Abstract
Intercellular amino acid transport is essential for the growth of all multicellular organisms, and its dysregulation is implicated in developmental disorders. By an unknown mechanism, amino acid efflux is stimulated in plants by overexpression of a membrane-localized protein (GLUTAMINE DUMPER 1 (GDU1)) that requires a ubiquitin ligase (LOSS OF GDU 2 (LOG2). Here we further explore the physiological consequences of the interaction between these two proteins. LOG2 ubiquitin ligase activity is necessary for GDU1-dependent tolerance to exogenous amino acids, and LOG2 self-ubiquitination was markedly stimulated by the GDU1 cytosolic domain, suggesting that GDU1 functions as an adaptor or coactivator of amino acid exporter(s). However, other consequences more typical of a ligase-substrate relationship are observed: disruption of the LOG2 gene increased the in vivo half-life of GDU1, mass spectrometry confirmed that LOG2 ubiquitinates GDU1 at cytosolic lysines, and GDU1 protein levels decreased upon co-expression with active, but not enzymatically inactive LOG2. Altogether these data indicate LOG2 negatively regulates GDU1 protein accumulation by a mechanism dependent upon cytosolic GDU1 lysines. Although GDU1-lysine substituted protein exhibited diminished in vivo ubiquitination, overexpression of GDU1 lysine mutants still conferred amino acid tolerance in a LOG2-dependent manner, consistent with GDU1 being both a substrate and facilitator of LOG2 function. From these data, we offer a model in which GDU1 activates LOG2 to stimulate amino acid export, a process that could be negatively regulated by GDU1 ubiquitination and LOG2 self-ubiquitination.
Keywords: amino acid transport, E3 ubiquitin ligase, protein degradation, ubiquitin, ubiquitin ligase, ubiquitylation (ubiquitination), glutamine dumper, Loss of GDU1 2, amino acid resistance
Introduction
Ubiquitination (or ubiquitylation) is a versatile post-translational protein modification conserved in eukaryotes in which the protein ubiquitin (Ub)6 is covalently linked to intracellular protein substrates on nucleophilic amino acids, typically lysines (1). Different fates await ubiquitinated substrates depending on their location and whether the linked Ub is itself ubiquitinated to form polyubiquitin chains. Monoubiquitination can drive internalization of cell surface transporters (2) and non-proteolytically curtail enzyme activity (3). Lysine 63-linked polyubiquitin chains facilitate lysosomal degradation of transmembrane substrates (4), although lysine 63-polyubiquitinated cytosolic proteins can nucleate signal transduction scaffolds (5). Lysine 48-linked chains usually destine cytosolic, nuclear, and nascent endoplasmic reticulum (ER) proteins to the proteasome (6). In all cases, ubiquitination requires a tripartite enzyme cascade comprising E1, E2, and E3 enzymes. E1s couple ATP hydrolysis with thioesterification of the Ub C terminus to the E1 active site cysteine, then transfer Ub to a corresponding cysteine in the E2 active site. Ubiquitin E3 ligases bind both E2-Ub thioesters and specific protein substrates to catalyze Ub transfer to a substrate nucleophile. Many E3s contain really interesting new gene (RING) domains, which enable interaction with E2-Ub thioesters (7). RING domains coordinate two zinc atoms necessary for activity via positionally conserved cysteines and/or histidines; alanine substitution at these sites abolishes E3 activity (8, 9).
Nitrogen transport between plant organs is necessary for optimal biomass production. However, nutritional excess of one amino acid can feedback-inhibit biosynthesis of shared precursors of other amino acids, which in turn restricts plant growth (10–12). Overexpression of Arabidopsis thaliana GLUTAMINE DUMPER 1 (AtGDU1) enhances non-selective efflux of a subset of proteinaceous amino acids and enables growth amid exogenous amino acid concentrations that ordinarily hinder plant germination and greening (13–15). The term “Gdu1D phenotype” describes the consequences of GDU1 overexpression, which also include decreased height and leaf size (9). A. thaliana GLUTAMINE DUMPER overexpression also causes a Gdu1D phenotype in Nicotiana tabacum (15), suggesting functional conservation in plants. GDUs are intrinsically disordered predicted single-pass transmembrane proteins defined by a five-amino acid motif (Val-Ile-Met-Ala-Gly (VIMAG)) in the cytosolic domain that exhibit high rates of phylogenetic divergence and lack any identified functional motif (16).
Multiple strategies to identify GDU1-interacting proteins uncovered the membrane-localized ubiquitin ligase LOSS OF GDU2 (LOG2) (13). LOG2 was shown to interact with and ubiquitinate the GDU1 cytosolic domain (cGDU1). Notably, the GDU1G100R/log1-1 mutation in the GDU1 VIMAG domain suppresses the Gdu1D phenotype (15) and strongly reduces the LOG2-GDU1 interaction (13). All aspects of the Gdu1D phenotype were lost in log2-reduced and loss-of-function backgrounds (13), indicating the LOG2-GDU1 interaction is necessary for GDU1-mediated amino acid tolerance. Because the Gdu1D phenotype evidently requires LOG2, a simple model in which LOG2 ubiquitinates and down-regulates GDU1 abundance and function was deemed untenable. LOG2 is homologous to MAHOGUNIN RING FINGER 1 (MGRN1), a gene that antagonizes melanocortin signaling and regulates neurogenesis in mammals (17). Whereas GDU orthologs are unknown in mammals, rat MGRN1 ubiquitinated the cytosolic region of GDU1 in vitro and partially complemented a log2 loss-of-function mutation in vivo (18), demonstrating an analogous function between these ubiquitin ligases.
Here, we report that GDU1 stimulates LOG2 enzymatic activity and that LOG2 activity is necessary for GDU1-activated amino acid tolerance. Although both LOG2 and GDU1 are unstable in vivo, they are degraded by distinct proteolytic pathways and have different half-lives. GDU1 abundance is regulated in part by LOG2, and LOG2 ubiquitinates GDU1 at specific lysines in vitro and in planta. Ubiquitination of GDU1 lysines is unexpectedly dispensable for the Gdu1D phenotype, although LOG2 is required in all cases. In conjunction with previous findings, these observations suggest that GDU1 is an adaptor or coactivator that promotes amino acid export by bolstering the E3 activity of LOG2. In this model, LOG2 turnover may be a form of negative feedback regulation.
Results
LOG2 Ubiquitin Ligase Activity, but Not N-Myristoylation, Is Necessary for the Gdu1D Phenotype
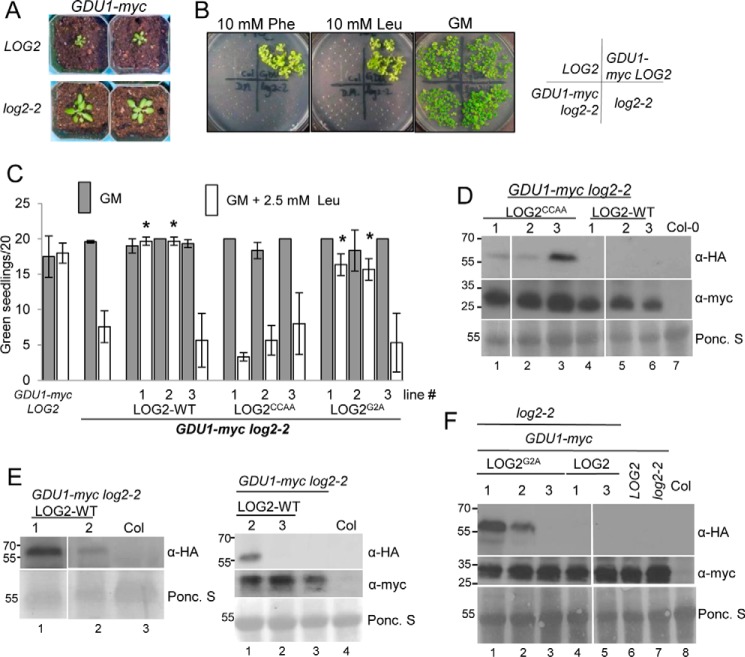
We generated plants that constitutively express GDU1-myc and crossed them to plants homozygous for the log2-2 allele that suppresses detectable LOG2 mRNA accumulation (13). Expression of GDU1-myc in the LOG2 background causes the same phenotypic changes compared with wild-type plants previously observed with the original enhancer-tagged gdu1–1D plants (13, 16), such as small rosette size (Fig. 1A) and resistance to exogenous amino acids (Fig. 1B) in germination media (GM). GDU1-myc log2-2 double homozygotes were significantly larger than GDU1-myc LOG2 plants at an equivalent age (Fig. 1A) and were unable to grow on solid GM supplemented with leucine or phenylalanine, although all lines grew equally well on nonsupplemented solid GM (Fig. 1B). Because the myc epitope enabled measurement of protein accumulation in the presence of LOG2-HA, GDU1-myc plants were used in subsequent experiments.
FIGURE 1.
Wild-type and myristoylation-defective LOG2 proteins restore the Gdu1D amino acid resistance phenotype in GDU1-myc log2-2 plants, but a catalytically inactive LOG2 protein does not. A, 3-week GDU1-myc log2-2 soil-grown plants are larger than age-matched GDU1-myc LOG2 siblings. B, GDU1-myc log2-2, GDU1-myc LOG2, and the non-GDU1-myc lines: control wild-type LOG2 (Col-0) or log2-2 alone seed were plated on GM (right) or GM supplemented with 10 mm phenylalanine (Phe) or leucine (Leu) and photographed after 2 weeks of growth. C, LOG2CCAA-HA, LOG2G2A-HA, and wild-type LOG2-HA transgenes were introduced into the GDU1-myc log2-2 background, triple homozygous seed was plated on GM (filled) or Leu-supplemented GM (open), and the number of green seedlings with expanded true leaves after 10 days of growth at RT were counted. 20 seeds per line were plated in 3 independent experiments. Asterisks indicate significantly different numbers of green seedlings compared with the progenitor GDU1-myc log2-2 line on GM + Leu as assessed by one-way analysis of variance (p < 0.05); all others on GM + Leu were not significantly different from the progenitor. Error bars represent twice the S.E. Seed from GDU1-myc LOG2 (left) and the GDU1-myc log2-2 progenitor (second from left) served as controls. D--F, immunoblots detecting wild-type and mutant LOG2-HA (top panels, anti-HA) and GDU1-myc (middle panels, anti-Myc) protein levels in 10-day-old seedlings grown on GM. Ponc. S (bottom panels), Ponceau stain loading controls. Molecular weight markers in kDa are shown on the left of each blot. Col is wild-type non-transformed control (name of ecotype for Columbia). Genetic background is indicated with bars above the blots. D, protein in LOG2CCAA lines. E, wild-type LOG2 lines. F, LOG2G2A lines. Immunoblots for LOG2CCAA and LOG2G2A lines (D and F) represent signal from 150 μg of total protein. Visualization of LOG2-HA signal (E) required a more sensitive ECL detection kit, longer exposure times, and more total protein compared with D and F (200 or 300 μg for the left and right blot, respectively). White lines in D and F denote removal of uninformative lanes from the same blot.
