Abstract
The 26S proteasome is an ATP-dependent dynamic 2.5 MDa protease that regulates numerous essential cellular functions through degradation of ubiquitinated substrates. Here we present a near-atomic-resolution cryo-EM map of the S. cerevisiae 26S proteasome in complex with ADP-AlFx. Our biochemical and structural data reveal that the proteasome-ADP-AlFx is in an activated state, displaying a distinct conformational configuration especially in the AAA-ATPase motor region. Noteworthy, this map demonstrates an asymmetric nucleotide binding pattern with four consecutive AAA-ATPase subunits bound with nucleotide. The remaining two subunits, Rpt2 and Rpt6, with empty or only partially occupied nucleotide pocket exhibit pronounced conformational changes in the AAA-ATPase ring, which may represent a collective result of allosteric cooperativity of all the AAA-ATPase subunits responding to ATP hydrolysis. This collective motion of Rpt2 and Rpt6 results in an elevation of their pore loops, which could play an important role in substrate processing of proteasome. Our data also imply that the nucleotide occupancy pattern could be related to the activation status of the complex. Moreover, the HbYX tail insertion may not be sufficient to maintain the gate opening of 20S core particle. Our results provide new insights into the mechanisms of nucleotide-driven allosteric cooperativity of the complex and of the substrate processing by the proteasome.
Keywords: high-resolution cryo-EM, proteasome, activated state, asymmetric nucleotide occupancy
Introduction
The ubiquitin-proteasome system (UPS) is the main protein degradation pathway in eukaryotic cells1,2. Dysfunction of the UPS is implicated in various diseases, including cancer and neurodegenerative diseases, making it a vital drug target2,3,4,5. In this pathway, the 26S proteasome plays a key role in the degradation of proteins marked by polyubiquitin chains2,6,7. This giant, 2.5 MDa, ATP-dependent macromolecular machine contains at least 33 distinct subunits, comprising a barrel-shaped 20S core particle (CP), which contains the internal proteolytic sites, and a 19S regulatory particle (RP) capping either side of the 20S CP8,9. The 19S RP functions to recognize and bind to the ubiquitinated substrates, and consequently to deubiquitinate, unfold, and translocate them into 20S CP for further degradation7,10,11,12.
The 19S RP is further divided into two sub-complexes: the base and the lid13. The base contains an AAA-ATPase, two largest subunits (Rpn1 and Rpn2), and the ubiquitin receptor Rpn13. The AAA-ATPase, a heterohexamer comprising subunits Rpt1-6, is responsible for substrate unfolding and translocation driven by cycles of ATP binding and hydrolysis14,15. Each AAA-ATPase subunit has a C-terminal AAA domain that assembles together into the AAA-ring. The six N-terminal oligonucleotide-binding (OB) domains form the N-ring, and three heterodimeric α-helical coiled coils radiate outwards from the N-ring. The lid, consisting of Rpn3, Rpn5-9, Rpn11, Rpn12, and Sem1 subunits in yeast, sits on the side of the proteasome and interacts with both the base and the 20S CP16,17,18. The lid subunits (except Rpn8, Rpn11 and Sem1) contain a PCI (Proteasome-CSN-eIF3) domain19 and their N-termini diverge into a horseshoe shape16,20,21,22,23, while the C-termini of all the lid subunits (except Sem1) form a helical bundle21,23,24. Rpn11, the catalytically active deubiquitinating enzyme, is indispensable for ubiquitin-dependent substrate degradation25. Rpn10, another resident ubiquitin receptor26, is positioned at the periphery of the lid.
With constant efforts of over two decades, the remarkable structures of 26S proteasome in the presence of ATP16,18,20,21,22, ATPγS18,27 and substrate17 have been determined to intermediate or relatively high resolution by single-particle cryo-EM, including a yeast proteasome structure in the presence of ATP at 4.6-6.1 Å resolution23. Previous studies reveal the existence of two states for yeast 26S proteasome in the presence of ATP23, including a resting state for substrate-free ATP structure and an activated state resembling the ATPγS structure28. Additionally, the isolated lid sub-complex29 as well as the human proteasome in the resting state30,31,32 has recently been determined to near-atomic resolution. Still, a near-atomic-resolution structure of proteasome in the activated transition state remains to be elucidated, in which state proteasome could utilize the energy provided by ATP hydrolysis to allosterically coordinate the entire macromolecular machine to perform substrate deubiquitination, unfolding, translocation, and degradation33.
Here we present an overall 4.2-Å-resolution cryo-EM map of the S. cerevisiae single-capped 26S proteasome in the presence of ADP-AlFx, which is shown to be in an activated state. This map reveals a new conformational configuration of the AAA-ATPase ring, especially in the pore loop region, with the overall conformation of the holoenzyme similar to that of the substrate-engaged state17. Our map also discloses that among the six AAA-ATPase subunits, four consecutive subunits have their nucleotide binding pockets occupied. We additionally present a cryo-EM map of the yeast proteasome in the resting state revealing new structural features. Our results provide new insights into the structural mechanism of nucleotide-driven proteasome allosteric coordination, correlative to the substrate unfolding and translocation by proteasome.
Results
Near-atomic-resolution cryo-EM structure of proteasome-ADP-AlFx
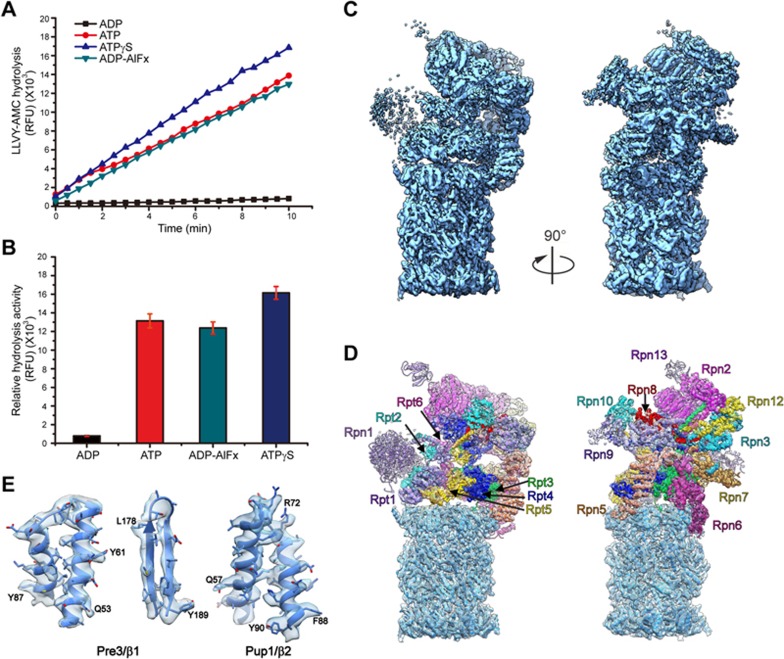
Our in vitro proteolytic activity experiment toward a fluorogenic peptide Suc-LLVY-AMC shows that the activity of proteasome in the presence of ADP-AlFx is comparable to that of proteasome in the presence of ATP and peptide substrate (Figure 1A and 1B). ADP-AlFx is the nucleotide analog usually used to mimic the transition state in ATP hydrolysis reaction34 in structural studies of other ATPase systems such as chaperonin35 and hexameric helicase36. Moreover, this proteolytic activity experiment also indicates that proteasome is more active in the presence of ATPγS, consistent with previous biochemical analysis37,38, but almost loses its activity when bound with ADP (Figure 1A and 1B, Supplementary information, Figure S1), which has been shown to be a poor stabilizer to associate 19S with 20S33 (Supplementary information, Figure S1C). Altogether, these data imply that proteasome remains functionally active when bound with ADP-AlFx.
