Abstract
In eukaryotes, the conjugation of the ubiquitin-like protein NEDD8 onto protein targets is an important post-translational modification. The best understood neddylation targets are the cullins, scaffold subunits of E3 ubiquitin ligases, where neddylation as well as deneddylation, facilitated by the protease activity of the CSN (COP9 signalosome), are required to control ubiquitin ligase assembly, function, and ultimately substrate degradation. Little is known about the role of other deneddylating enzymes besides CSN and the role of neddylation and deneddylation of their substrates. We previously characterized Arabidopsis thaliana mutants with defects in the conserved NEDD8-specific protease DEN1 (DENEDDYLASE1). These mutants display only subtle growth phenotypes despite the strong accumulation of a broad range of neddylated proteins. Specifically, we identified AXR1 (AUXIN-RESISTANT1), a subunit of the heterodimeric NAE (E1 NEDD8-ACTIVATING ENZYME), as highly neddylated in den1 mutants. Here, we examined the mechanism and consequences of AXR1 neddylation in more detail. We find that AXR1 as well as other neddylation enzymes are autoneddylated at multiple lysines. NAE autoneddylation can be linked to reduced NCE (E2 NEDD8-CONJUGATING ENZYME) NEDD8 thioester levels, either by critically reducing the pool of free NEDD8 or by reducing NAE activity. In planta, increasing NEDD8 gene dosage is sufficient to suppress den1 mutant phenotypes. We therefore suggest that DEN1 serves to recover diverted NEDD8 moieties from autoneddylated NAE subunits, and possibly also other neddylated proteins, to maintain NEDD8 pathway activity toward other NEDD8-dependent processes such as cullin E3 ligase regulation.
Keywords: Arabidopsis, Arabidopsis thaliana, deubiquitylation (deubiquitination), ubiquitin, ubiquitin-dependent protease, ubiquitylation (ubiquitination)
Introduction
The reversible attachment of the ubiquitin-related protein NEDD8 (NEURAL PRECURSOR CELL-EXPRESSED DEVELOPMENTALLY DOWN-REGULATED8) is an essential post-translational protein modification in most eukaryotes (1–5). NEDD8 conjugation is essentially analogous to the conjugation of ubiquitin and other UBLs2 (UBIQUITIN-LIKE PROTEINS) but is catalyzed by specific NEDD8 E1-activating enzymes (NAE) and E2-conjugating enzymes (NCE) and E3 ligases (6, 7). The heterodimeric NAE activates NEDD8 in an ATP-dependent reaction, first forming an adenylate and subsequently a high energy NAE∼NEDD8 thioester (8, 9). The activated NEDD8 moiety is then transferred to the active site Cys (cysteine) of the NCE, forming a NCE∼thioester (9). In the final step, NEDD8 is covalently attached to its substrate through an isopeptide bond involving the C-terminal Gly (glycine) of NEDD8 and an ε-amino group of a Lys (lysine) in the target protein (8). Cleavage of this bond and removal of the modification is achieved through the activities of specific deubiquitinating iso-peptidases, so-called DUBs (10).
Cullins are currently the best understood neddylation targets in all eukaryotes (11–13). Cullins are scaffold subunits of CRLs (cullin-RING E3 ubiquitin ligases), which regulate ubiquitin-dependent and substrate-specific protein degradation (14). Cullin neddylation is mediated by the above-described neddylation pathway, whereas cullin deneddylation is promoted by CSN5, a subunit of the CSN (COP9 signalosome) complex (15–18). Cullin neddylation and deneddylation are part of a recurring cycle that regulates CRL activity, substrate exchange, and thereby ultimately protein degradation (19–21).
In line with the importance of CRL-mediated protein degradation for many different cellular pathways, NEDD8 as well as neddylation and deneddylation pathway mutants are severely affected in growth and development (1–5). In the model plant Arabidopsis thaliana, the combined knock-out of RUB1 (RELATED TO UBIQUITIN1) and RUB2, two of the three genes coding for NEDD8 in Arabidopsis, results in embryonic growth arrest at the two-cell stage (22). Similarly, double mutants defective in the two paralogous NAE regulatory subunits AXR1 (AUXIN-RESISTANT1) and AXL (AXR1-LIKE) show defects in cell division and pattern formation as well as growth arrest during embryogenesis (23). Concurrently, loss of CSN5 activity and consequently cullin deneddylation leads to growth arrest at the seedling stage (15, 24).
Besides CSN5, also DEN1 (DENEDDYLASE1) has been identified as a deneddylating enzyme (25, 26). Like ubiquitin, NEDD8 requires maturation by C-terminal proteolytic processing of NEDD8 precursor forms. DEN1 was originally isolated as a NEDD8-specific C-terminal processing enzyme based on its in vitro activity (26). However, NEDD8 processing is not detectably impaired in den1 mutants from fission yeast (Schizosaccharomyces pombe), fungi (Aspergillus nidulans), Arabidopsis, Drosophila, or mice, suggesting that the in vivo activity of DEN1 may be different from its reported in vitro activity, or that other functionally redundant proteases can process NEDD8 in den1 mutants (27–31). Because den1 mutants from different species accumulate high levels of neddylated substrates of a broad molecular range, it was instead concluded that DEN1 serves in vivo to deconjugate NEDD8 from neddylated substrates (27–31).
Our previous comparative analyses of den1 and csn5 mutants from Arabidopsis thaliana showed that, at least in Arabidopsis, DEN1 is specifically required to remove NEDD8 from neddylated substrates other than cullins (31). We further identified AXR1 and, indirectly, also the paralogous AXL as prominent NEDD8-modified proteins that depend on DEN1 for deneddylation (31). AXR1 and AXL share ∼80% amino acid identity, and both can form a complex with ECR1 (E1 C-TERMINAL RELATED1) (23, 32). While axl single mutants display no prominent phenotype, axr1 single mutants have apparent growth defects. Adult axr1 plants are dwarfed, have reduced apical dominance, curled leaves, and reduced fertility (11, 33, 34). While den1 mutants show no obvious growth defects, the den1 mutation has a clear effect on plant growth in an AXR1-sensitized background, specifically, an axr1-30 Myc:AXR1 complementation line where the expression of a Myc-tagged AXR1 transgene only partially rescues the axr1-30 mutant phenotype (31, 32). This suggests that AXR1 neddylation negatively affects the neddylation pathway in den1 mutant backgrounds. It is, however, not yet understood what mechanisms lead to AXR1 neddylation in the first place, which steps of the neddylation cascade are affected by AXR1 neddylation, and whether this modification serves a regulatory function.
Here, we examine the molecular mechanism and consequences of AXR1 neddylation. We show that AXR1 and other neddylation enzymes are autoneddylated in an E2- and E3-independent manner in vitro and that AXR1 autoneddylation impairs NEDD8 E2∼thioester formation in the den1 mutant background in vivo. We further show that increasing NEDD8 gene dosage is sufficient to suppress den1 mutant phenotypes suggesting that the non-covalent NEDD8 pool may be limiting in den1. Taken together, our data indicate that DEN1 functions to maintain NEDD8 pathway activity by recovering diverted NEDD8 moieties from proteins of the NEDD8 conjugation machinery.