We previously demonstrated that alanine substitution of two RING domain zinc-chelating cysteines inhibited LOG2 ubiquitin E3 ligase activity (13). To test whether GDU1-mediated amino acid tolerance requires LOG2 enzymatic activity, GDU1-myc log2-2 plants were transformed with wild-type (LOG2-HA) or RING-mutated LOG2C354A/C357A (LOG2CCAA-HA) expression constructs. Homozygous T3 individuals from multiple independent lines were then tested for their ability to grow on GM supplemented with leucine. Two of three LOG2-HA GDU1-myc log2-2 lines (1 and 2) regained the Gdu1D phenotype, producing the equivalent number of true green leaves on GM alone or in the presence of 2.5 mm leucine (Fig. 1C). This leucine concentration was sufficient to significantly decrease growth of progenitor GDU1-myc log2-2 plants (Fig. 1C, left). In contrast, a LOG2CCAA-HA transgene did not restore amino acid resistance for GDU1-myc log2-2 in three independent lines (Fig. 1C) despite LOG2CCAA-HA protein accumulation exceeding that of the wild-type LOG2-HA complementation lines 1 and 2 (Fig. 1D, compare lanes 1–3 with 4 and 5). Notably, longer Western blot exposures were necessary to visualize wild-type LOG2-HA (Fig. 1E), and LOG2-HA protein was not detectable in the LOG2-HA line that failed to restore growth on leucine (line 3, Fig. 1E, right panel), which explains the failure of this line to restore the Gdu1D phenotype. The sensitivities to exogenous leucine seen in GDU1-myc log2-2 and GDU1-myc log2-2 LOG2CCAA-HA lines were not due to loss of GDU1-myc expression (Fig. 1, D–F, middle panels). These results showed that a functional LOG2 RING domain is required for the Gdu1D phenotype and that the C-terminal HA epitope tag does not block in vivo function.
Because LOG2 N-myristoylation facilitates localization to the plasma membrane, one major location of GDU1 (13), GDU1-myc log2-2 plants were also transformed with an N-myristoylation-inhibited construct (LOG2G2A-HA) (13). Two LOGG2A-HA GDU1-myc log2-2 lines were resistant to leucine and expressed detectable LOGG2A-HA protein, whereas a leucine-sensitive line lacked detectable protein (Fig. 1C, right set; protein in Fig. 1F, lanes 1–3). As with LOG2CCAA-HA plants, LOG2G2A-HA accumulated to higher levels than wild-type LOG2-HA (Fig. 1F, compare lanes 1–3 with 4 and 5). These results suggest that LOG2G2A can promote amino acid resistance, but differences in LOG2-HA protein among the lines obtained prevent direct comparisons of the efficacies of wild-type and myristoylation-inhibited LOG2.
GDU1 Protein Stability Is Higher in Plants with Reduced LOG2 Expression
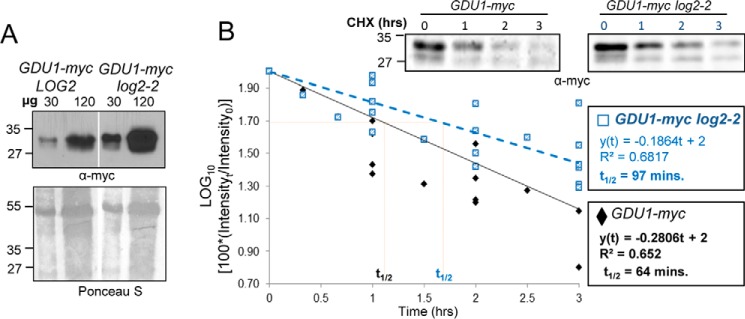
GDU1-myc protein was more abundant in log2-2 than in wild-type (LOG2) backgrounds when directly compared (Fig. 2A), suggesting LOG2 may negatively regulate GDU1-myc abundance in vivo. To determine whether LOG2 affected degradation of GDU1-myc, the half-life of GDU1-myc was determined in F3 seedlings descended from homozygous wild-type (LOG2) and log2-2 F2 siblings expressing GDU1-myc. GDU1-myc was more stable in LOG2-deficient plants (half-life of 64 versus 97 min in LOG2 and log2-2 backgrounds, respectively; Fig. 2B). A one-way analysis of covariance (ANCOVA) statistical test confirmed that the regression lines were significantly different from one another and that data distributions were homogeneous between regressions (supplemental Table 1). GDU1 thus has a longer half-life in the absence of LOG2, suggesting that LOG2 helps to promote GDU1 degradation in vivo.
FIGURE 2.
GDU1-myc steady state levels are higher and degradation is slowed in plants lacking LOG2. A, immunoblot of total protein extracts from GDU1-myc LOG2 (left) and GDU1-myc log2-2 (right) seedlings. Shown is Ponceau S staining for the total protein; molecular weight markers in kDa are on the left. B, semi-log plot of GDU1-myc immunoreactivity in total extracts as a function of time after the addition of the protein synthesis inhibitor CHX to 7-day seedlings from 7 independent experiments. Representative Western blots are shown in the upper right. Linear regressions were drawn in Excel and evaluated by a one-way ANCOVA (supplemental Table 1).
C-terminal Ubiquitination of GDU1 Is Not Sufficient to Bypass the LOG2 Dependence of the Gdu1D Phenotype
Because LOG2 ubiquitin ligase activity is required for the Gdu1D phenotype (Fig. 1) and LOG2 ubiquitinates GDU1 in vitro (13), we asked whether ubiquitination of GDU1 is sufficient for the phenotypic changes observed upon GDU1 overexpression. The requirement for an E3 ligase can be bypassed in some cases if a substrate protein is “ubiquitinated” by virtue of a genetically encoded attached ubiquitin rendering the requirement for post-translational ubiquitin addition by a ligase superfluous (19). For this test we generated transgenic plants expressing GDU1-HA-Ub, a translational fusion of full-length GDU1 with an HA epitope followed by a complete ubiquitin open reading frame. Detection of anti-HA protein from two independent GDU1-HA-Ub lines indicated the fusion protein was expressed, and the presence of high molecular weight forms suggested it is ubiquitinated (Fig. 3A). These two independent lines were then crossed to log2-2, and double homozygous sibling log2-2 and LOG2 lines expressing the GDU1-HA-Ub transgene were generated. The GDU1-HA-Ub fusion was biologically active, as expression conferred resistance to 10 mm leucine in LOG2 seedlings. However, leucine resistance was completely lost in the log2-2 siblings (Fig. 3B). All plants grew equally well on media without amino acid supplementation (Fig. 3B). Thus, a translational fusion of ubiquitin to the GDU1 C terminus is not sufficient to circumvent the role of LOG2 in GDU1-mediated resistance to exogenous amino acids.
FIGURE 3.
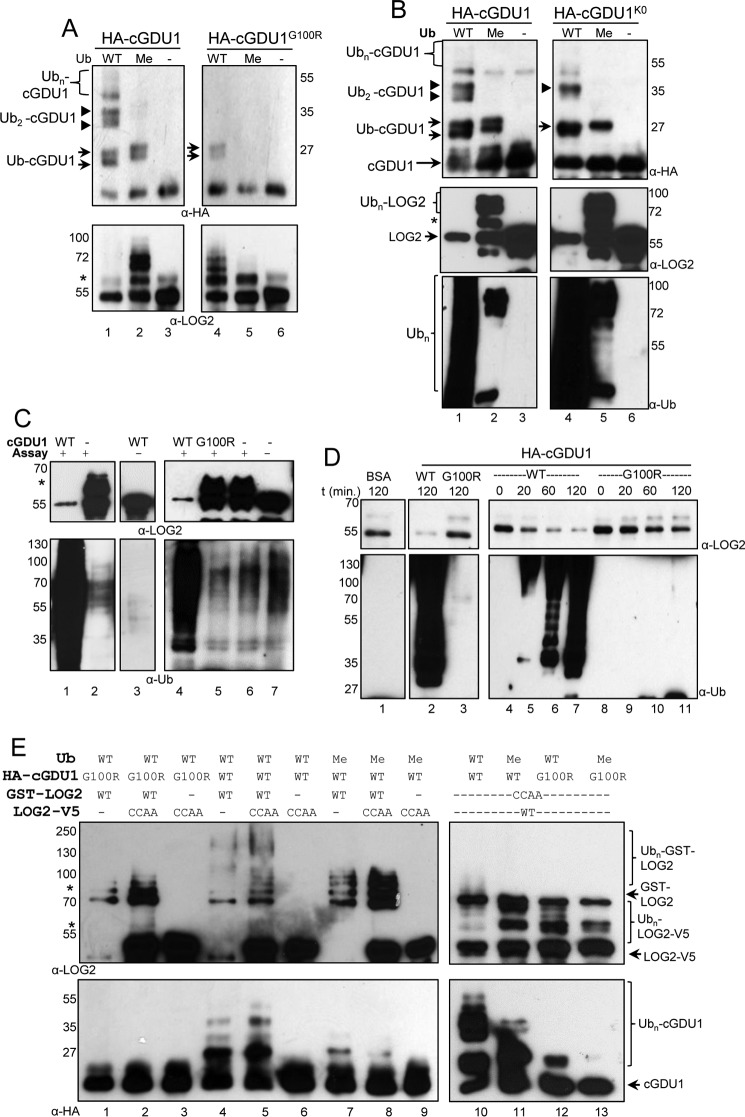
Genetically encoded ubiquitination failed to bypass a requirement for the E3 LOG2 in conferring amino acid resistance. A, two independent GDU1-HA-Ub lines (1 and 2) were crossed to log2-2, and analysis was performed with homozygous GDU1-HA-Ub LOG2 and GDU1-HA-Ub log2-2 F3 siblings. Shown is Western blot of ∼30 μg of total protein from 7-day-old seedlings separated by SDS-PAGE and probed with anti-HA antibodies. The arrow denotes unmodified GDU1-HA-Ub. Top panel, equal exposure indicating different levels of expression between the two independent lines; middle panels, separate exposures for each line for optimal visualization; bottom panel, Ponceau (Ponc. S) loading control. B, same assay as described in Fig. 1C, with the GDU1-HA-Ub expressing transgenic lines in LOG2 and log2-2 backgrounds. Wild-type (Col) without the GDU1-HA-Ub transgene served as the controls. Analysis was conducted as in Fig. 1C.
