Abstract
Mitophagy, the selective autophagic elimination of mitochondria, is a conserved cellular process critical for maintaining normal cellular physiology, and defects in this process are associated with certain pathophysiologies. In a recently published paper, Wei et al. describe their discovery of a hitherto unexplored mechanism of marking mitochondria for degradation.
Damaged and superfluous mitochondria are specifically removed from the cell by a process of selective autophagy called mitophagy1. Mitophagy plays critical roles in maintaining cellular homeostasis and in embryonic development by regulating the number and function of mitochondria. In their recently published work, Wei et al.2 demonstrate that the inner mitochondrial membrane (IMM) protein PHB2 (prohibitin 2) acts as a receptor for the mitophagic machinery.
Mitochondria are autonomous double-membrane organelles, which execute multiple important processes in the cell including oxidative phosphorylation, amino acid biogenesis, fatty acid catabolism and apoptosis. Mitochondrial quality control is, therefore, important for cell survival3. Mitophagy contributes to mitochondrial quality control by removal of faulty mitochondria, and neurodegenerative disorders related to mitochondrial dysfunction often develop under conditions of mitophagic deficiency1. Mitophagy has also been proposed to mediate degradation of paternal mitochondria during embryonic development4.
The loss of mitochondrial membrane potential facilitates the differentiation between functional and non-functional/dysfunctional mitochondria to mediate selective elimination. In mammalian cells this may be sensed by the kinase PINK1. In healthy mitochondria, PINK1 is transported into the mitochondria and degraded. Loss of membrane potential leads to the stabilization of PINK1 on the mitochondrial membrane. PINK1 then recruits and phosphorylates PARK2/parkin5, an E3 ligase, that ubiquitinates a number of outer mitochondrial membrane (OMM) proteins such as VDAC1. The ubiquitination of these OMM proteins promotes binding of receptors such as SQSTM1/p62 that link these proteins to LC3 and promote subsequent incorporation into phagophores, the precursors to autophagosomes6.
Recently, the PARK2-dependent generation of mitochondria-derived vesicles (MDVs) has been proposed as a parallel means for the selective loss of damaged components from individual mitochondria7. Mitophagy is DNM1L/Drp1 mediated and PARK2 independent in cardiomyocytes8 and BNIP3L/NIX mediated during programmed mitochondrial degradation in reticulocytes9. These findings illustrate that the cell may have developed different pathways for maintaining mitochondrial integrity.
Although the role of OMM components in mitophagy have been partially characterized, IMM dynamics during mitophagy have not been extensively studied. Wei et al. show that the IMM protein PHB2 can function as a mitophagy receptor. PHB2 is a component of the mitochondrial prohibitin complex along with PHB/PHB1 (prohibitin). Wei et al. utilize an immunoprecipitation-coupled mass spectrometry screen to show that in HeLa cells stably expressing PARK2 (HeLa-PARK2 cells), PHB and PHB2 coimmunoprecipitate with LC3-II upon CCCP administration. Treating these cells with a combination of oligomycin and antimycin (OA) yields a similar coimmunoprecipitation profile. Interestingly, this interaction is not observed in HeLa cells that do not stably express PARK2, indicating that PARK2 activity facilitates this interaction. Using in vitro binding assays they tease out this interaction to demonstrate that PHB2 directly binds LC3-II, whereas PHB associates with LC3-II via PHB2. The authors then investigate whether PHB2 regulates mitophagy in these cells. Upon induction of mitochondrial damage by OA treatment in HeLa-PARK2 cells, the number of mitochondria decreases rapidly as a result of mitochondrial clearance. Knockdown or conditional knockout of PHB2 in HeLa cells, however, leads to the retention of mitochondria in OA-treated cells, indicating a defect in mitochondrial clearance. The phenotype corroborates strongly with an ATG7 knockdown phenotype suggesting that mitophagy is specifically affected.
The authors also identify the LC3-interacting region (LIR) motif of PHB2 and show that mutations in the critical LIR residues (mutant LIR/mLIR) preclude LC3 binding by PHB2. mLIR-PHB2 mouse embryonic fibroblasts show reduced mitochondrial clearance upon OA treatment, a phenotype similar to OA-treated PHB2-knockdown cells. Cristae formation, mitochondrial organization and processing of the mitochondrial dynamin-like protein OPA1 in mLIR-PHB2 cells are comparable to those in wild-type PHB2 cells, indicating that the mLIR mutations in PHB2 do not compromise other known functions of the prohibitin complex in the mitochondria.
