Abstract
The yeast PHO5 promoter is a model system for the role of chromatin in eukaryotic gene regulation. Four positioned nucleosomes in the repressed state give way to an extended DNase I hypersensitive site upon induction. Recently this hypersensitive site was shown to be devoid of histone DNA contacts. This raises the mechanistic question of how histones are removed from the promoter. A displacement in trans or movement in cis, the latter according to the well established nucleosome sliding mechanism, are the major alternatives. In this study, we embedded the PHO5 promoter into the context of a small plasmid which severely restricts the space for nucleosome sliding along the DNA in cis. Such a construct would either preclude the chromatin transition upon induction altogether, were it to occur in cis, or gross changes in chromatin around the plasmid would be the consequence. We observed neither. Instead, promoter opening on the plasmid was indistinguishable from opening at the native chromosomal locus. This makes a sliding mechanism for the chromatin transition at the PHO5 promoter highly unlikely and points to histone eviction in trans.
All DNA-related processes like replication, transcription, recombination, and repair are mediated by protein factors and machines which use DNA as their substrate. It is therefore crucial that these proteins have access to the DNA for direct binding interaction, often in a sequence-specific manner. Restriction of this access is one mechanism by which cellular functions involving DNA are regulated. One way to achieve this goal is the packaging of the eukaryotic genome into a complex DNA-protein structure called chromatin, and modulation of the structure of chromatin is recognized as a key step in the cascade of DNA-related processes.
The basic unit of chromatin is the nucleosome, which is made up of a core of eight histone proteins with about 1.7 turns of DNA wound around this octamer. The nucleosomes form a beads-on-a-string structure along the DNA and are further organized into a so-far ill-defined hierarchy of higher order structures involving several nonhistone chromatin proteins. The influence of higher order structure on DNA-related processes is not well understood so far. At the level of the nucleosome, however, the tight interaction of the DNA with the surface of the histone octamer imposes a significant barrier for the access of proteins to their binding sites.
Our laboratory has been interested in the influence of chromatin structure on gene regulation for a long time. We employ the PHO genes in yeast as a model system which shows striking chromatin transitions upon the activation of several of the genes related to phosphate metabolism. In particular, under repressive conditions (with Pi [+Pi]) the promoter region of the PHO5 gene is organized into four clearly positioned nucleosomes with a short hypersensitive site of about 60-bp length between nucleosomes −2 and −3 (34) (Fig. 1A). Upon PHO5 activation by phosphate starvation (without Pi [−Pi]), these four nucleosomes become remodeled, leading to an extended hypersensitive site of about 600-bp length (4).
FIG. 1.
(A) Schematic of the chromatin organization at the PHO5 promoter in the repressed (+Pi) and induced state (−Pi). The positioned nucleosomes (circles) are numbered from −5 to +1 with respect to the open reading frame (broad black arrow). The open circles denote the nucleosomes which become remodeled upon induction. Upon induction, the short hypersensitive site (HS; asterisk) becomes extended (HS; dashed line) and the two Pho4 binding sites (black dots) bind Pho4 (striped arches). Remodeling of nucleosomes −1 and −4 is often less complete, as symbolized by stippled circles in the induced state. The position of the TATA box (T) is shown, and the buckled arrow (RNA Pol) symbolizes transcription of the PHO5 gene. The positions of relevant restriction sites are indicated by arrows as well as the relative position of the upstream ApaI-Sau3AI fragment which constitutes probe 1b. (B) Map of plasmid pTAP5C (2,481 bp). The genetic elements (CEN6, TRP1, ARS1, and the PHO5 promoter) are marked by boxes or arrows on the plasmid. The positions of nucleosomes along the plasmid are shown as circles. Nucleosomes are labeled with arabic numbers for the PHO5 promoter region (−5 to +1, as in panel A), with roman numerals for the remainder of the UNF region (36), and alphanumerically for the TRP1 region (T1-T4) and the CEN6 element (C6). The nucleosomes shaded in gray become remodeled upon induction of the PHO5 promoter. Asterisks mark hypersensitive sites. Restriction sites used as markers are shown and the probes used for indirect end labeling (Trp up, Trp down, 63, and 162) are indicated by curved arrows above the plasmid circle. The sites used for secondary cleavage for indirect end labeling are in bold.
It has been a long standing question what hypersensitive sites look like in molecular terms. Recently, our lab and others could show that the nucleosomes of the PHO5 promoter region not only become altered in their structure such that the DNA becomes accessible but that the nucleosomes are completely disassembled (7, 28, 29). In the induced state, histones are no longer in contact with this DNA region. This immediately raised the question: where do the histones go?
There are two major possibilities for how histones can be displaced from the promoter. Histones might leave the PHO5 promoter region either in cis or in trans with regard to the underlying DNA. Especially displacement in cis would correspond to the well documented sliding mechanism of chromatin remodeling. All chromatin remodeling complexes have been shown in vitro to catalyze the sliding of nucleosomes along DNA and especially for the so-called ISWI class there is also in vivo evidence (15, 20-22). Such remodeling reactions leave the histone octamer intact and change only its location relative to the DNA sequence. It is conceivable that the PHO5 promoter adopts the accessibility of naked DNA upon induction because the four nucleosomes slide up- and/or downstream into neighboring DNA regions.
In this study, we have tested the succinct predictions derived from such a sliding mechanism. To this end, we integrated the PHO5 promoter region into a small plasmid environment. The small size and circular nature of this environment severely restricts the possibility of accommodating additional nucleosomes in cis on the same DNA molecule and generating a histone-free PHO5 promoter region at the same time. Therefore a sliding mechanism would either preclude histone loss at the promoter altogether or would lead to gross changes in the chromatin structure around the plasmid.