LOG2 Ubiquitinates GDU1 on Lysines in Vitro and in Planta
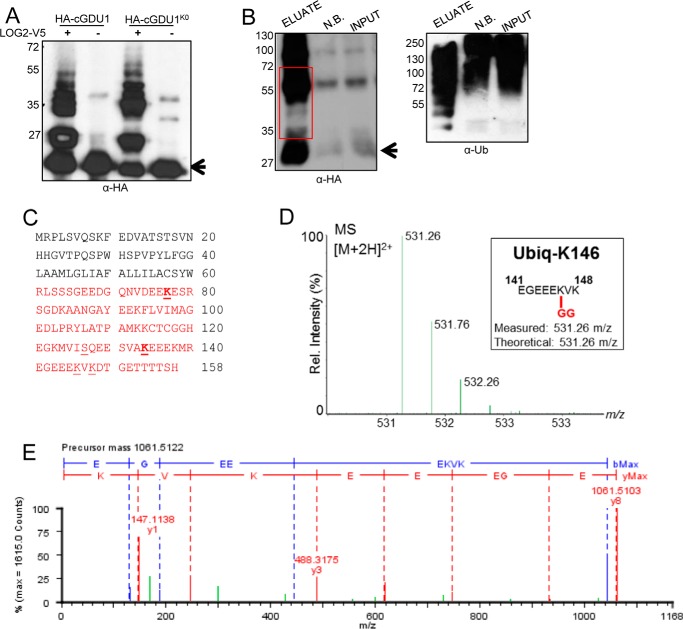
LOG2 was previously shown to ubiquitinate the cytosolic domain of GDU1 (cGDU1) in vitro (13). Because genetically encoded ubiquitination was not sufficient to confer LOG2 independence (Fig. 3), we hypothesized that the position of GDU1 ubiquitination might be critical for its biological activity. To address this possibility, we sought to identify ubiquitination sites on GDU1. In vitro ubiquitination assays were conducted in the presence or absence of V5-tagged LOG2, with recombinant, purified N-terminal His6-HA-tagged cGDU1 as the substrate. The HA epitope was chosen for its lack of lysines to ensure that the only lysine acceptor sites in the recombinant protein were those encoded by the native coding sequence. Control reactions contained HA-cGDU1K0, a mutant in which all lysines were substituted with arginine, an amino acid that cannot be ubiquitinated. Surprisingly, LOG2 exhibited ubiquitin ligase activity toward both wild-type and lysine-free cGDU1 (Fig. 4A), suggesting that LOG2 may ubiquitinate cGDU1 at non-lysine sites. Reactions were then analyzed by electrospray ionization tandem mass spectrometry (ESI-MS/MS). Proteomic analysis of ubiquitination assay milieus achieved 100% sequence coverage of both proteins. Ubiquitinated peptides were recovered corresponding to four ubiquitination sites in the wild-type protein (lysines 77, 134, 146, and 148 in the GDU1 coding sequence) (Fig. 4C, representative spectra in Fig. 4, D and E). A single residue (serine 127) was ubiquitinated in the lysine-free mutant (Fig. 4C, underlined), and no ubiquitin modification was detected at the N-terminal amino group or of any other amino acid R-group of HA-cGDU1. These data indicate that cGDU1 is ubiquitinated by LOG2 at specific residues in vitro. Moreover, modification at serine 127 in the lysine-free, but not wild-type HA-cGDU1 (Fig. 4C), suggests that LOG2 preferentially transfers ubiquitin to lysine residues but can utilize serine 127 when lysines are absent.
FIGURE 4.
GDU1 is ubiquitinated at lysines in vitro and in planta. A, in vitro ubiquitination of HA-cGDU1-His6 (wild-type or the K0 mutant lacking all cytosolic lysines) by V5-tagged LOG2. − lanes contain all ubiquitination assay components except LOG2. B, nickel-NTA affinity purification of His6-HA3-GDU1 from transiently transformed N. benthamiana leaves co-infiltrated with LOG2-HA. Input, cell-free supernatant after tissue lysis. NB, not bound (protein that did not bind nickel beads). Eluate, protein eluted from nickel-NTA beads with 350 mm imidazole and heat. The red box in B refers to the region on the corresponding SDS-polyacrylamide gels subjected to in-gel trypsinization and subsequent electrospray ionization-MS/MS tandem mass spectrometry to identify ubiquitination sites. Arrows in A and B denote un-modified HA-cGDU1-His6 and His6-HA3-GDU1, respectively. Numbers to the left of Western blots indicate protein mass in kDa. C, GDU1 amino acid sequence with mass spectrometry-identified ubiquitination sites indicated with underlines (identified in in vitro assays only) or bold underlines (identified in both in vitro and in planta assays). Red text demarcates the cGDU1 region in the GDU1 protein sequence. Serine 127 (underlined) was identified as the sole in vitro ubiquitination site in HA-cGDU1K0-His6. D and E, mass spectra for the parent ion (D) and MS/MS fragmentation (E) indicating in vitro ubiquitination of Lys-148 (ubiquitinated peptides in green).
To substantiate the in vitro ubiquitination assay, we next asked whether LOG2 ubiquitination of GDU1 also occurs at specific sites in plants. LOG2 and full-length His6-HA3-tagged GDU1 were transiently expressed in Nicotiana benthamiana leaves. The hexahistidine tag allowed for efficient precipitation of GDU1 from crude lysates of infiltrated leaves with nickel-conjugated (NTA) Sepharose beads, as evidenced by the enrichment of GDU1-HA immunoreactivity in Western blots (Fig. 4B). Subsequent proteomic analysis achieved 79% sequence coverage (the missing 21% comprising the His6-HA3 tag, which does not contain any lysines) and uncovered two of the same ubiquitinated sites found in the in vitro assays: GDU1 lysines 77 and 134 (Fig. 4C, bold). No additional modified residues were detected.
GDU1 Lysines Are Not Necessary for the LOG2-dependent Gdu1D Phenotype
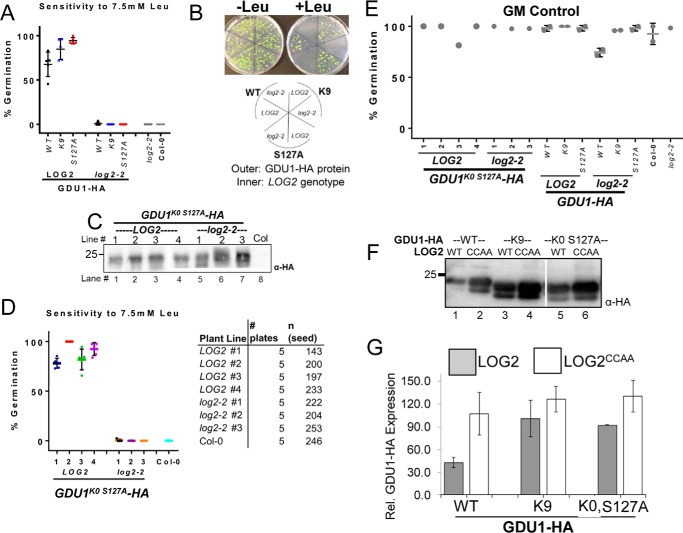
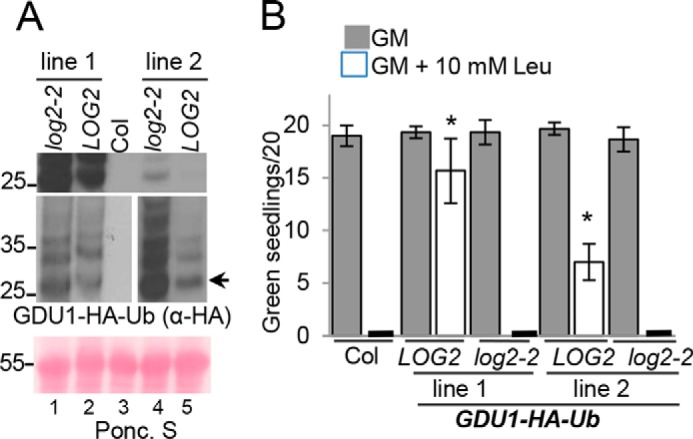
After demonstrating that GDU1 is ubiquitinated, we wanted to test whether ubiquitination is important for the Gdu1D phenotype. Multiple transgenes encoding C-terminal HA-tagged GDU1 proteins with lysine to arginine and/or serine 127 to alanine substitutions were generated and stably expressed in transgenic plants, and multiple independent lines were tested for their ability to confer amino acid resistance during germination as performed previously (Fig. 1, B and C, and Fig. 3B; Ref. 13). The proteins tested were: 1) GDU1 with all cytosolic lysines substituted, retaining only extracellular lysine 9 (GDU1K9-HA); 2) GDU1 with serine 127 substituted (GDU1S127A-HA); 3) the combination of these substitutions (GDU1K9 S127A-HA); 4) GDU1 with all lysines and serine 127 substituted (GDU1K0, S127A-HA). Homozygous T3 seedlings from two independent lines per construct were assessed for their resistance to supplemental leucine. Surprisingly, all lines were resistant to 5.0 and 7.5 mm leucine (Fig. 5, B and C), much like GDU1-myc plants, whereas control wild-type progenitor (Col-0) was completely sensitive (Fig. 5B). One GDU1S127A-HA line (line 1) exhibited <100% growth, but this also occurred on GM control plates and was hence unrelated to amino acid sensitivity.
FIGURE 5.
