Abstract
Tree shrews have a close relationship to primates and have many advantages over rodents in biomedical research. However, the lack of gene manipulation methods has hindered the wider use of this animal. Spermatogonial stem cells (SSCs) have been successfully expanded in culture to permit sophisticated gene editing in the mouse and rat. Here, we describe a culture system for the long-term expansion of tree shrew SSCs without the loss of stem cell properties. In our study, thymus cell antigen 1 was used to enrich tree shrew SSCs. RNA-sequencing analysis revealed that the Wnt/β-catenin signaling pathway was active in undifferentiated SSCs, but was downregulated upon the initiation of SSC differentiation. Exposure of tree shrew primary SSCs to recombinant Wnt3a protein during the initial passages of culture enhanced the survival of SSCs. Use of tree shrew Sertoli cells, but not mouse embryonic fibroblasts, as feeder was found to be necessary for tree shrew SSC proliferation, leading to a robust cell expansion and long-term culture. The expanded tree shrew SSCs were transfected with enhanced green fluorescent protein (EGFP)-expressing lentiviral vectors. After transplantation into sterilized adult male tree shrew's testes, the EGFP-tagged SSCs were able to restore spermatogenesis and successfully generate transgenic offspring. Moreover, these SSCs were suitable for the CRISPR/Cas9-mediated gene modification. The development of a culture system to expand tree shrew SSCs in combination with a gene editing approach paves the way for precise genome manipulation using the tree shrew.
Keywords: tree shrew, spermatogonial stem cells, Wnt/β-catenin signaling, Sertoli cells, gene editing
Introduction
The tree shrew (Tupaia belangeri) is widely distributed in South Asia, Southeast Asia, and Southwest China. It belongs to the order Scandentia and shares a high degree of similarity to primates in many of its molecular and physiological aspects1,2,3. Moreover, it has a small adult body size, a high brain-to-body mass ratio, a short reproductive cycle and life span, and a low cost of maintenance. Because of these advantageous features, the tree shrew has long been considered as a promising experimental animal in biomedical research and drug safety testing3,4. In particular, the tree shrew shows numerous advantages over rodents in studying hepatitis C virus5,6,7 and hepatitis B virus infections8,9, coxsackievirus A16 infection10, Mycobacterium tuberculosis infection11, visual system12,13,14, myopia15,16, stress response17, social stress and depression18,19, drug addiction20,21, learning behaviors22,23, and aging24. The tree shrew is also used to study cancers3,25 and metabolic diseases26,27. Importantly, recent release of a high-quality tree shrew genome has underscored its close relationship to primates1 and the potential as a useful alternative to high-order non-human primates such as old-world monkeys.
Despite the tree shrew having been used in biomedical research for several decades, it is not used as widely as once expected. One reason lies in the lack of practical gene manipulation techniques. In mammals, germline gene manipulation can be achieved by editing the genome in embryonic stem cells with germline transmission competence, in one-cell embryos or in spermatogonial stem cells (SSCs). To date, little information is available on the reproductive biology and assisted reproductive technologies in the tree shrew28; and gene editing methods using one-cell embryos or embryonic stem cells have hitherto been unsuccessful.
SSCs maintain spermatogenesis throughout the reproductive lifespan of males via life-long self-renewal and differentiation in vivo29. Under suitable conditions, SSCs can be stably expanded for many generations while maintaining stem cell properties of self-renewal and differentiation into sperm following transplantation into recipients29. This in vitro propagation of SSCs has been achieved in the mouse30, the rat31, and the human32. SSCs have also been used for sophisticated gene editing in the mouse and the rat33,34,35,36. Here, we report for the first time the development of a culture conditions for the propagation of tree shrew SSCs in vitro and the generation of transgenic tree shrew using these SSCs. The establishment of a SSC-based tree shrew transgenic platform will boost the wider application of the tree shrew in biomedical research and thus increase our understanding of human diseases by utilizing transgenic tree shrew as an animal model.
