Abstract
The addition of ubiquitin to a target protein has long been implicated in the process of degradation and is the primary mediator of protein turnover in the cell. Recently, however, many non-proteolytic functions of ubiquitination have emerged as key regulators of cellular homeostasis. In this review, we will describe the various non-traditional functions of ubiquitination, with particular focus on how they can be used as signaling entities in cancer formation and progression. Elaboration of this topic can lead to a better understanding of oncogenic mechanisms, as well as the discovery of novel druggable proteins within the ubiquitin pathway.
Keywords: cancer, deubiquitylation (deubiquitination), polyubiquitin chain, signal transduction, ubiquitylation (ubiquitination), monoubiquitination, non-degradative pathways, therapeutic intervention, ubiquitination, deubiquitination
Introduction
Post-translational modifications can alter the functional ability of a protein and are essential for their physiological actions within the eukaryotic cell (1). Ubiquitination, a post-translational modification involving the addition of a ubiquitin (Ub)2 moiety to a protein substrate, is the primary mechanism of protein turnover in the cell and is recognized as the “traditional” function of Ub tagging. Ub-mediated proteolysis has been extensively investigated for decades, and indeed, the Nobel Prize in Chemistry was awarded to Aaron Ciechanover, Avram Hershko, and Irwin Rose in 2004 for the discovery of this mechanism (2). Only in recent years has light been shed on the non-traditional functions of ubiquitination, that is, proteasome-independent functions of ubiquitin tagging.
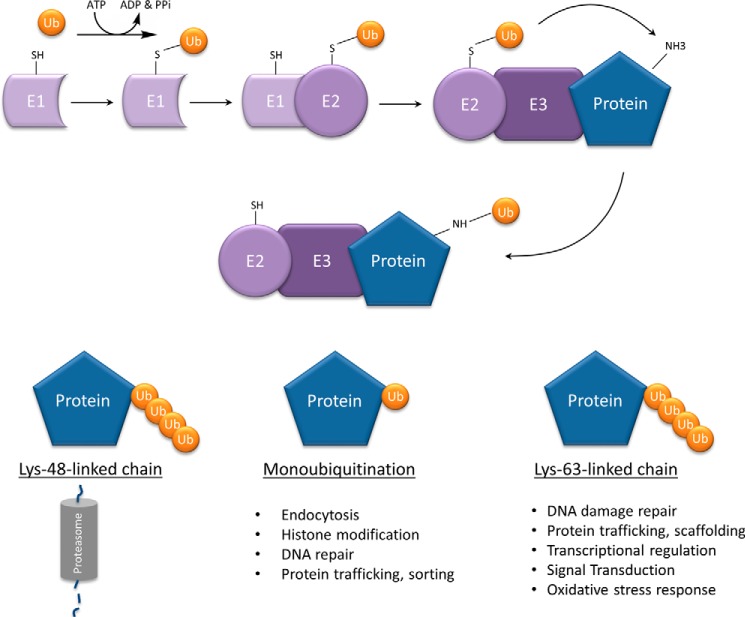
The addition of a Ub moiety to a target protein is a multistep, ATP-dependent process, illustrated in Fig. 1 (3, 4). In most cases, the first Ub molecule is added to an ϵ-amino group on a substrate lysine, but can also be added to the α-NH2 group at the N terminus of the substrate, named N-terminal ubiquitination (5). Ubiquitination of non-lysine sites such as cysteine, serine, and threonine has also been reported and may be a way in which cells adapt to the absence or the shielding of a lysine residue (6, 7). Overall, the end result of Ub tagging is a mono- or polyubiquitinated protein. Polyubiquitin chains can be generated using any of the seven lysine residues on the Ub molecule, with different chain topologies resulting in different functional consequences (8). Polyubiquitin chains linked at lysine residue 48 (Lys-48) shuttle the tagged protein to the 26S proteasome; this degradative form of ubiquitination is often termed the “molecular kiss of death” (9, 10). In contrast, monoubiquitination and Ub chains of different topologies result in altered target protein functionality (Table 1).
FIGURE 1.
Functional consequence of different Ub chain topologies. The addition of a Ub moiety to a target protein is a multistep, ATP-dependent process catalyzed by three key enzymes: Ub-activating enzyme (E1), Ub-conjugating enzyme (E2), and Ub ligase (E3). Initially, the C-terminal carboxylate group of Ub is linked to E1 via a high energy thioester bond. E1 adenylates Ub and transfers it to one of its own cysteine residues, which is then shifted to a sulfhydryl group on one of the many E2 enzymes. Finally, E3 specifically catalyzes the transfer of ubiquitin from E2 to a lysine residue on the substrate protein; really interesting new gene (RING) E3 enzymes catalyze the direct transfer of Ub to the substrate, whereas homologous to the E6-AP C terminus (HECT) E3 enzymes transiently bind Ub via a thioester intermediate before the transfer of Ub. The end result is a mono- or polyubiquitinated protein, with different chain topologies engaging in different functional consequences.
TABLE 1.
Functional consequences of different ubiquitination chain topologies
| Chain formation | Functions |
|---|---|
| Lys-48 | Proteasomal degradation (84) |
| Lys-11 | Proteasomal degradation (85) |
| Regulation of cell division (73) | |
| Lys-63 | DNA damage repair (13) |
| Protein trafficking (14) | |
| Protein sorting (86) | |
| Regulation of transcription (15) | |
| Kinase signaling (16) | |
| Protein scaffolding (16) | |
| Oxidative stress response (87) | |
| Lys-27 | DNA damage response (79) |
| Mitochondrial clearance (88) | |
| Lys-29 | Regulation AMPK-related kinases (74) |
| Lysosomal degradation (89) | |
| Lys-6 | BRCA1-mediated DNA damage repair (75) |
| Lys-33 | Regulation AMPK-related kinases (74) |
| DNA damage response (78) | |
| Monoubiquitination | Endocytosis (11) |
| Intracellular localization (90) | |
| Protein trafficking (91) | |
| Histone modification (12) | |
| DNA damage repair (42) |
Monoubiquitination, the addition of a single Ub molecule, is heavily involved in many proteasome-independent functions of ubiquitination such as endocytosis (11) and histone modification (12). Additionally, Lys-63-linked polyubiquitination is one of the only non-degradative chain formations with a firmly established series of non-traditional functions, for example DNA damage repair (13), protein trafficking (14), transcriptional regulation (15), and kinase signaling (16).
Spence et al. (17) were the first to demonstrate that different Ub chain linkages engage in different functional roles by substituting each individual lysine residue on Ub for arginine in Saccharomyces cerevisiae and observing defects in DNA damage repair. Amino acid substitutions at Lys-63 had no notable effect on protein turnover, indicating a role for Lys-63-linked ubiquitination in non-degradative pathways. Despite this discovery, the rationale behind this differential functional role was initially unknown. It has been since established over the last two decades that the functional diversity of Ub linkages is largely dependent on the structural conformation of the polyubiquitin chain (18). Lys-48-linked chains have a closed, compacted confirmation due to the position of Lys-48 relative to the C terminus of Ub, allowing linking Ub molecules to form a hydrophobic interface that is critical for recognition by the 26S proteasome. Conversely, Lys-63-linked chains have an extended conformation where the hydrophobic patches on the surface of the Ub molecule cannot interact with each other, leading to non-degradative roles (18, 19).
