Abstract
The mitogen-activated protein kinases (MAPKs) have been shown to regulate skeletal muscle function. Previously, we showed that MAPK phosphatase-5 (MKP-5) negatively regulates myogenesis and regeneration of skeletal muscle through inhibition of p38 MAPK and c-Jun N-terminal kinase (JNK). However, the identity and contribution of MKP-5-regulated MAPK targets in the control of skeletal muscle function and regenerative myogenesis have not been established. To identify MKP-5-regulated MAPK substrates in skeletal muscle, we performed a global differential phospho-MAPK substrate screen in regenerating skeletal muscles of wild type and MKP-5-deficient mice. We discovered a novel MKP-5-regulated MAPK substrate called guanine nucleotide exchange factor for Rab3A (GRAB) that was hyperphosphorylated on a phospho-MAPK motif in skeletal muscle of MKP-5-deficient mice. GRAB was found to be phosphorylated by JNK on serine 169. Myoblasts overexpressing a phosphorylation-defective mutant of GRAB containing a mutation at Ser-169 to Ala-169 (GRAB-S169A) inhibited the ability of C2C12 myoblasts to differentiate. We found that GRAB phosphorylation at Ser-169 was required for the secretion of the promyogenic cytokine interleukin 6 (IL-6). Consistent with this observation, MKP-5-deficient mice exhibited increased circulating IL-6 expression as compared with wild type mice. Collectively, these data demonstrate a novel mechanism whereby MKP-5-mediated regulation of JNK negatively regulates phosphorylation of GRAB, which subsequently controls secretion of IL-6. These data support the notion that MKP-5 serves as a negative regulator of MAPK-dependent signaling of critical skeletal muscle signaling pathways.
Keywords: cytokine, dual specificity phosphoprotein phosphatase, mitogen-activated protein kinase (MAPK), myogenesis, phosphoproteomics, skeletal muscle
Introduction
MAPK phosphatases (MKPs)2 are members of the dual specificity protein-tyrosine phosphatase family that specifically dephosphorylate the MAPKs (1). We have shown that the MKPs can both positively and negatively regulate myogenesis through coordinate MAPK dephosphorylation (2–5). In particular, we have shown that loss of MKP-5 expression in mice results in enhanced satellite cell proliferation and differentiation, which contribute to the increased capacity of skeletal muscle to regenerate (3). Furthermore, genetic ablation of MKP-5 in a model of dystrophic muscle disease abrogates the dystrophic phenotype, suggesting an important role for MKP-5 in skeletal muscle disease progression (3). Thus, understanding the signaling pathway(s) regulated by MKP-5 in skeletal muscle homeostasis might provide new insight into the mechanisms of both skeletal muscle growth and the progression of dystrophic muscle disease.
The maintenance of skeletal muscle function is a dynamic process that requires the interplay between skeletal muscle and trophic factors. An imbalance in the equilibrium between the factors that positively and negatively regulate skeletal muscle function is associated with diseases such as atrophy, cancer cachexia, muscular dystrophies, and neuromuscular disorders. Skeletal muscle produces and secretes a variety of cytokines and growth factors that control skeletal muscle growth and regeneration. In particular, interleukin-6 (IL-6) is the most prominent and has been defined as a myokine because of its production from the myofiber (6). IL-6 is induced in response to exercise and acts as a promyogenic cytokine that stimulates skeletal muscle regeneration and increases skeletal muscle mass (7–9).
Many of the MKPs have been shown to regulate a plethora of signaling events through MAPK-dependent mechanisms (1). However, little is known about the subsets of MAPK substrates that are controlled by a specific MKP/MAPK module(s). To identify the MKP-5-regulated MAPK substrates that contribute to the ability of MKP-5 to negatively regulate skeletal muscle growth and regeneration, we performed a differential MAPK substrate proteomic screen in regenerating skeletal muscles of MKP-5-deficient mice. We identified the guanine nucleotide exchange factor for Rab3A (GRAB) (10) as a novel MAPK substrate in regenerating skeletal muscle. We uncovered a mechanism whereby MKP-5-regulated JNK phosphorylation of GRAB is required for the secretion of IL-6 and subsequently myogenesis. Collectively, these data provide insight in to how IL-6 secretion is regulated in skeletal muscle and provide further evidence for a critical role for MKP-5 in the maintenance of skeletal muscle mass and coordination of regenerative myogenesis.
Results
Identification of GRAB as a MKP-5-regulated MAPK Substrate in Skeletal Muscle
To identify MKP-5-regulated MAPK substrates that contribute to the ability of MKP-5 to negatively control skeletal muscle growth and regeneration, we performed an unbiased differential phosphoproteomic MAPK substrate screen in skeletal muscle from mkp-5+/+ and mkp5−/− mice. Skeletal muscle lysates were collected from non-damaged mkp-5+/+ and mkp-5−/− mice and from mkp-5+/+ and mkp-5−/− mice 2 days after injection of cardiotoxin to induce skeletal muscle regeneration. Muscle homogenates were prepared and enriched for peptides containing phosphorylated MAPK substrates using a phospho-MAPK substrate motif antibody, which recognizes the conserved MAPK substrate motif (PXS*P or S*PX(R/K) where S* indicates phosphorylated serine) (Fig. 1A). The relative abundance of phosphorylated MAPK substrates was assessed by MS/MS spectrometry as described previously (11–13) (Fig. 1A). We focused on hyperphosphorylated phospho-MAPK peptides because those likely represented MAPK substrates that were regulated by MKP-5. In contrast, phospho-MAPK peptides that were hypophosphorylated were unlikely to be dependent upon the desired MKP-5/MAPK pathway. Our analysis revealed that 118 proteins represented those that were hyperphosphorylated exclusively (Fig. 1B). Additionally, 20 peptides were identified to be highly hyperphosphorylated (log2 >1.33) on the MAPK substrate motif in regenerating skeletal muscle from mkp-5−/− mice as compared with wild type controls (Fig. 1B). The highest hyperphosphorylated peptide was identified in regenerating skeletal muscle of mkp-5−/− mice to be GRAB (also known as Rab3A-interacting protein (Rabin3)-like 1 (Rab3il1) (10)) (Fig. 1, B and C). GRAB was hyperphosphorylated on the MAPK substrate motif by ∼8-fold in regenerating skeletal muscles from mkp-5−/− mice (Fig. 1, B and C). In contrast, GRAB was not found to be hyperphosphorylated in undamaged muscle. The most abundantly hyperphosphorylated protein in undamaged skeletal muscle of mkp-5−/− mice was the glycogen-targeting subunit of protein phosphatase 1 encoded by the gene PPP1R3A (14). Because the focus of this study was specifically on MKP-5 in regenerating skeletal muscle, we chose to focus on GRAB.
FIGURE 1.