Results
NEDD8 NAE and NCE Enzymes Can Be Autoneddylated
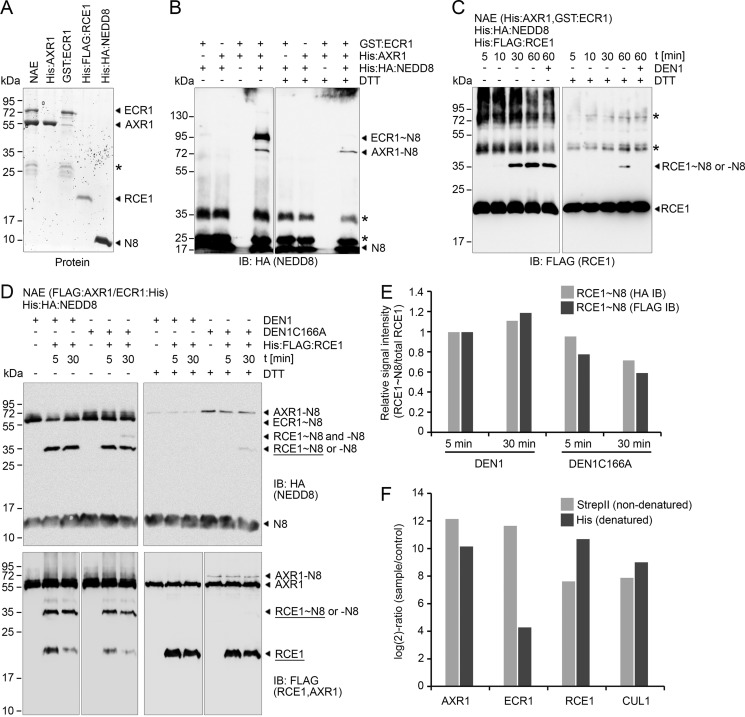
In Arabidopsis, the heterodimeric NAE is composed of the subunits ECR1 and AXR1 or the paralogous AXL. During NEDD8 activation, a thioester is formed between NEDD8 (NEDD8∼) and ECR1. The activated NEDD8 is then passed on, through a trans-thioester reaction, to the NCE enzyme RCE1 (RUB1-CONJUGATING ENZYME1) (5, 35). We have previously identified AXR1 as a neddylated protein from Arabidopsis wild type extracts and shown that AXR1-NEDD8 accumulates as a prominent neddylated protein in protein extracts from den1 mutants (31). To understand the molecular mechanism underlying this modification, we performed in vitro neddylation experiments with purified recombinant NEDD8 (His:HA:NEDD8), AXR1 (His:AXR1), and ECR1 (GST:ECR1) (Fig. 1A). Following incubation with both NAE subunits, NEDD8 was, as expected, conjugated to ECR1 but unexpectedly also to AXR1 (Fig. 1B). However, when each NAE subunit was examined individually, neither AXR1 nor ECR1 was NEDD8-modified, indicating that NAE activity was required for these modifications to occur (Fig. 1B). To characterize the biochemical nature of the observed NEDD8 conjugation, the reactions were performed in the presence or absence of DTT, which reduces thioester linkages but has no effect on isopeptide bonds. In the presence of the reducing agent, NEDD8 could be removed from ECR1 but not from AXR1 (Fig. 1B). We thus concluded that AXR1 is NEDD8-modified in vitro, concurrent with our previous report that AXR1 is NEDD8-modified in vivo and that this reaction can occur in an NCE-independent manner (31).
FIGURE 1.
NAE and NCE enzymes can be autoneddylated in vitro. A, Coomassie staining of purified recombinant proteins NAE (His:AXR1, GST:ECR1), His:AXR1, GST:ECR1, His:FLAG:RCE1, and His:HA:NEDD8. B, anti-HA immunoblot (IB) for the detection of free NEDD8 (His:HA:NEDD8), DTT-sensitive NEDD8 thioesters (∼N8), and DTT-insensitive NEDD8 isopeptide linkages (−N8) after incubation of ECR1 (GST:ECR1) and AXR1 (His:AXR1) with NEDD8. Asterisks indicate unspecific bands. C, anti-FLAG IB for the detection of the in vitro trans-thioester reaction of RCE1 (His:FLAG:RCE1). Reactions were stopped after incubation times as specified with non-reducing or reducing DTT-containing buffers. Asterisks indicate unspecific bands. D, anti-HA (NEDD8) and anti-FLAG (RCE1) IB for the detection of the effects of active DEN1 or inactive DEN1C166A on AXR1 neddylation and the trans-thioester reaction on the NCE RCE1. NAE reactions were incubated for 16 h before RCE1 addition. Reactions were stopped after an additional incubation time as specified with non-reducing buffer or reducing DTT-containing buffer. Protein bands used for quantification as shown in E are underlined. E, quantification of signal intensities of RCE1 thioester (RCE1∼N8) abundance in anti-HA (NEDD8) and anti-FLAG (RCE1). RCE1∼N8 signals from the anti-HA and anti-FLAG immunoblots were normalized to the total RCE1 protein input as detected in the reducing gel (D, right panel). F, graph of the log2 ratios of relative raw protein abundance (sample/control) measured by mass spectrometry. Enrichment of AXR1, ECR1, RCE1, and CUL1 (cullin1) after purification of His:StrepII:NEDD8-conjugates extracted from 7-day-old seedlings under non-denaturating (StrepII) conditions followed by a purification under denaturating conditions (His).
To examine whether the recombinant NAE heterodimer consisting of separately expressed and purified His:AXR1 and GST:ECR1 was formed correctly and functional, we repeated the neddylation reaction and included the NCE conjugation enzyme RCE1 (Fig. 1C). RCE1 was efficiently NEDD8-conjugated in this experiment, thus confirming the functionality of the NAE (Fig. 1C). However, not all of this conjugate was DTT-sensitive suggesting that a fraction of the NEDD8 modification was bound by an isopeptide bond rather than by a thioester (Fig. 1C). Because we had previously shown that the deneddylating enzyme DEN1 could cleave isopeptide bonds, we added DEN1 to the reaction. Although DEN1 treatment only weakly reduced the RCE1∼NEDD8 signal under non-reducing conditions, a complete deneddylation of RCE1 was achieved when DEN1 was combined with the DTT treatment (Fig. 1C). In a subsequent experiment, we pre-incubated recombinant NAE (FLAG:AXR1/ECR1:His) with proteolytically active or inactive DEN1 before the addition of RCE1 and an additional incubation for 5 or 30 min, respectively (Fig. 1D). A quantitative analysis of RCE1∼NEDD8 thioester formation, based on the signal intensities measured from the anti-HA (NEDD8) and anti-FLAG (RCE1) immunoblots, showed only a slight difference in NAE-NCE trans-thiolation efficiency in the presence of active or inactive DEN1 (Fig. 1E). Although we observed reduced signal intensities after the 30-min reaction with DEN1C166A, this could be explained by the covalent neddylation of RCE1 (REC1-NEDD8) because only the RCE1∼NEDD8 thioester band was used for the quantitative analysis (Fig. 1, D and E). This result thus suggests that the covalent NAE-NEDD8 modification has no strong effect on NAE enzymatic activity under these experimental conditions. However, only a small fraction of AXR1 was neddylated in our in vitro reactions, which could potentially lead to an underestimation of its in vivo effect (Fig. 1, D and E).
Because our in vitro experiments suggested that RCE1 may also become autoneddylated in vivo, we re-examined mass spectrometric data from a previously published study of a two-step protein purification performed under denaturing conditions of His:StrepII-tagged NEDD8 conjugates purified from Arabidopsis den1 mutant plants (31). Similar to AXR1 and CUL1 (cullin 1), RCE1 was enriched during both purification steps suggesting that DEN1-sensitive and DTT-insensitive NEDD8 modifications of RCE1 were present not only in vitro but also in vivo (Fig. 1F).