Results
Thymus cell antigen 1 cell surface marker can be used to enrich tree shrew SSCs
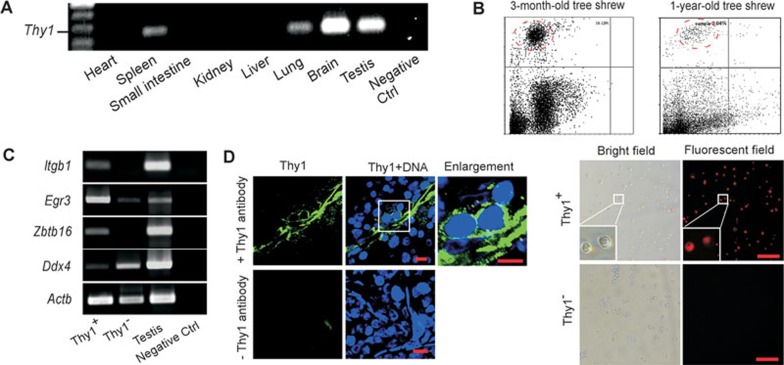
Previous studies have reported the expression of several cell surface markers in undifferentiated spermatogonia is conserved between rodents and human37,38. We therefore looked to see if one of them, thymus cell antigen 1 (Thy1) (also known as Cd90), is expressed in tree shrew SSCs and could be used to enrich SSCs. We designed PCR primers to amplify a fragment of Thy1 transcript according to the genome sequence of tree shrew1 and found that expression of Thy1 transcript could be detected in mRNA sample of tree shrew testis (Figure 1A). We then obtained a commercial antibody, which recognizes Thy1 in both rodents and primates. With this antibody, a small proportion of Thy1+ cells was reproducibly (five repeats) isolated from single-cell suspensions prepared from either pre-pubertal (about 3-month old) or adult (about 1-year old) tree shrew testicular tissues by fluorescence-activated cell sorting (FACS) (Figure 1B). These Thy1+ cells have a diameter of about 10 μm, similar to mouse SSCs (Figure 1B). RT-PCR analysis of gene expression in Thy1+ and Thy1− cell populations revealed that Egr3, Zbtb16, and Itgb1, several SSC markers38, were prominently expressed in Thy1+ cells, whereas the germ cell differentiation marker Ddx4 was predominantly expressed in Thy1− cells (Figure 1C). Also, immunostaining of the frozen testicular tissue sections showed that Thy1+ cells were localized at the basement membrane of the seminiferous tubules (Figure 1D), where the SSCs reside. Collectively, these results suggest that a tree shrew Thy1+ population contains SSCs and Thy1 can be used to prepare an enriched SSC population.
Figure 1.
Cell surface marker Thy1 can be used to enrich tree shrew spermatogonial stem cells (SSCs). (A) RT-PCR detected Thy1 mRNA expression in tree shrew testis, brain, lung, and spleen. No amplification was detected in the heart, small intestine, kidney, liver, and negative control (negative Ctrl) in which template cDNA was omitted. (B) Thy1+ cell population was reproducibly obtained by FACS of a testicular tissue single-cell suspension from prepubertal (3 month, ∼16%, n = 5) and adult (1 year, ∼3%, n = 5) tree shrews (upper panel). Lower panel show the morphology of sorted Thy1+ and Thy1− cells in fluorescent and bright fields (scale bar, 100 μm). (C) mRNA of several SSC markers (Itgb1, Egr3, and Zbtb16) was prominently expressed in Thy1+ cells, whereas Ddx4 was highly expressed in Thy1− cells. Negative Ctrl was included in which template cDNA was omitted. Experiments were repeated three times with independent biological samples. (D) Immunostaining with Thy1 antibody detected the presence of Thy1+ cells at the basement membrane of the seminiferous tubules. DNA was counterstained with DAPI. Immunostaining by omitting Thy1 antibody was used as negative Ctrl (scale bar, 10 μm). Experiments were repeated three times with independent biological samples.
RNA-sequencing analysis reveals Wnt/β-catenin signaling potentially regulates tree shrew SSC self-renewal
To culture SSC-enriched Thy1+ cells, we initially adopted a mouse SSC in vitro propagation system30,39 and made slight modifications (see the details in Materials and Methods). However, the initial culture conditions were unable to support tree shrew SSC self-renewal and survival. Thy1+ cells could only be sustained in culture for a short period and without obvious proliferation (about 2-3 passages, 1 week for each passage). In order to optimize the culture conditions, we attempted to understand by high-throughput RNA sequencing the molecular properties of tree shrew SSCs. We set up two experiments to compare the gene expression profiles between SSCs and differentiated germ cells (Supplementary information, Figure S1). In the first experiment, gene expression profiles were compared between SSC-enriched Thy1+ cells and differentiating germ cell-enriched Thy1− cells freshly isolated from the testis. In the second experiment, the comparisons were made among the successive passages of SSC-enriched cells cultured under inefficient conditions.