Furthermore, the functional diversity of Ub chains has also been associated with the combinatorial effect of E2 and E3 enzymes. The human genome encodes for ∼30 E2 enzymes within a single evolutionary family, while four distinct E3 families exist, accounting for an estimated 500–1000 ligases. These can form thousands of specific E2-E3 combinations, the pairing of which plays a crucial role in the type of Ub chain formed and the overall fate of the substrate. For example, the E2 enzyme UbcH5c creates a Lys-48-linked chain when combined with E6-AP; however, the same E2 enzyme generates a Lys-6-linked chain when combined with BRCA1/BARD1 (20). Christensen et al. (21) then later established a series of interactions between BRCA1/BARD1 and various E2 enzymes, each combination resulting in a different form of autoubiquitination on BRCA1.
Ubiquitination and Cancer
Because ubiquitination affects so many biochemical processes within the cell, it is not surprising that modifications in this system play a vital role in oncogenesis. The role of ubiquitination in cancer has gained a high degree of interest in recent years, mainly due to the unanticipated efficacy of proteasome inhibition in the clinic (22). Bortezomib, the first proteasome inhibitor to be evaluated in human clinical trials, was approved by the U. S. Food and Drug Administration (FDA) in 2003 for the treatment of relapsed multiple myeloma and in 2006 for mantle cell lymphoma. Acting in a dose-dependent and reversible manner, bortezomib binds to and represses the activity of the 26S proteasome, leading to the accumulation of cell cycle regulatory proteins, and as a result, apoptosis (23, 24). A second generation of proteasome inhibitors has followed, including carfilzomib, which was approved in 2012 to treat multiple myeloma patients who have failed on prior therapies (25), and more recently, ixazomib, the first oral proteasome inhibitor (26). Although inhibition of the proteolytic functions of the Ub system has been a success, these therapies are mostly limited to the treatment of hematological malignancies. With increased reports of drug resistance to add to their limitations, there is a growing demand for the identification of novel druggable components of the Ub system. The therapeutic potentials of the non-degradative functions of ubiquitination, specifically the enzymes that catalyze the addition/removal of these Ub chains, have begun to attract much attention. Here, we will explore the role of these non-traditional functions of ubiquitination and their role in oncogenic pathways.
Monoubiquitination
Histone Modification
Monoubiquitination, the attachment of a single Ub molecule to one or several lysines on a protein, has been widely implicated in oncogenesis largely due to its role in histone modification. Comprising the main structure of the nucleosome, histones are heavily subjected to many post-translational modifications, including methylation, phosphorylation, and indeed, ubiquitination. The post-translationally modified histone is a significant component of the epigenome and controls the accessibility of transcription factors to chromatin, the dysregulation of which can contribute to gene expression alteration and subsequent malignant initiation (12, 27).
Monoubiquitination of histone H2B at position Lys-120 (H2Bub1) can regulate many key cellular mechanisms, including the DNA damage response (DDR) and transcriptional regulation, by physically disrupting chromatin to allow access for DNA repair proteins and transcription factors (12). For example, Prenzel et al. (28) have demonstrated that proteasomal inhibition with bortezomib reduces monoubiquitination of H2B, and in turn, estrogen-driven gene expression. Although proteasomal inhibition was previously shown to alter estrogen receptor α (ERα) transcriptional activity, the mechanism by which it exerts this effect was unknown (29). A decrease in H2Bub1 resulted in defective transcriptional elongation on ERα target genes, and to support this, knockdown of the H2B E3 ligase RNF40 reduced ERα transcriptional activity (28). As 70% of breast cancers depend on ERα for growth and differentiation, elucidation of novel mechanisms that control this transcriptional machinery are vital for breast cancer treatment and management (30). It must be noted, however, that proteasomal inhibition in breast cancer may only benefit a subset of patients, such as those with a more differentiated, ERα/H2Bub1-positive tumor. The authors also demonstrated for the first time that H2Bub1 levels decrease during breast tumorigenesis, and may play a role in estrogen-independent proliferation. Many advanced tumors display decreased H2Bub1, which may correlate with the progressive stages of oncogenesis (12, 31). In this study, H2Bub1 was found to be reduced in primary and metastatic breast cancers, whereas ubiquitinated levels remain uniform in benign breast tissue (28, 32).
Further supporting this, a number of studies have found H2Bub1 to be reduced in gastric, parathyroid, and colorectal tumors (12). Immunohistochemical staining of a 129 colon cancer patient cohort displayed weaker staining in high grade colon cancer cases, when compared with low grade and normal tissue samples (33). Furthermore, cell division cycle 73 (CDC73), a tumor suppressor protein that is altered and/or mutated in the aforementioned tumor types, was found to regulate H2B monoubiquitination through interaction with the RNF20/40 E3 ligases. H2Bub1 was significantly down-regulated in CDC73 mutant parathyroid tumors, indicating a role for CDC73 in H2Bub1 maintenance and possibly a mechanism by which mutant CDC73 applies its oncogenic role (34). Finally, in inflammation-associated colon cancer, down-regulation of RNF20 and H2Bub1 led to enhanced recruitment of NF-κB dimers and transcription of NF-κB target genes. RNF20-deficient mice were prone to inflammation-associated colorectal cancer as a result, with an excess of myeloid-derived suppressor cells that suppress T-cell activity (35). This compelling evidence suggests that monoubiquitination of H2B acts as a tumor suppressor and a reduction in H2Bub1 correlates with a poor prognosis, and may represent a valid area of therapeutic intervention. Monoubiquitination of H2B could be restored by inhibition of its associated deubiquitinating enzymes (DUBs), USP22, USP44, and USP7 (12). Up-regulation of USP22 has been reported in many aggressive cancers including hepatocellular carcinoma (36) and non-small cell lung carcinoma (NSCLC) (37), supporting its potential as a druggable target. A number of studies have recently outlined the identification and use of USP DUB inhibitors in cancer biology, which have been extensively reviewed by D'Arcy et al. (38).
