Abstract
Loss and/or degradation of small molecules during sampling, sample transportation and storage can adversely impact biological interpretation of metabolomics data. In this study, we performed in vivo sampling using solid-phase microextraction (SPME) in combination with non-targeted liquid chromatography and high-resolution tandem mass spectrometry (LC-MS/MS) to capture the fish tissue exposome using molecular networking analysis, and the results were contrasted with molecular differences obtained with ex vivo SPME sampling. Based on 494 MS/MS spectra comparisons, we demonstrated that in vivo SPME sampling provided better extraction and stabilization of highly reactive molecules, such as 1-oleoyl-sn-glycero-3-phosphocholine and 1-palmitoleoyl-glycero-3-phosphocholine, from fish tissue samples. This sampling approach, that minimizes sample handling and preparation, offers the opportunity to perform longitudinal monitoring of the exposome in biological systems and improve the reliability of exposure-measurement in exposome-wide association studies.
Since genetic factors typically account for only about 18% of chronic disease risks1 exposome-wide association studies (EWAS) that measure entire classes of small molecules in biospecimens (resulting from myriad of environmental exposures) are being conducted to discover unknown causes of chronic diseases2. In biological systems, small molecules can be either substrates or end products of cellular metabolism and can originate from exogenous sources via a myriad of exposures, or from endogenous processes including host and microbial metabolism. Most EWAS rely on untargeted metabolomics analysis of biospecimens from incident disease cases and matched controls to measure hundreds or thousands of features that then generate candidate discriminating biomarkers3. The choice of sampling, sample collection, and sample preparation strategy plays an important role in the quality of metabolomics data4. Issues of incomplete metabolism quenching, ionization suppression, and metabolite instability have been well-documented and can adversely impact data interpretation. One way to circumvent these issues is to use in vivo sample preparation techniques that provides a true(r) representation of the exposome by eliminating some of the variability introduced in sample depletion and multistep sample handling5,6,7,8.
An example of such sample preparation technique is the sorbent-coated device, solid-phase microextraction (SPME). Over the past years, in vivo SPME has been successfully applied for targeted and untargeted metabolomics analysis of various biospecimens9 owing to the biocompatibility of SPME coatings. In vivo SPME combine sampling, metabolite extraction, and metabolism quenching in one step, limiting loss and/or degradation of metabolites9. We have recently demonstrated that it is possible to extract hundreds of chemicals with a single coated fiber from plasma, including short-lived and unstable metabolites not detected and/or quantified accurately with other techniques10,11.
Here we performed in vivo SPME in combination with liquid chromatography (LC) and data-dependent tandem high-resolution mass spectrometry (MS/MS) to capture the fish tissue exposome, and examined molecular differences obtained with ex vivo SPME analysis from the same biospecimens using molecular networking analysis. In vivo extraction of metabolites from fish tissue (n = 60, white sucker) was achieved by inserting a PAN-C18 coated SPME blade into the dorsal-epaxial muscle for 20 min. Ex vivo SPME sampling was conducted using the same procedure but from tissue samples collected after fish euthanasia and being stored frozen. Extracted molecules were desorbed in an 80% v/v acetonitrile solution, and analyzed with reversed-phase LC-MS/MS using a pentafluorophenyl column and a Q-Exactive Quadrupole-Orbitrap MS in positive ionization mode (Detailed description of experimental conditions can be found in Supplementary Information).
Materials and Methods
Study design
Adult white sucker (Catostomus commersonii) (40.8 ± 3.6 cm, 969.7 ± 303.3 g, n = 60) were collected by boat electrofishing from the Athabasca River in the Alberta oil sands region (Northern Alberta, Canada). As part of a larger sampling effort, twelve fish were collected in September 2013 from: 2 sites outside of the deposit, M0 (Athabasca) and M1, which are both downstream of a pulp and paper mill discharge; 1 site upstream of the oil sands development but within the deposit around Northlands Sawmill (Downstream of M3); 1 site adjacent to the oil sands development (Upstream of M4); and 1 site downstream of the Muskeg River within the deposit and downstream of the development (Downstream M4) (Fig. 1). In total, a subset of 6 males and 6 females were selected at each site and held briefly (<1 h) in cages in the river until sampling.