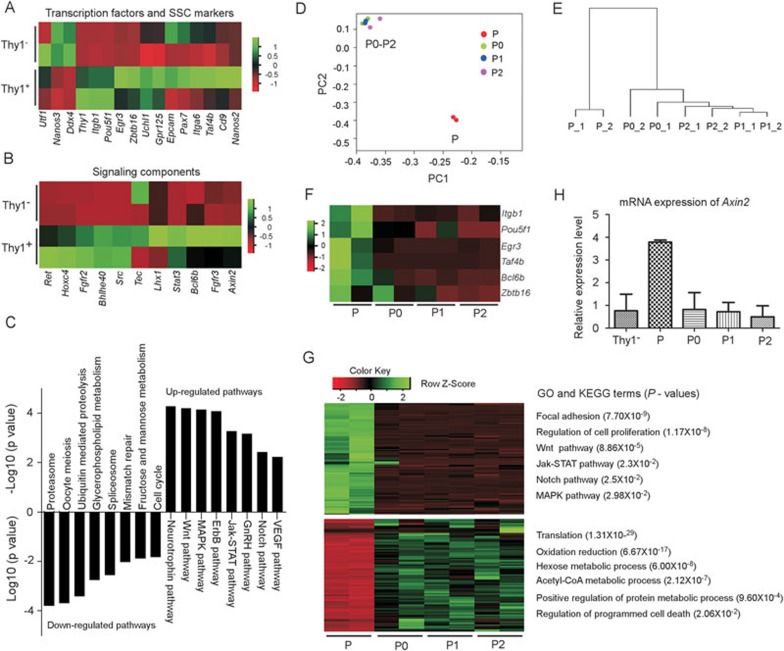
Single-cell suspensions of tree shrew testis were sorted into Thy1+ and Thy1− populations, in which SSCs and differentiating germ cells were enriched, respectively. Using the Illumina sequencing platform, we generated 48.66 million reads with high quality and an average of 12.17 million reads per sample. Genes (number of alignments over 10) were subject to differential gene expression analysis (|log2 (fold change)| ≥ 1 and FDR ≤ 0.05). A total of 2 024 and 3 870 differentially expressed genes (DEGs) were upregulated and downregulated in Thy1+ cells compared with Thy1− cells, respectively (Supplementary information, Table S1). Consistent with the stem cell identity, Thy1+ cells expressed higher levels of several well-known SSC markers (e.g., Thy1, Itgb1, Egr3, Uchl1, Gpr125, Cd9, and Pou5f1), transcription factors essential for SSC self-renewal (Zbtb16, Taf4b)29,38 (Figure 2A), and the key components of several signaling pathways (e.g., GDNF, FGF, Wnt, and Jak-Stat pathways) that were required for SSC proliferation and self-renewal29,30 (Figure 2B). Conversely, the differentiating germ cell-expressed gene Ddx440 was upregulated in Thy1− cells (Figure 2A). However, the undifferentiated spermatogonia markers reported in other species (e.g., Utf1, Nanos2, Nanos3, Epcam, and Itga6)29,37,38 were unable to mark the tree shrew SSCs (Figure 2A). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs uncovered eight signaling pathways that were significantly (P-value 0.01) enriched for genes highly expressed in Thy1+ cells (Figure 2C). The top pathways were the neurotrophin signaling pathway (GDNF signaling pathway) followed by the canonical Wnt (Wnt/β-catenin) pathway. GDNF signaling is well recognized as a highly conserved pathway necessary for SSC self-renewal in rodents and primates29,41. Consistently, key components of these pathways were predominantly expressed in Thy1+ cells (Figure 2B).
Figure 2.