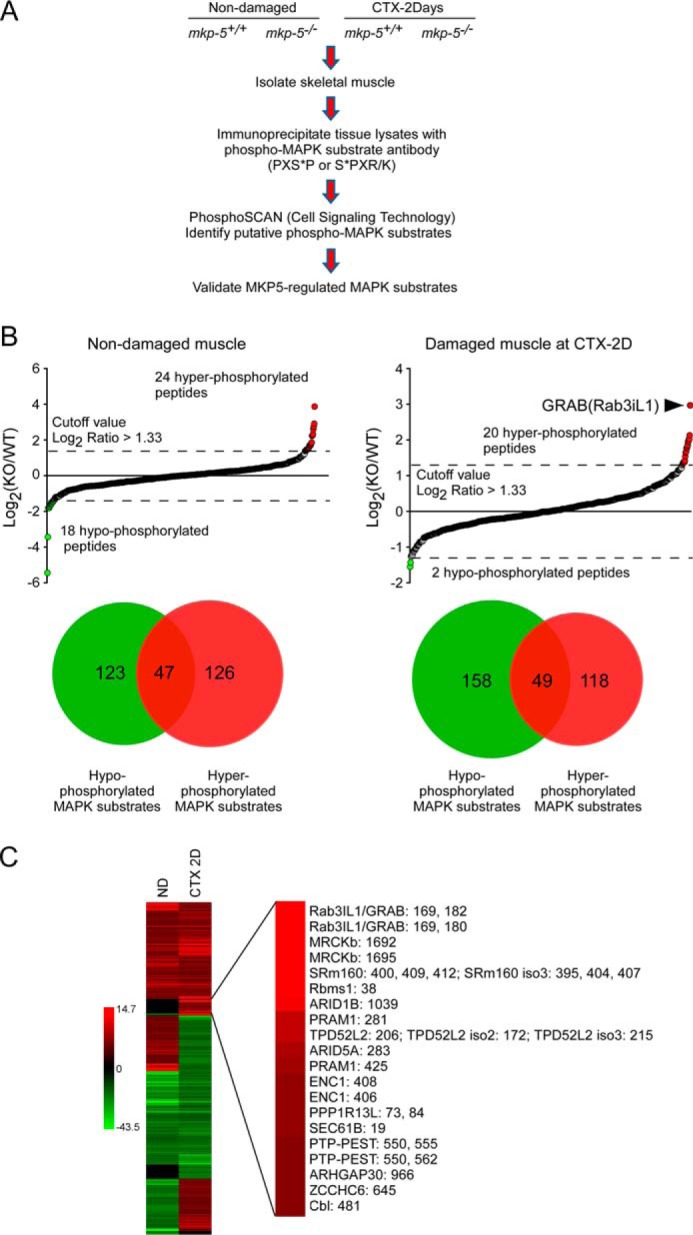
Phospho-MAPK substrate proteomic screen. A, schematic representation of phospho-MAPK substrate proteomic screen in skeletal muscle. B, phospho-MAPK substrate proteomic analysis of differentially phosphorylated proteins in skeletal muscle of mkp-5+/+ and mkp-5−/− mice. Upper graphs show the log2-transformed values for the ratio of each phosphopeptide in mkp-5+/+ (WT) and mkp-5−/− (KO) skeletal muscles. ND, non-damaged muscle; CTX 2D, damaged muscle 2 days after cardiotoxin injection. The Venn diagrams represent the number of hyper- (red) and hypophosphorylated (green) proteins in mkp-5−/− skeletal muscles as compared with mkp-5+/+ muscles. Overlapping proteins represent those containing more than one phosphorylation site identified to be hyper- and hypophosphorylated. C, heat map of hyper- (red) and hypophosphorylated (green) peptides showing the ratio of differentially phosphorylated peptides in skeletal muscles between mkp-5+/+ and mkp-5−/− mice. The right panel indicates the ranking of hyperphosphorylated proteins based on their relative level of phosphorylation of each peptide compared between regenerating skeletal muscles of mkp-5+/+ and mkp-5−/− mice.
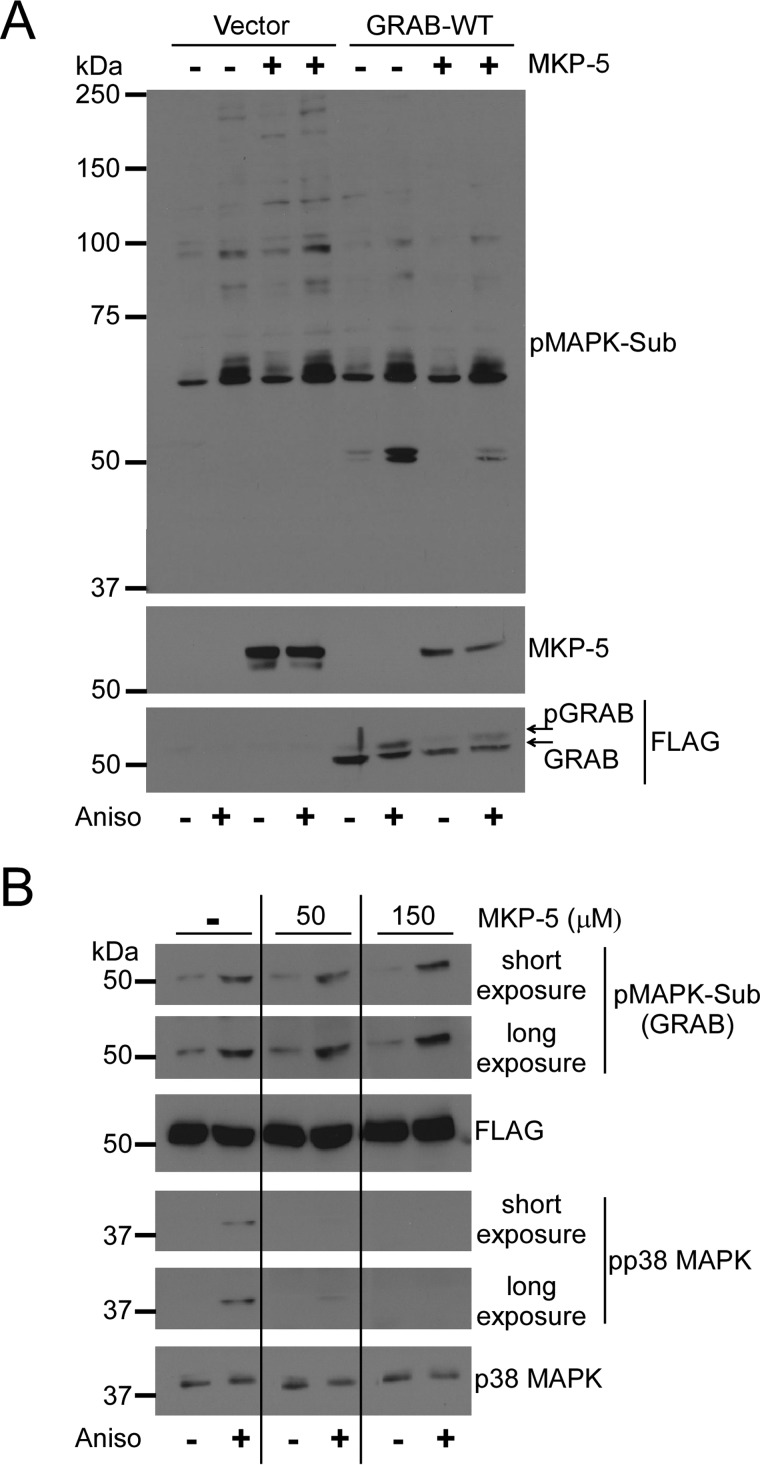
To validate GRAB as a MKP-5-regulated MAPK substrate, we asked whether MKP-5 overexpression reduced GRAB phosphorylation. These experiments utilized 293 cells because endogenous levels of GRAB are undetectable in these cells, allowing for the indiscriminate detection of exogenously expressed phosphorylated FLAG-tagged GRAB. First, 293 cells were stimulated with anisomycin to increase the levels of phosphorylated MAPK substrates (Fig. 2A). As anticipated, in vector control-expressing 293 cells, anisomycin-induced GRAB phosphorylation was undetectable (Fig. 2A). When MKP-5 was co-overexpressed along with FLAG-GRAB, a reduction in phosphorylation levels of the MAPK substrate motif on GRAB was observed as detected using anti-phospho-MAPK substrate motif antibody (Fig. 2A). Further evidence that MKP-5 reduced GRAB phosphorylation was revealed by the observation that GRAB underwent a mobility shift in response to anisomycin stimulation that was attenuated upon MKP-5 overexpression (Fig. 2A). It remained formally possible that GRAB might be a direct MKP-5 substrate. Far Western immunoblotting showed that purified MKP-5, when incubated with either immobilized phosphorylated p38 MAPK or phosphorylated GRAB, effectively dephosphorylated the MKP-5 substrate p38 MAPK but failed to dephosphorylate GRAB under identical conditions (Fig. 2B). These results suggest that MKP-5 does not act directly on GRAB but rather indirectly affects MAPK-mediated phosphorylation of GRAB.
