Abstract
Cell signaling affects gene expression by regulating the activity of transcription factors. Here, we report that mitogen-activated protein kinase (MAPK) phosphorylation of Ets-1 and Ets-2, at a conserved site N terminal to their Pointed (PNT) domains, resulted in enhanced transactivation by preferential recruitment of the coactivators CREB binding protein (CBP) and p300. We discovered this phosphorylation-augmented interaction in an unbiased affinity chromatography screen of HeLa nuclear extracts by using either mock-treated or ERK2-phosphorylated ETS proteins as ligands. Binding between purified proteins demonstrated a direct interaction. Both the phosphoacceptor site, which lies in an unstructured region, and the PNT domain were required for the interaction. Minimal regions that were competent for induced CBP/p300 binding in vitro also supported MAPK-enhanced transcription in vivo. CBP coexpression potentiated MEK1-stimulated Ets-2 transactivation of promoters with Ras-responsive elements. Furthermore, CBP and Ets-2 interacted in a phosphorylation-enhanced manner in vivo. This study describes a distinctive interface for a transcription factor-coactivator complex and demonstrates a functional role for inducible CBP/p300 binding. In addition, our findings decipher the mechanistic link between Ras/MAPK signaling and two specific transcription factors that are relevant to both normal development and tumorigenesis.
The connection of cell signaling to changes of gene expression represents a central step in many types of biological regulation. The Ras/mitogen-activated protein kinase (MAPK) pathway exemplifies the relevance of signaling to both normal development and disease. In this pathway, Ras, a GTPase, transmits extracellular signaling from receptor tyrosine kinases to two serine/threonine kinases (Raf and MEK) and, finally, to the activation of MAPKs. Upon nuclear import, MAPKs phosphorylate many different transcription factors, modulating DNA binding affinity, nuclear localization, stability, and interactions with coregulators (20, 58), thereby regulating gene expression. Genetic studies of Drosophila (54) and Caenorhabditis elegans (48) demonstrate that Ras/MAPK signaling plays a role in normal development. Furthermore, ∼15 to 20% of all human tumors have an activating mutation in one of three ras genes (N-, K-, or H-ras), and oncogenic mutations in the B-raf gene have been identified for a wide variety of cancers, with 66% of all melanomas being affected (6, 8). Although many of the components of Ras/MAPK signaling have been characterized, the full array of transcription factors affected is not known. Furthermore, the detailed mechanisms by which phosphorylation modulates transcription factors remain unclear in many cases.
Multiple members of the ETS family of transcription factors are phosphorylated upon activation of the Ras/MAPK signaling pathway. For example, Elk-1 phosphorylation results in recruitment of the mediator complex via its Sur-2 (MED23) subunit (49), enhanced interaction with the coactivator p300 (27), and increased DNA binding (62). Phosphorylation of the mammalian ERF and the Drosophila Yan, on the other hand, leads to their cytoplasmic export (26, 50). For vertebrate Ets-1, Ets-2, and their Drosophila ortholog Pointed P2 (PNT P2), which constitute one subclass within the ETS family of transcription factors, Ras/MAPK signaling stimulates transcriptional activity but does not affect in vitro DNA binding, nuclear localization, or stability of the proteins (5, 40, 55, 60). The effect of phosphorylation on ETS protein function differs among the family members, thus providing routes to specificity within this gene family.
Functional studies demonstrate a role for Ras/MAPK signaling in the Ets-1/Ets-2/PNT P2 subgroup of ETS proteins. Specific MAPKs (Rolled or extracellular signal-regulated kinases [ERKs], respectively) and a single threonine residue (Thr 151 or Thr 38/Thr 72) are implicated in Drosophila and mammalian cells, respectively (2, 33, 35, 40, 60). ERK-mediated Ets-1/Ets-2 phosphorylation results in persistent, not transient, activation of a distinct set of Ras-responsive element (RRE)-containing genes (9, 11). Mutation of the site of phosphorylation compromises Ras-dependent superactivation in transient expression assays in both Drosophila and mammalian systems (35, 36, 56, 60). In addition, mutation in PNT P2 (T151A) blocks R7 photoreceptor development in a dominant negative manner (2). Loss of the phosphoacceptor site in the sea urchin ortholog affects the epithelial-mesenchymal transition in the normal embryo (43). Introduction of the phosphoacceptor site mutation into the ets-2 locus in the mouse does not affect normal development, but it restricts mammary tumors promoted by a variety of different transgenic oncogenes. Furthermore, this restriction correlates with decreased MMP-3 and MMP-9 mRNA levels in macrophages of the Ets-2T72A mutant mouse (31). In spite of this extensive evidence for a role for the Ras/MAPK signaling in Ets-1, Ets-2, and PNT P2 functions, the mechanism(s) responsible for MAPK phosphorylation-enhanced Ets-1/Ets-2 activity had not been determined previously.
A structural framework for investigation of this problem is available for Ets-1 and, by homology, Ets-2 (Fig. 1). Based on a nuclear magnetic resonance (NMR) approach, the N-terminal PNT domain, which is conserved in Ets-1, Ets-2, and PNT P2, forms a five-helix globular structure that resembles a more broadly observed protein fold known as the SAM domain (Fig. 1B) (22, 29, 47). The site of MAPK phosphorylation is located at a conserved distance ∼20 amino acids N terminal to the PNT domain in Ets-1, Ets-2, and PNT P2 (45). This region displays no secondary structure and appears to be highly flexible. Furthermore, there is no structural change upon phosphorylation (47). A less-well-conserved transcriptional activation domain (TAD) lies between the PNT and C-terminal DNA binding domain, termed the ETS domain (7, 14) (Fig. 1A). Ets-2 appears to have a second TAD N terminal to the PNT domain (44). Transactivation studies suggest that the PNT domain might synergize with the central TAD (44). One route by which the PNT domain augments transactivation is by recruitment of MAPK. The PNT domain of Ets-1 and Ets-2 provides a docking site for the MAPK ERK2, which is conserved only in Ets-1, Ets-2, and PNT P2 (45). Additional functions for the domain are reasonable in light of the variety of functions attributed to this domain by other ETS proteins (22, 29).
FIG. 1.
Structural and functional domains of murine Ets-1 and Ets-2. (A) Schematic of full-length and C-terminal truncation mutants of Ets-1 and Ets-2, with MAPK phosphorylation sites T38 and T72 (asterisks), PNT and ETS domains (gray and black boxes, respectively), and TADs as illustrated. (B) Ribbon display of the NMR-based structure of Ets-129-138. The five α-helices (H1 to H5) of the PNT domain (residues 54 to 132) and unstructured N terminus bearing the MAPK substrate site are illustrated; the black sphere indicates the Cα of T38 in the space-filling model. The lowest-energy model from the ensemble of NMR-derived structures of Ets-1 PNT domain (1BQV) was drawn with WebLab ViewerPro 4.0 (Accelrys, San Diego, Calif.) (47).
We hypothesized that ERK2 phosphorylation might create a binding site for a ligand that would mediate transcriptional activation or that phosphorylation might disrupt a binding partner that represses the activation function. To search for proteins that may preferentially bind unphosphorylated or phosphorylated Ets-1 and Ets-2, we performed affinity chromatography of HeLa nuclear extracts with either mock-treated or ERK2-phosphorylated Ets-1 or Ets-2 as ligands. We discovered that CREB binding protein (CBP) and p300 displayed enhanced binding to phosphorylated Ets-1 and Ets-2. Both an intact phosphoacceptor and the PNT domain were required for the interaction. Functional in vivo tests, including transient expression assays and coimmunoprecipitation, were consistent with the biochemical approaches. We propose that CBP/p300 recruitment is the basis for Ras/MAPK-dependent superactivation by Ets-1 and Ets-2. Our findings suggest a unique interface for a phosphorylated transcription factor-CBP/p300 complex. The interface requires the well-structured PNT domain of Ets-1/Ets-2 as well as an unstructured region with a phosphate moiety. These findings establish the route to activation of Ets-1 and Ets-2 by Ras/MAPK signaling.
MATERIALS AND METHODS
Bacterial expression constructs.
FLAG-heart muscle kinase (HMK)-Ets-11-440in pET3a was previously described (5). The T38A mutant replaces a 279-bp XhoI fragment containing the wild-type (WT) codon (ACT) with a fragment encoding the mutated codon (GCT). 6xHis-FLAG-HMK-Ets-11-138 WT/T38A and 6xHis-FLAG-HMK-Ets-21-172 WT/T72A were constructed by cloning a FLAG/HMK cassette (5) into the NdeI site of the respective previously described pET28a(+) Ets-1/Ets-2 constructs (45). 6xHis-FLAG-HMK-Ets-11-52 and 6xHis-FLAG-HMK-Ets-21-86 were made by PCR amplification of the above Ets-11-138 and Ets-21-172 vectors, with products cloned into the unique NcoI and HindIII sites of pET33b(+) (Novagen).