RNA sequencing analyses reveal the potential signaling pathways activated in tree shrew undifferentiated SSCs. (A) Relative expression of some SSC marker genes and transcription factors in Thy1+ and Thy1− cells isolated by FACS. (B) Relative expression of some key components of signaling pathways including GDNF, Wnt/β-catenin, FGF2, and Jak-STAT in Thy1+ and Thy1− cells sorted by FACS. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed the signaling pathway enrichments for genes that were upregulated and downregulated in Thy1+ cells. (D) Principle component analysis (PCA) revealed that primary Thy1+ cells (P) were separated far from Thy1+ cells cultured for different passages (P0-P2), whereas P0-P2 cells were clustered together. (E) Unbiased clustering analysis of primary (P) and cultured Thy1+ cells at differential passages (P0-P2). (F) Relative expression of several marker genes and transcription factors in primary Thy1+ cells (P) and Thy1+ cells cultured for different passages (P0-P2). (G) Gene Ontology (GO) terms and KEGG analyses revealed the terms and signaling pathways enriched for genes downregulated or upregulated during culture of Thy1+ cells (P and P0-P2). (H) Quantitative RT-PCR revealed the relative mRNA expression levels of Axin2 in primary Thy1+ (P) and Thy1− cells, and in Thy1+ cells cultured for different passages (P0, P1, and P2). Experiments were repeated three times with independent biological samples.
We next investigated the gene expression changes along with the loss of SSC identity in culture. We collected P0 (1-week culture, passage 0), P1 (2-week culture, passage 1), and P2 (3-week culture, passage 2) cells and performed whole-genome RNA sequencing as described above. DEGs among the four cell types (fresh Thy1+ cells without culture (P), P0, P1, and P2) (Supplementary information, Table S2) were subject to principle component analysis (PCA) and unsupervised clustering. Both analyses revealed that P0-P2 samples clustered together, whereas P sample was separated far from the P0-P2 (Figure 2D and 2E). This pattern showed that the stem cell identity of tree shrew SSCs was lost shortly upon the initial culture of Thy1+ cells (P0 stage). The transcription factors required for SSC self-renewal (e.g., Zbtb16, Taf4b, and Bcl6b) and some tree shrew SSC markers (e.g., Itgb1, Egr3, and Pou5f1) were sharply downregulated from P to P0 (Figure 2F). Gene Ontology (GO) and KEGG term analyses revealed that many downregulated genes during culture were significantly enriched in the regulation of cell proliferation, Wnt/β-catenin, Jak-STAT, Notch, and MAPK signaling pathways, whereas upregulated genes were enriched in the apoptosis signaling pathway (Figure 2G). Collectively, these analyses suggested that several important signaling pathways including the Wnt/β-catenin were active in undifferentiated spermatogonia cells. Of note, real-time PCR validated that Axin2, which is a Wnt target gene and serves as a reporter of ongoing Wnt/β-catenin signaling42,43, was expressed at a higher level in primary Thy1+ cells (P) than in Thy1− cells or Thy1+ cells cultured in sub-optimal conditions (P0-P2) (Figure 2H). Because the Wnt/β-catenin signaling is able to activate the STAT and Notch signaling pathways44,45, we proposed that the Wnt/β-catenin pathway might have a primary role in maintaining tree shrew SSC identity. Consistently, in vivo self-renewal of mouse SSCs requires Wnt/β-catenin signaling and the presence of niche Sertoli cells, which can secrete Wnt6 to activate this pathway46.
Long-term propagation of tree shrew SSCs in culture
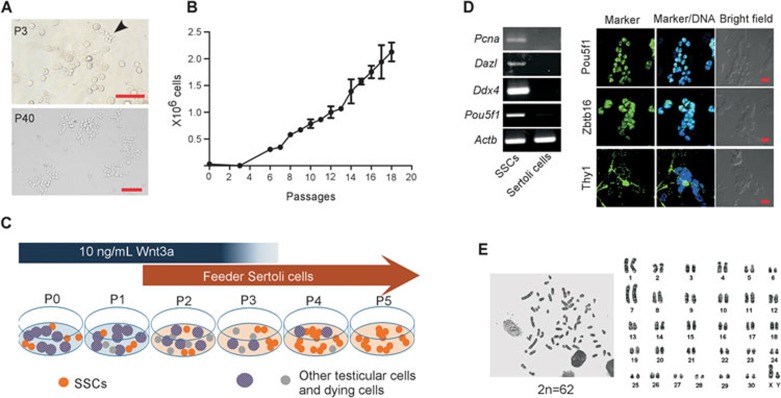
