Abstract
In adult skeletal muscle, β myosin heavy chain (βMyHC) gene expression is primarily restricted to slow type I fibers; however, its expression is down-regulated in response to muscle inactivity. Little is known about the signaling pathways and transcription factors that mediate this important functional response. This study demonstrates that increased binding of Sp3 to GC-rich elements in the βMyHC promoter is a critical event in down-regulation of βMyHC gene expression under non-weight-bearing conditions. Conversely, binding of Sp3 to these elements decreased while Sp1 binding increased with nuclear extracts from plantaris muscle exposed to mechanical overload, a stimulus that increases βMyHC gene expression. In addition, these experiments revealed the existence of an Sp4-DNA binding complex when using adult skeletal muscle nuclear extract was used but not when nuclear extracts from cultured myotubes were used. Sp3 proteins are competitive inhibitors of Sp1-mediated βMyHC reporter gene transactivation in both Drosophila SL-2 and mouse C2C12 myotubes. Sp4 is a weak activator of βMyHC gene expression in SL-2 cells, which lack endogenous Sp1 activity, but does not activate βMyHC gene expression in C2C12 myotubes, which have high levels of Sp1. These results suggest that competitive binding of Sp family proteins regulate βMyHC gene transcription in response to altered neuromuscular activity.
Skeletal muscle is a highly organized tissue that performs the specialized function of force development. In the adult mouse, four major myosin heavy chain (MyHC) isoforms [fast IIb, IIx/d, and IIa and slow type I (β)] are expressed in a manner that defines four primary fiber types, termed fast type IIb, IIx/d, and IIa and slow type I (37). Each of these distinct fiber types displays unique functional properties with respect to size, metabolism, fatigability, and intrinsic contractile properties. For example, slow type I fibers primarily populate slow-twitch muscles such as the soleus, rely on oxidative metabolism, have increased resistance to fatigue, and express high levels of the slow type β-isoform of MyHC (βMyHC) which is particularly efficient in energy utilization while maintaining tension. Thus, slow-twitch muscles like the soleus are primarily used in chronic activities such as postural maintenance and for sustained low-force locomotor activities. However, under non-weight-bearing (NWB) conditions, as would be encountered in the microgravity environment of space flight, an inactive lifestyle, injury, or disease, slow-twitch muscles undergo a marked degree of muscle atrophy accompanied by a slow-to-fast phenotypic change characterized by decreased βMyHC mRNA and protein expression (2, 5, 27, 28, 37, 41, 42). Because MyHC is a major determinant of the maximum unloaded shortening velocity (Vmax) of skeletal muscle fibers, the documented decrease in βMyHC protein has important physiological implications for skeletal muscles exposed to NWB conditions (2). While the phenotypic adaptations that occur in response to NWB have been well documented, identification of the precise transcriptional control mechanism(s) that mediates decreased βMyHC gene expression has been elusive since adult-stage muscle phenotypes, fiber-type transitions, and the effects of altered muscle loading conditions cannot be adequately duplicated in cultured muscle cells.
Skeletal muscles composed primarily of slow type I fibers, such as the soleus muscle, are more susceptible to atrophy during periods of reduced muscle activity (5). Our previous work has utilized the soleus muscle as a model system to understand the mechanistic basis underlying decreased βMyHC gene expression in response to NWB conditions (27, 28, 42). Our transgenic expression analysis of both the mouse and human βMyHC promoters delineated a 600-bp region of the βMyHC promoter that was sufficient to mimic endogenous βMyHC down-regulation which occurs in response to NWB (27). Further deletion of this 600-bp promoter region identified a strong negative regulatory element (dβNRE-S; −332/−311) and multiple positive-acting elements, including a distal MCAT (dMCAT; −290/−284), proximal MCAT (pMCAT; −210/−203), and an E-box/nuclear factor of activated T cells (E-box/NFAT; −183/−172) (28, 42). MCAT and NFAT elements are frequently found in the control region of muscle-specific genes and have been reported to function as inducible, muscle- or fiber-specific response elements (9, 17, 22, 24, 33, 35, 44, 45). However, when we examined each of these elements in detail, none was found to be solely responsible for NWB responsiveness, indicating the presence of additional elements required for down-regulation of the βMyHC under NWB conditions (42).
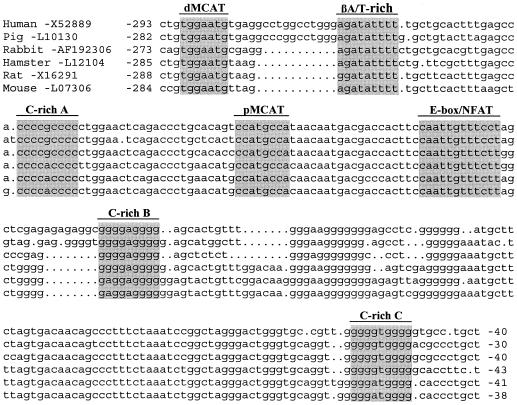
A computer-assisted analysis of the sequence located downstream from the dMCAT element identified three closely spaced GC-rich (GT/CACC) elements, termed C-richA (−248/−225), C-richB (−160/−140), and C-richC (−61/−41) in this paper. These three elements are highly conserved in sequence and location across species: an arrangement suggesting an important role in βMyHC regulation (Fig. 1). GC/GT elements are frequently found in the transcriptional control region of genes encoding proteins critical to a broad range of subcellular systems, including striated muscle. These elements are known to interact with members of the Sp family of transcription factors. In mammals, there are eight structurally similar Sp protein family members that are expressed in an overlapping fashion. Each contains a nearly identical DNA-binding domain consisting of three Cys2His2 zinc fingers, which likely accounts for their binding to the same GC/GT elements with variable affinity. Ubiquitously expressed Sp1 was the first family member identified, followed by Sp2, Sp3, and Sp4 (6, 30, 39). Targeted mutation of either Sp1, SP3, or SP4 results in early lethality due to a variety of different cellular defects indicating that each Sp protein can serve a distinct physiological role (7, 11, 12, 26, 32, 38). Recently, a subgroup of Sp protein family members comprising Sp5 to -8 has been isolated, and mice carrying targeted mutations of either Sp5, -7, or -8 also display distinct phenotypes (4, 16, 31).
FIG. 1.
βMyHC GC-rich elements are highly conserved in sequence and position across species. The nucleotide sequence comparison of the βMyHC proximal promoter of various species reveals high conservation of the GC-rich elements (shading). See the work of Vyas et al. (44, 45) for accession numbers and references of each sequence.
A recent study has implicated multiple CACC elements in regulation of the sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA1) gene under NWB conditions (29). However the proteins which bound to the CACC elements were not identified. In this study, we have identified and characterized the biochemical and functional properties of three βMyHC proximal promoter GC-rich elements. Mutagenesis analysis revealed the importance of each the GC-rich element for βMyHC reporter gene expression in C2C12 myotubes. Methylation interference footprinting delineated a binding site involving strong and weak protein-DNA interactions at the C-richA element that were identical for Sp1, Sp3, and Sp4. Electrophoretic mobility shift assay (EMSA) analysis revealed enriched binding of Sp3 proteins (115, 78, and 80 kDa) to these elements when nuclear extracts from soleus muscle exposed to NWB conditions were used. Binding of Sp3 to these elements was decreased while Sp1 binding increased with nuclear extracts from plantaris muscle following mechanical overload (MOV), a stimulus that induces a fast-to-slow phenotypic transition and increased βMyHC gene expression. Transient expression assays showed that Sp3 functioned as a competitive inhibitor of Sp1-mediated transactivation of a 293-bp βMyHC reporter gene in Drosophila SL-2 cells and C2C12 myotubes. In addition, a protein-DNA complex containing Sp4 was specifically detected when nuclear extracts from adult skeletal muscle were used. This Sp4-containing complex could not be detected with nuclear extracts isolated from a variety of cell lines, including HeLa, C2C12, and SOL8. These results provide clear evidence that Sp3 proteins play a functional role in mediating adult-stage skeletal muscle phenotype transitions in response to altered neuromuscular conditions. We suggest that the differential expression of Sp proteins will contribute to phenotypic adaptations that occur in all tissues affected by loading conditions imposed by disease, an inactive lifestyle, or space travel.
MATERIALS AND METHODS
Preparation of nuclear protein extract from adult skeletal muscle.
Nuclear extract was isolated from adult rat control soleus (CS), NWB soleus (NWB-S), control plantaris (CP), and MOV plantaris (MOV-P) muscles as described previously (28, 44). Protein concentration was determined with a commercial protein assay (Bio-Rad) according to the method of Bradford (8).
Animal care and MOV and NWB procedures.
The MOV and NWB procedures used in this study were approved by the Animal Care Committee for the University of Missouri—Columbia, and the MOV mice were housed in an Association for the Assessment and Accreditation of Laboratory Animal Care International-accredited animal facility. Rats and mice were prepared for the NWB experiment by an inexpensive modification of the noninvasive tail traction procedure, as described previously (27). The imposition of MOV on the fast-twitch plantaris muscle was accomplished as described by us previously (43). All animals were provided with food and water ad libitum and were housed at room temperature (24°C) with a 12-h light-dark cycle in either standard filter top cages (control and MOV mice) or cages designed for head-down tilt suspension (hind limb suspension).
Plasmids and constructs.
βMyHC promoter constructs contained 293 bp of human βMyHC promoter sequence, and 120 bp of 5′ untranslated region (UTR) was cloned into the HindIII site of the luciferase reporter vector, p0Luc. The βMyHC GC-rich elements (C-richA, C-richB, and C-richC) within the 293-bp human βMyHC promoter were mutated within the plasmid p0Luc-β293, using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Complementary oligonucleotide primers containing the desired mutations were designed to meet the length and melting temperature requirements, as listed below (mutated sites in boldface lowercase; core elements underlined): C-richAmut (5′-GCTGCACTTTGAGCCACCCtaCatgCTGGAACTCAGACCCTGCAC-3′), C-richBmut (5′-GGGACTGGGTGCCGTTGaGGacaGaGaTGCCCTGCTGCCCC-3′), and C-richCmut (5′-CCCTAGCTGGAcAGGCtGGtctGtGAGCACTGTTTGGGAAGGGGG-3′).
Unintentional transcription factor recognition sites were not created by these mutations as assessed by cross-referencing the mutated primers against the Eukaryotic Transcription Factor database (tfsites.dat) available from Genetics Computer Group.
The Drosophila expression vectors for Sp proteins, pPacSp1, pPacSp3, and pPacSp3/M1, in which the Sp1, Sp3, or a truncated Sp3 cDNA is fused with the Drosophila actin 5C promoter to drive expression in Drosophila Schneider line 2 (SL2) cells, were kindly provided by J. M. Horowitz (North Carolina State University, Raleigh). Sp4 expression vector (pPacSp4) was kindly provided by G. Suske (Klinikum der Philipps-Universität Marburg, Marburg, Germany) and the Copia-β-galactosidas expression vector and empty pPac0 were kindly provided by R. Tijian (Howard Hughes Medical Institute, Berkeley, Calif.). The C2C12 expression vectors for Sp proteins, pRc/CMV-Sp1 and Prc/cmv-Sp3 or pRc/CMV-Sp4, were also given to us by J. M. Horowitz and G. Suske, respectively. The in vitro transcribed and translated (TNT) expression plasmids for Sp1, Sp3, and Sp4 were constructed by inserting each cDNA into pCITE4 (Novagen) vectors in frame with the internal translation start site provided by the vector, creating new vectors pCITE4a-Sp1, pCITE4a-Sp3, and pCITE4c-Sp4.
