Abstract
Cell division in most bacteria is mediated by the tubulin-like FtsZ protein, which polymerizes in a GTP-dependent manner to form the cytokinetic Z ring. A diverse repertoire of FtsZ-binding proteins affects FtsZ localization and polymerization to ensure correct Z ring formation. Many of these proteins bind the C-terminal domain (CTD) of FtsZ, which serves as a hub for FtsZ regulation. FtsZ ring-associated proteins, ZapA–D (Zaps), are important FtsZ regulatory proteins that stabilize FtsZ assembly and enhance Z ring formation by increasing lateral assembly of FtsZ protofilaments, which then form the Z ring. There are no structures of a Zap protein bound to FtsZ; therefore, how these proteins affect FtsZ polymerization has been unclear. Recent data showed ZapD binds specifically to the FtsZ CTD. Thus, to obtain insight into the ZapD-CTD interaction and how it may mediate FtsZ protofilament assembly, we determined the Escherichia coli ZapD-FtsZ CTD structure to 2.67 Å resolution. The structure shows that the CTD docks within a hydrophobic cleft in the ZapD helical domain and adopts an unusual structure composed of two turns of helix separated by a proline kink. FtsZ CTD residue Phe-377 inserts into the ZapD pocket, anchoring the CTD in place and permitting hydrophobic contacts between FtsZ residues Ile-374, Pro-375, and Leu-378 with ZapD residues Leu-74, Trp-77, Leu-91, and Leu-174. The structural findings were supported by mutagenesis coupled with biochemical and in vivo studies. The combined data suggest that ZapD acts as a molecular cross-linking reagent between FtsZ protofilaments to enhance FtsZ assembly.
Keywords: cell division, cytokinesis, electron microscopy (EM), protein-protein interaction, X-ray crystallography, FtsZ, ZapD
Introduction
FtsZ is a highly conserved prokaryotic tubulin homolog that mediates cell division in most bacteria, chloroplasts, many archaea, as well as the mitochondria of primitive eukaryotes (1–38). It is composed of a short N-terminal region, a globular core that binds GTP, a long and flexible linker of variable sequence and length (ranging from ∼40 to 257 residues), and a short C-terminal domain (CTD)3 of ∼14 residues. The FtsZ CTD region can be further divided into an N-terminal conserved region (CTC) and a short C-terminal variable region (CTV) (19). GTP binding to the globular domains between different FtsZ protomers leads to the formation of linear protofilaments. The FtsZ protofilaments assemble at the cell center to create the Z ring, which then serves as the platform for the assembly of the divisome (9). There is still debate regarding the organization of FtsZ protofilaments within the Z ring. Super-resolution analyses on multiple bacteria indicate that the Z ring is composed of dispersed protofilament assemblages (10–15). However, other data suggest that FtsZ protofilaments form a continuous ring around the cell (18).
Although the specific organization of the Z ring is unclear, data show that its assembly is effected by regulatory proteins (19). Indeed, the intracellular levels of FtsZ in Escherichia coli and most bacteria remain largely unchanged throughout the cell cycle and exceed the critical concentration required to form the Z ring (17). Therefore, the regulation of Z ring formation occurs at the level of FtsZ filament assembly. FtsZ-interacting proteins modulate FtsZ polymerization and thus play key roles in this process (19–38). An important family of positive FtsZ regulatory factors is the FtsZ ring-associated proteins (Zap), ZapA–D. These proteins facilitate Z ring formation by enhancing the formation of protofilament assemblages. ZapA–D are recruited early during cytokinesis and have overlapping functions in stabilizing Z ring formation at midcell (20–24, 26–29, 35–38). Indeed, although individual Zap proteins are not essential for the assembly of the divisome, it is suggested that their collective functions in the stabilization of FtsZ protofilaments and their assembly is critical for optimal cell division.
The widely conserved ZapA interacts directly with FtsZ, and a zapA knock-out leads to cell elongation (21). ZapB does not bind FtsZ but enhances the role of ZapA in FtsZ bundling (22, 23). Like ZapA, both ZapC and ZapD interact directly with FtsZ and also promote FtsZ protofilament assembly (20, 27, 28). Structures have recently been obtained for all the Zap proteins and show that, interestingly, despite their overlapping functions, they are structurally distinct (22, 26, 35–38). ZapA and ZapB are coiled-coil proteins that form dimeric or tetrameric structures; ZapB consists almost entirely of an extended coiled coil, whereas ZapA contains N-terminal globular domains in addition to its coiled-coil region (22, 26). Binding studies revealed that the ZapA globular domains mediate FtsZ binding (26). ZapC is composed of an N-terminal α/β region and a C-terminal twisted β-barrel-like domain (35, 36). Residues located within two surface-exposed pockets of ZapC appear to be involved in FtsZ binding (35). Multiple studies showed that ZapC binds the FtsZ GTPase core (27, 28, 35). However, one analysis suggested that ZapC might bind the FtsZ CTD, albeit with low affinity (36). The ZapD protein contains a helical domain and a β-strand domain and is dimeric (37, 38). Two studies reporting on putative FtsZ-binding residues generated conflicting results, and hence the FtsZ binding pocket in ZapD remains unknown (37, 38).
Like the majority of FtsZ regulatory proteins, ZapD binds the FtsZ CTD (19, 39–45). Thus, the CTD has been called the regulatory hub of FtsZ (19). In addition to ZapD, proteins that have been shown to interact with the FtsZ CTD in E. coli include ZipA, FtsA, ClpX, SlmA, and the C-terminal domain of MinC (39–45). To date, however, only three structures of FtsZ regulatory protein-CTD complexes have been obtained; the E. coli FtsZ binding domain of ZipA bound to the CTD, the E. coli SlmA-DNA-CTD complex, and the Thermotoga maritima FtsA-CTD structure (40–42). FtsA, which has an actin-like structure, and ZipA, with a split βαβ-fold, both bind the FtsZ CTD as a helix, whereas the TetR family member and nucleoid occlusion protein, SlmA, binds the CTD as a fully extended strand. FtsA and ZipA are both membrane-associated proteins and function in tethering FtsZ to the membrane, whereas the nucleoid occlusion factor SlmA acts as a negative regulator of FtsZ assembly by preventing the Z ring from forming through chromosomal DNA. Interestingly, FtsZ CTD binding proteins show weak interactions with a single CTD. Studies from the Lutkenhaus laboratory provided an explanation for these data as they revealed that for regulatory proteins to bind the CTD with high affinity they must bind CTDs on an FtsZ protofilament, which exposes them as a multivalent ligand (39). Methods such as bio-layer interferometry and surface plasmon resonance, which partially mimic the multivalency of the CTD found in FtsZ protofilaments, have been used to obtain relative Kd values for these proteins that are in the micromolar range (38, 40, 41).
Although the ZipA, FtsA, and SlmA structures revealed how tethering and negative FtsZ regulatory proteins interact with the FtsZ CTD, to date there are no structures of FtsZ cytosolic positive regulators, such as the Zaps, bound to FtsZ, leaving unclear how these proteins may facilitate FtsZ protofilament assembly. Thus, to gain insight into this question, we determined the structure of the E. coli ZapD-FtsZ CTD complex to 2.67 Å resolution. The structure reveals a pocket on the ZapD surface that serves as a docking site for the CTD. The CTD conforms optimally to this pocket by adopting an unusual conformation composed of two turns of helix separated by a proline-induced kink. The sharp proline kink allows the deep insertion into the ZapD pocket of Phe-377, which serves as an anchoring residue. Complementary in vivo and in vitro studies support this structural model. Collectively, the data provide molecular insight into how ZapD may enhance FtsZ assembly.
