Abstract
We have analyzed the pathway networks of ischemia-affected and remote myocardial areas after repetitive ischemia/reperfusion (r-I/R) injury without ensuing myocardial infarction (MI) to elaborate a spatial- and chronologic model of cardioprotective gene networks to prevent left ventricular (LV) adverse remodeling. Domestic pigs underwent three cycles of 10/10 min r-I/R by percutaneous intracoronary balloon inflation/deflation in the mid left anterior descending artery, without consecutive MI. Sham interventions (n = 8) served as controls. Hearts were explanted at 5 h (n = 6) and 24 h (n = 6), and transcriptomic profiling of the distal (ischemia-affected) and proximal (non-affected) anterior myocardial regions were analyzed by next generation sequencing (NGS) and post-processing with signaling pathway impact and pathway network analyses. In ischemic region, r-I/R induced early activation of Ca-, adipocytokine and insulin signaling pathways with key regulator STAT3, which was also upregulated in the remote areas together with clusterin (CLU) and TNF-alpha. During the late phase of cardioprotection, antigen immunomodulatory pathways were activated with upregulation of STAT1 and CASP3 and downregulation of neprilysin in both zones, suggesting r-I/R induced intrinsic remote conditioning. The temporo-spatially differently activated pathways revealed a global myocardial response, and neprilysin and the STAT family as key regulators of intrinsic remote conditioning for prevention of adverse remodeling.
Pre-infarction angina, termed “warm-up angina”, increases myocardial resistance to subsequent ischemia, thereby reducing infarct size and mortality1. Exposure of the myocardium to single or repetitive brief episodes of ischemia and reperfusion (I/R), before sustained ischemia, induces cardioprotection, which enhances the ability of the myocardium to withstand the next ischemic insult2,3. Most ischemic preconditioning (IPC) protocols incorporate a sustained coronary occlusion following the IPC-stimulus, causing acute myocardial infarction (AMI). Problematically, the extensive myocardial damage incurred can mask changes to the genomic, proteomic, or metabolic profiles, which would be attributable solely to I/R-linked cardioprotection. Only few reports have employed repetitive I/R (r-I/R) without ensuing AMI to reveal the impact of r-I/R alone, using small animal models with open-chest procedures, or Langendorff perfused hearts in vitro4. Despite extensive research, translational breakthroughs have not yet been reached5, mainly due to the recognition that the experimentally proven powerful IPC-related cardioprotective substances are effective before infarction, and logically should be applied prior to the onset of infarction in the clinical scenario. Therefore, to date, IPC could not achieve practical clinical relevance.
The mechanisms underlying the early (up to 3 hours) and late (from 24 hours to several days post i-I/R) phase of IPC seem diverse: rapidly released and activated transmitter molecules confer beneficial effects in the acute phase, whereas cardioprotection in second windows of protection (SWOP) is instigated by a more complex response that requires de novo synthesis of effector proteins6. In contrast to IPC, postconditioning (PostIC) and remote ischemic conditioning (RIPC) are already used in clinical settings, albeit recent meta-analyses revealed a lack of definitive success for both in human procedures. This is partially explained by the application of potentially confounding cardioprotective medications7.
Since the term IPC is closely associated with subsequent AMI, we use here the expression r-I/R injury for clear separation of the two distinct phenomena. While IPC (with AMI) represents the warm-up angina followed by ST-segment elevation myocardial infarction (STEMI), r-I/R corresponds to a stable angina in humans.
To date, high-throughput gene analyses have not yet been used for dissecting the effects of IPC or r-I/R in a closed-chest, catheter-based setting in a clinically relevant large animal model, without the masking effect of infarction7,8, even if this approach is useful for identifying single genes or pathways, or entire pathway networks, and is thus well suited for exploring the complexity of the cardioprotection mechanism9.
Here we analyzed the effects of the r-I/R stimulus without subsequent infarction-related injury in a porcine model using next generation sequencing (NGS) combined with pathway network analysis. Our aim was to reveal the complex transcriptomic response that is involved in cardioprotection in ischemic and ischemia-unaffected regions of the heart, and to identify novel genes and networks that are responsible for protection of the heart against I/R. We analyzed the relevant genes and pathways at an early time point after r-I/R injury (early window, trigger of cardioprotection) and 24 h later (second window, performing cardioprotection), and elucidated the time-sequence of the activation of pathways and genes with conditioning function in the ischemic and the ischemia non-affected region, termed “intrinsic remote conditioning” responsible for prevention of LV adverse remodeling.
Results
A translational model of r-I/R and IPC
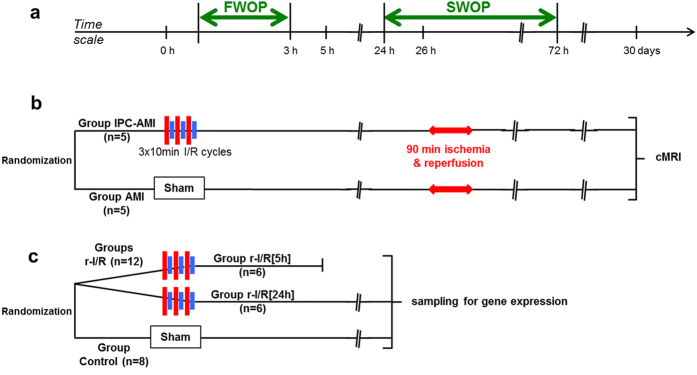
In contrast to our translational model, small animal open-chest models, with high heart rates differ substantially from the biological scenario presented by the human transient ischemic heart attack. Furthermore, in vitro models lack the pathophysiological complexity of the organism. Our assumption, that the typical 30 sec myocardial r-I/R cycles used in rodents for mimicking r-I/R or IPC, would be inadequate in provoking a sustained ischemic response in either pigs or humans, was evidence-based from our previous work showing that indications of adaptive mechanisms can be observed after a minimum of 5 min coronary occlusion in pigs10. We found that three 10 min cycles of r-I/R failed to provoke irreversible myocyte injury, as assessed by echocardiography, left ventricular (LV) hemodynamic measurements, necroenzyme release, or microscopic imaging, but induced substantial transcriptomic changes not only within the ischemic injury, but also in the remote myocardial area. Therefore, we induced r-I/R in pigs by using 3 × 10 min I/R by percutaneous balloon inflation/deflation of the mid left anterior descending coronary artery (LAD), followed by recovery of the animals. Sham-operated animals served as control (group control). At the pre-specified (5 h and 24 h) follow-ups (groups r-I/R [5 h] and r-I/R [24 h], respectively), the heart was explanted and the transcriptomics of the ischemia-affected distal anterior, border mid anterior and ischemia-non-affected proximal anterior parts of the heart were determined (Fig. 1). In order to prove the cardioprotective effect of the 3 × 10 min r-I/R stimulus, we have additionally induced AMI 26 h post r-I/R (SWOP phase) (group IPC-AMI) or sham I/R (group AMI) (Fig. 1).
Figure 1. Study design.
(a) Time scale of the experiments. FWOP: first window of protection; SWOP: second window of protection. (b) Efficacy study randomizing domestic pigs into two groups without (Group AMI) or with ischemic preconditioning (IPC; Group IPC-AMI) 26 h before acute myocardial infarction (AMI). Cardiac magnetic resonance imaging (cMRI) at 30 days. (c) Gene expression analysis study randomizing pigs into groups with repetitive ischemia/reperfusion (r-I/R) and sacrificing at 5 h (upstream phase of SWOP) and 24 h (SWOP phase) or control.
The r-I/R stimulus induces transient alterations in LV-hemodynamics without persistent myocardial dysfunction
The results of hemodynamic measurements, both invasive and non-invasive, at baseline, directly after the r-I/R stimulus and at follow-up are shown in Supplementary Table S2. As a direct response to the r-I/R stimulus, there was a significant decrease in LV systolic pressures, and an increase in LV end-diastolic pressure and isovolumic relaxation time with normalization at 5 h or 24 h follow-ups.