We show that chromatin opening of the plasmid-borne copy of the PHO5 promoter is indistinguishable from what we observe at the native chromosomal locus. Upon induction, the same extended hypersensitive site is generated, and the histones again lose contact with the DNA. As the chromatin structure of the remainder of the plasmid remains unchanged, we conclude that the loss of histones from the PHO5 promoter does not occur by nucleosome sliding in cis but that histones are evicted in trans upon induction.
MATERIALS AND METHODS
Yeast strains and media.
Yeast strains YS18 (MATα his3-11 his3-15 leu2-3 leu2-112 canR ura3Δ5) and YS31 (YS18 pho80::HIS3) were disrupted in the TRP1 gene by linear transformation with the trp1::URA3 disruption plasmid and YS188 (YS18 pho5::URA3; the disruption comprises the region between the BamHI site in the promoter and the second KpnI site in the coding region of the PHO5 gene) was disrupted with the trp1::LEU2 disruption plasmid, yielding strains YS1801, YS3101, and YS18018, respectively. pTAP5C- or pTAP5CΔTATA-containing strains were grown under selection conditions (without Trp) either in high-phosphate medium (0.67% [wt/vol] yeast nitrogen base without amino acids [Difco], supplemented with 2% [wt/vol] glucose and the necessary amino acids, uracil and adenine) or in no-phosphate medium to induce PHO5 as described by Almer et al. (4).
Plasmid construction.
DNA manipulation and cloning followed standard procedures (30). The plasmid pTAP5C was constructed from three PCR products. Template for all three PCRs was plasmid pCA/wt (12), which is a derivative from pCB/wt (14) with the PHO5 region extended beyond the BamHI up to the ApaI site. The primer pairs used were the following: P5-5Xho, CCGCTCGAGAAGAAAACAAGAGAC, and P5+1Sph, GACTTGCATGCATAGTCGCCAGGGAAAG, giving the PHO5 promoter fragment; MLCEN6Ecofor, CGGAATTCGACTATATTTCTTTTCATCAC, and MLCEN6Ecorev, CGGAATTCTTTCAACCTATTTTACATC, giving the CEN6 element; and TrpArsIIISph, GACTTGCATGCTATTATCTTCTACGC, and TrpArsIXho, CCGCTCGAGCGTATGCGCCTGTGAAC, giving the TRP1ARS1 circle fragment. In the latter case, pCA/wt was digested with EcoRI and religated, and the ligation reaction was used directly as template for the PCR. The PHO5 promoter fragment and the TRP1ARS1 circle fragment were ligated via XhoI and inserted via SphI into a pUC19 vector in which the EcoRI site was removed by EcoRI digestion, Klenow fill in, and blunt ligation. The third PCR product with the CEN6 element was ligated into this first pUC19 derivative in a second round of ligation via the unique EcoRI site of the TRP1ARS1 circle fragment. The identity of this second pUC19 derivative was confirmed by DNA sequencing, and the orientation of the CEN6 element was found to yield the DraI site closer to the SphI site (data not shown). pTAP5C corresponds to the 2.5-kb SphI fragment of the second pUC19 derivative and was cut out, ligated, and transformed into trp yeast strains with selection for the TRP1 marker. For the construction of pTAP5CΔTATA, the SphI-XhoI insert of the pUC19 derivative was swapped for the SphI-XhoI insert generated by PCR using pCAΔ26 (12, 14) as a template and the primers P5- 5Xho and P5+1Sph. The TATA box deletion was confirmed by DNA sequencing (data not shown).
The TRP1 disruption plasmid was generated from two PCR products using genomic yeast DNA as template and the following two primer pairs: TRP1 N-term ClaI for, CCATCGATCTTTCCTGCTTTGAATTAG, and TRP1 N-term HindIII rev, GTCAAAGCTTCATACTCCAAGCTGCCTTTG; and TRP1 C-term HindIII for, GTCAAAGCTTGCTAAGAAATAGGTTATTAC, and TRP1 C-term BamHI rev, CGGGATCCGTTTGTATTCATACTATGTG. The two PCR products were ligated via the HindIII site and inserted via the ClaI and the BamHI sites into pBR322. Either a URA3 or LEU2 marker cassette with HindIII linkers was ligated into the HindIII site of this pBR322 derivative. The ARS1 element was removed from this plasmid by digestion with BglII and NheI, Klenow fill in, and blunt religation in order to prevent episomal propagation of the transformed disruption plasmid in yeast. In the case of the URA3 marker, a ClaI/BamHI fragment (using a dam mutant Escherichia coli strain) of the ARS-free pBR322 derivative, and in the case of the LEU2 marker, a MluI/BamHI fragment was used for linear transformation.
Chromatin analysis.
Nuclei preparation and chromatin analysis by restriction enzymes or DNase I as well as indirect end labeling, gel electrophoresis, and blotting procedures were as described previously (4, 16).
Chromatin immunoprecipitation.
Yeast cultures of a density of 1 × 107 to 2 × 107 cells/ml were treated with 1% formaldehyde for 20 min at room temperature. Cross-linking was quenched by adding glycine to a final concentration of 125 mM, and then cells were sedimented and washed twice in ice-cold 0.9% NaCl. They were resuspended in HEG150 buffer (150 mM NaCl, 50 mM HEPES, pH 7.6, 10% glycerol, 1% Triton X-100, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) and treated with a French press three times at a pressure of 1,100 lb/in2. In this step cells were broken, and simultaneously the chromatin was sheared to an average fragment size of 450 bp. Immunoprecipitation was performed as described by Strahl-Bolsinger et al. (32). Antibodies against the C termini of histones H2B, H3, and H4 were gifts from A. Verreault. Immunoprecipitated DNA was quantitatively measured in duplicates by the ABI PRISM 7000 sequence detection system using the following amplicons: PHO5 UASp2-A, 5′-GAATAGGCAATCTCTAAATGAATCGA-3′; PHO5 UASp2-B, 5′-GAAAACAGGGACCAGAATCATAAATT-3′; PHO5 UASp2-probe, 5′-FAM-ACCTTGGCACTCACACGTGGGACTAGC-3′; TEL-A, 5′-TCCGAACGCTATTCCAGAAAGT-3′; TEL-B, 5′-CCATAATGCCTCCTATATTTAGCCTTT-3′; TEL-probe, 5′-FAM-TCCAGCCGCTTGTTAACTCTCCGACA-3′; TRP1-A, 5′-TGTTTTGGCTCTGGTCAATGATT-3′; TRP1-B, 5′-TGGTATTCTTGCCACGACTCATC-3′; and TRP1-probe, 5′-FAM-CGGCATTGATATCGTCCAACTGCATG-3′.