Figure 1. Map of different sites investigated generated from Google. (n.d.).
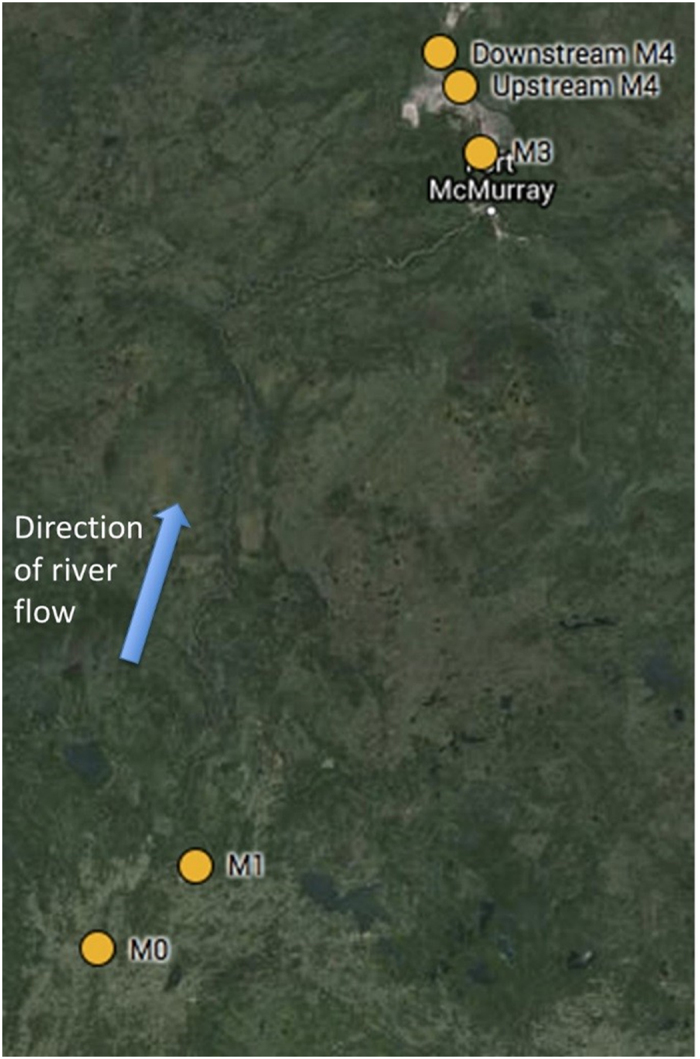
[Google Maps of fish collection sites in Alberta, Canada]. Map data ©2016 Google. Retrieved September 17, 2016, from https://goo.gl/HBAeBS.
SPME blade coating preparation
PAN-C18 SPME blades were prepared as previously described12 by immobilization of particles on the surface of stainless steel blades. Briefly, 5 μm particles (Supelco, PA) were immobilized (60 μm coating thickness) using a polyacrylonitrile (PAN) solution which acted as a bonding agent.
In vivo and ex vivo SPME sampling procedure of fish tissue
All experimental protocols were in accordance with and as approved by the University of Waterloo Animal Care Committee (AUPP #10–17). In vivo extraction of metabolites from fish tissue was conducted by inserting a PAN-C18 SPME coated blade into the dorsal-epaxial muscle (near the dorsal fin) of fish immobilized using a large foam bed13,14. The blade remained in place for 20 min while fish were held in an aerated, 28 L covered bucket. After 20 min, the blade was removed, rinsed with nanopure water to remove matrix components, and frozen in liquid nitrogen. Fish were then sacrificed and a small part of the dorsal-epaxial muscle was cut out, placed in aluminum foil and frozen in liquid nitrogen on-site and shipped to our laboratory. In the laboratory, ex-vivo SPME sampling was performed by inserting a PAN-C18 coated blade into a thawed, non-homogenized tissue sample for 20 min without agitation. Desorption of metabolites from the SPME coating of the blades was done by immersing them for 60 min in 1 mL of acetonitrile/water (80/20, v/v) with vortex agitation at 1000 rpm. Extracts were stored at −80 °C until analysis.