Biomarkers and histology
The r-I/R stimulus failed to change plasma levels of troponin I, myoglobin or CK (Fig. 2a). Histologic images of the proximal, mid and distal anterior wall regions showed no signs of morphological myocardial injury (Fig. 2b).
Figure 2. Laboratory data, histology and fluorescence microscopy of the animals with repetitive ischemia/reperfusion (r-I/R) [5 h] and r-I/R [24 h]) or without (group control) r-I/R.
(a) Serum troponin I (TnI), plasma myoglobin, and serum creatine kinase (CK) at baseline, immediately after r-I/R (post-r-I/R) or sham-r-I/R, at 5 h and 24 h. Animals of groups r-I/R [5 h] and r-I/R [24 h] were pooled as group r-I/R at baseline (n = 12) and post r-I/R (n = 12). Group control: n = 8, 5 h post-r-I/R: n = 6 and 24 h post-r-I/R n = 6. No significant differences between the groups were observed at any time point. Each data point represents the result of one animal, and mean ± s.d. is indicated for each group. (b) Haematoxylin-eosin staining (20x magnification) and alpha-actin (green) with DAPI (blue) staining (scale bar 10 μM) of myocardial samples of the proximal, mid and distal anterior regions. No structural changes of the myocardium after r-I/R without subsequent infarction were observed.
r-I/R leads to time-dependent changes in gene expression compared to non-conditioned hearts
Summary of retrieved genes
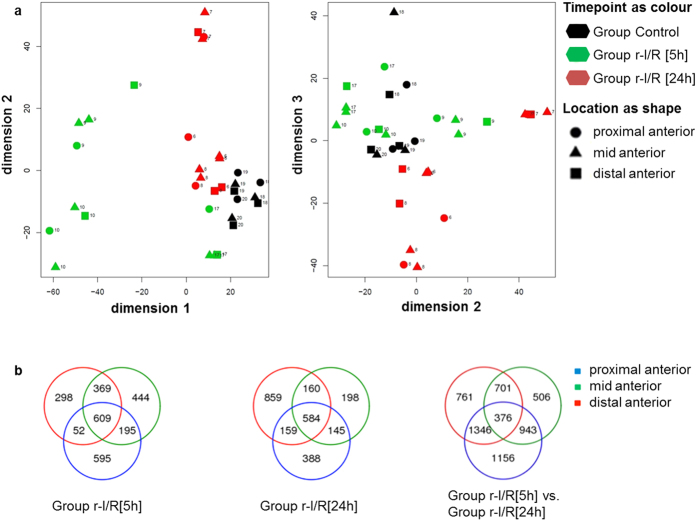
The comparison of the groups r-I/R [5 h] and r-I/R [24 h] with the control group showed that a total of 13758 genes were retrieved by NGS. PCA showed good discriminatory power between the three experimental groups with adequate similarity within the r-I/R groups even though originating from different anterior wall myocardial regions (Fig. 3a). The expressional differences between the r-I/R [5 h] group and controls were greater than between the r-I/R [24 h] group and controls, suggesting more pronounced alterations from normal cell function at 5 h than at 24 h after r-I/R.
Figure 3. Principal component analysis (PCA) and Venn diagrams of the gene expression analysis.
(a) Isomaps of 13948 analyzed genes of the myocardial samples from the proximal, mid and distal anterior wall regions from control animals (black) and animals 5 h (r-I/R [5 h], green) or 24 h (r-I/R [24 h], red) after the repetitive ischemia/reperfusion (r-I/R) stimulus. The distal anterior wall was regarded as ischemia/reperfusion-affected area. (b) Overlap of the altered gene expression in the proximal, mid and distal anterior regions following the r-I/R stimulus. Venn diagrams show genes with significantly altered expression in the proximal (blue), mid (green) and distal anterior wall (red) regions in the groups r-I/R [5 h] and r-I/R [24 h] compared to controls, and r-I/R [5 h] compared to r-I/R [24 h].
Venn diagrams showed significantly altered gene expression between control and r-I/R animals (regardless of the time) and between r-I/R -induced early and late changes for proximal (ischemia-unaffected), mid (border) and distal (ischemia-affected) anterior regions (Fig. 3b). Remarkably, also the proximal region, which was not directly affected by ischemia, showed distinct changes in gene expressions already after 5 h. A total of 5789 genes were differentially expressed between the myocardial samples of the groups r-I/R [5 h] and r-I/R [24 h], equally affecting all three regions.
In the distal anterior wall region, a total of 1328 and 1762 genes were differentially expressed at 5 h and at 24 h, respectively. In the mid anterior region (border zone of ischemia/reperfusion), 1617 and 1087 genes were deregulated 5 h and 24 h post r-I/R, respectively. Regarding the proximal anterior wall region, 1451 and 1276 genes showed differential expression at 5 h and 24 h post r-I/R.
Activated pathways and pathway networks of ischemia-affected and non-affected myocardial regions
Table 1 shows the results of the pathway analyses of the retrieved genes according to the different regions at 5 h and 24 h post r-I/R stimulus compared to controls. Results are presented as SPIA plots with false discovery rates of 5% and 10% for the corresponding pathways (Supplementary Fig. S1). Pathway analysis revealed that there were multiple networks involved at 5 h in the ischemia-affected area, primarily including the calcium signaling, the adipocytokine and insulin signaling pathways. At 24 h (during the late phase of cardioprotection), the immunomodulatory antigen processing and presentation, focal adhesion and extracellular matrix-receptor interaction pathways all were involved in the distal anterior area. The Ca-signaling pathway and the antigen processing and presentation pathways were also activated in the remote myocardial areas at 5 h and 24 h, respectively, indicating decisive roles in intrinsic remote conditioning (Table 1 and Supplementary Fig. S2). Table 2 and Fig. 4 display significantly deregulated genes, which are part of these relevant pathways.
Table 1. Significantly activated pathways in distal (ischemia-affected), mid (border zone of ischemia) and proximal (non-ischemia affected) anterior wall regions of the heart at 5 h (group r-I/R [5 h]) or 24 h (group r-I/R [24 h]) after repetitive ischemia/reperfusion (r-I/R) without subsequent myocardial infarction.
| Location | Name of pathway | ID of pathway | p- value* |
|---|---|---|---|
| Group r-I/R [5 h] | |||
| Distal anterior | Calcium signaling pathway | 4020 | 0.000 |
| Adipocytokine signaling pathway | 4920 | 0.008 | |
| Insulin signaling pathway | 4910 | 0.009 | |
| Mid anterior | Calcium signaling pathway | 4020 | 0.002 |
| Adipocytokine signaling pathway | 4920 | 0.022 | |
| Proximal anterior | Calcium signaling pathway | 4020 | 0.001 |
| Group r-I/R [24 h] | |||
| Distal anterior | Antigen processing and presentation | 4612 | 0.005 |
| Focal adhesion | 4510 | 0.044 | |
| Extracellular matrix-receptor interaction | 4512 | 0.044 | |
| Mid anterior | Antigen processing and presentation | 4612 | 0.000 |
| Complement and coagulation cascades | 4610 | 0.002 | |
| Graft-versus-host disease | 5332 | 0.004 | |
| Allograft rejection | 5330 | 0.004 | |
| Type I diabetes mellitus | 4940 | 0.004 | |
| Viral myocarditis | 5416 | 0.041 | |
| Cytosolic DNA-sensing pathway | 4623 | 0.042 | |
| Proximal anterior | Antigen processing and presentation | 4612 | 0.000 |
| Complement and coagulation cascades | 4610 | 0.010 | |
*Multiplicity correction at false discovery rate 5%.