Topological analysis.
Total DNA was prepared from logarithmically growing cultures by proteinase K-sodium dodecyl sulfate treatment and phenol extraction and resolved on 1.3% agarose gels with 33 μM chloroquine (Sigma) in both gel and electrophoresis buffer. The gels were run at 1.2 V/cm for 26 h, blotted, and probed specifically for the plasmid pTAP5CΔTATA with either probe Trp down or Trp up. The supercoil bands were quantified using a phosphorimager (Fuji FLA3000, AIDA software) and the distribution of intensities was fitted with a Gaussian model using the software CurveExpert 1.3 (Daniel Hyams). The trace profiles shown in Fig. 4B were generated from the phosphorimager screen with the AIDA software.
FIG. 4.
A change in superhelicity confirms the loss of nucleosomes from the plasmid upon induction. (A) Genomic DNA from strain YS1801 (wt) and YS3101 (pho80) both carrying the plasmid pTAP5CΔTATA was analyzed in an agarose gel containing 33 μM chloroquine. Three independent but parallel cultures were grown logarithmically in high-phosphate media for each strain. The arrowheads mark each maximum of the Gaussian distribution as averaged over the three lanes for each strain (closed arrowhead, YS1801; open arrowhead, YS3101). The difference in linking number between the repressed and induced states was 2.5 ± 0.4. Lanes 1 and 8 contain linearized plasmid as a marker and the position of the nicked circular and the linear form of the plasmid is indicated on the right side of the gel. (B) Trace profiles of lane 3 (top) and lane 5 (bottom) are shown and the respective maxima of the Gaussian fit indicated by arrowheads. The position of the nicked circular form was used as the origin for the trace profile.
RESULTS
Experimental design and plasmid construction.
In order to study chromatin opening at the PHO5 promoter in an environment where movement of nucleosomes in cis would be highly restricted, we inserted a promoter fragment of the PHO5 gene, comprising the region from nucleosome −5 to nucleosome +1, into the classical TRP1ARS1 plasmid (Fig. 1B). The TRP1ARS1 circle is one of the smallest stably propagated yeast plasmids known, and its chromatin structure is well characterized (36). The coding region of the TRP1 marker gene is organized into four loosely positioned nucleosomes; the ARS1 element and the region of the EcoRI site, where the circle is closed, are located in hypersensitive sites, and the region between the ARS1 element and the EcoRI site, called UNF (unknown function; now known to be part of the upstream region of GAL3), is packaged into three stably positioned nucleosomes numbered I to III starting at the ARS1 element. Our insertion of the PHO5 promoter fragment replaces nucleosome II, and the direction of transcription from the PHO5 promoter is away from the ARS1 element towards the TRP1 gene in order not to interfere with ARS function. Additionally, we inserted a minimal CEN6 element (11) into the EcoRI site, thereby keeping the copy number low. A high-copy-number plasmid might titrate out the activator Pho4, which is strictly necessary for PHO5 induction and the chromatin transition (13, 39), and thus might lead to mixed plasmid populations in the cell with variable extents of chromatin remodeling upon induction. The resulting plasmid was called pTAP5C (for TRP1, ARS1, PHO5, and CEN6) and comprises 2,481 bp.
In summary, we set out to construct a plasmid context for the PHO5 promoter where a chromatin transition by a sliding mechanism would either be impossible or lead to gross changes of the chromatin structure in the remainder of the plasmid. This can easily be tested by analysis of the chromatin structure around the plasmid before and after PHO5 induction as we show in the following results.
The PHO5 promoter fragment is an autonomous nucleosome positioning module for nucleosomes −4 to +1.
A prerequisite for this project was to establish whether the PHO5 promoter fragment in the new TRP1ARS1 plasmid context would be organized into the same nucleosomal structure under repressive conditions (+Pi) as the PHO5 promoter at the native chromosomal locus. Figure 2A shows that this is the case. We analyzed chromatin at both loci by limited DNase I digestion and by indirect end labeling in the strain YS1801 carrying the pTAP5C plasmid. This strain contains the PHO5 promoter in two locations: at the native position on chromosome II and on the pTAP5C plasmid in the form of the PHO5 promoter fragment. By using appropriate restriction enzymes for secondary cleavage and probing the same DNA blot with probes for either the plasmid or the chromosomal locus, we show directly that the positions of nucleosomes −4 to +1 are virtually superimposable.
FIG. 2.