On the basis of above analyses, we tested the possible roles of the Wnt/β-catenin signaling pathway on tree shrew SSC self-renewal in culture. To reduce the harm caused to cells by FACS, we used immunomagnetic isolation, which is gentler and can efficiently harvest Thy1+ cells. Because some somatic cells exist in the Thy1+ population, we plated Thy1+ cells in high density (105 cells/cm2) without feeder in a 24-well plate. Cells were treated with vehicle or 10 ng/mL recombinant Wnt3a, a ligand for the canonical Wnt signaling, and passaged once a week. At P0 and P1, no obvious cell proliferation or cell clumping was observed with or without Wnt3a exposure. At P2, we started to plate round germ cells (5 × 104 cells/cm2) on feeder cells. Different types of feeder cells are employed in culture of mammalian SSCs. For instance, mouse embryonic fibroblasts (MEFs) can support mouse SSC growth and propagation39, whereas the expansion of rat SSCs in culture is better supported by Sertoli cells31, the only neighboring cells of SSCs and able to function as a niche47. We then examined which feeder cell type is suitable for tree shrew SSC expansion in culture. Tree shrew Sertoli cells were prepared according to a previous report48 and the cell identity was verified by the expression of several marker genes including Vim and Sgp147,49 (Supplementary information, Figure S2). Beginning at P2, SSCs were plated on either mitotically inactivated MEFs or tree shrew Sertoli cells for continuous culture in the presence or absence of 10 ng/mL Wnt3a (Table 1). At P3, some cell clumps containing 5-9 cells with typical morphology of undifferentiated spermatogonia cells (round with ∼10 μm in diameter) appeared in the culture supplied with 10 ng/mL Wnt3a and Sertoli cells (Figure 3A and Supplementary information, Figure S3). Meanwhile, most germ cells enlarged, floated, and degenerated at the end of P3. After passage, these stem cell-like cells continued to proliferate and could be expanded and passaged for more than 50 generations (Figure 3A). Their propagation dynamics is shown in Figure 3B. In sharp contrast, the other culture conditions failed to efficiently support the propagation of SSCs in vitro, although success was occasionally obtained in the six trials in culture supplied with Sertoli cells but without Wnt3a (Table 1 and Supplementary information, Figure S3). Of note, we found that Wnt3a was unnecessary to the culture once the SSCs had shown robust proliferation from P4, and later. Collectively, these data demonstrate that Wnt3a is necessary at the initial stage of culture to facilitate the survival of SSCs, and tree shrew Sertoli cells are critical for the support of SSC expansion.
Table 1. Influence of different culture conditions on tree shrew SSC proliferation.
| Feeder cells | Success rate1 |
||
|---|---|---|---|
| Vehicle | Wnt3a (10 ng/mL) | Wnt3a (20 ng/mL) | |
| MEFs | 0/12 | 0/3 | N.A.2 |
| Sertoli cells | 2/6 | 8/8 | 0/3 |
1Definition of success rate: numbers of success/numbers of total test. Observation of obvious SSC colony formation by passage 5 is considered as success.
2N.A. = Not applicable.
Figure 3.
Establishment of tree shrew spermatogonial stem cell lines. (A) Thy1+ cells were cultured in the presence of 10 ng/mL Wnt3a and Sertoli cell feeder. Images show the appearance of germ cell clumps at passage 3 (P3, upper panel, arrow head), and stem cell colonies at P40 (lower panel) (scale bar, 100 μm). (B) Propagation dynamics of three lines of tree shrew SSCs in culture. Data are represented as mean ± SEM. (C) Schematic illustration of the culture system for tree shrew SSC expansion. (D) RT-PCR and immunostaining revealed the expression of several germ cell markers and SSC markers in long-term expanded tree shrew SSCs (scale bar, 20 μm). (E) Long-term expanded SSCs contain normal chromosome number (2n = 62).
We next examined whether higher dose of Wnt3a can boost SSCs to initiate proliferation earlier or to form more colonies. We compared the effects of 10 and 20 ng/mL Wnt3a on the initial culture of SSCs (P0-P5). Surprisingly, 20 ng/mL Wnt3a did not show any beneficial effect on SSC survival or initiation of colony formation. Cells cultured in 20 ng/mL Wnt3a degenerated at P3-P4 and no obvious SSC proliferation was observed (Table 1).
In summary, we developed culture conditions for tree shrew SSC expansion in vitro (Figure 3C) and established three tree shrew SSC lines, which have been passaged for more than 50 generations and retain the expression of several typical germ cell markers and SSC markers (Figure 3D). Tree shrew cells have 62 chromosomes (2n = 62)50 and G-band karyotyping analysis showed that cultured SSCs maintain normal karyotype after >30 passages of propagation (Figure 3E). Moreover, these SSCs can proliferate normally after cryopreservation and thawing.