GDU1 lysines and serine 127 are not required for amino acid resistance. A–C, 7-day-old seedlings expressing different GDU1 proteins with lysine and/or serine 127 substitutions to arginine and alanine, respectively, were grown on GM alone (A) or GM supplemented with 5 mm leucine (B) or 7.5 mm leucine (C) to test amino acid tolerance. The experiment was performed as described in Fig. 1C, except here is expressed as the percentage of green seedlings from 5–7 plantings of ∼25 seedlings from 2 representative independent experiments. Error bars denote twice the S.E. Numbers directly below the graphs denote transgenic line number. GDU1-myc and Col-0 seed served as positive and negative controls, respectively. Italicized abbreviations refer to plants that express the following GDU1-HA transgenes. WT, wild-type GDU1-HA. K9, GDU1-HA in which all lysine codons have been mutated to arginine codons except lysine 9. S127A, GDU1-HA in which the serine 127 codon has been mutated encode alanine. K9 S127A, GDU1-HA in which all lysine codons have been mutated to arginine codons and the serine 127 codon has been mutated to encode alanine. K0 S127A, GDU1-HA in which all lysine codons have been mutated to arginine codons and the serine 127 codon has been mutated encode alanine. D, effect of the lysine/serine substitutions on GDU1 electrophoretic behavior. Extracts of T3 seedlings (50 μg of protein) expressing the indicated GDU1-HA proteins in the wild-type background, visualized with an anti-HA antibody. The thin white line between lanes 1 and 2 represents removal of a non-informative band. Short exposure (right panel) is a shorter exposure of lanes 2 and 3 to demonstrate the difference in laddering between GDU1K9-HA and GDU1S127A-HA.
To verify that these substitutions affected in vivo GDU1 ubiquitination, protein extracts were prepared from seedlings expressing these proteins and immunoblotted for GDU1-HA. Although wild-type GDU1-HA and GDU1S127A-HA extracts exhibited electrophoretic “ladders” consistent with ubiquitination, GDU1K9-HA did not (Fig. 5D, compare lanes 1 and 2* with lane 3). In agreement with mass spectrometry experiments, these data suggest GDU1 is ubiquitinated on cytosolic lysines in planta and that serine 127 ubiquitination is not extensive in Arabidopsis.
Plants that express wild-type GDU1-HA, GDU1K9-HA, or GDU1S127A-HA were crossed to log2-2 to determine whether these altered GDU1 proteins required LOG2 to confer amino acid resistance. Homozygous LOG2 and log2-2 F3 seedlings were obtained from each cross. Resistance to leucine was lost in the log2-2 background even though LOG2 siblings from the same cross were resistant (Fig. 6, A and B). These results indicate that lysines in GDU1 and their ubiquitination are not necessary for amino acid resistance.
FIGURE 6.
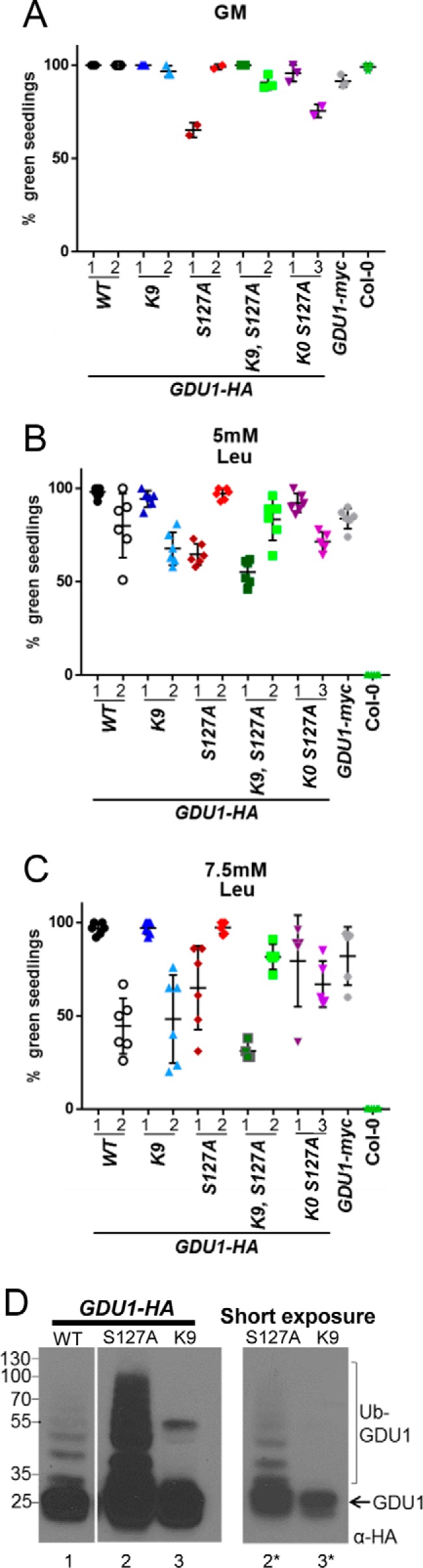
Substitution of GDU1 lysines did not affect amino acid resistance or its dependence on LOG2. A, germination percentages of F3 seed homozygous for various HA-tagged GDU1 variants on GM supplemented with 7.5 mm leucine in either the homozygous LOG2 (wild-type) or log2-2 background. GDU1 protein designations are as in Fig. 5A. The graph represents three biological replicates (each replicate is a percent germination from a plating of 20–40 seedlings with 1–3 technical replicates) of progeny of double homozygous F2 siblings that express the same GDU1-HA transgene in either the wild-type (LOG2) or the log2-2 background. B, representative plates (top) and legend (bottom) used to generate the data in A. C, GDU1K0 S127A-HA expression levels in 7 independent transgenic lines (4 in the wild-type (LOG2) and 3 in the log2-2 backgrounds) from 10 μg total protein from 14-day-old seedlings visualized with anti-HA antibody. The thin white line between lanes 3 and 4 indicates removal of a non-informative lane; all samples are from the same gel and exposure. D, germination percentage of multiple independent lines (shown in C) expressing GDU1K0 S127A-HA in the wild-type (LOG2) or log2-2 background on 7.5 mm leucine-supplemented media (left). Data represent five replicates, each replicate consisting of 20–50 seed (right). E, germination rates on GM for plant lines in A and D. F, co-expression of wild-type or mutant GDU1-HAs and wild-type or enzymatically inactive (CCAA) LOG2 in N. benthamiana leaves. G, densitometry of immunoreactivities in F. Intensities of wild-type and mutant GDU1-HA bands were quantified with a CCD camera and normalized to the average HA immunoreactivity for each blot. Data are representative of three infiltrations. Error bars correspond to twice the S.E.
To test whether serine 127 or the predicted extracellular lysine could substitute for the mutagenized cytosolic lysines, GDU1K0 S127A-HA was expressed in the log2-2 background. Expression of the transgene was confirmed by Western blotting and multiple lines expressing approximately equivalent or more GDU1K0 S127A-HA in log2-2 than in LOG2 (Fig. 6C) were chosen for further testing. Four GDU1K0 S127A-HA lines in LOG2 background exhibited robust leucine resistance, whereas three lines in the log2-2 background were indistinguishable from Col-0 (Fig. 6D). Importantly, all plants germinated well on GM (Fig. 6E). In summary, GDU1 lysines and serine 127 were found dispensable for the Gdu1D phenotype, but LOG2 was necessary in all cases.
LOG2-promoted GDU1 Instability and Protein Modification Partially Depended on GDU1 Lysines
If ubiquitination of GDU1 is dispensable for the Gdu1D phenotype, then what purpose does it serve? Because LOG2 affects GDU1 stability in vivo (Fig. 2), we tested the hypothesis that lysine ubiquitination and stability are causally linked. GDU1-HA, GDU1K9-HA, and GDU1K9 S127A-HA expression constructs were transiently co-expressed in N. benthamiana with untagged active LOG2 or the catalytically inactive LOG2CCAA as control to assess in vivo accumulation. Wild-type GDU1-HA abundance was significantly higher when co-expressed with LOG2CCAA than with active LOG2 (Fig. 6F, lanes 1 and 2, quantitated from three independent experiments in Fig. 6G), suggesting that increased degradation contributed to lower steady state levels. GDU1K9-HA abundance was not as sensitive to LOG2 catalytic activity (Fig. 6, F, lanes 3 and 4, and G). Further substitution of lysine 9 and serine 127 (GDU1K0 S127A-HA) (Fig. 6F, lanes 5 and 6, and 6G) did not affect the relative accumulation. These data suggest GDU1 stability is partially regulated by LOG2-dependent lysine ubiquitination.
LOG2 Ubiquitinates Itself and GDU1 with Different Degrees of Selectivity in Vitro
Our preceding results indicated that LOG2 enzymatic activity and the interaction between LOG2 and GDU1 are both necessary for the Gdu1D phenotype, whereas GDU1 ubiquitination is not. To better understand the biochemistry of the LOG2-GDU1 protein-protein interaction, in vitro ubiquitination assays were conducted with either wild-type ubiquitin or methylated ubiquitin (Me-Ub), which cannot form polyubiquitin chains. Consistent with prior results (13), electrophoretic laddering of HA-tagged cGDU1 was observed with wild-type ubiquitin, which could indicate ubiquitination at multiple lysines and/or polyubiquitination at one site (Fig. 7, A and B, lane 1, upper panels). In contrast, Me-Ub only permitted monoubiquitination of cGDU1 (Fig. 7, A and B, lane 2, upper panels). Ubiquitination at different lysines affords similar but not identical electrophoretic mobilities; closer inspection of Western blots revealed that ubiquitinated cGDU1 species comprised doublets, likely composed of cGDU1 ubiquitinated at either lysine 77 or 134 (Fig. 7, A and B, lanes 1 and 2, upper panels, highlighted by arrows). A monoubiquitinated doublet was also observed in reactions that contained HA-tagged cGDU1G100R (Fig. 7A, lane 4, upper panel), which is partially impaired in its capacity to interact with LOG2 (13). Lysine-free cGDU1K0 exhibited a single monoubiquitinated form with Me-Ub (Fig. 7B, lanes 4 and 5, upper panel), likely corresponding to serine 127. These results suggest that LOG2 selectively ubiquitinates GDU1 at one lysine per GDU1 protein molecule.
FIGURE 7.