FIGURE 2.
Validation of GRAB as a MKP-5-regulated MAPK substrate. A, FLAG-tagged GRAB was co-expressed with MKP-5 in 293 cells. Cells were serum-starved for 6 h and stimulated with 5 μm anisomycin (Aniso). Lysates were immunoblotted with pMAPK substrate (Sub) antibody and reprobed with anti-FLAG and anti-MKP-5 antibodies. B, FLAG-tagged GRAB was expressed in 293 cells and cells were serum-starved for 6 h and stimulated with 5 μm anisomycin. Whole cell lysates were transferred to membranes, and the blots were incubated with the indicated amount of purified MKP-5 protein for 3 h. Immunoblots were probed with phospho-p38 (pp38) MAPK or pMAPK substrate (Sub) antibodies and reprobed with p38 MAPK or FLAG antibody.
Identification of Phosphoserine 169 on GRAB as a Major Phosphorylation Site
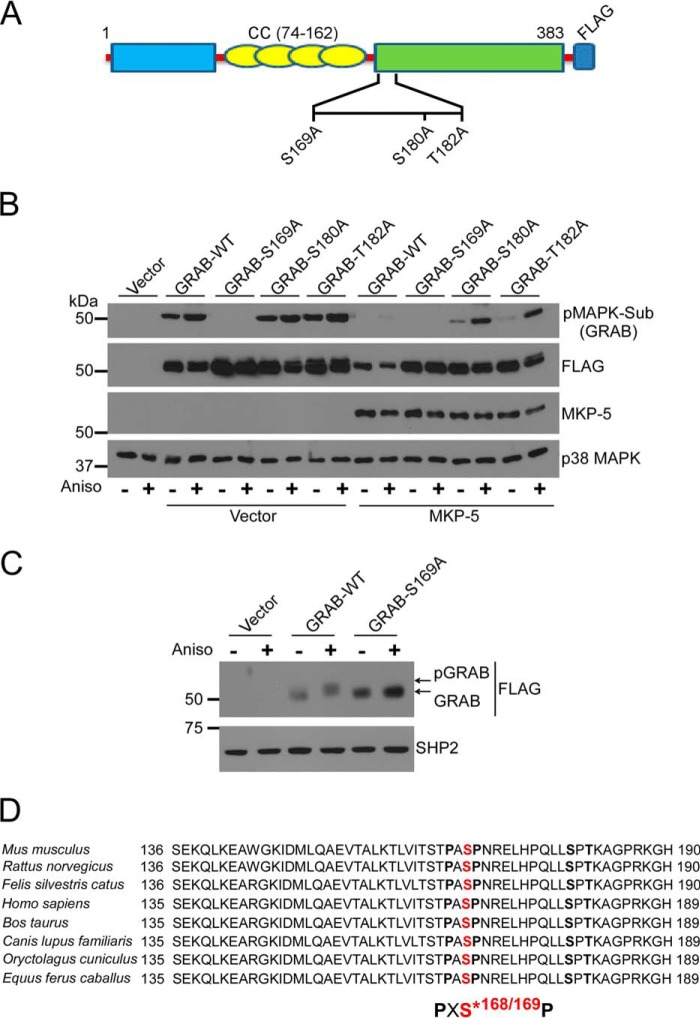
Three putative GRAB phosphorylation sites in the context of the MAPK substrate motif (PXS*P or S*PX(R/K)) were identified, serine 169 (Ser-169), serine 180 (Ser-180), and threonine 182 (Thr-182) (Figs. 1C and 3A). To further validate these phosphorylation sites on GRAB, we mutated Ser-169, Ser-180, and Thr-182 to alanine (Fig. 3A). We found that GRAB-S169A-expressing cells showed undetectable levels of phosphorylation either in the absence or presence of anisomycin stimulation, whereas GRAB-T182A- and GRAB-S180A-expressing cells were readily detectable as phosphorylated (Fig. 3B). Moreover, when co-expressed with MKP-5, wild type GRAB phosphorylation was reduced either in the absence or presence of anisomycin stimulation to levels comparable with that of GRAB-S169A (Fig. 3B). Importantly, both GRAB-S180A and -T182A mutants were still detectably phosphorylated even when MKP-5 was overexpressed, demonstrating that these sites marginally contribute to the net level of MKP-5-regulated GRAB phosphorylation (Fig. 3B). These results suggest the possibility that GRAB Ser-180 and/or Thr-182 sites might be regulated in a MKP-5/MAPK-independent manner. Notably, the GRAB-S169A mutant failed to exhibit a comparable mobility shift following anisomycin stimulation as compared with wild type GRAB, further supporting Ser-169 as a site of stoichiometric GRAB phosphorylation (Fig. 3C). Finally, inspection of the surrounding GRAB Ser-169 amino acid sequence across a variety of species revealed that Ser-169 of GRAB is highly conserved (Fig. 3D). Thus, we conclude that Ser-169 represents a major GRAB phosphorylation site.
FIGURE 3.
Identification of phosphoserine 169 on GRAB as a major phosphorylation site. A, schematic representation of GRAB showing identified MAPK phosphorylation sites. CC, coiled coil. B, wild type GRAB or the indicated GRAB phosphorylation site mutants were co-expressed with vector alone or MKP-5 in 293 cells. Cells were serum-starved for 6 h and stimulated with 5 μm anisomycin (Aniso). Total cell lysates were immunoblotted with pMAPK substrate (Sub) antibody and reprobed with FLAG, MKP-5, or p38 MAPK antibody. C, 293 cells were transfected with FLAG-tagged GRAB, and cells were serum-starved for 6 h and stimulated with 5 μm anisomycin for 30 min. Whole cell lysates were immunoblotted with anti-FLAG antibody and reprobed with anti-SHP2 antibody as a control. D, comparison of amino acid sequences of GRAB serine 169 with different species. The PXSP motif indicates the phospho-MAPK substrate motif.
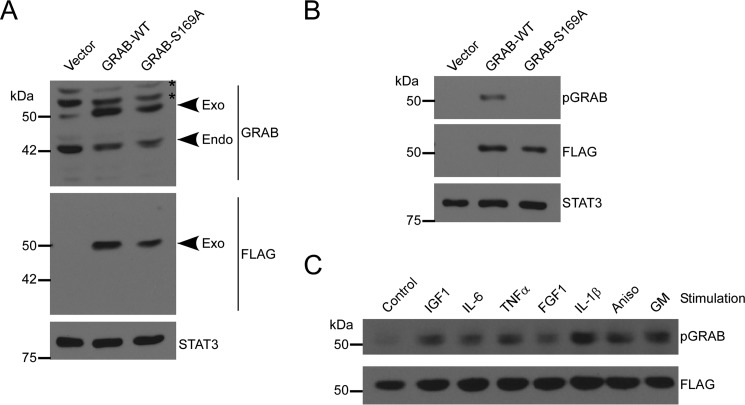
To further examine Ser-169 phosphorylation, we established stable C2C12 myoblasts expressing either FLAG-tagged GRAB-WT, GRAB-S169A, or vector control (Fig. 4A). C2C12 myoblasts express endogenous GRAB, and the exogenously expressed FLAG-GRAB migrated with a slightly higher molecular weight and at expression levels that were ∼2-fold above that of endogenous GRAB (Fig. 4A). Additionally, we generated a phospho-Ser-169-specific GRAB antibody and showed that this antibody could readily detect phosphorylated GRAB in GRAB-WT cells but not in GRAB-S169A mutant-expressing cells (Fig. 4B). Furthermore, stimulation of GRAB-WT-overexpressing myoblasts with IGF-1, IL-1β, FGF-1, TNF-α, and IL-6 demonstrated that growth factors and cytokines are capable of inducing GRAB Ser-169 phosphorylation (Fig. 4C). Hence, GRAB is a target for phosphorylation at Ser-169 in response to a variety of cytokines and growth factors.
FIGURE 4.