Mammalian expression vectors.
Plasmids encoding fusion proteins with the GAL4 DNA binding domain (1-147), GAL4:Ets-11-331, GAL4:Ets-11-243, GAL4:Ets-11-138, GAL4:Ets-11-52, GAL4:Ets-21-172, and GAL4:Ets-21-86, were created by cloning PCR-amplified coding sequences with an additional C-terminal FLAG tag into pFA-CMV (Stratagene) at the unique Asp718 and EcoRI sites. The Ets-11-138 T38A and Ets-21-172 T72A mutants were made by using a QuikChange Site-Directed Mutagenesis Kit (Stratagene). pMLC1 ΔN3 S218E, S222D MEK1, also known as CA-MEK1, was previously described (45). The corresponding empty vector was made by removal of the MEK1 open reading frame. pRc/RSV-mCBP-HA-RK (a kind gift of Dimitris Thanos, Columbia University), which expresses hemagglutinin-tagged full-length mouse CBP, has been described previously (46). The empty vector, pRc/RSV, was from Invitrogen. Cytomegalovirus (CMV) vectors expressing FLAG-tagged full-length mouse Ets-2 or Ets-2T72A were previously described (45).
Luciferase reporter vectors.
pFR-Luc, which has five GAL4 sites upstream of a TATA box driving firefly luciferase, was from Stratagene. 4xGAL4-TK-Luc, which has four GAL4 binding sites upstream of a thymidine kinase promoter, was a kind gift of Don Ayer, University of Utah. The artificial double ETS site RRE-Luc reporter was constructed by cloning annealed oligonucleotides 5′CGCGTAGGCCAGACCGGAAGCGTACTTCCGGTGCAATCGG 3′ and 5′CTAGCCGATTGCACCGGAAGTACGCTTCCGGTCTGGCCTA 3′, which display two inverted ETS binding sites (underlined), into the MluI and NheI sites of a minimal prolactin promoter (45). The MMP-9 RRE-Luc reporter has been previously described (45). The Renilla internal control, pRL-null, was from Promega.
Antibodies.
Anti-Ets-1 (UT2), which was raised in rabbits against the C-terminal 13 amino acids of murine Ets-1, has been described previously (15). Anti-Ets-1 (UT101A) against the Ets-1 PNT domain (residues 29 to 138) was raised in rabbits. Anti-CHD4 was a kind gift of Weidong Wang (National Institutes of Health). Commercial rabbit antibodies include anti-PCAF (Upstate Biotechnology), anti-BRG1 (Active Motif), anti-Daxx (Affinity BioReagents), and Santa Cruz Biotechnology antibodies anti-CBP/p300 (sc-1211), anti-CBP (A-22, sc-369), anti-p300 (N-15, sc-584), anti-RNA pol II LS (sc-899), anti-TRAP230 (sc-5372), anti-SRC-1 (sc-8995), anti-His (sc-804), anti-total ERK2 (sc-154), and anti-Ets-2 (C-20, sc-351). Commercial mouse monoclonal antibodies include anti-phos-Thr-Pro and anti-phospho-p44/42 MAPK E10 (Cell Signaling Technologies), anti-Sur-2 (BD Biosciences), anti-FLAG M2 Affinity Gel (Sigma), and anti-TAFII250 (sc-735) plus anti-GAL4 DNA binding domain (anti-GAL4DBD) (sc-510) (Santa Cruz Biotechnology). Secondary antibody-horseradish peroxidase conjugates were from Santa Cruz Biotechnology (goat) or Amersham (donkey).
Silver staining and Western blotting.
Silver staining of 4 to 15% or 4 to 20% gradient gels (Bio-Rad) was performed essentially as described previously (23). Western blotting was performed on protein-immobilized polyvinylidene difluoride (PVDF) membranes (Schleicher & Schuell) according to the ECL Plus detection kit (Amersham) protocol. Blots were stripped by an ECL Plus kit protocol.
Expression and purification of recombinant proteins.
Full-length FLAG-HMK-Ets-11-440 WT/T38A was expressed in bacteria and purified as described in reference 5. All 6xHis-FLAG-HMK-Ets-1/Ets-2 proteins were expressed in bacteria and purified as described previously (45), with the following exceptions. Ni-NTA agarose (QIAGEN) columns were employed, and His-tagged proteins were step eluted in 50 mM sodium phosphate (pH 7.8), 500 mM NaCl, and 250 mM imidazole (Sigma). Appropriate fractions, as determined by Coomassie blue staining, were pooled, dialyzed into buffer A (45) supplemented with 0.2 mM phenylmethylsulfonyl fluoride (PMSF), snap-frozen on liquid nitrogen, and stored at −80°C.
Baculoviruses encoding full-length human p300-6xHis (kind gift of W. Lee Kraus, Cornell University; 24) and 6xHis-murine CBP (kind gift of Dimitris Thanos; see reference 3) were used to infect Sf9 insect cells grown in liquid SFM II medium (Invitrogen) at 25°C with gentle shaking. Cell lysis was accomplished by Dounce homogenization in a previously described buffer (24). Soluble protein was then loaded onto a 5-ml HiTrap chelating column (Amersham), and His-tagged protein was eluted with a linear imidazole gradient (15 to 500 mM). Fractions were run on sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis (SDS-PAGE) gels, and appropriate fractions were pooled and stored at −80°C.
Phosphorylation of Ets-1 and Ets-2.
Phosphorylation of Ets-1 and Ets-2 species was performed with purified rat activated ERK2 (45). Complete phosphorylation was confirmed by mobility shifts of ETS proteins with SDS-PAGE gels or by incorporation of 32P by inclusion of [γ-32P]ATP (ICN Biomedicals, Inc.) in kinase reactions (C. E. Foulds, unpublished data). Kinase reactions were performed with ERK2 buffer, 25 mM Tris (pH 7.2), 1 mM dithiothreitol (DTT), 10 mM MgCl2, 1 mM ATP (Pharmacia), 12 mM β-glycerophosphate (B-GP) (Sigma or Calbiochem), 0.5 mM Na3VO4 (Sigma), and 0.2 mM PMSF with glycerol and KCl provided by the ETS and ERK2 protein solutions. A roughly 35 μM concentration of Ets-11-440, Ets-11-138, Ets-21-172, or the corresponding T38A/T72A mutants was incubated with ERK2 at a 10:1 molar ratio at 30°C for 1 h. Optimal phosphorylation of a ∼140 μM concentration of Ets-11-52 and Ets-21-86 was accomplished at a 100:1 molar ratio.
α-FLAG bead binding assay.
HeLa S3 cell pellets were obtained from the National Cell Culture Center, and nuclear extract was prepared as described previously (23). The concentration of protein in extract was determined by UV spectroscopy (16) and Bradford assay (Bio-Rad). The anti-FLAG bead binding assay was adapted from a previously described glutathione S-transferase affinity binding assay (41). FLAG-tagged Ets-1 or Ets-2 protein which was previously either mock treated or phosphorylated by ERK2 was immobilized to 60 μl of anti-FLAG M2 Affinity Gel in BB180 buffer, 20 mM HEPES (pH 7.9), 180 mM KCl, 10% glycerol, 0.2 mM EDTA, 0.5 mM PMSF, 12 mM Β-GP, 0.5 mM Na3VO4, and 1× complete mini-protease inhibitor cocktail (Roche) containing ∼0.25 mM DTT by gentle rocking at 4°C for 1 h. Beads were washed three times with 1.2 ml of the above buffer, and then preincubated with 1.2 ml of blocking buffer BB180 with 0.25 mM DTT, 0.05% NP-40, and 1 mg of bovine serum albumin (fraction V; Sigma)/ml at 4°C for 1 h with rotation. The blocking buffer was removed, and ∼2 mg of nuclear extract was added to the beads. Binding of proteins to beads occurred with gentle rotation at 4°C for at least 12 h. Beads were washed four times with 1.2 ml of BB180 supplemented with 0.25 mM DTT and 0.1% NP-40 to remove unbound proteins. FLAG-tagged Ets-1 and Ets-2 proteins and associated proteins were eluted competitively from the anti-FLAG beads with 0.1 mg of 1× FLAG peptide (Sigma)/ml in the above wash buffer. Eluates (∼600 μl) were trichloroacetic acid precipitated by a standard protocol (23), and dried protein pellets were resuspended in SDS sample buffer for SDS-PAGE. Western blotting, with the antibodies indicated in the figure legends, was used to detect CBP, p300, and other components present in the nuclear extract. Subsequent probing with anti-Ets-1 or anti-His showed a similar level of Ets-1/Ets-2 bound to the beads.
Direct protein-protein interaction assays.