Results
Structure Determination and Overall Structure of E. coli ZapD-FtsZ CTD Complex
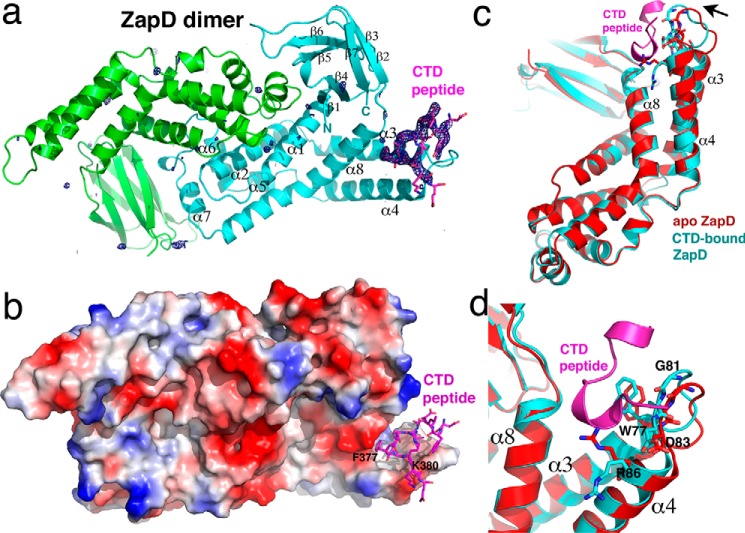
Crystals of the E. coli ZapD protein bound to the E. coli FtsZ CTD peptide, DYLDIPAFLRKQAD, were obtained, and the structure was solved by molecular replacement using the apo E. coli ZapD structure as a search model (Protein Data Bank code 5DKO). The structure was refined to final Rwork/Rfree values of 23.0%/26.5% (Table 1) (46). The structure contains a ZapD dimer in the crystallographic asymmetric unit (ASU). Each ZapD protomer is composed of two domains, a helical domain with eight α-helices (residues 12–179) and a β-strand domain with seven strands (residues 3–11; 180–247). The structure has the following topology: β1(residues 3–10)-α1(13–32)-α3(37–55)-α4(60–80)-α5(85–105)-α6(109–115) and α7(135–145)-α8(147–178)-β2(178–187)-β3(189–194)-β4(200–206)-β5(210–218)-β6(221–228)-β7(234–246) (Fig. 1, a and b). Formation of the ZapD dimer buries a considerable 1861 Å2 of protein surface from solvent, consistent with it being a functional dimer (47). Indeed, the structures of apo E. coli and the apo Vibrio parahemeolyticus ZapD homolog (2OEZ) revealed the identical dimer; no other shared dimer or oligomer states were identified in these structures. Superimpositions of the dimer from the ZapD-FtsZ CTD structure onto the apo E. coli ZapD and apo V. parahaemolyticus ZapD dimers resulted in root mean squared deviations (r.m.s.d.) of 1.4 and 1.5 Å, respectively.
TABLE 1.
Data collection and refinement statistics for E. coli ZapD-FtsZ CTD structure
| Data collection | |
| Space group | P21 |
| Cell constants (Å) | a = 71.3, b = 51.9, c = 79.1 |
| β = 90.7 | |
| Resolution (Å) | 79.1–2.67 |
| Rsym(%)a | 7.5 (48.0)b |
| Rpim (%) | 5.0 (40.9) |
| Overall I/σ(I) | 10.7 (1.6) |
| No. of unique reflections | 15,706 |
| No. of total reflections | 47,044 |
| % complete | 94.1 (73.1) |
| CC(1/2) | 0.997 (0.716) |
| Refinement statistics | |
| Rwork/Rfree(%)c | 23.0/26.5 |
| MolProbity score | 2.11 |
| B-Factors (average) (Å2) | |
| ZapD | 48.4 |
| CTD peptide | 80.0/69.2d |
| Solvent | 38.0 |
| r.m.s.d. | |
| Bond angles (°) | 0.445 |
| Bond lengths (Å) | 0.002 |
| Ramachandran analysis | |
| Favored (%) | 94.6 |
| Disallowed (%) | 0.0 |
aRsym = ΣΣ|Ihkl − Ihkl(j)|/ΣIhkl, where Ihkl(j) is observed intensity and Ihkl is the final average value of intensity.
bValues in parentheses are for the highest resolution shell.
cRwork = Σ‖Fobs| − |Fcalc‖/Σ|Fobs| and Rfree = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|, where all reflections belong to a test set of 5% randomly selected data.
dB-factor (average) for all CTD residues/B-factor (average) for CTD residues minus disordered side chains of residues 270, 381.
FIGURE 1.
Crystal structure of E. coli ZapD-FtsZ CTD complex. a, ribbon diagram of the E. coli ZapD-FtsZ CTD complex. The ZapD subunits in the dimer are colored green and cyan, and secondary structural elements are labeled for one subunit. The FtsZ CTD is shown as magenta sticks. Included in the structure is the Fo − Fc map contoured at 4.0 σ calculated after initial refinement of the structure, before the addition of the FtsZ CTD. This figure and Figs. 2, a and b, 4, and 7 were generated using PyMOL (51). b, electrostatic surface representation of the ZapD dimer shown in the same orientation as in a with the FtsZ CTD shown as magenta sticks. Blue and red regions represent electropositive and -negative regions, respectively. Also labeled are CTD residues Phe-377, which inserts into the ZapD hydrophobic cavity, and Lys-380, which interacts with electronegative regions of ZapD. c, comparison of apo ZapD subunit (red) and ZapD complexed to the FtsZ CTD (cyan). The CTD peptide is shown as a magenta ribbon. The figure shows an overlay of the subunits, which reveals that except for the region composed of ZapD residues 73–86, which shift inward upon CTD binding, the rest of the structure remains unchanged. d, close-up of c revealing the adjustment of key binding residue Trp-77 and the relocation of Arg-86 out of the pocket upon binding the CTD.
After initial refinement of the ZapD dimer, clear density was observed for residues 370–381 of the FtsZ CTD peptide in one subunit of the ZapD dimer (Fig. 1a). The structure shows that the CTD peptide binds within a pocket composed of α3, α4, and α8 located at the edge of the ZapD helical domain (Fig. 1c). Formation of the ZapD-CTD complex buries a significant 462 Å2 of complex surface from solvent. The other subunit of the ZapD dimer does not contain a bound peptide indicating that FtsZ CTD binding by ZapD is not cooperative. Although overlays of apo-ZapD subunits result in r.m.s.d. of <0.5 Å, comparison of the FtsZ CTD-bound ZapD subunit with the unbound subunit resulted in an r.m.s.d. of 1.1 Å indicating modest structural alterations take place upon CTD binding. Indeed, analysis of the overlaid structures revealed that ZapD residues 73–86, forming the α3-loop-α4 region of the CTD binding pocket, move inward upon peptide binding (Fig. 1, c and d). This shift removes Arg-86 from the pocket and orients hydrophobic residues for CTD contact (Fig. 1d). By contrast, α8, which includes the other edge of the CTD interaction pocket, is unchanged upon peptide binding (Fig. 1, c and d).