UPLC-Q-Exactive Orbitrap HRMS analysis
Metabolite profiling was conducted using an LC-MS system consisting of a ThermoAccela autosampler, pumps and a Q-Exactive Orbitrap System (Thermo Fisher Scientific, CA, USA). Metabolites were separated by a reversed-phase method using a pentafluorophenyl column (Kinetex Phenomenex, 2.1 mm × 100 mm, 1.7 μm particle size) at a flow rate of 300 μL/min. Mobile phase A consisted of water/formic acid (99.9/0.1, v/v) and mobile phase B consisted of acetonitrile/formic acid (99.9/0.1, v/v). The starting mobile phase conditions were 90% A from 0 to 1.0 min, followed by a linear gradient to 10% A from 1.0 to 9.0 min and an isocratic hold at 10% A until 12.0 min. The total run time was 18 min per sample, including a 6 min re-equilibration time. The injection volume was 10 μL. Analyses were performed in positive ionization mode in the mass range of m/z 50–750. To maintain a mass accuracy better than 5 ppm, we used the following lock mass: m/z 391.2843. Instrument parameters were set as follows: sheath gas (Nitrogen) flow rate, 35 arbitrary units; capillary voltage, 3.1 kV; ion source temperature 280 °C; full MS automatic gain control (AGC), 1.106; spectra rate acquisition, 3.7 spectra/s; full MS resolution, 70,000. MS/MS fragmentation of the ten most intense ions per spectrum was performed using a normalized collision energy (NCE) of 50; MS/MS resolution, 17,500; MS/MS AGC, 2.105; precursor ion mass isolation window, 1 ppm. MS/MS exclusion list was set after 3 analyses of blank SPME samples.
UPLC-MS/MS data processing and molecular networking
Molecular networking of LC-MS/MS data was performed using the Global Natural Products Social Molecular Networking (GNPS) software15. For MS and MS/MS spectral library search and molecular networking, we used an ion mass tolerance of 1 and 0.01 Da for precursor ions and fragment ions, respectively. A minimum cosine similarity score of 0.7 was used for MS/MS spectral library matching. A minimum cosine similarity score of 0.7 and a minimum number of 6 matched fragment ions were used to form a network of two consensus MS/MS spectra. Resulting molecular networks were built and visualized using Cytoscape 3.2.116. Precursor ion m/z was used as node attribute and cosine similarity score was used as edge attribute.
Results and Discussion
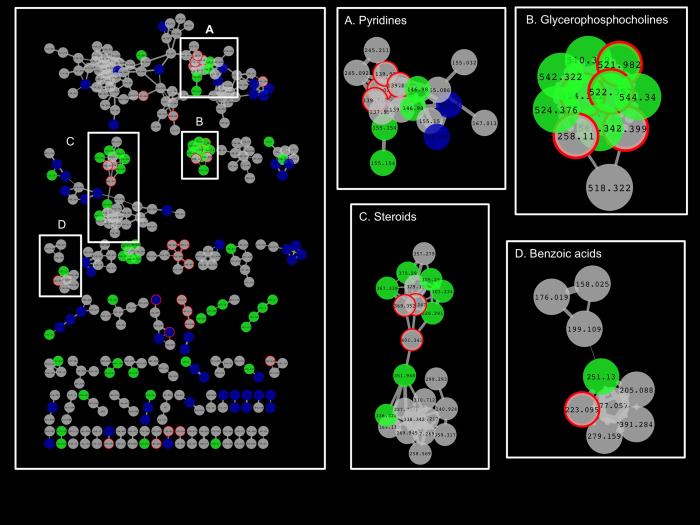
After molecular networking of fish tissue LC-MS/MS data, we found 494 nodes representing consensus of at least two or more MS/MS spectra (Fig. 2). The majority of the nodes matched chemicals detected in fish tissue using both sampling approaches. Only 7% of them could be identified by matching LC-MS/MS libraries with spectral similarity ≥0.7 (Table 1), suggesting that the vast majority of chemicals in fish tissue are unknown. However, mapping the chemical similarity between unknown and identified molecules can help uncover chemical classes of the uncharacterized molecules. Although the majority of identified chemicals were endogenous molecules, four molecules including 4-methoxycinnamic acid, 1-hydroxybenzotriazole, diethylphthalate, and phenoxybenzamine used in the formulation of sunscreen products17 dishwasher cleaning products18 and plasticizers19 or as active ingredients in pharmaceutical products20 respectively, were detected in fish tissues. These chemicals probably originated from external exposures (i.e. water contamination), confirming that fish tissue offers opportunity to reconstruct past environmental exposures due to bioaccumulation of persistent organic toxicants, and can be used to monitor aquatic ecosystem health.