Purified CBP or p300 protein (Coomassie stain estimate versus the bovine serum albumin standard) at a final concentration of ∼20 nM was incubated with ∼240 pmol of FLAG-tagged Ets-1/Ets-2 previously bound to 20 μl of anti-FLAG M2 Affinity Gel in BB180 buffer, as described above. FLAG-tagged ETS proteins were present in a molar excess of ∼50- or ∼120-fold over CBP or p300, respectively. Washing conditions to remove unbound protein were the same as for the bead binding assay with HeLa extracts. Bound proteins were eluted by boiling in SDS sample buffer, subjected to SDS-PAGE, transferred to PVDF membrane, and then probed with anti-CBP or anti-p300. Subsequent probing with anti-Ets-1 or anti-His showed that similar levels of Ets-1/Ets-2 had been bound to the beads.
Transfections and luciferase assays.
NIH3T3 mouse fibroblasts were grown, transfected by a calcium phosphate precipitation method, and serum starved as previously described (45). Firefly luciferase activities were normalized to Renilla luciferase activity from cotransfected pRL-null according to a published protocol (45). Each transfection was balanced for the same amount of total DNA (by adding sheared salmon sperm DNA where needed) and for promoter effects (by adding appropriate empty vectors). Transfection mixes shown in Fig. 4 contained 5 μg of total DNA, composed of 2.5 μg of luciferase reporter, 0.6 to 1 μg of pRL-null, and 100 ng of CA-MEK1 cotransfected with 100 ng of the indicated CMV-GAL4 fusion. In Fig. 5, 13 μg of DNA, composed of 1.9 μg of luciferase reporter, 0.9 μg of pRL-null, 100 ng of CA-MEK1, 100 ng of full-length Ets-2 or Ets-2T72A, and a total of 10 μg of Rous sarcoma virus vector (empty plus CBP expression vector; see figure legend), was transfected.
FIG. 4.
MAPK-enhanced transcriptional activation of Ets-1 and Ets-2 requires ERK2 phosphoacceptor sites and PNT domains. Transient expression assays in NIH3T3 cells are reported as relative luciferase activity (RLA), the ratio of firefly luciferase activity to Renilla luciferase activity (means and standard errors are shown). (A) Upper panel, minimal GAL4 dependent reporter (pFR-Luc) transfected with expression vectors for GAL4DBD fusions to Ets-1 C-terminal truncations (GAL4DBD:Ets-1) and CA-MEK1, as indicated. Lower panel, Western analysis of cell lysates transfected in parallel and probed with anti-GAL4. Arrowheads indicate the size expected for each full-length GAL4 fusion protein. Smaller fragments may represent degradation products. (B) 4xGal4-TK-Luc reporter transfected with expression vectors for GAL4DBD:Ets-1/Ets-2 species and CA-MEK1, as indicated. Left, schematic of GAL4DBD:Ets-1/Ets-2 fusion proteins with GAL4DBD (hatched), PNT (gray), and ETS domains (black). Wild-type (T38 and T72) or mutated MAPK phosphorylation sites are indicated. Right, reporter levels in the presence of CA-MEK1 (RLA × 10−3) as in panel A. Lower panel, Western analysis of GAL4 fusion protein levels in the presence of CA-MEK1 as in panel A.
FIG. 5.
CBP potentiates MAPK-enhanced Ets-2 transactivation of RREs. NIH3T3 cells were transfected as for Fig. 4, with a reporter driven by either the RRE of the MMP-9 promoter (upper panel) or two ETS protein binding sites (lower panel) and expression vectors for CA-MEK1, FLAG-tagged full-length Ets-2 or Ets-2T72A, and CBP (in increasing amounts [in micrograms], as indicated).
To determine steady-state GAL4 fusion protein levels with anti-GAL4DBD, 3T3 cells were transfected by Lipofectamine, according to Invitrogen's protocol, to obtain higher transfection efficiency. One microgram of GAL4:ETS fusion vector (or GAL4DBD only) and 1 μg of CA-MEK1 (or empty vector) was mixed with 10 μl of Lipofectamine. After transfection, cells were serum starved exactly as done previously. Lysates were prepared by boiling cells in 3× SDS sample buffer for Western blotting. Ponceau S staining confirmed that equal protein levels were transferred onto PVDF membranes.
ΔB-Raf:ER* cell assays.
ΔB-Raf:ER* NIH3T3 cells, which stably express a mouse B-Raf kinase domain-mouse ER fusion protein that is activated by 4-hydroxy-tamoxifen (4-HT), were a kind gift of Martin McMahon, University of California, San Francisco, Calif. (59). Cells were incubated in phenol red-free Dulbecco's modified Eagle medium containing 0.5% fetal bovine serum (HyClone) to serum starve prior to the addition of 4-HT. To monitor the effect of Ras/MAPK signaling on transcription activity, ∼2.5 × 105 ΔB-Raf:ER* cells were plated in six-well dishes (Falcon), transfected by a calcium phosphate precipitation method, serum starved for 20 to 24 h, and harvested for dual luciferase activities after indicated times of exposure to 1 μM 4-HT (Sigma). Transfection mixes contained 5 μg of total DNA composed of 2.5 μg of luciferase reporter, 1 μg of pRL-null, 100 ng of the indicated Ets-2 expression vector, and 1.4 μg of salmon sperm DNA. To assay endogenous ERK1/ERK2 activation, ∼2.5 × 105 ΔB-Raf:ER* cells were serum starved for 42 h. After receiving starvation medium lacking 4-HT or supplemented with 1 μM 4-HT for listed times, cells were harvested by direct lysis in 100 μl of 1.5× SDS sample buffer. One-fourth of the whole cell protein lysate was loaded onto a 10% SDS-PAGE gel and was assayed by Western blotting. For coimmunoprecipitations, ∼1.5 × 106 ΔB-Raf:ER* cells were plated in 100-mm dishes and transfected with 12 μg of CMV-FLAG-Ets-2 or -Ets-2T72A vector and 60 μl of Lipofectamine. After 42 h of serum starvation, starvation media either lacking 4-HT or containing 1 μM 4-HT was added for 4 h. Cells were washed once with ice-cold phosphate-buffered saline, and then scraped off dishes in 1-ml lysis buffer, 50 mM Tris-Cl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 1× complete mini-protease inhibitors (Roche), 1 mM PMSF, 50 mM NaF, 20 mM Β-GP, 0.2 mM activated Na3VO4, and 2.5 mM sodium pyrophosphate. Cells were sonicated as described previously (65) and clarified at 20,879 × g for 15 min at 4°C. Lysates were precleared with 40 μl of 50% slurry of protein A-agarose (Pierce) at 4°C for at least 30 min. Forty microliters of 50% slurry of anti-FLAG M2 Affinity Gel was added to the cleared lysates, and incubation was continued at 4°C for between 1.5 to 20 h. Following three washes in lysis buffer, 3× SDS sample buffer was added to the anti-FLAG beads and eluted proteins were analyzed by 4 to 15% gradient SDS-PAGE and Western blotting.
RESULTS
CBP/p300 interact preferentially with ERK2-phosphorylated Ets-1 and Ets-2.
ERK1/ERK2-dependent phosphorylation of Ets-1 and Ets-2 enhances transcriptional activity; thus, the modification may induce dissociation of a corepressor and/or recruitment of a coactivator. We searched for these putative interacting proteins by using bead-immobilized Ets-1 that was phosphorylated in vitro with purified ERK2 or mock treated (Fig. 2). Numerous proteins from HeLa nuclear extract bound both phosphorylated and mock-treated Ets-1; however, a single major species that was greater than 200 kDa bound preferentially to phosphorylated Ets-1 (Fig. 2A). Based on antibody detection of phosphorylated or mock-treated Ets-1 in eluate, we determined that this preference was not due to the uneven loading of beads. For identification of the phosphorylation-dependent binding species, the bead eluate was screened by Western analysis with antibodies against known transcription factors of high molecular weight. Candidates included CHD4, TAFII250 (TAF1), TRAP230 (MED12), the large subunit of RNA pol II, and CBP/p300. Only antibodies against CBP and/or p300 recognized a high-molecular-weight species with phosphorylation dependence (Fig. 2B) (Table 1). Consistent with this identification, the largest reactive species migrated on an SDS-PAGE gel with a mobility similar to that of purified human p300 (Foulds, unpublished). Antibody screening identified one of the phosphorylation-independent binding species as the mediator subunit Sur-2 (MED23) (Fig. 2B; Table 1). This phosphorylation-independent binding activity confirmed the equivalence of the Ets-1 species on the two types of affinity beads except for the addition of the phosphate moiety. Beads loaded with Ets-1T38A, which bears a mutation in the phosphoacceptor site, displayed severely reduced binding to CBP/p300, but retained binding to Sur-2 (Fig. 2B). These initial findings suggested that Ras/MAPK-directed phosphorylation of Ets-1 enhances the recruitment of coactivators CBP/p300.