Fanconi Anemia (FA) Disorder
Aside from its involvement in histone modification, accumulating evidence has highlighted a key role for monoubiquitination in FA disorder. FA disorder is a rare autosomal recessive condition that is associated with bone marrow failure, developmental abnormalities, and a lifelong susceptibility to cancer: in particular, acute myeloid leukemia and head and neck cancer (39). FA occurs when a mutation is present in any one of the 20 FA genes, all of which are critical in the repair of DNA interstrand cross-links (ICLs) (40, 41). When cells are exposed to a DNA cross-linking agent, the FA core complex of proteins is activated and initiates the DNA repair process. A key step in this activation includes FANCD2 and FANCI monoubiquitination, which results in dimerization and formation of the ID2 complex. This complex then interacts with RAD51 and BRCA1/2 at the point of DNA damage to coordinate repair (42). In FA patients, aberrations in the core complex will lead to the failure of FANCD2 and FANCI monoubiquitination and consequent abnormal DNA signaling. To further highlight the significance of ubiquitination in the FA pathway, a recent case study has identified an individual with a deficiency in UBE2T, the E2-conjugating enzyme responsible for monoubiquitination of FANCD2 and FANCI, with clinical symptoms of FA, identifying UBE2T as an additional FA gene (43). Monoubiquitination of FANCD2 is therefore thought to be an essential player in DNA repair and response to DNA cross-linking agents (44). The study of the FA pathway has been important in the understanding of how DNA damage repair defects contribute to oncogenesis, although the disorder itself is rare. As a result, elucidating the mechanisms that control ubiquitination within the FA pathway has a wide ranging applicability.
Several emerging studies have identified additional regulators of monoubiquitination of FANCD2 and expanded our understanding of the FA pathway. For example, Chun et al. (46) recently demonstrated a role for AMP-activating protein kinase (AMPK), a key regulator of cellular growth and metabolism (45), in FA activation. AMPK was shown to interact with both FANCA and FANCG, and AMPK knockdown reduced FANCD2 monoubiquitination and enhanced sensitivity to the DNA cross-linking agent mitomycin-C (MM-C) in the U2OS cell line (46). Furthermore, miR-302, a small non-coding RNA, was found to disrupt FANCD2 monoubiquitination and may be a novel regulator of the FA pathway (44). Overexpression of the miR-302 cluster reduced monoubiquitination of FANCD2 in the human embryonic carcinoma cell line NCCIT. miR-302-overexpressing cells were also more sensitive to MM-C and displayed a higher degree of chromosomal breakage (44). Finally, the DUB USP1 reversed the monoubiquitination of FANCD2 by removing Ub and inactivating the FA pathway. USP1 was found to co-localize with FANCD2 following DNA damage, whereas USP1 knockdown resulted in the accumulation of FANCD2-Ub (47). The autocleavage of USP1 was required for down-regulation of the DUB and sufficient monoubiquitination of FANCD2 and repair of ICLs (48). These studies highlight the importance of FA pathway interactors and may represent novel therapeutic targets. Not only could inhibition of these proteins restore FA pathway activation in FA patients, but these pathways could be manipulated to sensitize cancer patients to interstrand cross-linking agents such as MM-C and cisplatin.
On a final note, a recent study by Raghunandan et al. (49) indicated that the FA pathway is not as linear as we once assumed. Interestingly, non-ubiquitinated FANCD2 was found to accumulate on chromatin, and in concert with BRCA2 and FANCJ, it promoted aphidicolin-stalled replication fork recovery, independent of the FA core complex. The E3 ligase activity of the core complex was not required for this event to occur, indicating that the core complex members are dispensable in replication fork restart. Enhancement of replication fork recovery via this ubiquitin-independent mechanism may explain the milder FA phenotypes associated with mutations in the FA core complex (as opposed to downstream members), representing a backup mechanism when monoubiquitination of FANCD2 is impaired. The importance of monoubiquitination in the FA pathway, however, should not be dismissed, although this study provides a new angle from which we should study FA disorder.
Lys-63-linked Ubiquitination
NF-κB Activation
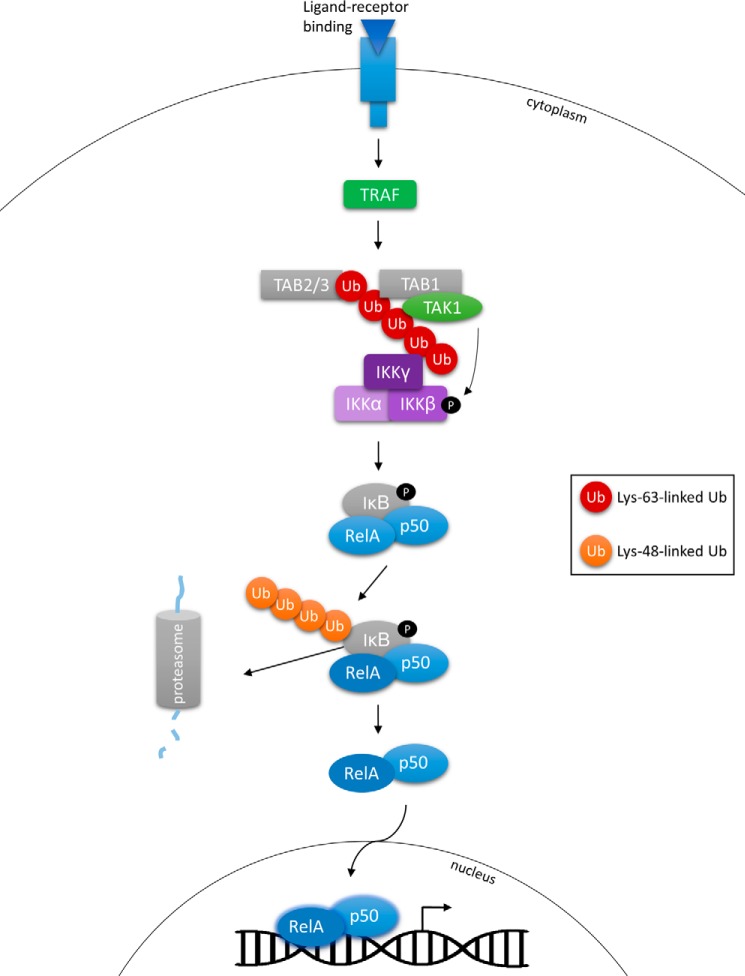
As outlined previously, Lys-63-linked polyubiquitin chains are not involved in protein degradation via the proteasome and carry alternative functional roles. Such functions of this chain formation are well characterized and are continuously emerging as key players in cell proliferation, survival, and cancer development (16). NF-κB, a central complex of transcription factors, plays a pivotal role in immune activation and oncogenesis and is heavily regulated by Lys-63-linked polyubiquitination (50). In the canonical NF-κB pathway, ligand-receptor binding (e.g. IL-1) can recruit members of the TNF receptor-associated factor (TRAF) family of E3 ligases, which mediate Lys-63-linked ubiquitination and recruitment of TGF-β-activated kinase 1 (TAK1). TAK1 phosphorylates and activates the inhibitor of κB kinase (IKK) complex, which in turn phosphorylates IκB, promoting proteasomal degradation and subsequent activation of NF-κB (16, 51). Of key interest here is Lys-63-linked polyubiquitination within the NF-κB pathway, which is responsible for the scaffolding and interaction of key components of the signaling network, allowing them to work together to initiate NF-κB activation (Fig. 2). Aberrations in this network can lead to constitutive NF-κB signaling and consequent cancer formation.
