Abstract
As senescence develops, cells sequentially acquire diverse senescent phenotypes along with simultaneous multistage gene reprogramming. It remains unclear what acts as the key regulator of the collective changes in gene expression at initiation of senescent reprogramming. Here we analyzed time series gene expression profiles obtained in two different senescence models in human diploid fibroblasts: replicative senescence and H2O2-induced senescence. Our results demonstrate that suppression of DNA methyltransferase 1 (DNMT1)-mediated DNA methylation activity was an initial event prior to the display of senescent phenotypes. We identified seven DNMT1-interacting proteins, ubiquitin-like with PHD and ring finger domains 1 (UHRF1), EZH2, CHEK1, SUV39H1, CBX5, PARP1, and HELLS (also known as LSH (lymphoid-specific helicase) 1), as being commonly down-regulated at the same time point as DNMT1 in both senescence models. Knockdown experiments revealed that, among the DNMT1-interacting proteins, only UHRF1 knockdown suppressed DNMT1 transcription. However, UHRF1 overexpression alone did not induce DNMT1 expression, indicating that UHRF1 was essential but not sufficient for DNMT1 transcription. Although UHRF1 knockdown effectively induced senescence, this was significantly attenuated by DNMT1 overexpression, clearly implicating the UHRF1/DNMT1 axis in senescence. Bioinformatics analysis further identified WNT5A as a downstream effector of UHRF1/DNMT1-mediated senescence. Senescence-associated hypomethylation was found at base pairs −1569 to −1363 from the transcription start site of the WNT5A gene in senescent human diploid fibroblasts. As expected, WNT5A overexpression induced senescent phenotypes. Overall, our results indicate that decreased UHRF1 expression is a key initial event in the suppression of DNMT1-mediated DNA methylation and in the consequent induction of senescence via increasing WNT5A expression.
Keywords: cellular senescence, DNA methylation, gene expression, gene regulation, microarray
Introduction
Cell senescence is a cellular fate in which mitotic cells progressively lose their replicative potential, eventually ceasing cell division. It has been proposed that cell senescence is an etiology of aging itself as well as of various aging-associated diseases, including cancer, Alzheimer's disease, and cardiovascular disease (1–3). Senescent cells also exhibit various deterioration phenotypes, including an enlarged and flattened cellular morphology with low elasticity, increased reactive oxygen species (ROS)3 generation, increased senescence-associated (SA) β-gal activity, and senescence-associated secretory phenotypes (1, 4).
In our previous study of replicative senescence (RS) in human diploid fibroblasts (HDFs), we demonstrated that individual phenotypes develop at different stages of senescence rather than appearing simultaneously (5). Moreover, time series gene expression profiles of RS indicate that stage-specific gene expression reprogramming is essential for stage-by-stage development of the senescent phenotype (5). Interestingly, the initial gene expression reprogramming in RS occurs at a fairly early stage, prior to the appearance of the typical senescence marker SA-β-gal. Little attention has focused on the initially reprogrammed genes because of their uncertain link to senescence. However, the initial reprogramming phase includes differential expression of over 1200 genes, with about 800 down-regulated and 500 up-regulated genes (5). This may include the causal event that triggers the presenescent condition, preparatory to senescence, and is likely controlled by master regulators.
In eukaryotic cells, DNA methylation is a key epigenetic modification that regulates gene expression and silencing. It most often occurs at the C5 position of cytosine, typically among the abundant CpG islands of a gene promoter region. CpG island hypermethylation is frequently linked to gene silencing, whereas hypomethylation is associated with gene transcription activation. DNA methylation patterns can vary throughout the life span of an organism, often changing to adapt to environmental conditions (6). Aged tissues show global loss of DNA methylation in all genomic compartments (promoter, intergenic, intronic, and exonic regions) (7, 8). However, hypermethylation on various tumor suppressor genes and Polycomb target genes has also been reported with age (9). This suggests that aberrant DNA hypomethylation may be closely linked with a massive induction of diverse genes that accelerates the aging process. Correspondingly, the reversibility of such hypomethylation may be essential for maintaining the vitality of an organism.
Humans exhibit two classes of DNA methylation activities executed by DNA methyltransferases (DNMTs): de novomethylation and maintenance methylation (10). Executed by DNMT3a and DNMT3b, de novo methylation forms the initial DNA methylation patterns during embryogenesis. On the other hand, maintenance methylation restores and preserves the DNA methylation patterns after each cellular DNA replication cycle. DNMT1 copies the DNA methylation patterns to daughter strands during DNA replication (10–12) and, thus, may be linked with senescence-associated epigenetic modifications.
Although DNMT1 possesses an enzymatic functional group that is critical for maintenance methylation, DNMT1 activity is delicately controlled by physical interactions with diverse proteins and posttranslational modifications (13). Binding of PCNA to DNMT1 increases methylation activity (14), whereas binding of ubiquitin-like with PHD and ring finger domains 1 (UHRF1) to DNMT1 facilitates recognition of hemimethylated DNA (15). DNMT1 is acetylated by KAT5, deacetylated by HDAC1 (16), phosphorylated by casein kinase 1δ/ϵ and AKT1 (17, 18), and methylated by SETD7 (17, 19). The overall integrated actions of these DNMT1-interacting proteins (DIPs) on DNMT1 contribute to maintenance methylation activity and the associated cellular phenotype. However, we do not yet clearly understand how DNMT1 and its interacting proteins are involved in senescence-associated gene reprogramming.
In this study, we analyzed time series gene expression profiles of 53 known DIPs in two different cell senescence model systems: replicative senescence and hydrogen peroxide (H2O2)-induced senescence (HS) of human diploid fibroblasts. We further evaluated how these commonly regulated DIPs were related to DNMT1 expression and to DNMT1-associated senescent processes.
Results
Decreased DNMT1 Expression Leads to Loss of Maintenance DNA Methylation, Inducing Cell Senescence
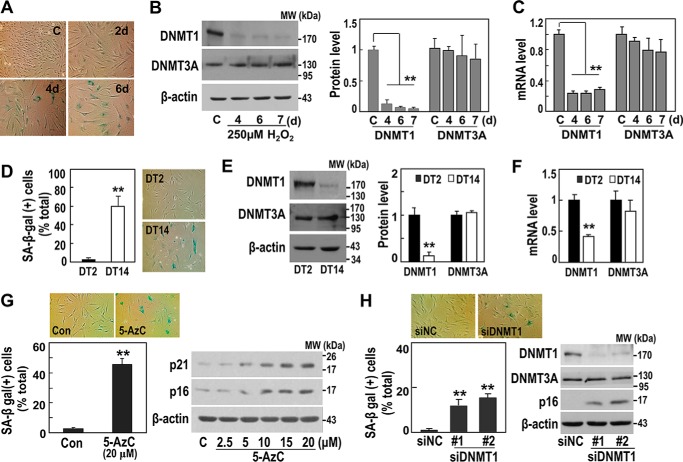
We first induced senescence in HDFs using a subcytotoxic dose (250 μm) of H2O2 (Fig. 1A) and examined the corresponding changes in the expression of DNMT1, a key DNA maintenance methyltransferase. Both DNMT1 protein and mRNA levels were significantly decreased in the HS HDF model. In contrast, there were no detectable changes in the protein or mRNA levels of DNMT3A, a methyltransferase that is critical in de novo DNA methylation (Fig. 1, B and C). Similar results were obtained in the RS HDF model (Fig. 1, D–F), suggesting that cell senescence is specifically linked to the loss of maintenance DNA methylation because of decreased DNMT1 mRNA expression. We then exposed young HDFs (with a doubling time of 2 days, DT2) to 5-aza-2′-deoxycytidine (5-AzC) for 5 days to block cellular DNA methylation activity—mainly maintenance methyltransferase activity. Subsequently, the HDFs exhibited senescent phenotypes that included gain of SA-β-gal and induction of the major cell cycle inhibitory regulators p21 and p16 (Fig. 1G). Moreover, siRNA-mediated knockdown of DNMT1 alone was sufficient to induce HDF senescence (Fig. 1H). These results support the importance of specific loss-of-maintenance DNA methylation by DNMT1 suppression in senescence induction.