FIG. 2.
Enhanced binding of CBP/p300 from HeLa nuclear extract to Ets-1 and Ets-2 requires ERK2 phosphorylation and PNT domain. Anti-FLAG agarose beads were loaded with different FLAG-Ets-1/Ets-2 species, as indicated, and then incubated with HeLa nuclear extract. After washing, bound species were eluted with FLAG peptide. Fifty percent of the sample was analyzed by stained gel, and 50% was used for Western analysis. NE, 2% of input nuclear extract; B, eluate from beads not loaded with Ets-1 species but incubated with extract; M, molecular mass standards (in kilodaltons). (A) Upper panel, silver-stained SDS-PAGE gradient gel of FLAG peptide eluate from beads loaded with Ets-11-440, treated or mock treated with ERK2. Arrowhead, >200 kDa phosphorylation-dependent binding species. Lower panels, Western blots of FLAG eluate for bead loading control: anti-Ets-1 (UT2) detects Ets-11-440 (the apparent doublet is due to saturation of film in enhanced chemiluminescence detection); anti-phos-Thr-Pro antibody confirms that ERK2 phosphorylation of Ets-1 is retained during the experiment. (B) Ets-11-440 with the wild-type or mutant phosphoacceptor site tested as in panel A. Western blotting of FLAG peptide eluates with antisera as indicated. Upper panels, antibody with reactivity for CBP and p300 as well as antibody specific for CBP detects phosphorylation-dependent species in the >200-kDa range. The smaller species detected by anti-CBP/p300 are assumed to be CBP/p300 fragments. Lower panel, anti-Ets-1 (UT101A) antibody detects Ets-11-440 species as bead loading control. (C) An assay of Ets-11-138 and Ets-21-172, as in panels A and B, demonstrated that N-terminal fragments bearing the PNT domains are sufficient for phosphorylation-dependent binding to CBP/p300. Upper panels, silver-stained SDS-PAGE gel. Middle panel, Western blot with anti-CBP/p300. Lower panel, anti-His detects Ets-1/Ets-2 species in loading controls. (D) Assay of Ets-11-52 and Ets-21-86, as in panel C, showed no phosphorylation-dependent binding. Anti-phos-Thr-Pro showed that Ets-11-52 and Ets-21-86 phosphorylation by ERK2 is retained during the experiment.
TABLE 1.
Survey of potential phosphorylation-dependent binding proteins of Ets-1 and Ets-2a
| Antibodyb | Ets-11-440
|
Ets-11-138
|
Ets-21-172
|
Activityc | |||
|---|---|---|---|---|---|---|---|
| −P | +P | −P | +P | −P | +P | ||
| p300/CBP | − | + | − | + | − | + | Coactivator/HAT |
| p300 | − | + | ND | ND | ND | ND | Coactivator/HAT |
| CBP | − | + | − | + | ND | ND | Coactivator/HAT |
| Sur-2 (MED23)d | + | + | + | + | + | + | Part of mediator |
| PCAFe | ND | ND | + | + | + | + | Coactivator/HAT |
| SRC-1e | ND | ND | − | − | + | + | Coactivator/HAT |
| Daxxf | ND | ND | − | − | − | − | Repressive in PODs |
| BRG-1f | − | − | − | − | − | − | ATPase of Swi/Snf |
| RNA pol II | − | − | − | − | ND | ND | Catalytic subunit |
| TAFII250 (TAF1) | − | − | ND | ND | ND | ND | Part of TFIID/HAT |
| TRAP230 (MED12) | − | − | ND | ND | ND | ND | Part of mediator |
| CHD4 | − | − | ND | ND | ND | ND | ATPase of NuRD |
−P, mock-treated; +P, phosphorylated protein; − or +, negative or positive binding status to ETS-immobilized beads; ND, not determined; HAT, histone acetyltransferase.
See Materials and Methods for details. All antibodies detected a polypeptide of the mass expected for the full-length protein in the input nuclear extracts.
Known activities of the various candidates. PODs, PML oncogenic domains.
Sur-2 bound Ets-1/Ets-2 independent of phosphorylation. This result was interesting, given that Ets-1/Ets-2 phosphorylation-enhanced transactivation is reduced in cells that lack Sur-2 (49).
Because both Ets-1 and the highly related Ets-2 are activated by Ras/MAPK signaling, we speculated that a domain conserved in Ets-1 and Ets-2 would be essential for the binding (Fig. 1A). Indeed, fragments of Ets-1 and Ets-2 that comprise both the conserved MAPK site (threonines 38 and 72, respectively) and PNT domain were sufficient for phosphorylation-enhanced CBP/p300 binding. Again, phosphoacceptor site mutants showed severely reduced binding (Fig. 2C). Interestingly, based on equimolar bead loading, phosphorylated Ets-21-172 showed higher binding activity for CBP/p300 than Ets-11-138. Phosphorylated N-terminal regions that flank the PNT domains and bear the phosphoacceptor sites Ets-11-52 and Ets-21-86 were not sufficient for CBP/p300 binding (Fig. 2D). Thus, in addition to the region bearing the phosphoacceptor, the PNT domain is necessary for the phosphorylation-dependent binding to CBP/p300.
Consistent with our previous report that the PNT domain of Ets-1 and Ets-2 provides a docking site for ERK2 (45), we detected both endogenous and added recombinant ERK2 bound to Ets-1/Ets-2-immobilized beads (Foulds, unpublished). Controls in which ATP addition or omission was used to produce phosphorylated or unphosphorylated species were performed. Phosphorylation-dependent binding of CBP/p300 was observed (Foulds, unpublished), thus demonstrating that ERK2 was not serving as a bridging factor in these bead-binding assays.
Phosphorylated Ets-1 and Ets-2 directly interact with CBP/p300.
Because nuclear extract may include auxiliary factors, we next tested for direct binding with CBP and p300 that was partially purified from an insect cell expression system (Fig. 3A) (M. L. Nelson, unpublished data). CBP and p300 bound unphosphorylated Ets-1, but this interaction was enhanced upon ERK2 phosphorylation (Fig. 3B) (Foulds, unpublished). Furthermore, phosphorylated Ets-11-138 and Ets-21-172, but not phosphoacceptor site mutants, bound CBP and p300 in an enhanced manner (Fig. 3B to D) (Foulds, unpublished). The minimal N-terminal fragments, Ets-11-52 and Ets-21-86, did not bind CBP/p300 at detectable levels (Fig. 3C and D) (Foulds, unpublished). In contrast to experiments with nuclear extract, the assays with purified CBP and p300 detected binding to unphosphorylated species. This binding may be inhibited in nuclear extracts by proteins that mask phosphorylation-independent interfaces on the ETS species. In conclusion, Ets-1 and Ets-2 interacted directly with CBP/p300 and, consistent with findings with extracts, the phosphorylation enhancement mapped to the PNT domain and N-terminal extensions.
FIG. 3.
Direct phosphorylation-enhanced binding of Ets-1 and Ets-2 to CBP. Anti-FLAG agarose beads were loaded with different FLAG-Ets-1/Ets-2 species, as indicated, and then incubated with CBP from a baculovirus expression system. After washing, bound CBP and Ets-1/Ets-2 species were eluted with SDS-PAGE sample buffer and subjected to electrophoresis and Western blotting for detection of CBP binding or for bead loading controls. M, molecular mass standards (in kilodaltons); B, eluate from beads not loaded with Ets-1/Ets-2 species but incubated with CBP. Western blots for loading controls detected Ets-1/Ets-2 fragments with anti-His, except for Ets-11-440 species, which were identified by anti-Ets-1 (UT2). (A) Coomassie-stained SDS-PAGE gel of His-tagged CBP after Ni column purification from insect cells. (B) Direct interaction between CBP and Ets-1 or Ets-2 species, as indicated, was detected in bead binding assay. Upper panel, Western blot of 67% of bead eluate probed with anti-CBP (A-22). Input lane, 4% of total CBP in binding reaction. Lower panel, Western loading control. (C) CBP binding to Ets-1 species analyzed as for panel B. Upper panel, Western blot of 67% of bead eluate probed with anti-CBP. Input lane, 4% of total CBP in binding reaction. Lower panel, Western loading control. (D) CBP binding to Ets-2 species, analyzed as for panel C.
Functional tests of phosphorylation-enhanced recruitment of CBP/p300.