FIGURE 2.
Ubiquitination is a key player in the activation of the NF-κB pathway. IκB is ubiquitinated with a Lys-48-linked chain, allowing for proteasomal degradation and transfer of NF-κB in to the nucleus. Lys-63-linked polyubiquitination, catalyzed by TRAF, acts as a docking site for TAB1-TAK1 activation. The IKK complex is also recruited, allowing for phosphorylation and activation by TAK1.
The IKK complex (or NEMO (NF-κB essential modulator)) is responsible for the maintenance of NF-κB signaling, the IKKβ subunit of which is also polyubiquitinated by a Lys-63-linked chain. Interestingly, mutations in IKKβ at Lys-171 (K171E and K171R), which have been identified in multiple myeloma and mantle cell lymphoma cancers, result in hyperactive kinase signaling and increased Lys-63-linked ubiquitination at this residue. This in turn leads to IL-6-dependent activation of STAT3, which can stimulate the expression of a variety of genes involved in cell growth and is also a positive regulator of Ubc13, the E2 Ub-conjugating enzyme that mediates the Lys-63-linked ubiquitination of TRAF6 (52). Continuous STAT3 activation may contribute to uncontrolled proliferation and evasion of apoptosis in cancers with IKKβ mutations and aberrant NF-κB signaling (53). Receptor-interacting protein 1 (RIP1), which plays a role in NF-κB activation following TNF binding, was found to be constitutively ubiquitinated with a Lys-63-linked poly-Ub chain by the inhibitor of apoptosis (IAP) proteins in a series of cancer cell lines. The poly-Ub chain linked to RIP1 also acts as a docking site for TAK1, which, as mentioned previously, activates NF-κB. Deubiquitination of RIP1 induces apoptosis, indicating that Lys-63-linked ubiquitination elicits a prosurvival function, possibly by constitutive NF-κB activation (54). Interestingly, Lys-63-linked ubiquitination and activation of the NF-κB pathway were also found to promote resistance to epidermal growth factor receptor (EGFR) inhibition in lung adenocarcinoma. Treatment with afatinib or erlotinib induced Lys-63-linked ubiquitination of TRAF2 and subsequent complex formation with RIP1 and the IKK complex, resulting in NF-κB activation and a mechanism by which cancer cells evade death following tyrosine kinase inhibition (55).
In the search for novel regulators of NF-κB activation, Golgi phosphoprotein 3 (GOLPH3) has been identified as a key driver of Lys-63-linked ubiquitination of TRAF2, RIP, and the IKK complex in hepatocellular carcinoma, an aggressive malignancy that is heavily dependent on NF-κB signaling. GOLPH3, an oncoprotein responsible for tubule and vesicle formation, accelerated levels of Lys-63-linked ubiquitination when overexpressed, although the mechanism by which this occurs remains unidentified (56). In addition, recent evidence has suggested a role for human T-cell leukemia virus type 1 (HTLV-1) Tax protein, a constituent activator of NF-κB activity and key player in the formation of adult T-cell leukemia, in the Lys-63-linked ubiquitination of anti-apoptotic protein MCL-1 through its interaction with TRAF6. This interaction caused TRAF6 to relocate to the mitochondria and ubiquitinate MCL-1 with a Lys-63-linked chain. This prevents degradation of MCL-1 and a poor response to chemotherapeutic agents in adult T-cell leukemia (57). Likewise, the T-cell IRAK1/4 signaling pathway was also found to activate TRAF6 and stabilize MCL-1, highlighting the clinical potential of TRAF6 inhibition and Lys-63-linked ubiquitination in T acute lymphoblastic leukemia (T-ALL) (58).
The TRAF family of proteins is of particular interest here due to their dysregulation in many cancer types. For example, TRAF6 was found to be up-regulated in colon tumor tissue and was associated with tumor grade. Sun et al. (59) also demonstrated that knockdown of TRAF6 slows proliferation in the RKO colon cancer cell line. Likewise, knockdown of TRAF6 in a series of lung cancer cell lines inhibited the invasive potential of the cells and promoted apoptotic cell death. In the same study, Lys-63-linked ubiquitination of TRAF6 was also observed alongside constitutive NF-κB signaling in the SPC-A1 human lung cancer cell line (60). Furthermore, TRAF2 has been identified as an oncogene that is amplified and rearranged in 15% of epithelial cancers. Knockdown of TRAF2 in multiple epithelial cancer cell lines harboring a TRAF2 copy number gain resulted in decreased cell proliferation and NF-κB activation (61). This cumulative evidence indicates that the TRAF family of E3 ligases may be the most viable therapeutic target in NF-κB-driven malignancies.
Deubiquitination within the NF-κB Pathway
Many DUBs have been identified as tumor suppressors due to their ability to remove Lys-63-linked Ub chains within the NF-κB pathway. Dysregulation of these tumor suppressor DUBs has been shown to confer a malignant phenotype. For example CYLD, which can negatively regulate NF-κB signaling by specifically deubiquitinating Lys-63-linked Ub chains on TRAF2 and NEMO, is the primary susceptibility gene in familial cylindromatosis, an autosomal dominant genetic disorder that leads to the formation of multiple tumors on the scalp (62). Knockdown of CYLD was found to be anti-apoptotic, possibly the mechanism by which loss of CYLD is oncogenic (63). Furthermore, the Ub-modifying enzyme A20 can remove Lys-63-linked Ub chains from RIP and subsequently ubiquitinate the protein for degradation using its dual DUB/E3 ligase activity (64). This has been identified as one of the key mechanisms by which A20 exerts an inhibitory effect on NF-κB activation, and is recognized as a key tumor suppressor in many cancers as a result (65). In contrast, USP2 has been identified as a positive regulator of NF-κB signaling. When USP2 was knocked down, nuclear translocation of NF-κB was inhibited, along with downstream target gene expression. The authors also demonstrate that USP2 is differentially regulated in breast cancer and tends to localize in the cytoplasm only in tumor tissue. This indicates that USP2 may play a role in breast cancer tumorigenesis, and as aberrant NF-κB signaling is common in this cancer, its therapeutic potential should be further explored (66). This evidence illustrates the importance of Lys-63-linked ubiquitination in aberrant NF-κB activation and identifies Lys-63-linked ubiquitinated proteins (and the E3 ligases driving their formation) as potential drug targets in cancers that are driven by NF-κB signaling.