There is no difference between the plasmid and the chromosome locus regarding the chromatin structure of the PHO5 promoter nucleosomes −4 to +1 both under repressed (+Pi) and induced (−Pi) conditions. (A) Nuclei of strain YS1801 carrying pTAP5C were subjected to limited DNase I digestion (wedges on top of the upper panel blot denote increasing DNase I concentration, dashes denote no DNase I addition) and secondary cleavage with HindIII and ApaI for indirect end labeling. The same blot membrane was probed with probe Trp down and reprobed with probe 1b for analysis of the plasmid and the chromosome locus, respectively (see also Fig. 1). The relevant genetic regions as well as schematics of the chromatin structure (labeled and stippled circles, asterisks, and dashed lines for the extended hypersensitive site HS, as in Fig. 1) are indicated at the sides of the blots. The asterisks labeled HS1 and HS3 refer to the hypersensitive sites upstream of the PHO5 promoter and between the PHO5 and the PHO3 gene (3), respectively. (B) Restriction enzyme analysis of nuclei from repressed (+Pi) and induced cells (−Pi) as in panel A with the indicated enzymes at 0.3 and 1.2 U/μl (left and right lane, respectively, for each enzyme) and secondary cleavage with HindIII and ApaI. Probes as in panel A.
The only difference was the region of nucleosome −5, which appears to be cramped in between nucleosome −4 and a hypersensitive site that is newly generated next to the strongly positioned UNF nucleosome I from the TRP1ARS1 circle. The chromosomal locus shows the hypersensitive site HS1 (3) instead of nucleosome I and therefore may provide different positioning information for nucleosome −5 compared to the plasmid. The chromatin interpretation of this plasmid region is tentative at the moment but is of no significance for the further argument of this study.
Further downstream (i.e., higher up in the lane in Fig. 2A) the chromatin structures of the plasmid and the chromosomal locus are necessarily different due to the differences in DNA sequence. The plasmid shows hypersensitive sites flanking the CEN6 element here and the chromosomal locus shows the typical hypersensitive site (HS3) between the PHO5 and PHO3 genes (3).
It appears that the PHO5 promoter fragment which we selected for these experiments contains sufficient nucleosome positioning information to position the nucleosomes −4 to +1 into the native structure even in a foreign sequence context and on a topologically constrained plasmid.
Chromatin opening of the PHO5 promoter region is identical at the plasmid and the chromosomal locus.
After confirming that both the plasmid and the chromosome locus started off with the same relevant chromatin structure under repressing conditions, we wished to determine if there would be any differences upon inducing conditions (−Pi). Quite strikingly, the response of the PHO5 promoter region on the plasmid and on the chromosome was indistinguishable (Fig. 2A). We induced cells overnight after shifting them into phosphate-free medium and analyzed chromatin at both loci with DNase I digestion and indirect end labeling and by probing and reprobing the same DNA blot. The PHO5 promoter became hypersensitive to DNase I over an extended region in the same way both on the plasmid and on the chromosome. This showed unambiguously that the PHO5 promoter fragment contained the relevant information not only for proper nucleosome positioning but also for the complete chromatin transition upon PHO5 induction.
In addition to DNase I digestion, we also probed the chromatin structure by restriction enzyme digestion (Fig. 2B) (4). We show the typical up and down in accessibility for the enzymes BamHI, ClaI, and BstEII along the PHO5 promoter which represent linker, nucleosomal, and again linker regions, respectively, in the repressed state (Fig. 1A). For monitoring the chromatin transition upon induction we routinely use the accessibility of the ClaI site in the −2 nucleosome of the PHO5 promoter where low and high accessibilities correspond to the repressed and the induced state, respectively. By the choice of probe and secondary cleavage, we again distinguished the plasmid from the chromosomal locus and again observed no significant difference between the two. At both loci the ClaI accessibility of the induced state was about 50%, which is somewhat lower than the usual 90% (4). We have, however, noticed in the past that depending on the strain background there are differences to what extent the ClaI site will become accessible upon PHO5 induction. Especially the YS series of yeast strains does not always reach full accessibility (J. Svaren and W. Hörz, unpublished data). In any case, for the purpose of the present study it is only important to confirm that there is no difference between plasmid and chromosomal location regarding nucleosome positioning and chromatin opening at the PHO5 promoter.
Histones are lost from the PHO5 promoter region on the plasmid upon chromatin opening.
The chromatin opening of the PHO5 promoter on the plasmid appeared indistinguishable from the opening on the chromosome as assayed by nuclease accessibility. It was vital, however, to confirm not only that the nucleosomes over the PHO5 promoter changed their nuclease protection properties for the DNA but also that their constituent histones were indeed displaced from the DNA of this region. In analogy to prior experiments (28), we performed chromatin immunoprecipitation analyses of the PHO5 promoter region of the plasmid. An amplicon in the coding region of the TRP1 gene region served as an internal control on the plasmid. In order to specifically probe only for the plasmid, we transformed the pTAP5C plasmid into strain YS18018, which contains a deletion of the chromosomal PHO5 locus in addition to the deletion of the TRP1 locus which is the prerequisite for the stable propagation of the plasmid. Therefore both the amplicon UASp2 of the PHO5 promoter as well as the amplicon TRP1 have no counterpart on the chromosome in this strain. As control for the overall amount of DNA after the immunoprecipitation step, we normalized to a telomeric region. The presence of histones was probed with three different antibodies directed against the C termini of histones H2B, H3, and H4. All three of them showed a specific decrease in histone abundance at the PHO5 promoter under inducing conditions (Fig. 3). The histone abundance at the TRP1 locus varied somewhat upon induction as well (Fig. 3A), but histone abundance at the PHO5 promoter dropped much more than at the TRP1 locus (see Fig. 3B for values relative to the TRP1 gene).
FIG. 3.
Chromatin immunoprecipitation analysis of the PHO5 promoter region on the plasmid shows reduced histone occupancy upon induction. (A) Immunoprecipitated DNA from strain YS18018 carrying pTAP5C using antibodies against the C terminus of H2B, H3, or H4 was quantified by real-time PCR with amplicons in the PHO5 promoter (UASp2) and in the open reading frame (ORF) of the TRP1 gene (TRP1). The PCR signals were controlled for amplicon efficiency with input DNA (T. Luckenbach, data not shown) and normalized versus the signal of an amplicon at the telomere. These normalized values represent the relative histone occupancy in the respective region and were determined for repressed (+Pi) and induced (−Pi) conditions. (B) Data as in panel A, but the values of amplicon UASp2 are divided by the values of amplicon TRP1 for each antibody and condition.