Generation of C2C12 myoblasts stably expressing GRAB. A, C2C12 myoblasts stably expressing GRAB-WT, GRAB-S169A, or empty vector were generated as described under “Experimental Procedures.” Cell lysates were immunoblotted with GRAB antibody and reprobed with FLAG or STAT-3 antibody as controls. Exo indicates exogenous FLAG-tagged GRAB, and Endo indicates endogenous GRAB. The asterisk indicates a nonspecific peptide. B, cell lysates were prepared and immunoblotted with pGRAB antibody and reprobed with FLAG or STAT-3 antibody. C, C2C12 myoblasts stably expressing wild type GRAB were serum-starved overnight and incubated with the indicated stimulus for 15 min, and then cells were lysed. Total lysates were immunoblotted with pGRAB and reprobed with FLAG antibody. Aniso, anisomycin; GM, growth medium.
Regulation of GRAB Phosphorylation by MAPK Signaling
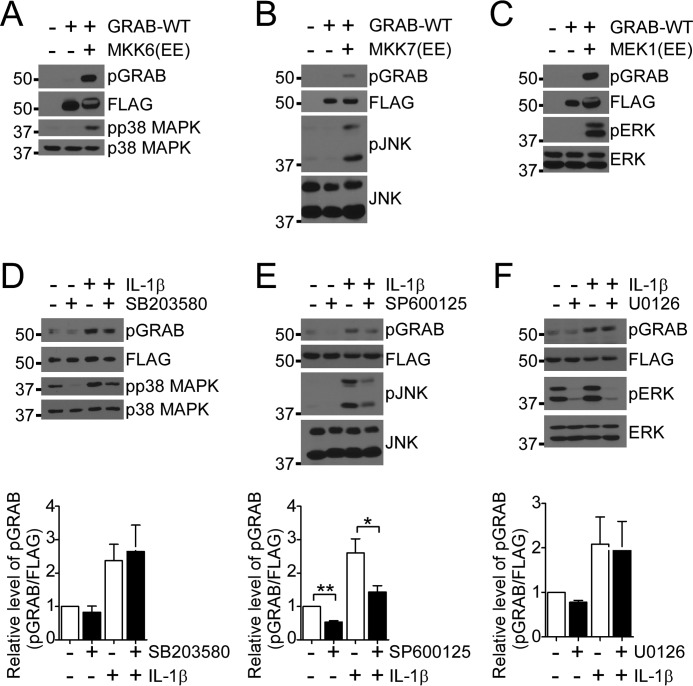
To examine which MAPK could phosphorylate GRAB, GRAB-WT was co-expressed in 293 cells with constitutively active mutants of MKK6, MKK7, and MEK1, which activate their downstream cognate MAPK targets p38 MAPK, JNK, and ERK1/2, respectively. Each activated MKK or MEK induced the phosphorylation of GRAB-WT on Ser-169 as determined using the phosphoserine 169-specific GRAB (pGRAB) antibodies (Fig. 5, A–C). These results demonstrate the sufficiency of the MAPKs to phosphorylate GRAB on Ser-169. Next, we tested which MAPK is required for the phosphorylation of GRAB. It has been shown that infiltration of macrophages within the damaged muscle releases proinflammatory cytokines such as IL-1β that stimulate the expression of cytokines such as IL-6 (15, 16). IL-6 subsequently acts to promote the repair process because mice lacking IL-6 fail to undergo regenerative myogenesis (7). When GRAB-WT myoblasts were stimulated with IL-1β, we found that GRAB underwent phosphorylation on Ser-169 (Fig. 5, D–F). Pretreatment of myoblasts with inhibitors of p38 MAPK, JNK, and ERK1/2 revealed that only JNK inhibition significantly reduced basal GRAB Ser-169 phosphorylation and attenuated IL-1β-induced pGRAB as compared with cells treated with either p38 MAPK or JNK inhibitors (Fig. 5, D–F). These data suggest that JNK plays a predominately greater role than either p38 MAPK or ERK toward the phosphorylation of GRAB on Ser-169.
FIGURE 5.
JNK-regulated phosphorylation of GRAB Ser-169. A–C, GRAB-WT was co-expressed with vector alone or a constitutively active mutant of MKK6 (A), MKK7 (B), or MEK1 (C) in 293 cells. Total lysates were immunoblotted with pGRAB or the indicated phosphospecific antibodies and reprobed with FLAG or the indicated total antibodies. D–F, GRAB-WT-expressing C2C12 myoblasts were serum-starved overnight and incubated with the indicated MAPK inhibitors (10 μm) for 1 h prior to stimulation with IL-1β (10 ng/ml) for 15 min. Total lysates from these cells were immunoblotted with pGRAB, phospho-p38 (pp38) MAPK, phospho-JNK (pJNK), and phospho-ERK (pERK) antibodies and reprobed with FLAG, p38 MAPK, JNK, and ERK antibodies, respectively. The graphs at the bottom of each panel represent quantitation of phosphorylated GRAB normalized to FLAG-GRAB. Data represent the mean ± S.E. from three independent experiments. Error bars indicate S.E. *, p < 0.05; **, p < 0.01.
Phosphorylation of GRAB at Serine 169 Is Required for Myogenesis
To examine the functional significance of GRAB Ser-169 phosphorylation, we initially analyzed the proliferation and migration of C2C12 myoblasts stably expressing either GRAB-WT or GRAB-S169A mutant. No differences in either proliferation or migration were observed between GRAB-WT- and GRAB-S169A-expressing myoblasts (Fig. 6, A and B). Remarkably, when GRAB-WT and GRAB-S169A myoblasts were induced to undergo differentiation, we found that GRAB-S169A myoblasts were severely impaired in their ability to differentiate as compared with GRAB-WT myoblasts (Fig. 6C). The fusion index in GRAB-S169A-expressing myoblasts was significantly reduced as compared with GRAB-WT myoblasts, and this correlated with reduced expression levels of myosin heavy chain and myogenin (Fig. 6, D and E). These results strongly suggest that the appropriate phosphorylation of GRAB at Ser-169 is required for myogenesis.
FIGURE 6.
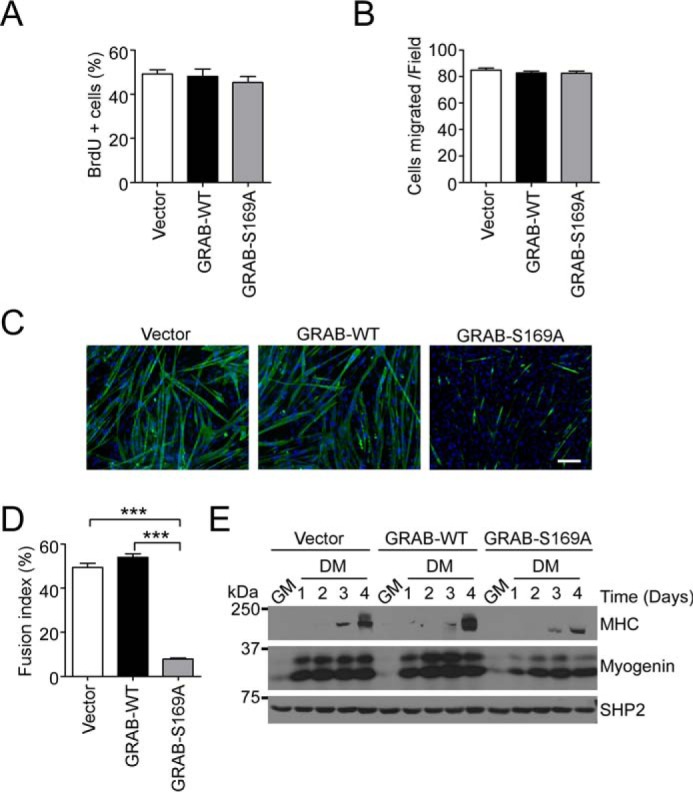
GRAB Ser-169 phosphorylation is required for myogenesis. A, stable C2C12 myoblasts were incubated in growth medium for 24 h, labeled with BrdU, and fixed for staining with BrdU and DAPI. Quantification of the percentage of positive DAPI-stained cells for BrdU is shown. Data represent the mean ± S.E. from three independent experiments. B, stable C2C12 myoblasts were analyzed for migration for 5 h. The number of cells migrated per 20× field is shown. Data represent the mean ± S.E. from three independent experiments, and error bars indicate S.E. C–E, stable C2C12 myoblasts expressing GRAB-WT, GRAB-S169A, or vector control were induced to differentiate for 6 days. Differentiated myoblasts were stained with myosin heavy chain (green) and DAPI (blue) (C) and quantified for myogenic fusion index (D). Scale bar, 100 μm. E, cell lysates prepared from differentiating GRAB-overexpressing myoblasts were immunoblotted with anti-myosin heavy chain (MHC), myogenin, and SHP2 antibodies. ***, p < 0.001. GM, growth medium; DM, differentiation medium.