FIGURE 1.
Decreased DNMT1-mediated DNA methylation activity is involved in senescence of HDF. A–C, HS of primary HDFs was developed as described under “Experimental Procedures.” A, representative images of the SA-β-gal assay. C, control; d, days. B, Western blotting analyses for DNMT1 and DNMT3A (left) and their quantifications (right). MW, molecular weight. C, messenger RNA levels by qRT-PCR. **, p < 0.01 versus control primary HDFs by Student's t test. D–F, RS of primary HDFs was generated as described under “Experimental Procedures,” and HDFs of DT2 (black columns) and DT14 (white columns) were used. D, quantifications (left) and representative images (right) of the SA-β-gal assay. E, Western blotting analysis (left) and their quantifications (right). F, messenger RNA levels by qRT-PCR. **, p < 0.01 versus DT2 by Student's t test. G, HDFs (DT2) were exposed to the indicated concentrations of 5-AzC (Sigma) for 5 days. Representative images (top panel) and quantifications (bottom left panel) of the SA-β-gal assay are shown. Also shown is a Western blotting analysis (bottom right panel). Con, control. **, p < 0.01 versus Con by Student's t test. H, HDFs (DT2) were transfected with siRNA for DNMT1 for 5 days. Representative images (top panel) and quantification data (bottom left panel) of the SA-β-gal assay are shown. Also shown is a Western blotting analysis (right bottom panel). **, p < 0.01 versus siRNA for negative control (siNC) or control by Student's t test.
Expressions of Seven Common DIPs Decrease at the Same Early Time Point as DNMT1 during Senescence Development
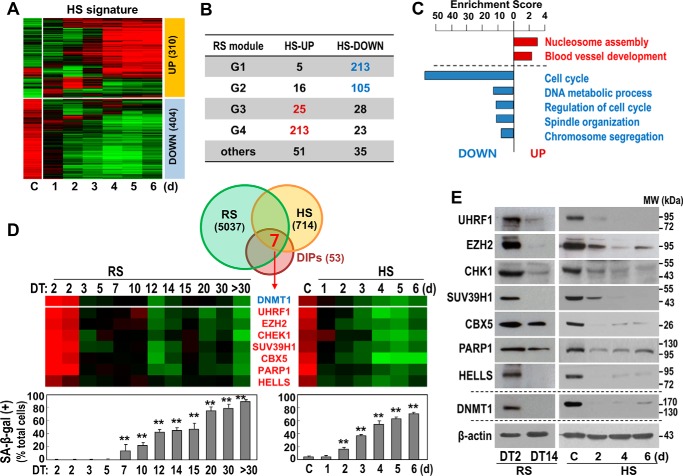
To evaluate when DNMT1 expression began to decrease during senescence development, we established a detailed time series in the HS model and performed a gene expression profiling analysis. Gene expression changes were normalized by subtracting the expression levels from the control experiment, and variable genes with a median absolute deviation of greater than 0.3 were filtered. Genes that were aberrantly expressed with a greater than 2-fold difference at any time point after H2O2 treatment were considered to be differentially expressed with H2O2 treatment (HS signature). This definition yielded 714 HS signature genes, including 310 that were up-regulated (HS_UP) and 404 that were down-regulated (HS_DOWN) (Fig. 2A). We next compared these signature genes with the four previously reported RS signature gene modules (5): G1, genes down-regulated genes after the early stage (earlier than DT3); G2, genes down-regulated after middle stage (DT3-DT7); G3, genes up-regulated at middle and advanced stages (DT10-DT20); and G4, genes up-regulated genes at advanced and very advanced stages (over DT30). Overall, 77.8% of the HS signature genes were matched with RS signature genes. Of the 310 HS_UP genes, most were found in the G3 (25) or G4 (213) RS modules. Of the 403 HS_DOWN genes, most were found in the G1 (213) or G2 (105) RS modules (Fig. 2B). The similarity of these gene expression profiles between the two different cell senescence models supports the appropriateness of these gene profiles for mechanistic investigation of senescence. Gene set enrichment analysis of the HS signature revealed substantial enrichment of cell cycle-related genes among the HS_DOWN genes (Fig. 2C).
FIGURE 2.
Expression of DNMT1-interacting proteins commonly regulated in both RS and HS of HDFs. A, time series HDFs obtained from the HS model were subjected cDNA microarray. A heat map of the time series gene expression profile is shown. C, control; d, days. B, progressively up-regulated (HS_UP, 310 genes) and down-regulated genes (HS_DOWN, 404 genes) were matched with four different modular genes (G1∼G4) identified from the time series RS model in our previous report (5). C, enrichment score indicating the −log10-transformed p values calculated from the gene set enrichment analysis. D, Venn diagram showing the number of the overlapping genes among the gene signatures for DIPs and RS and HS models. Seven genes were identified to be commonly regulated in the progress of the two HDF senescence models. Also shown are heat maps of time series gene expression profiles of the seven DIPs (top panels) and SA-β-gal assay (bottom panels) in the two HDF senescence models. **, p < 0.01 versus DT2 (left graph) or C (control, right graph) by Student's t test. E, the protein expression levels of the seven DIPs were validated by Western blotting analysis. MW, molecular weight.
Interestingly, DNMT1 expression began decreasing at the earliest time point, prior to the development of any clear senescent phenotype such as SA-β-gal (Fig. 2D, top of the heat map and bottom panels). We next analyzed the expression levels of previously reported DIPs (n = 53) (13). Among these DIP genes, seven were commonly found in both RS and HS signature genes: UHRF1, EZH2, CHEK1, SUV39H1, CBX5, PARP1, and HELLS. Unexpectedly, each of these seven DIPs showed progressive down-regulation in a pattern similar to DNMT1 (Fig. 2D). In further experiments, we validated their protein expression levels (Fig. 2E). Collectively, these findings indicate that the decrease in DNMT1-mediated DNA methyltransferase activity observed during senescence development likely results from suppressed expressions of DNMT1 itself and of DIPs that probably play essential or auxiliary roles in promoting optimal DNMT1 activity.