EMSAs.
All oligonucleotide probes used in this study are listed in Table 1. EMSAs were carried out as previously described (28). The annealed C-rich oligonucleotide probes were labeled by end labeling with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]dATP (Perkin-Elmer) and gel purified. Binding reactions were performed with 4 μg of CS, NWB-S, CP, or MOV-P nuclear extract and 20,000 cpm of labeled probe for 20 min at room temperature in a 25-μl total volume in binding buffer (50 mM Tris-HCl, pH 7.9, 50 mM KCl, 2.5 mM ZnCl2, 0.5 mM dithiothreitol, 5% [wt/vol] glycerol). Where indicated, either in vitro TNT Sp1, Sp3, and Sp4 or HeLa, C2C12, or Sol8 nuclear extract was used in place of adult skeletal muscle nuclear extract (see figure legends for specific amounts). Supershift assays were performed by including 2 μl of preimmune (PI) serum, anti-Sp1 (Santa Cruz Biotechnology, Inc.; catalog no. PEP2), anti-Sp3 (Santa Cruz Biotechnology, Inc., catalog no. D-20), anti-Sp4 (Santa Cruz Biotechnology, Inc.; catalog no. V-20), or anti-Egr1 (Santa Cruz Biotechnology, Inc., catalog no. 588) antibody in the binding reaction mixture prior to the addition of probe. Protein-DNA complexes were electrophoretically resolved from unbound oligonucleotide probe on a 5% nondenaturing polyacrylamide gel at 220 V for 2.5 h at 4°C.
TABLE 1.
Oligonucleotides used in EMSA and methylation interference analysisa
| Designation | Oligonucleotide (sequence 5′ to 3′) | Position | Reference |
|---|---|---|---|
| C-richA | TGAGCCACCCCGCCCCCTGGAACT | −248 to −255 | This study |
| C-richB | AGAGGCGGGGAGGGGAGCACT | −160 to −140 | This study |
| C-richC | GCCGTTGGGGGTGGGGGTGCCTG | −63 to −41 | This study |
| Sp1 consensus | ATTCGATCGGGGCGGGGCGAGC | Promega, catalog no. E323A | |
| C-richMT1 | TGAGCCACaCtaCatgCTGGAACT | This study | |
| C-richMI | TGCACTTTGAGCCACCCCGCCCCCTAGAACTCAGAC | −225 to −220 | This study |
In each case, the species was human. Lowercase represents mutations; underlining represents core elements.
DMS interference assay.
Dimethyl sulfate (DMS) DNA-footprinting assays were performed as previously described (44). 32P-labeled C-richA probe was modified by 0.7% DMS for 15 min at 25°C. The probe was used for a preparative EMSA as described above, except the reactions were scaled-up 10-fold. Bands corresponding to in vitro-synthesized Sp1/C-richA, Sp1/Sp4/C-richA, and Sp3/C-richA protein-DNA complexes and free probe were excised from the EMSA gel and recovered by electroelution. Base-specific cleavage of the recovered DNA was carried out in 100 μl of 1 M piperidine incubation for 30 min at 90°C, which was followed by repeated rounds of lyophilization to remove the piperidine. Equivalent amounts (1,000 cpm/lane) of free and bound cleaved probe were resolved on a 20% polyacrylamide-8 M urea denaturing sequencing gel. Gels were autoradiographed for 24 h.
Western blots and antibodies.
Western blotting was carried out as previously described (18). Herein 0.8 μl of in vitro-synthesized Sp protein or 30 to 50 μg of CS, NWB-S, CP, or MOV-P nuclear extract was fractionated on NuPage 4 to 12% polyacrylamide Bis-Tris gels (Invitrogen) according to the manufacturer's instructions and transferred to a polyvinylidene difluoride membrane (Bio-Rad Laboratories). The blots were incubated with either anti-Sp1 (1:1,000), anti-Sp3 (1:1,000), or anti-Sp4 (1:200) rabbit polyclonal immunoglobulin G (IgG). The blots were then washed with Tris-buffered saline-Tween and further incubated with goat anti-rabbit IgG conjugated with horseradish peroxidase (Cell Signal Technology; 1:2,000). After washing, horseradish peroxidase activity was detected with an enhanced chemiluminescence detection SuperSignal substrate (Pierce) and subjected to autoradiography.
In vitro TNT.
In vitro TNT reactions were performed with 1 μg of pCITE-Sp1, -Sp3, or -Sp4 expression plasmid in the T7 TNT rabbit reticulocyte lysate system, according to the manufacturer's instructions (Promega). Parallel TNT reactions were performed in the presence of [35S]methionine (Perkin-Elmer). Efficient translation and expected molecular weights of the protein products were verified by resolving the radiolabeled reaction products on NuPage 4 to 12% polyacrylamide Bis-Tris gel (Invitrogen). Parallel reactions of lysate not programmed with plasmid DNA served as negative controls and are called unprogrammed lysate (UL).
Cell culture, transfection, and reporter assays.
Drosophila Schneider SL2 cells (American Type Culture Collection) were grown at 25°C in Schneider's Drosophila medium (Gibco BRL) supplemented with 10% (vol/vol) heat-inactivated insect-tested fetal bovine serum (FBS) (Sigma), 100-U/ml penicillin, 100-μg/ml streptomycin (Sigma), and 2 mM l-glutamine (Sigma). Cells used for transfection were plated at a density of 5 × 106 cells per 60-mm-diameter cell culture plate in Drosophila Schneider medium supplemented with only 10% FBS. Cells were transfected by a calcium phosphate-DNA coprecipitation method according to the manufacturer's manual (Invitrogen). Each plate was cotransfected with 0.2 μg of Copia-β-galactosidase expression vector as an internal normalization control to estimate variations in transfection efficiency. The ratios and the amounts of other reporter and expression plasmids used in transfection experiments were experimentally determined and are listed in the respective figure legends. The total amount of DNA was kept constant at 8 μg by compensation with the addition of plasmid pPac0. Cells were washed free of Ca2+ by transferring suspended cells to sterile tubes, pelleted by centrifugation for 4 min at 25°C at 500 × g, and then resuspended in 1× phosphate-buffered saline (PBS). The washed cell pellets were pelleted and resuspended in fresh propagation media, transferred to fresh 60-mm plates, and incubated at 25°C for 24 h. Cells were lysed with the addition of 200 μl of 1× lysis buffer (Promega Corp.) to the cell pellets, following two washes with 1× PBS as described above, and incubated for 15 min at room temperature. The lysed cells were centrifuged at 125,000 rpm for 2 min, and the supernatant was removed and then stored at −80°C until analyzed.
Mouse skeletal muscle C2C12 myoblasts (American Type Culture Collection) were maintained in Dulbecco's modified Eagle's medium (Gibco) containing 10% (vol/vol) FBS at 37°C supplemented with 10% CO2. Transfection experiments were carried out as previously described (18). Cells were transfected with FuGene6 (Roche Applied Science) according to the manufacturer's manual, including cotransfection of 0.2 μg of pRSV-β-galactosidase expression vector (Promega) as the internal control. The ratios of other reporter and expression plasmids were experimentally determined and are listed in the respective figure legends. The total amount of DNA was kept constant by compensation with the addition of plasmid pPac0 as nonspecific.
Protein concentrations for all cell culture experiments were determined by the method of Bradford (8). Reporter assays were done as described previously with the following modifications. For SL2 cell extract, the following total amounts of protein were used per assay: luciferase assay, 80 μg; and β-galactosidase, 200 μg incubated for 70 min at 37°C. For the C2C12 myotube extract, the total amounts of protein were used per assay: luciferase assay, 15 μg; and β-galactosidase, 15 μg incubated for 70 min at 37°C. Bioluminescent assays were performed with a Turner Designs Model TD-20/20 luminometer with dual auto injectors. β-Galactosidase assays (Promega Corp.) were performed with a Beckman DU640 spectrophotometer at 420 nm. Normalized relative light units (RLU) were obtained by dividing luciferase RLU by β-galactosidase concentrations (milligrams per milliliter).
Statistical analysis.
Statistical analyses were performed by using the SPSS Graduate Pack 10.0 program (SPSS, Chicago, Ill.). A Levene's test for equality of variances was performed, followed by a two-tailed independent-sample t test to assess differences between group means. Where the Levene's test was rejected (significance of ≤0.05), the separate variance t test for means was used, where equal variances were not assumed. The lowest significance level accepted was P < 0.05. All data are reported as the mean ± standard error.
RESULTS
The human βMyHC promoter contains three GC-rich regulatory elements, two that were previously unreported.
Our previous transgenic and protein-DNA interaction analyses of dβNRE-S (−332/−311), dMCAT (−290/−284), βA/T-rich (−269/−258), pMCAT (−210/−203), and E-box/NFAT (−183/−172) revealed that these elements were not sufficient to account for complete NWB responsiveness, suggesting the presence of unidentified elements within the βMyHC promoter that were required for complete βMyHC transgene NWB responsiveness (27, 28, 42). A computer-assisted sequence analysis of the βMyHC proximal promoter located three closely spaced GC-rich elements [C-richA (−248/−225), C-richB (−160/−140), and C-richC (−61/−41)] that are highly conserved in sequence and location across species (Fig. 1). Interestingly, a previous transgenic mutagenesis analysis revealed that the C-richA element is not required for high-level muscle-specific expression; however, this result may have been due to the presence of the C-richB and C-richC elements which have previously been unrecognized (22). Of importance to this study is the finding that closely spaced GC/GT-rich elements favor stable binding of Sp3 isoproteins versus Sp1, a binding pattern that favors decreased and/or repressed gene transcription (49).
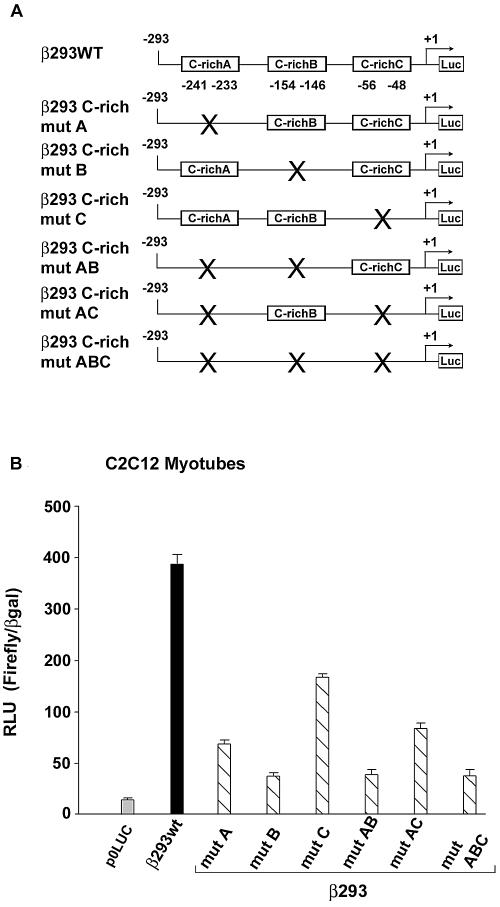
Mutation of the C-rich elements significantly reduces βMyHC promoter activity in C2C12 myotubes.