ZapD-FtsZ CTD Interactions Are Mediated by an Unusual CTD Conformation
The ZapD-FtsZ CTD structure reveals that ZapD interacts with CTD residues 374–381, where residues 374–379 are part of the conserved CTC and residues 380–381 are from the CTV. This finding is consonant with previous studies that showed that removal of N-terminal CTD residues 367–371 resulted in only a 2-fold reduction in ZapD binding (38). Indeed, although CTD residues 370–373 are visible in the structure, only one residue in this region, Tyr-371, contacts ZapD. In this interaction the phenyl ring atoms of the Tyr-371 side chain make van der Waals interactions with ZapD residue Gly-82. The ZapD pocket bound by the FtsZ CTD is primarily hydrophobic but is surrounded on its edges by acidic residues (Fig. 1b). FtsZ residue Lys-380 makes electrostatic contacts to ZapD residue Asp-84. This interaction along with hydrogen bonds between the peptide backbone atoms of Lys-380 and Gln-381 to ZapD residues 81–83 anchor the CTD peptide onto ZapD (Fig. 2a). Electron density is not visible for the Gln-381 side chain, consistent with the fact that only its peptide backbone makes contacts with ZapD. FtsZ CTV residues 380–381 make key contacts with ZapD, which explains the in vivo data showing that these residues are required for ZapD to localize with FtsZ during cell division (45). Bio-layer interferometry analyses also showed that truncating FtsZ to residue 379 reduced ZapD binding from 4 μm to >2 mm (38).
FIGURE 2.
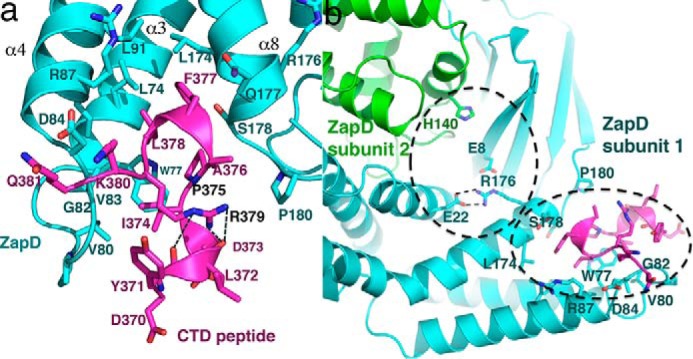
Interactions between ZapD and the FtsZ CTD. a, ribbon diagram showing zoomed-in view of the contacts between the FtsZ CTD and ZapD. The CTD binds to the ZapD helical domain between α3, α4, and α8. The inner region of this pocket is hydrophobic, although the surrounding ledges of the pocket are primarily electronegative. The CTD is colored magenta, and ZapD is cyan. Interacting residues are shown as sticks. CTD residues Pro-375 and Arg-379, labeled in black, play key roles in the formation of the unusual CTD conformation that allows it to dock within the pocket. b, ribbon diagram of the ZapD-CTD complex showing the relative locations of a putative pocket identified by Choi et al. (38) and the CTD binding pocket shown in the structure. The pockets are outlined by dashed lines, which underscores that they are adjacent to each other. Mutation of Glu-22 to an arginine impacted CTD binding likely by destabilizing the CTD binding pocket revealed in the crystal structure. One ZapD subunit is green, and the other is cyan. The CTD peptide is colored magenta.
The anchoring contacts from FtsZ CTD residues 380–381 permits docking of CTD residues 374–379 deep into the ZapD pocket (Fig. 2a). When bound in the ZapD pocket, CTD residues 371–379 adopt an unusual structure consisting of two turns of the helix separated by a proline-induced kink. CTD residue Arg-379 stabilizes this unique conformation by hydrogen bonding with the carbonyl oxygens located at the C terminus of the first turn of the CTD helix (Fig. 2a). The hydrophobic side of the CTD helix-kink-helix inserts into the ZapD pocket. A key hydrophobic interaction is provided by CTD residue Phe-377, which inserts deeply into the ZapD crevice (Fig. 1b). This contact provides another anchor for CTD binding. The inserted Phe-377 side chain interacts with ZapD residues Leu-74, Leu-91, Leu-174, and the Cβ and Cγ atoms of the Gln-177 side chain. The side chains of FtsZ residues Leu-378 and Pro-375, which are also located on the hydrophobic face of the CTD, interact with ZapD residues Leu-74 and Trp-77. Finally, CTD residue Ile-374 makes van der Waals contacts with Trp-77 as well as the side chains of Val-80 and Val-83 (Fig. 2a).
In contrast to residues in the α3-loop-α4 region, which undergo a conformational shift upon FtsZ binding, α8 located at the other side of the binding pocket appears to serve as a rigid scaffolding for the CTD interaction (Fig. 1, c and d). Some support for this notion comes from a recent study by Choi et al. (38). Based on studies examining FtsZ CTD binding to the ZapD mutants, E22R and H140R, Choi et al. (38) hypothesized that Glu-22, Arg-176, Glu-8, and His-140′ (where ′ indicates other subunit of the dimer) may form a putative CTD binding pocket in ZapD (Fig. 2b) (38). However, although the ZapD(E22R) substitution led to a 15-fold reduction in CTD binding, the His-140′ had a negligible effect on CTD binding (38). The ZapD-FtsZ CTD structure provides an explanation for these data as the putative pocket containing His-140′ and Glu-22 is adjacent to the FtsZ CTD binding site revealed in the ZapD-CTD complex (Fig. 2b). In the structure, Glu-22 makes bifurcated hydrogen bonds with α8 residue Arg-176 that would be predicted to stabilize this region (Fig. 2b). An E22R substitution would not only abrogate this contact, but the proximal placement of two arginine residues would also lead to significant electrostatic repulsion.
As hydrophobic contacts are not specific, they do not demand complete identity among homologs to retain contacts to partner proteins. Multiple sequence alignments of ZapD proteins show that CTD-interacting residues are either completely conserved, such as Trp-77, Val-83, Asp-84, and Leu-174, or have conservative substitutions that would maintain the interactions with the CTD (Fig. 3). Interestingly, the sequence alignments reveal that several arginine residues on the face of the ZapD dimer opposite the CTD binding site are also conserved. The importance of these residues is currently unclear; however, one possibility is that they could be involved in making secondary interactions with the FtsZ core, which is highly electronegative. The FtsZ-binding protein SlmA provides a precedent for such binding. The FtsZ CTD interacts with SlmA with high affinity and allows the formation of secondary interactions to the FtsZ core (42). The possible function of these secondary interactions is currently unclear, but it has been suggested that it may be to effect lateral contacts between FtsZ protofilaments. This could also be the case for positive regulators of FtsZ assembly, such as ZapD. Indeed, this type of binding model could explain data from Roach et al. (37) that showed cross-linking between ZapD and FtsZ core residues. Alternatively, the results from these cross-linking experiments could be due to nonspecific contacts between ZapD and FtsZ resulting from ZapD induced bundling and aggregation of FtsZ molecules.
FIGURE 3.