Figure 2. Chemical-similarity maps of small molecules (n = 494 with MS/MS spectrum) in fish tissue (Tanimoto coefficient ≥0.7).
Green and blue nodes represent small molecules observed only after in vivo SPME sampling and ex vivo SPME sampling, respectively. Grey nodes represent small molecules detected using both sampling methods. Nodes with red border indicates annotated molecule by matching LC-MS/MS libraries with spectral similarity ≥0.7 Edge represents similarity between MS/MS spectra. Thickness of the edges indicates the level of similarity (the thicker is an edge, the more similar are MS/MS spectra).
Table 1. Small molecules identified in fish tissue using solid-phase microextraction combined with LC-MS/MS with a Q-Exactive Quadrupole-Orbitrap mass spectrometer.
| Metabolite | Family | Precursor m/z | Precursor adduct | RT (min) | Sample class | Similarity cosinea |
|---|---|---|---|---|---|---|
| 4-methoxycinnamic acid | Cinnamic acids | 179.070 | [M + H]+ | 8.48 | G2 | 0.78 |
| Phenoxybenzamine | Phenylmethylamines | 304.147 | [M + H]+ | 6.98 | G2 | 0.82 |
| Nicotinamide | Pyridinecarboxylic acids | 123.055 | [M + H]+ | 0.86 | G2 | 0.83 |
| 1-Oleoyl-sn-glycero-3-phosphocholine | Glycerophosphocholines | 522.355 | [M + H]+ | 7.5 | G2 | 0.87 |
| 1-Palmitoleoyl-glycero-3-phosphocholine | Glycerophosphocholines | 494.324 | [M + H]+ | 6.78 | G2 | 0.70 |
| 4-Chlorophenol | Chlorophenols | 129.010 | [M + H]+ | 0.70 | G1 | 0.74 |
| Hydroxyproline | Carboxylic acids and derivates | 132.065 | [M + H]+ | 0.88 | G1 | 0.83 |
| 6-Hydroxynicotinate | Pyridinecarboxylic acids | 140.034 | [M + H]+ | 0.97 | G1 | 0.78 |
| 2-Heptyl-3-hydroxy 4-quinolone | Quinolones | 260.165 | [M + H]+ | 8.30 | G1 | 0.78 |
| Phenylalanine | Phenylpropanoic acids | 166.087 | [M + H]+ | 1.38 | G1, G2 | 0.96 |
| Tyrosine | Phenylpropanoic acids | 182.081 | [M + H]+ | 0.85 | G1, G2 | 0.97 |
| Deoxycarnitine | Fatty acids and conjugates | 146.118 | [M + H]+ | 0.99 | G1, G2 | 0.99 |
| 1-Hydroxybenzotriazole | Benzotriazoles | 136.049 | [M + H]+ | 0.90 | G1, G2 | 0.70 |
| Creatine | Carboxylic acids and derivates | 132.077 | [M + H]+ | 0.88 | G1, G2 | 0.97 |
