Dear Editor,
The lifespans of model animals, such as Caenorhabditis elegans, have been demonstrated to be extended when they are subjected to hypoxic conditions in the laboratory1,2. In addition, wild mammals with hypoxic habitats, such as the naked mole rat that lives in burrow, and whales that must hold their breath as they dive, seem to live longer3,4,5. In stark contrast with observations in the laboratory, humans living under natural hypoxic environment of the Tibetan Plateau have generally been thought to have much shorter lifespans compared with people living at lower altitudes. No scientific ascertainment exists for this paradox, neither has the underlying genetic basis been resolved.
In an effort to explore this issue, we first examined published Chinese census data (year 2010, available online) from the National Bureau of Statistics of the People's Republic of China. Indeed, the proportion of the Tibetan population > 60 years of age was significantly lower than that for the Han population (Supplementary information, Figure S1A). The harsh environment and the limited health and medical resources found in the Tibetan Plateau, compared with other regions of China, likely contribute to the differences in lifespan. For example, chronic hypoxic atmosphere theoretically increases the load of oxygen transport. It has been shown that death cases related to cerebrovascular and circulatory diseases are the highest in Tibet compared with other regions. Particularly, the rate of hypertensive heart disease is more than twice higher in Tibet than in the province with the second highest rate6.
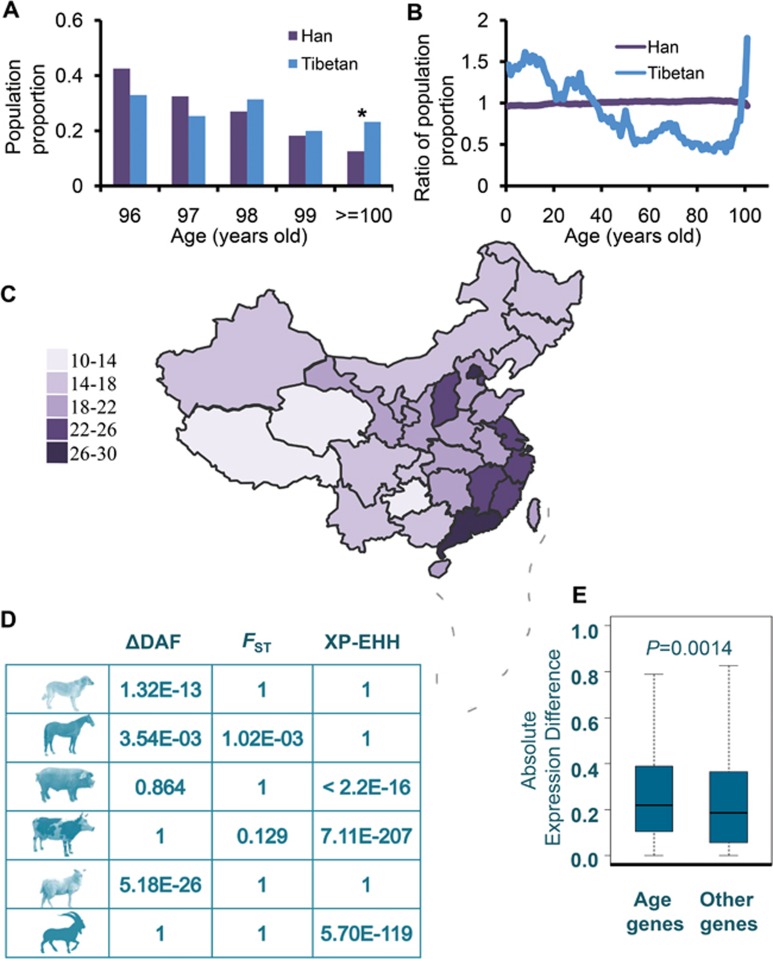
However, we interestingly noted that the proportion of individuals older than 100 years of age was higher for male (but not female) Tibetans than for Hans (P = 1.23E-07 by χ2-test, Figure 1A, Supplementary information, Figure S1B). We then compared the age distributions of the Tibetan and Han Chinese populations. Here, ratios were calculated as the proportion of Tibetan or Han population at each age divided by the proportion of the whole Chinese population at the same age (Figure 1B). In comparison with the Han population, a dramatic rise in the ratio of proportion of populations older than 91 years of age was seen in the Tibetans (Figure 1B). The pattern remained the same when males and females were considered separately (Supplementary information, Figure S1C). We then examined data of the age at death for each province of China based on the year 2010 census data. Proportion of deaths was defined as the number of deaths in an age group divided by the total number of people for the age group. We examined two different age populations, i.e., < 60 years and ≥ 60 years. The relative proportion of deaths in the old population (ab. RPDO) (≥ 60 years old) was defined as the proportion of deaths in old population (≥ 60 years old) divided by the proportion of deaths in the other population (< 60 years). The RPDO value is the lowest for the Tibetan Plateau, stretching across Tibet and Qinghai provinces (Figure 1C). We also calculated the ratio of deaths for old people, defined as the number of deaths of old people divided by the total number of deaths, which was also found to be the lowest for the Tibetan Plateau (Supplementary information, Figure S1D). Our findings strongly suggest that old people of the Tibetan Plateau have a longer lifespan than old people in other regions of China.