ZapD sequence alignments. Sequence alignments of ZapD proteins and the corresponding FtsZ CTD sequence from the same bacteria (E. coli (ZapD/FtsZ accession code: NP_414644.1/AJF44961.1); Vibrio cholerae (APF68703.1/CSA82682.1); Aeromonas hydrophilia (WP_065475370.1/YP_858319.1); Klebsiella pneumoniae (WP_004145941.1/CDI24806.1); Enterobacter cloacae (CQR78072.1/KGB11780.1); Shigella flexneri (WP_001194727.1/KFZ98273.1); Salmonella enterica (WP_000557441.1/EDZ24255.1); Serratia marcescens (WP_015670929.1/KFL05802.1); Yersinia pestis (KXF93325.1/KNX88873.1); and Vibrio parahemeolyticus (KKZ07905.1/ESV67294.1). ZapD residues that make contact with the FtsZ CTD in the crystal structure are highlighted in cyan, and the Glu-22–Arg-176 residues that make a salt bridge are highlighted in gray. Secondary structural elements of ZapD are indicated above the sequence alignments. Residues in the FtsZ CTDs that are visible in the crystal structure are highlighted in magenta.
FtsZ CTD Can Adopt Multiple Structures When Binding FtsZ Regulatory Proteins
The majority of FtsZ-binding proteins that have been characterized to date bind the FtsZ CTD. As a result, the FtsZ CTD has been called the FtsZ landing pad (19). The consequences of these binding protein-CTD interactions are varied. Some regulators enhance the formation of protofilaments by FtsZ and some prevent it, whereas others mediate FtsZ recruitment (19, 25, 40–42). Structures that have been solved of regulatory protein-CTD complexes include the E. coli ZipA-CTD complex, the T. maritima FtsA-CTD complex, and the E. coli SlmA-DNA-CTD complex. ZipA and FtsA both function to recruit FtsZ to the membrane, whereas SlmA-DNA antagonizes FtsZ protofilament formation and bundling (40–42). The structures of ZipA and FtsA bound to the CTD were obtained first and revealed the CTD bound as helices, which led to the suggestion that the CTD adopts a helical state when complexed with binding proteins (40, 41). However, the helices bound in the structures differed. Although both helices initiated with the conserved CTD proline, the FtsA-bound helix is distorted due to a glycine not present in the E. coli CTD, and T. maritima FtsA contacts the last CTV C-terminal residue, which is not found in the E. coli CTD. Indeed, the E. coli and T. maritima CTD share only five residues in common, and the T. maritima CTD contains a much longer CTV (T. maritima CTD is PEGDIPAIYRYGLEGLL compared with the E. coli FtsZ CTD, DYLDIPAFLRKQAD; conserved residues underlined). Thus, differences in these helices might be due to the dissimilarity in the E. coli and T. maritima CTD sequences. However, when the E. coli SlmA-DNA-CTD structure was obtained with the same CTD peptide as utilized in the ZipA-CTD structure, it revealed a fully extended and non-helical conformation for the CTD, demonstrating that at least the E. coli CTD is a flexible region that can adopt multiple conformations that depend on its binding partners. The ZapD-FtsZ CTD structure confirms this supposition as it reveals yet a different conformation for the E. coli CTD than those observed in the SlmA-CTD and ZipA-CTD structures (Fig. 4). Thus, these data indicate that deducing the binding mode of the E. coli FtsZ CTD by a given regulatory protein will require structural information.
FIGURE 4.
Comparison of E. coli FtsZ CTD complexes with regulatory proteins. Ribbon diagrams are shown for the E. coli regulatory proteins ZapD, ZipA, and SlmA bound to the CTD. The regulatory proteins are colored light cyan and the CTDs are magenta. For reference, Phe-377 is shown in the same orientation, which underscores the very different structures adopted by the CTD when bound to these proteins.
Single Mutants of Potential FtsZ-interacting Residues on ZapD Rescue Viability Defects and Filamentation
The ZapD-FtsZ CTD structure is largely consistent with and explains previous biochemical data (35, 38). However, to further probe the structural model, we next made mutations in ZapD residues that the structure indicated are key for CTD binding. In particular, ZapD residues Trp-77, Asp-84, Leu-91, and Leu-174 were mutated to alanines, and the effects of these substitutions were analyzed (Tables 2–3). As described previously, increased expression of wild type (WT) ZapD leads to lethal filamentation and reduced cell viability (20). Therefore, we tested whether overexpression of the constructed ZapD mutants displayed altered viability and cell morphology phenotypes (Fig. 5a). In contrast to WT ZapD, overexpression of ZapD single mutants ZapD(W77A), ZapD(L91A), and ZapD(L174A) were viable and displayed reduced cell elongation even at high concentrations of the inducer (Fig. 5b and Table 4). The ZapD mutant D84A, which makes long range electrostatic contacts to CTD residue Lys-380, showed partially increased viability and a modest reduction in cell elongation compared with WT ZapD and was not used in further studies. Immunoblotting of FtsZ and ZapD mutant proteins revealed that these phenotypes were not due to changes in expression levels or instability of mutant proteins (Fig. 5c).
TABLE 2.
Bacterial strains and plasmid used in this study
| Strains and plasmids | Relevant genotype | Source or Ref.a |
|---|---|---|
| Strains | ||
| MG1655 | F− λ− ilvG rfb50 rph1 | Laboratory collection |
| C41 (DE3) | F− ompT hsdSB (rB− mB−) gal dcm (DE3) | 49 |
| KHH72 | C41 (DE3) pET28b-h10-smt3-zapD | |
| KHH441 | MG1655 pTrc99a | |
| KHH442 | MG1655 pTrc99a-zapD | |
| KHH443 | MG1655 pTrc99a-zapD D84A | |
| KHH445 | MG1655 pTrc99a-zapD L91A | |
| KHH448 | MG1655 pTrc99a-zapD (W77A) | |
| KHH478 | MG1655 pTrc99a-zapD (L174A) | |
| KHH488 | C41 (DE3) pET28b-h10-smt3-zapD (L174A) | |
| KHH490 | C41 (DE3) pET21b-ftsZ | |
| KHH491 | C41 (DE3) pET28b-h10-smt3-zapD (W77A) | |
| KHH492 | C41/DE3 pET28b-h10-smt3-zapD (L91A) | |
| Plasmids | ||
| pAM4 | pTrc99a-zapD (L174A) | |
| pET28b-his10-smt3-zapD | pBR322 ori PT7 h10-smt3-zapD, KanR | 20 |
| pKHH45 | pTrc99a zapD (W77A) | |
| pKHH46 | pTrc99a zapD (D84A) | |
| pKHH47 | pTrc99a zapD (L91A) | |
| pKHH48 | pET28b h10-smt3-zapD (L174A) | |
| pKHH49 | pET28b h10-smt3-zapD (L91A) | |
| pKHH50 | pET28b h10-smt3-zapD (W77A) | |
| pTrc99a | ColE1 ori Ptac AmpR | Laboratory collection |
a This work was used unless otherwise noted.
TABLE 3.