AXR1 and ECR1 Carry NEDD8 Isopeptide Linkages at Multiple Lysine Residues
When increasing the NAE input in the in vitro reaction, we detected not only one but two DTT-insensitive bands after an extended incubation time (Fig. 2A). These bands could be explained by the expected molecular weight shift of neddylated AXR1 or ECR1, respectively (Fig. 2A). To confirm the protein identity of the designated bands and to map the residues of AXR1 and ECR1 carrying NEDD8 modifications, we analyzed the excised gel pieces, band A and band B, by mass spectrometry. AXR1 could be identified as the most highly abundant protein in band A and ECR1 as the most highly abundant protein in band B. For AXR1, we detected the typical Gly-Gly (glycine-glycine) footprint that remains after tryptic digest of a NEDD8 isopeptide-conjugated Lys on five different AXR1 Lys residues (Lys-21, -222, -262, -350, and -383) (Figs. 2B and 3A). For ECR1, we detected in total seven different Lys carrying the Gly-Gly footprint (Lys-20, -99, -101, -185, -243, -412, and -424) (Figs. 2B and 3B). Because we observed a discrete band of the modified subunits, we reasoned that AXR1 and ECR1 are interchangeably mononeddylated at different Lys residues rather than being polyneddylated at multiple sites at the same time. We positioned these modified Lys on the predicted structure of AXR1-ECR1, which we had modeled on the structure of the human NAE composed of APPBP1 and UBA3 (AMYLOID PRECURSOR PROTEIN-BINDING PROTEIN1 and UBIQUITIN-LIKE MODIFIER ACTIVATING ENZYME3; Fig. 2B). There, we found many of the modified Lys to be surface-exposed and close to, but not in absolute vicinity of, the NEDD8-reactive Cys-215 of ECR1. In AXR1, the modified Lys were on average closer to Cys-215 (about 30 Å) than the non-modified Lys (about 45 Å; Fig. 2C). This was not observed in ECR1, but in ECR1 all Lys residues were on average closer to Cys-215 (35–40 Å) than the non-modified Lys found in AXR1 (Fig. 2C). We also examined whether these Lys modifications may interfere with NAE and NCE function. Two of the neddylated Lys of ECR1 (Lys-20 and Lys-185) were positioned at the interface of ECR1 with AXR1 and may therefore disturb the AXR1-ECR1 interaction. In turn, ECR1 Lys-412 was positioned on the UBL domain, which facilitates the interaction with the NCE, and the modification on this residue may thus potentially interfere with the NAE-NCE interaction (8, 9). However, because we have no evidence for the occurrence of ECR1 neddylation in vivo, we cannot exclude that these in vitro ECR1 neddylations are artifacts of the thioester assay that may not necessarily be of functional relevance in vivo (31).
FIGURE 2.
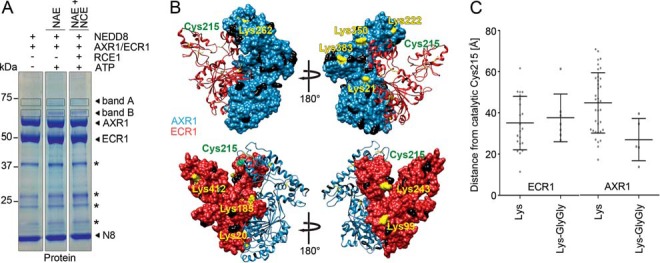
Identification of modified lysines in NEDD8 NAE subunits. A, SDS-PAGE of thioester reactions of NAE (FLAG:AXR1/ECR1:His), NAE and NCE (His:FLAG:RCE1), and NEDD8 (His:HA:NEDD8) reduced with DTT and stained for total protein with Coomassie Brilliant Blue (Protein). Bands A and B were excised from each lane and analyzed by LC-MS/MS. B, Swiss-Model structure prediction of AXR1 (blue) and ECR1 (red) based on the structure of human NAE (APPBP1/UBA3; Protein Data Bank accession 1YOV (8)). The model was analyzed using the Chimera software. Lys residues with a Gly-Gly modification as identified by mass spectrometry are shown in yellow; non-modified Lys residues are shown in black, and the catalytically active Cys-215 of ECR1 is shown in green. C, graph displaying the direct physical distances between the Cα positions of the catalytically active Cys-215 of ECR1 and the Lys residues of AXR1 or ECR1. Graphs show the average distance and S.D. for all unmodified Lys residues or Lys residues carrying a GlyGly-modification (LysGlyGly). * indicates background bands.
FIGURE 3.
AXR1 and ECR1 alignment and their Gly-Gly modification sites. A, ClustalW alignment of A. thaliana NAE subunits AXR1 (At_AXR1) and AXL (At_AXL) together with their human counterpart Hs_APPBP1. B, ClustalW alignment of A. thaliana NAE subunit ECR1 (At_ECR1) and the human catalytic subunit UBA3 (Hs_UBA3). Lys residues carrying a Gly-Gly modification after autoneddylation as identified in this study; in the case of APPBP1 and UBA3, Lys residues reported to carry a Gly-Gly modification in PhosphositePlus are marked by an arrow. An asterisk indicates the position of the catalytically active Cys in ECR1 and UBA3.
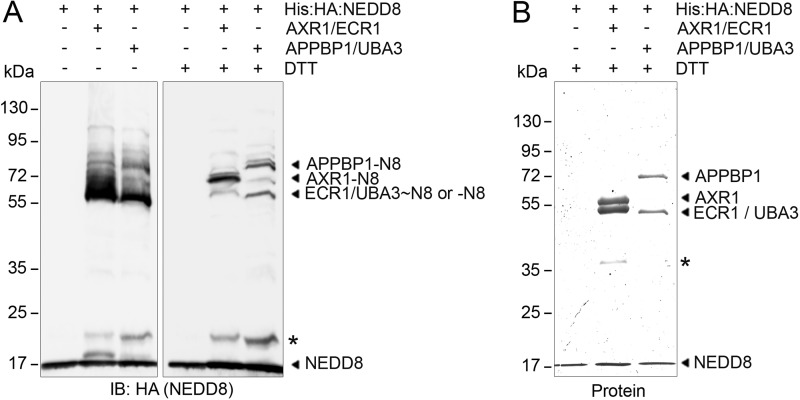
Each of the NEDD8-modified Lys residues in AXR1 was conserved in its closely related paralog AXL. Interestingly, none of them were conserved between AXR1/AXL and human APPBP1 or between ECR1 and human UBA3 (Fig. 3, A and B). Thus, if these modifications were to represent regulatory modifications, they were likely specific for the Arabidopsis proteins. Proteomics data annotated in PhosphositePlus indicated that human APPBP1 and UBA3, similarly to AXR1 and ECR1, carried Gly-Gly modifications at multiple Lys residues (Fig. 3, A and B) (36). Unfortunately, the sequence identity between the C terminus of NEDD8 and ubiquitin does not allow us to make conclusive statements about the identity of these modifications, because ubiquitin or NEDD8 modifications result in the same mass footprint after tryptic digest. We reasoned, however, that the human proteins may undergo similar autoneddylation reactions as the plant NAE and performed the corresponding experiment with recombinant human APPBP1 and UBA3. There, we detected indeed DTT-resistant neddylation of both subunits in addition to the UBA3∼NEDD8 thioester confirming that a similar modification to the plant NAE can also occur on the human counterpart (Fig. 4, A and B).
FIGURE 4.
Conservation of NAE vitro autoneddylation in human and Arabidopsis. A, IB with anti-HA (NEDD8) of 16 h in vitro neddylation reactions with Arabidopsis NAE (FLAG:AXR1/ECR1:His) and human NAE (APPBP1/His:UBA3) with NEDD8 (N8) (His:HA:NEDD8) under non-reducing and reducing (DTT) conditions. Asterisks indicate background cross-reactive bands. B, protein staining of in vitro neddylation reactions from A under reducing conditions. Asterisks indicate unspecific background.
Neddylation Has No Apparent Effect on AXR1 Stability or Interactions
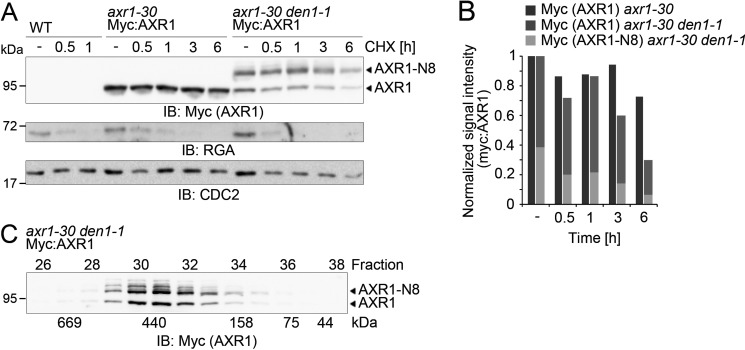
AXR1 accumulates in its neddylated form in Arabidopsis den1 mutants, and low levels of neddylated AXR1 can also be detected in the wild type after AXR1 immunoprecipitation (31). We reasoned that a comparative analysis between wild type and den1 mutants could provide an indication of the effects of neddylation on AXR1 protein behavior and function. For these analyses, we made use of a previously described Myc:AXR1 den1 transgenic line, where about half of the Myc:AXR1 protein accumulated in its neddylated form (Fig. 5). We first performed a cycloheximide (CHX) chase experiment where the addition of the protein biosynthesis inhibitor allows us to follow changes in protein abundance over time, for example due to protein degradation. We found that the unmodified and the NEDD8-modified form of AXR1 were similarly stable in the wild type and the den1 background for up to 3 h following the CHX treatment (Fig. 5, A and B). Following longer incubation times, however, non-neddylated as well as neddylated AXR1 were less stable in den1 than AXR1 in the wild type (Fig. 5, A and B). Thus, although the absence of DEN1 and, consequently, the presence of AXR1 neddylation do not have a strong effect on AXR1 stability, it is noteworthy that there may be a direct or indirect effect on AXR1 protein turnover. The dynamics of the reaction make it difficult to judge whether this effect is specific only for neddylated AXR1 or for both protein pools in the den1 mutant.