Topological analysis confirms the loss of nucleosomes upon induction.
As we worked with a circular plasmid, we had the opportunity to confirm the loss of nucleosomes upon induction by a topological analysis. Boeger et al. (7) reported such an analysis comparing the linking number of chromatin circles which were excised in vivo from the PHO5 locus in wild-type and pho80 cells representing the repressed and induced state, respectively. In their construct the PHO5 promoter was truncated at the BamHI site (Fig. 1A) and therefore encompassed only three of the four nucleosomes which become remodeled upon induction. In this system, they found a linking number difference of 1.85 upon induction, which correlated well with a loss of about 1.9 nucleosomes as measured by limit nuclease digestion. It was concluded that on average two out of three nucleosomes are lost from the PHO5 promoter upon induction. As an important technical detail, Boeger et al. found that transcription from the induced PHO5 promoter led to topological changes by itself and therefore obscured the determination of the linking number difference solely due to the chromatin transition.
Therefore, we introduced a TATA box deletion into plasmid pTAP5C, yielding pTAP5CΔTATA. Our lab characterized this deletion previously (14) and showed that transcription is abolished whereas the chromatin transition upon induction is not affected. We report here that the generation of the extended hypersensitive site proceeds indistinguishably (see Fig. 6A) and confirmed in addition, using the antibodies directed against the C termini of histones H3 and H4, that histones are lost from the promoter region in the same way as with plasmid pTAP5C (data not shown). When we compared the linking number of pTAP5CΔTATA in wild-type and pho80 cells under high-phosphate conditions, we observed a linking number difference of 2.5 ± 0.4 (Fig. 4). We expected a larger linking number difference than those in the study of Boeger et al., as our system includes all four nucleosomes which become remodeled.
FIG. 6.
The chromatin structures of regions adjacent to the PHO5 promoter on plasmid pTAP5CΔTATA remain unchanged upon induction. Chromatin analysis was done by using DNase I digestion with indirect end labeling for the plasmid pTAP5CΔTATA in strain YS18018 under repressive and inducing conditions. The labeling is analogous to Fig. 2A and 5. Compare panel A with the “Plasmid” panel in Fig. 2A and panels B, C, and D with panel A, B, and C of Fig. 5, respectively. The marker bands in panel B are rather weak and are therefore indicated by dashes. The marker for probe 63 in panel C was overloaded and a shorter exposure of this lane is shown in panel C, whereas the lane was cut out in panel D.
The chromatin structure of the plasmid environment remains the same upon PHO5 induction.
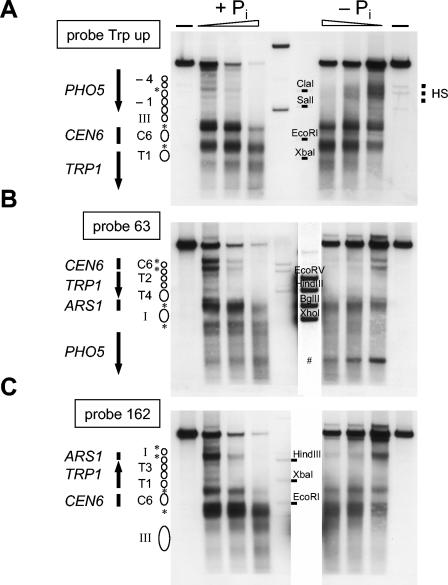
Now that the characteristic opening of the PHO5 promoter on the small plasmid, including the loss of histones, was established, the question of where these histones went became more poignant. We sought to distinguish between movement in cis or in trans by assaying the effect of the chromatin transition at the PHO5 promoter on the chromatin structure of the residual plasmid. We chose appropriate secondary cleavage sites and probes to analyze DNase I sensitivities around the whole plasmid (Fig. 5). Again, this analysis needed to be specific for the plasmid and was therefore done again in the pho5 deletion strain YS18018, as we had to use probes within the PHO5 promoter to achieve good resolution of the TRP1 region. Figure 5A to C shows unambiguously that there is no difference in chromatin structure between repressing and inducing conditions in the non-PHO5 part of the plasmid which could account for an accommodation of the nucleosomes from the PHO5 promoter. Especially the hypersensitive sites flanking the CEN6 element as well as the hypersensitive site of the ARS1 element remain unchanged. This was important to show because such nucleosome-free regions might have served as acceptors for nucleosomes if displacement were to occur in cis. We have also confirmed that the copy number of the plasmid remains unchanged upon induction (data not shown). A change in copy number could have been indicative of some interference with ARS function due to putative sliding nucleosomes pushing into this region.
FIG. 5.
The chromatin structure of regions adjacent to the PHO5 promoter on plasmid pTAP5C remain largely unchanged upon induction. Nuclei of strain YS18018 carrying pTAP5C and grown under repressive (+Pi) or inducing (−Pi) conditions were analyzed by DNase I digestion and indirect end labeling (wedges and dashes, as in Fig. 2A). HindIII was used for secondary cleavage in panel A and ClaI in panels B and C, and probes were as indicated on the left of the blots (see also Fig. 1B). The relevant genetic regions as well as schematics of the chromatin structure (labeled and stippled circles, asterisks, and dashed lines for the extended hypersensitive site HS, as in Fig. 1A and B) are indicated at the sides of the blot. The blot of panel A was also probed with probe Trp down with the same result as in the “Plasmid” panel in Fig. 2A (P. Korber and D. Blaschke, data not shown). The two middle lanes of all blots contain markers for different probes. Only one of the two lanes is relevant for a particular probe and is labeled accordingly. The marker lane for probe Trp up (A) was underloaded and the position of the restriction fragments as indicated by dashes was deduced from a longer exposure. Panels B and C show the same blot which was probed with two different probes. The marker lane for probe 63 (B) was overloaded; therefore, a shorter exposure of this lane is shown in panel B and it was cut out in panel C. The pound sign in panel B denotes an artifact band.