Akt Signaling
Recent studies have also indicated a role for Lys-63-linked ubiquitination in the regulation of Akt signaling. An imperative member of the phosphoinositide 3-kinase (PI3K) signal transduction pathway, Akt has been recognized as a hub of cell signaling and is involved in cell growth, proliferation, and survival, and has been identified as a key player in the initiation and development of many human malignancies (67, 68). The post-translational regulation of Akt is widely recognized and has been extensively reviewed by Chan et al. (69); however, only in recent years has the regulation of Akt signaling by ubiquitination come to light. Several studies have demonstrated that Akt is targeted by multiple E3 ligases for Lys-48-linked ubiquitination and degradation by the proteasome. However, the studies mentioned below have outlined that Akt is also a substrate for Lys-63-liked ubiquitination and is a key mechanism by which mutated or hyperactive Akt provokes an oncogenic phenotype. For example, mutant Akt E17K, which is found in a subset of human cancers including breast, ovarian, and colorectal (70), has a higher basal level of Lys-63-linked Ub when compared with wild type. Blocking this hyperubiquitination prevents Akt localization to the membrane, indicating that E17K Akt applies its oncogenic role via this mechanism (16).
It was initially assumed that phosphorylation by PDK1 and mTORC2 was the primary mechanism by which Akt is hyperactivated in oncogenesis, but this did not indicate why cancers with normal PI3K pathway activity continued to display aberrant Akt signaling. It was then later established that Lys-63-linked ubiquitination of Akt is responsible for recruitment of the kinase to the cytoplasmic membrane, where it is activated by phosphorylation. This modification may induce a conformational change, allowing Akt to interact with a putative chaperone protein and aid in its membrane recruitment. TRAF6 was identified as the E3 ligase mediating Lys-63-linked ubiquitination of Akt, with overexpression resulting in increased phosphorylation at Thr-308 and Akt activity in primary mouse embryonic fibroblasts (71). Alongside TRAF6, the Skp2 SCF E3 ligase complex was also found to induce Lys-63-linked ubiquitination of Akt in response to EGF signaling via the human epidermal growth factor receptors (HER). Interestingly, Skp2 overexpression correlated with activation of Akt and breast cancer progression, identifying Skp2 as a novel prognostic marker in HER2-positive patients. Chan et al. (72) demonstrated that Skp2 deficiency sensitizes HER2-positive tumors to trastuzumab treatment, highlighting the clinical potential of targeting E3 ligases, and in turn, Lys-63-linked ubiquitination.
Atypical Ubiquitination
Overview
Although the non-degradative functions of monoubiquitination and Lys-63-linked ubiquitination are well established, the functions associated with atypical chain formations have been elusive for a long time. Some of these functions have been characterized in recent years; for example, Lys-11-linked ubiquitination has been identified as a regulator of cell division (73), whereas Lys-29- and Lys-33-linked Ub chains have been found to regulate AMPK-related kinases (74). Nonetheless, information on the role of these atypical Ub chains in oncogenesis is sparse, with the focus primarily falling on the deregulation of the DDR.
BRCA Signaling
The BRCA1 tumor suppressor protein is a widely recognized key player in the DDR. Patients with mutations in the BRCA1 gene produce a truncated BRCA1 protein, leading to impaired DNA damage repair and increased risk of developing early onset breast and ovarian cancer. BRCA1 contains a RING finger domain within the N terminus of the protein that catalyzes the formation of Ub chains when it forms a heterodimer with BARD1: an interaction essential for the E3 ligase activity of BRCA1 (75, 76). Although the majority of BRCA1 mutations affect the C terminus of the protein, a number of missense mutations affect the RING finger coding region within the N terminus. Patients with the C61G missense mutation in the RING finger domain displayed reduced levels of E3 ligase activity, indicating that the BRCA1/BARD1 heterodimer is important to the tumor suppressor function of BRCA1 (20, 77). A comparative mutational analysis involving the BRCA1/BARD1 heterodimer and the E3 ligase E6-AP revealed that BRCA1/BARD1 catalyzes the formation of Lys-6-linked Ub chains and has a preference for this form of chain linkage. It was later demonstrated by Morris and Solomon (75) that BRCA1/BARD1 catalyzes the formation of Lys-6-linked Ub chains during S phase after treatment with hydroxyurea, an agent that inhibits ribonucleotide reductase, and in turn, DNA replication. Relocation of BRCA1 to points of double strand breaks following ionizing radiation treatment was also observed, as determined by γH2AX and BrdU incorporation. Ubiquitination was also observed at these sites, demonstrating a role for BRCA1 at DNA damage foci. Mutational analysis on a series of lysine residues, whereby lysine was converted to arginine (K11R, K33R, K29R, K48R, and K63R), indicated that the majority of Ub conjugation was taking place at Lys-6. Transfection of K6R in the MCF7 cell line resulted in a reduction of conjugated Ub in both hydroxyurea-treated and non-treated cells.
To further support these findings, Elia et al. (78) demonstrated for the first time that endogenous Lys-6-linked ubiquitination (as well as Lys-33-linked ubiquitination) is increased in response to DNA damage induced by UV radiation. Remarkably, there is also evidence for the involvement of Lys-27-linked ubiquitination in the DDR. The E3 ligase RNF168 was found to catalyze the formation of Lys-27-linked Ub chains on histones H2A and H2A.X following DNA damage. These specific Ub chain topologies are recognized by several of the DDR proteins, including 53BP1, Rap80, and BRCA1, and are crucial for the normal DDR activation (79). This strongly suggests a role for atypical polyubiquitination in DNA damage repair, although the exact mechanisms by which these chains can elicit this function should be further explored. Importantly, elucidation of the mechanisms by which BRCA partakes in the DDR is critical to the field and will advance our understanding of the role of BRCA in both familial and sporadic breast and ovarian cancer.
Concluding Remarks
Up to now, the role of non-traditional ubiquitination in cancer has been relatively understudied, with focus falling primarily on the degradative functions and how these can be targeted in the clinic. With the recent emergence of many high profile studies in this topic, the role of non-traditional ubiquitination within key oncogenic pathways has come to light. The Ub system now represents an attractive area for therapeutic intervention, and the development of novel inhibitors to act within this biomolecular pathway may be a feasible task. The inhibition of E3 ligases may be one of the most attractive approaches, as these enzymes target a limited number of substrates and contain an enzymatic catalytic site for which a small molecule inhibitor can be specifically designed (80). DUB inhibition is also an emerging area of interest, with many large pharmaceutical companies now investing in this class of drugs (81). Several DUBs have been described as key players in tumorigenesis, and as cysteine proteases, they also represent a very “druggable” class of proteins (38). To support this, there are a number of drug candidates in preclinical development with several DUB inhibitors already validated at an in vitro level (82, 83).
Here, we have highlighted the emerging role of non-traditional ubiquitination in oncogenic pathways, and although more research is required to determine the effectiveness of targeting such mechanisms, this form of ubiquitination may represent a novel area of therapeutic interest in the oncology arena.
This work was supported by BREAST-PREDICT, The Irish Cancer Society's Collaborative Cancer Research Centre (Grant CCRC13GAL), and Science Foundation Ireland, in particular the OPTi-PREDICT project, which was funded under the Investigator Programme (Grant 15/IA/3104). The authors declare that they have no conflicts of interest with the contents of this article.