To determine the functional significance of the three human βMyHC GC-rich elements (C-richA, -B, and -C), we used site directed mutagenesis to generate six β293 mutant reporter constructs (Fig. 2A). Nucleotides chosen for mutations were based on our methylation interference footprinting results (see below). In transient transfection assays using C2C12 myotubes, independent mutation of each C-rich element (β293mutA, β293mutB, or β293mutC) significantly reduced promoter activity compared to wild-type 293-bp βMyHC reporter gene activity (Fig. 2B). The simultaneous mutation of the C-richA and -B elements (β293mutAB) reduced expression to a greater degree than when elements C-richA and C-richC were simultaneously mutated (β293AC). The expression level obtained when all three GC-rich elements were simultaneously mutated (β293mutABC) was indistinguishable from those obtained when elements C-richA and -B were simultaneously mutated, indicating that elements C-richA and C-richB play a more significant role in conferring βMyHC promoter activity than element C-richC (Fig. 2B). Although the independent or combinatorial mutation of the three GC-rich elements significantly reduced the activity of the 293-bp βMyHC reporter gene (to as low as 16% of wild-type levels), these mutations did not completely eliminate promoter activity. The residual expression following mutation of the GC-rich elements is likely driven by other elements, including dMCAT, βA/T-rich, pMCAT, and NFAT, within the β293wt construct (Fig. 1). Nevertheless, these data show that each GC-rich element participates in the expression of the wild-type 293-bp βMyHC reporter gene.
FIG. 2.
Mutations of the βMyHC GC-rich elements significantly decrease βMyHC promoter activity. (A) Schematic representation of the wild-type β293wt reporter gene (293-bp βMyHC promoter) and those that harbor mutations to individual or combinatorial GC-rich elements (C-richA, C-richB, or C-richC). Site-directed mutations involved those nucleotides shown to be involved in protein-DNA interaction by methylation interference footprint (Fig. 4). (B) Promoter activity of wild-type and GC-rich mutant reporter genes (2 μg) determined in C2C12 myotubes. All data were normalized to β-galactosidase (β-Gal) to accommodate variations in transfection efficiency. Data are reported as luciferase-normalized RLU (RLU/β-Gal ratio) and are expressed as the mean ± standard error (n = 10 for each reporter gene).
The C-richA element binds zinc-dependent nuclear proteins.
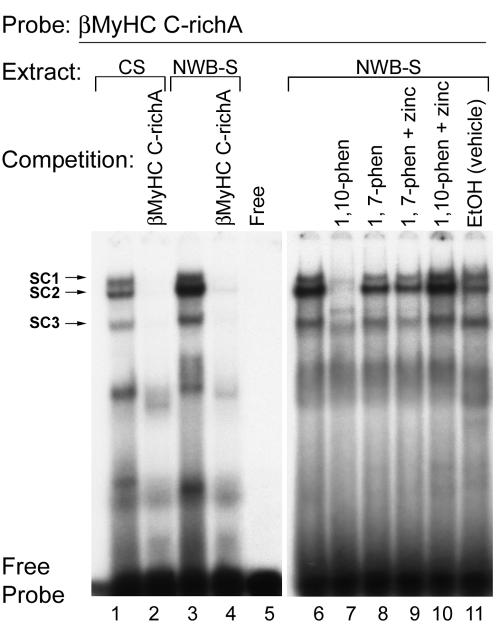
Having demonstrated the functional importance of each GC-rich element in conferring basal expression of the 293-βMyHC reporter gene in C2C12 myotubes, we wished to examine their binding properties. To determine the binding properties of the human βMyHC C-richA element, we performed an EMSA analysis using nuclear extracts prepared from intact adult-stage soleus muscles under control conditions (CS) and NWB conditions (NWB-S). Incubation of the double-stranded 32P-labeled human βMyHC C-richA probe (−248/−225; Table 1) with 4 μg of CS nuclear extract revealed the formation of three specific low-mobility binding complexes termed SC1, SC2, and SC3 (Fig. 3, lane 1). Notably, complexes SC2 and SC3 were highly enriched when 4 μg of NWB-S nuclear extract was used, whereas the intensity of complex SC1 revealed an insignificant change (Fig. 3, lane 1 versus 3). The addition of 100-fold molar excess cold C-richA element to binding reaction mixtures containing either CS or NWB-S nuclear extract abolished complex formation (Fig. 3, lanes 2 and 4).
FIG. 3.
EMSA assessment of sequence-specific protein-DNA interaction at the βMyHC C-richA element. The 32P-labeled βMyHC C-richA oligonucleotide probe was incubated with 4 μg of either CS (lanes 1, 2, and 6) or NWB-S (lanes 3, 4, and 7 to 11) nuclear extract. For competition assays, nonradioactive C-richA competitor oligonucleotide was added to the reaction mixture at a 100-fold molar excess prior to the addition of the 32P-labeled C-richA probe (lanes 2 and 4). Three specific complexes (SC1, SC2, and SC3) were resolved when using CS or NWB-S nuclear extract. To characterize the properties of the protein(s) in the binding complex, 4 mM zinc chelating agent 1,10-phenanthroline (1, 10-phen) or nonchelating agent 1,7-phenanthroline (1, 7-phen) was added to the binding reaction mixture. 1,10-Phenanthroline abolished the protein-DNA complex (lane 7), which was rescued with the supplement of 0.5 mM zinc (lane 10), while the addition of 1,7-phenanthroline or 1,7-phenanthroline plus zinc did not affect the binding pattern (lanes 8 and 9), confirming the presence of zinc finger proteins in the binding complex. Lane 9 shows that the phenanthroline vehicle (ethanol [EtOH]) did not affect the binding complex obtained when using NWB-S nuclear extracts. The Free lane represents the binding reaction mixture minus nuclear extract. EMSAs were repeated with three independent batches of nuclear extract.
Because GC- or GT/CACC elements reportedly interact with several transcription factors (Sp proteins, ZBP-89, Erg-1, and Kruppel-like factors) that contain a DNA binding domain comprising three highly conserved Cys2His2 zinc fingers (6, 34, 39, 40, 50), we examined whether the addition of a zinc ion chelator (1,10-phenanthroline) to binding reaction mixtures containing NWB-S nuclear extract would abolish binding complex formation at the βMyHC C-richA element. The addition of 4 mM 1,10-phenanthroline to binding reactions abolished formation of complexes SC1, SC2, and SC3, which were reconstituted by the addition of 0.5 mM ZnCl2 (Fig. 3, lane 6 versus 7 and lane 7 versus10). Addition of 1,7-phenanthroline, 1,7-phenanthroline plus 0.5 mM ZnCl2, or vehicle (ethanol) to binding reaction mixtures did not alter binding complex formation (Fig. 3, lane 6 versus 8, 9, and 11). These data demonstrate that binding complexes SC1 to -3 formed at the βMyHC C-richA element are specific and contain zinc-dependent DNA-binding nuclear proteins. Moreover, formation of SC2 and SC3 at the C-richA element is highly enriched only when NWB-S nuclear extract is used.
Methylation interference footprinting identified a nucleotide binding site that was identical for both the zinc-dependent nuclear protein(s) and in vitro-synthesized Sp1.
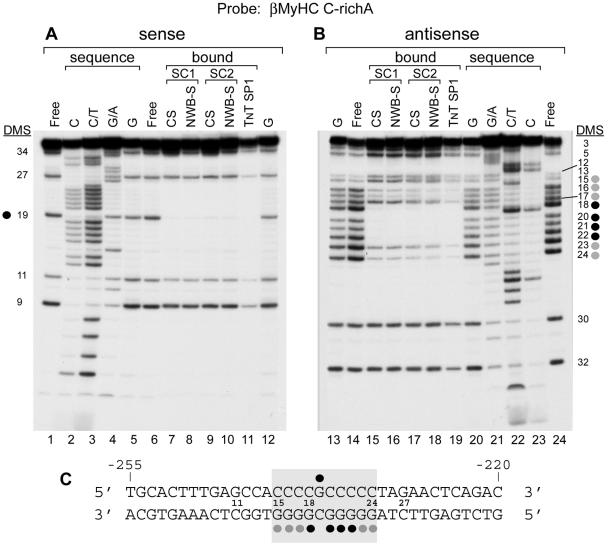
To identify the nucleotides within the human βMyHC C-richA element that interact with the zinc-dependent nuclear protein(s) comprising binding complexes SC1 and SC2 when either control- or NWB-S nuclear extract is used, we performed DMS interference footprinting analysis. DMS footprinting delineated a binding site on the sense strand of the 36-bp C-richA oligonucleotide that involved methylation of a single guanine residue (position 19) that strongly interfered with nuclear protein binding (Fig. 4A, lanes 7 to 11, and 3C). In contrast, DMS modification of the antisense strand distinguished a binding site that encompassed nucleotides −241 to −232, wherein methylation of guanine residues either partially (positions 15 to 17, 23, and 24) or strongly (positions 18 and 20 to 22) interfered with nuclear protein binding (Fig. 4B, lanes 15 to 19, and 3C). Our DMS interference footprint identified a binding site within the human βMyHC C-richA element that conforms to a consensus Sp protein element and revealed that the zinc-dependent nuclear protein(s) comprising SC1 and SC2 contacts the same nucleotides as in vitro-synthesized Sp1 protein whether CS or NWB-S nuclear extract was used. Thus, these data provide strong evidence in support of the notion that Sp proteins correspond to the zinc-dependent nuclear protein(s) comprising SC1 and SC2.
FIG. 4.
Methylation interference analysis delineated the specific nucleotides involved in protein-DNA interactions at the βMyHC C-richA element. (A and B) DMS interference footprinting of the C-richA element sense or antisense strand. Parallel reaction mixtures with the labeled probe were partially methylated with DMS. The modified probes were incubated in the presence of CS or NWB-S nuclear extracts or in vitro-synthesized Sp1 and resolved by preparative EMSA as described in Materials and Methods. The cleavage patterns of the bound (B) and free (F) probe are shown for the sense (lanes 6 to 12) and antisense (lanes 14 to 19) strands. Control sequences C, C/T, G/A, and G (lanes 2 to 5, 12, 13, and lanes 20 to 23) represent base-specific chemical cleavage of the unbound probe. The positions of the modified guanine (G) residues which either completely (black circles) or partially (gray circles) interfere with NWB-S factor(s) binding are shown on the side of each panel. (C) Summary of the DMS footprint formed by in vitro-synthesized Sp1 protein-, CS-, or NWB-S-interacting nuclear protein(s) at the C-richA element. The oligonucleotide numbering begins at the 5′ end of the sense strand. The probe extends from nucleotide −255 to −220 of the human βMyHC promoter. Black circles depict complete interference; gray circles depict partial interference.
Competition EMSA analysis provides evidence of SP protein binding to the C-richA element.