In Fig. 5B and C, the −Pi patterns appear weaker and less distinct than the +Pi patterns; the four positioned nucleosomes over the TRP1 gene become especially less discernible. We found that this is due to the induced transcription from the strong PHO5 promoter under −Pi conditions and did not observe this difference anymore when repeating the same kind of analysis with the pTAP5CΔTATA plasmid (Fig. 6B to D).
DISCUSSION
In this study, we address the fate of the histones that are lost from the PHO5 promoter upon induction. In this process, four positioned nucleosomes give way to an extended hypersensitive site (4), which has recently been shown to lack histone-DNA contacts (7, 28). This implies that the nucleosomes have not just adopted an altered state with higher accessibility to nucleases but that the nucleosomes became disassembled and that the histones actually moved away from the PHO5 promoter region. By transferring the PHO5 promoter to a small plasmid, we are making an important step towards understanding the mechanism by which they do so.
Changes of nucleosome structure have been shown both in vitro and in vivo to be catalyzed by chromatin remodeling complexes (6). Depending on the type of remodeler, different mechanisms and different end products of remodeling reactions have been reported, although there may be a common underlying principle (21). With respect to the chromatin transition at the PHO5 promoter, we focus on possible remodeling mechanisms that can lead to histone-free DNA as the final outcome. This narrows down the variety of observed and discussed remodeling mechanisms to two alternatives: either the transfer of histones away from the promoter region in trans onto some kind of acceptor molecules or the movement of nucleosomes in cis, i.e., the sliding of whole histone octamers along the DNA into adjacent regions.
The sliding mechanism is well documented both in vitro and in vivo for the ISWI and the SWI/SNF class of chromatin remodeling complexes (15, 20, 21, 41). In the case of the induced PHO5 promoter, it is conceivable that the positioned nucleosomes would slide away into the neighboring up- and/or downstream regions. As we have shown in the past that nucleosomes −5 and +1, bordering the region of the chromatin transition, remain largely unchanged (4), the arrival of additional nucleosomes up- and/or downstream would have to be diluted over extended stretches of chromatin along the chromosome. Therefore, the result of sliding would not necessarily be detectable within the actual acceptor regions but just lead to accessible, histone-free DNA at the vacated promoter.
In this work we present data which make such a scenario highly unlikely. We embedded a PHO5 promoter fragment into a very small circular plasmid context. That way the space for nucleosomes sliding away from the promoter region in cis is very restricted, as is the possibility of the neighboring regions to maintain an undisturbed chromatin structure while accommodating additional nucleosomes. We show here that both the initial positioning of the promoter nucleosomes as well as their remodeling upon induction occur in this plasmid context in the same way as in the native chromosomal context. Importantly, the generation of the extended hypersensitive site upon induction again leads to the loss of histone-DNA contacts, implying that histones have moved to a new location. The plasmid construct harbors several regions of DNase I hypersensitivity which could have been thought to provide space for accommodation of nucleosomes in cis. Nonetheless, we find that the chromatin structure of the remainder of the plasmid including the hypersensitive sites is completely unaffected by the chromatin transition at the PHO5 promoter. Therefore the possibility of nucleosome sliding in cis becomes highly unlikely as the mechanism of chromatin opening at the PHO5 promoter.
By exclusion of this alternative, we are drawn to conclude that the histones leave the PHO5 promoter in trans. Support for such a mechanism comes from a number of in vitro remodeling assays with the SWI/SNF or the RSC complex (23, 26, 27, 41). In these assays, histone movement in trans was observed either as the eviction of histones during disassembly of mononucleosomes leading to free DNA or the transfer of histone octamers from unlabeled mononucleosomes or polynucleosomal arrays onto labeled competitor DNA yielding labeled mononucleosomes. This mode of remodeling has never been observed so far for remodeling complexes of the ISWI family (20, 38), and chromatin remodeling at the PHO5 promoter points—as far as we are aware—to the first clear in vivo case of histone eviction in trans. Importantly, the chromatin transition at the PHO5 promoter is independent of replication and transcription (14, 31). The eviction of histones here may therefore represent a case different from the disassembly of nucleosomes observed during replication or the replication-independent histone exchange at actively transcribed loci (2). It remains to be seen whether this disassembly in all cases is catalyzed only by remodeling machines, as seems to be true for the PHO5 promoter, or whether the polymerases play an important role in that process during replication, transcription, and repair (33).
We consider the state of the induced PHO5 promoter that is generated by such a trans mode of remodeling action not as a static state but as the steady state of a competition between dis- and reassembly of nucleosomes and agree here with the concept of Boeger et al. (7) of the equilibrium of removal and reformation of nucleosomes. In this regard we want to comment on the apparent discrepancy between four remodeled nucleosomes and the quantitation of nucleosome loss by topological analysis as 2.5 out of 4 or 1.9 out of 3 (7), which we observe to represent the same proportion of 63% in both cases. It was shown early on (4) using nuclease digestion techniques that all four nucleosomes −4 to −1 are affected in their accessibility upon induction, but the flanking nucleosomes −4 and −1 less completely so (50% versus ca. 100% restriction enzyme accessibility). In keeping with this, Fig. 1A and 6A show again that nucleosome −1 and also some upstream part of the region of nucleosome −4 are still rather protected in the DNase I digestion analysis, and Fig. 2B shows that even the accessibility for ClaI, monitoring nucleosome −2, can be less than maximal in some strain backgrounds. Therefore it comes as no surprise that the topological analysis detects a linking number difference which is considerably smaller than the number of remodeled nucleosomes. This technique measures how many nucleosomes are completely lost on time average but not which ones at a given time point in a given cell. Over time and sampled over a cell population, any of the four nucleosomes −4 to −1 can be affected as shown by nuclease techniques, but on time average only roughly two thirds will be completely disassembled.