Figure 1.
Proteasome proteolytic activity and cryo-EM structure of the single-capped 26S proteasome-ADP-AlFx. (A) In vitro proteolytic activity of proteasome toward a fluorogenic peptide substrate Suc-LLVY-AMC in different nucleotide states. RFU, relative fluorescence units. (B) Relative hydrolysis ability of proteasome toward the fluorogenic substrate was analyzed for the same samples as in A. Standard deviations were calculated from three independent experiments. (C) The cryo-EM map of proteasome-ADP-AlFx in different views (dodger blue). (D) The corresponding pseudo-atomic model fitted into the proteasome-ADP-AlFx map, with the models for different 19S RP subunits in color and that for 20S CP in dodger blue. Different subunits in 19S RP are labeled. (E) Representative high-resolution α helix and β strand features of the 20S β subunits (β1 and β2) in the proteasome-ADP-AlFx map. The β strand separation can be well resolved. The selected residues with bulky side chain are labeled.
To capture the structure of proteasome in the presence of ADP-AlFx (denoted as “proteasome-ADP-AlFx”) at high resolution, we collected 13 871 micrographs of the yeast single-capped 26S proteasome in complex with ADP-AlFx. Data collection was carried out on a Titan Krios transmission electron microscope equipped with a K2 Summit direct electron detector (Supplementary information, Figure S2A and Table S1). Its cryo-EM structure was determined (from 178 576 cleaned-up particles) at an overall near-atomic resolution of 4.2 Å (Figure 1C, Supplementary information, Figures S2G and S3). In this map, 20S displays a resolution of up to ∼4.0 Å, and the majority of 19S shows a resolution of 5.0-6.5 Å due to its highly dynamic and heterogeneous nature39,40 (Figure 1E and Supplementary information, Figure S2E). This allowed us to build a pseudo-atomic model for the complex (Figure 1D and Supplementary information, Figure S4).
We also resolved a cryo-EM map of the yeast single-capped 26S proteasome in the resting state (the substrate-free ATP structure; Supplementary information, Figure S5) at an overall resolution of 6.3 Å from 26 000 particles, with 20S at a resolution of 5.5-6.0 Å and 19S at a resolution of 6.0-7.0 Å (Supplementary information, Figure S2B, S2F-S2G and Table S1). Although this map is in a similar conformation to that of the yeast 26S proteasome also in the resting state23 (Supplementary information, Figure S5B), new features have been revealed by our structure. With these structure details, we were able to build a pseudo-atomic model into our resting state map (Supplementary information, Figure S5A).
Here we chose the single-capped 26S proteasome since a previous sub-tomogram averaging of the in situ proteasome in hippocampal neurons has suggested the single-capped 26S proteasome to be more prevalent (73%) than the double-capped 26S proteasome (27%)41. Moreover, the two 19S RPs in the double-capped 26S proteasome are functionally independent41. Thus, choosing the single-capped 26S proteasome in our cryo-EM study may have also reduced the compositional heterogeneity40 and been beneficial for the high-resolution reconstruction.
Proteasome-ADP-AlFx represents a new conformational state
The AAA-ATPase is the most critical motor for the ATP-driven conformational change of this gigantic macromolecular machine. Our proteasome-ADP-AlFx map represents a new AAA-ATPase ring conformational configuration revealed by comparisons with previous proteasome cryo-EM structures in distinct chemical or functional states17,18,23, although the overall conformation of the proteasome-ADP-AlFx is similar to that of the substrate-engaged proteasome17 (Supplementary information, Figure S6 and Movie S1).
We first compared the AAA-ATPase ring between the proteasome-ADP-AlFx and the resting state maps, where the AAA-ATPase ring of the resting state is often used as a standard to measure the conformational switches in this region of other states17,27. Aligning the least varied 20S portions of the two maps together revealed that the AAA-ATPase alters its relative position to 20S and its internal topology dramatically (Supplementary information, Figure S7). The Rpt3-Rpt4-Rpt5 side of the AAA-ATPase ring in the proteasome-ADP-AlFx structure was found to be displaced away from 20S (by up to 14.5 Å in the vertical direction and tilted about 22°, Supplementary information, Figure S7A and S7B), leading to a 37% reduction in the interaction interface between 19S and 20S (Supplementary information, Figure S7A). Furthermore, we also observed a continuous widened channel for the substrate to the peptidase active sites in the ADP-AlFx state (Supplementary information, Figure S7C), which is consistent with the ATPγS and substrate-engaged state maps17,18,27, but different from the resting state map16,21. This widened channel combined with the Rpn11 position right above the N-ring pore (discussed below) and its proteolytic activity in vitro (Figure 1A and 1B, Supplementary information, Figure S1C) suggests that proteasome-ADP-AlFx is in an activated state, which may represent one of the transition states of proteasome along the enzymatic pathway for ATP hydrolysis and substrate unfolding and translocation.
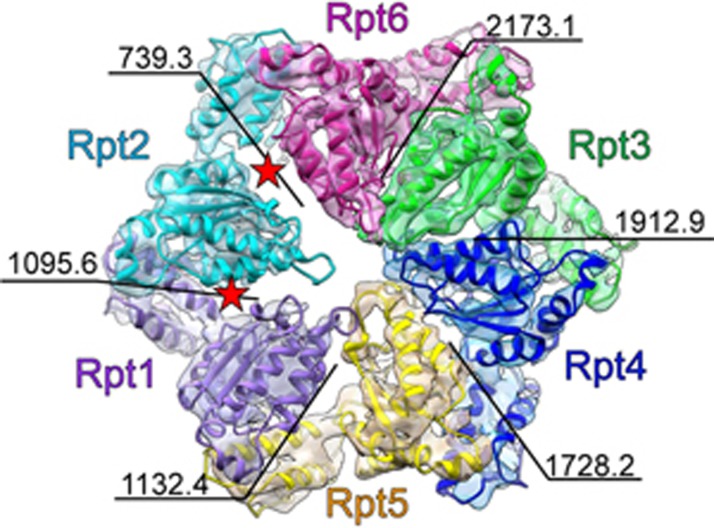
Surprisingly, our proteasome-ADP-AlFx map revealed two sizable gaps next to the Rpt2 subunit (Figure 2), which was not observed in the proteasome maps of the resting, ATPγS, and substrate-engaged states. These gaps lead to an ∼50% reduction in the interaction interface with its neighboring subunits Rpt1 and Rpt6 (Figure 2 and Supplementary information, Figure S8A). We then compared the AAA-ATPase structure of proteasome-ADP-AlFx to that of proteasome-ATPγS (S3 state, PDB ID: 4CR4)18, which revealed a pronounced 37.6° inter-domain movement of the Rpt2 subunit between its large and small AAA domains, further delineating the origin of the gaps (Supplementary information, Figure S8B-S8E and Movie S2). Basically, the large AAA domain of Rpt2 showed a large outward tilt away from the central pore, and an anticlockwise twist towards the neighboring Rpt6 subunit but away from the other neighboring subunit, Rpt1 (Supplementary information, Movie S2). Consequently, these domain motions led to the noticeable gap between Rpt2 and Rpt1. In this process, the twist of the large AAA domain of Rpt2 was accompanied by a clockwise rotation of the Rpt6 subunit away from Rpt2, generating another gap between Rpt2 and Rpt6. This coordinated motion resulted in a slight expansion of the AAA-ring accompanied by an ∼3° rotation of the entire base (Supplementary information, Figure S8B and Movie S2). Moreover, the N-ring was observed to be positioned a little closer to the AAA-ring, meanwhile, the large AAA domain of Rpt2 was elevated up slightly along with Rpn1, leading to a reduced distance between the N-ring and the AAA-ring (Supplementary information, Movie S2). In summary, proteasome-ADP-AlFx is in an activated state, which also depicts a new conformational configuration of proteasome, especially in the AAA-ATPase motor region.