Table 2. Deregulated genes and their expression (fold changes), which are involved in the activated pathway networks in the distal (ischemia-affected), mid (border zone of ischemia) and proximal (not ischemia-affected) anterior wall regions of the heart at 5 h group r-I/R [5 h] or 24 h group r-I/R [24 h] after repetitive ischemia/reperfusion (r-I/R) without subsequent myocardial infarction.
| Gene code | Gene name | Group r-I/R [5 h] | Group r-I/R [5 h] | Group r-I/R [5 h] | Group r-I/R [24 h] | Group r-I/R [24 h] | Group r-I/R [24 h] |
|---|---|---|---|---|---|---|---|
| Distal | Mid | Proximal | Distal | Mid | Proximal | ||
| NCOA1 | nuclear receptor coactivator 1 | 0.13 | 0.28 | 0.35 | −0.11 | −0.01 | 0.12 |
| CPT1A | carnitine O-palmitoyltransferase 1 | 0.57 | 0.54 | 0.65 | 0.04 | 0.25 | 0.24 |
| IKBKG | nuclear factor kappa-B essential modulator | 0.94 | 0.77 | 0.99 | 0.46 | 0.46 | 0.48 |
| PPARA | peroxisome proliferator-activated receptor alpha | 1.24 | 1.19 | 0.89 | −0.33 | −0.45 | −0.09 |
| SLC2A4 | Solute carrier family 2 | 0.82 | 0.55 | 0.38 | −0.92 | −0.55 | −1.04 |
| STAT3 | Signal transducer and activator of transcription 3 | 0.65 | 0.64 | 0.66 | −0.33 | 0.03 | 0.05 |
| TNFRSF1A | tumor necrosis factor receptor superfamily member 1 A | 0.50 | 1.30 | 0.23 | 0.21 | 1.33 | 0.04 |
| HK2 | hexokinase-2 | 3.36 | 1.64 | 1.09 | −0.86 | −1.43 | −0.49 |
| ALB | albumin | −1.96 | −1.35 | −1.59 | −0.22 | −0.7 | −0.5 |
| TRAF6 | TNF receptor-associated factor 6 | 0.31 | 0.41 | 0.35 | −0.46 | −0.03 | −0.11 |
| CAMKK2 | calcium/calmodulin dependent protein kinase kinase 2 | −0.58 | −0.51 | −0.45 | −0.8 | −0.66 | −0.77 |
| MET | hepatocyte growth factor receptor precursor | 0.74 | 0.58 | 0.33 | 0.26 | 0.16 | 0.01 |
| TNFRSF1B | tumor necrosis factor receptor superfamily member 1B | 0.48 | 0.33 | 0.35 | 0 | 0.21 | 0.22 |
| IKBKB | inhibitor of nuclear factor kappa-B kinase subunit beta | −0.83 | −0.8 | −0.89 | −0.66 | −0.33 | −0.51 |
| NEK6 | serine/threonine-protein kinase Nek6 | 0.21 | 0.18 | 0.31 | −0.47 | −0.19 | −0.35 |
| BMP6 | bone morphogenetic protein 6 | 0.53 | 0.27 | 0.57 | 0.10 | 0.40 | 0.33 |
| MAPK14 | mitogen-activated protein kinase 14 | 0.11 | 0.22 | 0.40 | −0.23 | −0.03 | 0.14 |
| IL4R | interleukin-4 receptor subunit alpha | 0.54 | 0.50 | 0.61 | 0.41 | 0.41 | 0.33 |
| CXCL10 | C-X-C motif chemokine 10 | 0.99 | 1.13 | 1.23 | 1.90 | 2.21 | 2.37 |
| HMGB1 | high mobility group protein B1 | 0.14 | 0.17 | 0.28 | −0.08 | −0.02 | 0.15 |
| CLU | clusterin | 2.48 | 2.3 | 2.14 | 3.67 | 3.71 | 3.69 |
| FAS | tumor necrosis factor receptor superfamily member 6 precursor | 0.54 | 0.43 | 0.66 | 0.05 | 0.17 | 0.37 |
| CAT | catalase | −0.8 | −0.62 | −0.8 | −0.5 | −0.2 | −0.2 |
| ARNTL | aryl hydrocarbon receptor nuclear translocator-like protein | 1.10 | 1.29 | 0.94 | −0.10 | −0.09 | 0.16 |
| JAK3 | tyrosine-protein kinase | 0.35 | 0.51 | 0.72 | 0.33 | 0.64 | 0.51 |
| IRF1 | interferon regulatory factor 1 | 0.60 | 0.53 | 0.76 | 0.77 | 0.97 | 0.88 |
| CD14 | monocyte differentiation antigen CD14 | 1.11 | 1.21 | 1.14 | 0.41 | 0.82 | 0.53 |
| STAT1 | signal transducer and activator of transcription 1 | 0.29 | 0.5 | 0.57 | 0.68 | 0.83 | 0.98 |
| CAV1 | caveolin-1 | 0.40 | 0.38 | 0.48 | −0.25 | −0.05 | 0.17 |
| F2R | proteinase-activated receptor 1 precursor | 0.51 | 0.40 | 0.56 | −0.09 | 0.22 | 0.32 |
| CXCL9 | C-X-C motif chemokine 9 | 0.50 | 0.66 | 1.22 | 1.50 | 1.93 | 2.00 |
| CCR2 | chemokine (C-C motif) receptor 2 | 1.61 | 2.03 | 1.89 | 0.83 | 1.40 | 1.17 |
| TGFBR2 | TGF-beta receptor type-2 | −0.21 | −0.02 | 0.07 | −0.73 | −0.15 | −0.27 |
| DPP4 | dipeptidyl peptidase 4 | 0.52 | 0.69 | 0.82 | −0.47 | 0.01 | 0.22 |
| COL5A1 | collagen alpha-1 (V) chain | 0.58 | 0.62 | 0.23 | 0.38 | 0.4 | 0.4 |
| SOX9 | transcription factor SOX-9 | 0.29 | −0.05 | 0.11 | 0.91 | 1.03 | −0.37 |
| SPARC | SPARC precursor | 0.02 | −0.15 | 0.12 | −0.63 | −0.25 | −0.32 |
| TIMP1 | metalloproteinase inhibitor 1 precursor | 0.32 | 0.05 | 0 | 1.08 | 0.55 | 0.24 |
| CCL2 | C-C motif chemokine 2 | 0.07 | 0.09 | 0.35 | 0.94 | 0.77 | 0.76 |
| CXCL12 | stromal cell-derived factor 1 | 0.01 | −0.25 | 0.35 | −0.68 | −0.22 | −0.05 |
| NOS2 | nitric oxide synthase, inducible | −0.73 | −0.86 | −0.44 | −0.83 | −0.6 | −0.68 |
| CASP3 | caspase-3 subunit | 0.42 | 0.49 | 0.41 | 0.64 | 0.58 | 0.85 |
| NFKB | NF-kappa-B inhibitor | 0.31 | −0.32 | 0.8 | 0.3 | 0.33 | 0.34 |
| TYK2 | non-receptor tyrosine-protein kinase | −0.74 | −0.57 | −0.48 | −0.48 | −0.22 | −0.28 |
| KIT | mast/stem cell growth factor receptor precursor | 1 | 1 | 0.89 | −0.78 | −0.6 | −0.44 |
| MMP2 | 72 kDa type IV collagenase | 0.08 | −0.11 | 0.09 | −0.65 | −0.12 | −0.25 |
| APOE | apolipoprotein E precursor | 0.06 | 0.31 | 0.35 | 1.45 | −0.54 | 1.22 |
| GPC3 | glypican 3 | −0.19 | −0.13 | −0.44 | −0.74 | −0.2 | −0.52 |
| MME | neprilysin | 1.15 | 0.92 | 1.07 | −0.81 | −0.09 | −0.03 |
| ACE | angiotensin-converting enzyme | 0.15 | 0.25 | 0.08 | 0.4 | −0.57 | 0.01 |
| ND1 | NADH-ubiquinone oxidoreductase chain 1 | 0.05 | 0.1 | −0.14 | −0.5 | −0.63 | −0.6 |
| SOD1 | superoxide dismutase | 0.18 | 0.04 | 0.05 | −0.56 | −0.46 | 0.28 |
| TGFBI | transforming growth factor, beta-induced | −0.29 | −0.20 | −0.17 | −0.07 | 0.15 | −0.09 |
| CRYAB | alpha-crystallin B chain | 0.61 | 0.38 | 0.18 | 0.54 | 0.13 | 0.0 |
| COL1A1 | collagen, type I, alpha 2 | 0.11 | −0.25 | 0.23 | −0.38 | −0.20 | −0.3 |
| COL5A2 | collagen, type 2, alpha 2 | −0.08 | −0.07 | 0.25 | −0.86 | −0.18 | −0.13 |
| TUBA4A | tubulin | −1.47 | −1.44 | −1.73 | −1.39 | −1.61 | −1.87 |
| SOX9 | transcription factor SOX-9 | 0.29 | −0.05 | 0.11 | 0.91 | 1.03 | 1.05 |
| NME1 | nucleoside diphosphate kinase A | 0.05 | −0.25 | −0.03 | 1.03 | 0.14 | 0.81 |
| PCNA | proliferating cell nuclear antigen | −0.02 | −0.11 | 0.30 | −0.40 | −0.28 | −0.27 |
| THBD | thrombomodulin | 0.50 | 0.48 | 0.64 | 0.67 | 0.80 | 0.74 |
| PLAU | urokinase-type plasminogen activator | 0.42 | 0.50 | 0.48 | 0.45 | 0.63 | 0.56 |