LOG2 ubiquitinates itself and GDU1 by different mechanisms, and GDU1 stimulates LOG2 activity in vitro. A, immunoblots of cGDU1 (top, anti-HA) or LOG2 (bottom, anti-LOG2) from ubiquitination assays of HA-cGDU1 and HA-cGDU1G100R (encoded by the log1-1 allele) by LOG2-V5 in the presence of wild-type ubiquitin (WT) or methylated ubiquitin (Me) lacking acceptor lysines for ubiquitin chain formation. Small arrows and arrowheads in A and B indicate mono- and di-ubiquitinated cGDU1, respectively. B, ubiquitination of HA-cGDU1 and HA-cGDU1K0 (lysine-less) by LOG2-V5 in the presence of wild-type ubiquitin (WT) or methylated ubiquitin (Me) and immunoblotted for cGDU1 or LOG2 as in A (top and middle panels). The same reactions were immunoblotted for ubiquitin (bottom panel, anti-ubiquitin antibody). The asterisk indicates monoubiquitinated LOG2. C, LOG2 E3 ligase activity is stimulated by the binding of cGDU1. The plus sign indicates inclusion of E1, E2, Mg-ATP, and Ub. WT, FLAG-cGDU1; log1-1, FLAG-cGDU1G100R. D, time course of cGDU1-activated LOG2 ubiquitination activity. BSA, reaction contained 1 mg·ml−1 bovine serum albumin. E, LOG2 self-ubiquitination occurs in cis but not in trans. Assays contained both wild-type and enzymatically inactive (CCAA) LOG2 (with either a GST or a -V5 tag to allow for adequate electrophoretic separation of the two forms of LOG2) in the presence of HA-cGDU1 (wild-type or the G100R mutant). For all immunoblots, an aliquot of the reaction mixture was fractionated by SDS-PAGE, and proteins were visualized by immunoblotting with the indicated antibody. In all instances, higher migrating forms of the active ligase (but not the inactive ligase) can be observed, indicating LOG2 can promote intramolecular self-ubiquitination (cis-ubiquitination) but not intermolecular (trans-) self-ubiquitination. Single and double asterisks highlight monoubiquitinated LOG2-V5 and GST-LOG2, respectively.
In addition to facilitating ubiquitin transfer to substrates, E3 ligases can also self-ubiquitinate (20, 21). LOG2 self-ubiquitination was assessed with an antibody raised against LOG2 peptides. Unlike cGDU1, LOG2 formed multiple higher molecular mass conjugates in the presence of Me-Ub (Fig. 7A, lane 2, lower panel; Fig. 7B, lanes 2 and 5, middle panel). This indicates that LOG2 self-ubiquitination is less selective than for GDU1 and occurs at multiple sites per LOG2 molecule.
LOG2 cis-Autoubiquitination Is Stimulated by GDU1 in Vitro
LOG2 self-ubiquitination is strikingly robust in the presence of cGDU1 such that unmodified LOG2 is depleted from the reaction milieu (Fig. 7B, middle panel, compare lanes 1 and 3; Fig. 7, C and D, top panels). Compared with wild-type cGDU1, interaction-impaired cGDU1G100R triggers much lower total ubiquitination activity of LOG2 (assessed with anti-Ub blots) and self-ubiquitination (inferred by depletion of unmodified LOG2) (Fig. 7C, compare lanes 4 and 5). The same result was observed in time course assays; unmodified LOG2 was rapidly depleted from reactions containing wild-type cGDU1 but not cGDU1G100R (Fig. 7D, top panel, compare lanes 4–7 with 8-11). The observed LOG2 depletion likely reflects epitope masking by polyubiquitination and poor transfer of high molecular weight proteins to PVDF membranes (22). Bovine serum albumin had no further stimulatory effect on LOG2 E3 ligase activity than did cGDU1G100R (Fig. 7D, lane 1). These data indicate that LOG2 autoubiquitination is specifically stimulated by cGDU1 binding and does not result from a protein crowding effect.
Autoubiquitination can occur both in cis (an E3 ligase intramolecularly ubiquitinates itself) and in trans (an E3 ligase intermolecularly modifies another molecule of the same E3 ligase) (23, 24). To distinguish between these mechanisms, in vitro ubiquitination assays were conducted with GST-tagged wild-type LOG2 and V5-tagged LOG2CCAA in the same reaction. The molecular mass difference between these two tags allowed for simultaneous visualization of both proteins. Wild-type GST-LOG2 displayed, respectively, moderate and robust self-ubiquitination in the presence of cGDU1G100R and wild-type cGDU1 (Fig. 7E, top panel, compare lanes 1 and 2 with 4 and 5). Me-Ub permitted GST-LOG2 multi-monoubiquitination (Fig. 7E, top panel, lanes 7 and 8). In contrast, LOG2CCAA-V5 modification was not apparent under any condition (Fig. 7E, upper panel, compare lane 2 with 3, lane 5 with 6, and lane 8 with 9). Similar results were obtained when the V5 and GST tags were swapped between the active and inactive proteins (Fig. 7E, top right panel). Because electrophoretic laddering of LOG2CCAA was never observed, we conclude that LOG2 does not autoubiquitinate in trans. Instead, these data indicate that LOG2 catalyzes intramolecular, cis autoubiquitination and that this activity is stimulated by interaction with GDU1.
LOG2 and GDU1 Are Degraded by Distinct Proteolytic Pathways in Vivo
Ubiquitin ligase self-ubiquitination and subsequent proteasomal degradation are known mechanisms for feedback inhibition (23, 24). Given the rate of self-ubiquitination in vitro, we hypothesized that LOG2 is rapidly degraded in plants. To measure LOG2 stability, the protein synthesis inhibitor cycloheximide (CHX) (25, 26) was applied to 7-day-old T3 seedlings of two homozygous LOG2-HA complementation lines in the GDU1-myc log2-2 background (described in Fig. 1). LOG2-HA was undetectable in whole plant lysates 1 h after CHX treatment, whereas pretreatment with the proteasomal inhibitor MG132 dramatically slowed LOG2 degradation (Fig. 8A, upper panels). In contrast, GDU1-myc degradation was more gradual and was not stabilized by MG132 (Fig. 8A, lower panels). In a second experiment, seedlings were treated with DMSO or MG132 for 4 h or with CHX for 1 h, after which microsomes membranes were harvested and LOG2-HA levels were examined. As in whole plant lysates, MG132 pretreatment promoted membrane-localized LOG2 accumulation relative to solvent control, whereas LOG2 was undetectable 1 h after CHX treatment (Fig. 8B). These data indicate LOG2 is rapidly degraded in vivo by the proteasome, whereas GDU1 degradation occurs more slowly and by a different mechanism.
FIGURE 8.
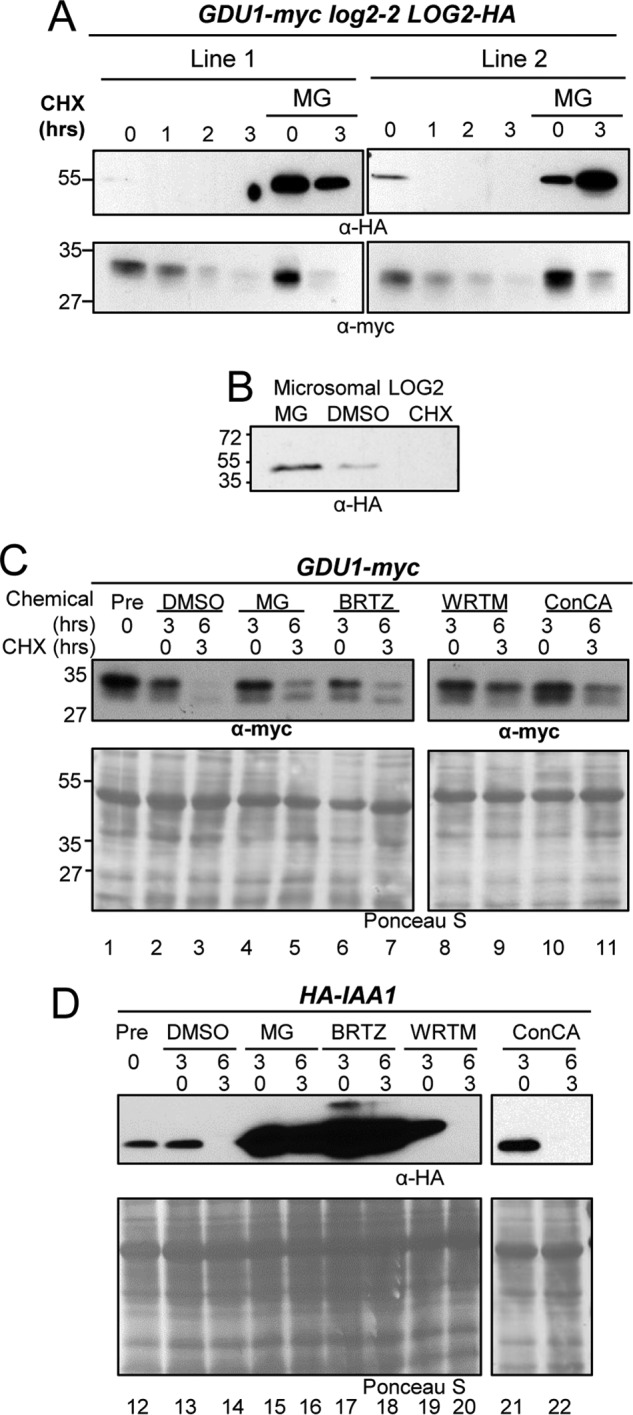
LOG2 and GDU1 are degraded by different proteolytic pathways in vivo. A, LOG2-HA expressed in GDU1-myc log2-2 LOG2-HA complementation lines (visualized from total lysates from seedlings after 7 days in liquid GM media by immunoblotting) after treatment with protein synthesis inhibitor CHX or with a 2-h pretreatment with 100 μm MG132. B, microsomal LOG2-HA from 7-day-old seedlings stable transgenic Arabidopsis were treated with 100 μm MG132 or 1% (v/v) DMSO for 4 h or 200 μg/ml CHX for 1 h before microsomal purification. Equal total microsomal protein was fractionated by SDS-PAGE, and LOG2-HA was visualized with anti-HA antibodies. C, 7-day-old seedlings grown in liquid GM media were treated with solvent control (DMSO), proteasome inhibitors (MG132 (MG) or bortezomib IBRTZ)), wortmannin (WRTM, a vacuolar maturation inhibitor), or concanamycin A (ConCA, which inhibits vacuolar acidification) for 3 h before the addition of CHX. D, same as C with seedlings that express HA-IAA1, an HA-tagged auxin response transcriptional repressor IAA1, a known proteasomal target. Equal total protein was loaded, and immunoblot analysis was performed using anti-myc (for GDU1) or anti-HA (for HA-IAA1). Ponceau S staining indicates total protein.