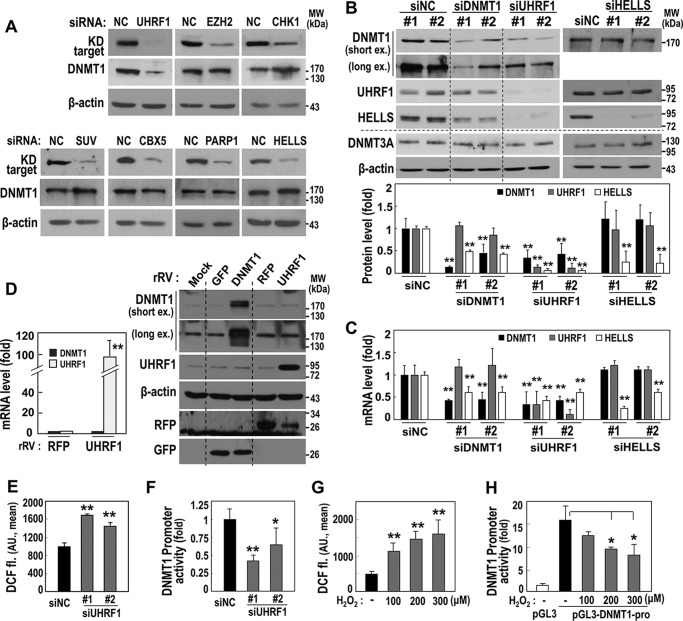
UHRF1 Is an Upstream Regulator of DNMT1 Expression
DIPs reportedly control DNMT1 activity through physical protein interactions (13). Here we further investigated whether any DIP contributed to regulating DNMT1 expression. Using siRNA-mediated knockdown, the seven DIPs were individually suppressed in HDF (DT2). Only UHRF1 knockdown led to decreased DNMT1 protein expression (Fig. 3A). Although UHRF1 suppression significantly down-regulated both mRNA and protein expression of DNMT1, DNMT1 knockdown did not alter UHRF1 expression (Fig. 3, B and C), indicating the role of UHRF1 as an upstream regulator of DNMT1 transcription. To verify the specificity of the effect of UHRF1 on DNMT1 expression, HELLS knockdown was used as a control (Fig. 3, B and C). Interestingly, cDNA microarray analysis after UHRF1 knockdown in young HDFs showed down-regulation of six DIPs (all except CHEK1) (data not shown), suggesting that UHRF1 could potentially be a master regulator of DNMT1 and the DIPs. However, UHRF1 overexpression in HDF (DT5, stage with decreased UHRF1 expression) did not increase DNMT1 mRNA and protein expression (Fig. 3D). These findings indicate that, as an upstream regulator, UHRF1 is essential but not sufficient for DNMT1 expression. To date, there is no clear evidence to support the role of UHRF1 as a direct transcription regulator. However, DNMT1 promoter activity was significantly decreased by siRNA-mediated UHRF1 suppression (Fig. 3E), further supporting regulation of DNMT1 transcriptional activity by decreased UHRF1 expression. Interestingly, siRNA-mediated UHRF1 suppression increased the intracellular ROS level (Fig. 3F), and DNMT1 promoter activity was decreased by the augmented intracellular ROS upon exposure to exogenous H2O2 (Fig. 3, G and H). These results suggest that decreased UHRF1 expression indirectly down-regulates DNMT1 transcription via ROS generation.
FIGURE 3.
UHRF1 is an upstream regulator of DNMT1 expression. A–C, HDFs (DT2) were transfected with siRNAs for the indicated targets for 3 days. A, Western blotting analyses. The bands of knockdown (KD) targets were obtained at the same position as shown in Fig. 2E. NC, negative control; MW, molecular weight. B, Western blotting analyses (top panel) and their quantification (bottom panel). ex, exposure. **, p < 0.01 versus siNC. C, messenger RNA levels by qRT-PCR. **, p < 0.01 versus siNC. D, an HDF (DT7) was infected with a recombinant retrovirus (rRV) harboring the indicated target cDNA for 3 days. Shown are messenger RNA levels by qRT-PCR (left panel) and Western blotting analyses (right panel). **, p < 0.01 versus RFP by Student's t test. AU, arbitrary unit. **, p < 0.01 versus siNC. E, HDFs (DT2) were transfected with siUHRF1 for 5 days. Intracellular ROS levels were monitored by flow cytometric analysis after staining cells with DCF-DA fluorescence dye (DCF fl). *, p < 0.05 versus siNC; **, p < 0.01 versus siNC. F, after an HDFs (DT2) was transfected with siRNA for UHRF1 (siUHRF1) for 24 h, the cells were transfected again with the pGL3-DNMT1-pro plasmid for 2 days and then subjected to a promoter assay. G, HDFs (DT2) were exposed to the indicated dose of H2O2 for 2 days, and intracellular ROS levels were monitored. **, p < 0.01 versus no H2O2 treatment. H, after an HDF (DT2) was transfected with the pGL3-DNMT1-pro or pGL3-basic plasmid (pGL3) for 24 h, the cell was exposed to the indicated dose of H2O2 for 2 days and then subjected to intracellular ROS level analysis using DCF-DA fluorescence dye. *, p < 0.05 versus siNC or no H2O2 treatment by Student's t test.
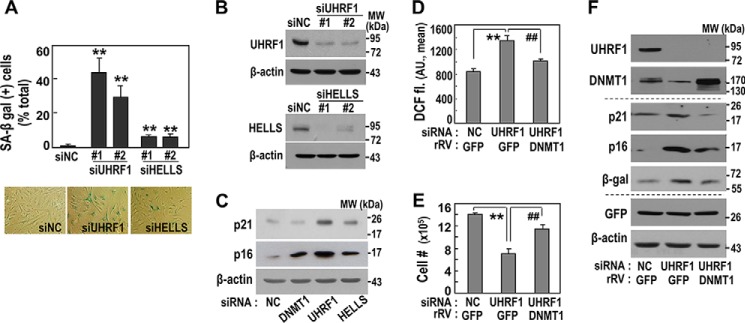
UHRF1 Suppression Effectively Induces Senescence through DNMT1 Down-expression
We next confirmed that UHRF1 knockdown in young HDFs (DT2) induced senescent phenotypes, including SA-β-gal, p21 and p16 induction, and intracellular ROS increase (Fig. 4, A–D). Compared with DNMT1 knockdown (Fig. 1D), UHRF1 knockdown more effectively led to the gain of senescent phenotypes (Fig. 4A). Unexpectedly, HELLS knockdown also induced senescent phenotypes but only slightly, implying that HELLS-mediated regulation of DNMT1 activity may partially contribute to senescence induction. Restoring the cellular DNMT1 level by overexpression effectively attenuated the senescence phenotypes acquired by UHRF1 knockdown, although not completely (Fig. 4, D–F). These findings suggest that UHRF1 is an effective upstream regulator of DNMT1 expression and, consequently, of senescence control.
FIGURE 4.
Knockdown of UHRF1 effectively induces senescence via DNMT1 suppression. A–C, an HDF (DT2) was transfected with siRNAs for the indicated targets for 5 days. A, quantification (top panel) and representative images (bottom panel) of the SA-β-gal assay are shown. B, knockdown levels of the targets were confirmed by Western blotting analysis. **, p < 0.01 versus siNC by Student's t test. MW, molecular weight. C, Western blotting analysis. NC, negative control. D–F, an HDF (DT2) was transfected with siRNAs for the indicated targets and on the next day infected with an rRV harboring the indicated target cDNA for 4 days. DCF-fl, DCF-DA fluorescence dye. D, intracellular ROS levels using DCF-DA fluorescence dye (DCF fl). AU, arbitrary unit. **, p < 0.01 versus siNC/GFP; ##, p < 0.01 versus siUHRF1/GFP. E, cell growth rate by counting cell numbers. **, p < 0.01 versus siNC/GFP; ##, p < 0.01 versus siUHRF1/GFP. F, Western blotting analysis.