MKP-5 Negatively Regulates IL-6 Secretion in Skeletal Muscle
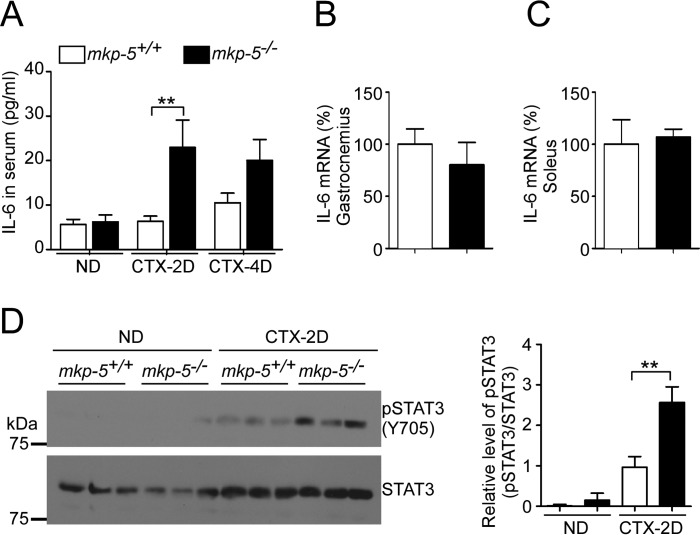
In an effort to further understand the pathways through which MKP-5/MAPK negatively regulates regenerative myogenesis (3), we noted that during skeletal muscle regeneration circulating IL-6 levels in mkp-5−/− mice were significantly increased as compared with mkp-5+/+ mice (Fig. 7A). Although mkp-5−/− mice had enhanced levels of secreted IL-6 during skeletal muscle regeneration compared with mkp-5+/+ mice, we found no difference in IL-6 mRNA expression between regenerating skeletal muscle of mkp-5+/+ and mkp-5−/− mice (Fig. 7, B and C). Consistent with the increased secretion of IL-6 during regeneration, skeletal muscles of mkp-5−/− mice showed enhanced levels of STAT-3 Tyr-705 phosphorylation (Fig. 7D). These data demonstrate that MKP-5 negatively regulates the levels of circulating IL-6 during regenerative myogenesis.
FIGURE 7.
MKP-5 negatively regulates IL-6 secretion during regeneration of skeletal muscle. A, serum IL-6 levels in non-damaged (ND) and at 2 days (CTX-2D) and 4 days (CTX-4D) after cardiotoxin damaged skeletal muscle (n = 5 per genotype). B and C, IL-6 mRNA expression 2 days after cardiotoxin-induced damage (n = 5 per genotype). D, lysates from muscle in non-damaged (ND) and 2 days after cardiotoxin-damaged (CTX-2D) were immunoblotted with phospho-STAT-3 (Tyr-705) antibody and reprobed with STAT-3 antibody. The graph represents quantitation of phospho-STAT-3 (pSTAT3) normalized to total STAT-3 (n = 3 per genotype). Open bars, mkp-5+/+; closed bars, mkp-5−/− mice. Data represent the mean ± S.E., and error bars indicate S.E. **, p < 0.01. CTX-4D, 4-day cardiotoxin-damaged.
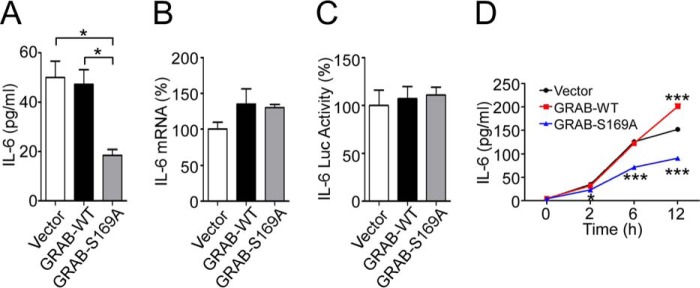
GRAB functions as a guanine nucleotide exchange factor for the Rab3A small GTPase that is involved in cytokine, growth factor, neurotransmitter, and hormonal secretion (10, 17–21). Given the functional relevance of GRAB to the secretory pathway, we examined the functional significance of GRAB Ser-169 phosphorylation specifically on IL-6 secretion. Myoblasts expressing GRAB-S169A exhibited significantly reduced levels of IL-6 secretion (Fig. 8A) in the absence of any apparent change in either IL-6 mRNA expression or transcriptional activity of the proximal IL-6 promoter (Fig. 8, B and C). When GRAB-S169A myoblasts were stimulated with IL-1β, the levels of secreted IL-6 were significantly reduced as compared with GRAB-WT myoblasts (Fig. 8D). Collectively, these data demonstrate that GRAB Ser-169 phosphorylation plays an essential role in the secretion of IL-6 from myoblasts.
FIGURE 8.
GRAB Ser-169 phosphorylation is required for IL-6 secretion in myoblasts. A, C2C12 myoblasts stably expressing GRAB-WT, GRAB-S169A, or empty vector were cultured in growth medium, and the amount of secreted IL-6 was measured by ELISA. B, IL-6 mRNA expression in stable C2C12 myoblasts was determined by real time quantitative PCR. C, levels of relative IL-6 promoter activity in C2C12 myoblasts co-transfected with pGL3-IL-6 and the indicated GRAB constructs. D, stable C2C12 myoblasts were serum-starved for 24 h and stimulated with IL-1β for the indicated times. Secreted IL-6 was measured by ELISA. Data represent the mean ± S.E., and error bars indicate S.E. *, p < 0.05; ***, p < 0.001. Luc, luciferase.
Discussion
Using a non-biased approach to identify putative MKP-5-regulated MAPK substrates, we performed differential phosphoproteomic analyses targeting the phospho-MAPK substrate motif in regenerating skeletal muscles. We identified GRAB as a major hyperphosphorylated MAPK substrate in regenerating skeletal muscle of mkp-5−/− mice. GRAB (Rab3il1) has been shown to be a GEF for Rab3A (10). Rab3A controls vesicular transport, membrane trafficking, and exocytosis (22). Based upon our analysis of other potential top hit substrates (Fig. 1C), we identified the following potential MKP-5-regulated MAPK substrates: 1) MRCKβ, 2) SRm160, 3) ARID1B, and 4) RBMS1. Interestingly, MRCKβ, which is known as the myotonic dystrophy kinase-related Cdc42 binding kinase β, is involved in actin-cytoskeleton signaling and is required for myosin light chain 2 and MYPT2 phosphorylation (23, 24). SRm160 is an RNA-binding protein (25), ARID1B is a Brahma-associated factor-associated protein involved in chromatin remodeling (26), and RBMS1 is a transcription factor RNA-binding motif that plays a role in regulating DNA replication, transcription, apoptosis, and cell cycle progression by interacting with c-Myc (27). It is conceivable that these potential MAPK substrates also play additional roles as downstream targets that are regulated through MKP-5/MAPK in the control of skeletal muscle regeneration. Further work will be required to validate and subsequently investigate these MKP-5/MAPK targets for their role in regenerative myogenesis.