FIGURE 5.
AXR1 neddylation does not affect AXR1 stability or apparent protein interactions in vivo. A, IB analyses with anti-Myc, anti-RGA, and anti-CDC2 following a CHX chase experiment with wild type (WT) seedlings and axr-30 or axr-30 den1-1 mutant seedlings expressing Myc:AXR1. Seedlings were grown on growth medium for 7 days and subsequently transferred to liquid growth medium with 50 μm CHX for up to 6 h. The anti-CDC2 IB serves as a loading control; the anti-RGA IB against the unstable RGA protein serves as a positive control for the CHX treatment. B, graph depicting the normalized signal intensities of Myc:AXR1 in its unmodified (AXR1) and NEDD8-modified form (AXR1-N8) as detected in axr-30 or axr-30 den1-1 backgrounds. In the case of the axr-30 den1-1 measurements, the results from AXR1 and AXR1-N8 were summed up to provide a measure for the overall AXR1 abundance. C, IB with anti-Myc of 20 μl eluate per fraction following a Superose 6 gel filtration of total protein extract from 7-day-old Myc:AXR1 axr-30 den1-1 seedlings. Fraction numbers and corresponding molecular weights are specified accordingly. N8, NEDD8.
When we performed gel filtration experiments to examine the association of AXR1 with other proteins, we found that AXR1 neddylation did not affect the stable association or interaction of AXR1 with other proteins because both the neddylated and unneddylated forms had the same elution profiles (Fig. 5C). Thus, AXR1 neddylation did not lead to apparent changes in AXR1 protein stability or stable protein interactions.
RCE1 NEDD8∼Thioester Formation Is Deficient in den1 Mutants
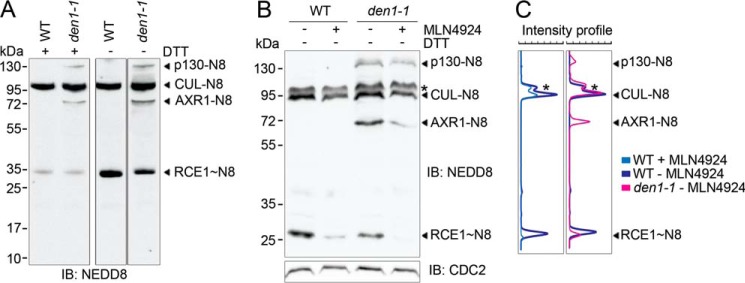
In vivo, the neddylation of AXR1 and ECR1 may interfere with the downstream trans-thioester reaction from NAE to NCE. To evaluate the neddylation status of RCE1 in vivo, we compared NEDD8 immunoblots from wild type and den1 mutant plant extracts using reducing and non-reducing conditions. Under non-reducing conditions, we observed a neddylated protein at around 30 kDa, which should corresponds to the RCE1∼NEDD8 thioester according to its calculated molecular weight (Fig. 6A). To rule out background staining visible only under non-reducing conditions, we included samples grown on the neddylation inhibitor MLN4924 and could show a clear reduction of the NEDD8 signal for both cullin-NEDD8 as well as RCE1∼NEDD8 (Fig. 6, B and C) (37). When subsequently comparing the protein levels in den1 and wild type, we noted a less severe but still substantial reduction for RCE1∼NEDD8 under these conditions, whereas steady-state cullin neddylation was not apparently affected (Fig. 6, B and C). We therefore concluded that RCE1∼NEDD8 thioester formation is impaired in den1 mutants, either due to reduced NAE activity or because free NEDD8 monomers are consumed in NEDD8-modified proteins that accumulate in den1 mutants.
FIGURE 6.
AXR1 neddylation correlates with reduced RCE1∼NEDD8 thioester formation. A, anti-NEDD8 IB analysis of crude protein extracts (60 μg) after non-reducing or reducing SDS-PAGE. NEDD8 conjugates (−N8) and NEDD8 thioesters (∼N8) are labeled accordingly. B, anti-NEDD8 immunoblot analysis of crude protein extracts (60 μg) after non-reducing SDS-PAGE. Seedlings were grown for 7 days on growth medium with or without 10 μm MLN4924. The CDC2 IB serves as a loading control. C, signal intensity profile comparison of B for treated (light blue) and untreated wild type (WT) (dark blue) and untreated WT and den1-1 mutant (pink) samples generated with the MultiGauge analysis software.
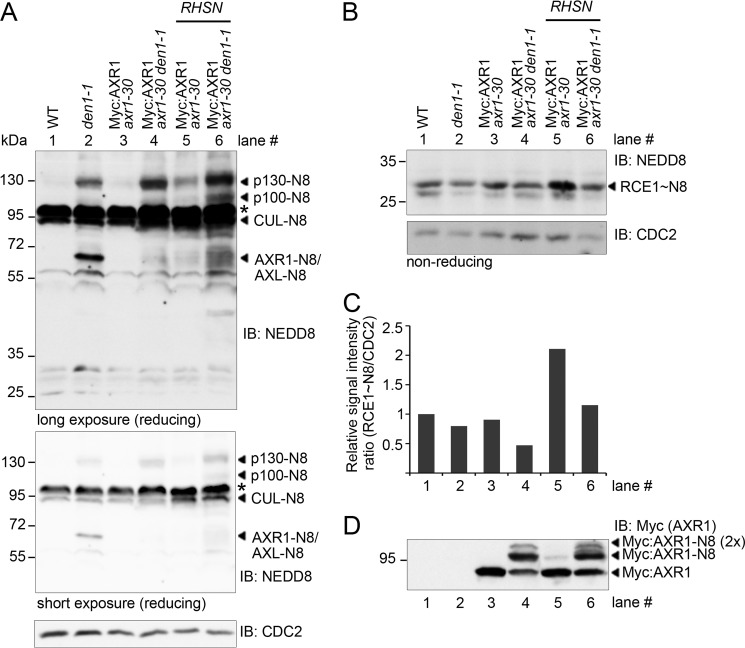
We next examined the effects of increasing NEDD8 gene dosage on neddylation reactions in the den1 background. To this end, we introduced a transgene designated RSHN for the expression of a His:StrepII-tagged NEDD8 under the control of a 2-kb RUB1 (an Arabidopsis NEDD8 gene) promoter fragment into Myc:AXR1 axr1-30 and Myc:AXR1 axr1-30 den1 (31). Increasing NEDD8 concentrations in this manner resulted in the increased neddylation of p130, a protein of unknown identity that we had previously shown to accumulate in its neddylated form in den1 (Fig. 7A) (31). Additionally, we noted an increase in cullin neddylation as well as the increased neddylation of a protein of similar molecular weight to AXR1, which we concluded to be AXL based on its increased abundance in axr1 mutants (Fig. 7A) (31, 32). Furthermore, we noted the appearance of an additional neddylated protein of unknown identity in Myc:AXR1 axr1-30 den1 that was not detectable in den1 or Myc:AXR1 axr1-30 but could correspond to hyperneddylated Myc:AXR1 as suggested by an anti-Myc immunoblot (Fig. 7, A and D, lane 6).
FIGURE 7.