WNT5A Is a Downstream Target of the UHRF1/DNMT1 Axis
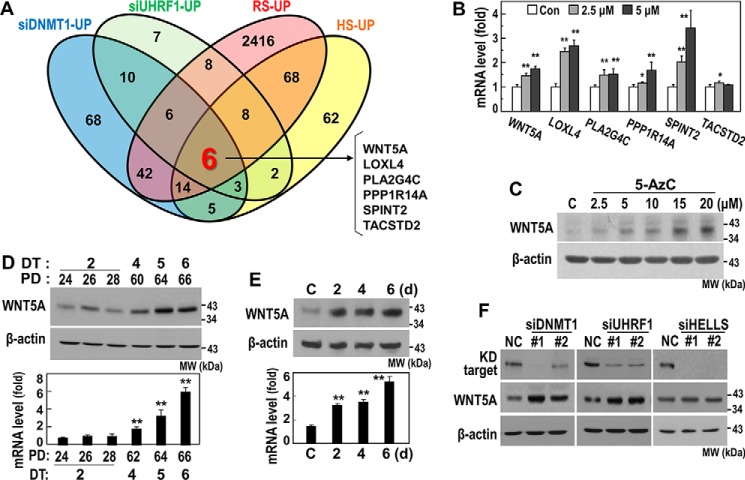
Hypomethylation induced by DNMT1 suppression in a gene promoter may activate transcription of certain effectors to induce senescence. To screen for downstream effectors of the UHRF1/DNMT1 axis, we performed cDNA microarrays after knockdown of UHRF1 or DNMT1, analyzed the commonly up-regulated genes, and finally matched the identified genes with the commonly up-regulated signature genes in the RS and HS models. This process identified the following six genes: WNT5A, LOXL4, PLA2G4C, PPP1R14A, SPINT2, and TACSTD2 (Fig. 5A).
FIGURE 5.
WNT5A is a downstream effector of the UHRF1/DNMT1 axis. A, an HDF (DT2) was transfected with siRNAs for DNMT1 or UHRF1 for 3 days, and total cellular RNAs were subjected to a cDNA microarray and bioinformatics analysis. Up-regulated genes by DNMT1 knockdown and UHRF1 knockdown were matched with the up-regulated genes in the two HDF senescence models (RS and HS). The Venn diagram shows the number of the overlapping genes among DNMT1_UP, UHRF1_UP, RS_UP, and HS_UP. Six commonly up-regulated genes were identified. B and C, an HDF (DT2) was exposed to the indicated concentration of 5-AcZ for 5 days. B, messenger RNA levels were monitored by qRT-PCR. Con, control. *, p < 0.05 versus Con; **, p < 0.01 versus Con. C, Western blotting analysis for WNT5A protein expression. MW, molecular weight. D, Western blotting analysis (top panel) and qRT-PCR (bottom panel) for WNT5A in the progress of RS. **, p < 0.01 versus PD24 by Student's t test. E, Western blotting analysis (top panel) and qRT-PCR (bottom panel) for WNT5A in the progress of HS. **, p < 0.01 versus control (C). d, days. F, an HDF (DT2) was transfected with siRNAs for the indicated targets for 4 days and subjected to Western blotting analysis. The bands of knockdown (KD) targets were obtained at the same position as shown in Fig. 2E.
We next examined whether expression of these six putative targets was truly regulated by DNA methylation. In young HDFs (DT2), we inhibited DNA methylation with 5 days of exposure to 5-AzC. This treatment significantly induced the mRNA levels of five genes (WNT5A, LOXL4, PLA2G4C, PPP1R14A, and SPINT2), whereas TACSTD2 was slightly induced with 2.5 μm 5-AzC (Fig. 5B), implying their transcriptional regulation by DNA methylation. Among the six tested genes, we focused on WNT5A because of its previously reported potential link to senescence (20). As expected, the WNT5A protein level was dose-dependently induced by blocking DNA methylation using 5-AzC (Fig. 5C). We additionally validated the increases in both WNT5A mRNA and protein in the senescent cells using a time series of the RS and HS models (Fig. 5, D and E). Knockdown of either DNMT1 or UHRF1 clearly induced WNT5A protein expression, whereas HELLS knockdown showed a minor effect (Fig. 5F). Overall, these results indicate that WNT5A is a downstream target of the UHRF1/DNMT1 axis.
Hypomethylation from −1569 bp to −1363 bp of the WNT5A Promoter Is Potentially Linked to Senescence-associated WNT5A Induction
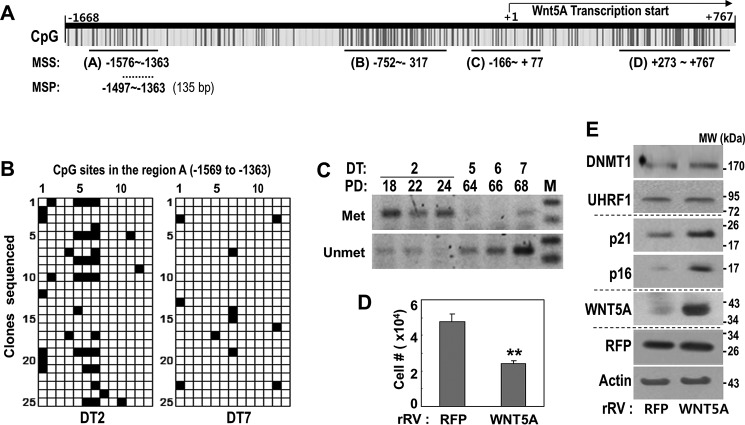
Finally, to examine the DNA methylation status of the WNT5A promoter, we performed methylation-specific sequencing of four major CpG-rich regions, A to D. These regions were estimated by using the MethPrimer program to analyze the WNT5A promoter sequence from −1668 bp to +767 bp from the WNT5A transcription start (NC_000003.12) (Fig. 6A). Unexpectedly, young HDFs (DT2) showed no significant cytosine methylation within the indicated CpG-rich regions B, C, and D (data not shown). Only region A, which is relatively distal from the transcription start, showed abundant cytosine methylation in young HDFs as well as decreased methylation in senescent HDFs (DT7) (Fig. 6B). DNA methylation hot spots included three CpG dinucleotides located at −1490, −1483, and −1476 bp (marked as CpG sites 5, 6, and 7 in Fig. 6B) from the WNT5A transcription start (Fig. 6B). To further monitor the time series methylation status on this hot spot region of the WNT5A promoter, we performed methylation-specific PCR using both methylation-specific primers and non-methylation-specific primers. During the RS process, the methylation status gradually decreased, whereas non-methylation increased (Fig. 6C). As expected, WNT5A overexpression in young HDFs (DT2) induced senescence phenotypes without altered DNMT1 or UHRF1 expression, as evidenced by delayed cell growth and p21 and p16 induction (Fig. 6, D and E). Altogether, our results suggest that hypomethylation of CpG islands between −1569 and −1363 bp upstream of the WNT5A promoter increases WNT5A expression, eventually inducing senescence phenotypes.
FIGURE 6.