| 3-Methylhistidine | Carboxylic acids and derivates | 170.093 | [M + H]+ | 0.92 | G1, G2 | 0.82 |
| Cholesterol | Steroids | 369.352 | [M−H2O + H]+ | 10.1 | G1, G2 | 0.95 |
| Histidine | Carboxylic acids and derivates | 156.078 | [M + H]+ | 0.90 | G1, G2 | 0.99 |
| Tryptophan | Indoles | 205.097 | [M + H]+ | 2.90 | G1, G2 | 0.93 |
| Arginine | Carboxylic acids and derivates | 175.119 | [M + H]+ | 0.94 | G1, G2 | 0.95 |
| 2-amino-2-methylpropanoate | Carboxylic acids and derivates | 106.050 | [M+H]+ | 0.74 | G1, G2 | 0.98 |
| Proline | Carboxylic acids and derivates | 116.071 | [M+H]+ | 0.76 | G1, G2 | 0.87 |
| Betaine | Carboxylic acids and derivates | 118.086 | [M+H]+ | 0.75 | G1, G2 | 0.95 |
| Threonine | Carboxylic acids and derivates | 120.066 | [M+H]+ | 0.74 | G1, G2 | 0.91 |
| L-Glutamine | Carboxylic acids and derivates | 147.076 | [M+H]+ | 1.01 | G1, G2 | 0.95 |
| L-Methionine | Carboxylic acids and derivates | 150.058 | [M+H]+ | 0.84 | G1, G2 | 0.81 |
| Carnitine | Fatty acids and conjugates | 162.112 | [M+H]+ | 0.95 | G1, G2 | 0.94 |
| O-Acetylcarnitine | Fatty acids and conjugates | 204.123 | [M+H]+ | 1.25 | G1, G2 | 0.96 |
| Diethylphthalate | Benzoic acids | 223.096 | [M+H]+ | 6.22 | G1, G2 | 0.90 |
| Inosine | Purine nucleosides | 269.088 | [M+H]+ | 0.80 | G1, G2 | 0.94 |
| Adenosine | Purine nucleosides | 268.103 | [M+H]+ | 0.89 | G1, G2 | 0.97 |
| Guanosine | Purine nucleosides | 284.099 | [M+H]+ | 0.80 | G1, G2 | 0.91 |
| Inosinic acid | Purine nucleotides | 349.054 | [M+H]+ | 0.72 | G1, G2 | 0.88 |
| Desmosterol | Steroids | 385.346 | [M+H]+ | 7.98 | G1, G2 | 0.75 |
| 3β-Hydroxy-5-cholestenal | Steroids | 401.341 | [M+H]+ | 8.88 | G1, G2 | 0.80 |
| sn-Glycero-3-phosphocholine | Glycerophosphocholines | 258.110 | [M+H]+ | 0.70 | G1, G2 | 0.94 |
| 1-Palmitoyl-sn-glycero-3-phosphocholine | Glycerophosphocholines | 496.339 | [M+H]+ | 7.3 | G1, G2 | 0.89 |
| 1-Palmitoyl-sn-glycero-3-phosphocholine | Glycerophosphocholines | 518.322 | [M+Na]+ | 7.3 | G1, G2 | 0.89 |
aCosine similarity value between experimental MS/MS spectrum and MS/MS spectrum from public LC-MS/MS libraries.
G1: ex-vivo SPME sampling; G2: in-vivo SPME sampling.