Figure 2.
A new AAA-ATPase ring conformational configuration in proteasome-ADP-AlFx map. The AAA-ATPase ring structure of proteasome-ADP-AlFx elucidates two obvious gaps beside Rpt2 subunit (indicated by two red stars). The map is rendered as transparent surface and the model is in color. The interaction interface (in Å2) between the neighboring AAA domains is labeled. The color scheme for the map and model of individual AAA-ATPase subunit is followed throughout. The map here is low-pass filtered to 5.5-Å resolution (local resolution in this region) for better visualization.
Interestingly, our resting state map clearly revealed a solid piece of extra density, which belongs to the Rpt1 C-terminus and is near the 20S gate, indicating a close proximity between the Rpt1 C-terminus and the N-terminus of the 20S α6 subunit (Supplementary information, Figure S7E). This extra density was not seen in our proteasome-ADP-AlFx map, suggesting that it might be activated and more dynamic in the ADP-AlFx state. Still, the functionality of this Rpt1 C-terminal tail remains unclear.
A distinct pore loop configuration
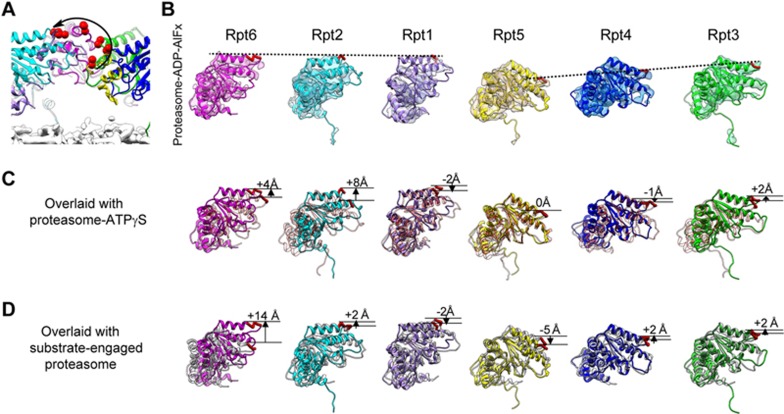
The central pore of AAA-ATPase is formed by the so-called “pore loops”, which line the channel within the RP protruding from each AAA-ATPase subunit to interact with substrate and move axially to direct vectorial motion of the substrate towards the CP, therefore playing an essential role in substrate unfolding and translocation42,43,44. Our proteasome-ADP-AlFx map reveals a new asymmetric vertical configuration of the pore loops, forming a right-handed “lockwasher” (Figure 3A). In this configuration, the pore loop of Rpt5 locates the lowest down forming the beginning of the lockwasher, and that of Rpt6 positions the highest up within this AAA-ATPase ring; the pore loops of the following Rpt2 and Rpt1 subunits locate slightly lower than that of Rpt6, with Rpt1 forming the end of the lockwasher (Figure 3A and 3B). This pore loop configuration is distinct from that of the previously reported ones16,17,27, further substantiating the notion that with ATP binding and hydrolysis, the AAA-ATPase ring of proteasome especially the pore loops varies its vertical configuration, which could play a key role in substrate unfolding and translocation42,43,44.
Figure 3.
A distinct configuration of pore loops in the AAA-ATPase ring of proteasome-ADP-AlFx. (A) Cutaway side view of the AAA-ATPase ring of proteasome-ADP-AlFx, with the upper location of each of the pore loops denoted by two red spheres. A noticeable “lockwasher-like” configuration of the pore loops can be observed (indicated by a black curve with an arrow head), with Rpt5 and Rpt1 forming the beginning and the end of the “lockwasher”, respectively. (B) The AAA domain of individual AAA-ATPase subunit shown in the same orientation. The pore loop is shown as thick red loop and the height of the pore loops is denoted by the dashed black line. Same low-pass filtered map as in Figure 2 is used here. (C) The overlaid AAA domains from proteasome-ADP-AlFx (colored) and proteasome-ATPγS (S3, PDB ID: 4CR4, transparent salmon)18, with the vertical differences between their positions in the two structures indicated. (D) The overlaid AAA domains from proteasome-ADP-AlFx (colored) and substrate-engaged proteasome (grey), with the vertical differences between their positions indicated. The model of the substrate-engaged proteasome structure was generated from PAN (PDB ID: 3H4M)80.
We further inspected the structures to decipher the motion of the pore loops. During the transformation from the ATPγS state (PDB ID: 4CR4)18 to our ADP-AlFx state, the pore loop of Rpt2 elevates upwards most pronouncedly (∼8 Å) in the vertical direction (Figure 3C); meanwhile, this upwards motion is associated with the largest rigid-body rotation (about 19.6°) of Rpt2 (Supplementary information, Figure S8D). Furthermore, as described above, Rpt2 exhibits a prominent inter-domain movement between its large and small AAA domains in this transformation (Supplementary information, Figure S8C, S8E and Movie S2), indicating that Rpt2 may play an essential role in the conformational transformation between these two states in yeast proteasome. Additionally, the pore loop of the neighboring Rpt6 subunit also exhibits a noticeable upwards elevation (∼4 Å, Figure 3C) and the entire Rpt6 subunit rotates about 9.4°, more pronouncedly than the remaining four consecutive subunits in the ring (Supplementary information, Figure S8D).
In addition, we compared the structures between our proteasome-ADP-AlFx and the substrate-engaged proteasome17. From the substrate-engaged state to the ADP-AlFx state, the pore loop of Rpt6 exhibits the largest vertical elevation of 14 Å (Figure 3D), which is accompanied by a 22.2° rigid-body rotation of Rpt6 (Supplementary information, Figure S9C); meanwhile, Rpt2 displays an obvious inter-domain rotation (∼35.7°), which is not observed in the other five AAA-ATPase subunits (Supplementary information, Figure S9B and S9D). Taken together, the pore loops of Rpt2 and Rpt6 exhibit rather substantial movements in the transformations from the ATPγS state or the substrate-engaged state to the ADP-AlFx state; in the meantime, Rpt2 also shows a pronounced inter-domain movement. Therefore, the neighboring subunits Rpt2 and Rpt6 display pronounced conformational variations responding to ATP hydrolysis in the AAA-ATPase ring, and may play an indispensable role in substrate processing of yeast proteasome.
ATP hydrolysis in proteasome could be a dynamic process and there may exist multiple potential intermediate states, especially in the AAA-ATPase ring and the pore loop regions. However, only one conformational configuration was resolved from our ADP-AlFx dataset. This configuration may be the most dominant population in this nucleotide state, representing either the lowest energy configuration or the rate-limiting state in the conformational landscape. Similarly, in previous work on substrate-engaged proteasome, only one conformation was resolved, which may be a snapshot of a dwell state in the substrate processing procedure17. Overall, the state that we captured here may stand for a rate-limiting snapshot in the dynamic ATP hydrolysis process.