To test the functional relevance of phosphorylation-enhanced CBP/p300 binding, we compared the transcriptional activities of Ets-1 and Ets-2 under conditions of MAPK activation in the mouse fibroblast cell line NIH3T3. To facilitate tests of ETS protein fragments, GAL4 DNA binding domain fusions and reporter plasmids with multiple GAL4 binding sites were used in transient expression assays. Controls showed that all GAL4 fusion proteins were expressed at roughly similar levels (Fig. 4). To ensure maximal MAPK (presumably ERK1/ERK2) activation, an expression plasmid for constitutively active MEK1 (CA-MEK1) was coexpressed. Cells were serum starved after transfection to enhance the stimulatory effect of CA-MEK1. We first addressed the minimal region required for MEK1 induction by using Ets-11-331, Ets-11-243, and Ets-11-138 fragments fused to GAL4 (Fig. 4A). Although basal levels of these fusion proteins differed due to the presence of the TAD between 138 and 243 (Fig. 1A), the 1.5- to 3.5-fold induction by CA-MEK1 was similar. These mapping results are similar to those obtained by the use of oncogenic Ras with GAL4 fusions to an alternative form of Ets-1 in chickens (55) and with a GAL4-Ets-21-172 fusion (10). To compare CBP/p300 binding to transcriptional activity more directly, we tested additional GAL4 fusion proteins from Ets-1 and Ets-2. Tests with Ets-11-138 and Ets-21-172 demonstrated the expected requirement for an intact phosphoacceptor for maximal transcription (Fig. 4B). Interestingly, Ets-21-172 had stronger activation activity than Ets-11-138, consistent with the stronger CBP/p300 binding activity of Ets-21-172. Also, consistent with binding studies, the transcriptional activity of Ets-11-52 and Ets-21-86 fusions were similar to the activity of the GAL4 DNA binding domain alone (Fig. 4B). This result is consistent with the failure of phosphorylated Ets-11-52 and Ets-21-86 to bind CBP/p300 in vitro. Alternatively, the requirement for the PNT domain may reflect the role of this domain in efficient ERK phosphorylation of Ets-1 and Ets-2 (45). In conclusion, the phosphoacceptors and PNT domains of Ets-1 and Ets-2 are required for Ras/MAPK enhancement of both transcription and CBP/p300 binding.
The binding and transcription assays predict that CBP/p300 are phosphorylation-enhanced coactivators of Ets-1 and Ets-2. To test directly for this synergism, we added CBP to Ets-2 transient expression assays in NIH3T3 cells. Native Ras-responsive elements (RREs) that represented either the composite Ets/AP-1 site from the murine MMP-9 promoter or an arrangement of two inverted ETS binding sites that mimicked those found in the MMP-3 promoter were utilized. Consistent with previous reports (45, 49), Ets-2, in the presence of CA-MEK1, synergistically activated both reporters (Fig. 5). CBP coexpression further enhanced both the MMP-9 and double ETS site reporters. The phosphoacceptor site mutant, Ets-2T72A, was compromised in both superactivation and, more importantly, CBP coactivation (Fig. 5). This result was not due to differences in the levels of wild-type and mutant Ets-2 as determined by quantitative immunoprecipitations (45). The use of the double ETS site reporter demonstrated that an AP-1-type binding factor was not necessary for Ets-2:CBP cooperation. In conclusion, CBP binding data correlated with enhanced transcription of Ras-responsive promoters.
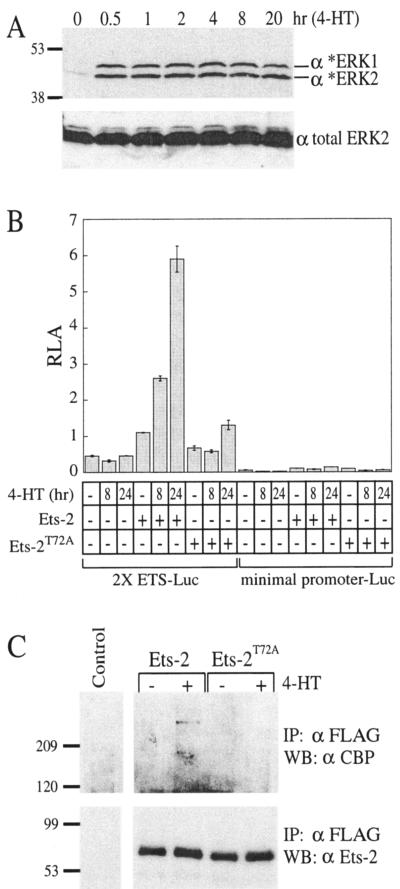
To further investigate the functional significance of the phosphorylation-enhanced binding of CBP, we tested for an effect of Ras/MAPK signaling on the in vivo association of CBP and Ets-2. We used an NIH3T3 cell line with a conditional B-raf allele (ΔB-Raf:ER*) in which B-Raf activity can be induced by 4-hydroxy-tamoxifen (4-HT) treatment (59). As previously reported, the ΔB-Raf:ER* fusion protein is rapidly activated in a sustained manner, as detected by ERK1/ERK2 activation (Fig. 6A). Transfection of a double ETS site reporter, but not the minimal promoter lacking ETS sites, along with FLAG-Ets-2 resulted in transcriptional activation in response to 4-HT treatment that was reduced in the Ets-2T72A mutant (Fig. 6B). These results are consistent with previous work with a 3T3 cell line expressing a Raf-1:ER fusion, in which Ets-2 phosphorylation was directly correlated with reporter activation (33). This inducible system was then used to test whether endogenous CBP associated with Ets-2. By coimmunoprecipitation, we observed a 4-HT-dependent binding of endogenous CBP with FLAG-Ets-2, but not with FLAG-Ets-2T72A. This result was not due to differences in the amounts of Ets-2 immunoprecipitated by anti-FLAG beads or by different CBP levels in the lysates (Fig. 6C) (C. E. Foulds and A. G. Blaszczak, unpublished data). In sum, these findings support the proposal that Ras/MAPK signaling leads to enhanced recruitment of CBP/p300 by Ets-1 and Ets-2 to augment their transcriptional activation.
FIG. 6.
Signaling-dependent association of Ets-2 and CBP correlates with activated transcription. NIH3T3 cells with a conditional allele of B-raf (ΔB-Raf:ER* cells) were used to detect in vivo interactions between recombinant Ets-2 and CBP. (A) To assay activation of B-Raf signaling induced with 4-HT, cell lysates were probed for the presence of dually phosphorylated, activated endogenous ERK1/ERK2 (asterisks, upper panel) or for total ERK2 (lower panel) by Western analysis. (B) B-Raf signaling enhances transcriptional activation mediated by Ets-2. Cells were transfected with a luciferase reporter with two ETS protein binding sites or a minimal reporter lacking binding sites and expression vectors for either FLAG-tagged full-length Ets-2 or Ets-2T72A. After serum starvation, cells were harvested either immediately for dual luciferase assays or after incubation with 4-HT for 8 or 24 h. A ∼2-fold increase in 2X ETS-Luc reporter activity was observed after 8 h of 4-HT treatment, upon cotransfection with Ets-2. (A similar result was seen with 4 h of 4-HT treatment [A. G. Blaszczak, unpublished data]). (C) Coimmunoprecipitation with anti-FLAG beads detected a phosphorylation-dependent interaction between endogenous CBP and FLAG-tagged Ets-2. Cells were transfected with expression vectors for FLAG-Ets-2 or FLAG-Ets-2T72A, followed by 4-HT addition for 4 h or mock treatment. After SDS-PAGE, immunoprecipitated proteins were analyzed by Western blotting with anti-CBP (A-22) (upper panel). Lower panel, anti-Ets-2 (C-20) detected equivalent levels of recombinant Ets-2 in lysates.
DISCUSSION
Model for how MAPK phosphorylation activates Ets-1/Ets-2 by CBP/p300 recruitment.
This study demonstrated the mechanism by which Ras/MAPK-dependent phosphorylation enhances the transcriptional activity of Ets-1 and Ets-2. We show that CBP and p300 preferentially bind phosphorylated Ets-1 and Ets-2 in cell extracts and with purified proteins. Importantly, we have shown that phosphorylation at the MAPK site of Ets-1/Ets-2 and the presence of a PNT domain are both required for maximal CBP/p300 interaction. To test the functional significance of the phosphorylated Ets-1/Ets-2-CBP/p300 interaction, we showed that the Ets-1/Ets-2 residues required for maximal binding in vitro are also critical for MAPK-induced transcription in vivo, that CBP augmented MAPK-induced Ets-2 transactivation, and, finally, that endogenous CBP and recombinant Ets-2 displayed phosphorylation-enhanced interaction in vivo. We predict that the apparent ortholog of vertebrate Ets-1 and Ets-2, Drosophila PNT P2, which has been genetically implicated to function downstream of Ras signaling, also displays this interaction.