| ITGAV | integrin alpha-V | 0.30 | 0.37 | 0.68 | 0.58 | 0.59 | 0.99 |
| F3 | tissue factor | −0.56 | 0.20 | 0.82 | 1.00 | 1.19 | 1.32 |
| HMOX1 | heme oxygenase 1 | −0.31 | −0.67 | −0.38 | −0.62 | −0.97 | −0.92 |
| TNFSF10 | tumor necrosis factor ligand superfamily member 10 | 0.07 | −0.03 | 0.58 | 0.37 | 0.77 | 0.91 |
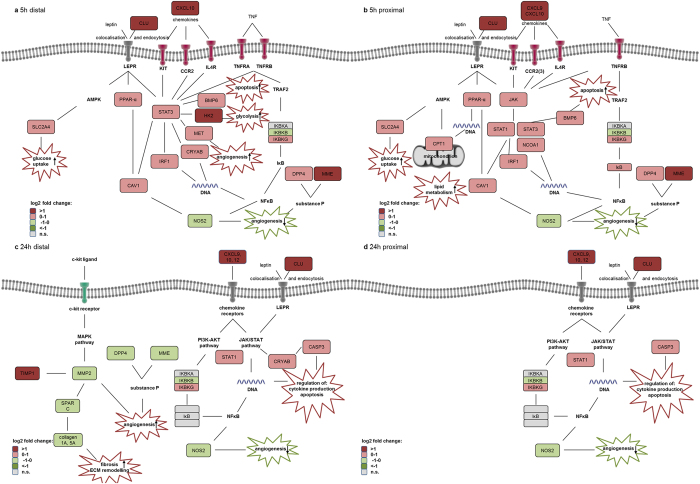
Figure 4. Significantly deregulated gene- and pathway networks at 5 h (group r-I/R [5 h)] and 24 h (group r-I/R [24 h]) after repetitive ischemia/reperfusion (I/R) in the I/R-affected (distal anterior) and –non-affected (proximal anterior) myocardial areas.
(a) 5 h after r-I/R in the myocardial area subjected to r-I/R. A strong activation of cytokines, their receptors, and downstream signaling molecules of the JAK/STAT and other pathways were found. In particular, strong up regulation of hexokinase 2 (HK2), the hepatocyte growth factor nuclear receptor (MET), interferon regulatory factor 1 (IRF1), alpha-crystallin B chain (CRYAB), and the glucose transporter Glut4 (SLC2A4) resulted, all of which are enhancing either angiogenesis or energy consumption (glucose uptake and glycolysis). Upregulation of peptidases neprilysin (MME) and dipeptidyl peptidase 4 (DPP4), both mediating substance P hydrolysis and inactivation, also indirectly regulates angiogenesis through NO production. (b) 5 h after r-I/R in the non-ischemic area. The genes with differential expression in the non-ischemic area are somewhat similar to those found in the ischemic area. A stronger activation of chemokines (CXCL9) and members of the JAK/STAT pathway (JAK3, STAT1) was detected, but no significant regulation of downstream modulators of angiogenesis and glycolysis. This indicates a weaker effect in the remote area, which is not directly affected by ischemia. (c) 24 h after r-I/R in the myocardial area subjected to r-I/R. At this time point, a strong up regulation of chemokines was encountered, with a less pronounced differential expression of components of the JAK/STAT pathway (upregulation of STAT1, but not STAT3 or JAK3) or of the NF-κB pathway. Downregulation of critical components of the regulation of collagen production was found, and reduced expression of peptidases MME and DPP4, which enhance angiogenesis through derepression of substance P. (d) 24 h after r-I/R in the non-ischemic area. In the non-ischemic area, lower regulatory effects were detected. The alterations in chemokines and the JAK/STAT pathway is similar to the ischemic area at this time point, but no effect on genes that are essential for collagen production or substance P inhibition was present.
Table 3 and Supplementary Table S3 show a list of relevant selected genes of additional pathways that are significantly involved, showing expressional changes at 5 h and 24 h in the different regions. The genes are listed according to functional classes or established pathways, and changes in expression are indicated according to time points and regions.
Table 3. Genes with significantly altered expression according to functional groups in distal (ischemia-affected), mid (border zone of ischemia) and proximal (not ischemia-affected) anterior wall regions of the heart at 5 h (group r-I/R [5 h]) or 24 h (group r-I/R [24 h]) after repetitive ischemia/reperfusion (r-I/R) without subsequent myocardial infarction.
| Functional group | Gene code | Gene name | Function | Group r-I/R[5 h] | Group r-I/R[5 h] | Group r-I/R[5 h] | Group r-I/R[24 h] | Group r-I/R[24 h] | Group r-I/R[24 h] |
|---|---|---|---|---|---|---|---|---|---|
| Distal | Mid | Proximal | Distal | Mid | Proximal | ||||
| Apoptosis/survival | FAIM | Fas apoptotic inhibitory molecule | Protects against death receptor-triggered apoptosis | −1.7 | −1.6 | −1.7 | −1.7 | −1 | −1.2 |
| SHISA2 | Shisa family member | Attenuates both FGF and WNT signaling | 2.7 | 2.5 | 2.7 | 1.8 | 2.4 | 1.9 | |
| TNFRSF12A | Tumor necrosis factor receptor superfamily, member 12 A | Promotes angiogenesis and the proliferation of endothelial cells | 1.8 | 1.3 | 1.8 | 2 | 1.2 | 1.2 | |
| CLU/APOJ | Clusterin | Secreted chaperone. suggested role in cell death | 2.5 | 2.3 | 2.5 | 3.6 | 3.7 | 3.7 | |
| CASP3 | Caspase 3, apoptosis-related cysteine peptidase | Apoptosis. necrosis. and inflammation pathway | 0.5 | 0.6 | 0.5 | 0.6 | 0.6 | 0.9 | |
| ADRA 1B | Adrenoceptor alpha 1B | Regulates growth and proliferation | 2.2 | 2.1 | 2.2 | 0.7 | 0.5 | 0.3 | |
| Oxidative stress | NOS2 | Nitric oxide synthase | Generates nitric oxide. high affinity for Ca2+/calmodulin | −0.7 | −0.8 | −0.7 | −0.8 | −0.6 | −0.7 |
| GSS | Gluthathione synthethase | Protects cells from oxidative damage by free radicals | −0.8 | −0.6 | −0.8 | 0 | 0.1 | 0.1 | |
| HSF4 | Heat shock transcription factor 4 | Activates heat-shock response genes | −1 | −0.9 | −1 | −0.2 | 0 | −0.1 | |
| TRAP1/HSP75 | TNF receptor-associated protein 1 | Modulates the balance between oxidative phosphorylation and aerobic glycolysis | −2.1 | −2.1 | −2.1 | 0 | 0.1 | 0 | |
| HIF1-α | Hypoxia inducible factor 1-α subunit | Master transcriptional regulator of the adaptive response to hypoxia | 0.1 | 0.3 | 0.1 | −0.5 | −0.1 | 0.2 | |
| TXN | Thioredoxin | Thioredoxin reductase. glutaredoxin and glutathione reductase activities. inhibits caspase-3 | −0.3 | 0.3 | −0.4 | 0.3 | −0.4 | 0.2 | |
| DNA damage/repair | ERCC4/XPF | Excision repair cross-complementation group 4 | Catalytic component of a structure-specific DNA repair endonuclease | 1.7 | 1.7 | 1.7 | 0.1 | 0 | 0.1 |
| LIG1 | DNA ligase I, ATP-dependent | Integral role in DNA repair and replication | −0.8 | −0.8 | −0.8 | −0.5 | −0.2 | −0.4 | |
| Ca-signaling | CNN1 | Calponin 1, basic, smooth muscle | Implicated in the regulation and modulation of smooth muscle contractions | 2 | 1.8 | 2 | 2 | 1.3 | 0.8 |
| MYL1 | Myosin, light chain 1, alkali, skeletal, fast | ATPase cellular motor protein | 1.5 | 1.6 | 1.5 | 0.4 | 0.5 | −0.3 | |