In addition to the proteasome, eukaryotic cells possess another means of regulated proteolysis: the vacuole (or lysosome). Because vacuolar proteolysis is more common among endomembrane-resident proteins, we hypothesized that GDU1 is degraded by the vacuole (4, 27, 28). To test this hypothesis, GDU1-myc and GDU1-myc log2-2 seedlings were treated with proteasomal and vacuolar protease inhibitors before protein synthesis inhibition with CHX (Fig. 8C). Seedlings expressing HA-tagged IAA1, a well characterized proteasome substrate (29), were treated in parallel (Fig. 8D). GDU1-myc degradation was substantially slowed by treating plants with wortmannin, which inhibits trans-Golgi network (TGN)-to-vacuole trafficking (30), and concanamycin A, which inhibits vacuolar acidification (31) (Fig. 8C, lanes 8 and 9 and lanes 10 and 11). HA-IAA1 was not detectably stabilized (Fig. 8D, lanes 19–22). In contrast, pretreatment with proteasome inhibitors MG132 (32) and bortezomib (33) markedly stabilized HA-IAA1 (Fig. 8D, lanes 15–18) but only marginally slowed GDU1-myc degradation (Fig. 8C, lanes 4–7). Neither protein was stabilized by the DMSO solvent control (Fig. 8, C and D, lanes 2–3 and 13–14 for GDU1-myc and HA-IAA1, respectively). These results suggest GDU1 is primarily degraded by the vacuole/lysosome in vivo.
Discussion
Amino Acid Export May Be Controlled by a GDU1-activated LOG2 E3 Ligase Complex
Enzymatically inactive LOG2CCAA cannot promote the GDU1 overexpression phenotype in the GDU1-myc log2-2 background despite being present at higher levels than wild-type LOG2 (Fig. 1). This highlights the importance of LOG2 ubiquitin ligase activity in regulating amino acid export and homeostasis upon GDU1 overexpression. LOG2 clearly facilitates GDU1 degradation in vivo (Fig. 2 and Fig. 6, F and G) via ubiquitination, although GDU1 ubiquitination was not required for amino acid tolerance (Fig. 3 and Fig. 5, A--C).
Based on the data presented here we propose a model in which additional GDU1 expression recruits and over-activates LOG2 activity toward an unknown target protein substrate (represented as X), the ubiquitination of which leads to enhanced amino acid export by an unknown mechanism (Fig. 9). It should be noted that the Gdu1D phenotype is not completely suppressed by LOG2 knock-out; amino acid content and uptake are not back to wild-type levels (13).7 This suggests that other proteins, possibly the other LOG2-family members (13) that interact with GDU1, also ubiquitinate the target protein X. Because these proteins play a minor role relative to LOG2, they have been omitted from the model.
FIGURE 9.
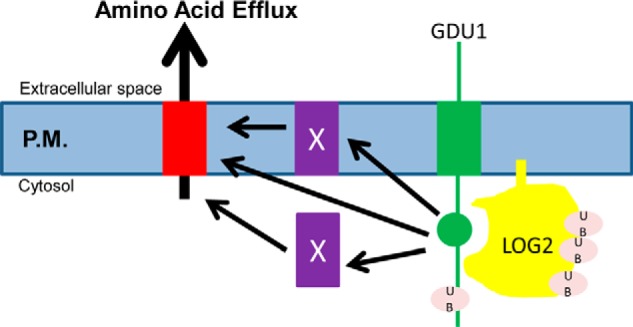
A model for the dependence of the Gdu1D phenotype on LOG2 ubiquitin ligase activity. In the presence of LOG2 (yellow), GDU1 (green) overexpression activates an amino acid transport system either by directly associating with an amino acid exporter (red), by functioning through as yet-undiscovered facilitator(s) (X), or by down-regulating an inhibitor of the transporter. This process depends in part on GDU1-activated LOG2 ubiquitination activity. GDU1 may also be an adaptor protein that aids in the recognition of amino acid exporters or unknown inhibitors/facilitators. P. M., plasma membrane.
In this model, GDU1 would function similarly to arrestins, which mediate endocytosis in yeast by recruiting E3s to integral membrane protein substrates (34). GDU1 is then an “incidental” LOG2 substrate that is stochastically ubiquitinated and degraded due to its association with the ubiquitin ligase even if this is not its major role. Multiple lines of evidence support this hypothesis. GDU1 ubiquitination appears to play no role in conferring resistance to the toxic effects of exogenous leucine. First, genetically encoded ubiquitination is not sufficient for the Gdu1D phenotype in the absence of LOG2 (Fig. 3). Furthermore, GDU1 variants that are not detectably or are weakly ubiquitinated still require LOG2 for amino acid resistance (Figs. 5 and 6).
LOG2 self-ubiquitination markedly increased upon binding the GDU1 cytosolic domain, suggesting that GDU1 modulates LOG2 activity (Fig. 7). Characterized adaptor proteins are known to enhance ubiquitin ligase activity of their cognate E3s. In metazoans, binding of small mothers against decapentaplegic 7 (Smad7) to the HECT-type E3 ligase SMad-specific ubiquitin ring finger 2 (Smurf2) relaxes an autoinhibitory domain, thereby promoting stronger binding of ubiquitin conjugating enzymes (35). A similar mechanism was demonstrated for activation of the E3 ligase atrophin-1-Interacting protein 4 (AIP4) by the adapter protein SPARTIN (36).
Robust LOG2 self-ubiquitination, evidenced by multi-monoubiquitination in the presence of Me-Ub (Fig. 7) and rapid in vivo degradation (Fig. 8A), may serve to prevent pathway overstimulation. Such negative feedback would be biologically useful if GDU1 is a LOG2 coactivator or substrate adaptor. Indeed, CULLIN-type E3 ligases ubiquitinate both their substrates and themselves more efficiently in the presence of substrate adaptors (23). Although GDU1 appears to be a vacuolar protease substrate, LOG2 is probably turned over by the proteasome (Fig. 8). This is reasonable given that GDU1 is an integral membrane protein, whereas LOG2 is a soluble protein anchored to the membrane via a myristyl modification (13, 16).
GDU1 is a member of a multigene family of intrinsically disordered proteins whose primary sequences vary considerably across taxa (15). Although GDU homologs are not found in mammals (18), analogous intrinsically disordered proteins such as polyQ protein inclusions have been shown to regulate MGRN1 function (37). Identification of additional in vivo LOG2 ubiquitination targets will help reveal the scope of processes regulated by LOG2, particularly the extent of conservation between LOG2 and MGRN1 and the breadth of the GDU1-LOG2 pathway for amino acid export in plants.
Experimental Procedures
Cloning and Constructs
Primer sequences are available upon request. Construction of cauliflower mosaic virus 35S promoter (35S)-GDU1-myc and HA-tagged LOG2 transgenes including LOG2G2A (G2A, myristoylation-inhibited) and LOG2C354A/C357A (CCAA) mutants was described previously (13). pET-DEST 42_LOG2CCAA-V5-His6 was made from Gateway-cloning of pDONR201_LOG2CCAA. Sequences encoding full-length GDU1 cytosolic lysine (retaining Lys-9) and serine mutants and lysine mutants of the cytosolic GDU1 domain (cGDU1, comprising amino acids 61–158) were constructed by the Kunkel mutagenesis method (38). Constructs to express N-terminally HA-tagged cGDU1, cGDU1K0 (lacking all cytosolic lysines), and cGDU1G100R were made with primers adding 5′ Shine-Dalgarno and HA epitope-encoding sequences using the respective pDONR-Zeo constructs (13) as templates. Resulting products were Gateway-cloned into pDONR201 and pET-DEST42 (Thermo Fisher Scientific). Full-length GDU1 ORFs with serine 127 codon substituted with an alanine codon or the above additionally with all lysine codons substituted with arginine codons were purchased from Genewiz.
The Arabidopsis ubiquitin 10 (UBQ10) promoter-driven His6-HA3-GDU1 construct (used to identify in planta ubiquitination by mass spectrometry) was made with primers that added a 5′ NdeI site in-frame with the start codon and a 3′ BamHI site after the stop codon. Amplification products were cloned into a modified pGREENII plasmid (p3756) containing a 5′ proUBQ10:: His6-HA3 cassette (39). The GDU1-HA C-terminal ubiquitin fusion construct (GDU1-HA-Ub) was made by overhang extension PCR using pEARLEYGATE_HA-Ub derived from an HA4-Ub plasmid (40) as template. The ORF was subcloned from pDONR-Zeo (13) into the plant expression vector pSWNkan,8 under the CsVMV promoter (41). For transient expression experiments, wild-type and mutant GDU1 was cloned by restriction digestion-ligation into pPHTkan, whereas untagged LOG2 was cloned by Gateway cloning into pPWThyg (vector sequence and information available upon request); in both vectors, the constructs are under the control of the cauliflower mosaic virus 35S promoter.
A Gateway-type vector with an Arabidopsis UBQ10 promoter (pGWUBQ10) was made from pGWB14 (42) as follows. The UBQ10 promoter (1003 bp of genomic DNA upstream of the first intron in the 5′-UTR of the UBIQ10 gene) was PCR-amplified from p2543 (43) to include 5′ and 3′ HindIII and NheI sites. The amplicon was cloned into pGWB14 (excised of the 35S promoter). UBQ10 promoter-driven LOG2-HA, LOG2G2A-HA, and LOG2CCAA-HA plant expression vectors were made by Gateway cloning LOG2 coding sequences into pGWUBQ10.
Crosses and Transformations of A. thaliana and N. benthamiana
GDU1-myc log2-2 Arabidopsis were generated from a cross between log2-2 and 35S::GDU1-myc plants (13). These plants were transformed with wild-type LOG2, LOG2G2A, and LOG2CCAA expression constructs described above by the floral dip method (44) using Agrobacterium tumefaciens strain AGL1 (45). Col plants were similarly transformed with wild-type and mutant GDU1-HA constructs and were crossed to log2-2. Homozygosity was confirmed by PCR and antibiotic selection. F3 and T3 material was used for all analyses. The genotype of the plants was confirmed by LOG2 PCR genotyping (13) and sequencing the GDU1 ORF obtained by PCR from plant genomic DNA.
Transient expression assays were conducted with leaves of 4- to 6-week N. benthamiana. Plants were co-transformed with suspensions of one or more AGL1 or GV3101 (pMP90) Agrobacterium strains harboring a particular construct (46) and a strain expressing the p19 of viral silencing repressor to enhance protein expression (47), according to previous methods (48).