Primers used in this study
F is forward, and R is reverse.
| Name | Sequence 5′–3′ | Plasmid |
|---|---|---|
| SUMO-5 YacF-BamHI | CGCGGATCCATGCAGACCCAGGTCCTTTTTG | pET28b-his10x-smt3-zapD |
| SUMO-3 YacF-HindIII | CCCAAGCTTTTAGCAACAGGCCAGTTCGAA | pET28b-his10×-smt3-zapD |
| ZapD-BamHI F | GGTAGGATCCATGCAGACCCAGGTCCTTTTTGA | pKHH48–50 |
| ZapD-SalI R | GGCGGTCGACTTAGCAACAGGCCAGTTCGAAAT | pAM4, pKHH44–50 |
| ZapD-W77A F | GCTAAACTCCAGACCGCGATTGGCGTGCCTGG | pKHH45 |
| ZapD-W77A R | CCAGGCACGCCAATCGCGGTCTGGAGTTTAGC | pKHH45 |
| ZapD-D84A F | CGTGCCTGGCGTGGCCCAGAGCCGTATTGA | pKHH46 |
| ZapD-D84A R | TCAATACGGCTCTGGGCCACGCCAGGCACG | pKHH46 |
| ZapD-L91A F | CCGTATTGAAGCAGCAATTCAGCAGTTAAA | pKHH47 |
| ZapD-L91A R | TTTAACTGCTGAATTGCAGCTTCAATACGG | pKHH47 |
| ZapD-L174A F | CATGGTGCTGGATGCAATTCGCCAGTCGGC | pAM4 |
| ZapD-L174A R | CGGCACTGGCGAATTGCATCCAGCTCCATG | pAM4 |
FIGURE 5.
Cell viability and morphology phenotypes upon overexpression of WT ZapD or ZapD mutant variants. a, spot viability assays of wild type (MG1655) strain bearing pTrc99a vector alone, WT ZapD, ZapD(W77A), ZapD(D84A), ZapD(L91A), or ZapD(L174A). The 10-fold dilutions of cells cultured as described under “Experimental Procedures” were spotted on LB plates with ampicillin at the indicated concentrations of IPTG and grown at 37 °C for ∼16 h. The experiment was repeated three times, and a representative image is shown. b, cell morphology phenotypes of wild type (MG1655) strain bearing pTrc99a vector alone (panel i), WT ZapD (panel ii), ZapD(W77A) (panel iii), ZapD(D84A) (panel iv), ZapD(L91A) (panel v), or ZapD(L174A) (panel vi). Cells were grown and imaged under conditions described under “Experimental Procedures.”. Bar, 3 μm. c, quantitative immunoblots of FtsZ, WT ZapD, and ZapD mutant protein levels. Whole cell proteins were harvested from cells at mid-log grown under the same conditions described in the legend to b. Samples were normalized to optical densities and separated by SDS-PAGE. Western analysis was conducted using anti-FtsZ rabbit polyclonal antibody (GenScript) at 1:10,000 and anti-ZapD rabbit polyclonal antibody (GenScript) at 1:1000. Protein bands were normalized to total proteins (BLOT-FastStain, G-Biosciences) as loading and transfer controls. Bands were visualized using a LI-COR Odyssey CLx imager, and intensities were measured using ImageStudio software (LI-COR). Three independent experiments were conducted, and a representative blot with average band intensities ± S.D. is shown. Std indicates molecular size standards, and na indicates not available.
TABLE 4.
Cell lengths of strains overexpressing either WT ZapD or a ZapD mutant in trans
| Plasmida | Average ± S.D.b | Nc |
|---|---|---|
| μm | ||
| Vector | 3.0 ± 0.7 | 331 |
| ZapD | 46.6 ± 17.9 | 71 |
| W77A | 6.4 ± 3.3 | 278 |
| D84A | 21.2 ± 13.7 | 102 |
| L91A | 7.0 ± 4.8 | 252 |
| L174A | 3.4 ± 1.1 | 374 |
a Wild type or ZapD mutant plasmids were expressed in trans in the pTrc99a vector backbone in a wild type strain (MG1655) background.
b Cell lengths reported are for cells sampled at OD = ∼0.6–0.8 grown in LB with 25 μm IPTG at 37 °C. S.D. = standard deviation.
c N represents the number of individual cells measured for each strain. In case of filamentous strains, fewer numbers of cells were present within each field of view.
Mutant ZapD Proteins Fail to Increase FtsZ Polymerization in Vitro
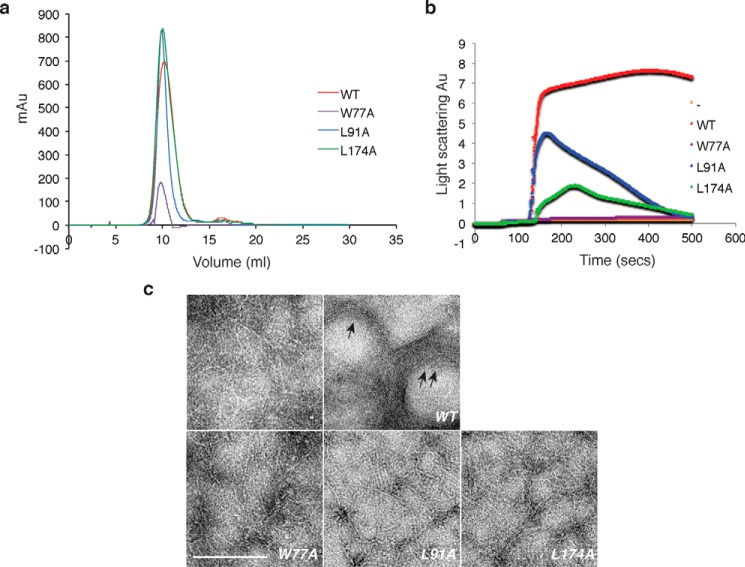
To investigate the effects of the ZapD mutants, W77A, L91A, and L174A, on FtsZ polymerization in vitro, we purified the mutant proteins and examined FtsZ assembly in a 90° light scattering assay. We also visualized the effects of these mutants on the morphological properties of FtsZ polymers by TEM. All mutant ZapD proteins eluted with a near identical profile (mostly dimeric) to the WT protein suggesting that the mutants were likely folded (Fig. 6a). When incubated in the presence of GTP alone, WT FtsZ showed an increase in light scattering signal, and single protofilaments of FtsZ were visualized by electron microscopy (Fig. 6, b and c). Upon addition of ZapD, the light scattering signal was further enhanced, and large bundled polymeric assemblies were observed by TEM (Fig. 6, b and c). In contrast, the ZapD mutants either failed to promote (W77A) or promoted moderate increases (L91A, L174A) in light scattering signal followed by a rapid reduction in the signal (Fig. 6b). Consistent with these data, visualization of FtsZ polymerization in the presence of ZapD mutant proteins via TEM showed single FtsZ protofilaments, or 2–3 protofilament bundles, similar to WT FtsZ alone (Fig. 6c). Taken together, the in vitro results are consistent with Trp-77, Leu-91, and Leu-174 residues of ZapD mediating critical interactions with the FtsZ CTD.
FIGURE 6.