Increasing NEDD8 gene dosage results in increased neddylation and increased RCE1∼thioester formation in planta. A, anti-NEDD8 (long and short exposures) and anti-CDC2 IB of crude protein (60 μg) extracts from 7-day-old seedlings after reducing SDS-PAGE. RHSN is a transgene for the expression of the Arabidopsis NEDD8 gene RUB1 under control of a RUB1 promoter fragment. NEDD8 conjugates (−N8) are labeled accordingly. An asterisk indicates a background cross-reactive band. The short exposure of the anti-NEDD8 immunoblot serves to show differences in cullin (CUL) NEDD8 modification. The CDC2 IB serves as a loading control. B, anti-NEDD8 and anti-CDC2 IB from non-reducing SDS-PAGE of the same protein extracts as shown in A. The RCE1∼N8 thioester, identified based on its molecular weight and NEDD8 immunoreactivity, is marked. C, graph displaying the signal intensities of RCE1∼N8 relative to the respective anti-CDC2 signal and normalized to the wild type signal ratio. D, anti-Myc immunoblot of the same protein extracts as shown in A.
To understand whether elevated NEDD8 levels had an effect on RCE1∼NEDD8 thioester formation, we additionally examined RCE1∼NEDD8 abundance in immunoblots from non-reducing SDS-PAGE (Fig. 7B). Quantification of the RCE1 thioester signal showed reduced thioester levels when comparing den1 with wild type (Fig. 7C). We also observed a clear increase in RCE1∼NEDD8 thioester formation that correlated with elevated NEDD8 expression and that was particularly strong in Myc:AXR1 axr1-30 (Fig. 7, B and C). Because Myc:AXR1 cannot be efficiently separated from neddylated cullins in anti-NEDD8 immunoblots, we examined Myc:AXR1 abundance and neddylation using anti-Myc immunoblots. There, we found that increases in the NEDD8 gene dosage also led to increased multi- or polyneddylation of AXR1 (Fig. 7D, AXR1-N8 (2×), lane 6). Importantly, we did not detect apparent differences in AXR1 abundance in den1 as may have been expected following our observation that AXR1 has a slightly reduced stability in den1 mutants (Fig. 5, A and B). Thus, the thioester formation defect in den1 mutant backgrounds might be suppressed by increasing NEDD8 availability. The fact that increasing the abundance of NEDD8 was sufficient to enhance protein neddylation and that DEN1 mutation correlated with reduced RCE1∼NEDD8 thioesters indicated that the control of NEDD8 levels, activation, and transfer are key regulatory steps of the neddylation pathway that are impaired in den1.
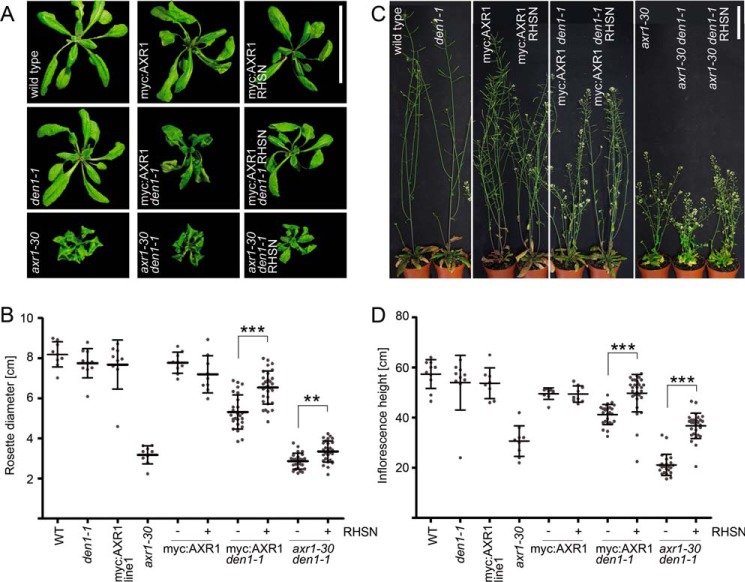
Increased NEDD8 Levels Can Suppress den1 Mutant Phenotypes
When compared with the wild type, den1 mutants do not display any obvious growth defects in standard growth conditions (31). In the sensitized axr1-30 or Myc:AXR1 axr1-30 backgrounds, however, den1 mutants are smaller than plants with the wild type DEN1 allele (Fig. 8). In den1, this phenotype is accompanied by an increase in non-cullin NEDD8-conjugated formation and reduced NCE thioester levels (Fig. 7). Because the biochemical analyses had shown that increasing NEDD8 gene dosage could rescue the den1 defects in RCE1∼NEDD8 formation, we reasoned that this may also lead to a suppression of the DEN1-dependent growth defects in axr-30 and Myc:AXR1 axr-30. Indeed, the rosette growth defects as well as the plant height defects of the axr-30 den1 and Myc:AXR1 axr-30 den1 mutants were at least partially suppressed in the presence of the RHSN transgene (Fig. 8). We concluded that the deneddylation of NAE subunits and other neddylated proteins is one functional role of DEN1 as a NEDD8-specific deneddylase. This could serve to restore NAE function and possibly replenish the pool of free NEDD8 that is available for productive neddylation reactions.
FIGURE 8.
Increased NEDD8 levels suppress den1 phenotypes. A–D, representative photographs of 28-day-old (A) and 42-day-old (C) plants and quantitative analysis of rosette size (B) and plant height (D) of mutant and transgenic plants expressing RUB1::His:Strep:NEDD8 (RHSN) as specified in the figure. Scale bars, 5 cm. The experiment was conducted twice under the same growth conditions with similar outcome; one representative experiment is shown. Mean and S.D. are shown (n = 7–29). Two-sided Mann-Whitney U test was performed for Myc:AXR1 den1-1 (−) versus (+) and axr-30 den1-1 (+) versus (−) respectively: ***, p < 0.001; **, p < 0.01.
Discussion
We have previously reported that Arabidopsis AXR1 can be neddylated in the wild type and that AXR1 neddylation is strongly increased in the den1 mutant. In this study, we examined the molecular mechanisms leading to the NEDD8 modification of the NAE subunit AXR1 (31). Our analyses revealed that the NAE subunit ECR1 as well as the NCE enzyme RCE1 can be NEDD8-modified in a manner similar to AXR1 in vitro, although with a different efficiency in vivo. Because these modifications require an active NAE but can take place in the absence of the NCE, they are best explained by autoneddylation reactions that occur when the activated NAE∼NEDD8 thioester on the catalytically active ECR1 Cys-215 comes in proximity to Lys residues in AXR1 and ECR1.
The presence of the NEDD8 modification on multiple AXR1 Lys residues in the vicinity of the active site NEDD8 thioester indirectly supports the hypothesis that this modification is not targeted but is the result of a general reactivity of the activated NEDD8. NEDD8 activation as an adenylate, thioester formation, and thioester transfer to the NCE require an active site remodeling of the NAE enzyme that might promote the nucleophilic attack of different internal Lys residues on the NEDD8 thioester. This could also provide an explanation for our observation that different Lys can be neddylated in vitro (9). Until now, we were not able to identify the modification site or modification sites of AXR1 in vivo. However, the hypothesis of an unspecific modification reaction is further supported by the fact that the Lys residues found to be NEDD8- modified in the Arabidopsis NAE are not conserved when aligned with the highly similar human counterparts APPBP1 and UBA3, even though our in vitro analysis showed that the human NAE subunits are likewise prone to autoneddylation. Proteomics data provide evidence for Lys modifications of the human enzymes also in vivo. However, the fact that NEDD8 and ubiquitin leave identical mass footprints after tryptic digestion does not allow unequivocal conclusions about the identity of these modifications in vivo (36, 38–40).
The neddylation pathway as well as the other ubiquitin and ubiquitin-like pathways depend on the reactivity of thioester intermediates that can be readily transferred along the conjugation cascade. When considering the possibility of autoneddylation, as demonstrated in our in vitro experiments, it is not surprising that Arabidopsis AXR1, as a protein physically close to NEDD8 due to its enzymatic activity, is also one of the most prominent substrates of this modification in vivo. Both our in vitro and in vivo experiments suggested a preference for NEDD8 to form an isopeptide bond with AXR1 and not ECR1. Even though we could identify several modification sites on ECR1 in vitro, AXR1 appears as the more prominent neddylated NAE subunit in vivo. Thus, NEDD8 modifications on ECR1 detected in vitro that may potentially interfere with NAE activity may not be of functional relevance in vivo.