| KCNT2 | Potassium channel. subfamily T, member 2 | Ca2+-activated potassium channel | 3 | 3.3 | 3 | 1.7 | 2.6 | 3 | |
| CAMTA2 | Calmodulin binding transcription activator 2 | Calmodulin-binding transcription activator | 0 | 0 | 0 | −0.5 | −0.4 | −0.4 | |
| ATP2B3 | ATPase, Ca2+ transporting, plasma membrane 3 | Critical role in intracellular calcium homeostasis | 0.8 | 0.5 | 0.8 | 1.3 | 0.3 | 0.5 | |
| CAMKK2 | Calcium/calmodulin-dependent protein kinase kinase 2, beta | Phosphorylates the downstream kinases in the calcium/calmodulin-dependent (CaM) kinase cascade | −0.6 | −0.5 | −0.6 | −0.8 | −0.7 | −0.8 | |
| AGT | Angiotensinogen | Essential component of the renin-angiotensin system (RAS) | 1.3 | 1.3 | 1.3 | 0.3 | 1.1 | 1.2 | |
| Cell structure | MYOC | Myocilin | Role in cytoskeletal function | −2.4 | −2.5 | −2.4 | −0.5 | 0.4 | −0.7 |
| COL4A2 | Collagen, type IV, alpha 2 | Major structural component of basement membranes | 0.6 | 0.4 | 0.6 | 0.3 | 0.4 | 0.4 | |
| GATA4 | GATA binding protein | Myocardial differentation and function | 0 | 0 | 0 | 0.2 | 0.4 | 0.1 | |
| MEF2c | Myocyte Enhancer Factor 2 C | Role in myogenesis | −0.2 | −0.2 | −0.2 | −0.4 | −0.2 | 0 | |
| PKC | Protein kinase C | Cell signaling. cell adhesion. cell transformation. | 1 | 1.1 | 1 | 0.2 | 0.1 | 0.7 | |
| Immunomodulation | SELL/LECAM 1 | Selectin L | Cell surface adhesion protein | 1.6 | 1.5 | 1.6 | 0.2 | 0.7 | 0.8 |
| CXCL10 | Chemokine (C-X-C Motif) ligand 10 | Chemotactic for monocytes and T-lymphocytes | 0.9 | 1.1 | 0.9 | 1.9 | 2.2 | 2.4 | |
| Protein turnover | CAPN2 | Calpain 2, (M/II) large subunit | Calcium-activated neutral protease | −3.4 | −3.9 | −3.4 | −1.6 | −1.2 | −1.6 |
| Energy metabolism | HK2 | Hexokinase 2 | Couples extramitochondrial glycolysis to intramitochondrial oxidative phosphorylation | 3 | 2.6 | 3 | −0.9 | −1.4 | −0.5 |
| APOD | Apolipoprotein D | Closely associated with the enzyme lecithin:cholesterol acyltransferase | 2.3 | 2.3 | 2.3 | 0.1 | 0.4 | 0.5 | |
| Cell signaling | MSTN | Myostatin | Negative regulator of skeletal muscle growth | −1.9 | −1.1 | −1.9 | −2.3 | −1 | −1.7 |
| FGF16 | Fibroblast growth factor 16 | Required for normal cardiomyocyte proliferation and heart development | −1.9 | −1.8 | −1.9 | 0.8 | 0.8 | 1.3 | |
| NFKBIB | Nuclear Factor Of Kappa Light Polypeptide Gene Enhancer In B Cells Inhibitor, Beta | Inhibits NF-kappa-B by complexing with and trapping it in the cytoplasm | 0.4 | −0.1 | 0.8 | 0.3 | 0.8 | 0.2 | |
| CTGF | Connective tissue growth factor | Mitoattractant secreted by vascular endothelial cells | 1.9 | 1.5 | 1.9 | 0.8 | 1.1 | 0.9 | |
| TYK2 | Tyrosine kinase 2 | Involved in the initiation of type I IFN signaling | −0.7 | −0.6 | −0.7 | −0.5 | −0.3 | −0.3 | |
| CREB | CAMP responsive element binding protein 1 | Synchronization of circadian rhythmicity and the differentiation of adipose cells | 0.9 | 0.6 | 0.9 | 0.6 | 0.4 | 0.6 | |
| MAPK1/ERK2 | Mitogen-activated protein kinase 1 | Transduces signals from growth factors and phorbol esters | 0.3 | 0.3 | 0.3 | −0.3 | −0.1 | 0 | |
| STAT1 | Signal transducer and activator of transcription 1 | Transcription of IFN-stimulated genes | 0.3 | 0.5 | 0.3 | 0.7 | 0.8 | 1 |
Major pathways with known role in ischemia/reperfusion
The key regulator STAT3 is overexpressed in the ischemic, but also in the border and non-ischemic areas at 5 h, and mediates the Ca-signaling, adipocytokine and insulin signaling pathways. It is involved in both the PI3K/Akt and the JAK-STAT pathways. Moreover, SHISA2 showed a striking upregulation in both the early and late phases, suggesting the involvement of Wnt-signaling in ischemic preconditioning-induced cardioprotection11.
Activation of PI3K and its downstream targets Akt and glycogen synthase kinase 3β (GSK3B), has been demonstrated to be essential for ischemic preconditioning-induced cardioprotection involved in the RISK pathway12. Briefly, CREB-1 shows significant upregulation in the early phase, inducing transcriptional regulation of the cAMP pathway. Transcriptional changes in either GSK3A or GSK3B could not be detected in r-I/R-induced injury without subsequent AMI, neither at the early nor at the late phase, suggesting that the involvement of these proteins in cardioprotection does not depend on enhanced transcription. Indeed, most studies imply a post-translational regulation of GSK3B via phosphorylation at serine 913. The PI3K-Akt pathway with the genes found deregulated at 5 h and 24 h post r-I/R is displayed in Supplementary Fig. S3. Our data show that CREB, RTK and ITGA were up- and c-Myb and GF were downregulated. At 24 h, RTK and ITGA showed stable upregulation, while c-Myb displayed baseline levels. We found no modulation of anti-apoptotic Bcl-2, and the transcription and cell survival factor NF-κB showed modest upregulation at 5 h and at 24 h (Supplementary Fig. S3).
The recruitment of STAT3 and the activation of the JAK-STAT pathway play a major role in upregulation of cardioprotective and anti-apoptotic proteins14. Other STAT proteins (STAT-1, 5 and 6) have been shown to be upregulated as well, but their precise functional role is not yet fully elucidated15. Expression of STAT3 is upregulated in the early phase of r-I/R-induced changes, along with altered expression of several associated proteins, such as TNFα, connective tissue growth factor, osteopontin, tenascin, and several paracrine factors and cytokines16. STAT3-KO mice developed cardiac fibrosis, LV remodeling and heart failure16. Activation of STAT3 with TNF and its receptors is referred to as the survivor activating factor enhancement (SAFE) pathway17. Supplementary Table S4 lists significantly up- or downregulated genes in the RISK and SAFE pathways that have been shown to be upregulated in different phases of IPC (I/R injury with subsequent AMI). Interestingly, r-I/R without subsequent infarction did not result in significant activation of these pathways as a whole, even though some components were deregulated.