Asymmetric nucleotide occupancy in proteasome-ADP-AlFx
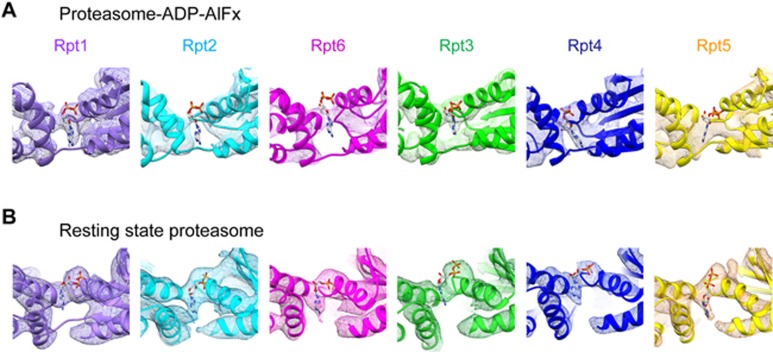
We further inspected the nucleotide density in each of the AAA-ATPase subunits in our proteasome-ADP-AlFx map and found relative strong density in the nucleotide-binding pockets of four consecutive AAA-ATPase subunits (Rpt3-Rpt4-Rpt5-Rpt1), no obvious extra density in Rpt2 pocket, and rather weak density for the potential nucleotide in Rpt6 pocket (Figure 4A). Since cryo-EM single-particle analysis is an averaging technique, such weak nucleotide density implies a rather low nucleotide occupancy in Rpt6. Collectively, our ADP-AlFx state map demonstrates an asymmetric nucleotide occupancy pattern in the AAA-ATPase ring. Still, the resolution in this region did not allow us to accurately distinguish ADP-AlFx from ADP in the occupied pockets. Previous analyses on analogous AAA-ATPase systems (including ClpX45, HslU46, and PAN47) have suggested that a maximum of four subunits can bind nucleotide simultaneously in the functional states, which is mostly consistent with our observation. Moreover, this asymmetric nucleotide occupancy pattern in consecutive subunits is to some extent in line with the recent biochemical analysis of the PAN system48, but different from the ClpX system in which the two unloaded subunits locate on the opposite positions of the ring49,50.
Figure 4.
Distinct nucleotide occupancy statuses between proteasome-ADP-AlFx and resting state proteasome. (A) Nucleotide occupancy status in the nucleotide-binding pocket of individual AAA-ATPase subunit in the proteasome-ADP-AlFx map. We used ADP to locate the approximate position of potential nucleotide. The map reveals relative strong density in the nucleotide-binding pockets of four consecutive subunits (Rpt3-Rpt4-Rpt5-Rpt1), no obvious extra density in the Rpt2 pocket, and rather weak density for potential nucleotide in the Rpt6 pocket. Same low-pass filtered map as in Figure 2 is used here. (B) Our yeast resting state structure reveals that the nucleotide-binding pockets of all the six AAA-ATPase subunits are occupied. In both maps, due to resolution limitation in 19S RP region, we are not able to distinguish different nucleotide forms (ADP-AlFx, ADP, or ATP) in the individual pocket.
Interestingly, in our yeast proteasome resting state map all the six nucleotide-binding pockets are occupied (Figure 4B), which was not reported in previous study of yeast proteasome in the same state, but is in line with recent cryo-EM studies of human proteasome in the resting state30,31,32. Although similar excessive amount of nucleotide was present in both the resting state and the activated ADP-AlFx state, the nucleotide occupancy pattern is distinct, indicating that the proteasome nucleotide occupancy status could be related to the nucleotide state and the activation status of the complex. Still, we should mention that in the cryo-EM maps of human proteasome in the resting state30,31, the nucleotide binding forms were reported to be distinct among the six AAA-ATPase subunits.
The 20S gate remains closed in the proteasome-ADP-AlFx map
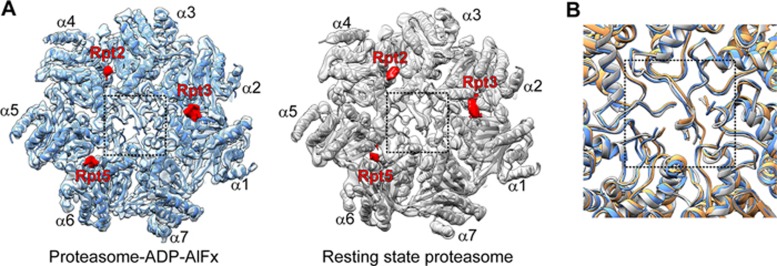
The established notion for the 20S gate opening is that, upon ATP binding, the C-terminal HbYX (hydrophobic-tyrosine-any residue) motifs of subunits Rpt2, Rpt3, and Rpt5 would insert into the defined pockets on the surface of 20S to trigger gate opening51,52. Both of our yeast proteasome-ADP-AlFx and resting state maps clearly depict pieces of density belonging to the C-termini of Rpt2, Rpt3, and Rpt5 in the corresponding 20S pockets (Figure 5A), which is consistent with the functional or biochemical experiments51,52. However, the gate remains in the closed conformation in both our maps (Figure 5A). Moreover, a good match was observed between our two corresponding pseudo-atomic models and the crystal structure of eukaryotic 20S with a closed gate (PDB ID: 1RYP)53 (Figure 5B). This is in agreement with cryo-EM studies of the 26S proteasome in the presence of ATP21,30,31; and by comparing with the proteasome-ATPγS map18, we also noticed a similar feature. It is possible that the gate is conformationally heterogeneous, which may be in the closed conformation at most of the time and only transiently show the open conformation. This is supported by a recent study on human proteasome, showing that the dominant population of the particles (∼92%) is in the closed gate configuration32.
Figure 5.
Insertion of the AAA-ATPase subunit C-terminal HbYX motif into 20S is not sufficient to trigger 20S gate opening. (A) End-on view of 20S with the inserted C-terminal tails of Rpt2, Rpt3, and Rpt5 (red pieces of density) in the proteasome-ADP-AlFx (dodger blue) and resting state (grey) maps. The gate region is indicated by a dash line square. (B) Superimposed models showing only the gate region, with that of resting state in grey, that of proteasome-ADP-AlFx state in dodger blue, and that of the gate closed eukaryotic 20S crystal structure (PDB ID: 1RYP)53 in sandy brown.
Our data suggest that in yeast proteasome insertion of the C-terminal HbYX motif of Rpt2, Rpt3, and Rpt5 is not sufficient to maintain an open 20S gate during ATP-dependent substrate degradation. Other factors such as a contact between the unstructured tail of the substrate and the gate region may serve as an additional signal to open the 20S gate54,55,56, or the C-terminal tail of other AAA-ATPase subunits (in addition to Rpt2, Rpt3, and Rpt5) may also play a role in this process32,57. The previous atomic force microscopy analysis revealed that substrate, including short peptide, could induce a shift toward the open gate conformation for the 20S proteasome58. In summary, our study indicates that eukaryotic 20S gate opening by 19S is more selective and intricate than previously thought.
Discussion
Here we present the first near-atomic-resolution cryo-EM structure of the yeast 26S proteasome in the activated ADP-AlFx state, which reveals a new conformational state of proteasome with a distinct AAA-ATPase ring configuration. Our biochemical and structural analyses further demonstrate that proteasome-ADP-AlFx is functionally active and may represent one of the transition states along the enzymatic pathway. Additionally, the proteasome-ADP-AlFx map also depicts an asymmetric nucleotide occupancy pattern in the AAA-ATPase ring, which is observed for the first time for yeast proteasome in the activated state.