ZapD mutant derivatives do not support FtsZ assemblies in vitro. a, overlaid elution profiles of WT ZapD (red), ZapD(W77A) (purple), ZapD(L91A) (blue), and ZapD(L174A) (green) are shown. Calculated molecular masses indicate that WT and mutant proteins elute mostly as dimers. A calibrated Superdex10-75 (10/300 GL) column was equilibrated in buffer (50 mm MOPS/KOH, pH 6.5, 50 mm KCl, 2.5 mm MgCl2, 0.2 mm DTT, and 0.2% glycerol), loaded with 500 μl of purified protein, and run at 0.5 ml/min with equilibration buffer. b, 90° light scattering data of FtsZ assembly alone (orange), in the presence of WT ZapD (red), ZapD(W77A) (purple), ZapD(L91A) (blue), and ZapD(L174A) (green). Data from FtsZ (5 μm) and WT ZapD or ZapD mutant proteins (5 μm) are shown. Reaction conditions are described under “Experimental Procedures.” c, electron micrographs of FtsZ assemblies in reaction conditions as described in b legend. – = FtsZ alone. Arrows point to FtsZ bundles in the presence of WT ZapD. Bar, 300 nm.
Discussion
Bacterial cytokinesis is initiated by formation of an unstable proto-ring by FtsZ. FtsZ regulatory factors such as the Zap proteins contribute to the stabilization of the proto-ring into the Z ring. Although the precise arrangement of FtsZ protofilaments in the Z ring is unclear, studies have indicated that individual FtsZ protofilaments in this structure are ∼60–120 Å apart (12, 41, 48). Therefore, it is unlikely that there are tight contacts between the FtsZ core regions of multiple protofilaments assembled in the Z ring. Hence, regulatory factors such as the Zap proteins may be involved in mediating FtsZ polymer assembly. Indeed, studies indicate that ZapA, ZapC, and ZapD greatly enhance FtsZ protofilament assembly (20–24, 26–29). However, the molecular mechanisms utilized by these proteins to facilitate FtsZ protofilament bundling is not clear as there have been no structures reported of a Zap protein bound to FtsZ.
Data have clearly demonstrated that the ZapD protein binds the FtsZ CTD (20). Thus, to gain insight into how a positive Zap protein assembly factor may bind FtsZ and promote Z ring formation, here we determined the structure of the ZapD-FtsZ CTD complex. The ZapD structure reveals an elongated dimer, and the FtsZ CTD was shown to bind at the end of a ZapD subunit in a pocket formed by α3, α4, and α8. When bound to ZapD, the FtsZ CTD adopts an unusual conformation composed of two turns of helix separated by a proline kink. The CTD Arg-379 side chain inserts between the helical turns to hydrogen bond to the carbonyl oxygens of the first helical turn and stabilize this unusual structure. The resulting CTD structure makes numerous hydrophobic contacts with ZapD.
Because ZapD is a dimer, two FtsZ CTDs could possibly bind to each end of the dimer, although this remains to be demonstrated. This would suggest a mechanism by which ZapD could function as a molecular cross-linking reagent, aligning between FtsZ polymers to stabilize the formation of laterally arranged FtsZ protofilaments (Fig. 7). A similar cross-linking mechanism may be at play for other Zaps. It is not clear how ZapA interacts with FtsZ. However, data show that ZapA forms dimers and tetramers (26). Hence, ZapA could interact with several FtsZ molecules and provide proximal linkage of several protofilaments. Multiple studies indicate that ZapC is monomeric (27, 28, 35). However, recent analyses suggesting that one monomer may harbor multiple FtsZ interacting pockets (35) indicate that it too could function as a cross-linker by using these pockets to link together FtsZ molecules from multiple protofilaments.
FIGURE 7.

Model of how ZapD dimers (with subunits colored magenta and cyan) could function as cross-linking reagents between FtsZ protofilaments. The FtsZ protofilaments are shown as gray surfaces.
In a ZapD-FtsZ cross-linking model, the long linker connecting the FtsZ GTPase core domain with the FtsZ CTD would enable flexibility between the interacting proteins and allow distances between ZapD-connected protofilaments on the order of 60–120 Å, which as noted is similar to the observed distances between protofilaments in the Z ring (12, 41, 48,). In one model of the Z ring, FtsZ protofilaments form continuous bands around the cell and slide past each other (18). The properties of the ZapD-FtsZ CTD interaction, including non-cooperative, weak contacts with the FtsZ CTD, would facilitate such a model as one ZapD subunit could release its CTD, whereas the other subunit retained CTD binding, allowing the sliding of adjacent protofilaments while maintaining their juxtaposed organization. Possible secondary interactions between FtsZ core regions to ZapD-conserved arginine residues could provide further stability in FtsZ assembly. However, a better understanding of the mechanism by which Zap proteins facilitate FtsZ assembly necessitates a clearer picture of the organization and the dynamics of FtsZ protofilaments within the Z ring.
Experimental Procedures
ZapD-FtsZ CTD Crystallization and Data Collection
The ZapD protein was purified as described previously (20), and the His10×-Smt3 tag was removed prior to crystallization. The purified protein was buffer exchanged into 25 mm Tris, pH 7.5, 150 mm NaCl, 5% glycerol, 1 mm dithiothreitol (DTT) for crystallization. The ZapD protein was then concentrated to 20 mg/ml, and the 14-mer E. coli FtsZ CTD peptide (DYLDIPAFLRKQAD) (GenScript) was added to a final concentration of 2 mm. The ZapD-FtsZ CTD peptide solution was crystallized using the hanging drop vapor diffusion method at room temperature by mixing it 1:1 with a solution of 27% PEG 1500, 0.1 m MES, pH 6.5. Crystals took 1–5 days to grow and were cryo-preserved by dipping them in a solution consisting of the crystallization reagent supplemented with 10% glycerol for 1–2 s prior to placement in the cryo-stream.
Structure Determination and Refinement of the ZapD-FtsZ CTD Structure
The ZapD-FtsZ CTD crystals take the space group P21, with a = 71.3 Å, b = 51.9 Å, c = 79.1 Å and β = 90.7°. X-ray intensity data were collected to 2.67 Å at Advanced Light Source beamline 8.3.1, and the data were processed with MOSFLM. The structure was solved by molecular replacement using the apo E. coli ZapD structure (5DKO) as a search model. This apo ZapD structure contains one subunit in the crystallographic ASU. Hence a multisearch procedure in Phaser was employed to find the two subunits in the ZapD-FtsZ-CTD ASU. After initial refinement of the starting model, electron density was observed for one FtsZ CTD. CTD peptide residues 370–381 and 62 solvent molecules were added at this point, and the structure was refined to convergence using Phenix (46). CTD residues 382–383 and the side chains of Asp-370 and Gln-381 were not visible, and only weak density was observed for the Lys-380 side chain (Table 1). With the exception of Met-1, all ZapD residues were visible and included in the model. The final structure has a MolProbity score of 2.11, which places it in the 97th percentile range of crystal structures solved at a similar resolution. Final X-ray data collection and refinement statistics are presented in Table 1.
Strains and Growth Conditions
Bacterial strains were grown in LB (0.5% NaCl) broth or agar plates with ampicillin at 100 μg/ml or kanamycin at 50 μg/ml at 37 °C unless noted otherwise. All strains and plasmids are derivatives of MG1655. Strains, plasmids, and primers used in the study are listed in Tables 2 and 3.