Regulation of further pathways and genes
In the late phase (24 h), expression of STAT1 was upregulated together with CASP3, both of which have a central role in the activated pathways and are promoting apoptosis. Transcription of CLU (a secreted chaperone that is protective against apoptosis) is significantly elevated both at 5 h and 24 h post r-I/R in ischemic and non-ischemic regions, with an even more pronounced increase at the later time point. CLU has been found to induce ischemic tolerance in neurons18, but its role in cardiac r-I/R injury is currently obscure. CLU is secreted in response to cellular stress, promotes cell survival, modulates matrix metalloproteinase expression, and stimulates angiogenesis. Together with leptin, it binds to the leptin receptor, which is subsequently internalized and activates transcriptional pathways including the JAK/STAT pathway19. These roles and the marked increase after r-I/R are indicative of an important role in restoring cell function post I/R, and/or indicate a surfeit of CLU to ensure future cardioprotection.
Similarly to a previous experiment by El-Adawi et al.20, the intrinsic angiotensin II autocrine loop was activated early after r-I/R involving neprilysin (MME), which is recently acknowledged as playing a significant role in LV adverse remodeling. However, without subsequent deep and long-lasting ischemia leading to AMI, the expression of this endopeptidase was almost reduced to baseline levels at 24 h, while even being significantly down regulated in the distal area. MME and the peptidase DPP4 are both upregulated 5 h after r-I/R, but down regulated in the distal zone after 24 h. Among other effects, substance P, which is cleaved by both peptidases21, enhances angiogenesis and vasodilatation through release of nitric oxide22. The activation of MME and DPP4 at the 5 h time point possibly indicates repression of angiogenesis, followed by de-repression of substance P and angiogenesis in the distal area after 24 h. In addition, the inducible nitric oxide synthase (iNOS2) was reduced at all time points. This indicates a potential cross talk between the MME/DPP4 system and the JAK/STAT pathway (Fig. 4).
Supplementary Table S5 demonstrates a selection of well-documented genes with currently unknown function in the cardiovascular system. The upregulation of expression of some of these genes were stronger than the known cardioprotection-associated genes and further characterization may identify these gene as new potential biomarkers and/or targets.
Confirmation of gene expression changes of the anterior wall regions after the r-I/R-stimulus
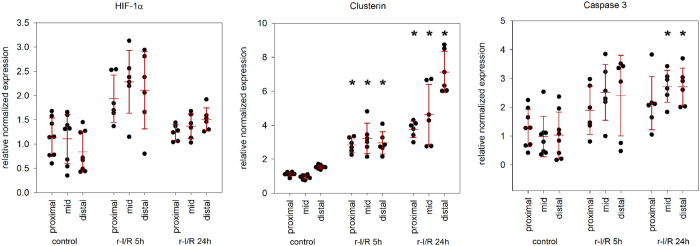
Quantitative RT-PCR of selected targets confirmed the findings of the NGS analysis (Fig. 5). The r-I/R stimulus resulted in a significant increase of clusterin (CLU) at 5 h with further upregulation at 24 h, and similar, but less pronounced increase in CASP3. Both results are well in line with the sequencing data. We also examined the expression of HIF-1α, the master regulator of cellular oxygen homeostasis. Interestingly, both NGS and qPCR failed to identify a significantly altered transcriptional regulation of HIF-1α in response to the r-I/R stimulus, even though a trend towards increase in HIF-1α expression at 5 h was observed. This could be attributed to short I/R cycles and to the fact that activity of HIF-1α is upregulated primarily by stabilization of the encoded protein, via reduced hydroxylation (mediated by HIF prolyl-hydroxylases), rather than de novo protein synthesis.
Figure 5. Real-time polymerase chain reaction (RT-PCR) of selected candidate genes in the control (C), and repetitive ischemia/reperfusion groups (r-I/R [5 h] and r-I/R [24 h]) in the different anterior wall regions.
A significant increase in clusterin (CLU) was found 5 h after the r-I/R stimulus with a further increase at 24 h. Similar, but less pronounced changes of expression of CASP3 resulted. A trend towards increase in hypoxia-inducible factor-alpha (HIF-1α) resulted. These results verify the NGS data. Each data point represents the result of one animal, and mean ± s.d. is indicated for each group. *p < 0.05 between Group r-I/R [5 h] vs group control in the corresponding myocardial regions.
IPC reduces myocardial scar tissue area leading to significantly higher LV EF 30 days after MI during SWOP
In order to prove the cardioprotective effect of the 3 × 10 min r-I/R stimulus, we have additionally induced AMI 26 h post r-I/R (SWOP phase) (group IPC-AMI) or sham I/R (group AMI). The AMI was induced by 90 min catheter-based occlusion of the mid left anterior descending coronary artery followed by reperfusion. The results of cardiac MRI measurements at 30 days are summarized in Table 4. IPC treatment before MI in SWOP resulted in a significant reduction of myocardial scar tissue area, smaller LV ESV and higher LV EF, as well as better segmental wall motion of the ischemia-affected and remote areas, confirming the cardioprotective and reverse remodeling effect of IPC in the late window in this pig model.
Table 4. Cardiac magnetic resonance imaging (cMRI) data collected 30 days after reperfused acute myocardial infarction (AMI) in pigs with and without ischemic preconditioning (IPC), induced by 3 times 10 min ischemia/reperfusion.
| cMRI parameter | group IPC-AMI (n = 5) | group AMI (n = 5) | p-value* |
|---|---|---|---|
| LVEF, % (IQR) | 44.02 (42.31–48.75) | 38.60 (37.80–39.80) | 0.016 |
| CO, l/min (IQR) | 3.31 (2.96–3.56) | 3.00 (2.80–3.00) | 0.346 |
| LVEDV, ml (IQR) | 62.29 (59.59–73.74) | 75.60 (75.10–78.30) | 0.117 |
| LVESV, ml (IQR) | 39.97 (30.43–42.75) | 47.00 (46.10–47.10) | 0.047 |
| LV scar tissue, % (IQR) | 5.15 (3.83–8.33) | 16.20 (14.10–17.70) | 0.028 |
| LVM, mg (IQR) | 69.59 (62.09–76.26) | 73.80 (70.40–80.80) | 0.251 |
| Infarction transmurality, % (IQR) | 55.35 (54.83–61.97) | 69.80 (63.50–74.10) | 0.175 |
| MO, % (IQR) | 0.14 (0.08–0.25) | 0.90 (0.50–1.80) | 0.053 |
| RVEF, % (IQR) | 44.35 (32.53–44.81) | 40.70 (39.20–41.50) | 0.917 |
| Segmental contraction velocity of ischemia–affected anterior area | 14.45 (12.63–16.76) | 12.24 (9.98–14.68) | 0.044 |
| Remote anterior area | 22.54 (20.39–23.97) | 20.12 (18.77–23.55) | 0.086 |
AMI was induced 26 h after IPC (group IPC-AMI) or sham-IPC procedure (group AMI). Data are given in median and interquartile ranges (IQR).
Fonts in bold indicate statistical significance (p < 0.05). *p-values were calculated by the Mann-Whitney-U-test.
LVEF indicates left ventricular ejection fraction; CO, cardiac output; LVEDV, left ventricular end-diastolic volume; LVESV, left ventricular end-systolic volume; LV, left ventricle; LVM, left ventricular mass; MO, microvascular obstruction; RVEF, right ventricular ejection fraction.