Our proteasome-ADP-AlFx map reveals that the pore loop heights are related to the nucleotide binding status. For instance, for Rpt2 and Rpt6 (with empty or only weakly occupied nucleotide pocket), their pore loops stay in the relative higher positions compared to those of the other four consecutive subunits with bound nucleotide (Figures 3A, 3B and 4A). The correlative relationship between the heights of the substrate-binding pore loops and the nucleotide binding states was also observed in the comparable Rho helicase system59.
Additionally, our data reveal a pronounced inter-domain movement of Rpt2 and an associated noticeable motion of Rpt6 during the transition from the ATPγS state to the ADP-AlFx state, which may represent a collective result of allosteric cooperativity of all the AAA-ATPase subunits responding to ATP hydrolysis. Rpt2 with mutation abrogating its hydrolysis ability has been indicated to function virtually identically to wild-type proteasome in ATP hydrolysis and substrate degradation ability in reconstituted system57; however, introducing mutation in the pore loop of Rpt2 leads to severe defects in substrate degradation57, indicating the important role of Rpt2 pore loop in substrate processing. Interestingly, the aforementioned pronounced inter-domain movement of Rpt2 brings its pore loop to the top ranking position of the lockwasher, which is accompanied by a pore loop elevation of the neighboring Rpt6. This Rpt2 motion-induced pore loop repositioning may delineate the important role of Rpt2 in substrate processing. Moreover, other biochemical analyses have shown that mutation of the invariant lysine residue K229 of Rpt2, in the nucleotide-binding pockets and essential for ATP hydrolysis, is lethal in the endogenous yeast system, which is not the case for other Rpt proteins60. Further studies substantiate the notion that Rpt2 plays a major role in regulating the peptidase activity of the CP60,61. Taken together, our data imply that the discharged energy from ATP hydrolysis can be converted through allosteric cooperativity into mechanical forces associated with the dramatic conformational changes of Rpt2 and noticeable motion of Rpt6 and the consequent pore loop elevation, which may facilitate the substrate processing of proteasome.
Noteworthy, although incubated with over excess ADP-AlFx, the nucleotide occupancy pattern in the six AAA-ATPase subunits is asymmetric, suggesting that, in the activated proteasome-ADP-AlFx, it is unlikely that proteasome consumes nucleotide in a concerted manner. Interestingly, previous studies have shown that the pore loop of Rpt1 stays in the highest position in the AAA-ATPase ring of proteasome-ATPγS18, and Rpt1 could probably trigger the ATP hydrolysis progressively around the ring57. Furthermore, in our proteasome-ADP-AlFx map, the particular inter-domain motion of Rpt2 may accommodate its Arg-finger projecting towards the ATP in the nucleotide-binding pocket of the adjoining Rpt1, which may facilitate ATP hydrolysis27,48. Putting together, it is possible that Rpt1 initiates ATP hydrolysis under the aid of Rpt2. The following three consecutive AAA-ATPase subunits (Rpt5-Rpt4-Rpt3) all have their nucleotide-binding pockets occupied (Figure 4A) and prepared for the following ATP hydrolysis, which therefore may operate in a counterclockwise sequential manner. The ATP hydrolysis would rapidly induce dramatic conformational changes around the ring, facilitating the unfolding and translocation of substrate through the ATPase channel.
Interestingly, the AAA-ATPase subunit Rpt2 is adjacent to the dynamic Rpn1 subunit, a platform to recruit ubiquitinated substrate and deubiquitinating enzyme Ubp662. The close proximity between Rpt2 and Rpn1 could link the ATPase cycle to the positioning of Rpn1, which may contribute to substrate positioning and translocation.
Previous data have shown that in the transition from the resting state to the ATPγS state, the horseshoe-like configuration of PCI domains dramatically rotates along the base to reposition Rpn11 to the top of the AAA-ATPase pore, which is beneficial for efficient substrate deubiquitination18,27. Here our data reveal that, in the following transition from the ATPγS state to the ADP-AlFx state, the relative position of Rpn11 remains on the top of the AAA-ATPase pore trapped by the coiled coils of Rpt3/6 and Rpt4/5 (Supplementary information, Figure S10), but with a rotation of 13.4° (due to the rotation of the lid, discussed below). The relative position of Rpn11 to the pore further corroborates the notion that proteasome-ADP-AlFx is in an activated state. Collectively, the factors discussed above may allosterically regulate and maintain the activity of Rpn11.
In our proteasome-ADP-AlFx structure, the ubiquitin receptor Rpn10 contacts the coiled coil of the AAA-ATPase dimer Rpt4/5, similar to that observed in the proteasome-ATPγS structure18,27 (Supplementary information, Figure S10A and S10B). However, Rpn10 rotates about 18° clockwise along with the coiled coil of Rpt4/5 due to the rotation of the entire AAA-ATPase ring triggered by ATP hydrolysis (Supplementary information, Figures S8B and S10C). The conformational change in Rpt2 and Rpt6 and the subsequent motion in the entire base could be propagated to the lid through the rotation of the coiled coils of Rpt3/6 and Rpt4/5 (Supplementary information, Movie S2). These results demonstrate that, in the nucleotide cycle, conformational changes in the ATPase ring could initiate the allosteric coordination of the lid and subsequently the entire complex so as to exhibit its substrate processing function.
Our high-resolution cryo-EM structure of yeast proteasome in one of its possible activated transition states demonstrates that the ATP hydrolysis-driven collective motion of Rpt2 and Rpt6 could lead to an elevation of their pore loops, which may play a special role in substrate processing. Our results provide new insights into the structural mechanism of proteasome allosteric coordination and take a step forward towards elucidating the structural mechanism of substrate degradation by the proteasome.
Materials and Methods
Yeast strain and plasmid construction
Single-capped 26S proteasomes were purified from S. cerevisiae by using Rpn11-3×FLAG63,64. To integrate the 3×FLAG tag into the C-terminus of Rpn11, the respective plasmid was created as follows: first, the sequence encoding the 3×FLAG epitope, the stop codon of the Rpn11 gene, and the URA3 gene were amplified and fused by sequential PCR (cassette I). Next, the 3′-segment (478 bp) of the ORF of the Rpn11 gene (without the stop codon), cassette I and 500 bp downstream of Rpn11 were cloned into a pUC19 vector by using the ClonExpress MultiS One Step Cloning Kit (Vazyme). The 3′-segment of the Rpn11-cassette I-downstream was amplified by PCR from the resultant plasmid and introduced to the BY4741 background yeast strain (MATa, his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) by a chromosomal homologous recombination.
Purification of the single-capped 26S proteasome
To purify the endogenous single-capped 26S proteasome, yeast cells cultured in YPD medium at 30 °C were harvested, washed twice with water and stored at −80 °C. The frozen yeast cells were lysed with an ultra-high pressure cell disrupter in lysis buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 4 mM ATP, 10% glycerol, 0.2% NP-40). The lysate was centrifuged at 20 000× g for 30 min at 4 °C. The clarified lysate was filtered through cheesecloth and then incubated with anti-FLAG M2 agarose beads (Sigma) for 2 h at 4 °C. The beads were recovered and washed twice with wash buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 2 mM ATP, 10% glycerol, 0.1% NP-40) before eluting with 500 μg/ml 3×FLAG peptide (Shanghai Bootech BioScience & Technology). For separation of the single-capped 26S proteasome, the eluted samples were further subjected to a 10%-40% glycerol density gradient centrifugation, fractionated, and analyzed by activity assay, SDS-PAGE and negative stain EM.