Subcellular Fractionation, Genomic DNA Analysis, Immunoblots, and Recombinant Protein Expression
Arabidopsissubcellular fractionation for membrane enrichment, genomic DNA isolation, PCR-based genotyping, and recombinant fusion protein expression and purification were performed as described previously (13). Arabidopsis seedling proteins were extracted with buffer PLB (50 mm Tris, 150 mm NaCl, 10% (v/v) glycerol, 1% (v/v) Nonidet P-40, pH 7.5, with freshly added 1 mm PMSF and 0.5× Complete protease inhibitor mix (Roche Applied Science). Protein concentration was assessed by Bradford or BCA assays. Western blots were performed as described previously (13) and either visualized with film or with ImageQuant (GE Healthcare) with the following addenda: antibodies against the LOG2 peptides LKKESLRLEPDPDNPG and FSVFEDVELFKAAADTEI (amino acids 138–153 and 216–233, respectively) raised in rabbit (21st Century Biochemicals). Quantitative protein expression comparisons were made with IRDye800-labeled secondary antibodies and an Odyssey flatbed scanner (LiCoR Biosciences P/N 929-70020, used in Fig. 2) or CCD camera-detected ECL with HRP-labeled mouse anti-HA antibodies (Sigma H6533, used for Fig. 4, A and B). Both of these approaches enable linear sensitivity >4 orders of magnitude (49, 50).
Phenotypic Assays
Arabidopsis seedlings were grown on GM agar (1× Murashige and Skoog salts, 1% sucrose) ± amino acids from the same master media stock for 7–14 days using the same batch of surface-sterilized seed as described previously (13). Leucine or phenylalanine was added to GM at concentrations between 2.5 and 10 mm as indicated in figure legends. Data from two or more experiments are shown (all experiments performed at least 3 times) with at least 20 seeds per plate per genotype analyzed by a one-way analysis of variance with Tukey post-hoc tests for multiple comparisons with the VassarStats online calculator.
In Vitro Ubiquitination Assays
LOG2 Ub ligase activity was measured essentially as described previously (51); 30-μl reactions contained 4 μg of bovine Ub (Sigma) or Me-Ub (Boston Biochem) and were quenched with 10 μl of 5× Laemmli sample buffer. To distinguish cis from trans autoubiquitination, glutathione-Sepharose (GE Healthcare)-bound wild-type or enzymatically inactive GST-LOG2 was added directly to reactions.
Proteolytic Inhibitor Treatments and Half-life Measurements
Chemical treatments to assess rates and modes of protein degradation were performed on 7-day, liquid-grown Arabidopsis seedlings. Solutions of cycloheximide (Sigma) were prepared in liquid GM (13) and used immediately. Wortmannin (Millipore), bortezomib (LC Laboratories), concanamycin A (Millipore), and MG132 (Millipore) were reconstituted in DMSO and applied to liquid-grown plants at final concentrations of 100 μm, 100 μm, 1 μm, and 100 μm, respectively, for 2 h before the addition of cycloheximide (200 μg/ml). Wortmannin treatments were conducted in the dark to prevent photo-degradation. Seedlings were flash-frozen in liquid nitrogen at various time points after cycloheximide addition, after which protein was extracted in PLB buffer and assessed by Western blot. To determine the in vivo half-life of GDU1-myc in wild-type and log2-2 backgrounds, IRDye800 fluorescence intensity was quantified for GDU1-myc-specific bands and normalized to the zero time point, and the common logarithms of each intensity were graphed as a function of time. Linear regressions of best fit were drawn using Microsoft Excel, and a one-way analysis of covariance (ANCOVA) test for independent samples was performed with the VassarStats online calculator. Time points from seven independent assays were included in the statistical analysis.
Protein Immunoprecipitation and Preparation of Samples for Mass Spectrometry
In vitro ubiquitination reactions for mass spectrometry were prepared identically to before but scaled up 20-fold in volume. To assess in planta GDU1ubiquitination, N. benthamiana leaves were infiltrated with AGL1 cultures harboring p19, p3756_GDU1 + pSOUP, or pGWB14_LOG2 mixed at a ratio of 9.5:9.5:1 (v/v/v). Three days after infiltration ∼10 g of leaf tissue was homogenized in 50 mm Na2HPO4 + 300 mm NaCl (pH 7.5) + 0.2% Nonidet P-40 + 1 mm PMSF + 0.5× Complete Protease Inhibitors. This suspension was ground 5 × 20 s in a Waring blender. CHAPS detergent was added to 0.25%, 0.5%, and 0.75% (w/v) on the 3rd, 4th, and 5th cycle, respectively. The crude homogenate was filtered through 4 layers of miracloth and centrifuged for 10 min at 10,000 × g at 6 °C. The recovered supernatant (“input”) was brought to 20 mm imidazole, combined with equilibrated Ni-NTA-agarose beads (GE Healthcare), and rotated at 4 °C for 45 min, after which the mixture was centrifuged at 1000 × g to recover the nickel beads. Beads were washed 3 times with seven column volumes lysis buffer, with 30 min/wash. Protein was eluted after a 15-min incubation with the homogenization buffer above plus 350 mm imidazole at 4 °C. Upon imidazole eluate recovery, the beads were boiled in 5× Laemmli buffer. The 5× Laemmli buffer eluate was then added to the imidazole eluate. The sample was loaded onto an 8-cm, 10% NuPAGE Novex Bis-Tris Mini Gel (Thermo Fisher Scientific) and run with NuPAGE MOPS-SDS buffer (Thermo Fisher Scientific). The gel was then stained with Imperial Protein Stain (Thermo Fisher Scientific) and destained in water. Gel bands were sliced into small portions followed by standard in-gel trypsin digestion (52). After digestion, peptides were lyophilized and analyzed by nano-RPLC-MS/MS. Some samples were prepared by the on-filter tryptic digestion via filter-aided sample preparation (FASP) procedure (53). Briefly, the combined Laemmli buffer-imidazole eluate was buffer-exchanged to 100 mm ammonium bicarbonate and loaded into an Amicon Ultra-0.5-ml centrifugal filter with a 10,000 molecular weight cutoff membrane (Millipore) and flushed with 400 μl of urea 4 times, after which the retentate was reduced, alkylated, and suspended in 100 mm ammonium bicarbonate. After overnight tryptic digestion, peptides were lyophilized and analyzed by nano-RPLC-MS/MS.
Nano-flow Reverse Phase Liquid Chromatography-Tandem Mass Spectrometry (Nano-RPLC-MS/MS) and Database Search
Tryptic peptides were dissolved in 0.5% trifluoroacetic acid, 2% acetonitrile, 97.5% water (v/v/v) before LC-MS/MS analysis. Nano-RPLC tandem mass spectrometric analysis was performed on both a LTQ-OrbitrapXL mass spectrometer (Thermo Fisher Scientific) equipped with an ADVANCE nanospray ion source (Michrom Bioresources) and a XEVO-G2-QTOF mass spectrometer equipped with a NanoAcquity-UPLC-TRIZAIC source (Waters). For the analysis on the LTQ-OrbitrapXL system, a house-packed C18AQ column was used (54). For the analysis on the XEVO-G2-QTOF, an equilibrated TRIZAIC nanoTile (Waters) with a 180-μm × 20-mm trapping column and an 85 μm × 100-mm analytical column (HSS T3, 1.8 μm, 100 Å) was used. The sample was loaded onto the trapping column for 3 min at 2% solvent B (0.1% (v/v) formic acid in acetonitrile) and 98% solvent A (0.1% (v/v) formic acid in water) at a flow rate of 5 μl/min. Peptides were resolved by the following gradient at a flow rate of 450 nl/min: 3–40% solvent B for 40 min, 40–85% solvent B for 2 min, and 85% B for 4 min. The XEVO-G2-QTOF mass spectrometer was operated in the data-dependent acquisition mode or MSe mode to automatically switch between MS and tandem mass spectrometric acquisition. Mass spectra were processed by MassLynx V4.1 (Waters), and MSe data were processed and searched by PLGS V2.5 (Waters). All tandem mass spectra from data-dependent acquisition on either the LTQ-OrbitrapXL or the XEVO-G2-QTOF system were searched through MASCOT V2.1 (Matrix Science). The mass spectrometry analysis was performed twice with independent samples, and the same Ub sites were identified in both experiments.
Author Contributions
D. G. and J. C. conceived the study. G. P. and J. C. supervised the research. Along with all other authors, R. P. and S. Y. contributed to the design, execution, and interpretation of the research. W. J. and J. L. performed the mass spectrometry analysis. D. G., S. M. C., and J. C. wrote the manuscript with editing by all authors.
Supplementary Material
Acknowledgments
We thank all members of the Callis laboratory for helpful discussions and Khin Kyaw for expert technical assistance. We are grateful to Daniel Guerra for critical advice in choosing a title for our manuscript. We are also indebted to Jennifer Gilda and Aldrin Gomes for advice in the use of bortezomib, the UC Davis Campus Controlled Environment Facility for growth of plants, the UC Davis Campus Mass Spectrometry Facility, and our funding sources.
This work was supported by the National Science Foundation (MCB-0929100 and IOS-1557760 (to J. C.) and MCB-1052048 (to G. P.)). This work was also supported, in whole or in part, by the National Institutes of Health (Predoctoral Training Grant fellowship GM0007377l to J. Trimmer (principal investigator) (to D. G.). and Grant 1S10RR023558-01 (to J. L.)), the Hatch Program of the National Institute of Food and Agriculture (VA-135908 (to G. P.) and CA-D-MCB-5554-H (to J. C.)), and the Virginia Agricultural Experiment Station (to G. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table 1.
S. Yu and G. Pilot, unpublished data.
R. Pratelli and G. Pilot, unpublished results.
- Ub
- ubiquitin
- Me-Ub
- methylated ubiquitin
- ER
- endoplasmic reticulum
- RING
- really interesting new gene
- GDU1
- glutamine dumper 1
- cGDU1
- cytosolic domain of GDU1
- LOG2
- loss of GDU 2
- VIMAG
- valine-isoleucine-methionine-alanine-glycine motif in GDU1
- GM
- germination media
- ANCOVA
- analysis of covariance
- NTA
- nickel-nitrilotriacetic acid
- CHX
- cycloheximide
- RPLC
- reverse phase liquid chromatography.