Figure 1.
Analysis of the age composition of Tibetans, and rapid evolution and differential expression of genes associated with aging. (A) Proportion of the population (×10−4) with male individuals > 95 years of age, Han and Tibetans. *P = 1.23E-07. (B) Ratio of the population proportion computed as the proportion of Tibetan or Han population divided by the proportion for the whole Chinese population for different ages. (C) Heatmap of RPDO across China. The relative proportion of deaths in the old population (ab. RPDO) (≥ 60 years old) was defined as the proportion of deaths in old population (≥ 60 years old) divided by the proportion of deaths in the other population (< 60 years). Data for Taiwan are not available. (D) Significantly higher values of ΔDAF, FST and XP-EHH for SNPs in genes associated with aging versus SNPs in other genes. P-values according to Mann-Whitney U-test are presented. (E) Higher level of gene expression difference was found for aging-related genes than for other genes. FPKM, as expression value, was calculated by Cufflinks for each gene15. Expression difference for each gene was calculated by the absolute difference of log2 (FPKM+1) values of the gene in lungs of Tibetan and Min pigs.
We went a step further and investigated potential contributing factors to the observed longer lifespan of old people in Tibet. It may be argued that Tibet was very poor, particularly that Tibet was governed by slave holders 100 years ago and few people had birth certificates during that time, which might induce an age bias in older Tibetans. To exclude this potential confounding factor, we analyzed census data in the city and countryside (village) of each province to examine the potential influence of economic development status. We did not find a correlation between the proportion of old people (> 100 years old) and the proportion of countryside people across the provinces (Supplementary information, Figure S1E), suggesting that the age bias cannot explain our observation. In addition, we computed a ratio, defined as the proportion of old people (> 100 years old) in the countryside divided by the proportion of old people (> 100 years old) in the city, for each province. The ratio in Tibet does not significantly differ from values in other provinces.
A down-regulated basal metabolic rate, due to the lower temperature, could contribute to an extended lifespan. However, it has been demonstrated that the Tibetan population has a normal basal metabolic rate expected for their age, sex and body weight, as do Andeans from another highland region7. Therefore, the lower average temperature at the highland is unlikely to be a critical factor for the extended human longevity, particularly in Tibet.
On the other hand, hypoxia might induce longevity in Tibetans, as has been stated for model animals under laboratory experimentation1,2, as well as wild mammals that live in natural hypoxic environments, such as the naked mole rat and whales3,4,5. Considering the strongly significant greater frequency of interaction between aging-associated genes and hypoxia response genes (Supplementary information, Figure S1F and Data S1), and the fact that 18 genes were shared between these two groups of genes, it is likely that hypoxia stress would accelerate the evolution of aging-associated genes. In this case, we would expect that the biological function of genes associated with aging might have changed after these people moved to the Tibetan Plateau, and that these genes might have evolved rapidly under positive selection. To test this hypothesis, we used genome-wide single-nucleotide polymorphisms (SNPs), genotyped by the Affymetrix Genome-Wide Human SNP 6.0 Array, from 31 unrelated Tibetans from a previous study8 and compared them with genotype data for Han people from HapMap (phase II, http://hapmap.ncbi.nlm.nih.gov/). We found that SNPs at genes associated with aging harbored significantly higher levels of population differentiation, as evaluated by FST9, between Tibetan and Han (mean FST value is 0.060), than SNPs at other genes (mean FST value is 0.0585) (P = 1.39×10−4, by Mann-Whitney U-test). Using other parameters, locus-specific branch length (LSBL)10 and ΔDAF (the difference of the derived allele frequencies between Tibetan and Han), SNPs at genes associated with aging also gave higher values compared with SNPs at other genes, although the difference did not reach statistical significance. In contrast, genes involved in metabolism did not show rapid evolution than other genes among Tibetans by virtue of FST, LSBL and ΔDAF, suggesting that metabolism might not be a critical factor for the extended longevity in Tibetans.