- Ub
- ubiquitin
- DDR
- DNA damage response
- ERα
- estrogen receptor α
- FA
- Fanconi anemia
- DUB
- deubiquitinating enzyme
- ICL
- interstrand cross-link
- USP
- ubiquitin-specific protease
- AMPK
- AMP-activating protein kinase
- MM-C
- mitomycin-C
- TRAF
- TNF receptor-associated factor
- IKK
- inhibitor of κB kinase
- RIP
- receptor-interacting protein.
References
- 1. Wang Y. C., Peterson S. E., and Loring J. F. (2014) Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 24, 143–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hershko A., and Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 3. Haas A. L., and Siepmann T. J. (1997) Pathways of ubiquitin conjugation. FASEB J. 11, 1257–1268 [DOI] [PubMed] [Google Scholar]
- 4. Ciechanover A., Orian A., and Schwartz A. L. (2000) Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays 22, 442–451 [DOI] [PubMed] [Google Scholar]
- 5. Ciechanover A., and Ben-Saadon R. (2004) N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 14, 103–106 [DOI] [PubMed] [Google Scholar]
- 6. Kravtsova-Ivantsiv Y., and Ciechanover A. (2012) Non-canonical ubiquitin-based signals for proteasomal degradation. J. Cell Sci. 125, 539–548 [DOI] [PubMed] [Google Scholar]
- 7. Cadwell K., and Coscoy L. (2005) Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 309, 127–130 [DOI] [PubMed] [Google Scholar]
- 8. Suryadinata R., Roesley S. N., Yang G., and Sarčević B. (2014) Mechanisms of generating polyubiquitin chains of different topology. Cells 3, 674–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pickart C. M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 10. Pickart C. M., and Eddins M. J. (2004) Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 [DOI] [PubMed] [Google Scholar]
- 11. Haglund K., Di Fiore P. P., and Dikic I. (2003) Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci. 28, 598–603 [DOI] [PubMed] [Google Scholar]
- 12. Cole A. J., Clifton-Bligh R., and Marsh D. J. (2015) Histone H2B monoubiquitination: roles to play in human malignancy. Endocr.-Relat. Cancer 22, T19–T33 [DOI] [PubMed] [Google Scholar]
- 13. Messick T. E., and Greenberg R. A. (2009) The ubiquitin landscape at DNA double-strand breaks. J. Cell Biol. 187, 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Erpapazoglou Z., Walker O., and Haguenauer-Tsapis R. (2014) Versatile roles of K63-linked ubiquitin chains in trafficking. Cells 3, 1027–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vallabhapurapu S., and Karin M. (2009) Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27, 693–733 [DOI] [PubMed] [Google Scholar]
- 16. Wang G., Gao Y., Li L., Jin G., Cai Z., Chao J. I., and Lin H. K. (2012) K63-linked ubiquitination in kinase activation and cancer. Front. Oncol. 2, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spence J., Sadis S., Haas A. L., and Finley D. (1995) A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 15, 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Varadan R., Walker O., Pickart C., and Fushman D. (2002) Structural properties of polyubiquitin chains in solution. J. Mol. Biol. 324, 637–647 [DOI] [PubMed] [Google Scholar]
- 19. Varadan R., Assfalg M., Haririnia A., Raasi S., Pickart C., and Fushman D. (2004) Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J. Biol. Chem. 279, 7055–7063 [DOI] [PubMed] [Google Scholar]
- 20. Wu-Baer F., Lagrazon K., Yuan W., and Baer R. (2003) The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J. Biol. Chem. 278, 34743–34746 [DOI] [PubMed] [Google Scholar]
- 21. Christensen D. E., Brzovic P. S., and Klevit R. E. (2007) E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 14, 941–948 [DOI] [PubMed] [Google Scholar]
- 22. Moreau P., Richardson P. G., Cavo M., Orlowski R. Z., San Miguel J. F., Palumbo A., and Harousseau J. L. (2012) Proteasome inhibitors in multiple myeloma: 10 years later. Blood 120, 947–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bold R. (2004) “Development of the proteasome inhibitor Velcade (Bortezomib)” by Julian Adams, Ph.D., and Michael Kauffman, M.D., Ph.D. Cancer Invest. 22, 328–329 [DOI] [PubMed] [Google Scholar]
- 24. Pellom S. T. Jr., and Shanker A. (2012) Development of proteasome inhibitors as therapeutic drugs. J. Clin. Cell. Immunol. S5, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fostier K., De Becker A., and Schots R. (2012) Carfilzomib: a novel treatment in relapsed and refractory multiple myeloma. Onco Targets Ther. 5, 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moreau P., Masszi T., Grzasko N., Bahlis N. J., Hansson M., Pour L., Sandhu I., Ganly P., Baker B. W., Jackson S. R., Stoppa A. M., Simpson D. R., Gimsing P., Palumbo A., Garderet L., et al. (2016) Oral Ixazomib, Lenalidomide, and Dexamethasone for multiple myeloma. N. Engl. J. Med. 374, 1621–1634 [DOI] [PubMed] [Google Scholar]
- 27. Chervona Y., and Costa M. (2012) Histone modifications and cancer: biomarkers of prognosis? Am. J. Cancer Res. 2, 589–597 [PMC free article] [PubMed] [Google Scholar]
- 28. Prenzel T., Begus-Nahrmann Y., Kramer F., Hennion M., Hsu C., Gorsler T., Hintermair C., Eick D., Kremmer E., Simons M., Beissbarth T., and Johnsen S. A. (2011) Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B. Cancer Res. 71, 5739–5753 [DOI] [PubMed] [Google Scholar]
- 29. Fan M., Nakshatri H., and Nephew K. P. (2004) Inhibiting proteasomal proteolysis sustains estrogen receptor-α activation. Mol. Endocrinol. 18, 2603–2615 [DOI] [PubMed] [Google Scholar]
- 30. Oosterkamp H. M., Hijmans E. M., Brummelkamp T. R., Canisius S., Wessels L. F., Zwart W., and Bernards R. (2014) USP9X downregulation renders breast cancer cells resistant to tamoxifen. Cancer Res. 74, 3810–3820 [DOI] [PubMed] [Google Scholar]
- 31. Cao J., and Yan Q. (2012) Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu Y., Chen P., Jing Y., Wang C., Men Y. L., Zhan W., Wang Q., Gan Z., Huang J., Xie K., Mi J., Yu C., Yu X., Chen P. C., Chang J. F., et al. (2015) Microarray analysis reveals potential biological functions of histone H2B monoubiquitination. PLoS One 10, e0133444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Z., Zhu L., Guo T., Wang Y., and Yang J. (2015) Decreased H2B monoubiquitination and overexpression of ubiquitin-specific protease enzyme 22 in malignant colon carcinoma. Hum. Pathol. 46, 1006–1014 [DOI] [PubMed] [Google Scholar]
- 34. Hahn M. A., Dickson K. A., Jackson S., Clarkson A., Gill A. J., and Marsh D. J. (2012) The tumor suppressor CDC73 interacts with the ring finger proteins RNF20 and RNF40 and is required for the maintenance of histone 2B monoubiquitination. Hum. Mol. Genet. 21, 559–568 [DOI] [PubMed] [Google Scholar]