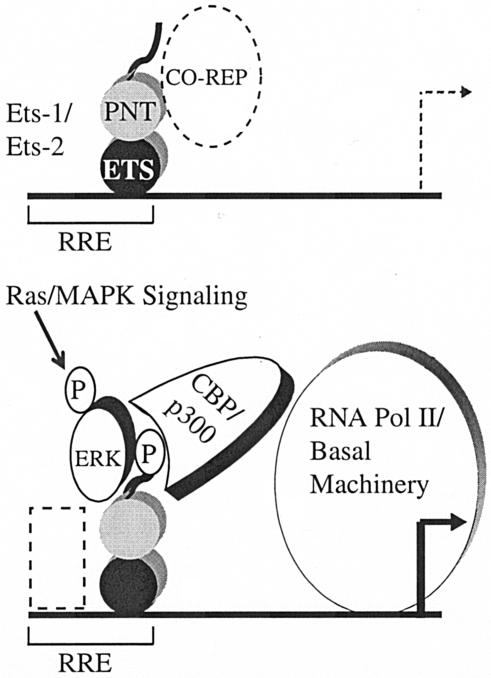
We propose a working model for how MAPK-phosphorylated Ets-1/Ets-2 activates transcription at a subset of RREs by recruitment of CBP/p300 (Fig. 7). For simplicity, we have modeled this interaction on Ets-1/Ets-2 prebound to target genes in the nucleus. Corepressor binding, documented for unphosphorylated Ets-1 and Ets-2 via their PNT domains (1, 21, 28, 38, 57) is added to the uninduced state. Upon stimulation of the Ras/MAPK signaling pathway, activated ERK1/ERK2 bind Ets-1/Ets-2 through the docking site on their PNT domains. Next, ERK phosphorylation in the flexible N terminus stimulates the binding of CBP/p300, which may recruit RNA polymerase II and/or the basal transcriptional machinery. MAPK-stimulated Ets-1 and Ets-2 functions impinge on RREs that are composed of an ETS site juxtaposed to either an AP-1, another ETS site, a Pit-1, or a vitamin D receptor binding site (12, 66). Because Ras/MAPK signaling also affects the activity of DNA binding partners of Ets-1/Ets-2 (e.g., c-Jun/ATF-2 at an overlapping AP-1 site) (20, 58), these proteins are added to the induced state. This mechanism is different from Ras/MAPK signaling to the well-characterized ETS protein Elk-1 that functions at serum response elements with SRF. Elk-1, which lacks a PNT domain, recruits both p300 and the mediator complex by a phosphorylation-dependent mechanism (27, 49).
FIG. 7.
Model of MAPK phosphorylation-enhanced Ets-1/Ets-2-CBP/p300 interaction. (Upper panel) In the absence of Ras/MAPK signaling, unphosphorylated Ets-1 and Ets-2 are bound with associated corepressors (CO-REP) to RREs. (Lower panel) Upon Ras pathway stimulation, ERKs become activated by phosphorylation and translocate to the nucleus. Activated ERKs are recruited by docking with the PNT domain to phosphorylate Ets-1 and Ets-2 at T38 and T72, respectively. We speculate that this association happens at the RRE and that CBP/p300 binding then ensues. We also illustrate that recruited CBP/p300 then increases transcription in part by stabilizing preinitiation complex formation (i.e., RNA pol II and the basal machinery) at the promoter (reviewed in reference 53). Additional DNA binding factors, such as AP-1, Pit-1, and vitamin D receptors (dashed box), may also bind the activated promoter.
Binding interface for the phosphorylated Ets-1/Ets-2-CBP/p300 complex.
The finding that phosphorylation enhances the CBP/p300 interaction with Ets-1/Ets-2 extends previous findings (18, 32, 59a, 61). Specifically, the CBP/p300 CH1/TAZ1 domain is reported to interact with Ets-11-210 and Ets-21-290 (18, 61), the CBP/p300 CH3 domain associates with full-length Ets-1, Ets-21-290, or Ets-2289-469 (18, 61), and the CBP C-terminal SRC1 interaction domain binds the Ets-2 PNT domain (residues 60 to 170) (32). Our data provide new insight into these interactions, as we show that Ets-11-138 and Ets-21-172, upon ERK2 phosphorylation, have increased affinity for CBP/p300. We provide evidence that phosphorylation at T38/T72 and the PNT domain of Ets-1/Ets-2 are both required for the binding interface. The residues 153 to 210 of the Ets-1 central TAD, not the PNT domain, were reported to be necessary for CH1/TAZ1 binding (61), and the effect of phosphorylation of Ets-2 on SRC1 interaction domain binding was not tested. Based on comparing CBP/p300 binding to that of Ets-11-440 and Ets-11-138, we hypothesize that there are both constitutive and inducible CBP/p300 binding regions in full-length Ets-1, with residues 1 to 138 providing the phosphorylation enhancement and residues 139 to 440 potentially providing constitutive contacts. This postulate may explain why Ets-1T38A retains some transcriptional activity (see references 37 and 45). Finally, based on the interface of the phosphorylated Ets-1/Ets-2-CBP/p300 binary complex, we can suggest a possible mechanism for functional specificity in the ets family of transcriptional regulators. Approximately 40% of ETS proteins have a PNT domain, yet only the subgroup with vertebrate Ets-1/Ets-2 and Drosophila PNT P2 has a juxtaposed MAPK substrate site (45). Therefore, MAPK and subsequent CBP/p300 docking provides a distinct regulatory pathway for this subgroup of ETS proteins.
The phosphorylated Ets-1 and Ets-2 interaction with CBP/p300 is distinct from that observed for other phosphorylated transcription factors, e.g., CREB (42), NF-κB p65 subunit (68), Elk-1 (27), GATA-4 (51), Smad3 (17), E2F-5 (34), forkhead box M1B (30), Kruppel-like factor 5 (67), Stat1 (52), p53 (25), HIF-like factor (13), and microphthalmia transcription factor (39). The phospho-CREB:CBP interaction was the first discovered (4) and remains the best structurally characterized example of phosphorylation-dependent CBP binding. The unstructured, phosphorylated kinase-inducible domain of CREB adopts structure upon CBP binding (42). The interfaces of other transcription factors also have been proposed to employ unstructured regions, although no structural data are available. We have shown that an NMR-defined unstructured region of Ets-1 is important for CBP/p300 interaction; however, the juxtaposed well-folded PNT domain is also required for maximal CBP/p300 binding. The unstructured region may be induced to fold in the complex or function in the flexible state. Further biochemical and structural studies are required to determine whether structural transitions play a role in the specificity of the interface.
A biological role for Ets-1/Ets-2 phosphorylation-enhanced CBP/p300 recruitment.
To date, constitutive Ras/MAPK signaling can result from activating mutations in either ras or B-raf genes (6, 8). The resultant oncogenic proteins can affect cellular processes associated with malignancy, such as stimulated proliferation, inhibited apoptosis, induced angiogenesis, and increased invasiveness (8). Targets of the Ras/MAPK pathway include multiple members of the ETS family of transcription factors (14, 66). Ets-1 and Ets-2 proteins were the focus of this study because their phosphorylation at T38 and T72, respectively, correlates with increased transcription of a subset of RRE-containing genes. Several of these genes, which are candidate Ets-1/Ets-2 targets, regulate remodeling of the extracellular matrix, an essential step in tumor growth and invasion, including the serine protease uPA and several matrix metalloproteinases (e.g., MMP-1, MMP-3, and MMP-9) (7, 31, 56, 60, 66). A better understanding of the phosphorylated Ets-1/Ets-2-CBP/p300 interface will facilitate the development of potential cancer therapeutics aimed at disrupting the interaction and potentially reducing the transcription of some RRE-containing genes.
In conclusion, our results provide an example of how functional specificity can be achieved in a transcription factor family by the use of a phosphorylation site and a juxtaposed protein interaction domain common to only a few members. Our findings demonstrate how the Ras/MAPK signaling pathway activates Ets-1/Ets-2 by promoting a unique binding interface with a coactivator. The binding mechanism further implicates a physiological role in converting extracellular proliferation signals to transcriptional activation of Ets-1- and Ets-2-dependent target genes.
Acknowledgments
The National Institutes of Health supported this work (grant R01 GM38663 to B.J.G., training grant support for C.E.F. [T32 DK07115 and T32 CA09602] and M.L.N [GM08537], and a cancer center support grant [CA-24014] to the Huntsman Cancer Institute). We also gratefully acknowledge support from the Huntsman Cancer Foundation.
We thank those cited within the text for reagents. We thank members of the Graves lab for advice and Karen Davis for aid in manuscript preparation.
Footnotes
This study is dedicated by C.E.F. to the memory of his father, Jon M. Foulds.