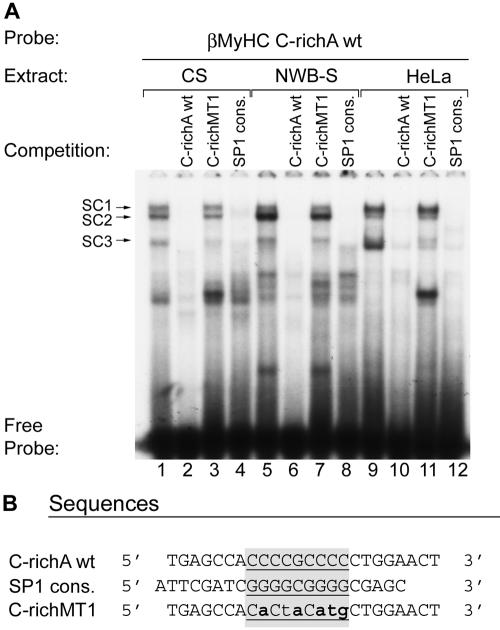
To further verify the specificity of binding complexes SC1 to -3, we performed competition EMSA analysis. Incubation of the double-stranded 32P-labeled human βMyHC C-richA probe with 4 μg of CS or NWB-S nuclear extract or 0.75 μg of HeLa nuclear extract revealed the formation of three specific complexes (SC1, SC2, and SC3) that were completely abolished by the addition of either 100-fold excess cold wild-type C-richA element or a consensus Sp1 element to the binding reaction mixture (Fig. 5A, lane 1 versus 2 and 4, lane 5 versus 6 and 8, and lane 9 versus 10 and 12; and B). In contrast, addition of 100-fold excess cold mutant C-richMT1 probe to binding reaction mixtures did not abolish the formation of complexes SC1 to -3 (Fig. 5A, lanes 3, 7, and 11). These data demonstrate that the binding complexes (SC1 to -3) formed at the human βMyHC C-richA element are specific whether CS, NWB-S, or HeLa nuclear protein is used and further suggest that these complexes comprise Sp proteins.
FIG. 5.
Competition EMSA analysis of sequence-specific protein-DNA interactions at the C-richA element. (A) Four micrograms of either CS (lanes1 to 4) or NWB-S (lanes 5 to 8) nuclear extract or 0.75 μg of HeLa nuclear extract was incubated in the presence of 20,000 cpm of the 32P-labeled C-richA wild type (lanes 1 to 12). For competition assays, the following nonradioactive competitor oligonucleotides were added to the reaction mixture at a 100-fold molar excess prior to the addition of the 32P-labeled C-richA probe: C-richAwt (lanes 2, 6, and 10), C-richMT1 (lanes 3, 7, and 12), and Sp1 consensus (cons.; lanes, 4, 8, and 12). (B) Alignment of sequences used in competition assays. The core elements are underlined and highlighted in gray. The mutated nucleotides are in bold lowercase. EMSAs were repeated with three independent batches of nuclear extract.
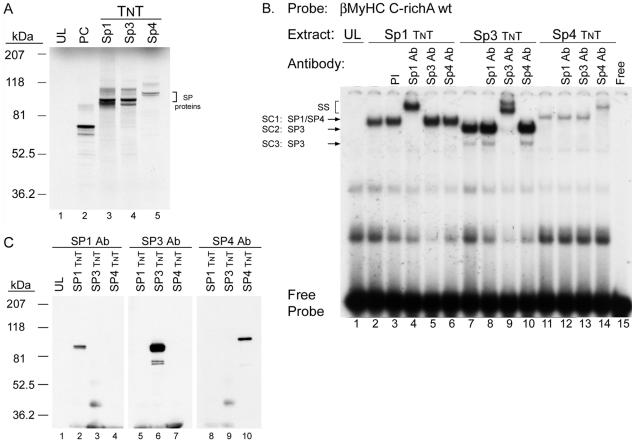
EMSA and Western blot analyses using in vitro-synthesized Sp proteins demonstrate specificity of Sp antibodies.
To determine whether the C-richA element binds Sp proteins, as indicated by our competition EMSA and footprinting analyses, we performed EMSA and Western analyses using in vitro-synthesized Sp1, Sp3, and Sp4 proteins; the most extensively studied Sp family members with commercially available antibodies (Fig. 6). In addition, given the high structural similarity between these Sp family members, this analysis allowed us to assess if cross-reactivity between anti-Sp1, anti-Sp3, and anti-Sp4 antibodies exists. A detectable binding complex did not form when the 32P-labeled human βMyHC C-richA element was incubated with 2 μl of UL, whereas a specific complex formed when reacted with 2 μl of either in vitro-synthesized Sp1, Sp3, or Sp4 (Fig. 6B, lane 1 versus 2, 7, and 11). The addition of PI serum did not alter specific binding complex formation when added to reaction mixtures containing the 32P-labeled human βMyHC C-richA element and 2 μl of in vitro-synthesized Sp1 (Fig. 6B, lane 2 versus 3). Importantly, a supershift formed only when anti-Sp1, anti-Sp3, or anti-Sp4 antibodies were added to binding reaction mixtures containing their cognate protein (Fig. 6B, lanes 4, 9, and 14). To further test the specificity of the Sp antibodies, we performed a Western blot analysis using in vitro-synthesized Sp1, Sp3, and Sp4 proteins. A detectable band was only observed by anti-Sp1, anti-Sp3, or anti-Sp4 antibodies in those lanes, wherein in vitro-synthesized Sp1, Sp3, or Sp4 protein had been electrophoretically fractionated, respectively (Fig. 6C, lanes 2, 6, and 10). The absence of antibody cross-reactivity observed in these analyses clearly demonstrates the specificity of each Sp antibody for its corresponding target protein.
FIG. 6.
EMSA analysis of the specific binding of in vitro-synthesized (TNT) Sp1, Sp3 and Sp4 proteins to C-richA binding. (A) One microliter of [35S]methionine-labeled TNT Sp1 (lane 3) and Sp3 (lane 4) and 2 μl of Sp4 (lane 5) were fractionated on 4 to 20% polyacrylamide Bis-Tris polyacrylamide gel electrophoresis gel to confirm the correct size of the proteins produced. Twice the amount (2 μl) of TNT Sp4 was loaded in lane 5 to demonstrate the low in vitro transcription and translation efficiency for Sp4, a common result observed for Sp4 expression, which is suspected to be an intrinsic property of the Sp4 protein sequence (13). The luciferase cDNA was programmed as a positive control (PC; lane 2), and the UL was used as a negative control (lane 1). (B) Antibody (Ab) EMSA analysis verifies specific binding of in vitro-synthesized Sp1 (lanes 2 to 7), Sp3 (lanes 7 to 10), and Sp4 (lanes 11 to 14) proteins to 32P-labeled C-richA oligonucleotide. A control reaction was performed with PI serum (lane 3). The Free lane represents the binding reaction mixture minus nuclear extract. SS, supershift. (C) Western blot analysis of in vitro-synthesized Sp1, Sp3, and Sp4 proteins using rabbit polyclonal Sp1 (1:1,000), Sp3 (1:1,000), and Sp4 (1:200) antibody to show their specificity. Anti-Sp1, -Sp3, and -Sp4 antibody only recognized in vitro-synthesized Sp1 (lane 2), Sp3 (lane 6), or Sp4 (lane 10), respectively. UL (lane 1) served as a control to show the absence of nonspecific cross-reactivity of Sp1, Sp3, and Sp4 antibody, respectively.
Sp protein family members Sp1, Sp3, and Sp4 represent the zinc-dependent nuclear protein(s) that interacts at the human βMyHC C-richA, -B, and -C elements.
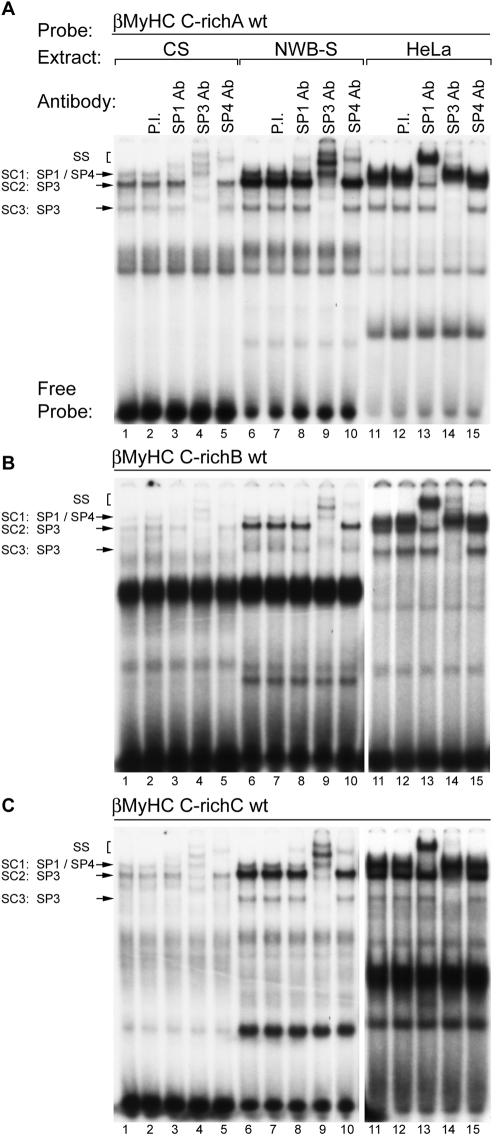
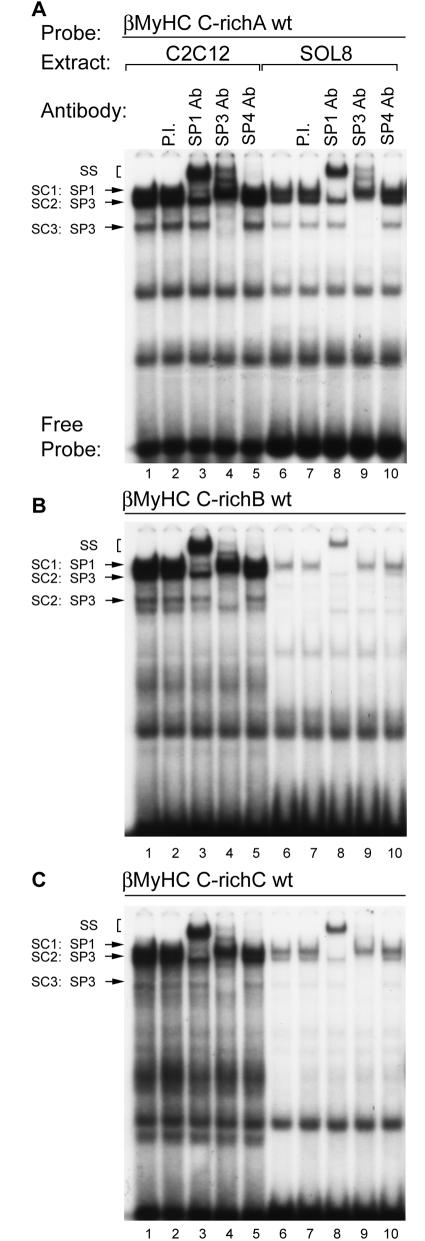
To determine if the Sp proteins comprise the zinc-dependent nuclear protein binding complexes formed at the human βMyHC C-richA element, we performed antibody EMSA analyses using polyclonal antibodies that specifically recognized either Sp1, Sp3, or Sp4 (Fig. 6 and 7A). Incubation of 32P-labeled human βMyHC C-richA oligonucleotide with 4 μg of CS nuclear extract revealed the formation of the three distinct low-mobility binding complexes, SC1, SC2, and SC3 (Fig. 7A to C, lane 1). A similar binding pattern was observed when 4 μg of NWB-S nuclear extract was used, except that binding complexes SC2 and SC3 were highly enriched (Fig. 7A, lane 1 versus 6). Binding complexes when either CS or NWB-S nuclear extract was used were not altered by addition of PI serum, while the addition of anti-Sp1 antibody to binding reaction mixtures produced a very-low-intensity supershift from complex SC1 (Fig. 7A, lane 2 versus 3 and lane 7 versus 8). The addition of increasing amounts of Sp1 antibody (1 to 4 μl) did not change this result (data not shown). Furthermore, the addition of Sp2, Erg-1, or ZBP-89 antibodies to binding reaction mixtures did not immunodeplete or supershift any of the binding complexes formed at either the C-richA, -B, or -C elements when CS or NWB-S nuclear extract was used (data not shown). Surprisingly, the addition of anti-Sp4 antibody produced a nearly complete supershift of complex SC1 at the C-richA element when CS or NWB-S nuclear extract was used (Fig. 7A, lane 1 versus 5 and lane 6 versus 10). The addition of anti-SP3 antibody to binding reaction mixtures containing CS or NWB-S nuclear extract resulted in a complete supershift or immunodepletion of only SC2 and SC3 (Fig. 7A, lane 1 versus 4 and lane 6 versus 9). It should be noted that a single 4.2-kb mRNA encodes three distinct Sp3 isoforms (110, 80, and 70 kDa); the two smaller Sp3 proteins are derived from two adjacent internal translation initiation sites. Therefore, in our EMSA analysis, it is likely that SC2 represents that full-length Sp3 protein while SC3 is likely to comprise both small Sp3 isoforms.