Access of LC3 to PHB2 requires prior disruption of the OMM. This may be achieved by PARK2-mediated ubiquitination of OMM proteins and recruitment of the proteasome to mitochondria, promoting subsequent degradation of the OMM proteins and rupture of the OMM10. Using electron microscopy, Wei et al. demonstrate that the OMM is degraded in HeLa-PARK2 cells upon OA treatment. This membrane rupture can be prevented by treating the cells with the proteasomal inhibitor epoxomicin. Consistent with their hypothesis, epoxomicin treatment also abrogates the co-immunoprecipitation and colocalization of PHB2 with LC3-II indicating that OMM degradation precedes PHB2-LC3-II interaction. OMM degradation is independent of PHB2 function as PHB2 knockdown drastically reduces mitochondrial clearance but does not prevent OMM rupture. Using super-resolution microscopy the authors show that OA treatment compromises mitochondrial membrane integrity in HeLa-PARK2 cells and in the same cells with a knockdown of PHB2 but not in epoxomicin-treated cells. However, whereas the phagophore localizes to the mitochondria in OA-treated HeLa-PARK2 cells, it is localized away from the mitochondria in both PHB2-knockdown and epoxomicin-treated cells.
Selective mitochondrial degradation during development facilitates exclusive inheritance of maternal mitochondrial DNA. In C. elegans, the paternal mitochondria are selectively eliminated soon after fertilization by an autophagy-dependent process4. Wei et al. show that in C. elegans, knockdown of PHB2 in males leads to the retention of paternal mitochondrial DNA in the subsequent generation. Labeled paternal mitochondria are also retained in embryos derived from PHB2-knockdown males at later stages of embryonic development. Furthermore, this phenotype of paternal mitochondria retention recapitulates the phenotype of embryos derived from ATG7-knockdown females.
The findings presented by Wei et al. indicate a conserved role for PHB2 as a mitochondrial receptor for mitophagy (Figure 1). While they establish that PHB2 interacts with LC3-II, it remains to be seen whether the PHB2-LC3 interaction is an important determinant of mitophagy in cells expressing “wild-type” levels of PARK2. Similarly, the role of PHB2 and other IMM proteins in PARK2-independent mitophagy may be an exciting area for further investigation. In conclusion, their findings affirm that the regulation of mitophagy in mammals may be mediated by multiple mechanisms and is context dependent.
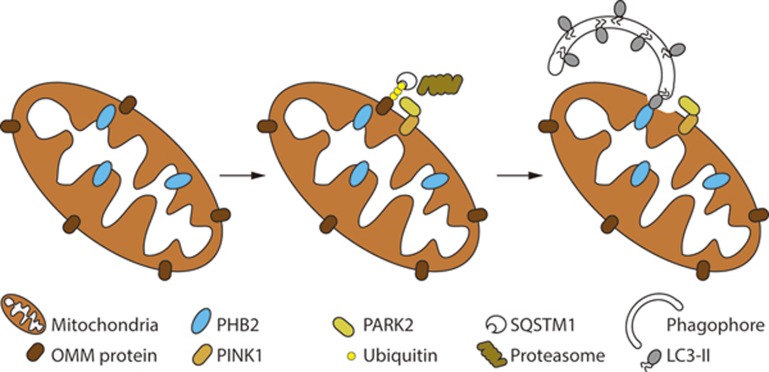
Figure 1.
Depolarized mitochondria express PINK1 on the outer membrane, resulting in recruitment of PARK2 and subsequent ubiquitination and degradation of mitochondrial outer membrane proteins. The resulting rupture of the outer membrane allows LC3-II on a phagophore to bind the mitochondrial inner membrane protein PHB2, leading to mitophagy.
References
- Wang K, Jin M, Liu X, et al. Autophagy 2013; 9:1828–1836. [DOI] [PMC free article] [PubMed]
- Wei Y, Chiang WC, Supmter Jr R, et al. Cell 2017; 168:224–238. [DOI] [PMC free article] [PubMed]
- Mao K, Klionsky DJ. Autophagy 2013; 9:1900–1901. [DOI] [PMC free article] [PubMed]
- Sato M, Sato K. Science 2011; 334:1141–1144. [DOI] [PubMed]
- Shiba-Fukushima K, Imai Y, Yoshida S, et al. Sci Rep 2012; 2:1002. [DOI] [PMC free article] [PubMed]
- Geisler S, Holmström KM, Skujat D, et al. Nat Cell Biol 2010; 12:119–131. [DOI] [PubMed]
- McLelland G-L, Lee SA, McBride HM, et al. J Cell Biol 2016; 214:275–291. [DOI] [PMC free article] [PubMed]
- Kageyama Y, Hoshijima M, Seo K, et al. EMBO J 2014; 33:2798–2813. [DOI] [PMC free article] [PubMed]
- Schweers RL, Zhang J, Randall MS, et al. Proc Natl Acad Sci USA 2007; 104:19500–19505. [DOI] [PMC free article] [PubMed]
- Yoshii SR, Kishi C, Ishihara N, et al. J Biol Chem 2011; 286:19630–19640. [DOI] [PMC free article] [PubMed]