Several ubiquitin and ubiquitin-like pathway enzymes have previously been reported to likewise carry automodifications. In the case of ubiquitination and SUMOylation, the molecular consequences of these modifications included changes in substrate specificity, protein localization, or reactivity (41–47). We could not detect a strong impact of the NEDD8 modification on AXR1 stability or protein interactions, and our in vivo analyses do not suggest that AXR1 abundance is affected in den1. We did, however, observe a decreased transfer efficiency of activated NEDD8 from the NAE to the NCE. This can be explained by a competition between the NAE and the NCE for a limited NEDD8 monomer pool or by a progressive reduction of NAE activity due to its neddylation. Although we could observe neddylation modifications on ECR1 that could potentially interfere with NAE function, we consider it unlikely that these are limiting because increased expression of NEDD8 alone could suppress the den1-associated phenotypes.
DEN1 is highly conserved in most eukaryotes, with the notable exception of Saccharomyces cerevisiae, where the neddylation pathway is not essential. Although den1 mutants from several species accumulate neddylated proteins of a broad molecular range, there are only very subtle consequences of the loss of DEN1 in most of these organisms (27–29, 31). Our results suggest that Arabidopsis DEN1 serves to deconjugate NEDD8 from substrates that have become neddylated in concert with or independent of an E3 reaction to maintain the functionality of the neddylation machinery for targeted neddylation reactions, most prominently the neddylation of the cullin subunits of the CRL-type E3 ligases. Such a role for DEN1 would indicate that the pool of free and activated NEDD8 can become limiting in a physiological context. Our observation that increasing NEDD8 gene dosage can partially suppress molecular and growth defects of den1 mutants supports this hypothesis. Similar situations have been reported for ubiquitin where it was found that the DUBs Doa4 or USP14/Upb6 are essential for the recycling of ubiquitin for ubiquitin-dependent degradation by the 26S proteasome (41, 48–50). Loss-of-function of these DUBs leads to a depletion of the free ubiquitin pool, and the corresponding phenotypes can be rescued by increasing ubiquitin levels. For NEDD8, controlling the levels of free, thioester-linked, and conjugated forms may also be important in regard to its interaction with the ubiquitin system. As shown previously, increases in NEDD8 abundance can result in the ubiquitin conjugation machinery using NEDD8 rather than ubiquitin as a substrate (51, 52). Because we have observed autoneddylation for the human NAE subunits, it is tempting to speculate that the reactions examined here in detail for the plant proteins may also serve to control NEDD8 abundance and neddylation in other organisms. It remains to be elucidated whether the proposed role of DEN1 to recycle NEDD8 from autoneddylated proteins of the NEDD8 conjugation machinery applies to all NEDD8-modified substrates retrieved in den1 mutants and whether these modifications are merely side reactions or have regulatory functions. The regulation of NAE and NCE activity or interaction poses a potential regulatory checkpoint at the start of ubiquitin and UBL conjugation cascades. Recently, an example of ubiquitin NCE regulation was reported where the family of MEMBRANE-ANCHORED UBIQUITIN FOLD PROTEINS competes with the ubiquitin-activating enzyme for a binding surface on the ubiquitin-conjugating enzyme (53). Given the established function of NEDD8 in the control of CRL activity and subsequently its impact on protein ubiquitylation, the NEDD8 pathway would be well suited to allow a global regulation of diverse CRL-dependent protein degradation processes. In this regard, it is tempting to speculate that NAE autoneddylation and its control by DEN1 form a regulatory feedback loop to attenuate protein neddylation.
Experimental Procedures
Biological Material
All recombinant protein expression was performed in the Escherichia coli strain Rosetta(DE3) pLysS (Madison, WI). The mutants and transgenic lines axr-30, den1-1, Myc:AXR1, Myc:AXR1 den1-1, axr-30 den1-1, and RUB1:His:Strep:NEDD8 (RHSN) were previously described (31). The transgene was introgressed from one original line into the above-listed mutant backgrounds. For each genotype, three lines were confirmed by phenotypic and PCR-based analysis and used for the growth analysis experiment. The experiment was conducted twice under the same growth conditions (22 °C, continuous light) with a similar outcome.
Protein Purification
Protein purification of recombinant proteins was performed using standard approaches. The constructs for the expression of recombinant GST:DEN1, GST:DEN1C166A, His:AXR1, GST:ECR1, and His:FLAG:RCE1 were described previously (31, 32). His:HA:NEDD8 was cloned by amplification of the coding sequence for the processed NEDD8 protein (RUB1 Thr-77–Gly-152) with the primers 1 and 2 into pET28a (Merck, Darmstadt, Germany). Constructs for FLAG:AXR1 and ECR1:His were obtained by insertion of the AXR1 and ECR1 cDNA sequences into pET21a and pET28a, respectively, following amplification of the cDNAs with primers 3 and 4 (AXR1) or 5 and 6 (ECR1). FLAG:AXR1 and ECR1:His were coexpressed in E. coli and purified as a heterodimer. All primer sequences are listed in Table 1. Recombinant human NAE enzyme was purchased from UBPBio (Aurora, CO).
TABLE 1.
Primer sequences
fw indicates forward, and rv indicates reverse.
| No. | Construct | Name | 5′ |
|---|---|---|---|
| 1 | His:HA:NEDD8 | HAN8 NdeI fw | AATATCATATGTATCCATACGATGTTCCAG |
| ATTATGCTGTCGGTGGAGGAGGTGGTACT | |||
| ATGATTAAGGTG | |||
| 2 | His:HA:NEDD8 | N8 EcoRI rv | ATGAATTCTTAACCACCCCTAAGGGCAAG |
| 3 | FLAG:AXR1 | FLAG-AXR1 NdeI fw | ATCCATATGGACTACAAAGACGATGACGAC |
| AAGGGTATGCAAGCAGTAAAAAGATC | |||
| 4 | FLAG:AXR1 | AXR1 NotI rv | ATGCGGCCGCCTACAATTTCAATAACTGAG |
| 5 | ECR1:His | ECR1 NcoI fw | ATCCATGGCTGATCTCGATGTTC |
| 6 | ECR1:His | ECR1 XhoI rv | ATCTCGAGTGCTCCAATGGCTGTATC |
Biochemical Experiments
In vitro neddylation experiments were performed for 50 min at 23 °C with 1 μg of recombinant NAE components (15 pmol) and NEDD8 (80 pmol) in a reaction buffer (RB) containing 20 mm Tris-HCl, pH 8.5, 150 mm NaCl, 10 mm MgCl2, 0.1 mm DTT, and 2 mm ATP. The in vitro trans-thioester reaction was performed for 60 min at 23 °C with 10 pmol of His:AXR1, GST:ECR1, and His:FLAG:RCE1, 100 pmol of His:HA:NEDD8, and 1 pmol of DEN1 in RB at pH 6.8. The effect of DEN1 in the in vitro reactions was tested by incubating 50 pmol of NAE (FLAG:AXR1/ECR1:His), 500 pmol of His:HA:NEDD8, and 10 pmol of DEN1 or DEN1C166A in RB at pH 8 for 16 h at 23 °C. 50 pmol of FLAG:His:RCE1 was subsequently added for 5 or 30 min. Reactions were stopped with non-reducing or reducing (100 mm DTT) SDS-buffer containing 4% SDS, 10% glycerol, and 0.01% bromphenol blue in 20 mm Tris-HCl, 150 mm NaCl, pH 8. To increase the amount of automodified protein for LC-MS/MS analysis, 100 pmol of recombinant NAE (FLAG:AXR1/ECR1:His) and 100 pmol of His:HA:NEDD8 were incubated for 16 h at 23 °C. The control reaction was performed in the absence of ATP. To study the effect of NAE NCE interaction, 100 pmol of His:FLAG:RCE1 was added to the reaction. For the comparison of Arabidopsis NAE (AXR1/ECR1) and human NAE (APPBP1/UBA3), 50 pmol of the respective NAE was incubated with 100 pmol of His:HA:NEDD8 in RB at pH 8.5 for 16 h at 23 °C. NEDD8 conjugates were detected with an anti-HA-peroxidase antibody (1:1000, Roche Diagnostics, Penzberg, Germany). His:FLAG:RCE1 and FLAG:AXR1 were detected using an anti-FLAG antibody (1:3000, Sigma, Taufkirchen, Germany). Gels were stained with Coomassie Brilliant Blue to control for protein loading. Immunoblot signal intensities were quantified with the Fuji Film MultiGauge software analysis tool.