Nucleotide exchange and native gel electrophoresis
In the proteasome purification process, oversaturated amount of ATP is used, which needs to be consumed before exchanging to other ATP analogues. Apyrase was used to deplete all these ATP33. To ensure the activity of apyrase, the buffer for the purification of 26S proteasome was exchanged to the aforementioned wash buffer (except for the exchange of NP-40 to DTT) with reduced glycerol to 5%. The proteasome was treated with apyrase for 1 min to deplete ATP, and incubated with 1 mM ATP analogues (including ADP-AlFx, ATPγS, and ADP) at 30 °C for 1 h following the protocol reported previously33. The samples were subsequently analyzed by native PAGE. The PAGE was prepared with 4% acrylamide gels containing 1 mM ATP and 1 mM DTT, as described previously65. About 1 μg of proteasome was loaded on the gel with the native loading buffer. The gel electrophoresis was then carried out at 100 V at 4 °C for 3 h. Afterwards, the gel was either stained for protein with coomassie blue or overlaid with Suc-Leu-Leu-Val-Tyr-AMC (100 μM) for assessment of proteasome activity.
Proteasome activity measurement
Overall, after cleaving the fluorogenic peptide Suc-Leu-Leu-Val-Tyr-AMC by the proteasome, the proteasome activity can be determined by measuring the resulting fluorescence due to free AMC66. In more details, after the glycerol density gradient centrifugation, we carried out the activity assay in a 96-well plate to quickly define the fractions containing proteasome. In addition, we can also perform native PAGE to accurately distinguish each forms of proteasome including double-capped proteasome, single-capped proteasome, and 20S. Afterwards, to detect the fluorescence, the samples either in the 96-well plate or from the native PAGE were incubated in buffer containing 100 μM fluorogenic peptide for 10 min at 30 °C. The reaction was stopped by addition of 1% SDS65. The fluorescence of the product was visualized by using a gel image system (Tanon-1600, Shanghai, China). For the continuous measurement of the proteasome proteolytic activity, the sample was buffer exchanged and incubated at 30 °C for 1 h, and the fluorescence was detected at an excitation of 380 nm and an emission of 460 nm every 30 s for a total of 10 min using a multimode microplate reader (BioTek).
Cryo-EM sample preparation and data collection
Except for the nucleotide exchange step, the following sample preparation procedure was identical for the resting state and proteasome-ADP-AlFx samples. 200 mesh holey carbon Quantifoil R2/1 grids were plasma cleaned using a Solarus plasma cleaner (Gatan), followed by grid pretreatment with polylysine16 to obtain more random orientations of proteasome particles. The glycerol concentration in the purified sample was reduced to 2% in the same wash buffer (except for the exchange of NP-40 to DTT) before cryo-sample preparation. A volume of 2 μl of the sample was placed onto a grid, followed by blotting for 1 s with −1 offset in 100% humidity utilizing Vitrobot Mark IV (FEI company), and then flash frozen in liquid ethane.
Images were taken by using a Titan Krios transmission electron microscope (FEI) operated at 300 kV and equipped with a Cs corrector. We collected the data cross several months in six sessions with the Cs values ranging from 0.001 to 0.006 mm. Images for proteasome-ADP-AlFx were collected by using a K2 Summit direct electron detector (Gatan) in counting mode with a pixel size of 1.32 Å. Each movie was dose-fractioned into 38 frames. The exposure time was 7.6 s with 0.2 s for each frame, generating a total dose of ∼38 e−/Å2. The resting state proteasome data were also recorded on the Titan Krios but with a Falcon II detector. The pixel size was 1.09 Å, and each movie was dose-fractioned into 18 frames with a total dose of ∼25 e−/Å2. Defocus values for both datasets varied from −1.8 to −2.5 μm. All the images were collected by utilizing the SerialEM automated data collection software package67.
Cryo-EM data processing
A total of 13 871 and 3 499 micrographs were used for the proteasome-ADP-AlFx and resting state proteasome structure determinations, respectively. All images were aligned and summed using MotionCorr whole-image motion correction software68. Unless otherwise specified, single-particle analysis was mainly executed in RELION 1.369. After CTF parameter determination by CTFFIND370, particle auto-picking, manual particle checking, and reference-free 2D classification, 1 107 189 and 89 000 particles remained for the proteasome-ADP-AlFx and resting state datasets, respectively. For both datasets, the initial model was a single-capped 26S proteasome, derived from the previous double-capped 26S resting state cryo-EM map (EMDB ID: 1992)16 but was segmented out as a single-capped 26S proteasome and low-pass filtered to 60 Å using proc3d program in EMAN 1.971.
For proteasome-ADP-AlFx reconstruction, one round of 3D classification was carried out to extract 331 012 good single-capped proteasome particles. Then an auto-refine procedure was performed in RELION to generate a 4.6-Å-resolution map. Afterwards, the 3-34 frames were used for the first round of per-particle motion correction (also known as particle polishing72) without B factor weighting. These polished particles were further three-dimensionally classified into four classes, among which one class with better features (containing 178 576 particles) was subjected to the auto-refine procedure, resulting in a 4.4-Å-resolution map. To further improve the map quality, we chose 3-14 frames (with a lower dose of 14 e−/Å2) to perform a second round of per-particle motion correction without B factor weighting. These newly polished particles were subjected to one more round of auto-refinement, which gave rise to a map with a resolution of 4.2 Å. This map was corrected for modulation transfer function of the detector and was sharpened by applying a negative B-factor (−127 Å2), estimated by an automated procedure in RELION. The resolution was accessed based on the gold-standard criterion with FSC at 0.143, and the local resolution was estimated by ResMap73.
For the resting state dataset with 89 000 particles, we performed an auto-refine procedure to obtain a 7.4-Å-resolution map. We then used 3-18 frames for the first round of per-particle motion correction72 without B factor weighting. These polished particles were then 3D classified into four classes. Among them, one class with 26 000 particles showed better structural details and showed a conformation similar to those of the previous resting state structures16,18,20,21,22,23, and was hence referred to as the resting state. Considering the extremely dynamic nature of Rpn1 and Rpn13 and the weak density in each of these two regions, we generated a mask without Rpn1 and Rpn13 and further auto-refined the data with this mask, which gave rise to a 6.3-Å-resolution map after post-processing. For the same dataset, we carried out another auto-refinement procedure without any masking. This refinement led to a 6.9-Å-resolution map, in which the density levels for Rpn1 and Rpn13 were comparable to that in the reconstruction with the mask. We then chose the masked map for further refinement. Afterwards, 3-11 frames were used for a second round of per-particle motion correction without B factor weighting, with a dose of 15 e−/Å2. After one round of auto-refinement, the final resolution of the 3D map was at 6.3 Å but with improved features, and was used for model building. This map was corrected for modulation transfer function of the detector and was sharpened by applying a negative B-factor of −168 Å2, automatically determined by RELION.
We should mention that based on the ATP presented dataset, we also reconstructed an 8.9-Å-resolution cryo-EM map from a class of 11 588 particles, which resembled the proteasome-ATPγS structure18,23,27. This confirmed the notion that in the presence of ATP, there exists a mixture of conformations, i.e., the resting state and activated state23. Since this map did not reveal new structural features, we did not include it in the analysis of this work.