Long-term expanded tree shrew SSCs retain full spermatogenesis potential
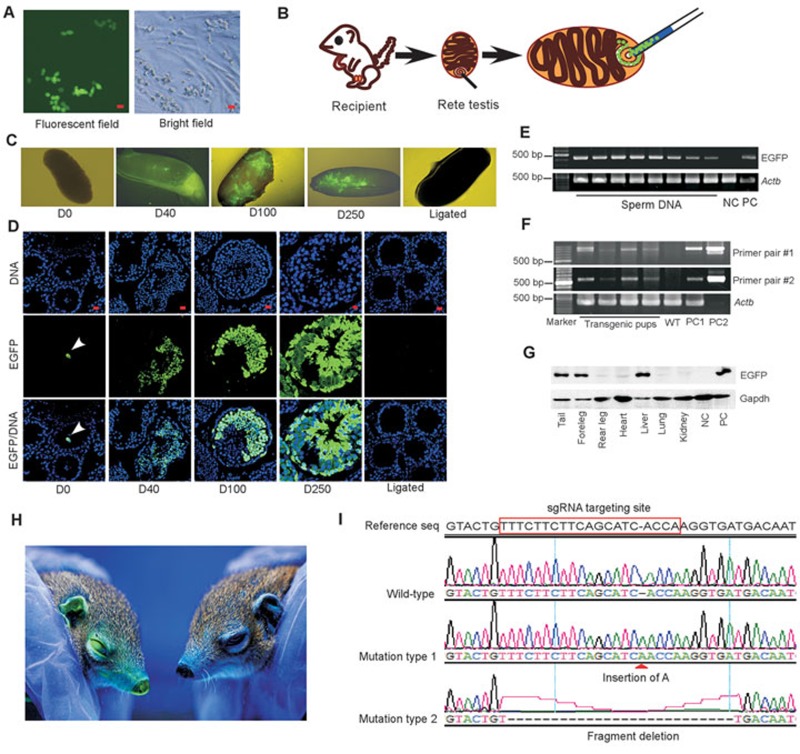
To test whether these propagated SSCs retain their stem cell characteristics, i.e., self-renewal and spermatogenesis potentials, we infected SSCs at P15 with EGFP-expressing lentivirus51. Following viral infection and 1-week culture, EGFP+ SSCs were purified by FACS and propagated to obtain cells stably expressing EGFP in culture (Figure 4A). These EGFP-SSCs were transplanted into seminiferous tubules of recipient adult males52 (Figure 4B), which had been sterilized by pre-treatment with busulfan (40 mg/kg body weight) to destroy pre-existing germ cells (Supplementary information, Figure S4). Recipient testes were collected and examined immediately after transplantation (day 0, D0), or at 40 days (D40), 100 days (D100), and 250 days (D250) post transplantation. At D0, no EGFP expression was observed in the entire testis under fluorescent dissection microscope. Examination of testicular tissue sections by immunostaining with GFP antibody showed that rare EGFP+ cells were suspended in the lumen of seminiferous tubules, but no EGFP+ cell was detected within the tubules. At D40, green seminiferous tubules were clearly visible. The colonization of EGFP-SSCs at the basement membrane of the seminiferous tubules and the presence of EGFP-tagged differentiating germ cells within the tubules were confirmed by confocal microscopy. At D100, more green seminiferous tubules were detected and all stages of green spermatogenic cells were observed. The spermatogenesis from EGFP-SSCs persisted at D250. In contrast, no green germ cell was detected in ligated control testis (Figure 4C and 4D). To verify that the transplanted EGFP-SSCs can generate transgenic sperm, we collected sperm from epididymis at D250 and examined the insertion of EGFP transgene in the sperm genome by PCR. Indeed, EGFP fragment was successfully amplified from DNA samples prepared from the sperm, and the sequence was confirmed by sequencing PCR products (Figure 4E). Collectively, these results demonstrate that the long-term expanded tree shrew SSCs retain self-renewal and spermatogenesis abilities after transplantation.
Figure 4.