FIG. 7.
Antibody EMSA analysis identified Sp proteins as specific binding proteins interacting at the βMyHC C-richA, C-richB, and C-richC elements when using CS, NWB-S, or HeLa nuclear extract. 32P-labeled βMyHC C-richA, -B, or -C oligonucleotide probes were incubated with 4 μg of either CS (lanes 1 to 5) or NWB-S (lanes 6 to 10) nuclear extract or 0.75 μg of HeLa nuclear extract (lanes 11 to 15). Addition of anti-Sp1 antibody (Ab) to the binding reaction mixture containing CS or NWB-S nuclear extract resulted in a partial supershift (SS) of SC1 (lanes 3 and 8); addition of anti-Sp3 antibody resulted in a supershift of SC2 and SC3 (lanes 4 and 9); addition of anti-Sp4 antibody resulted in a nearly complete supershift of SC1 (lanes 5 and 10). In contrast, when using HeLa cell nuclear extract, anti-Sp1 antibody completely supershifted SC1 while addition of Sp4 antibody had no effect. Control reactions were performed with PI serum (lanes 2, 7, and 12). EMSAs were repeated with three independent batches of nuclear extract.
These experiments also included an evaluation of the binding properties of the human βMyHC C-richB and C-richC elements (Fig. 7B and C). Incubation of 32P-labeled human βMyHC C-richB or C-richC oligonucleotides with 4 μg of CS nuclear extract also revealed the formation of the three distinct low-mobility binding complexes, SC1, SC2, and SC3, whose intensity was considerably reduced compared to that of the complexes formed at the C-richA element (compare lanes 1 in Fig. 7A to C). Although the intensity of complexes SC1 to -3 formed at the C-richB and C-richC elements was weak when CS nuclear extract was used, binding complexes SC2 and SC3 were highly enriched when 4 μg of NWB-S nuclear extract was used in binding reactions (Fig. 7B and C, lane 1 versus 6). The addition of PI serum to binding reaction mixtures containing either CS or NWB-S nuclear extract did not alter complex formation, whereas the addition of anti-Sp1 antibody produced a very-low-intensity supershift, presumably from complex SC1 (Fig. 7B and C, lane 2 versus 3 and lane 7 versus 8). As observed for the C-richA element, the addition of anti-Sp4 antibody produced a complete supershift of complex SC1 when CS or NWB-S nuclear extract was used (Fig. 7B and C, lane 1 versus 5 and 6 versus 10). The addition of anti-SP3 antibody to binding reaction mixtures containing CS or NWB-S nuclear extract resulted in a complete supershift or immunodepletion of only SC2 and SC3 (Fig. 7B and C, lane 1 versus 4 and lane 6 versus 9).
A similar, but more intense binding pattern for SC1 to -3 was obtained when the 32P-labeled human βMyHC C-richA, -B, or -C oligonucleotides were incubated with 0.75 μg of HeLa nuclear extract (Fig. 7A to C, lanes 11 to 15). The addition of PI serum did not alter binding complex formation (Fig. 7A to C, lane 11 versus 12). Importantly, the addition of anti-Sp1 resulted in a complete supershift of complex SC1, while addition of anti-Sp4 antibody did not supershift or immunodeplete any binding complex (Fig. 7A to C, lane 11 versus 13 and lane 13 versus 15). The addition of anti-Sp3 antibody to binding reaction mixtures supershifted or immunodepleted only complexes SC2 and SC3 (Fig. 7A to C, lane 11 versus 14). These experiments conclusively identify Sp1, Sp3, and Sp4 as the zinc-dependent nuclear proteins comprising SC1 to -3 when CS, NWB-S, or HeLa nuclear extract was used. In addition, these experiments are the first to reveal the formation of a specific Sp4 binding complex when adult-stage skeletal muscle nuclear extract, but not HeLa nuclear extract, was used.
MOV results in increased Sp1 and decreased Sp3 protein binding at the βMyHC C-richA element.
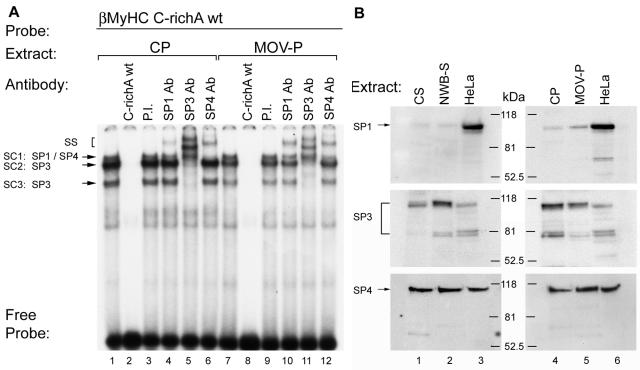
Our EMSA analysis has established increased binding of Sp3 isoproteins at the βMyHC C-richA, -B, and -C elements in response to NWB, a physiological stimulus associated with a slow-to-fast fiber-type shift and decreased βMyHC gene expression (5, 27, 28, 42). Thus, it was of interest to determine whether the Sp protein binding patterns at these three elements would differ in response to MOV, a stimulus associated with a fast-to-slow fiber-type switch and induced βMyHC expression (5, 41, 43-45). Incubation of 32P-labeled human βMyHC C-richA element with 4 μg of CP nuclear extract revealed the formation of specific complexes SC1, SC2, and SC3 (Fig. 8A, lane 1). Interestingly, when 4 μg of MOV-P nuclear extract was used, the intensity of complex SC1 did not appear to change; however, the intensity of specific complexes SC2 and SC3 markedly decreased compared to that of these complexes formed when CP nuclear extracts were used (Fig. 8A, lane 1 versus 7). Complex formation was completely abolished by the addition of 100-fold molar excess cold C-richA element to binding reaction mixtures containing either CP or MOV-P nuclear extract (Fig. 8A, lanes 2 and 8). The addition of PI serum did not alter binding complex formation when either CP or MOV-P nuclear extract was used (Fig. 8A, lanes 3 and 9). Interestingly, addition of anti-Sp1 antibody to binding reaction mixtures containing CP nuclear extract produced a very-low-intensity supershift which was enriched when MOV-P nuclear extract was used (Fig. 8A, lane 4 versus 10). Addition of anti-SP3 antibody to binding reaction mixtures containing CP or MOV-P nuclear extract produced a complete supershift or immunodepletion of complexes SC2 and SC3 (Fig. 8A, lanes 5 and 11). The addition of anti-Sp4 antibody produced a nearly complete supershift of complex SC1 when CP nuclear extract was used, whereas when MOV-P nuclear extract was used, only a partial supershift was formed (Fig. 8A, lane 6 versus 12). These data clearly demonstrate that in response to MOV, binding of Sp1 at the βMyHC C-richA element increased, while Sp3 binding decreased, a binding pattern that would favor increased βMyHC gene expression.
FIG. 8.
Antibody EMSA analysis of binding complexes formed at the βMyHC C-richA element with CP and MOV-P nuclear extract. (A) 32P-labeled βMyHC C-richA probe was incubated with 4 μg of either CP (lanes 1 to 6) or MOV-P (lanes 7 to 12) nuclear extract. For competition assays, nonradioactive competitor C-richA oligonucleotide was added to the binding reaction mixture at a 100-fold molar excess prior to the addition of the 32P-labeled C-richA probe (lanes 2 and 8). Addition of anti-Sp1 antibody (Ab) to the binding reaction mixture containing CP or MOV-P nuclear extract resulted in a partial supershift (SS) of SC1 (lanes 4 and 10); addition of anti-Sp3 antibody resulted in supershift of SC2 and SC3 (lanes 5 and 11); and addition of anti-Sp4 antibody resulted in a nearly complete supershift of SC1 (lanes 6 and 12). Control reactions were performed with PI serum (lanes 3 and 9). EMSAs were repeated with three independent batches of nuclear extract. (B) Sp protein expression pattern in CS, NWB-S, CP, and MOV-P. Western blot results are shown for rat CS and NWB-S nuclear extract (40 μg; lanes 1 and 2), CP and MOV-P nuclear extract (30 μg; lanes 4 and 5), and HeLa nuclear extract (30 μg; lanes 3 and 6), using rabbit polyclonal Sp1 (1:1,000), Sp3 (1:1,000), and Sp4 (1:200) antibody. HeLa extract served as a positive control for the presence of Sp1, Sp3, and Sp4 proteins.
Western analysis revealed increased differential regulation of Sp1 and Sp3 proteins in response to NWB and MOV.