References
- 1. Pickart C. M., and Eddins M. J. (2004) Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 [DOI] [PubMed] [Google Scholar]
- 2. Lucero P., Peñalver E., Vela L., and Lagunas R. (2000) Monoubiquitination is sufficient to signal internalization of the maltose transporter in Saccharomyces cerevisiae. J. Bacteriol 182, 241–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Uhrig R. G., She Y.-M., Leach C. A., and Plaxton W. C. (2008) Regulatory monoubiquitination of phosphoenolpyruvate carboxylase in germinating castor oil seeds. J. Biol. Chem. 283, 29650–29657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guerra D. D., and Callis J. (2012) Ubiquitin on the Move: the ubiquitin modification pathway plays diverse roles in the regulation of Er-and membrane-localized proteins. Plant Physiol. 160, 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kerscher O., Felberbaum R., and Hochstrasser M. (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 [DOI] [PubMed] [Google Scholar]
- 6. Jacobson A. D., Zhang N. Y., Xu P., Han K. J., Noone S., Peng J., and Liu C. W. (2009) The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26S proteasome. J. Biol. Chem. 284, 35485–35494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Metzger M. B., Pruneda J. N., Klevit R. E., and Weissman A. M. (2014) RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1843, 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lorick K. L., Jensen J. P., Fang S., Ong A. M., Hatakeyama S., and Weissman A. M. (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 96, 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brzovic P. S., Keeffe J. R., Nishikawa H., Miyamoto K., Fox D. 3rd, Fukuda M., Ohta T., and Klevit R. (2003) Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl. Acad. Sci. U.S.A. 100, 5646–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bonner C. A., Rodrigues A. M., Miller J. A., and Jensen R. A. (1992) Amino acids are general growth inhibitors of Nicotiana sylvestris in tissue culture. Physiol. Plant. 84, 319–328 [Google Scholar]
- 11. Pratelli R., and Pilot G. (2014) Regulation of amino acid metabolic enzymes and transporters in plants. J. Exp. Bot. 65, 5535–5556 [DOI] [PubMed] [Google Scholar]
- 12. Pratelli R., and Pilot G. (2007) Altered Amino Acid Metabolism in glutamine Dumper1 plants. Plant Signal. Behav. 2, 182–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pratelli R., Guerra D. D., Yu S., Wogulis M., Kraft E., Frommer W. B., Callis J., and Pilot G. (2012) The ubiquitin E3 ligase LOSS OF GDU1 2 is required for GLUTAMINE DUMPER1-induced amino acid secretion in Arabidopsis. Plant Physiol. 158, 1628–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pratelli R., Voll L. M., Horst R. J., Frommer W. B., and Pilot G. (2010) Stimulation of nonselective amino acid export by glutamine dumper proteins. Plant Physiol. 152, 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pratelli R., and Pilot G. (2006) The plant-specific VIMAG domain of glutamine Dumper1 is necessary for the function of the protein in Arabidopsis. FEBS Lett. 580, 6961–6966 [DOI] [PubMed] [Google Scholar]
- 16. Pilot G., Stransky H., Bushey D. F., Pratelli R., Ludewig U., Wingate V. P., and Frommer W. B. (2004) Overexpression of glutamine dumper1 leads to hypersecretion of glutamine from hydathodes of Arabidopsis leaves. Plant Cell 16, 1827–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Upadhyay A., Amanullah A., Chhangani D., Mishra R., Prasad A., and Mishra A. (2016) Mahogunin ring Finger-1 (MGRN1), a multifaceted ubiquitin ligase: recent unraveling of neurobiological mechanisms. Mol. Neurobiol. 53, 4484–4496 [DOI] [PubMed] [Google Scholar]
- 18. Guerra D. D., Pratelli R., Kraft E., Callis J., and Pilot G. (2013) Functional conservation between mammalian MGRN1 and plant LOG2 ubiquitin ligases. FEBS Lett. 587, 3400–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhadina M., and Bieniasz P. (2010) Functional interchangeability of late domains, late domain cofactors and ubiquitin in viral budding. PLoS Pathog. 6, e1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bourgeois-Daigneault M. C., and Thibodeau J. (2012) Autoregulation of MARCH1 expression by dimerization and autoubiquitination. J. Immunol. 188, 4959–4970 [DOI] [PubMed] [Google Scholar]
- 21. Amemiya Y., Azmi P., and Seth A. (2008) Autoubiquitination of BCA2 RING E3 ligase regulates its own stability and affects cell migration. Mol. Cancer Res. 6, 1385–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mimnaugh E. G., and Neckers L. M. (2005) Measuring ubiquitin conjugation in cells. In Ubiquitin-Proteasome Protocols (Patterson C., and Cyr D., eds) pp. 223–242, Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 23. de Bie P., and Ciechanover A. (2011) Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 18, 1393–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weissman A. M., Shabek N., and Ciechanover A. (2011) The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat. Rev. Mol. Cell Biol. 12, 605–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen Y.-T., Liu H., Stone S., and Callis J. (2013) ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J. 75, 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dreher K. A., Brown J., Saw R. E., and Callis J. (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18, 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen X., and Yin X.-M. (2011) Coordination of autophagy and the proteasome in resolving endoplasmic reticulum stress. Vet. Pathol. 48, 245–253 [DOI] [PubMed] [Google Scholar]
- 28. Ihara Y., Morishima-Kawashima M., and Nixon R. (2012) The ubiquitin-proteasome system and the autophagic-lysosomal system in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2, a006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramos J. A., Zenser N., Leyser O., and Callis J. (2001) Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome-dependent. Plant Cell 13, 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hansen S. H., Olsson A., and Casanova J. (1995) Wortmannin, an inhibitor of phosphoinositide 3-kinase, inhibits transcytosis in polarized epithelial cells. J. Biol. Chem. 270, 28425–28432 [DOI] [PubMed] [Google Scholar]
- 31. Woo J. T., Shinohara C., Sakai K., Hasumi K., and Endo A. (1992) Isolation, characterization and biological activities of concanamycins as inhibitors of lysosomal acidification. J. Antibiot. 45, 1108–1116 [DOI] [PubMed] [Google Scholar]
- 32. Amanso A. M., Debbas V., and Laurindo F. R. (2011) Proteasome inhibition represses unfolded protein response and Nox4, sensitizing vascular cells to endoplasmic reticulum stress-induced death. PLoS ONE 6, e14591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen D., Frezza M., Schmitt S., Kanwar J., and Dou Q. (2011) Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr. Cancer Drug Targets 11, 239–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nikko E., and Pelham H. R. (2009) Arrestin-Mediated Endocytosis of Yeast Plasma Membrane Transporters. Traffic 10, 1856–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Léon S., and Haguenauer-Tsapis R. (2009) Ubiquitin ligase adaptors: regulators of ubiquitylation and endocytosis of plasma membrane proteins. Exp. Cell Res. 315, 1574–1583 [DOI] [PubMed] [Google Scholar]
- 36. Hooper C., Puttamadappa S. S., Loring Z., Shekhtman A., and Bakowska J. C. (2010) Spartin activates atrophin-1-interacting protein 4 (AIP4) E3 ubiquitin ligase and promotes ubiquitination of adipophilin on lipid droplets. BMC Biol. 8, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chhangani D., Nukina N., Kurosawa M., Amanullah A., Joshi V., Upadhyay A., and Mishra A. (2014) Mahogunin ring finger 1 suppresses misfolded polyglutamine aggregation and cytotoxicity. Biochim. Biophys. Acta 1842, 1472–1484 [DOI] [PubMed] [Google Scholar]
- 38. Kunkel T. (1985) Rapid and efficient site-directed mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. U.S.A. 82, 488–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hellens R. P., Edwards E. A., Leyland N. R., Bean S., and Mullineaux P. M. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832 [DOI] [PubMed] [Google Scholar]
- 40. Nishikawa H., Ooka S., Sato K., Arima K., Okamoto J., Klevit R. E., Fukuda M., and Ohta T. (2004) Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J. Biol. Chem. 279, 3916–3924 [DOI] [PubMed] [Google Scholar]
- 41. Verdaguer B., de Kochko A., Fux C. I., Beachy R. N., and Fauquet C. (1998) Functional organization of the cassava vein mosaic virus (CsVMV) promoter. Plant Mol. Biol. 37, 1055–1067 [DOI] [PubMed] [Google Scholar]
- 42. Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., and Kimura T. (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41 [DOI] [PubMed] [Google Scholar]
- 43. Norris S. R., Meyer S. E., and Callis J. (1993) The intron of Arabidopsis polyubiquitin genes is conserved in location and is a quantitative determinant of chimeric gene expression. Plant Mol. Biol. 21, 895–906 [DOI] [PubMed] [Google Scholar]
- 44. Clough S. J., and Bent A. F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 45. Lazo G. R., Stein P. A., and Ludwig R. A. (1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology 9, 963–967 [DOI] [PubMed] [Google Scholar]
- 46. Batoko H., Zheng H.-Q., Hawes C., and Moore I. (2000) A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal golgi movement in plants. Plant Cell 12, 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lakatos L., Szittya G., Silhavy D., and Burgyán J. (2004) Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23, 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu S., Pratelli R., Denbow C., and Pilot G. (2015) Suppressor mutations in the glutamine Dumper1 protein dissociate disturbance in amino acid transport from other characteristics of the Gdu1D phenotype. Front. Plant Sci. 6, 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Loebke C., Sueltmann H., Schmidt C., Henjes F., Wiemann S., Poustka A., and Korf U. (2007) Infrared-based protein detection arrays for quantitative proteomics. Proteomics 7, 558–564 [DOI] [PubMed] [Google Scholar]
- 50. Hommema E., Jarrett N., Shiflett S., Hong S., Rangaraj P., and Webb B. (2013) Comparison of cooled-CCD digital imaging versus X-ray film for sensitivity, dynamic range and signal linearity with enhanced chemiluminescence (ECL). Thermo Fisher Scientific Application Note 1602635 [Google Scholar]
- 51. Hsia M. M., and Callis J. (2010) BRIZ1 and BRIZ2 Proteins form a heteromeric E3 ligase complex required for seed germination and post-germination growth in Arabidopsis thaliana. J. Biol. Chem. 285, 37070–37081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shevchenko A., Tomas H., Havlis J., Olsen J. V., and Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 53. Winiewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 54. Jia W., Shaffer J. F., Harris S. P, and Leary J. A. (2010) Identification of novel protein kinase A phosphorylation sites in the M-domain of human and murine cardiac myosin binding protein-C using mass spectrometry analysis. J. Proteome Res. 9, 1843–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.