REFERENCES
- 1.Baker, K. M., G. Wei, A. E. Schaffner, and M. C. Ostrowski. 2003. Ets-2 and components of mammalian SWI/SNF form a repressor complex that negatively regulates the BRCA1 promoter. J. Biol. Chem. 278:17876-17884. [DOI] [PubMed] [Google Scholar]
- 2.Brunner, D., K. Dücker, N. Oellers, E. Hafen, H. Scholz, and C. Klambt. 1994. The ETS domain protein Pointed-P2 is a target of MAP kinase in the Sevenless signal transduction pathway. Nature 370:386-389. [DOI] [PubMed] [Google Scholar]
- 3.Chen, C. J., Z. Deng, A. Y. Kim, G. A. Blobel, and P. M. Lieberman. 2001. Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol. Cell. Biol. 21:476-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chrivia, J. C., R. P. Kwok, N. Lamb, M. Hagiwara, M. R. Montminy, and R. H. Goodman. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365:855-859. [DOI] [PubMed] [Google Scholar]
- 5.Cowley, D. O., and B. J. Graves. 2000. Phosphorylation represses Ets-1 DNA binding by reinforcing autoinhibition. Genes Dev. 14:366-376. [PMC free article] [PubMed] [Google Scholar]
- 6.Davies, H., G. R. Bignell, C. Cox, P. Stephens, S. Edkins, S. Clegg, J. Teague, H. Woffendin, M. J. Garnett, W. Bottomley, N. Davis, E. Dicks, R. Ewing, Y. Floyd, K. Gray, S. Hall, R. Hawes, J. Hughes, V. Kosmidou, A. Menzies, C. Mould, A. Parker, C. Stevens, S. Watt, S. Hooper, R. Wilson, H. Jayatilake, B. A. Gusterson, C. Cooper, J. Shipley, D. Hargrave, K. Pritchard-Jones, N. Maitland, G. Chenevix-Trench, G. J. Riggins, D. D. Bigner, G. Palmieri, A. Cossu, A. Flanagan, A. Nicholson, J. W. Ho, S. Y. Leung, S. T. Yuen, B. L. Weber, H. F. Seigler, T. L. Darrow, H. Paterson, R. Marais, C. J. Marshall, R. Wooster, M. R. Stratton, and P. A. Futreal. 2002. Mutations of the BRAF gene in human cancer. Nature 417:949-954. [DOI] [PubMed] [Google Scholar]
- 7.Dittmer, J. 2003. The biology of the Ets1 proto-oncogene. Mol. Cancer 2:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Downward, J. 2003. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3:11-22. [DOI] [PubMed] [Google Scholar]
- 9.Fafeur, V., D. Tulasne, C. Queva, C. Vercamer, V. Dimster, V. Mattot, D. Stehelin, X. Desbiens, and B. Vandenbunder. 1997. The ETS1 transcription factor is expressed during epithelial-mesenchymal transitions in the chick embryo and is activated in scatter factor-stimulated MDCK epithelial cells. Cell Growth Differ. 8:655-665. [PubMed] [Google Scholar]
- 10.Foos, G., C. K. Galang, C. F. Zheng, and C. A. Hauser. 2001. Ras signaling to transcription activation: analysis with GAL4 fusion proteins. Methods Enzymol. 333:61-73. [DOI] [PubMed] [Google Scholar]
- 11.Fowles, L. F., M. L. Martin, L. Nelsen, K. J. Stacey, D. Redd, Y. M. Clark, Y. Nagamine, M. McMahon, D. A. Hume, and M. C. Ostrowski. 1998. Persistent activation of mitogen-activated protein kinases p42 and p44 and ets-2 phosphorylation in response to colony-stimulating factor 1/c-fms signaling. Mol. Cell. Biol. 18:5148-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galang, C. K., C. J. Der, and C. A. Hauser. 1994. Oncogenic Ras can induce transcriptional activation through a variety of promoter elements, including tandem c-Ets-2 binding sites. Oncogene 9:2913-2921. [PubMed] [Google Scholar]
- 13.Gradin, K., C. Takasaki, Y. Fujii-Kuriyama, and K. Sogawa. 2002. The transcriptional activation function of the HIF-like factor requires phosphorylation at a conserved threonine. J. Biol. Chem. 277:23508-23514. [DOI] [PubMed] [Google Scholar]
- 14.Graves, B. J., and J. M. Petersen. 1998. Specificity within the ets family of transcription factors, p. 1-55. In G. V. Woude and G. Klein (ed.), Advances in cancer research, vol. 75. Academic Press, San Diego, Calif. [DOI] [PubMed] [Google Scholar]
- 15.Gunther, C. V., and B. J. Graves. 1994. Identification of ETS domain proteins in murine T lymphocytes that interact with the Moloney murine leukemia virus enhancer. Mol. Cell. Biol. 14:7569-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Janknecht, R., N. J. Wells, and T. Hunter. 1998. TGF-β-stimulated cooperation of Smad proteins with the coactivators CBP/p300. Genes Dev. 12:2114-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jayaraman, G., R. Srinivas, C. Duggan, E. Ferreira, S. Swaminathan, K. Somasundaram, J. Williams, C. Hauser, M. Kurkinen, R. Dhar, S. Weitzman, G. Buttice, and B. Thimmapaya. 1999. p300/cAMP-responsive element-binding protein interactions with ets-1 and ets-2 in the transcriptional activation of the human stromelysin promoter. J. Biol. Chem. 274:17342-17352. [DOI] [PubMed] [Google Scholar]
- 19.Kamei, Y., L. Xu, T. Heinzel, J. Torchia, R. Kurokawa, B. Gloss, S. C. Lin, R. A. Heyman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1996. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85:403-414. [DOI] [PubMed] [Google Scholar]
- 20.Karin, M. 1994. Signal transduction from the cell surface to the nucleus through the phosphorylation of transcription factors. Curr. Opin. Cell Biol. 6:415-424. [DOI] [PubMed] [Google Scholar]
- 21.Kasten, M., and A. Giordano. 2001. Cdk10, a Cdc2-related kinase, associates with the Ets2 transcription factor and modulates its transactivation activity. Oncogene 20:1832-1838. [DOI] [PubMed] [Google Scholar]
- 22.Kim, C. A., and J. U. Bowie. 2003. SAM domains: uniform structure, diversity of function. Trends Biochem. Sci. 28:625-628. [DOI] [PubMed] [Google Scholar]
- 23.Kingston, R. E. 1993. Current protocols in molecular biology, vol. 1. Current Protocols, Brooklyn, N.Y.