Because our EMSA analysis revealed differential binding of the Sp proteins in response to NWB versus MOV, we performed a Western analysis to determine if these differences would be reflected by changes in nuclear Sp isoprotein levels (Fig. 8B). Using anti-Sp1 antibody, a 95- to 100-kDa band was detected in HeLa extract, which aligned with a barely detectable band when either CS or NWB-S nuclear extract was used (Fig. 8B, lanes 1 to 3). In contrast, anti-Sp1 antibody detected a low-intensity band with CP nuclear extract that was more intense than when MOV-P nuclear extract was used (Fig. 8B, lanes 4 to 6). Anti-Sp3 antibody detected bands at approximately 100, 80, and 78 kDa that were more intense when NWB-S versus CS nuclear extract was used (Fig. 8B, lanes 1 to 3). In contrast, these three bands reflected decreased intensity when comparing MOV-P versus CP nuclear extract (Fig. 8B, lanes 4 to 6). These data are significant because the greatest change in Sp3 isoprotein levels was found to occur for the 78- to 80-kDa isoforms, which have been shown to act as strong repressors of Sp protein-mediated gene transcription (20). When anti-Sp4 antibody was used, a band of approximately 110 kDa was detected with HeLa, CS, NWB-S, Cp, and MOV-P nuclear extract that did not reflect a difference in intensity between CS and NWB-S but did reflect a minor increase in intensity in MOV-P compared to that in CP (Fig. 8B, lanes 1 to 6). These data show clear changes in nuclear Sp isoprotein levels indicating that these proteins are regulated in response to NWB and MOV.
C2C12 and Sol8 nuclear extract show Sp1 but not SP4 binding at theβMyHC C-richA, -B, and -C elements.
Because C2C12 and Sol8 myotubes are often used as an in vitro model of muscle differentiation, most if not all studies concerned with Sp protein regulation of muscle-specific gene expression have been conducted with these cells. Since our current EMSA analyses using nuclear extract isolated from adult-stage skeletal muscle revealed Sp4 binding activity, it was important to determine whether nuclear extract isolated from these cells also contains Sp4 binding activity. Thus, we performed antibody EMSA analyses using polyclonal antibodies that recognized Sp1, Sp3, or Sp4 (Fig. 9). Incubation of 32P-labeled human βMyHC C-richA, C-richB, or C-richC elements with either C2C12 (1 μg) or Sol8 (1.5 μg) nuclear extract revealed the formation of complexes SC1, SC2, and SC3, which appeared as more intense bands when C2C12 nuclear extracts were used (Fig. 9A to C, lane 1 versus 6). The addition of PI serum to binding reaction mixtures did not alter complex formation (Fig. 9A to C, lanes 2 and 7), whereas addition of anti-Sp1 antibody resulted in the complete supershift of SC1 (Fig. 9A to C, lane 1 versus 3 and 8). Binding complexes SC2 and SC3 were supershifted or immunodepleted by the addition of anti-SP3 antibody to binding reaction mixtures containing either C2C12 or Sol8 nuclear extract (Fig. 9A to C, lane 1 versus 4 and 9). Importantly, the addition of anti-Sp4 antibody did not produce a detectable supershift or immunodepletion of complex SC1 when C2C12 or Sol8 nuclear extract was used (Fig. 9A to C, lane 1 versus 5 and 10). The absence of a specific Sp4 binding complex when C2C12 and Sol8 nuclear extract was used was not expected since our Western blot analysis detected the presence of nuclear Sp4 protein in nuclear extracts isolated from these cells (data not shown). Consistent with our EMSA analysis with CS nuclear extract, elements C-richB and C-richC did not interact strongly with the Sp proteins within nuclear extract isolated from Sol8 (a mouse soleus cell line) myotubes (Fig. 9B and C, lanes 6 to 10, and Fig. 7B and C, lanes 1 to 5).
FIG. 9.
Antibody EMSA analysis of binding complexes formed at the βMyHC C-richA element when using C2C12 or Sol8 nuclear extract. 32P-labeled βMyHC C-richA oligonucleotide probe was incubated with 0.75 μg of either C2C12 (lanes 1 to 5) or 1 μg of Sol8 (lanes 6 to 10) nuclear extract. Addition of anti-Sp1 antibody (Ab) to the binding reaction mixture containing C2C12 or Sol8 nuclear extract resulted in a complete supershift (SS) of SC1 (lanes 3 and 8), addition of anti-Sp3 antibody resulted in a supershift of SC2 and SC3 (lanes 4 and 9), and addition of anti-Sp4 antibody did not supershift any of the specific protein-DNA complex (lanes 5 and 10). Control reactions were performed with PI serum (lanes 2 and 7). EMSAs were repeated with three independent batches of nuclear extract.
Sp3 isoproteins act as competitive inhibitors of Sp1-mediated βMyHC reporter gene expression in Drosophila SL-2 and C2C12 myotubes.
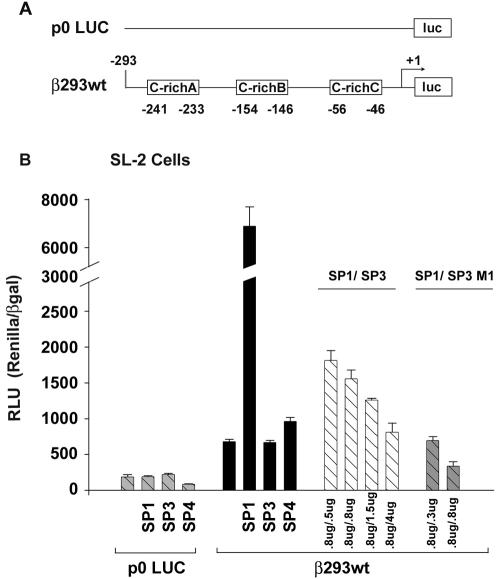
To determine the functional significance of Sp protein binding to the βMyHC C-richA, -B, and -C elements, we conducted transient expression assays in which Drosophila Sp protein expression vectors (pPacSp1, pPacSp3, and pPacSp4 [0.8 μg of each]) were cotransfected with the wild-type 293-bp βMyHC luciferase reporter gene (β293wt; 4 μg; Fig. 10). These experiments made use of Drosophila SL-2 cells since these cells lack Sp protein binding activity, thereby eliminating possible confounding effects as a result of endogenous SP proteins. The promoterless p0Luc plasmid (2 μg) was not highly expressed in Drosophila SL-2 cells, and it did not exhibit regulated expression in response to cotransfection with Sp1, Sp3, or Sp4 expression vectors (Fig. 10). The basal expression level of the wild-type β293wt reporter construct in Drosophila SL-2 cells was significantly higher (fourfold) than that of p0Luc (Fig. 10). Expression levels of the wild-type β293wt reporter construct increased 10- and 1.4-fold above basal levels when cotransfected with either Sp1 or Sp4 expression vectors, respectively, whereas cotransfection of Sp3 expression vector slightly decreased expression compared to basal levels (Fig. 10).
FIG. 10.
Transactivation of βMyHC promoter in Drosophila SL2 cells. The wild-type 293-bp βMyHC-Luc reporter gene (4 μg) was assayed with transient cotransfection assays in Drosophila SL2 cells with 0.8 μg each of Sp1, Sp3, Sp3/M1, and Sp4 expression vectors. All data were normalized to β-galactosidase (β-Gal) to accommodate variations in transfection efficiency. Data are reported as Renilla luciferase-normalized RLU (RLU/β-Gal ratio) and are expressed as the mean ± standard error (n = 10 for each reporter gene).
To determine if Sp1 transactivation of the wild-type β293wt reporter gene could be competitively inhibited by increasing amounts of Sp3 expression, we transfected Drosophila SL-2 cells with a constant amount of Sp1 expression vector (0.8 μg) and increasing amounts of either Sp3 (full length) or Sp3/M1 (an Sp3 isoform derived from an internal translation initiation site) (Fig. 10). As the amount of Sp3 was increased (from 0.5 to 4.0 μg), transcriptional activation of the wild-type β293wt reporter construct by Sp1 decreased approximately to basal expression levels (Fig. 10). Importantly, when Drosophila SL-2 cells were cotransfected with equivalent amounts of Sp1 (0.8 μg) and Sp3/M1 (0.8 μg) expression vectors, expression of the wild-type β293wt reporter gene decreased to approximately 50% of basal levels, demonstrating that Sp3/M1 acts as a potent negative regulator (Fig. 10).
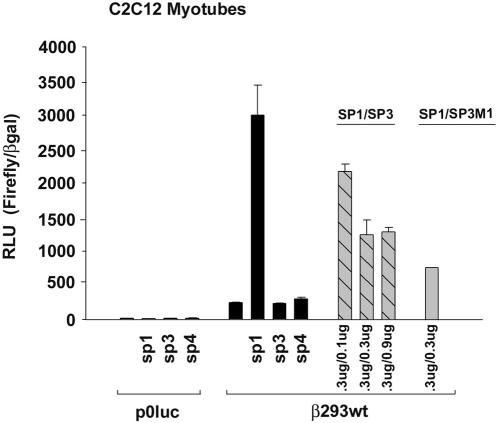
Because C2C12 myotubes express Sp1 and Sp3 isoproteins (data not shown), we used C2C12 cells as an in vitro muscle model system to determine whether altered levels of expressed Sp protein, as seen during NWB or MOV, would be effective in regulating the wild-type β293wt reporter gene. As seen when using Drosophila SL-2 cells, p0Luc (2 μg) expression in C2C12 myotubes was low and the cells did not exhibit regulated expression in response to cotransfection with Sp1, Sp3, or Sp4 (0.3 μg of each) expression vectors (Fig. 11). Basal expression of the wild-type β293wt reporter gene (2 μg) in C2C12 cells was 17-fold greater than that of p0Luc. Cotransfection with Sp1 increased wild-type β293wt expression 19-fold above basal levels, whereas cotransfection with either Sp3 or Sp4 expression vectors did not increase expression above basal levels (Fig. 11). When the wild-type β293wt reporter gene was cotransfected with a constant amount of Sp1 (0.3 μg) and increasing amounts of full-length Sp3 (0.1 and 0.9 μg) or an equivalent amount of Sp3/M1 (0.3 μg), Sp1-driven expression was significantly inhibited. Collectively, these data provide clear evidence that Sp3 isoproteins act as efficient competitive inhibitors of Sp1-mediated transcriptional activation of wild-type βMyHC reporter gene expression and support the notion that increased expression of Sp3 isoproteins under NWB conditions likely means these isoproteins function as negative regulators of βMyHC gene expression in adult skeletal muscle.
FIG. 11.
Transactivation of βMyHC promoter in C2C12 myotubes. The wild-type 293-bp βMyHC-Luc reporter gene was assayed with transient cotransfection assays in C2C12 myotubes with 0.3 μg each of the Sp1, Sp3, Sp3/M1, and Sp4 expression vectors. All data were normalized to β-galactosidase (β-Gal) to accommodate variations in transfection efficiency. Data are reported as luciferase-normalized RLU (Firefly/β-Gal ratio) and are expressed as the mean ± standard error (n = 10 for each reporter gene).
DISCUSSION
The inability to accurately mimic altered neuromuscular activity using cell lines has been a major limitation in understanding how skeletal muscles respond to changes in muscle activity. For example, although a discernible decrease in βMyHC gene expression represents an important functional alteration that occurs in skeletal muscle in response to decreased muscle activity, the mechanistic basis underlying this adaptation is poorly understood (5, 27, 28, 42). This study reports the novel finding that enriched binding of Sp3 proteins to three highly conserved GC-rich elements within the βMyHC proximal promoter is a critical event in down-regulation of βMyHC gene expression in response to NWB conditions. In addition, we provide the first evidence that Sp family proteins are differentially expressed in adult skeletal muscle as compared to permanent muscle cell lines, as exemplified by detection of an Sp4-DNA complex that forms when using adult skeletal muscle nuclear extract but not when using nuclear extracts obtained from various cell lines. These data suggest that the exact composition of Sp family members is an important determinant in modulating striated muscle phenotype, as well as the phenotype of other tissues influenced by conditions of altered weight bearing.