Shortly before submitting the manuscript for this study, Boeger et al. (8) published data that are equivalent to the study presented here, although using a different approach and a different major line of argument. Similar to our data, they find that promoter opening on a small and in vivo-excised PHO5 chromatin circle proceeds normally after shifting cells to phosphate-free medium. They also confirm the loss of histones by chromatin immunoprecipitation analysis and also find a linking number difference for the circle upon induction. Based on the argument that a sliding process would not have changed the overall topology, they come to the same conclusion as we did, i.e., that histones must have left in trans. This data set is fully compatible with our data, but our line of argument mainly rests on the observation that the chromatin structure in the remainder of the plasmid remains unchanged upon induction (Fig. 5 and 6).
The trans displacement of histones that leads to the disassembly of nucleosomes is catalyzed by only a subset of remodeler families, as opposed to the sliding reaction which can be performed by all remodelers tested, including the SWI/SNF complex (20, 41), and therefore seems to be a more difficult or special task. The stoichiometry of SWI/SNF complex to chromatin substrate had to be about 10-fold higher for moving nucleosomes in trans than for movement in cis and appeared the less-favored mode of action (41). Similarly, histone eviction in trans at the PHO5 promoter in vivo seems to be a slow process compared to nucleosome sliding in cis. Even though PHO5 is renowned as one of the most strongly induced genes in yeast (37), its induction at the chromatin level has a half time of about three hours (5) whereas nucleosome sliding has a half time of 60 min at the POT1 promoter (15).
In light of the many binding interactions stabilizing the nucleosome structure (24) it makes sense that the complete disassembly of a nucleosome should be less favorable than its mere relocation on the DNA which may proceed stepwise by a looping mechanism (21). At this point we cannot exclude the possibility that if the mechanism of the chromatin transition at the PHO5 promoter includes an initial sliding step which may help to start the process of histone eviction. The DNase I pattern of a gcn5 pho80 strain, which corresponds to a repressed state but exhibits randomized nucleosome positions, may capture such an intermediate state (17). Boeger et al. (8) have confirmed in their topological analysis our previous finding that nucleosomes are fully retained under these conditions.
Alternatively or in addition, the disassembly of a nucleosome can be stimulated if, e.g., a specific DNA binding factor competes with the histones for access to the DNA. Owen-Hughes et al. (26) demonstrated the eviction of histones in a SWI/SNF-catalyzed reaction from only that nucleosome in an array which contained five Gal4 binding sites and only in the presence of Gal4. Also the PHO5 promoter exhibits one intranucleosomal binding site in nucleosome −2 for the transactivator Pho4 (Fig. 1A), but the competition between histones and Pho4 for binding to the DNA cannot explain the remodeling of the remaining three nucleosomes.
Another way to favor the disassembly of nucleosomes consists of providing suitable acceptors for the displaced histones, like competitor DNA or histone chaperones. It has long been known in uncatalyzed systems that the presence of competitor DNA can lead to transfer of histone octamers in trans, although with very low efficiency at physiological ionic strength. This reaction can be again stimulated by, e.g., Gal4 binding to nucleosomal sites and even further by the presence of histone chaperones like nucleoplasmin or NAP1 (10, 40, 42). As a role for competitor DNA seems unlikely for the in vivo situation and as chromatin opening at the PHO5 promoter has to account for disassembled nucleosomes without internal factor binding sites, we speculate that histone chaperones play a major role, and studies addressing this hypothesis are currently under way (P. Korber, T. Luckenbach, and W. Hörz, unpublished data). Very recently, Adkins et al. (1) reported that the histone chaperone Asf1 appears to be essential for opening of the PHO5 promoter. In our own studies with asf1 strains we obtained conflicting data (S. Barbaric, P. Korber, T. Luckenbach, and W. Hörz, unpublished data). However, we agree that Asf1 plays a role in the chromatin transition and will discuss this point in a future publication.
The possible importance of a collaboration between remodeling complexes and carriers of histones has recently received attention in connection with the isolation of specific histone chaperone complexes for different variants of histone H3 (35) as well as the recognition of a specific histone exchange reaction catalyzed by the newly isolated SWR1 complex (18, 19, 25) and the exchange of H2A-H2B dimers during remodeling by the SWI/SNF or RSC but not ISWI complexes (9). We note, however, that the complete disassembly of nucleosomes goes an important step further than histone exchange. Still, it could be the same subset of chromatin remodeler families which is able to break up the histone octamer and therefore lead to exchange or eviction of histones.
Acknowledgments
We thank Johannes Hegemann for advice on and the sequence of the minimal CEN6 element. We are grateful to Alain Verreault for sharing histone antibodies.
This work was funded in part by Deutsche Forschungsgemeinschaft, Transregio 5, and Fonds der Chemischen Industrie.