Chemical Treatments
For NEDD8 inhibition, seedlings were grown for 7 days under continuous light on growth medium supplemented with 10 μm MLN4924 (Millennium Pharmaceuticals, Cambridge, MA). The protein stability experiments for Myc:AXR1 in the axr1 and axr1 den1 backgrounds were performed by incubating 7-day-old seedlings with 50 μm CHX in a liquid growth medium. Myc:AXR1 abundance was examined in immunoblots with anti-Myc antibodies (anti-c-Myc, 1:3000; Sigma). An anti-RGA antibody (1:1000) directed against the unstable DELLA repressor protein REPRESSOR-OF-ga1–3 was used as a control for the efficiency of the CHX treatment (54). Anti-CDC2 (1:5000, Santa Cruz Biotechnology, Heidelberg, Germany) was used as a protein loading control. Immunoblot signal intensities were quantified with the Fuji Film MultiGauge software analysis tool.
Gel filtration was performed using 1 mg of crude protein extract in a Superose 6 column (GE Healthcare, Freiburg, Germany) as described previously (55). 0.5-ml fractions were collected, and proteins of a known molecular weight were resolved on an SDS-polyacrylamide gel to determine protein sizes.
Mass Spectrometry
In vitro neddylation reactions were stopped with reducing LDS buffer (NuPAGE, Thermo Fisher Scientific, Waltham, MA) and stained with Coomassie Brilliant Blue for total protein loading. Reduction, alkylation with chloroacetamide, and tryptic in-gel digestion of bands corresponding to NEDD8-modified AXR1 and ECR1 were performed according to standard procedures. Nanoflow liquid chromatography-tandem mass spectrometry (MS) was performed by coupling an Eksigent nano LC-Ultra 1D+ (Sciex, Framingham, MA) to an Orbitrap Velos (Thermo Scientific, Waltham). After 10 min of loading on a trap column (100 μm × 2 cm, packed in-house with Reprosil-Pur C18-AQ 5-μm resin; Dr. Maisch, Ammerbuch, Germany) at a flow rate of 5 μl/min in 100% solvent A (0.1% formic acid in HPLC-grade water), peptides were transferred to an analytical column (75 μm × 40 cm, packed in-house with Reprosil-Pur C18-AQ 5 μm resin; Dr. Maisch, Ammerbuch, Germany) and separated using a 60-min gradient from 4 to 32% solvent B (0.1% formic acid and 5% DMSO in acetonitrile, solvent A: 0.1% formic acid, 5% DMSO in water) at 300 nl/min flow rate. The Orbitrap Velos was operated in data-dependent mode, automatically switching between MS and MS2. Full scan MS spectra (m/z 360–1300) were acquired in the Orbitrap at 30,000 (400 m/z) resolution. Tandem MS spectra were generated for up to 10 precursors in the multipole collision cell using higher energy collisional dissociation (AGC value 3 × 104, normalized collision energy 30%) and analyzed in the Orbitrap at a resolution of 7500 (400 m/z). Raw MS data files were processed with MaxQuant/Andromeda (version 1.5.3.8) and searched against TAIR10 (TAIR_pep_20101214_updated, download November 25, 2015), a corresponding decoy database (reversed protein sequences) and common contaminants for protein identification (56). The variable Lys modification (Gly-Gly) was included in the database search, and modified peptides were filtered using an Andromeda score cutoff of 40 and a localization probability greater than 0.75.
Lys modification sites were combined with a structural model of AXR1/ECR1. The model was obtained with Swiss Model using the structure of human APPBP1/UBA3 (Protein Data Bank identifier 1YOV (8)) and modified with the UCSF Chimera package (Version 1.10.2) (57). Distance measurements were performed with UCSF Chimera. The conservation of NEDD8-modified Lys between Arabidopsis AXR1, AXL, and human APPBP1 as well as between ECR1 and human UBA3 was examined based on ClustalW protein alignments (58). Gly-Gly-modified Lys residues for APPBP1 and UBA3 were derived from PhosphoSitePlus (36).
Statistical Analysis
Results for plant inflorescence height and rosette diameter are expressed as the mean ± S.D. Statistical relevance was obtained using the Mann-Whitney U test using Microsoft Excel 2013 with the Real Statistics Add-In.
Author Contributions
J. M. and C. S. conceived the project and planned the experiments. J. M. performed the experiments. B. K. discussed the project with J. M. and C. S. and directed the MS experiment. J. M. and C. S. wrote the paper. All authors contributed to revising the manuscript.
Acknowledgments
We thank Judy Callis (University of California, Davis) for providing the His:AXR1, GST:ECR1, and His:FLAG:RCE1 constructs as well as for providing the axr1 Myc:AXR1 transgenic line.
This work was supported by Deutsche Forschungsgemeinschaft Grant SCHW751/11-1 (to C. S.) as part of the Schwerpunktprogramm SPP1365 “Ubiquitin Family Proteins” and as part of Sonderforschungsbereich Grant 924 (to C. S. and B. K.). This work was also supported by the COST ACTION BM1307 PROTEOSTASIS in the European Union Framework Programme Horizon 2020. The authors declare that they have no conflicts of interest with the contents of this article.
- UBL
- ubiquitin-like
- NAE
- NEDD8 E1 activating enzyme
- NCE
- NEDD8 E2-conjugating enzyme
- CHX
- cycloheximide
- IB
- immunoblot
- DUB
- deubiquitinating.