Discussion
Time-dependent activation of adaptive genomic responses of the myocardium to ischemic stimulus
This is the first study investigating an r-I/R stimulus without subsequent MI in a translational large animal model by the means of NGS, able to retrieve all mRNA transcripts, to identify cardioprotective signaling and the de novo synthesized proteins mediating cardioprotection of the late window. Pathway network and gene expression analyses confirmed time-dependent reprogramming of the ischemic and ischemia non-affected LV gene expression in response to the r-I/R stimulus without MI, with activation of the Ca-signaling, adipocytokine and insulin signaling pathway in the ischemic region early after I/R, and the immunomodulatory pathways at the time of the induced cardioprotection, during the late window of protection. The pathway network analysis provides evidence of spatiotemporal changes in the activation of diverse pathways and complex gene expression profiles of ischemic and non-ischemia-affected zones of the LV. Our data demonstrate that the r-I/R stimulus triggers a distinct adaptive genomic response of the myocardium in a time-dependent manner. Spatiotemporal analysis suggested protective responses in remote, non-ischemia-affected myocardial regions, which might be a target for prevention of ischemic LV remodeling. In contrast to previous experiments, which primarily examined selected genes or groups of related genes, the methods used here, i.e. NGS and pathway network modeling, allowed us to identify the complex responses to I/R in a more comprehensive manner. We analyzed the expressional changes of over 13000 genes by NGS mapping. The results confirmed the reprogramming of the LV in response to the r-I/R injury stimulus. Our data revealed that numerous cardioprotective proteins are necessary for the late windows of protection, which relies on de novo protein synthesis.
Gene networks and pathways altered by r-I/R
We have identified different networks of pathways at the site of the ischemic injury that were activated sequentially, including Ca-signaling, adipocytokine and insulin signaling pathways at 5 h and several immunomodulatory pathways at 24 h. We found that altered gene expression and related pathway activation were more pronounced at 5 h than at 24 h after r-I/R, underlining the significance of early de-novo protein synthesis, reaching their full effectivity in the SWOP. STAT3 proved to be a key regulator of pathway networks in the early phase, while STAT1 and CASP3 are central nodes in the late phase of cardioprotection. We found an early upregulation of neprilysin both in the ischemic, but also in the non-ischemic region. This endopeptidase has recently been acknowledged as an important signaling molecule of heart insufficiency. Our data suggests also its active role in LV remodeling after I/R injury. In the remote myocardium, Ca-signaling and immunomodulatory pathways were activated at 5 h and at 24 h, indicating intrinsic remote conditioning, in contrast with the neurohumoral mechanism of the classic remote conditioning applying repetitive ischemia in the peripheral muscles. The complexity of the gene regulation patterns revealed in this study suggests that profound biological reactions are triggered by r-I/R and shows that powerful bioinformatics analyses of pathways, gene and protein interactions have the potential to reveal novel targets for therapeutic intervention for clinical relevance.
During conditioning r-I/R cycles, the myocardium activates adaptive mechanisms in an attempt to maintain cell function during actual and delayed hypoxic conditions, and to counterbalance apoptotic signaling by limiting DNA-damage. Unaware of how long the ischemic event will last, the myocardium initially responds in an identical manner to that seen during sustained ischemia. Once a conditioning response is confirmed, the question is to determine the threshold of ischemic burden that will trigger conditioning-induced signaling (instilling ischemic memory) towards irreversible damage.
With reference to previous studies, our NGS data confirms the involvement of pathways and networks already known to be implicated in I/R, such as Ca-signaling, energy metabolism including mitochondrial respiratory chain proteins, myocyte and matrix structure proteins, and stress response proteins including cell fate effectors12,17,23. The strength and novelty of our study lies in its comprehensive approach in identifying all interconnected transcripts and interacting pathways, and their regulation with respect to activation, in the ischemic and in the non-ischemic myocardium. The discovery of several activated pathways and upregulated genes in the ischemia-affected and non-affected regions with yet unknown cardiovascular function may serve as a starting point for further research on mitigation of I/R injury and post-ischemic left ventricular remodeling.
Implications for therapeutic development
Taken together, our data indicates that complex cellular and molecular mechanisms are responsible for cardioprotection through r-I/R, and further regulatory pathways than just the SAFE and RISK pathways are triggered. A number of genes with yet unknown role in I/R have been identified and their precise role and mechanism in cardioprotection may be characterized by further investigations. The profound alterations of several distinct pathways indicate that a multi-targeted therapeutic approach may be feasible. Regarding the concept of systems pharmacology, the precise role of all involved pathways should be taken into account, including the resilience of individual targets to interventional strategies, in order to develop effective therapies. Clearly, I/R is difficult to implement in the clinic. Instead, the elucidation of the functional outcomes may direct the development of novel pharmacologic or gene therapies based on the molecular changes that are responsible for cardioprotection. In this regard, the insight generated by this study facilitates further target characterization and selection to prevent ischemic injury and reverse remodeling of the human heart.
Study Limitation
Indeed, r-I/R without subsequent infarction did not cause significant myocardial damage and did not elevate the cardiac markers. The 10 min duration of ischemia was selected based on the guidelines on the management of the stable coronary artery disease24 defining the stable angina as ischemic pain lasting up to 15 min, and also on our previous observations in pigs10 that at least 5 min coronary occlusion is needed to manifest ischemia in intracardiac electrocardiograms. The transition of reversible to non-reversible myocardial ischemia has no clear time-threshold, and depends on a number of different factors, including comorbidities24. Our earlier experiments10 showed that 10 min r-I/R causes neither permanent low electrical signals nor the segmental wall motion disturbances typical for infarction. However, it results in an incomplete recovery of the myocardial electrical activity even at 24 h post I/R injury, which might also explain the trigger of cardioprotective protein synthesis as a possible mechanism of the second window of cardioprotection in pigs. Although de-novo synthesis of protein is crucial for cardioprotection in SWOP, a complete interpretation of the IPC-induced mechanisms necessitates integration of our NGS findings with proteomic, and other methods that can investigate and verify functional effects and post-translational modifications.
Here, IPC was performed on healthy animals, but atherosclerosis and other cardiovascular co-morbidities may attenuate cardioprotection by IPC25. Unfortunately, no animal model exists to simulate the complete palette of human atherosclerotic coronary artery disease. However, as the pig circulation and heart anatomy is very similar to humans, the porcine closed-chest catheter-based coronary intervention model is accepted as the most suitable translational model for human coronary ischemia and treatment.
Conclusion
We demonstrate for the first time that r-I/R stimuli provokes sequential changes in pathway networks and gene expression profiles not only in ischemic but also in the non-ischemia-affected regions of the myocardium; we introduce the term “intrinsic remote conditioning”, describing an intrinsic protective mechanism against adverse LV remodeling. This experimental approach, using the current, clinically relevant animal model, provides a useful tool for the identification of early and late r-I/R-induced gene expression networks, and may reveal relevant pathways for targeted drug intervention. Concerning the complexity of the response, it is likely that the simultaneous regulation of multiple targets (e.g. mechanisms optimizing cellular metabolism, contractility, inflammation, DNA-repair, and cell survival) is a viable strategy for induction of robust cardioprotection.
Methods
Porcine Model of Ischemic Preconditioning
Animal investigations were carried out in accordance with the “Position of the American Heart Association on Research Animal Use,” as adopted by the AHA on November 11, 1984. The study was approved by the Ethics Committee on Animal Experimentation at the University of Kaposvar, Hungary. The study design is displayed in Fig. 1.
Domestic pigs underwent cardiac catheterization (Supplementary Methods). The r-I/R protocol consisted of three repetitive cycles of 10 min I/R via percutaneous balloon occlusion and deflation in the mid left anterior descending coronary artery (LAD) under general anesthesia. Details are described in the Supplementary Methods.
(a) Effect of IPC on 30 days cardiac function after acute myocardial infarction (AMI) in SWOP
To prove the effect of IPC in SWOP ten domestic pigs were randomized into two groups with (n = 5, group IPC-AMI) and without (n = 5, group AMI) IPC stimulus (Fig. 1a,b and Supplementary Methods).