Hypomethylation in a specific region (from −1569 to −1363) of the WNT5A promoter in the process of senescence. A, schematic for potential CpG islands of WNT5A promoter regions and the selected regions used for the MSS and MSP. B, MSS of region A was performed with HDFs (DT2 and DT14) as described under “Experimental Procedures.” Fourteen CpG dinucleotides exist within the region, and methylated (Met) CpG dinucleotides are shown as black and non-methylated (Unmet) ones as white. C, MSP was performed using the primer set for the confirmed CpG methylation hot spots (−1490, −1483, and −1476 bp from the WNT5A transcription start) within the WNT5A promoter in the progress of replicative HDF senescence. M, DNA size marker. D and E, an HDF (DT2) was infected with the rRV encoding WNT5A cDNA for 6 days. D, cell growth rate by cell number counting. **, p < 0.01 versus RFP by Student's t test. E, Western blotting analysis.
Discussion
Compared with other degenerative cellular fates, including apoptosis and necrosis, the development of senescence shows relatively slower and more progressive acquisition. Moreover, senescence displays diverse cellular phenotypes, such as irreversible growth arrest (loss of cell division capacity), enlarged and flattened cellular morphology (increased cell mass and size), increased cell granularity, high ROS generation, gain of SA-β-gal, and senescence-associated secretory phenotype induction and release, that do not develop simultaneously but, rather, manifest sequentially (5). Although SA-β-gal is considered a key senescence marker, it develops at a later stage. Other senescence phenotypes appear earlier and are progressively amplified. However, these earlier phenotypes and the initial gene reprogramming have been scarcely investigated because of their ambiguous relationship with impending cellular senescence. We recently published bioinformatics data, including a detailed time series of the gene expression profiling of replicative senescence in HDF, which provides useful groundwork for further investigating this subject (5). In this study, we further developed a time series for a stress-induced (H2O2-triggered) senescence model with the aim of identifying a common master regulator for initial gene reprogramming. We found that DNMT1, which can simultaneously regulate diverse gene expressions, was down-regulated as part of the earliest gene alterations, suggesting its potential role as a master regulator for the initial reprogramming and consequent senescent phenotypes. Although prior studies have reported global hypomethylation and DNMT1 involvement in senescence (7, 8, 21, 22), this is the first investigation to pinpoint DNMT1 suppression as a primary event in senescence.
DNMT1 is essential for maintenance of DNA methylation activity, in which the gene expression program of the mother cell is imprinted to the daughter cell following cell division—a critical epigenetic modification. Thus, it is clear that loss of DNMT1 expression during senescence development could effectively reprogram gene expression patterns. Loss of DNMT1 expression occurs gradually, resulting in slow accumulation of hypomethylated regions on cellular DNA and generating progressive gene reprogramming. Hypomethylation of a gene promoter region allows gene transcription, suggesting that the hypomethylation resulting from DNMT1 loss is likely linked to the induction of certain genes that may control senescence. This raises the question of how stage-specific genes can be induced by random hypomethylation. Although DNMT1 possesses the key enzymatic activity for maintenance methylation, its DNA binding and methylation activities are exquisitely regulated by its protein-protein interaction with diverse proteins. The actions of these specified DIPs and their additional interacting regulators may help determine the sensitivity to hypomethylation and target gene transcription.
Recently, Qin et al. (13) thoroughly reviewed the actions of 53 DIPs on DNMT1 activity. Among these 53 DIPs, we identified seven (UHRF1, EZH2, CHEK1, SUV39H1, CBX5, PARP1, and HELLS) that decreased in expression at the same time point as DNMT1 in both the RS and HS models. Interestingly, we found no DIPs with increasing expression. EZH2 reportedly recruits DNMT1 to target genes, thereby mediating promoter methylation (23). However, EZH2 also antagonizes a subset of p53-regulated genes associated with cell senescence (24), implying its irrelevance to direct senescence induction. Chk1 recruits DNMT1 to DNA damage sites (25) and directs both senescence and mitotic catastrophe in HCT116 human colorectal cancer cells (26). SUV39H1 specifically interacts with both Dnmt1 and Dnmt3a in vitro (27), and SUV39H1 down-regulation is involved in senescence induced by ionizing radiation (28). Heterochromatin protein 1α (HP1α) is encoded from the CBX5 gene and specifically localizes to pericentric heterochromatin sites, whereas HP1β and HP1γ localize to both heterochromatic and euchromatic sites (29) and interact with DNA methyltransferase (27). PARP1 inhibits DNA methylation through the non-covalent interaction of its ADP-ribose polymers with Dnmt1 (30), and Parp1−/− mice exhibit moderately faster aging compared with wild-type control mice (31). HELLS (also named LSH1) forms a complex with DNMT1, DNMT3b, and HDACs, suggesting a role in transcriptional repression (32). Additionally, LSH (lymphoid-specific helicase) delays senescence through p16 repression (33). UHRF1 acts as an essential regulator of DNA methylation by binding to specific DNA sequences and recruiting DNMT1 (34). In zebrafish liver tissue, UHRF1 overexpression drives DNA hypomethylation by causing Dnmt1 mislocalization or destabilization, triggering a TP53-mediated senescence program (35). These seven DIPs reportedly have diverse and sometimes controversial effects on DNMT1 activity and senescence. Overall, the collective loss of these seven DIPs, together with DNMT1, may contribute to effectively attenuating maintenance DNA methylation activity, supporting the importance of the loss of maintenance methylation activity in the initial induction of senescence.
Another unanswered question is which factor acts as the key upstream regulator suppressing DNMT1 transcription at the initial stage of senescence. We investigated whether any of the presently examined seven DIPs were involved in senescence-associated DNMT1 suppression, although the DIPs are known to directly bind and modulate the methyltransferase of DNMT1. Unexpectedly, our results identified UHRF1 as an upstream regulator of DNMT1 transcription. UHRF1 belongs to a subfamily of RING finger-type E3 ubiquitin ligases and possesses ubiquitin ligase activity in addition to its activity in the direct recruitment of DNMT1 to specific DNA sequences. Although no clear evidence supports its direct role as a transcription regulator, UHRF1 could potentially ubiquitinate a transcription factor involved in DNMT1 expression, leading to the proteasomal degradation of the factor. Supporting this hypothesis, Ma et al. (36) recently reported that UHRF1 promotes non-degradative ubiquitination of p53 and suppresses p53-mediated transactivation. Another report demonstrated that p53 regulated DNMT1 expression by forming a complex with Sp1 on the DNMT1 promoter (37), implying that UHRF1 may control DNMT1 transcription via modulating p53/Sp1 activity. Thus, the loss of UHRF1 may have powerful dual effects on DNMT1 activity, both inactivating DNMT1 transcription and promoting inefficient DNMT1 recruitment on its specific DNA sequence. Notably, UHRF1 suppression showed a greater influence on senescence induction compared with DNMT1 suppression alone, implying that UHRF1 suppression has unknown additional action in triggering senescence beside its DNMT1 control and supporting its powerful action in senescence control. A detailed study of the involvement of UHRF1 in transcriptional regulation of DNMT1 is currently underway.
DNMT1 suppression leads to DNA hypomethylation that can activate the transcription of multiple genes. Although some of these genes may be involved in senescence induction, others may be active in protection against senescence. The additional loss of UHRF1 may help DNMT1 select senescence-inducing genes. Thus, it is critical to identify target genes of the UHRF1/DNMT1 axis that are essential in senescence induction. Our present efforts led to the identification of WNT5A as a true target of the axis and as a senescence trigger. UHRF1 was recently identified as a novel oncogene in primary liver cancer (35), suggesting its potential role as a switch molecule between senescence and cancer. Overall, we conclude that UHRF1 loss is a key initial event in the inactivation of DNMT1 transcription and activity, thereby triggering senescence-associated gene reprogramming and subsequent senescent phenotypes.