References
- 1. Osaka F., Saeki M., Katayama S., Aida N., Toh-E A., Kominami K., Toda T., Suzuki T., Chiba T., Tanaka K., and Kato S. (2000) Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 19, 3475–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jones D., and Candido E. P. (2000) The NED-8 conjugating system in Caenorhabditis elegans is required for embryogenesis and terminal differentiation of the hypodermis. Dev. Biol. 226, 152–165 [DOI] [PubMed] [Google Scholar]
- 3. Tateishi K., Omata M., Tanaka K., and Chiba T. (2001) The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J. Cell Biol. 155, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ou C. Y., Lin Y. F., Chen Y. J., and Chien C. T. (2002) Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 16, 2403–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dharmasiri S., Dharmasiri N., Hellmann H., and Estelle M. (2003) The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J. 22, 1762–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vierstra R. D. (2012) The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 160, 2–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Osaka F., Kawasaki H., Aida N., Saeki M., Chiba T., Kawashima S., Tanaka K., and Kato S. (1998) A new NEDD8-ligating system for cullin-4A. Genes Dev. 12, 2263–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walden H., Podgorski M. S., and Schulman B. A. (2003) Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature 422, 330–334 [DOI] [PubMed] [Google Scholar]
- 9. Huang D. T., Hunt H. W., Zhuang M., Ohi M. D., Holton J. M., and Schulman B. A. (2007) Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature 445, 394–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Komander D., Clague M. J., and Urbé S. (2009) Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 11. del Pozo J. C., Dharmasiri S., Hellmann H., Walker L., Gray W. M., and Estelle M. (2002) AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis Cullin AtCUL1 is required for auxin response. Plant Cell 14, 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pan Z. Q., Kentsis A., Dias D. C., Yamoah K., and Wu K. (2004) Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23, 1985–1997 [DOI] [PubMed] [Google Scholar]
- 13. Enchev R. I., Schulman B. A., and Peter M. (2015) Protein neddylation: beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 16, 30–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saha A., and Deshaies R. J. (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dohmann E. M., Kuhnle C., and Schwechheimer C. (2005) Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant Cell 17, 1967–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cope G. A., and Deshaies R. J. (2003) COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114, 663–671 [DOI] [PubMed] [Google Scholar]
- 17. Lyapina S., Cope G., Shevchenko A., Serino G., Tsuge T., Zhou C., Wolf D. A., Wei N., Shevchenko A., and Deshaies R. J. (2001) Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292, 1382–1385 [DOI] [PubMed] [Google Scholar]
- 18. Schwechheimer C., Serino G., Callis J., Crosby W. L., Lyapina S., Deshaies R. J., Gray W. M., Estelle M., and Deng X. W. (2001) Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292, 1379–1382 [DOI] [PubMed] [Google Scholar]
- 19. Pierce N. W., Lee J. E., Liu X., Sweredoski M. J., Graham R. L., Larimore E. A., Rome M., Zheng N., Clurman B. E., Hess S., Shan S. O., and Deshaies R. J. (2013) Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell 153, 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu S., Zhu W., Nhan T., Toth J. I., Petroski M. D., and Wolf D. A. (2013) CAND1 controls in vivo dynamics of the cullin 1-RING ubiquitin ligase repertoire. Nat. Commun. 4, 1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zemla A., Thomas Y., Kedziora S., Knebel A., Wood N. T., Rabut G., and Kurz T. (2013) CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nat. Commun. 4, 1641. [DOI] [PubMed] [Google Scholar]
- 22. Bostick M., Lochhead S. R., Honda A., Palmer S., and Callis J. (2004) Related to ubiquitin 1 and 2 are redundant and essential and regulate vegetative growth, auxin signaling, and ethylene production in Arabidopsis. Plant Cell 16, 2418–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dharmasiri N., Dharmasiri S., Weijers D., Karunarathna N., Jurgens G., and Estelle M. (2007) AXL and AXR1 have redundant functions in RUB conjugation and growth and development in Arabidopsis. Plant J. 52, 114–123 [DOI] [PubMed] [Google Scholar]
- 24. Gusmaroli G., Feng S., and Deng X. W. (2004) The Arabidopsis CSN5A and CSN5B subunits are present in distinct COP9 signalosome complexes, and mutations in their JAMM domains exhibit differential dominant negative effects on development. Plant Cell 16, 2984–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mendoza H. M., Shen L. N., Botting C., Lewis A., Chen J., Ink B., and Hay R. T. (2003) NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J. Biol. Chem. 278, 25637–25643 [DOI] [PubMed] [Google Scholar]
- 26. Gan-Erdene T., Nagamalleswari K., Yin L., Wu K., Pan Z. Q., and Wilkinson K. D. (2003) Identification and characterization of DEN1, a deneddylase of the ULP family. J. Biol. Chem. 278, 28892–28900 [DOI] [PubMed] [Google Scholar]
- 27. Christmann M., Schmaler T., Gordon C., Huang X., Bayram O., Schinke J., Stumpf S., Dubiel W., and Braus G. H. (2013) Control of multicellular development by the physically interacting deneddylases DEN1/DenA and COP9 signalosome. PLoS Genet. 9, e1003275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chan Y., Yoon J., Wu J. T., Kim H. J., Pan K. T., Yim J., and Chien C. T. (2008) DEN1 deneddylates non-cullin proteins in vivo. J. Cell Sci. 121, 3218–3223 [DOI] [PubMed] [Google Scholar]
- 29. Zhou L., and Watts F. Z. (2005) Nep1, a Schizosaccharomyces pombe deneddylating enzyme. Biochem. J. 389, 307–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vogl A. M., Brockmann M. M., Giusti S. A., Maccarrone G., Vercelli C. A., Bauder C. A., Richter J. S., Roselli F., Hafner A. S., Dedic N., Wotjak C. T., Vogt-Weisenhorn D. M., Choquet D., Turck C. W., Stein V., et al. (2015) Neddylation inhibition impairs spine development, destabilizes synapses and deteriorates cognition. Nat. Neurosci. 18, 239–251 [DOI] [PubMed] [Google Scholar]
- 31. Mergner J., Heinzlmeir S., Kuster B., and Schwechheimer C. (2015) DENEDDYLASE1 deconjugates NEDD8 from non-cullin protein substrates in Arabidopsis thaliana. Plant Cell 27, 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hotton S. K., Eigenheer R. A., Castro M. F., Bostick M., and Callis J. (2011) AXR1-ECR1 and AXL1-ECR1 heterodimeric RUB-activating enzymes diverge in function in Arabidopsis thaliana. Plant Mol. Biol. 75, 515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lincoln C., Britton J. H., and Estelle M. (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2, 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Leyser H. M., Lincoln C. A., Timpte C., Lammer D., Turner J., and Estelle M. (1993) Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364, 161–164 [DOI] [PubMed] [Google Scholar]
- 35. Pozo J. C., Timpte C., Tan S., Callis J., and Estelle M. (1998) The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science 280, 1760–1763 [DOI] [PubMed] [Google Scholar]
- 36. Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., and Skrzypek E. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., et al. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 [DOI] [PubMed] [Google Scholar]
- 38. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., and Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mertins P., Qiao J. W., Patel J., Udeshi N. D., Clauser K. R., Mani D. R., Burgess M. W., Gillette M. A., Jaffe J. D., and Carr S. A. (2013) Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods 10, 634–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., and Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Walz T., Ploegh H., and Finley D. (2002) Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 [DOI] [PubMed] [Google Scholar]
- 42. Banerjee A., Gregori L., Xu Y., and Chau V. (1993) The bacterially expressed yeast CDC34 gene product can undergo autoubiquitination to form a multiubiquitin chain-linked protein. J. Biol. Chem. 268, 5668–5675 [PubMed] [Google Scholar]
- 43. Bencsath K. P., Podgorski M. S., Pagala V. R., Slaughter C. A., and Schulman B. A. (2002) Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J. Biol. Chem. 277, 47938–47945 [DOI] [PubMed] [Google Scholar]
- 44. Arnold J. E., and Gevers W. (1990) Auto-ubiquitination of ubiquitin-activating enzymes from chicken breast muscle. Biochem. J. 267, 751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Banka P. A., Behera A. P., Sarkar S., and Datta A. B. (2015) RING E3-catalyzed E2 self-ubiquitination attenuates the activity of Ube2E ubiquitin-conjugating enzymes. J. Mol. Biol. 427, 2290–2304 [DOI] [PubMed] [Google Scholar]
- 46. Truong K., Lee T. D., and Chen Y. (2012) Small ubiquitin-like modifier (SUMO) modification of E1 Cys domain inhibits E1 Cys domain enzymatic activity. J. Biol. Chem. 287, 15154–15163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Truong K., Lee T. D., Li B., and Chen Y. (2012) Sumoylation of SAE2 C terminus regulates SAE nuclear localization. J. Biol. Chem. 287, 42611–42619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reyes-Turcu F. E., Ventii K. H., and Wilkinson K. D. (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Papa F. R., and Hochstrasser M. (1993) The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature 366, 313–319 [DOI] [PubMed] [Google Scholar]
- 50. Swaminathan S., Amerik A. Y., and Hochstrasser M. (1999) The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10, 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hjerpe R., Thomas Y., Chen J., Zemla A., Curran S., Shpiro N., Dick L. R., and Kurz T. (2012) Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes. Biochem. J. 441, 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hjerpe R., Thomas Y., and Kurz T. (2012) NEDD8 overexpression results in neddylation of ubiquitin substrates by the ubiquitin pathway. J. Mol. Biol. 421, 27–29 [DOI] [PubMed] [Google Scholar]
- 53. Lu X., Malley K. R., Brenner C. C., Koroleva O., Korolev S., and Downes B. P. (2016) A MUB E2 structure reveals E1 selectivity between cognate ubiquitin E2s in eukaryotes. Nat. Commun. 7, 12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Willige B. C., Ghosh S., Nill C., Zourelidou M., Dohmann E. M., Maier A., and Schwechheimer C. (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19, 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schwechheimer C., Serino G., and Deng X. W. (2002) Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14, 2553–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 57. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 58. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]