghu, an ancient nomadic group of North China. Hum. Biol. 84, 365–378 (2012). [DOI] [PubMed] [Google Scholar]
- Loogväli E. L. et al. Disuniting uniformity: a pied cladistic canvas of mtDNA haplogroup H in Eurasia. Mol. Biol. Evol. 21, 2012–21 (2004). [DOI] [PubMed] [Google Scholar]
- Murzin V. Y. Skifskaya arkhaika Severnogo Prichernomor’a (ed. Murzin V. Y.) (1984). [Google Scholar]
- Raevsky D. S. Mir Skifskoj Kultury (ed. Raevsky D. S.) (2006). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.