Footnotes
This paper is dedicated to Mark N. Hoover Thames and Viticia A. Thames on the occasion of their wedding.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191-1200. [DOI] [PubMed] [Google Scholar]
- 3.Almer, A., and W. Hörz. 1986. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 5:2681-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almer, A., H. Rudolph, A. Hinnen, and W. Hörz. 1986. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbaric, S., J. Walker, A. Schmid, J. Q. Svejstrup, and W. Hörz. 2001. Increasing the rate of chromatin remodeling and gene activation—a novel role for the histone acetyltransferase Gcn5. EMBO J. 20:4944-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker, P. B., and W. Hörz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 7.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. [DOI] [PubMed] [Google Scholar]
- 8.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14:667-673. [DOI] [PubMed] [Google Scholar]
- 9.Bruno, M., A. Flaus, C. Stockdale, C. Rencurel, H. Ferreira, and T. Owen-Hughes. 2003. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol. Cell 12:1599-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, H., B. Y. Li, and J. L. Workman. 1994. A histone-binding protein, nucleoplasmin, stimulates transcription factor binding to nucleosomes and factor-induced nucleosome disassembly. EMBO J. 13:380-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cottarel, G., J. H. Shero, P. Hieter, and J. H. Hegemann. 1989. A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:3342-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fascher, K. D. 1989. Der Einfluβ der Chromatinstruktur auf die Genregulation am Beispiel eines Phosphatase-Gens aus Hefe. Ph.D. thesis. Universität München, München, Germany.
- 13.Fascher, K. D., J. Schmitz, and W. Hörz. 1990. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 9:2523-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fascher, K. D., J. Schmitz, and W. Hörz. 1993. Structural and functional requirements for the chromatin transition at the PHO5 promoter in Saccharomyces cerevisiae upon PHO5 activation. J. Mol. Biol. 231:658-667. [DOI] [PubMed] [Google Scholar]
- 15.Fazzio, T. G., and T. Tsukiyama. 2003. Chromatin remodeling in vivo: evidence for a nucleosome sliding mechanism. Mol. Cell 12:1333-1340. [DOI] [PubMed] [Google Scholar]
- 16.Gregory, P. D., S. Barbaric, and W. Hörz. 1999. Restriction nucleases as probes for chromatin structure. Methods Mol. Biol. 119:417-425. [DOI] [PubMed] [Google Scholar]
- 17.Gregory, P. D., A. Schmid, M. Zavari, L. Lui, S. L. Berger, and W. Hörz. 1998. Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol. Cell 1:495-505. [DOI] [PubMed] [Google Scholar]
- 18.Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings, A. J. Link, H. D. Madhani, and J. Rine. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2:E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan, H. Ding, R. A. Haw, J. Pootoolal, A. Tong, V. Canadien, D. P. Richards, X. Wu, A. Emili, T. R. Hughes, S. Buratowski, and J. F. Greenblatt. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12:1565-1576. [DOI] [PubMed] [Google Scholar]
- 20.Längst, G., and P. B. Becker. 2001. Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J. Cell Sci. 114:2561-2568. [DOI] [PubMed] [Google Scholar]
- 21.Längst, G., and P. B. Becker. 2004. Nucleosome remodeling: one mechanism, many phenomena? Biochim. Biophys. Acta 1677:58-63. [DOI] [PubMed] [Google Scholar]
- 22.Längst, G., E. J. Bonte, D. F. Corona, and P. B. Becker. 1999. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97:843-852. [DOI] [PubMed] [Google Scholar]
- 23.Lorch, Y., M. Zhang, and R. D. Kornberg. 1999. Histone octamer transfer by a chromatin-remodeling complex. Cell 96:389-392. [DOI] [PubMed] [Google Scholar]
- 24.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 25.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 26.Owen-Hughes, T., R. T. Utley, J. Côté, C. L. Peterson, and J. L. Workman. 1996. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science 273:513-516. [DOI] [PubMed] [Google Scholar]
- 27.Phelan, M. L., G. R. Schnitzler, and R. E. Kingston. 2000. Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases. Mol. Cell. Biol. 20:6380-6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reinke, H., and W. Hörz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 29.Reinke, H., and W. Hörz. 2004. Anatomy of a hypersensitive site. Biochim. Biophys. Acta 1677:24-29. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schmid, A., K. D. Fascher, and W. Hörz. 1992. Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication. Cell 71:853-864. [DOI] [PubMed] [Google Scholar]
- 32.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 33.Studitsky, V. M., W. Walter, M. Kireeva, M. Kashlev, and G. Felsenfeld. 2004. Chromatin remodeling by RNA polymerases. Trends Biochem. Sci. 29:127-135. [DOI] [PubMed] [Google Scholar]
- 34.Svaren, J., and W. Hörz. 1997. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci. 22:93-97. [DOI] [PubMed] [Google Scholar]
- 35.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51-61. [DOI] [PubMed] [Google Scholar]
- 36.Thoma, F., L. W. Bergman, and R. T. Simpson. 1984. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J. Mol. Biol. 177:715-733. [DOI] [PubMed] [Google Scholar]
- 37.Toh-e, A., H. Nakamura, and Y. Oshima. 1976. A gene controlling the synthesis of non specific alkaline phosphatase in Saccharomyces cerevisiae. Biochim. Biophys. Acta 428:182-192. [DOI] [PubMed] [Google Scholar]
- 38.Varga-Weisz, P. D., M. Wilm, E. Bonte, K. Dumas, M. Mann, and P. B. Becker. 1997. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388:598-602. [DOI] [PubMed] [Google Scholar]
- 39.Vogel, K., W. Hörz, and A. Hinnen. 1989. The two positively acting regulatory proteins PHO2 and PHO4 physically interact with PHO5 upstream activation regions. Mol. Cell. Biol. 9:2050-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walter, P. P., T. Owen-Hughes, J. Côté, and J. L. Workman. 1995. Stimulation of transcription factor binding and histone displacement by nucleosome assembly protein 1 and nucleoplasmin requires disruption of the histone octamer. Mol. Cell. Biol. 15:6178-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitehouse, I., A. Flaus, B. R. Cairns, M. F. White, J. L. Workman, and T. Owen-Hughes. 1999. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400:784-787. [DOI] [PubMed] [Google Scholar]
- 42.Workman, J. L., and R. E. Kingston. 1992. Nucleosome core displacement in vitro via a metastable transcription factor nucleosome complex. Science 258:1780-1784. [DOI] [PubMed] [Google Scholar]