In this study, we have examined the role of three highly conserved and closely spaced GC/GT-rich elements in the βMyHC proximal promoter. Although classical GC/GT-rich elements have been shown to bind a variety of transcriptional regulators, Sp and XKLF-like family members, MNF, CBF-40, ZBP-89, HCB1, HCB2, and Erg-1 (3, 6, 10, 39, 40, 50), we provide multiple lines of evidence that, in adult skeletal muscle, these βMyHC GC-rich elements exclusively bind members of the Sp family of zinc finger transcription factors. First, our direct and competition EMSA results using control and NWB-S nuclear extracts revealed the formation of three specific binding complexes (SC1, SC2, and SC3) that were abolished in the presence of the metal ion chelator 1,10-phenanthroline. These protein-DNA complexes were reconstituted by the addition of ZnCl2. Second, the patterns of protection as determined by methylation interference footprinting analysis of the C-richA element were identical between adult skeletal muscle nuclear extract and in vitro-synthesized Sp1 protein. Third, in competition EMSAs, an oligonucleotide containing a consensus Sp1 binding site completely abolished complex formation at the C-richA element as efficiently as the wild-type C-richA element. Fourth, when using nuclear extracts from either control, NWB, or MOV muscle, antibody supershift analysis convincingly demonstrated that binding complexes SC1 to -3 were composed exclusively of Sp family proteins.
A novel finding in our results is that Sp family proteins are differentially expressed in adult skeletal muscle as compared to permanent muscle cell lines. Notably, our antibody EMSA analyses revealed the formation of an Sp4-specific binding complex when using adult skeletal muscle nuclear extract. The presence of an Sp4-specific complex was not anticipated since previous Northern blot and in situ hybridization analysis has shown that Sp4 is expressed at high levels primarily in the central nervous system (6, 12, 32, 38). Our results represent the first evidence in support of the notion that Sp4 regulates muscle-specific gene expression. In contrast, a specific Sp4 binding complex was not detected by EMSA analysis when using nuclear extract obtained from HeLa, C2C12, or Sol8 myotubes despite the fact that Western blot analysis detected the presence of Sp4 protein in these extracts (15; this work and data not shown). Because Sp1 levels are much higher in cell lines than adult skeletal muscle, the absence of a specific Sp4 binding complex when using extracts from cell lines likely reflects the ability of Sp1 to bind with higher affinity to GC/GT elements than Sp4. Our observation of a unique Sp4 binding complex cannot be attributed to Sp antibody cross-reactivity because anti-Sp4 antibody only recognized in vitro-synthesized Sp4 protein in EMSA and Western analysis, while anti-Sp1 and anti-Sp3 antibodies specifically recognized in vitro-synthesized Sp1 and Sp3, respectively. On the other hand, it is possible that other cell types (fibroblast, neural, smooth muscle, and epithelial) contained within adult skeletal muscle may have been the contributing source of Sp4 detected within our nuclear extract. While the latter is possible, it is unlikely since multinucleated myofibers constitute the majority of the cell mass comprising an adult skeletal muscle. In addition, Supp et al. (38) have shown Sp4 mRNA expression in adult striated muscle, a finding that is well matched with our detection of Sp4 protein in adult skeletal muscle. Thus, while numerous studies using permanent cell lines have attributed Sp family-mediated gene regulation to Sp1, our experimental findings using nuclear extracts from adult skeletal muscle indicate a role for Sp4 in muscle gene regulation (see reference 6 and the references cited within).
Consistent with the notion that Sp4 can activate gene expression in adult skeletal muscle, our transient expression assays using Drosophilia SL-2 cells, which are devoid of endogenous Sp protein activity, showed that Sp4 activated expression of the 293-bp βMyHC promoter. However, Sp4 did not activate the βMyHC promoter in C2C12 myotubes. Our findings showing differential cell-type-specific activation by Sp4 is similar to those of others (1, 13, 14, 23, 25, 48). In this regard, our Western blot analysis demonstrated that C2C12 myotubes express constitutively high levels of endogenous Sp1 and Sp3 proteins, a situation that favors binding site occupancy by Sp1 or Sp3 as opposed to Sp4 (6; data not shown). It may be that activation of gene expression by Sp4 will also require specific posttranslational modifications (phosphorylation, glycosylation, and sumoylation) that operate in adult skeletal muscle but are absent in cultured cells.
Another difference in Sp family proteins between adult skeletal muscle and myogenic cells in culture was increased levels of Sp3-specific complexes (SC2 and SC3) in nuclear extracts from adult skeletal muscle. Of particular importance to this study were our EMSA and Western blot analyses, which showed enriched binding of Sp3 proteins and an increase in the level of intensity of nuclear Sp3 protein, respectively, when nuclear extracts from NWB-S muscle were used. Mechanistically, it is noteworthy that there are three distinct Sp3 proteins (110, 80, and 70 kDa) that are encoded from a single 4.2-kb mRNA. The two smaller Sp3 proteins are derived from two adjacent internal translation initiation sites, and as a result, they display the same DNA binding site specificity as the full-length Sp3 isoform; however, because they lack the majority of the N-terminal transactivation domain, they display a greatly diminished transactivation potential and act primarily as potent competitive inhibitors of Sp-mediated gene transcription (20). Furthermore, full-length Sp3 has been reported to function as a negative transcriptional regulator in a promoter context-dependent manner and, in particular, when multiple adjacent GC-rich sites are present (14, 20, 21, 23, 49). This arrangement of GC-rich elements, which exist in the βMyHC proximal promoter studied herein, has been shown to support more stable binding of Sp3 and thus more efficient competition for binding site occupancy (49). In agreement with the latter notion, when Drosophila SL-2 cells and C2C12 myotubes were cotransfected with increasing amounts of either full-length Sp3 or the short Sp3 isoform (Sp3/M1), Sp1 transactivation of the 293-bp βMyHC reporter gene was significantly reduced. Collectively, these experiments provide compelling evidence supporting a role for Sp3 proteins in down-regulation of βMyHC gene expression in response to muscle inactivity.
To further examine the notion that Sp3 proteins mediate changes in muscle gene transcription in response to altered neuromuscular activity, we used the stimulus of MOV, which in contrast to NWB leads to increased βMyHC expression in both fast- and slow-twitch muscles (41, 43-45). Antibody EMSA analysis using nuclear extracts from overloaded plantaris muscles revealed a notable decrease in Sp3 protein binding activity, while the binding activity of Sp1 increased. This observation was mirrored by Western blot analysis, which showed a parallel decrease in Sp3 and increase in Sp1. The increase in Sp1 protein concurrent with decreased Sp3 protein would be expected to favor increased βMyHC expression. Consistent with this notion, our transient expression assays revealed that Sp1 was a potent transactivator of βMyHC transcription in both Drosophila SL-2 cells and C2C12 myotubes. The latter finding is consistent with a large body of evidence that establishes Sp1 as a potent activator of gene transcription and is consistent with findings that Sp1 plays an important regulatory role during hypertrophy of the heart, another striated muscle. For example, using a cell culture model of cardiomyocyte hypertrophy, an Sp1 element was shown to be a necessary requirement for the activation of a skeletal α-actin/reporter gene (19). Furthermore, in response to pressure overload-induced hypertrophy of the mouse heart, increases in both Sp1 binding activity and nuclear levels have been reported (36). Consistent with the latter report, we have also documented increased Sp1 binding activity at the C-richA element and an increase in nuclear Sp1 levels in the pressure-overloaded mouse heart (data not shown).
We propose that Sp family proteins function as part of a complex gene regulatory network involved in altered gene expression in response to altered skeletal muscle activity. In our studies, nuclear Sp3 protein levels and DNA binding activity increase in response to NWB conditions and are decreased in response to the MOV condition. Although our study is limited to the analysis of the βMyHC promoter, Sp3-mediated repression of gene expression may be more broadly relevant since Sp3 proteins are widely expressed across tissues and GC-rich elements appear in the control region of numerous genes. Additional work will be necessary to determine the scope of Sp3 protein participation in coordinate negative regulation of gene expression in response to various physiological stimuli. Mechanistically, it is conceivable that the participation of Sp proteins in the modulation of muscle phenotypes involves the selective recruitment of histone deacetylases (HDACs) and/or the release of histone acetyltransferases. This potential mode of regulation has been shown previously wherein the association of either Sp1 or Sp3 with HDACs resulted in transcriptional repression (6, 46, 47). Thus, in response to NWB activity, the increased levels of Sp3 protein would be expected to favor binding of Sp3 proteins and the likely recruitment of HDACs and other corepressors to form a multiprotein complex resulting in decreased βMyHC transcription. On the other hand, activation of βMyHC gene expression in response to MOV may involve the release of HDACs and the recruitment of histone acetyltransferases, Sp1, and other coactivator proteins. Our ongoing investigations of these mechanisms further characterize the role Sp family proteins play in directing muscle gene transcription and fiber-type conversions in response to various physiological stimuli.
Acknowledgments
This work was supported by Public Health Service grants AR41464 and AR47197 from the National Institute of Arthritis and Musculoskeletal and Skin Disease.
We thank Mark Hannink for critical review of the manuscript. We thank J. M. Horowitz, G. Suske, and R. Tijian for their generous sharing of all Sp family plasmids used in this study.
Footnotes
This article is dedicated to the memory of Gretchen L. Tsika.
REFERENCES
- 1.Ahlgren, R., G. Suske, M. R. Waterman, and J. Lund. 1999. Role of Sp1 in cAMP-dependent transcriptional regulation of bovine CYP11A gene. J. Biol. Chem. 274:19422-19428. [DOI] [PubMed] [Google Scholar]
- 2.Barany, M. 1967. ATPase activity of myosin correlated with speed of muscle shortening. J. Gen. Physiol. 5:197-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassel-Duby, R., M. D. Hernandez, Q. Yang, J. M. Rochelle, M. F. Seldin, and R. S. Williams. 1994. Myocyte nuclear factor, a novel winged-helix transcription factor under both developmental and neural regulation in striated myocytes. Mol. Cell. Biol. 14:4596-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, S. M., C. M. Schreiner, R. R. Waclaw, K. Campbell, S. S. Potter, and W. Scott. 2003. Sp8 is crucial for limb outgrowth and neuropore closure. Proc. Natl. Acad. Sci. USA 100:12195-12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booth, F. W., and K. M. Baldwin. 1996. Muscle plasticity: energy demand and supply processes, p. 1075-1123. In L. B. Rowell and J. T. Shepard (ed.), Handbook of physiology. Exercise: regulation and integration of multiple systems. American Physiological Society, Bethesda, Md.