Plasmid Construction
Plasmid pTrc99a expressing zapD under the control of an isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible promoter was constructed by amplifying zapD using the ZapD-SacI forward and ZapD-SalI reverse primers. The PCR product was digested by SacI and SalI endonucleases and ligated into the same sites of the pTrc99a vector. The resulting clones were verified by Sanger sequencing (Genewiz). Plasmid pTrc99a-zapD was used to create the zapD mutants by QuikChange mutagenesis (Agilent) using the appropriate zapD mutant primer pairs. After the correct clones were verified by sequencing, the mutagenized zapD region of the plasmid was excised using a SacI/SalI digest, and the resulting fragment was re-ligated into the same sites of the pTrc99a parent vector. Plasmid pET28b-his10×-smt3 expressing zapD or a zapD mutant was constructed by amplifying WT or mutant zapD fragments from pTrc99a-zapD or zapD mutant clones using the ZapD-BamHI forward and ZapD-HindIII (for WT) or ZapD-SalI (for mutants) reverse primer pairs. The PCR product was digested by BamHI and HindIII or SalI and ligated into the same sites of the pET28b-his10×-smt3 vector. The resulting clones were verified by Sanger sequencing (Genewiz).
Spot Viability Assays
For spot viability experiments, overnight cultures of MG1655 expressing the vector alone, WT ZapD, or ZapD mutant plasmids were grown in LB with ampicillin at 37 °C, centrifuged, and then resuspended in LB containing ampicillin. Cell suspensions were normalized to A600 = 1.0, serially diluted to 100,000-fold, and 3 μl from each dilution were spotted on LB plates containing ampicillin and varying concentrations of IPTG. The plates were incubated at 37 °C for ∼16 h, at which point they were imaged (Syngene Gel-Doc System).
Cell Length Measurements
Overnight cultures of MG1655 expressing vector alone, WT ZapD, or ZapD mutant plasmids were grown at 37 °C in LB media with ampicillin and 0.2% glucose. Cells were subcultured 1:100 and grown in LB ampicillin until A600 = ∼0.1–0.2, at which point IPTG at 25 μm was added, and cells were sampled at A600 = ∼0.6–0.8. Cells were imaged using a Nikon TiE microscope on 1% agarose pads on glass slides. ObjectJ (National Institutes of Health) was used to measure cell lengths from images as described (20).
Protein Purification
FtsZ, ZapD, and ZapD mutants were purified as described previously and used in light scattering assays and electron microscopy experiments (20, 45).
90° Light Scattering Assays
Light scattering assays were done as described (50) using a DM-45 spectrofluorimeter (Olis). Initial reaction mixtures contained 5 μm FtsZ in assembly buffer (50 mm MOPS/KOH, pH 6.5, 50 mm KCl, 2.5 mm MgCl2). Readings were taken every 0.25 s at room temperature, and a baseline was established for 1 min prior to the addition of 1 mm GTP. WT or mutant variants of ZapD were added at 5 μm, 1 min after addition of 1 mm GTP. Data were collected by SpectralWorks (Olis) for 10 min per reaction and exported into Microsoft Excel for processing.
Electron Microscopy
To visualize the morphology of the FtsZ polymers under similar conditions as those used for light scattering assays, reaction mixtures were assembled as follows: FtsZ and ZapD (at 5 μm each) were incubated in polymerization buffer (50 mm MOPS/KOH, pH 6.5, 50 mm KCl, 2.5 mm MgCl2). GTP was added to a final concentration of 1 mm to start FtsZ polymerization, and the samples were incubated for 10 min at room temperature. A 10-μl aliquot of each sample was applied onto a glow-discharged 400 mesh carbon-coated copper grid, incubated for 10 s, wicked with a filter paper, and negatively stained using 2% uranyl acetate for 5–10 s. The grids were visualized using a JEOL 2100 transmission electron microscope operated at 200 kV with 30 pA/cm2 current density and recorded on a 2k-by-2k CCD camera at 15,000 or 30,000 magnification.
Author Contributions
M. A. S. and A. J. designed the research and analyzed the data. M. A. S. crystallized and solved the ZapD-FtsZ CTD structure. W. Z. performed protein purifications. K.-H. H. carried out in vivo and biochemical experiments. M. A. S. wrote the paper with input from A. J. All authors reviewed the results and approved the final manuscript.
Acknowledgments
We acknowledge the Advanced Light Source beamlines 8.3.1 and 5.0.2 for data collection. The Berkeley Center for Structural Biology is supported in part by NIGMS, National Institutes of Health, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the United States Department of Energy under Contract No. DE-AC02-05CH11231. Beamline 8.3.1 at the Advanced Light Source is operated by the University of California Office of the President, Multicampus Research Programs and Initiatives Grant MR-15-328599 and Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation. We also thank Aaron Mychack for help in plasmid construction.
This work was supported by National Institutes of Health Grant GM115563 (to M. A. S.) and National Science Foundation Award IOS1615858 (to A. J.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (code 5KOA) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CTD
- C-terminal domain
- r.m.s.d.
- root mean squared deviation
- FtsZ
- filament temperature-sensitive mutant Z
- Zap
- FtsZ ring-associated protein
- TEM
- transmission electron microscopy
- ASU
- asymmetric unit
- CTV
- C-terminal variable region
- CTC
- C-terminal conserved region
- IPTG
- isopropyl β-d-1-thiogalactopyranoside.
References
- 1. Adams D. W., and Errington J. (2009) Bacterial cell division, assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7, 642–653 [DOI] [PubMed] [Google Scholar]
- 2. Bi E. F., and Lutkenhaus J. (1991) FtsZ ring structure associated with division in. Escherichia coli. Nature 354, 161–164 [DOI] [PubMed] [Google Scholar]
- 3. de Boer P. A. (2010) Advances in understanding E. coli cell fission. Curr. Opin. Microbiol. 13, 730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Erickson H. P., Anderson D. E., and Osawa M. (2010) FtsZ in bacterial cytokinesis, cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74, 504–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lutkenhaus J., Pichoff S., and Du S. (2012) Bacterial cytokinesis, from Z ring to Divisome. Cytoskeleton 69, 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Boer P., Crossley R., and Rothfield L. (1992) The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359, 254–256 [DOI] [PubMed] [Google Scholar]
- 7. RayChaudhuri D., and Park J. T. (1992) Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359, 251–254 [DOI] [PubMed] [Google Scholar]
- 8. Mukherjee A., and Lutkenhaus J. (1998) Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 17, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Söderström B., and Daley D. O. (2016) The bacterial divisome: more than a ring. Curr. Genet. 10.1007/s00294-016-0630-2 [DOI] [PubMed] [Google Scholar]
- 10. Rowlett V. W., and Margolin W. (2014) 3D-SIM super-resolution of FtsZ and its membrane tethers in Escherichia coli cells. Biophys. J. 107, L17–L20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Biteen J. S., Goley E. D., Shapiro L., and Moerner W. E. (2012) Three-dimensional super-resolution imaging of the midplane protein FtsZ in live Caulobacter crescentus cells using astimatism. ChemPhysChem 13, 1007–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Z., Trimble M. J., Brun Y. V., and Jensen G. J. (2007) The structure of FtsZ filaments in vivo suggest a force-generating role in cell division. EMBO J. 26, 4694–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buss J., Coltharp C., Huang T., Pohlmeyer C., Wang S. C., Hatem C., and Xiao J. (2013) In vivo organization of the FtsZ ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol. Microbiol. 89, 1099–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fu G., Huang T., Buss J., Colrharp C., Hensel Z., and Xiao J. (2010) In vivo structure of the E. coli FtsZ ring by photoactivated localization microscopy (PALM). PLoS ONE 5, e12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jennings P. C., Cox G. C., Monahan L. G., and Harry E. J. (2011) Super-resolution imaging of the bacterial cytokinetic protein FtsZ. Micron 42, 336–341 [DOI] [PubMed] [Google Scholar]