In addition to demonstrating by mass spectrometry that GRAB is hyperphosphorylated in regenerating skeletal muscles of mkp-5−/− mice, several additional lines of evidence support the conclusion that GRAB is a MKP-5-regulated MAPK substrate. First, we show that overexpression of MKP-5 results in the dephosphorylation of MAPK and a concomitant decrease in phosphorylated GRAB in cells. Second, either activation of MKKs or inhibition of the MAPKs pharmacologically stimulated and inhibited GRAB phosphorylation, respectively. Specifically, JNK inhibition reduced GRAB phosphorylation basally and in response to IL-1β. Our dephosphorylation assays do not support the interpretation that GRAB is a direct substrate for MKP-5. Finally, mutation of Ser-169 to a non-phosphorylatable residue on GRAB that exists within a highly conserved MAPK motif completely abrogated GRAB phosphorylation. These data argue that GRAB is phosphorylated in skeletal muscle by MAPK and that JNK is the likely MAPK responsible for this phosphorylation.
The ability of skeletal muscle to register changes in the extracellular environment and coordinately adapt is critical for the maintenance of skeletal muscle physiology. IL-6 can positively regulate regenerative myogenesis and skeletal muscle growth (28). However, understanding of the mechanisms of how the regulation of IL-6 expression is coupled to extracellular signals remains incomplete. In response to skeletal muscle damage, the systemic levels of IL-6 are elevated in MKP-5-deficient mice as compared with wild type mice, demonstrating that MKP-5 negatively regulates IL-6 production during muscle regeneration. Previous reports have demonstrated that IL-6 levels are negatively regulated by MKP-5 (29, 30). In those cases, IL-6 expression levels were ascribed to the commensurate increase in IL-6 mRNA expression mainly in macrophages (30) or in prostate cancer cells (29). Here, in response to muscle injury, we found that IL-6 mRNA expression levels were similar in regenerating skeletal muscles of mkp-5−/− mice, suggesting that in response to injury the increased circulating levels of IL-6 could, at least in part, be generated through mechanisms other than those that were solely transcriptional. Furthermore, under these conditions, IL-6 signaling to STAT-3 was elevated. Finally, cell-based experiments supported the notion of a myoblast autonomous mechanism because overexpression of the GRAB-S169A mutant impaired IL-6 secretion independently of changes in IL-6 mRNA. These data now demonstrate that in skeletal muscle MKP-5 is a negative regulator of IL-6.
Although there are studies indicating that the secretory pathways regulated by the Rabs are responsive to the MAPKs (31), much less is known about their GEFs and how they are coupled to extracellular responses. That MKP-5 negatively regulates GRAB phosphorylation at the site in which IL-6 secretion is promoted suggests that the actions of MKP-5 on skeletal muscle growth are mediated, at least in part, through this pathway. Consistent with this notion, we found that overexpression of the non-phosphorylatable GRAB mutant impaired myogenesis. These results demonstrate that MKP-5 negatively regulates myogenesis in a JNK/GRAB-dependent manner. Precisely how JNK phosphorylation of GRAB affects its ability to signal to the Rabs remains to be elucidated. Interestingly, it has been shown that the GEF for Rab8, Rabin8, is phosphorylated by ERK, and this promotes GTP exchange on Rab8 (32). It is conceivable that JNK phosphorylation of GRAB similarly promotes the activation and/or interactions of additional Rabs; however, this will need to be formally determined.
In summary, our data reveal that MKP-5 negatively regulates JNK-dependent phosphorylation of GRAB and signaling to the secretory machinery to control IL-6 secretion in skeletal muscle. Moreover, GRAB phosphorylation appears to play an integral role in the progression of skeletal muscle differentiation. Hence, MKP-5 plays an important role in coupling extracellular signals that trigger skeletal muscle regeneration and growth, to those of myokine secretory pathways.
Experimental Procedures
Phospho-MAPK Substrate Proteomic Screen
The PhosphoScan method was performed as described previously using services provided by Cell Signaling Technology (11–13). Non-damaged and damaged skeletal muscle from mkp-5+/+ and mkp-5−/− mice were homogenized, sonicated, and clarified by centrifugation at 20,800 × g for 20 min at 4 °C. Protein concentration was determined using BCA reagent according to the manufacturer's instructions (Pierce). Muscle homogenates were reduced with 4.5 mm DTT for 30 min at 55 °C and alkylated with iodoacetamide (0.095 g/5 ml of H2O) for 15 min at room temperature in the dark. The homogenates were digested overnight with 10 μg/ml trypsin, digested peptide lysates were acidified with 1% trifluoroacetic acid (TFA), and peptides were desalted over 360-mg Sep-Pak Classic C18 columns (Waters). Peptides were eluted with 40% acetonitrile in 0.1% TFA, dried under vacuum, and stored at −80 °C.
Lyophilized peptides were redissolved, and phosphopeptides were enriched by immunoprecipitation using a phospho-MAPK substrate motif antibody (2325, Cell Signaling Technology), which recognizes the conserved MAPK substrate motif (PXS*P or S*PX(R/K)). Peptides were loaded directly onto a 10-cm × 75-μm PicoFrit capillary column packed with Magic C18 AQ reversed-phase resin to remove non-peptide materials. The column was developed with a 72-min linear gradient of acetonitrile in 0.125% formic acid delivered at 280 nl/min. Each sample was split and run to generate analytical replicates and increase the number of tandem MS (MS/MS) identifications from each sample. Tandem mass spectra were collected with an LTQ-Orbitrap Elite mass spectrometer running XCalibur using a top 20 MS/MS method (run time, 96 min; MS1 scan range, 300.0–1500.00). MS/MS spectra were evaluated using SEQUEST 3G and the SORCERER 2 platform from Sage-N Research (Milpitas, CA; v4.0). Searches were performed against the most recent update of the NCBI Mus musculus database with mass accuracy of ±50 ppm for precursor ions and 1 Da for product ions. Results were filtered with a mass accuracy of ±5 ppm on precursor ions and the presence of the intended motif.
Label-free quantitation from phosphopeptide intensities was performed using Progenesis v4.1 (Waters Corp.) and Skyline version 3.1 and produced from either the apex peak height or peak area of the corresponding peptide assignment. Quantitation was based on a single peptide ion for each protein/site. Peptide abundance data were manually reviewed to ensure accurate quantitation in Skyline. Statistical analysis of the quantitative data was done using a two-tailed t test between two groups. The maximum negative log p value from three comparison pairs was used to indicate significance for abundance changes of a certain peptide between two groups. The false discovery rate for each binary comparison was further controlled by applying the Benjamini-Hochberg procedure. The -fold changes were normalized using a technique in which the median log2 ratio is set to 0 and all -fold changes are adjusted relative to the median.
Cell Culture, Differentiation, and Cytokine and Luciferase Assays
C2C12 mouse myoblasts were cultured and differentiated as described (33). Differentiated C2C12 myoblasts were stained with anti-myosin heavy chain antibody (Developmental Study Hybridoma Bank; 1:20 dilution) and with DAPI for nuclei. At least 10 fields for each sample were collected. The myogenic fusion index was calculated as the ratio of the numbers of nuclei in myosin-stained myotubes to the total number of nuclei in DAPI-stained cells. Serum IL-6 levels were measured using an enzyme-linked immunosorbent assay (ELISA) kit (mouse IL-6 Quantikine ELISA, R&D Systems). For measuring IL-6, culture medium was collected and spun at 1,000 × g for 10 min, and the supernatant was used to measure IL-6 by ELISA. C2C12 myoblasts were co-transfected with the IL-6 promoter fused to either firefly luciferase or Renilla luciferase and GRAB mutants. The activities of firefly luciferase and Renilla luciferase were examined using the Dual-Luciferase reporter system kit from Promega following the manufacturer's protocol, and luminescence was measured using a GloMax 20/20 luminometer (Promega Corp.).