- 35. Tarcic O., Pateras I. S., Cooks T., Shema E., Kanterman J., Ashkenazi H., Boocholez H., Hubert A., Rotkopf R., Baniyash M., Pikarsky E., Gorgoulis V. G., and Oren M. (2016) RNF20 links histone H2B ubiquitylation with inflammation and inflammation-associated cancer. Cell Rep. 14, 1462–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tang B., Tang F., Li B., Yuan S., Xu Q., Tomlinson S., Jin J., Hu W., and He S. (2015) High USP22 expression indicates poor prognosis in hepatocellular carcinoma. Oncotarget 6, 12654–12667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ding F., Bao C., Tian Y., Xiao H., Wang M., Xie X., Hu F., and Mei J. (2014) USP22 promotes NSCLC tumorigenesis via MDMX up-regulation and subsequent p53 inhibition. Int. J. Mol. Sci. 16, 307–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. D'Arcy P., Wang X., and Linder S. (2015) Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol. Ther. 147, 32–54 [DOI] [PubMed] [Google Scholar]
- 39. Mathew C. G. (2006) Fanconi anaemia genes and susceptibility to cancer. Oncogene 25, 5875–5884 [DOI] [PubMed] [Google Scholar]
- 40. Jo U., and Kim H. (2015) Exploiting the Fanconi anemia pathway for targeted anti-cancer therapy. Mol. Cells 38, 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Renaudin X., Koch Lerner L., Menck C. F., and Rosselli F. (2016) The ubiquitin family meets the Fanconi anemia proteins. Mutat. Res. Rev. Mutat. Res. 769, 36–46 [DOI] [PubMed] [Google Scholar]
- 42. Gregory R. C., Taniguchi T., and D'Andrea A. D. (2003) Regulation of the Fanconi anemia pathway by monoubiquitination. Semin. Cancer Biol. 13, 77–82 [DOI] [PubMed] [Google Scholar]
- 43. Rickman K. A., Lach F. P., Abhyankar A., Donovan F. X., Sanborn E. M., Kennedy J. A., Sougnez C., Gabriel S. B., Elemento O., Chandrasekharappa S. C., Schindler D., Auerbach A. D., and Smogorzewska A. (2015) Deficiency of UBE2T, the E2 ubiquitin ligase necessary for FANCD2 and FANCI ubiquitination, causes FA-T Subtype of Fanconi anemia. Cell Rep. 12, 35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Suresh B., Kumar A. M., Jeong H. S., Cho Y. H., Ramakrishna S., and Kim K. S. (2015) Regulation of Fanconi anemia protein FANCD2 monoubiquitination by miR-302. Biochem. Biophys. Res. Commun. 466, 180–185 [DOI] [PubMed] [Google Scholar]
- 45. Mihaylova M. M., and Shaw R. J. (2011) The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 13, 1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chun M. J., Kim S., Hwang S. K., Kim B. S., Kim H. G., Choi H. I., Kim J. H., Goh S. H., and Lee C. H. (2016) AMP-activated protein kinase is involved in the activation of the Fanconi anemia/BRCA pathway in response to DNA interstrand crosslinks. Oncotarget 7, 53642–53653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nijman S. M., Huang T. T., Dirac A. M., Brummelkamp T. R., Kerkhoven R. M., D'Andrea A. D., and Bernards R. (2005) The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell 17, 331–339 [DOI] [PubMed] [Google Scholar]
- 48. Kim M., and Kim J. M. (2016) The role of USP1 autocleavage in DNA interstrand crosslink repair. FEBS Lett. 590, 340–348 [DOI] [PubMed] [Google Scholar]
- 49. Raghunandan M., Chaudhury I., Kelich S. L., Hanenberg H., and Sobeck A. (2015) FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi Anemia core complex. Cell Cycle 14, 342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hayakawa M. (2012) Role of K63-linked polyubiquitination in NF-κB signalling: which ligase catalyzes and what molecule is targeted? J. Biochem. 151, 115–118 [DOI] [PubMed] [Google Scholar]
- 51. Lavorgna A., and Harhaj E. W. (2014) Regulation of HTLV-1 tax stability, cellular trafficking and NF-κB activation by the ubiquitin-proteasome pathway. Viruses 6, 3925–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang H., Hu H., Greeley N., Jin J., Matthews A. J., Ohashi E., Caetano M. S., Li H. S., Wu X., Mandal P. K., McMurray J. S., Moghaddam S. J., Sun S. C., and Watowich S. S. (2014) STAT3 restrains RANK- and TLR4-mediated signalling by suppressing expression of the E2 ubiquitin-conjugating enzyme Ubc13. Nat. Commun. 5, 5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gallo L. H., Meyer A. N., Motamedchaboki K., Nelson K. N., Haas M., and Donoghue D. J. (2014) Novel Lys63-linked ubiquitination of IKKβ induces STAT3 signaling. Cell Cycle 13, 3964–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bertrand M. J., Milutinovic S., Dickson K. M., Ho W. C., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., and Barker P. A. (2008) cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell 30, 689–700 [DOI] [PubMed] [Google Scholar]
- 55. Blakely C. M., Pazarentzos E., Olivas V., Asthana S., Yan J. J., Tan I., Hrustanovic G., Chan E., Lin L., Neel D. S., Newton W., Bobb K. L., Fouts T. R., Meshulam J., Gubens M. A., et al. (2015) NF-κB-activating complex engaged in response to EGFR oncogene inhibition drives tumor cell survival and residual disease in lung cancer. Cell Rep. 11, 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dai T., Zhang D., Cai M., Wang C., Wu Z., Ying Z., Wu J., Li M., Xie D., Li J., and Song L. (2015) Golgi phosphoprotein 3 (GOLPH3) promotes hepatocellular carcinoma cell aggressiveness by activating the NF-κB pathway. J. Pathol. 235, 490–501 [DOI] [PubMed] [Google Scholar]
- 57. Choi Y. B., and Harhaj E. W. (2014) HTLV-1 tax stabilizes MCL-1 via TRAF6-dependent K63-linked polyubiquitination to promote cell survival and transformation. PLoS Pathog. 10, e1004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li Z., Younger K., Gartenhaus R., Joseph A. M., Hu F., Baer M. R., Brown P., and Davila E. (2015) Inhibition of IRAK1/4 sensitizes T cell acute lymphoblastic leukemia to chemotherapies. J. Clin. Invest. 125, 1081–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sun H., Li X., Fan L., Wu G., Li M., and Fang J. (2014) TRAF6 is upregulated in colon cancer and promotes proliferation of colon cancer cells. Int. J. Biochem. Cell Biol. 53, 195–201 [DOI] [PubMed] [Google Scholar]