- 16. Romberg L., and Levin P. A. (2003) Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu. Rev. Microbiol. 57, 125–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weart R. B., and Levin P. A. (2003) Growth rate-dependent regulation of medial FtsZ ring formation. J. Bacteriol. 185, 2826–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Szwedziak P., Wang Q., Bharat T. A., Tsim M., and Löwe J. (2014) Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. Elife 3, e04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang K. H., Durand-Heredia J., and Janakiraman A. (2013) FtsZ ring stability, of bundles, tubules, crosslinks, and curves. J. Bacteriol. 195, 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Durand-Heredia J., Rivkin E., Fan G., Morales J., and Janakiraman A. (2012) Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J. Bacteriol. 194, 3189–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mohammadi T., Ploeger G. E., Verheul J., Comvalius A. D., Martos A., Alfonso C., van Marle J., Rivas G., and den Blaauwen T. (2009) The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry 48, 11056–11066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ebersbach G., Galli E., Møller-Jensen J., Löwe J., and Gerdes K. (2008) Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol. Microbiol. 68, 720–735 [DOI] [PubMed] [Google Scholar]
- 23. Galli E., and Gerdes K. (2012) FtsZ-ZapA-ZapB interactome of Escherichia coli. J. Bacteriol. 194, 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pacheco-Gómez R., Cheng X., Hicks M. R., Smith C. J., Roper D. I., Addinall S., Rodger A., and Dafforn T. R. (2013) Tetramerisation of ZapA is required for FtsZ bundling. Biochem. J. 449, 795–802 [DOI] [PubMed] [Google Scholar]
- 25. Dajkovic A., Lan G., Sun S. X., Wirtz D., and Lutkenhaus J. (2008) MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr. Biol. 18, 235–244 [DOI] [PubMed] [Google Scholar]
- 26. Roach E. J., Kimber M. S., and Khursigara C. M. (2014) Crystal structure and site-directed mutagenesis reveals key residues involved in Escherichia coli ZapA function. J. Biol. Chem. 289, 23276–23286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Durand-Heredia J. M., Yu H. H., De Carlo S., Lesser C. F., and Janakiraman A. (2011) Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J. Bacteriol. 193, 1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hale C. A., Shiomi D., Liu B., Bernhardt T. G., Margolin W., Niki H., and de Boer P. A. (2011) Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J. Bacteriol. 193, 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bhattacharya A., Ray S., Singh D., Dhaked H. P., and Panda D. (2015) ZapC promotes assembly and stability of FtsZ filaments by binding at a different site in FtsZ than ZipA. Int. J. Biol. Macromol. 81, 435–442 [DOI] [PubMed] [Google Scholar]
- 30. Camberg J. L., Hoskins J. R., and Wickner S. (2009) ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc. Natl. Acad. Sci. U.S.A. 106, 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hamoen L. W., Meile J.-C., de Jong W., Noirot P., and Errington J. (2006) SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol. Microbiol. 59, 989–999 [DOI] [PubMed] [Google Scholar]
- 32. Król E., van Kessel S. P., van Bezouwen L. S., Kumar N., Boekema E. J., and Scheffers D. J. (2012) Bacillus subtilis SepF binds to the C terminus of FtsZ. PLoS ONE 7, e43293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goley E. D., Dye N. A., Werner J. N., Gitai Z., and Shapiro L. (2010) Imaging based identification of a critical regulator of FtsZ protofilament curvature in Caulobacter. Mol. Cell 39, 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Potluri L.-P., Kannan S., and Young K. D. (2012) ZipA is required for FtsZ dependent pre-septal peptidoglycan synthesis prior to invagination during cell division. J. Bacteriol. 194, 5334–5342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schumacher M. A., Zeng W., Huang K. H., Tchorzewski L., and Janakiraman A. (2016) Structural and functional analyses reveal insights into the molecular properties of the Escherichia coli Z ring stabilizing protein, ZapC. J. Biol. Chem. 291, 2485–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ortiz C., Kureisaite-Ciziene D., Schmitz F., McLaughlin S. H., Vicente M., and Löwe J. (2015) Crystal structure of the Z-ring associated cell division protein ZapC from Escherichia coli. FEBS Lett. 589, 3822–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roach E. J., Wroblewski C., Seidel L., Berezuk A. M., Brewer D., Kimber M. S., and Khursigara C. M. (2016) Structure and mutational analyses of Escherichia coli ZapD reveal charged residues involved in FtsZ bundling. J. Bacteriol. 198, 1683–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Choi H., Min K., Mikami B., Yoon H. J., and Lee H. H. (2016) Structural and biochemical studies reveal a putative FtsZ recognition site on the Z-ring stabilizer ZapD. Mol. Cells 39, 814–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Du S., Park K. T., and Lutkenhaus J. (2015) Oligomerization of FtsZ converts the FtsZ tail motif (conserved carboxy-terminal peptide) into a multivalent ligand with high avidity for partners ZipA and SlmA. Mol. Microbiol. 95, 173–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mosyak L., Zhang Y., Glasfeld E., Haney S., Stahl M., Seehra J., and Somers W. S. (2000) The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 19, 3179–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Szwedziak P., Wang Q., Freund S. M., and Löwe J. (2012) FtsA forms actin-like protofilaments. EMBO J. 31, 2249–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schumacher M. A., and Zeng W. (2016) Structures of the nucleoid occlusion protein SlmA bound to DNA and the C-terminal domain of the cytoskeletal protein FtsZ. Proc. Natl. Acad. Sci. U.S.A. 113, 4988–4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Camberg J. L., Hoskins J. R., and Wickner S. (2011) The interplay of ClpXP with the cell division machinery in Escherichia coli. J. Bacteriol. 193, 1911–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Camberg J. L., Viola M. G., Rea L., Hoskins J. R., and Wickner S. (2014) Location of dual sites in the E. coli FtsZ important for degradation by ClpXP; one at the C terminus and one in the disordered linker. PLoS ONE 9, e94964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang K. H., Mychack A., Tchorzewski L., and Janakiraman A. (2016) Characterization of the FtsZ C-terminal variable (CTV) region in Z-ring assembly and interaction with the Z-ring stabilizer ZapD in E. coli cytokinesis. PLoS ONE 11, e0153337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 48. Mingorance J., Tadros M., Vicente M., González J. M., Rivas G., and Vélez M. (2005) Visualization of single Escherichia coli FtsZ filament dynamics with atomic force microscopy. J. Biol. Chem. 280, 20909–20914 [DOI] [PubMed] [Google Scholar]
- 49. Buske P. J., and Levin P. A. (2012) Extreme C terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. J. Biol. Chem. 287, 10945–10957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Arjes H. A., Lai B., Emelue E., Steinbach A., and Levin P. A. (2015) Mutations in the bacterial cell division protein FtsZ highlight the role of GTP binding and longitudinal subunit interactions in assembly and function. BMC Microbiol. 15, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Delano W. L. (2002) The PyMOL Molecular Graphics System, Version 1.6, DeLano Scientific, San Carlos, CA [Google Scholar]