We then evaluated the ability of in vivo SPME to capture unstable molecules. We observed that 16% of the nodes were only detected after in vivo SPME sampling, while 21% only found after ex vivo SPME sampling. Molecules only observed with in vivo SPME include cinnamic acids, glycerophosphocholines, phenylmethylamines, phenoxybenzamines and pyridincarboxylic acids originating from both exogenous and endogenous exposures (Table 1). For example, 1-oleoyl-sn-glycero-3-phosphocholine is a lipid-signaling molecule generated by phospholipase enzymes21. This compound contains an unsaturated acyl chain where the hydrogen atom on methylene groups adjacent to the double bounds has low carbon-hydrogen (C-H) bond energies, and is therefore a major target for modification under oxidative conditions after sample collection22. Five other glycerophosphocholines (Fig. 1B; m/z 542.322, m/z 524.376, m/z 568.342, m/z 544.340, and m/z 510.359) were only detected using in vivo SPME, but was not successfully identified due to the lack of adequate similarity (cosine ≥0.7) between their MS/MS spectra and those from public LC-MS/MS libraries. This result possibly indicates that these molecules have not been previously identified due to their chemical instability or experimental MS/MS spectra produced were a composite of two or more molecules. Similarly, cinnamic acids, phenylmethylamines, phenoxybenzamines and pyridincarboxylic acids only detected after in-vivo SPME are also highly reactive compounds prone to auto-oxidation during sample transportation and storage. However, glycerophosphocholines containing saturated acyl chains, such as 1-palmitoyl-sn-glycero-3-phosphocholine, which was detected after both in vivo and ex vivo SPME, are more resistant to oxidation22. Some molecules were only observed after ex-vivo SPME, including 6-hydroxynicotinate, hydroxyproline and 2-heptyl-3-hydroxy 4-quinolone. Previous studies have demonstrated that 6-hydroxynicotinate and 2-heptyl-3-hydroxy 4-quinolone are essentially produced from bacterial metabolism23,24 and possibly originated from tissue degradation during sample transportation and storage. Hydroxyproline, a major constituent of collagen, is also used as an indicator of tissue damage or degradation25.
Conclusion
In summary, we demonstrated that in vivo SPME sampling provided extraction and stabilization of highly reactive chemicals, not detected by ex vivo sample preparation techniques due to their auto-oxidation during sample collection, transportation and storage. In vivo SPME sampling provides extraction of molecules with a wide range of chemical and physical properties9 (balanced coverage), which originate from exogenous sources via environmental exposures or from endogenous processes including host and microbial metabolism. In vivo SPME sampling as a minimally invasive technique also offers the opportunity to perform repeated sampling over time on the same subject, limiting inter and intra-individual variability in levels of circulating molecules arising from changes in environmental factors (e.g. diet or sources of pollutants).
Additional Information
How to cite this article: Bessonneau, V. et al. In vivo microsampling to capture the elusive exposome. Sci. Rep. 7, 44038; doi: 10.1038/srep44038 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
This work was supported by Environment Canada through the Environmental Damages Fund (Grant EC-129114) and by funding provided to Environment Canada through the Joint Oil Sands Monitoring Program.
Footnotes
The authors declare no competing financial interests.
Author Contributions J.P., M.S., J.I. and V.B. designed experiments. M.M. and J.I. collected fish, performed in vivo SPME sampling and collected fish tissue samples. V.B. performed ex vivo SPME sampling, LC-MS/MS analysis, data processing and statistical analysis, and prepared the manuscript with help from all co-authors who also helped interpret the results.
References
- Rappaport S. M. Genetic factors are not the major causes of chronic diseases. PLoS ONE 11, e0154387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild C. P. Complementing the genome with an “exposome”: The outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomarkers Prev. 14, 1847–1850 (2005). [DOI] [PubMed] [Google Scholar]