Pseudo-atomic model building and validation
For 19S, the initial model for our resting state structure was based on the previous model in the same state (4CR2)18, and the initial model for our proteasome-ADP-AlFx structure was from that of the previous proteasome-ATPγS (4CR4)18 structure; and the model of 20S was based on the crystal structure of the S. cerevisiae 20S (1RYP)53. We carried out further flexible fitting against our cryo-EM maps of the resting state and proteasome-ADP-AlFx to build a pseudo-atomic model for each of these two maps. First, some of the missing structural information in the original models but well resolved in our maps was added to the model using COOT74, including a loop of 11 amino acids at the Rpt1 C-terminus in the resting state, and a helix of the Rpn5 N-terminus in both maps. For each of the two states, we fitted the model into the corresponding map as a rigid body utilizing Chimera75. COOT74 was also used for manual local refinement of the model. Following that, we carried out flexible fitting of the model, without Rpn1 and Rpn13 due to the weak density in these regions in both maps probably caused by their distal location and intrinsic plasticity76, against the corresponding map by using the real-space refinement software package Rosetta77. Still, for a more complete visualization of the structure, we used the previous models for these two subunits18 and manually placed them into the density.
Model validation was performed following the protocol of phenix.molprobity78,79. UCSF Chimera was used for map segmentation and figure generation75.
Accession numbers
EM maps have been deposited in the Electron Microscopy Data Bank under accession codes of EMD-6694 (proteasome-ADP-AlFx) and EMD-6693 (the resting state proteasome). Pseudo-atomic models for proteasome-ADP-AlFx and the resting state proteasome have been deposited in the Protein Data Bank under accession numbers of 5WVK and 5WVI, respectively.
Author Contributions
ZD and YC conceived and designed the experiments. ZD, ZF and YW designed constructs for structural studies. ZD, ZF, YW, JC and YFW performed protein purification. ZD, ZF and CX performed functional analysis. ZD, ZF and LK performed cryo-EM data collection and analysis. NL and RZ performed SAXS data collection and analysis. ZD, CX and JL performed structure reconstruction. ZD and YC analyzed the structure and wrote the paper.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Drs Jinqiu Zhou and Zhaocai Zhou (Institute of Biochemistry and Cell Biology, Shanghai) for their generous support. We thank Yingyi Zhang and Mi Cao from the Electron Microscopy facility and Min Xue from Data Base & Computation system at National Centre for Protein Science Shanghai (NCPSS) for their assistance with the EM instrument management and data storage and parallel computing. We are grateful to the NCPSS Protein Expression and Purification facility, Mass Spectrometry facility, and BL19U2 beamline at NCPSS and Shanghai Synchrotron Radiation Facility (SSRF) for instrument support and technical assistance. This work was supported by grants from the CAS Pilot Strategic Science and Technology Projects B (XDB08030201), the National Natural Science Foundation of China (31270771, 31670754), the National Basic Research Program of China (2013CB910401), the STS program of the CAS (KFJ-EW-STS-098), and the CAS-Shanghai Science Research Center (CAS-SSRC-YH-2015-01).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Biochemical analyses of the yeast proteasome.
Cryo-EM analysis of the proteasome.
Work flow for the processing of the high-resolution proteasome-ADP-AlFx cryo-EM data.
Pseudo atomic models of proteasome 19S RP subunits fitted into the cryo-EM map of proteasome-ADP-AlFx.
Cryo-EM maps of yeast proteasome in the presence of ATP and conformation comparison with related previous cryo-EM maps.
Conformation comparison of our proteasome-ADP-AlFx map with the previously reported cryo-EM proteasome maps.
The AAA-ATPase ring conformational change between proteasome-ADP-AlFx and the resting state structures.
AAA-ATPase subunit interaction interface analysis and conformational changes of the AAA-ATPase ring from proteasome-ATPγS to proteasome-ADP-AlFx.
Conformational changes of the AAA-ATPase subunits from the substrate-engaged state to the ADP-AlFx state.
Conformational changes of Rpn10 and Rpn11 between proteasome-ATPγS and proteasome-ADP-AlFx.
Statistics of cryo-EM data collection and refinement.
Comparison of our proteasome-ADP-AlFx map with the previous proteasome structures confirms that our proteasome-ADP-AlFx adopted a new conformation.
Base region conformational changes between proteasome-ATPγS and proteasome-ADP-AlFx, revealing the most prominent difference to be for Rpt2 in the AAA-ATPase ring and the origin of the gaps next to Rpt2.
References
- Schwartz AL, Ciechanover A. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev Pharmacol Toxicol 2009; 49:73–96. [DOI] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 2009; 78:477–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL. Nobel committee tags ubiquitin for distinction. Neuron 2005; 45:339–344. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 2002; 82:373–428. [DOI] [PubMed] [Google Scholar]
- Leroy E, Boyer R, Auburger G, et al. The ubiquitin pathway in Parkinson's disease. Nature 1998; 395:451–452. [DOI] [PubMed] [Google Scholar]
- Kish-Trier E, Hill CP. Structural Biology of the Proteasome. Annu Rev Biophys 2013; 42:29–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 1999; 68:1015–1068. [DOI] [PubMed] [Google Scholar]
- Saeki Y, Tanaka K. Assembly and function of the proteasome. Methods Mol Biol 2012; 832:315–337. [DOI] [PubMed] [Google Scholar]
- Peters JM, Cejka Z, Harris JR, Kleinschmidt JA, Baumeister W. Structural features of the 26 S proteasome complex. J Mol Biol 1993; 234:932–937. [DOI] [PubMed] [Google Scholar]
- Demartino GN, Gillette TG. Proteasomes: machines for all reasons. Cell 2007; 129:659–662. [DOI] [PubMed] [Google Scholar]
- Smith DM, Benaroudj N, Goldberg A. Proteasomes and their associated ATPases: a destructive combination. J Struct Biol 2006; 156:72–83. [DOI] [PubMed] [Google Scholar]
- Groll M, Bochtler M, Brandstetter H, Clausen T, Huber R. Molecular machines for protein degradation. Chembiochem 2005; 6:222–256. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 1998; 94:615–623. [DOI] [PubMed] [Google Scholar]
- Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem 2011; 80:587–612. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Hartmann MD, Habeck M, et al. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell 2009; 34:580–590. [DOI] [PubMed] [Google Scholar]
- Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature 2012; 482:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyskiela ME, Lander GC, Martin A. Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol 2013; 20:781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unverdorben P, Beck F, Sledz P, et al. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc Natl Acad Sci USA 2014; 111:5544–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci 1998; 23:204–205. [DOI] [PubMed] [Google Scholar]
- Lasker K, Forster F, Bohn S, et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci USA 2012; 109:1380–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck F, Unverdorben P, Bohn S, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci USA 2012; 109:14870–14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca PC, He J, Morris EP. Molecular model of the human 26S proteasome. Mol Cell 2012; 46:54–66. [DOI] [PubMed] [Google Scholar]
- Luan B, Huang X, Wu J, et al. Structure of an endogenous yeast 26S proteasome reveals two major conformational states. Proc Natl Acad Sci USA 2016; 113:2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrin E, Lopez-Blanco JR, Chacon P, Martin A. Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid. Structure 2013; 21:1624–1635. [DOI] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 2002; 298:611–615. [DOI] [PubMed] [Google Scholar]