To determine whether rapid evolution of genes associated with aging commonly occurs among other mammals inhabiting the highlands, we used whole-genome sequence data of sheep, goats, cattle, dogs, pigs and horses from the Tibetan Plateau (134 genomes), as well as those of domestic populations from lowland areas (193 genomes), to identify SNPs across the genome by a stringent pipeline implemented in the GATK program11. Differentiation of the SNPs was calculated using FST9, XP-EHH (cross population extended haplotype homozygosity)12, and ΔDAF (the difference of the derived allele frequencies) between the highland and lowland populations to evaluate their evolutionary rates. SNPs at genes associated with aging showed significantly higher differentiation compared with SNPs at all other genes (Figure 1D), supporting the proposed rapid evolution of the genes involved in aging.
Gene expression is the most fundamental level at which the genotype gives rise to the phenotype. Therefore, we reasoned that genes associated with aging might also exhibit changes in RNA expression levels in individuals living in highlands. To test this hypothesis, we profiled the transcriptomes of lung tissues from four Tibetan pigs and four lowland Min pigs by RNA sequencing. As expected, genes associated with aging showed significantly higher levels of mRNA expression difference as compared to other genes (Figure 1E, P = 0.0014 by Mann-Whitney U-test). Finally, we explored the potential association between changes in gene expression under hypoxia and changes in gene expression during aging. We retrieved and re-analyzed transcription data for human peripheral blood sampled at different ages from a previous study13, and found that the change in direction of gene expression under hypoxia is significantly negatively associated with the change in direction of gene expression during aging (P = 0.008, Pearson correlation coefficient is −0.202). This result suggests that genes downregulated during aging tend to be upregulated under hypoxia, and vice versa. Hypoxia, thus, might offset the effect of aging and therefore extend lifespan, as observed in model and wild animals1,2,3,4,5.
In conclusion, we found that older people living at the Tibetan Plateau tend to have a longer lifespan than older people living at lower altitudes, suggesting an association between hypoxia and longevity in Tibetans. Actually, linkage between longevity of people at the Andean region in America and high altitude has also been reported, with Ecuador being termed a “Valley of Longevity”14.
The caveat of our analysis is that it was based on the comparison of Han and Tibetan populations without stratifying these populations living at different regions. Our analyses suggest that hypoxia might offset the effect of aging and therefore extend lifespan. Hypoxia adaptation of aging-associated genes and hypoxia response genes, particularly genes involved in HIF pathways, could contribute to longevity of Tibetans. More generally, hypoxia might be a key driving force that has promoted human longevity under natural hypoxic conditions.
Acknowledgments
DDW was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13020600). YL was supported by the National Natural Science Foundation of China (31401083). DDW, WW, PS and YPZ were supported by the National Natural Science Foundation of China (31321002). NOO thanks the support of CAS-TWAS President's Fellowship Program for Doctoral Candidates.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Supplementary information, Figure S1A–S1F
Methods
References
- Mehta R, Steinkraus KA, Sutphin GL, et al. Science 2009; 324:1196–1198. [DOI] [PMC free article] [PubMed]
- Leiser SF, Fletcher M, Begun A, et al. J Gerontol A Biol Sci Med Sci 2013; 68:1135–1144. [DOI] [PMC free article] [PubMed]
- Fang X, Seim I, Huang Z, et al. Cell Rep 2014; 8:1354–1364. [DOI] [PMC free article] [PubMed]
- Kim EB, Fang X, Fushan AA, et al. Nature 2011; 479:223–227. [DOI] [PMC free article] [PubMed]
- Keane M, Semeiks J, Webb AE, et al. Cell Rep 2015; 10:112–122. [DOI] [PMC free article] [PubMed]
- Zhou M, Wang H, Zhu J, et al. Lancet 2016; 387:251–272. [DOI] [PubMed]
- Beall CM. Proc Natl Acad Sci USA 2007; 104 (Suppl 1):8655–8660. [DOI] [PMC free article] [PubMed]
- Simonson TS, Yang Y, Huff CD, et al. Science 2010; 329:72–75. [DOI] [PubMed]
- Akey JM, Zhang G, Zhang K, et al. Genome Res 2002; 12:1805–1814. [DOI] [PMC free article] [PubMed]
- Bigham AW, Mao X, Mei R, et al. Hum Genomics 2009; 4:79–90. [DOI] [PMC free article] [PubMed]
- McKenna A, Hanna M, Banks E, et al. Genome Res 2010; 20:1297–1303. [DOI] [PMC free article] [PubMed]
- Sabeti PC, Varilly P, Fry B, et al. Nature 2007; 449:913–918. [DOI] [PMC free article] [PubMed]
- Peters MJ, Joehanes R, Pilling LC, et al. Nat Commun 2015; 6:8570. [DOI] [PMC free article] [PubMed]
- Mazess RB. JAMA 1978; 240:1781.
- Trapnell C, Williams BA, Pertea G, et al. Nat Biotech 2010; 28:511–515. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Figure S1A–S1F
Methods