- 60. He Z., Huang C., Lin G., and Ye Y. (2016) siRNA-induced TRAF6 knockdown promotes the apoptosis and inhibits the invasion of human lung cancer SPC-A1 cells. Oncol. Rep. 35, 1933–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shen R. R., Zhou A. Y., Kim E., O'Connell J. T., Hagerstrand D., Beroukhim R., and Hahn W. C. (2015) TRAF2 is an NF-κB-activating oncogene in epithelial cancers. Oncogene 34, 209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kovalenko A., Chable-Bessia C., Cantarella G., Israël A., Wallach D., and Courtois G. (2003) The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature 424, 801–805 [DOI] [PubMed] [Google Scholar]
- 63. Brummelkamp T. R., Nijman S. M., Dirac A. M., and Bernards R. (2003) Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature 424, 797–801 [DOI] [PubMed] [Google Scholar]
- 64. Wertz I. E., O'Rourke K. M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D. L., Ma A., Koonin E. V., and Dixit V. M. (2004) De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 430, 694–699 [DOI] [PubMed] [Google Scholar]
- 65. Hymowitz S. G., and Wertz I. E. (2010) A20: from ubiquitin editing to tumour suppression. Nat. Rev. Cancer 10, 332–341 [DOI] [PubMed] [Google Scholar]
- 66. Metzig M., Nickles D., Falschlehner C., Lehmann-Koch J., Straub B. K., Roth W., and Boutros M. (2011) An RNAi screen identifies USP2 as a factor required for TNF-α-induced NF-κB signaling. Int. J. Cancer 129, 607–618 [DOI] [PubMed] [Google Scholar]
- 67. Testa J. R., and Tsichlis P. N. (2005) AKT signaling in normal and malignant cells. Oncogene 24, 7391–7393 [DOI] [PubMed] [Google Scholar]
- 68. Altomare D. A., and Testa J. R. (2005) Perturbations of the AKT signaling pathway in human cancer. Oncogene 24, 7455–7464 [DOI] [PubMed] [Google Scholar]
- 69. Chan C. H., Jo U., Kohrman A., Rezaeian A. H., Chou P. C., Logothetis C., and Lin H. K. (2014) Posttranslational regulation of Akt in human cancer. Cell Biosci. 4, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Carpten J. D., Faber A. L., Horn C., Donoho G. P., Briggs S. L., Robbins C. M., Hostetter G., Boguslawski S., Moses T. Y., Savage S., Uhlik M., Lin A., Du J., Qian Y. W., Zeckner D. J., et al. (2007) A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 448, 439–444 [DOI] [PubMed] [Google Scholar]
- 71. Yang W. L., Wang J., Chan C. H., Lee S. W., Campos A. D., Lamothe B., Hur L., Grabiner B. C., Lin X., Darnay B. G., and Lin H. K. (2009) The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 325, 1134–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chan C. H., Li C. F., Yang W. L., Gao Y., Lee S. W., Feng Z., Huang H. Y., Tsai K. K., Flores L. G., Shao Y., Hazle J. D., Yu D., Wei W., Sarbassov D., Hung M. C., et al. (2012) The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell 149, 1098–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wickliffe K. E., Williamson A., Meyer H. J., Kelly A., and Rape M. (2011) K11-linked ubiquitin chains as novel regulators of cell division. Trends Cell Biol. 21, 656–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Al-Hakim A. K., Zagorska A., Chapman L., Deak M., Peggie M., and Alessi D. R. (2008) Control of AMPK-related kinases by USP9X and atypical Lys29/Lys33-linked polyubiquitin chains. Biochem. J. 411, 249–260 [DOI] [PubMed] [Google Scholar]
- 75. Morris J. R., and Solomon E. (2004) BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum. Mol. Genet. 13, 807–817 [DOI] [PubMed] [Google Scholar]
- 76. McCarthy E. E., Celebi J. T., Baer R., and Ludwig T. (2003) Loss of Bard1, the heterodimeric partner of the Brca1 tumor suppressor, results in early embryonic lethality and chromosomal instability. Mol. Cell. Biol. 23, 5056–5063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hashizume R., Fukuda M., Maeda I., Nishikawa H., Oyake D., Yabuki Y., Ogata H., and Ohta T. (2001) The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 276, 14537–14540 [DOI] [PubMed] [Google Scholar]
- 78. Elia A. E., Boardman A. P., Wang D. C., Huttlin E. L., Everley R. A., Dephoure N., Zhou C., Koren I., Gygi S. P., and Elledge S. J. (2015) Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol. Cell 59, 867–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gatti M., Pinato S., Maiolica A., Rocchio F., Prato M. G., Aebersold R., and Penengo L. (2015) RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep. 10, 226–238 [DOI] [PubMed] [Google Scholar]
- 80. Shen M., Schmitt S., Buac D., and Dou Q. P. (2013) Targeting the ubiquitin-proteasome system for cancer therapy. Expert Opin. Ther. Targets 17, 1091–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Farshi P., Deshmukh R. R., Nwankwo J. O., Arkwright R. T., Cvek B., Liu J., and Dou Q. P. (2015) Deubiquitinases (DUBs) and DUB inhibitors: a patent review. Expert Opin. Ther. Pat 25, 1191–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kushwaha D., O'Leary C., Cron K. R., Deraska P., Zhu K., D'Andrea A. D., and Kozono D. (2015) USP9X inhibition promotes radiation-induced apoptosis in non-small cell lung cancer cells expressing mid-to-high MCL1. Cancer Biol. Ther. 16, 392–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Colland F., Formstecher E., Jacq X., Reverdy C., Planquette C., Conrath S., Trouplin V., Bianchi J., Aushev V. N., Camonis J., Calabrese A., Borg-Capra C., Sippl W., Collura V., Boissy G., et al. (2009) Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol. Cancer Ther. 8, 2286–2295 [DOI] [PubMed] [Google Scholar]
- 84. Pickart C. M. (1997) Targeting of substrates to the 26S proteasome. FASEB J. 11, 1055–1066 [DOI] [PubMed] [Google Scholar]
- 85. Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., and Peng J. (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lauwers E., Jacob C., and André B. (2009) K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185, 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Silva G. M., Finley D., and Vogel C. (2015) K63 polyubiquitination is a new modulator of the oxidative stress response. Nat. Struct. Mol. Biol. 22, 116–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., and Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 89. Chastagner P., Israël A., and Brou C. (2006) Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep. 7, 1147–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jura N., Scotto-Lavino E., Sobczyk A., and Bar-Sagi D. (2006) Differential modification of Ras proteins by ubiquitination. Mol. Cell 21, 679–687 [DOI] [PubMed] [Google Scholar]
- 91. Su Y. T., Gao C., Liu Y., Guo S., Wang A., Wang B., Erdjument-Bromage H., Miyagi M., Tempst P., and Kao H. Y. (2013) Monoubiquitination of filamin B regulates vascular endothelial growth factor-mediated trafficking of histone deacetylase 7. Mol. Cell. Biol. 33, 1546–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]