Animal Experiments
MKP-5 knock-out mice (mkp-5−/−) were described previously, and cardiotoxin-induced muscle damage was performed as described (3). The Yale University Institutional Animal Care and Use Committee approved all procedures.
Plasmids and Stable Cell Lines
FLAG-tagged mouse wild type GRAB (Rab3il1) in pCMV6-entry (Origen) and GRAB mutants were generated using a QuikChange XL site-directed mutagenesis kit (Stratagene). MKK6, MKK7, and MEK1 in pCDNA3 were obtained from Addgene. The various inserts of GRAB in pCMV6-entry were cloned into the pLenti-EF1a-GFP-2A-Puro lentiviral vector (Applied Biological Materials Inc.) using EcoRI/XbaI sites. Lenti-Combo Packing Mix plasmids (Applied Biological Materials Inc.) and each GRAB lentiviral vector were co-transfected into 293T cells using the Lentifectin transfection reagent (Applied Biological Materials Inc.). Supernatants were collected, purified, and quantitated by quantitative PCR (viral titer, ∼2 × 107 infection units/ml). To generate C2C12 myoblasts stably expressing GRAB, C2C12 cells were plated in 6-well plates, and Polybrene (Sigma) was added to a final concentration of 10 μg/ml with the virus. After overnight incubation, medium was replaced, and on the 2nd day of infection cells were selected with puromycin (Sigma; 10 μg/ml).
Cell Lysis, Immunoblotting, and Immunoprecipitation
Cells were lysed on ice in lysis buffer containing 150 mm NaCl, 50 mm Tris-HCl (pH 7.8), 1 mm EDTA, 1% Nonidet P-40, 1 mm Na3VO4, 10 mm NaF, 1 mm DTT, 1 mm benzamidine, 1 mm PMSF, 1 μg/ml pepstatin A, 5 μg/ml aprotinin, and 5 μg/ml leupeptin for 30 min and clarified by centrifugation at 20,800 × g at 4 °C for 20 min. Protein concentration was determined using BCA reagent according to the manufacturer's instructions (Pierce). For immunoprecipitation experiments, 200–500 μg of cell lysate was incubated with 2 μg of FLAG mouse monoclonal antibody (Sigma) overnight at 4 °C. Immune complexes were collected on either protein A- or G-Sepharose (GE Healthcare). Immune complexes were washed three times with lysis buffer and once with STE buffer (100 mm NaCl, 10 mm Tris-HCl (pH 8.0), and 1 mm EDTA), and Sepharose beads were heated to 95 °C in sample buffer (62.5 mm Tris (pH 6.8), 4% glycerol, 2% SDS, 5% β-mercaptoethanol, and 0.02% bromphenol blue) for 5 min.
For immunoblotting, lysates or immune complexes were resolved by SDS-PAGE and transferred onto either Immobilon-P membranes (Millipore) or nitrocellulose membranes (Bio-Rad). Membranes were first blocked with 5% nonfat dry milk or 5% BSA in Tris-buffered saline/Tween 20 (TBS-T) for 1 h at room temperature or overnight at 4 °C. Primary antibodies were diluted in 5% BSA or 5% nonfat dry milk in TBS-T. The following primary antibodies were used. MKP-5 (3484), phospho-p38 MAPK (Thr-180/Tyr-182) (9125), phospho-JNK1/2 (Thr-183/Tyr-185) (4668), phospho-ERK1/2 (Thr-202/Tyr-204) (9101), phospho-STAT-3 (Tyr-705) (9145), and phospho-MAPK substrate motif and STAT-3 (9139) antibodies were obtained from Cell Signaling Technology. ERK1/2 (sc-94), p38 MAPK (sc-535), SHP-2 (sc-280), and JNK (sc-571) antibodies were purchased from Santa Cruz Biotechnology. GRAB (SAB1400234) and FLAG (F3165) antibodies were purchased from Sigma. Myosin heavy chain (MF20) and myogenin (F5A) were purchased from Developmental Study Hybridoma Bank. After incubation with primary antibodies overnight at 4 °C, membranes were washed in TBS-T at least three times for 10 min. The membranes were incubated in secondary anti-rabbit IgG horseradish peroxidase (HRP)-linked goat antibody or anti-mouse IgG HRP-linked horse antibody (Cell Signaling Technology) for 1 h at room temperature at 1:5,000 dilution and washed in TBS-T three times for 10 min. Membranes were visualized using enhanced chemiluminescence (Amersham Biosciences).
For the generation of phosphospecific antibodies to Ser(P)-169 on mouse GRAB a Ser(P)-169-containing peptide surrounding Ser-169 was synthesized and used to immunize rabbits (Cell Signaling Technology). Anti-Ser(P)-169 rabbit antiserum (pGRAB) was affinity-purified and used for experiments. For immunoblotting, membranes were blocked in BSA and incubated overnight with pGRAB at a dilution of 1:1,000. Membranes were washed several times with TBS-T, and proteins were visualized using an anti-rabbit IgG HRP-linked goat antibody at a 1:5,000 dilution followed by enhanced chemiluminescence.
Cell Migration Assay
Migration assays were performed using a Transwell chamber (Costar) following the manufacturer's protocol as described previously (34). The Transwell insert membrane containing 8-μm size pores was coated with 10 μg/ml human fibronectin for 2 h at 25 °C. Cells (1 × 105 per insert in 250 μl of serum-free medium) were placed in the upper chamber, and growth medium was placed in the lower chamber. After 5-h incubation at 37 °C, the insert membrane was fixed with 99% methanol (Fisher Scientific) for 10 min, rinsed with PBS, stained with 0.2% crystal violet (Fisher Scientific) for 10 min, and washed twice with water. Non-migrated cells on the upper surface of the membrane were wiped off with a cotton swab, and the membrane was washed with distilled water twice. Images of migrated cells on the bottom of the membrane were collected on a Zeiss Axiovert S100 microscope (at 20× and 40×) using Axiovision software (Zeiss).
Cell Proliferation Assay
C2C12 proliferation assays were determined using 5-bromo-2′-deoxyuridine (BrdU). Briefly, cells were incubated with BrdU at 10 μm final concentration for 1 h and then fixed with 70% ethanol. After treatment with 1.5 m HCl, immunostaining with anti-BrdU antibodies (Developmental Study Hybridoma Bank G3G4) was carried out, and nuclei were stained by DAPI. Imaging was collected on a Zeiss Axiovert S100 microscope using Axiovision software (Zeiss).
RNA Isolation and Quantitative RT-PCR
Total RNA from cells or skeletal muscles was extracted using an RNeasy Kit (Qiagen) according to the manufacturer's instructions. Isolated RNA was reverse transcribed to generate cDNA using a reverse transcriptase PCR kit (Applied Biosystems). TaqMan IL-6 and 18S rRNA primers were used in combination with the TaqMan Gene Expression Master Mix (Applied Biosystems). A 7500 Fast-Real Time PCR system (Applied Biosystems) was used to quantify mRNA expression. IL-6 mRNA relative abundance was normalized to internal control 18S rRNA and expressed as percent change.
Statistical Analysis
Statistical analyses were performed using Prism software (GraphPad Software). Statistical analyses in three or more groups were performed using a one-way analysis of variance followed by the Bonferroni t test for multiple comparisons. Statistical differences between two groups were analyzed using an unpaired two-tailed t test.
Author Contributions
H. L. performed experiments with the assistance of K. M., J.-S. Y., H. S., W. C., and L. J. H. L., K. M., J.-S. Y., and A. M. B. analyzed data. H. L. and A. M. B. designed the research project and wrote the paper.
Acknowledgment
We thank Dr. Jungmin Choi for bioinformatics assistance.
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- MKP
- MAPK phosphatase
- GRAB
- guanine nucleotide exchange factor for Rab3A
- Rab3il1
- Rab3A-interacting protein (Rabin3)-like 1
- MKK
- MAPK kinase
- pGRAB
- phosphoserine 169-specific GRAB
- GEF
- guanine nucleotide exchange factor
- pMAPK
- phospho-MAPK.