- 6.Bouwman, P., and S. Philipsen. 2002. Regulation of the activity of Sp1-related transcription factors. Mol. Cell. Endocrinol. 195:27-38. [DOI] [PubMed] [Google Scholar]
- 7.Bouwman, P., H. Gollner, H. P. Elsasser, G. Eckhoff, A. Karis, F. Grosveld, S. Philipsen, and G. Suske. 2000. Transcription factor Sp3 is essential for post-natal survival and late tooth development. EMBO J. 19:655-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Carbtree, G. R., and E. N. Olson. 2002. NFAT signaling: choreographing the social lives of cells. Cell 109:S67-S79. [DOI] [PubMed] [Google Scholar]
- 10.Dellow, K. A., P. K. Bhavsar, N. J. Brand, and P. J. R. Barton. 2001. Identification of novel, cardiac-restricted transcription factors binding to a CACC-box within the human cardiac troponin I promoter. Cardiovasc. Res. 50:24-33. [DOI] [PubMed] [Google Scholar]
- 11.Gollner, H., C. Dani, B. Phillips, S. Philipsen, and G. Suske. 2001. Impaired ossification in mice lacking the transcription factor Sp3. Mech. Dev. 106:77-83. [DOI] [PubMed] [Google Scholar]
- 12.Gollner, H., P. Bouwman, M. Mangold, A. Karis, H. Braun, I. Rohner, A. D. Rey, H.-O. Besedovsky, A. Meinhardt, M. Van der Brock, T. Cutforth, F. Grosveld, S. Philipsen, and G. Suske. 2001. Complex phenotype of mice homozygous for a null mutation in the Sp4 transcription factor gene. Genes Cells 6:689-697. [DOI] [PubMed] [Google Scholar]
- 13.Hagen, G., J. Denning, A. Preiβ, M. Beato, and G. Suske. 1995. Functional analysis of the transcription factor Sp4 reveal properties distinct from Sp1 and Sp3. J. Biol. Chem. 270:24989-24994. [DOI] [PubMed] [Google Scholar]
- 14.Hagen, G., S. Muller, M. Beato, and G. Suske. 1994. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 13:3843-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagen, G., S. Muller, M. Beato, and G. Suske. 1992. Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res. 20:5519-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison, S. M., D. Houzelstein, S. L. Dunwoodie, and R. S. Beddington. 2000. Sp5, a new member of the Sp1 family, is dynamically expressed during development and genetically interacts with Brachyury. Dev. Biol. 227:358-372. [DOI] [PubMed] [Google Scholar]
- 17.Horsley, V., and G. K. Pavlath. 2002. NFAT: ubiquitous regulator of cell differentiation and adaptation. J. Cell Biol. 156:771-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karasseva, N., G. L. Tsika, J. Ji, A. Zhang, X. Mao, and R. W. Tsika. 2003. Transcription enhancer factor 1 binds multiple muscle MEF2 and A/T-rich elements during fast-to-slow skeletal muscle fiber type transitions. Mol. Cell. Biol. 23:5143-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karns, L. R., K. Kariya, and P. C. Simpson. 1995. M-CAT, CArG, and Sp1 elements are required for α1-adrenergic induction of the skeletal α-actin promoter during cardiac myocyte hypertrophy. J. Biol. Chem. 270:410-417. [DOI] [PubMed] [Google Scholar]
- 20.Kennett, S. B., A. J. Udvadia, and J. M. Horowitz. 1997. Sp3 encodes multiple proteins that differ in their capacity to simulate or repress transcription. Nucleic Acids Res. 25:3110-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennett, S. B., K. S. Moorefield, and J. M. Horowitz. 2002. Sp3 represses gene expression via the titration of promoter-specific transcription factors. J. Biol. Chem. 277:9780-9789. [DOI] [PubMed] [Google Scholar]
- 22.Knotts, S., H. Rindt, J. Neumann, and J. Robbins. 1994. In vivo regulation of the mouse β myosin heavy chain gene. J. Biol. Chem. 269:31275-31282. [PubMed] [Google Scholar]
- 23.Kwon, H.-S., M.-S. Kim, H. J. Edenberg, and M.-W. Hur. 1999. Sp3 and Sp4 can repress transcription by competing with Sp1 for the core cis-elements on the human ADH5/FDH minimal promoter. J. Biol. Chem. 274:20-28. [DOI] [PubMed] [Google Scholar]
- 24.Larkin, S. B., I. K. Farrance, and C. P. Ordahl. 1996. Flanking sequences modulate the cell specificity of M-CAT elements. Mol. Cell. Biol. 16:3742-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lerner, L. E., Y. E. Gribanova, L. Whitaker, B. E. Knox, and D. B. Faber. 2002. The rod cGMP-phosphodiesterase β-subunit promoter is a specific target for Sp4 and is not activated by other Sp proteins or CRX. J. Biol. Chem. 277:25877-25883. [DOI] [PubMed] [Google Scholar]
- 26.Marin, M., A. Karis, P. Visser, F. Grosveld, and S. Philipsen. 1997. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell 89:619-628. [DOI] [PubMed] [Google Scholar]
- 27.McCarthy, J. J., A. M. Fox, G. L. Tsika, L. Gao, and R. W. Tsika. 1997. β-MHC transgene expression in suspended and mechanically overloaded/suspended soleus muscle of transgenic mice. Am. J. Physiol. 272:R1552-R1561. [DOI] [PubMed] [Google Scholar]
- 28.McCarthy, J. J., D. R. Vyas, G. L. Tsika, and R. W. Tsika. 1999. Segregated regulatory elements direct β-myosin heavy chain expression in response to altered muscle activity. J. Biol. Chem. 274:14270-14279. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell-Felton, H., R. B. Hunter, E. J. Stevenson, and S. C. Kandarian. 2000. Identification of weight-bearing-responsive elements in the skeletal muscle sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA) gene. J. Biol. Chem. 275:23005-23011. [DOI] [PubMed] [Google Scholar]
- 30.Moorefield, K. S., S. J. Fry, and J. M. Horowitz. 2004. Sp2 DNA binding activity and trans-activation are negatively regulated in mammalian cells. J. Biol. Chem. 279:13911-13924. [DOI] [PubMed] [Google Scholar]
- 31.Nakashima, K., X. Zhou, G. Kunkel, Z. Zhang, J. M. Deng, R. R. Behringer, and B. Crombruggle. 2002. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108:17-29. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen-Tran, V. T., S. W. Kubalak, S. Minamisawa, C. Fiset, K. C. Wollert, A. B. Brown, P. Ruiz-Lozano, S. Barrere-Lemaire, R. Kondo, L. W. Norman, R. G. Gourdie, M. M. Rahme, G. K. Feld, R. B. Clark, W. R. Giles, and K. R. Chien. 2000. A novel genetic pathway for sudden cardiac death via defects in the transition between ventricular and conduction system cell lineage. Cell 102:671-682. [DOI] [PubMed] [Google Scholar]
- 33.Olson, E. N., and R. S. Williams. 2000. Calcineurin signaling and muscle remodeling. Cell 101:689-692. [DOI] [PubMed] [Google Scholar]
- 34.Rafty, L. A., and L. M. Khachigian. 1998. Zinc finger transcription factors mediated high constitutive platelet-derived growth factor-B expression in smooth muscle cells derived from aortae of newborn rats. J. Biol. Chem. 273:5758-5764. [DOI] [PubMed] [Google Scholar]
- 35.Rindt, H., J. Gulick, and J. Robbins. 1995. Segregation of cardiac and skeletal muscle-specific regulatory elements of the β-myosin heavy chain gene. Proc. Natl. Acad. Sci. USA 92:1540-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sack, M. N., D. L. Disch, H. A. Rockman, and D. P. Kelly. 1997. A role for Sp and nuclear receptor transcription factors in a cardiac hypertrophic growth program. Proc. Natl. Acad. USA 94:6438-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiaffino, S., and A. L. Serrano. 2002. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol. Rev. 76:371-432. [DOI] [PubMed] [Google Scholar]
- 38.Supp, D. M., D. P. Witte, W. W. Branford, E. P. Smith, and S. S. Potter. 1996. Sp4, a member of the Sp1-family of zinc finger transcription factors, is required for normal murine growth, viability, and male fertility. Dev. Biol. 176:284-299. [DOI] [PubMed] [Google Scholar]
- 39.Suske, G. 1999. The Sp-family of transcription factors. Gene 238:291-300. [DOI] [PubMed] [Google Scholar]
- 40.Swirnoff, A. H., and J. Milbrandt. 1995. DNA-binding specificity of NGFI-A and related zinc finger transcription factors. Mol. Cell. Biol. 15:2275-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsika, G. L., J. L. Wiedenman, L. Gao, J. J. McCarthy, K. Sheriff-Carter, I. D. Rivera-Rivera, and R. W. Tsika. 1996. Induction of β-MHC transgene in overloaded skeletal muscle is not eliminated by mutation of conserved elements. Am. J. Physiol. 271:C690-C699. [DOI] [PubMed] [Google Scholar]
- 42.Tsika, R. W., J. J. McCarthy, N. Karasseva, Y. Ou, and G. L. Tsika. 2002. Divergence in species and regulatory role of β-myosin heavy chain proximal promoter muscle-CAT elements. Am. J. Physiol. Cell Physiol. 283:C1761-C1775. [DOI] [PubMed] [Google Scholar]
- 43.Tsika, R. W., S. D. Hauschka, and L. Gao. 1995. M-creatine kinase gene expression in mechanically overloaded skeletal muscle of transgenic mice. Am. J. Physiol. 269:C665-C674. [DOI] [PubMed] [Google Scholar]
- 44.Vyas, D. R., J. J. McCarthy, and R. W. Tsika. 1999. Nuclear protein binding at the β-myosin heavy chain A/T-rich element is enriched following increased skeletal muscle activity. J. Biol. Chem. 274:30832-30842. [DOI] [PubMed] [Google Scholar]
- 45.Vyas, D. R., J. J. McCarthy, G. L. Tsika, and R. W. Tsika. 2001. Multiprotein complex formation at the β-myosin heavy chain distal muscle CAT element correlates with slow muscle expression but not mechanical overload responsiveness. J. Biol. Chem. 276:1173-1184. [DOI] [PubMed] [Google Scholar]
- 46.Won, J., J. Yim, and T. K. Kim. 2002. Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoter in normal human somatic cells. J. Biol. Chem. 277:38230-38238. [DOI] [PubMed] [Google Scholar]
- 47.Xiao, H., T. Hasegawa, and H.-I. Isobe. 2000. p300 collaborates with Sp1 and Sp3 in p21waf1/cip1 promoter activation induced by histone deacetylase inhibitor. J. Biol. Chem. 275:1371-1376. [DOI] [PubMed] [Google Scholar]
- 48.Yan, S., I. M. Berquin, B. R. Troen, and B. F. Sloane. 2000. Transcription of human cathepasin B is mediated by Sp1 and Ets family factors in glioma. DNA Cell Biol. 19:79-91. [DOI] [PubMed] [Google Scholar]
- 49.Yu, B., P. K. Datta, and S. Bagchi. 2003. Stability of the Sp3-DNA complex is promoter-specific: Sp3 efficiently competes with Sp1 for binding to promoters containing multiple Sp-sites. Nucleic Acids Res. 31:5368-5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, X., I. H. Diab, and Z. E. Zehner. 2003. ZBP-89 represses vimentin gene transcription by interacting with the transcriptional activator, Sp1. Nucleic Acids Res. 31:2900-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]