Experimental Procedures
Cell Culture and Development of Senescence
Primary HDFs isolated from the foreskin of a 5-year-old boy by a method described previously (5) were cultured in DMEM supplemented with 10% FBS (Invitrogen) and antibiotics (Invitrogen) at 37 °C in a humidified incubator with 5% CO2. To develop RS of HDFs, confluent cells were continuously subcultured by being transferred evenly into two new dishes, and the numbers of population doublings (PDs) as well as the DT were counted as described previously (5). To generate HS of HDFs, primary HDF cells (DT2) were treated with 150 μm H2O2 twice with a 12-h interval to induce stably senescence and further incubated for the indicated time periods.
Senescence-associated β-Galactosidase Activity Staining
SA-β-gal activity was assayed at pH 6.0 as described previously (38). The stain was visible for 12 h after incubation at 37 °C. By counting the numbers of the blue-colored and total cells under using ImageJ software (National Institutes of Health), the percentage of the cells stained blue was estimated to represent the population of senescent cells.
Introduction of siRNAs and Plasmids into Cells
HDFs were treated with the siRNA duplexes and plasmids using OligofectamineTM reagent (Invitrogen) and FuGENE HD (Roche Diagnostics), respectively, according to the instructions of the manufacturer. All siRNAs for targets, UHRF1 (#1, 5′-GCUGACCAUGCAGUAUCCATT; #2, 5′-ACUGCUUAGCGUCUGAGAUTT), DNMT1 (#1, 5′-CGAGUCUGGUUUGAGAGUTT; #2, 5′-GGAAUGGCAGAUGCCAACAGCTT), HELLS (#1, 5′-GUUGUUUAUCGCCUUGUUATT; #2, 5′-CAGCAAAUACUAUCGAUCATT), CBX5 (5′-AGGAAUGAACAUGAGACUUAATT), EZH2 (5′-GGAUAGAGAAUGUGGGUUUAUTT), SUV39H1 (5′-ACCACAGAAACUUUGUACUAGTT), PARP1 (5′-GGCAGAGGUGAAGGCAGAGCCTT), CHK1 (5′-GGUCUUUCCUUAUGGGAUACTT) and negative control (NC, 5′-CCUACGCCACCAAUUUCGUTT), were generated by Bioneer (Daejeon, Korea).
Construction of the Retroviral Vector and Production of Retrovirus
To generate a retroviral vector containing the DNMT1 gene, a cDNA fragment for DNMT1 was obtained from the pcDNA3/Myc-DNMT1 vector, which was provided by Prof. Tae-Young Roh (Postech), using EcoRI and NotI restriction enzymes and subcloned into the pMXs-IRES-GFP retroviral vector (Cell Biolabs, Inc., San Diego, CA). The cDNAs for UHRF1 and WNT5A were amplified by PCR using total cDNAs isolated from Chang cells and pcDNA-WNT5A (Addgene, Cambridge, MA), respectively. The primer sets (Bioneer) used for the cDNA amplification were as follows: UHRF1, 5′-CAGTTAACATGGGGGTTTTTGCTGTCCC and 5′-GACTCGAGTCACCGGCCATTGCCGTA; WNT5A, 5′-CAGTTAACATGAAGAAGTCCATTGG and 5′-GACTCGAGTCACTTGCACACAA. The amplified cDNA fragments were inserted into HpaI and XhoI sites of the pCMMP-MCS-IRES-RFP vector (Addgene).
To generate recombinant retrovirus, H29D cells, a modified 293T cell line, was cultured in DMEM (Invitrogen) as described previously (39) and transfected with retroviral plasmids containing target cDNAs using Lipofectamine reagent (Invitrogen). The virus-containing medium (supernatant) was harvested daily for 4 days. The harvested virus supernatant was filtered through a 0.45-μm filter unit (UFC 920008, EMD Millipore Corp., Darmstadt, Germany) and stored at 80 °C.
Construction of the Promoter-Luciferase Reporter Plasmid and Promoter Assay
The human DNMT1 promoter region of 657 bp (−344 to +313, NC_000019.10) was cloned by targeted PCR against total genomic DNA of Chang cells, an immortalized normal hepatocyte cell line (ATCC) using the primer set 5′-TTACGCGTCCCACACACTGGGTATAG and 5′-TGCAGATCTCTGCGGACATCGTCG. The amplified DNMT1 promoter region was inserted between the MluI and BglII sites of the pGL3-basic vector (Promega, Fitchburg, WI), generating the pGL3-DNMT1 promoter reporter plasmid (pGL3-DNMT1-pro). After construction, the inserted promoter was confirmed by DNA sequencing.
To monitor DNMT1 promoter activity, cells were transfected with a total of 1 μg of DNA (950 ng of the cloned reporter plasmid and 50 ng of thymidine kinase promoter-driven Renilla luciferase plasmid as an internal control) using FuGENE HD reagent. After 2 days, the luciferase activity of cell extracts was measured by Synergy 2 multimode reader (BioTek Instruments, Inc., Winooski, VT) according to the protocol provided with the Dual-Luciferase reporter assay system (Promega). The luciferase activities were normalized by the Renilla luciferase activity.
Gene Expression Profiling and Data Analysis
Total RNA was amplified and purified to yield biotinylated cRNA as described previously (5). Briefly, total RNA isolated from HDFs for microarray hybridization was amplified and purified using the Ambion Illumina RNA amplification kit (Ambion, Austin, TX). Labeling and hybridization to the human HT-12 v4 expression bead chip array was performed according to the instructions of the manufacturer (Illumina Inc., San Diego, CA). Raw data were obtained using GenomeStudio software (Illumina Inc.), filtered by detection p value (p < 0.05), and further processed by log2 transformation and quantile normalization. The gene set analysis of biological function was performed using David software (https://david.ncifcrf.gov/). All data preprocessing works and subsequent data analysis were performed using R/Bioconductor packages.
Estimation of Intracellular ROS Level
To estimate intracellular ROS level, cells were stained with 10 μm 2′,7′-dichlorodihydrofluorescein diacetate (Molecular Probes, Eugene, OR) for 15 min at 37 °C. Stained cells were washed, resuspended in PBS, and analyzed by flow cytometry (FACS Vantage, BD Biosciences). Mean values of arbitrary fluorescence units of 10,000 cells were expressed as a percentage of the negative control.