References
- 1. Caunt C. J., and Keyse S. M. (2013) Dual-specificity MAP kinase phosphatases (MKPs): shaping the outcome of MAP kinase signalling. FEBS J. 280, 489–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bennett A. M., and Tonks N. K. (1997) Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases. Science 278, 1288–1291 [DOI] [PubMed] [Google Scholar]
- 3. Shi H., Verma M., Zhang L., Dong C., Flavell R. A., and Bennett A. M. (2013) Improved regenerative myogenesis and muscular dystrophy in mice lacking Mkp5. J. Clin. Investig. 123, 2064–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shi H., Boadu E., Mercan F., Le A. M., Flach R. J., Zhang L., Tyner K. J., Olwin B. B., and Bennett A. M. (2010) MAP kinase phosphatase-1 deficiency impairs skeletal muscle regeneration and exacerbates muscular dystrophy. FASEB J. 24, 2985–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shi H., Gatzke F., Molle J. M., Lee H. B., Helm E. T., Oldham J. J., Zhang L., Gerrard D. E., and Bennett A. M. (2015) Mice lacking MKP-1 and MKP-5 reveal hierarchical regulation of regenerative myogenesis. J. Stem Cell Regen. Biol. 1, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pedersen B. K., and Febbraio M. A. (2008) Muscle as an endocrine organ: focus on muscle-derived interleukin-6. Physiol. Rev. 88, 1379–1406 [DOI] [PubMed] [Google Scholar]
- 7. Serrano A. L., Baeza-Raja B., Perdiguero E., Jardí M., and Muñoz-Cánoves P. (2008) Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab. 7, 33–44 [DOI] [PubMed] [Google Scholar]
- 8. Zhang C., Li Y., Wu Y., Wang L., Wang X., and Du J. (2013) Interleukin-6/signal transducer and activator of transcription 3 (STAT3) pathway is essential for macrophage infiltration and myoblast proliferation during muscle regeneration. J. Biol. Chem. 288, 1489–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tierney M. T., Aydogdu T., Sala D., Malecova B., Gatto S., Puri P. L., Latella L., and Sacco A. (2014) STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat. Med. 20, 1182–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luo H. R., Saiardi A., Nagata E., Ye K., Yu H., Jung T. S., Luo X., Jain S., Sawa A., and Snyder S. H. (2001) GRAB: a physiologic guanine nucleotide exchange factor for Rab3A, which interacts with inositol hexakisphosphate kinase. Neuron 31, 439–451 [DOI] [PubMed] [Google Scholar]
- 11. Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., et al. (2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 [DOI] [PubMed] [Google Scholar]
- 12. Stokes M. P., Farnsworth C. L., Moritz A., Silva J. C., Jia X., Lee K. A., Guo A., Polakiewicz R. D., and Comb M. J. (2012) PTMScan direct: identification and quantification of peptides from critical signaling proteins by immunoaffinity enrichment coupled with LC-MS/MS. Mol. Cell. Proteomics 11, 187–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gu H., Ren J. M., Jia X., Levy T., Rikova K., Yang V., Lee K. A., Stokes M. P., and Silva J. C. (2016) Quantitative profiling of post-translational modifications by immunoaffinity enrichment and LC-MS/MS in cancer serum without immunodepletion. Mol. Cell. Proteomics 15, 692–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang P. M., Bondor J. A., Swiderek K. M., and DePaoli-Roach A. A. (1991) Molecular cloning and expression of the regulatory (RG1) subunit of the glycogen-associated protein phosphatase. J. Biol. Chem. 266, 15782–15789 [PubMed] [Google Scholar]
- 15. Luo G., Hershko D. D., Robb B. W., Wray C. J., and Hasselgren P. O. (2003) IL-1β stimulates IL-6 production in cultured skeletal muscle cells through activation of MAP kinase signaling pathway and NF-κB. Am. J. Physiol. Regul. Integr. Comp. Physiol. 284, R1249–R1254 [DOI] [PubMed] [Google Scholar]
- 16. Otis J. S., Niccoli S., Hawdon N., Sarvas J. L., Frye M. A., Chicco A. J., and Lees S. J. (2014) Pro-inflammatory mediation of myoblast proliferation. PLoS One 9, e92363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 18. Barr F., and Lambright D. G. (2010) Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pfeffer S. R. (2013) Rab GTPase regulation of membrane identity. Curr. Opin. Cell Biol. 25, 414–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geppert M., Bolshakov V. Y., Siegelbaum S. A., Takei K., De Camilli P., Hammer R. E., and Südhof T. C. (1994) The role of Rab3A in neurotransmitter release. Nature 369, 493–497 [DOI] [PubMed] [Google Scholar]
- 21. Cazares V. A., Subramani A., Saldate J. J., Hoerauf W., and Stuenkel E. L. (2014) Distinct actions of Rab3 and Rab27 GTPases on late stages of exocytosis of insulin. Traffic 15, 997–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mizuno-Yamasaki E., Rivera-Molina F., and Novick P. (2012) GTPase networks in membrane traffic. Annu. Rev. Biochem. 81, 637–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wilkinson S., Paterson H. F., and Marshall C. J. (2005) Cdc42-MRCK and Rho-ROCK signalling cooperate in myosin phosphorylation and cell invasion. Nat. Cell Biol. 7, 255–261 [DOI] [PubMed] [Google Scholar]
- 24. Gomes E. R., Jani S., and Gundersen G. G. (2005) Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell 121, 451–463 [DOI] [PubMed] [Google Scholar]
- 25. Blencowe B. J., Issner R., Nickerson J. A., and Sharp P. A. (1998) A coactivator of pre-mRNA splicing. Genes Dev. 12, 996–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sim J. C., White S. M., and Lockhart P. J. (2015) ARID1B-mediated disorders: mutations and possible mechanisms. Intractable Rare Dis. Res. 4, 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takai T., Nishita Y., Iguchi-Ariga S. M., and Ariga H. (1994) Molecular cloning of MSSP-2, a c-myc gene single-strand binding protein: characterization of binding specificity and DNA replication activity. Nucleic Acids Res. 22, 5576–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muñoz-Cánoves P., Scheele C., Pedersen B. K., and Serrano A. L. (2013) Interleukin-6 myokine signaling in skeletal muscle: a double-edged sword? FEBS J. 280, 4131–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nonn L., Peng L., Feldman D., and Peehl D. M. (2006) Inhibition of p38 by vitamin D reduces interleukin-6 production in normal prostate cells via mitogen-activated protein kinase phosphatase 5: implications for prostate cancer prevention by vitamin D. Cancer Res. 66, 4516–4524 [DOI] [PubMed] [Google Scholar]
- 30. Zhang Y., Blattman J. N., Kennedy N. J., Duong J., Nguyen T., Wang Y., Davis R. J., Greenberg P. D., Flavell R. A., and Dong C. (2004) Regulation of innate and adaptive immune responses by MAP kinase phosphatase 5. Nature 430, 793–797 [DOI] [PubMed] [Google Scholar]
- 31. Farhan H., and Rabouille C. (2011) Signalling to and from the secretory pathway. J. Cell Sci. 124, 171–180 [DOI] [PubMed] [Google Scholar]
- 32. Wang J., Ren J., Wu B., Feng S., Cai G., Tuluc F., Peränen J., and Guo W. (2015) Activation of Rab8 guanine nucleotide exchange factor Rabin8 by ERK1/2 in response to EGF signaling. Proc. Natl. Acad. Sci. U.S.A. 112, 148–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kontaridis M. I., Liu X., Zhang L., and Bennett A. M. (2002) Role of SHP-2 in fibroblast growth factor receptor-mediated suppression of myogenesis in C2C12 myoblasts. Mol. Cell. Biol. 22, 3875–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee H., and Bennett A. M. (2013) Receptor protein tyrosine phosphatase-receptor tyrosine kinase substrate screen identifies EphA2 as a target for LAR in cell migration. Mol. Cell. Biol. 33, 1430–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]