- vanNocker S, Sadis S, Rubin DM, et al. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol 1996; 16:6020–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz P, Unverdorben P, Beck F, et al. Structure of the 26S proteasome with ATP-γS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc Natl Acad Sci USA 2013; 110:7264–7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Yu H, Mim C, Matouschek A. Regulated protein turnover: snapshots of the proteasome in action. Nat Rev Mol Cell Biol 2014; 15:122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambacher CM, Worden EJ, Herzik MA, Martin A, Lander GC. Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. eLife 2016; 5:e13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer A, Aufderheide A, Rudack T, et al. Structure of the human 26S proteasome at a resolution of 3.9 Å. Proc Natl Acad Sci USA 2016; 113:7816–7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Luan B, Wu J, Shi Y. An atomic structure of the human 26S proteasome. Nat Struct Mol Biol 2016; 23:778–785. [DOI] [PubMed] [Google Scholar]
- Chen S, Wu J, Lu Y, et al. Structural basis for dynamic regulation of the human 26S proteasome. Proc Natl Acad Sci USA 2016; 113:12991–12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CW, Li X, Thompson D, et al. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol Cell 2006; 24:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabre M. Aluminofluoride and beryllofluoride complexes: a new phosphate analogs in enzymology. Trends Biochem Sci 1990; 15:6–10. [DOI] [PubMed] [Google Scholar]
- Meyer AS, Gillespie JR, Walther D, Millet IS, Doniach S, Frydman J. Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell 2003; 113:369–381. [DOI] [PubMed] [Google Scholar]
- Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell 2012; 151:267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WH, de Poot SA, Lee JH, et al. Open-gate mutants of the mammalian proteasome show enhanced ubiquitin-conjugate degradation. Nat Commun 2016; 7:10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell 2011; 144:526–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Martin A, Nogales E. The proteasome under the microscope: the regulatory particle in focus. Curr Opin Struct Biol 2013; 23:243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster F, Unverdorben P, Sledz P, Baumeister W. Unveiling the long-held secrets of the 26S proteasome. Structure 2013; 21:1551–1562. [DOI] [PubMed] [Google Scholar]
- Asano S, Fukuda Y, Beck F, et al. Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science 2015; 347:439–442. [DOI] [PubMed] [Google Scholar]
- Wang J, Song JJ, Franklin MC, et al. Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure 2001; 9:177–184. [DOI] [PubMed] [Google Scholar]
- Yamada-Inagawa T, Okuno T, Karata K, Yamanaka K, Ogura T. Conserved pore residues in the AAA protease FtsH are important for proteolysis and its coupling to ATP hydrolysis. J Biol Chem 2003; 278:50182–50187. [DOI] [PubMed] [Google Scholar]
- Finley D, Chen X, Walters KJ. Gates, channels, and switches: elements of the proteasome machine. Trends Biochem Sci 2016; 41:77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch GL, Burton RE, Bolon DN, Baker TA, Sauer RT. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell 2005; 121:1017–1027. [DOI] [PubMed] [Google Scholar]
- Yakamavich JA, Baker TA, Sauer RT. Asymmetric nucleotide transactions of the HslUV protease. J Mol Biol 2008; 380:946–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz AA, Navon A, Groll M, Smith DM, Reis C, Goldberg AL. ATP-induced structural transitions in PAN, the proteasome-regulatory ATPase complex in Archaea. J Biol Chem 2007; 282:22921–22929. [DOI] [PubMed] [Google Scholar]
- Kim YC, Snoberger A, Schupp J, Smith DM. ATP binding to neighbouring subunits and intersubunit allosteric coupling underlie proteasomal ATPase function. Nat Commun 2015; 6:8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson BM, Nager AR, Glynn SE, Schmitz KR, Baker TA, Sauer RT. Nucleotide binding and conformational switching in the hexameric ring of a AAA+ machine. Cell 2013; 153:628–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell 2009; 139:744–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's α ring opens the gate for substrate entry. Mol Cell 2007; 27:731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell 2008; 30:360–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, et al. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 1997; 386:463–471. [DOI] [PubMed] [Google Scholar]
- Forster F, Lasker K, Beck F, Nickell S, Sali A, Baumeister W. An atomic model AAA-ATPase/20S core particle sub-complex of the 26S proteasome. Biochem Biophys Res Commun 2009; 388:228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bech-Otschir D, Helfrich A, Enenkel C, et al. Polyubiquitin substrates allosterically activate their own degradation by the 26S proteasome. Nat Struct Mol Biol 2009; 16:219–225. [DOI] [PubMed] [Google Scholar]
- Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol 2004; 11:830–837. [DOI] [PubMed] [Google Scholar]
- Beckwith R, Estrin E, Worden EJ, Martin A. Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nat Struct Mol Biol 2013; 20:1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmulski PA, Hochstrasser M, Gaczynska M. A tetrahedral transition state at the active sites of the 20S proteasome is coupled to opening of the α-ring channel. Structure 2009; 17:1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell 2009; 139:523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J 1998; 17:4909–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Cascio P, Leggett DS, Woo KM, Goldberg AL, Finley D. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol Cell 2001; 7:1143–1152. [DOI] [PubMed] [Google Scholar]
- Shi Y, Chen X, Elsasser S, et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 2016 Feb 19. doi: 10.1126/science.aad9421 [DOI] [PMC free article] [PubMed]
- Sone T, Saeki Y, Toh-e A, Yokosawa H. Sem1p is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. J Biol Chem 2004; 279:28807–28816. [DOI] [PubMed] [Google Scholar]
- Verma R, Chen S, Feldman R, et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell 2000; 11:3425–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett DS, Glickman MH, Finley D. Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol Biol 2005; 301:57–70. [DOI] [PubMed] [Google Scholar]
- Hough R, Pratt G, Rechsteiner M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J Biol Chem 1987; 262:8303–8313. [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 2005; 152:36–51. [DOI] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 2013; 10:584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 2003; 142:334–347. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 1999; 128:82–97. [DOI] [PubMed] [Google Scholar]
- Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife 2014; 3:e03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods 2014; 11:63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera — a visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Forster F, Villa E. Integration of cryo-EM with atomic and protein-protein interaction data. Methods Enzymol 2010; 483:47–72. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Song Y, Li X, et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods 2015; 12:361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB. Arendall WB 3rd, Headd JJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Hu M, Tian G, et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell 2009; 34:473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Biochemical analyses of the yeast proteasome.
Cryo-EM analysis of the proteasome.
Work flow for the processing of the high-resolution proteasome-ADP-AlFx cryo-EM data.
Pseudo atomic models of proteasome 19S RP subunits fitted into the cryo-EM map of proteasome-ADP-AlFx.
Cryo-EM maps of yeast proteasome in the presence of ATP and conformation comparison with related previous cryo-EM maps.
Conformation comparison of our proteasome-ADP-AlFx map with the previously reported cryo-EM proteasome maps.
The AAA-ATPase ring conformational change between proteasome-ADP-AlFx and the resting state structures.
AAA-ATPase subunit interaction interface analysis and conformational changes of the AAA-ATPase ring from proteasome-ATPγS to proteasome-ADP-AlFx.
Conformational changes of the AAA-ATPase subunits from the substrate-engaged state to the ADP-AlFx state.
Conformational changes of Rpn10 and Rpn11 between proteasome-ATPγS and proteasome-ADP-AlFx.
Statistics of cryo-EM data collection and refinement.
Comparison of our proteasome-ADP-AlFx map with the previous proteasome structures confirms that our proteasome-ADP-AlFx adopted a new conformation.
Base region conformational changes between proteasome-ATPγS and proteasome-ADP-AlFx, revealing the most prominent difference to be for Rpt2 in the AAA-ATPase ring and the origin of the gaps next to Rpt2.