Methylation-specific Sequencing and Methylation-specific PCR
Potential CpG islands within the WNT5A promoter region (from −1668 bp to +767 bp from WNT5A transcription start, NC_000003.12) were estimated using the MetPrimer program. To determine the methylation pattern, we converted cytosine residues of isolated genomic DNA (500 ng) to uracil with bisulfite, leaving 5-meyhylcytosine residues unaffected, according to the instruction of the EZ DNA methylation kit (Zymo Research, Orange, CA). For methylation-specific sequencing (MSS), bisulfite-modified DNA was amplified against four different CpG-rich regions (A to D) by PCR using the following primer sets, which were designed to anneal with the sequences containing thymine residues converted from non-CpG cytosine residues: region A (from −1576 bp to −1363 bp from the transcription start), 5′-GTGTTGGAGGTAG and 5′-TACATAATTACTCATCTAACTC; region B (from −752 bp to −317 bp), 5′-AGAGAGTAAGGTAGTTG and 5′-AATTAATCTCTCTTTTCCC; region C (from −166 bp to +77 bp), 5′-TAATTTGGGGTTGATTTTTGTAGTT and 5′-ATCTCCAACTCCTCCTCTCTAAATC); region D (from +273 bp to +767 bp), 5′-GGGTTGGAAAGTTTTAATTAT and 5′-ACTAAACACCTACCTTCATA. PCR products were cloned into the pGEM T-easy vector (Promega). Multiple clones (>15 clones) from each PCR product were subjected to DNA sequencing at Cosmogenetech Inc. (Seoul, Korea).
For methylation-specific PCR (MSP), the CpG-rich region (from −1496 to −1363 bp, 135 bp in full length) containing methylation hot spots (−1490, −1483, and −1476 bp from the WNT5A transcription start), which was identified by the above MSS, was amplified against the bisulfite-modified DNA using the following primers (Bioneer) for the non-CpG region: forward primer for methylated hot spots, 5′-AATTTCGGTGTTCGGAATTC; forward primer for unmethylated hot spots, 5′-GAAATTTTGGTGTTTGGAATTTG); common reverse primer, 5′-TACATAATTACTCATCTAACTC. PCR conditions were as follows: 95 °C for 5 min, followed by 40 cycles of 95 °C for 1 min, 53.4 °C (for methylated hot spots) or 54.8 °C (for unmethylated hot spots) for 1 min and 72 °C for 1 min and a final extension at 72 °C for 10 min.
Western Blotting Analysis
Western blotting analysis was performed using standard procedures. Antibodies for DNMT1 (sc-20701), DNMT3A (sc-20703), HELLS (sc-46665), CHK1 (sc-377231), p16 (sc-759), p21 (sc-397), GFP (sc-9996), and β-actin (sc-1616) were obtained from Santa Cruz Biotechnology (Dallas, TX). Antibodies against Suv39H1 (8729) and EZH2 (5246) were purchased from Cell Signaling Technology (Beverly, MA). Antibodies for PARP1 (ab32071) and RFP (ab62341) were purchased from Abcam (Cambridge, UK). Antibodies for UHRF1 (3A11) and CBX5 (05-689) were obtained from Calbiochem and Millipore, respectively. Antibodies for WNT 5A (MAB645) and human β-galactosidase (AF6464) were from R&D Systems (Minneapolis, MN).
Quantitative RT-PCR
Total RNA was isolated using TRIzol (Invitrogen), and cDNA was prepared using avian myeloblastosis virus (AMV) reverse transcriptase (Promega). PCR was performed using Thunderbird® SYBR® qPCR Mix (Toyobo Co. Ltd., Osaka, Japan) according to the protocol of the manufacturer. The primer sets were produced by Bioneer as follows: DNMT1, 5′-GAGCTACCACGCAGACATCA and 5′-CGAGGAAGTAGAAGCGGTTG; DNMT3A, 5′-TATTGATGAGCGCACAAGAGAGC and 5′-GGGTGTTCCAGGGTAACATTGAG; UHRF1, 5′-TGTGGACCATGGGAATTTTT and 5′-GCTATTCTTGCCACCCTTGA; HELLS, 5′-ACTGGAGGAGTGATGCGATG and 5′-GCCACAGACAAGAAAAGGTC; WNT5A, 5′-CAAGGGCTCCTACGAGAGTG and 5′-CTTCTCCTTCAGGGCATCAC; LOXL4, 5′-AACTGCCTCTCCAAGTCTGC and 5′-TGCTGTGGTAATGCCTGTGG; PLA2G4C, 5′-GACCCCGAAAGGAAAGGCTG and 5′-CGAGTGGGAAGGGAGTGTTG; PPP1R14A, 5′-GCTGGACGTGGAGAAGTGGA and 5′-CCTGGATGAAGTCCTCGACAG; SPINT2, 5′-TGTGTATGGGGGCTGTGACG and 5′-CTTCTGGGAGCACTTGGGACA; TACSTD2, 5′-GTCACGCTTCCTGATTCCTC and 5′-GGACCGAAAGGGGATACATT; β-actin, 5′-CCTTCCTGGGCATGGAGTCCTGT and 5′-GGAGCAATGATCTTGATCTTC.
Author Contributions
H. J., H. B., S. M., U. J., Y. L., and Y. S. performed the experiments. B. J. performed the bioinformatics analysis. H. W. supervised the bioinformatics analysis and prepared the manuscript. G. Y. designed and supervised the project and prepared the manuscript.
This work was supported by National Research Foundation of Korea Grants 2012R1A5A2048183 and 2015R1A2A1A10055038. The authors declare that they have no conflicts of interest with the contents of this article.
The raw data from the microarray experiments are available in the GEO database (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE80322.
- ROS
- reactive oxygen species
- RS
- replicative senescence
- HDF
- human diploid fibroblast
- DNMT
- DNA methyltransferase
- DIP
- DNMT1-interacting protein
- HS
- H2O2-induced senescence
- DT
- doubling time
- 5-AzC
- 5-aza-2′-deoxycytidine
- PD
- population doubling
- MSS
- methylation-specific sequencing
- MSP
- methylation-specific PCR
- qRT-PCR
- quantitative RT-PCR
- rRV
- recombinant retrovirus
- RFP
- red fluorescent protein
- DCF-DA
- 2′,7′–dichlorofluorescin diacetate.