(b) IPC-induced transcriptomic changes are essential for cardioprotection in SWOP
To assess the time-dependent gene expression alterations as well as pathways and pathway networks that are crucial for triggering cardioprotection and protection in the late window (SWOP), twelve pigs underwent r-I/R, while animals undergoing sham intervention (n = 8) served as controls (group Control). Baseline echocardiography and invasive LV hemodynamic measurements via a pigtail catheter (Cordis, a J&J Company, Fremont, CA) were performed before the preconditioning stimulus. Animals with r-I/R were randomized to groups r-I/R [5 h] (n = 6), or r-I/R [24 h] (n = 6) according to sacrificing time points. At 5 h and 24 h post r-I/R or sham-r-I/R, echocardiography and LV hemodynamics measurements were repeated. Animals were then sacrificed and pig hearts were explanted (Fig. 1c). Myocardial tissue samples from the distal (ischemia-affected), mid (border) and proximal (ischemia non-affected) anterior walls were harvested using biopsy kits (Acu-Punch, Acuderm, Fort Lauderdale, FL).
Additional Information
How to cite this article: Pavo, N. et al. Sequential activation of different pathway networks in ischemia-affected and non-affected myocardium, inducing intrinsic remote conditioning to prevent left ventricular remodeling. Sci. Rep. 7, 43958; doi: 10.1038/srep43958 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This study was funded by the Ludwig Boltzmann Institute Cluster Cardiovascular Research, The Hungarian Scientific Research Fund (OTKA ANN 107803) and the Austrian Science Fund (KPIO1277FWF). Zoltan Giricz holds a “János Bolyai Fellowship” from the Hungarian Academy of Sciences. Péter Ferdinandy is a Szentágothai Fellow of the Hungarian National Program of Excellence (TAMOP 4.2.4.A/2-11-1-2012-0001). Marta Sarkozy was supported by the UNKP-UNKP-6-4 IKT/147-1787/8/2016-ÖSZT-120 New National Excellence Program of the Ministry of Human Capacities. Derek J Hausenloy is funded by the British Heart Foundation (grant number FS/10/039/28270), the Rosetrees Trust, and is supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Footnotes
The authors declare no competing financial interests.
Author Contributions N.P. and M.G. conceived the study, N.P., and M.G. designed the study, N.P., D.Lu, K.Z., A.Z., D.P., K.A., R.G., and M.G. conducted the experiments N.P., D.Lu., K.Z., A.Z., D.Lo., G.G., J.W., D.P., K.A., H.J.A., Z.G., T.B., M.S., A.J., M.Y.E., S.P.H., D.J.L., P.F., and M.G. analysed and interpreted data, G.M. interpreted data, N.P., J.W., and M.G. drafted the manuscript. All authors reviewed and approved the manuscript.
References
- Reiter R., Henry T. D. & Traverse J. H. Preinfarction angina reduces infarct size in st-elevation myocardial infarction treated with percutaneous coronary intervention. Circ. Cardiovasc. Interv. 6, 52–58 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdinandy P., Schulz R. & Baxter G. F. Interaction of cardiovascular risk factors with myocardial ischemia/reperfusion injury, preconditioning, and postconditioning. Pharmacol. Rev. 59, 418–458 (2007). [DOI] [PubMed] [Google Scholar]
- Murry C. E., Jennings R. B. & Reimer K. A. Preconditioning with ischemia: A delay of lethal cell injury in ischemic myocardium. Circulation 74, 1124–1136 (1986). [DOI] [PubMed] [Google Scholar]
- Das D. K. & Maulik N. Cardiac genomic response following preconditioning stimulus. Cardiovasc. Res. 70, 254–263 (2006). [DOI] [PubMed] [Google Scholar]
- Ludman A. J., Yellon D. M. & Hausenloy D. J. Cardiac preconditioning for ischaemia: Lost in translation. Disease Models & Mechanisms 3, 35–38 (2010). [DOI] [PubMed] [Google Scholar]
- Hausenloy D. J. & Yellon D. M. The second window of preconditioning (swop) where are we now? Cardiovasc. Drugs Ther. 24, 235–254 (2010). [DOI] [PubMed] [Google Scholar]
- Ónody A. et al. Effect of classic preconditioning on the gene expression pattern of rat hearts: A DNA microarray study. FEBS Lett. 536, 35–40 (2003). [DOI] [PubMed] [Google Scholar]
- Varga Z. V. et al. Micrornas associated with ischemia-reperfusion injury and cardioprotection by ischemic pre- and postconditioning: Protectomirs. Am. J. Physiol. Heart Circ. Physiol. 307, H216–H227 (2014). [DOI] [PubMed] [Google Scholar]
- Heusch G. & Gersh B. J. The pathophysiology of acute myocardial infarction and strategies of protection beyond reperfusion: A continual challenge. Eur. Heart J. in press (2016). [DOI] [PubMed] [Google Scholar]
- Pavo N. et al. On-line visualization of ischemic burden during repetitive ischemia/reperfusion. JACC Cardiovasc. Imaging 7, 956–958 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barandon L. et al. Involvement of frza/sfrp-1 and the wnt/frizzled pathway in ischemic preconditioning. Circ. Res. 96, 1299–1306 (2005). [DOI] [PubMed] [Google Scholar]
- Bolli R. Preconditioning: A paradigm shift in the biology of myocardial ischemia. American journal of physiology. Heart and circulatory physiology 292, H19–H27 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Imahashi K., Steenbergen C. & Murphy E. Phosphorylation of glycogen synthase kinase-3β during preconditioning through a phosphatidylinositol-3-kinase–dependent pathway is cardioprotective. Circ. Res. 90, 377–379 (2002). [DOI] [PubMed] [Google Scholar]
- Bolli R. et al. A murine model of inducible, cardiac-specific deletion of stat3: Its use to determine the role of stat3 in the upregulation of cardioprotective proteins by ischemic preconditioning. J. Mol. Cell. Cardiol. 50, 589–597 (2011). [DOI] [PubMed] [Google Scholar]
- Knight R. A., Scarabelli T. M. & Stephanou A. Stat transcription in the ischemic heart. JAK-STAT 1, 111–117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghikia A., Ricke-Hoch M., Stapel B., Gorst I. & Hilfiker-Kleiner D. Stat3, a key regulator of cell-to-cell communication in the heart. Cardiovasc. Res. 102, 281–289 (2014). [DOI] [PubMed] [Google Scholar]
- Heusch G. Molecular basis of cardioprotection signal transduction in ischemic pre-, post-, and remote conditioning. Circ. Res. 116, 674–699 (2015). [DOI] [PubMed] [Google Scholar]
- Wiggins A. K., Shen P.-J. & Gundlach A. L. Delayed, but prolonged increases in astrocytic clusterin (apoj) mrna expression following acute cortical spreading depression in the rat: Evidence for a role of clusterin in ischemic tolerance. Mol. Brain Res. 114, 20–30 (2003). [DOI] [PubMed] [Google Scholar]
- Byun K. et al. Clusterin/apoj enhances central leptin through lrp2‐mediated endocytosis. EMBO reports 15, 801–808 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Adawi H. et al. The functional role of the jak–stat pathway in post-infarction remodeling. Cardiovasc. Res. 57, 129–138 (2003). [DOI] [PubMed] [Google Scholar]
- Pare G. et al. Genetic variants associated with angiotensin-converting enzyme inhibitor-associated angioedema. Pharmacogenet. Genomics 23, 470–478 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seegers H. C., Hood V. C., Kidd B. L., Cruwys S. C. & Walsh D. A. Enhancement of angiogenesis by endogenous substance p release and neurokinin-1 receptors during neurogenic inflammation. J. Pharmacol. Exp. Ther. 306, 8–12 (2003). [DOI] [PubMed] [Google Scholar]
- Yellon D. M. & Hausenloy D. J. Myocardial reperfusion injury. N. Engl. J. Med. 357, 1121–1135 (2007). [DOI] [PubMed] [Google Scholar]
- Montalescot G. et al. Esc guidelines on the management of stable coronary artery disease: The task force on the management of stable coronary artery disease of the european society of cardiology. Eur. Heart J. 34, 2949–3003 (2013). [DOI] [PubMed] [Google Scholar]
- Ferdinandy P., Hausenloy D. J., Heusch G., Baxter G. F. & Schulz R. Interaction of risk factors, comorbidities, and comedications with ischemia/reperfusion injury and cardioprotection by preconditioning, postconditioning, and remote conditioning. Pharmacol. Rev. 66, 1142–1174 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.