- Rappaport S. M. Implications of the exposome for exposure science. J. Expo. Sci. Environ. Epidemiol. 21, 5–9 (2011). [DOI] [PubMed] [Google Scholar]
- Ryan D. & Robards K. Metabolomics: The greatest omics of them all? Anal. Chem. 78, 7954–7958 (2006). [DOI] [PubMed] [Google Scholar]
- Bruce S. J., Tavazzi I., Parisod V., Kochlar S. & Guy P. A. Investigation of human blood plasma sample preparation for performing metabolomics using ultrahigh performance liquid chromatography/mass spectrometry. Anal. Chem. 81, 3285–3296 (2009). [DOI] [PubMed] [Google Scholar]
- Moco S., Vervoort J., Bino R. J., De Vos R. C. H. & Bino R. Metabolomics technologies and metabolite identification. Trends Anal. Chem. 26, 855–866 (2007). [Google Scholar]
- Theodoridis G., Gika H. G. & Wilson I. D. LC-MS-based methodology for global metabolite profiling in metabonomics/metabolomics. Trends Anal. Chem. 27, 251–260 (2008). [Google Scholar]
- Canelas A. B. et al. Quantitative evaluation of intracellular metabolite extraction techniques for yeast metabolomics. Anal. Chem. 81, 7379–7389 (2009). [DOI] [PubMed] [Google Scholar]
- Bojko B. et al. Solid-phase microextraction in metabolomics. Trends Anal. Chem. 61, 168–180 (2014). [Google Scholar]
- Vuckovic D. et al. In vivo solid-phase microextraction: Capturing the elusive portion of metabolome. Angew. Chem. Int. Ed. 50, 5344–5348 (2011). [DOI] [PubMed] [Google Scholar]
- Bessonneau V., Zhan Y., De Lannoy I. A. M., Saldivia V. & Pawliszyn J. In vivo solid-phase microextraction liquid chromatography-tandem mass spectrometry for monitoring blood eicosanoids time profile after lipopolysaccharide-induced inflammation in Sprague-Dawley rats. J. Chromatogr. A 1424, 134–138 (2015). [DOI] [PubMed] [Google Scholar]
- Mirnaghi F. S., Chen Y., Sidisky L. M. & Pawliszyn J. Optimization of the coating procedure for a high-throughput 96-blade solid-phase microextraction system coupled with LC-MS/MS for analysis of complex samples. Anal. Chem. 83, 6018–6025 (2011). [DOI] [PubMed] [Google Scholar]
- Ouyang G. et al. Sampling-rate calibration for rapid and nonlethal monitoring of organic contaminants in fish muscle by solid-phase microextraction. Environ. Sci. Technol. 45, 7792–7798 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang X., Oakes K. D., Wang S., Cui S. & Pawliszyn J. In vivo sampling of environmental organic contaminants in fish by solid-phase microextraction. Trend Anal. Chem. 32, 31–39 (2012). [Google Scholar]
- Wang M. et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nature Biotechnology 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. J. & Miller I. J. The effect of molecular environment on the photochemistry of p-methoxycinnamic acid and its esters. J. Photochem. Photobiol. A 118, 93–97 (1998). [Google Scholar]
- Janna H., Scrimshaw M. D., Williams R. J., Churchley J. & Sumpter J. P. From dishwasher to tap? Xenobiotic substances benzotriazole and tolyltriazole in the environment. Environ. Sci. Technol. 45, 3858–3864 (2011). [DOI] [PubMed] [Google Scholar]
- Sánchez-Avila J., Tauler R. & Lacorte S. Organic micropollutants in coastal waters from NW Mediterranean Sea: sources distribution and potential risk. Environ. Int. 46, 50–62 (2012). [DOI] [PubMed] [Google Scholar]
- Guzzetta N. A. Phenoxybenzamine in the treatment of hypoplastic left heart syndrome: A core review. Anesth. Analg. 105, 312–315 (2007). [DOI] [PubMed] [Google Scholar]
- Mustranta A., Forsell P., Aura A. M., Suortti T. & Poutanen K. Modification of phospholipids with lipases and phospholipases. Biocatalysis 9, 181–194 (1994). [Google Scholar]
- Reis A. & Spickett C. M. Chemistry of phospholipid oxidation. Biochim. Biophys. Acta 1818, 2374–2387 (2012). [DOI] [PubMed] [Google Scholar]
- Hurh B., Ohshima M., Yamane T. & Nagasawa T. Microbial production of 6-hydroxynicotinic acid, an important building block for the synthesis of modern insecticides. J. Ferment. Bioeng. 77, 382–385 (1994). [Google Scholar]
- Diggle S. P. et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 14, 87–96 (2007). [DOI] [PubMed] [Google Scholar]
- Nogueira Ade C., Vale R. G., Gomes A. L. & Dantas E. H. The effect of muscle actions on the level of connective tissue damage. Res. Sports Med. 19, 259–270 (2011). [DOI] [PubMed] [Google Scholar]