References
- 1. Campisi J., Andersen J. K., Kapahi P., and Melov S. (2011) Cellular senescence: a link between cancer and age-related degenerative disease? Semin. Cancer Biol. 21, 354–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Collado M., Blasco M. A., and Serrano M. (2007) Cellular senescence in cancer and aging. Cell 130, 223–233 [DOI] [PubMed] [Google Scholar]
- 3. Rodier F., and Campisi J. (2011) Four faces of cellular senescence. J. Cell Biol. 192, 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hwang E. S., Yoon G., and Kang H. T. (2009) A comparative analysis of the cell biology of senescence and aging. Cell. Mol. Life Sci. 66, 2503–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim Y. M., Byun H. O., Jee B. A., Cho H., Seo Y. H., Kim Y. S., Park M. H., Chung H. Y., Woo H. G., and Yoon G. (2013) Implications of time-series gene expression profiles of replicative senescence. Aging Cell 12, 622–634 [DOI] [PubMed] [Google Scholar]
- 6. Jaenisch R., and Bird A. (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254 [DOI] [PubMed] [Google Scholar]
- 7. Wilson V. L., and Jones P. A. (1983) DNA methylation decreases in aging but not in immortal cells. Science 220, 1055–1057 [DOI] [PubMed] [Google Scholar]
- 8. Gonzalo S. (2010) Epigenetic alterations in aging. J. Appl. Physiol. 109, 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maegawa S., Hinkal G., Kim H. S., Shen L., Zhang L., Zhang J., Zhang N., Liang S., Donehower L. A., and Issa J. P. (2010) Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 20, 332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Law J. A., and Jacobsen S. E. (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goll M. G., and Bestor T. H. (2005) Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 [DOI] [PubMed] [Google Scholar]
- 12. Yoder J. A., Soman N. S., Verdine G. L., and Bestor T. H. (1997) DNA (cytosine-5)-methyltransferases in mouse cells and tissues: studies with a mechanism-based probe. J. Mol. Biol. 270, 385–395 [DOI] [PubMed] [Google Scholar]
- 13. Qin W., Leonhardt H., and Pichler G. (2011) Regulation of DNA methyltransferase 1 by interactions and modifications. Nucleus 2, 392–402 [DOI] [PubMed] [Google Scholar]
- 14. Schermelleh L., Haemmer A., Spada F., Rösing N., Meilinger D., Rothbauer U., Cardoso M. C., and Leonhardt H. (2007) Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 35, 4301–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arita K., Ariyoshi M., Tochio H., Nakamura Y., and Shirakawa M. (2008) Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455, 818–821 [DOI] [PubMed] [Google Scholar]
- 16. Du Z., Song J., Wang Y., Zhao Y., Guda K., Yang S., Kao H.-Y., Xu Y., Willis J., Markowitz S. D., Sedwick D., Ewing R. M., and Wang Z. (2010) DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci. Signal. 3, ra80-ra80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Estève P.-O., Chang Y., Samaranayake M., Upadhyay A. K., Horton J. R., Feehery G. R., Cheng X., and Pradhan S. (2011) A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat. Struct. Mol. Biol. 18, 42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sugiyama Y., Hatano N., Sueyoshi N., Suetake I., Tajima S., Kinoshita E., Kinoshita-Kikuta E., Koike T., and Kameshita I. (2010) The DNA-binding activity of mouse DNA methyltransferase 1 is regulated by phosphorylation with casein kinase 1δ/ϵ. Biochem. J. 427, 489–497 [DOI] [PubMed] [Google Scholar]
- 19. Estève P. O., Chin H. G., Benner J., Feehery G. R., Samaranayake M., Horwitz G. A., Jacobsen S. E., and Pradhan S. (2009) Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 106, 5076–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bitler B. G., Nicodemus J. P., Li H., Cai Q., Wu H., Hua X., Li T., Birrer M. J., Godwin A. K., Cairns P., and Zhang R. (2011) Wnt5a suppresses epithelial ovarian cancer by promoting cellular senescence. Cancer Res. 71, 6184–6194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. So A. Y., Jung J. W., Lee S., Kim H. S., and Kang K. S. (2011) DNA methyltransferase controls stem cell aging by regulating BMI1 and EZH2 through microRNAs. PLoS ONE 6, e19503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng Q. H., Ma L. W., Zhu W. G., Zhang Z. Y., and Tong T. J. (2006) p21Waf1/Cip1 plays a critical role in modulating senescence through changes of DNA methylation. J. Cell. Biochem. 98, 1230–1248 [DOI] [PubMed] [Google Scholar]
- 23. Viré E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J. M., Bollen M., Esteller M., Di Croce L., de Launoit Y., and Fuks F. (2006) The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439, 871–874 [DOI] [PubMed] [Google Scholar]
- 24. Iannetti A., Ledoux A. C., Tudhope S. J., Sellier H., Zhao B., Mowla S., Moore A., Hummerich H., Gewurz B. E., Cockell S. J., Jat P. S., Willmore E., and Perkins N. D. (2014) Regulation of p53 and Rb links the alternative NF-κB pathway to EZH2 expression and cell senescence. PLoS Genet. 10, e1004642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Palii S. S., Van Emburgh B. O., Sankpal U. T., Brown K. D., and Robertson K. D. (2008) DNA methylation inhibitor 5-aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol. Cell. Biol. 28, 752–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poehlmann A., Habold C., Walluscheck D., Reissig K., Bajbouj K., Ullrich O., Hartig R., Gali-Muhtasib H., Diestel A., Roessner A., and Schneider-Stock R. (2011) Cutting edge: Chk1 directs senescence and mitotic catastrophe in recovery from G2 checkpoint arrest. J. Cell. Mol. Med. 15, 1528–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fuks F., Hurd P. J., Deplus R., and Kouzarides T. (2003) The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 31, 2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sidler C., Li D., Wang B., Kovalchuk I., and Kovalchuk O. (2014) SUV39H1 down-regulation induces deheterochromatinization of satellite regions and senescence after exposure to ionizing radiation. Front. Genet. 5, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maison C., and Almouzni G. (2004) HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5, 296–304 [DOI] [PubMed] [Google Scholar]
- 30. Zampieri M., Guastafierro T., Calabrese R., Ciccarone F., Bacalini M. G., Reale A., Perilli M., Passananti C., and Caiafa P. (2012) ADP-ribose polymers localized on Ctcf-Parp1-Dnmt1 complex prevent methylation of Ctcf target sites. Biochem. J. 441, 645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Piskunova T. S., Yurova M. N., Ovsyannikov A. I., Semenchenko A. V., Zabezhinski M. A., Popovich I. G., Wang Z. Q., and Anisimov V. N. (2008) Deficiency in poly(ADP-ribose) polymerase-1 (PARP-1) accelerates aging and spontaneous carcinogenesis in mice. Curr. Gerontol. Geriatr. Res. 754190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Myant K., and Stancheva I. (2008) LSH cooperates with DNA methyltransferases to repress transcription. Mol. Cell. Biol. 28, 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou R., Han L., Li G., and Tong T. (2009) Senescence delay and repression of p16INK4a by Lsh via recruitment of histone deacetylases in human diploid fibroblasts. Nucleic Acids Res. 37, 5183–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T. A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., Tajima S., Mitsuya K., Okano M., and Koseki H. (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912 [DOI] [PubMed] [Google Scholar]
- 35. Mudbhary R., Hoshida Y., Chernyavskaya Y., Jacob V., Villanueva A., Fiel M. I., Chen X., Kojima K., Thung S., Bronson R. T., Lachenmayer A., Revill K., Alsinet C., Sachidanandam R., Desai A., et al. (2014) UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell 25, 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ma J., Peng J., Mo R., Ma S., Wang J., Zang L., Li W., and Fan J. (2015) Ubiquitin E3 ligase UHRF1 regulates p53 ubiquitination and p53-dependent cell apoptosis in clear cell Renal Cell Carcinoma. Biochem. Biophys. Res. Commun. 464, 147–153 [DOI] [PubMed] [Google Scholar]
- 37. Lin R. K., Wu C. Y., Chang J. W., Juan L. J., Hsu H. S., Chen C. Y., Lu Y. Y., Tang Y. A., Yang Y. C., Yang P. C., and Wang Y. C. (2010) Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 70, 5807–5817 [DOI] [PubMed] [Google Scholar]
- 38. Seo Y. H., Jung H. J., Shin H. T., Kim Y. M., Yim H., Chung H. Y., Lim I. K., and Yoon G. (2008) Enhanced glycogenesis is involved in cellular senescence via GSK3/GS modulation. Aging Cell 7, 894–907 [DOI] [PubMed] [Google Scholar]
- 39. Kim J. H., Kim H. Y., Lee Y. K., Yoon Y. S., Xu W. G., Yoon J. K., Choi S. E., Ko Y. G., Kim M. J., Lee S. J., Wang H. J., and Yoon G. (2011) Involvement of mitophagy in oncogenic K-Ras-induced transformation: overcoming a cellular energy deficit from glucose deficiency. Autophagy 7, 1187–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]