- 24.Kraus, W. L., and J. T. Kadonaga. 1998. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 12:331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambert, P. F., F. Kashanchi, M. F. Radonovich, R. Shiekhattar, and J. N. Brady. 1998. Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273:33048-33053. [DOI] [PubMed] [Google Scholar]
- 26.Le Gallic, L., D. Sgouras, G. Beal, Jr., and G. Mavrothalassitis. 1999. Transcriptional repressor ERF is a Ras/mitogen-activated protein kinase target that regulates cellular proliferation. Mol. Cell. Biol. 19:4121-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Q. J., S. H. Yang, Y. Maeda, F. M. Sladek, A. D. Sharrocks, and M. Martins-Green. 2003. MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300. EMBO J. 22:281-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, R., H. Pei, D. K. Watson, and T. S. Papas. 2000. EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene 19:745-753. [DOI] [PubMed] [Google Scholar]
- 29.Mackereth, C. D., M. Scharpf, L. N. Gentile, S. E. MacIntosh, C. M. Slupsky, and L. P. McIntosh. 2004. Diversity in structure and function of the Ets family PNT domains. J. Mol. Biol. 342:1249-1264. [DOI] [PubMed] [Google Scholar]
- 30.Major, M. L., R. Lepe, and R. H. Costa. 2004. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol. Cell. Biol. 24:2649-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Man, A. K., L. J. Young, J. A. Tynan, J. Lesperance, M. Egeblad, Z. Werb, C. A. Hauser, W. J. Muller, R. D. Cardiff, and R. G. Oshima. 2003. Ets2-dependent stromal regulation of mouse mammary tumors. Mol. Cell. Biol. 23:8614-8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuda, S., J. C. Harries, M. Viskaduraki, P. J. Troke, K. B. Kindle, C. Ryan, and D. M. Heery. 2004. A conserved α-helical motif mediates the binding of diverse nuclear proteins to the SRC1 interaction domain of CBP. J. Biol. Chem. 279:14055-14064. [DOI] [PubMed] [Google Scholar]
- 33.McCarthy, S. A., D. Chen, B.-S. Yang, H. Cherwinski, C. A. Hauser, X.-R. Chen, M. L. Klagsbrun, M. C. Ostrowski, and M. McMahon. 1997. Rapid phosphorylation of Ets-2 accompanies mitogen-activated protein kinase activation and the induction of heparin-binding epidermal growth factor gene expression by oncogenic Raf-1. Mol. Cell. Biol. 17:2401-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris, L., K. E. Allen, and N. B. La Thangue. 2000. Regulation of E2F transcription by cyclin E-Cdk2 kinase mediated through p300/CBP co-activators. Nat. Cell Biol. 2:232-239. [DOI] [PubMed] [Google Scholar]
- 35.O'Neill, E. M., I. Rebay, R. Tjian, and G. M. Rubin. 1994. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78:137-147. [DOI] [PubMed] [Google Scholar]
- 36.Patton, S. E., M. L. Martin, L. L. Nelsen, X. Fang, G. B. Mills, R. C. Bast, Jr., and M. C. Ostrowski. 1998. Activation of the ras-mitogen-activated protein kinase pathway and phosphorylation of ets-2 at position threonine 72 in human ovarian cancer cell lines. Cancer Res. 58:2253-2259. [PubMed] [Google Scholar]
- 37.Paumelle, R., D. Tulasne, Z. Kherrouche, S. Plaza, C. Leroy, S. Reveneau, B. Vandenbunder, V. Fafeur, and D. Tulashe. 2002. Hepatocyte growth factor/scatter factor activates the ETS1 transcription factor by a RAS-RAF-MEK-ERK signaling pathway. Oncogene 21:2309-2319. [DOI] [PubMed] [Google Scholar]
- 38.Pei, H., J. S. Yordy, Q. Leng, Q. Zhao, D. K. Watson, and R. Li. 2003. EAPII interacts with ETS1 and modulates its transcriptional function. Oncogene 22:2699-2709. [DOI] [PubMed] [Google Scholar]
- 39.Price, E. R., H. F. Ding, T. Badalian, S. Bhattacharya, C. Takemoto, T. P. Yao, T. J. Hemesath, and D. E. Fisher. 1998. Lineage-specific signaling in melanocytes. C-kit stimulation recruits p300/CBP to microphthalmia. J. Biol. Chem. 273:17983-17986. [DOI] [PubMed] [Google Scholar]
- 40.Rabault, B., M. F. Roussel, C. T. Quang, and J. Ghysdael. 1996. Phosphorylation of Ets1 regulates the complementation of a CSF-1 receptor impaired in mitogenesis. Oncogene 13:877-881. [PubMed] [Google Scholar]
- 41.Rachez, C., Z. Suldan, J. Ward, C. P. Chang, D. Burakov, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1998. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 12:1787-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radhakrishnan, I., G. C. Perez-Alvarado, D. Parker, H. J. Dyson, M. R. Montminy, and P. E. Wright. 1997. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell 91:741-752. [DOI] [PubMed] [Google Scholar]
- 43.Rottinger, E., L. Besnardeau, and T. Lepage. 2004. A Raf/MEK/ERK signaling pathway is required for development of the sea urchin embryo micromere lineage through phosphorylation of the transcription factor Ets. Development 131:1075-1087. [DOI] [PubMed] [Google Scholar]
- 44.Schneikert, J., Y. Lutz, and B. Wasylyk. 1992. Two independent activation domains in c-Ets-1 and c-Ets-2 located in non-conserved sequences of the ets gene family. Oncogene 7:249-256. [PubMed] [Google Scholar]
- 45.Seidel, J. J., and B. J. Graves. 2002. An ERK2 docking site in the Pointed domain distinguishes a subset of ETS transcription factors. Genes Dev. 16:127-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheppard, K. A., K. M. Phelps, A. J. Williams, D. Thanos, C. K. Glass, M. G. Rosenfeld, M. E. Gerritsen, and T. Collins. 1998. Nuclear integration of glucocorticoid receptor and nuclear factor-κB signaling by CREB-binding protein and steroid receptor coactivator-1. J. Biol. Chem. 273:29291-29294. [DOI] [PubMed] [Google Scholar]
- 47.Slupsky, C. M., L. N. Gentile, L. W. Donaldson, C. D. Mackereth, J. J. Seidel, B. J. Graves, and L. P. McIntosh. 1998. Structure of the Ets-1 pointed domain and mitogen-activated protein kinase phosphorylation site. Proc. Natl. Acad. Sci. USA 95:12129-12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sternberg, P. W., and M. Han. 1998. Genetics of RAS signaling in C. elegans. Trends Genet. 14:466-472. [DOI] [PubMed] [Google Scholar]
- 49.Stevens, J. L., G. T. Cantin, G. Wang, A. Shevchenko, and A. J. Berk. 2002. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296:755-758. [DOI] [PubMed] [Google Scholar]
- 50.Tootle, T. L., P. S. Lee, and I. Rebay. 2003. CRM1-mediated nuclear export and regulated activity of the receptor tyrosine kinase antagonist YAN require specific interactions with MAE. Development 130:845-857. [DOI] [PubMed] [Google Scholar]
- 51.Tremblay, J. J., and R. S. Viger. 2003. Transcription factor GATA-4 is activated by phosphorylation of serine 261 via the cAMP/protein kinase a signaling pathway in gonadal cells. J. Biol. Chem. 278:22128-22135. [DOI] [PubMed] [Google Scholar]
- 52.Varinou, L., K. Ramsauer, M. Karaghiosoff, T. Kolbe, K. Pfeffer, M. Muller, and T. Decker. 2003. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-γ-dependent innate immunity. Immunity 19:793-802. [DOI] [PubMed] [Google Scholar]
- 53.Vo, N., and R. H. Goodman. 2001. CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 276:13505-13508. [DOI] [PubMed] [Google Scholar]
- 54.Wassarman, D. A., M. Therrien, and G. M. Rubin. 1995. The Ras signaling pathway in Drosophila. Curr. Opin. Genet. Dev. 5:44-50. [DOI] [PubMed] [Google Scholar]
- 55.Wasylyk, C., A. P. Bradford, A. Gutierrez-Hartmann, and B. Wasylyk. 1997. Conserved mechanisms of Ras regulation of evolutionary related transcription factors, Ets1 and Pointed P2. Oncogene 14:899-913. [DOI] [PubMed] [Google Scholar]
- 56.Watabe, T., K. Yoshida, M. Shindoh, M. Kaya, K. Fujikawa, H. Sato, M. Seiki, S. Ishii, and K. Fujinaga. 1998. The Ets-1 and Ets-2 transcription factors activate the promoters for invasion-associated urokinase and collagenase genes in response to epidermal growth factor. Int. J. Cancer 77:128-137. [DOI] [PubMed] [Google Scholar]
- 57.Wei, G., A. E. Schaffner, K. M. Baker, K. C. Mansky, and M. C. Ostrowski. 2003. Ets-2 interacts with co-repressor BS69 to repress target gene expression. Anticancer Res. 23:2173-2178. [PubMed] [Google Scholar]
- 58.Whitmarsh, A. J., and R. J. Davis. 2000. Regulation of transcription factor function by phosphorylation. Cell. Mol. Life Sci. 57:1172-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woods, D., D. Parry, H. Cherwinski, E. Bosch, E. Lees, and M. McMahon. 1997. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol. Cell. Biol. 17:5598-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59a.Xu, D., T. J. Wilson, D. Chan, E. De Luca, J. Zhou, P. J. Hertzog, and I. Kola. 2002. Ets1 is required for p53 transcriptional activity in UV-induced apoptosis in embryonic stem cells. EMBO J. 21:4081-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang, B.-S., C. A. Hauser, G. Henkel, M. S. Colman, C. Van Beveren, K. J. Stacey, D. A. Hume, R. A. Maki, and M. C. Ostrowski. 1996. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol. Cell. Biol. 16:538-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, C., L. H. Shapiro, M. Rivera, A. Kumar, and P. K. Brindle. 1998. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol. Cell. Biol. 18:2218-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang, S. H., P. Shore, N. Willingham, J. H. Lakey, and A. D. Sharrocks. 1999. The mechanism of phosphorylation-inducible activation of the ETS-domain transcription factor Elk-1. EMBO J. 18:5666-5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 64.Yao, T. P., G. Ku, N. Zhou, R. Scully, and D. M. Livingston. 1996. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl. Acad. Sci. USA 93:10626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yochum, G. S., and D. E. Ayer. 2001. Pf1, a novel PHD zinc finger protein that links the TLE corepressor to the mSin3A-histone deacetylase complex. Mol. Cell. Biol. 21:4110-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yordy, J. S., and R. C. Muise-Helmericks. 2000. Signal transduction and the Ets family of transcription factors. Oncogene 19:6503-6513. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, Z., and C. T. Teng. 2003. Phosphorylation of Kruppel-like factor 5 (KLF5/IKLF) at the CBP interaction region enhances its transactivation function. Nucleic Acids Res. 31:2196-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhong, H., R. E. Voll, and S. Ghosh. 1998. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell 1:661-671. [DOI] [PubMed] [Google Scholar]