Expanded tree shrew SSCs are capable of normal spermatogenesis and suitable for CRISPR-/Cas9-mediated gene editing. (A) Tree shrew SSCs transfected with lentiviral vectors stably expressed EGFP (scale bar, 20 μm). (B) Schematic illustration of SSC transplantation. (C) Recipient testes examined by fluorescent dissection microscopy at 0 day (D0), 40 days (D40), 100 days (D100), and 250 days (D250) post transplantation of EGFP-SSCs. Note that green seminiferous tubules were clearly visible at D40, D100, and D250. Testis ligated for 100 days was set as negative control (NC). (D) Recipient testicular tissue sections were examined by immunostaining for the expression of EGFP. At D0, no EGFP+ cells were detected within the seminiferous tubules and rare EGFP+ cells were found suspended in the lumen of seminiferous tubules (arrow head). At D40 through D250, EGFP+ germ cells at differential developmental stages were clearly detected within seminiferous tubules (scale bar, 20 μm). (E) Genomic DNA was prepared from sperm collected from epididymis of recipient tree shrew at D250 post transplantation. Nest-PCR analysis showed the insertion of EGFP transgene in sperm genome (∼200 sperm per sample), which was confirmed by sequencing PCR products. PCR reaction with genomic DNA from wide-type SSCs as templates was used as NC, whereas PCR reaction with genomic DNA from EGFP-SSCs as templates was set as positive control (PC). (F) Genomic DNA was prepared from offspring generated by recipient males. PCR amplification of integrated transgene was conducted with two different primer pairs and PCR products were sequenced to confirm the integration of transgene. Genomic DNA from wild-type (WT) tree shrew was used as NC. Genomic DNA from EGFP-SSCs was used as positive control 1 (PC1) and EGFP-lentiviral expressing plasmids were used as positive control 2 (PC2). (G) Immunoblotting analysis confirmed the expression of EGFP proteins in several tissues collected from one transgenic pup. Protein from WT SSCs was used as NC. Proteins from EGFP-SSCs were used as PC. (H) Fluorescent image showing the EGFP expression in the transgenic tree shrew (left) versus the control tree shrew (right). (I) CRISPR-/Cas9-mediated targeting of the App gene in tree shrew SSCs. DNA sequencing of PCR products confirmed the targeted mutations in the App gene. Mutation type 1 has an insertion of “A” at the target site, and mutation type 2 contains a 25-bp deletion at the target site.
Generation of EGFP-integrated transgenic tree shrews via in vitro expanded SSCs
To verify the normal fertilization function of the transgenic sperm and to evaluate the feasibility of generating transgenic tree shrew offspring using SSCs, 12 recipient males were mated with wild-type female tree shrews and their offspring were examined for the integration of EGFP transgene in their genome. Two different tissues (tail-tip and ear-tip) were collected from each pup at different times to prepare genome DNA samples. Insertion of virus-EGFP fragment in the genome was detected by PCR analysis with two different primer pairs and confirmed by sequencing PCR products. As shown in Supplementary information, Table S3, 7 out of 123 pups (5.7%) contained virus-EGFP transgene in their genome (Figure 4F). We also validated EGFP protein expression in different tissues of one transgenic pup by immunoblotting with GFP antibody (Figure 4G). In addition, a healthy transgenic pup emitted intense green fluorescence under the UV light (Figure 4H). Collectively, these results indicate that transgenic sperm has normal fertilization ability and SSC-based gene manipulation approach is workable to obtain gene-modified tree shrews for biomedical research.
Tree shrew SSCs are suitable for CRISPR/Cas9-mediated gene editing
CRISPR/Cas9 technology has been widely utilized in various cell types to generate sophisticated gene editing. We tested whether CRISPR/Cas9 can work in tree shrew SSCs. We designed an sgRNA targeting sequence to disrupt the gene encoding amyloid <0x8272>>β precursor protein (App) (Figure 4I). The targeting plasmids expressing sgRNA and Cas9 proteins were delivered into SSCs by electroporation and at about 20-24 h post transfection, individual transfected SSCs expressing mcherry (Supplementary information, Figure S5) were manually collected by mouth pipette and DNA was examined for the generation of mutations at the target site. Among the 70 single cells successfully sequenced for the App gene fragment, 17 were found to contain indel mutations and the example sequencing results were shown in Figure 4I. Thus, CRISPR-/Cas9-mediated gene editing can work in tree shrew SSCs.
Discussion
Due to its close phylogenetic relation to primates and the many advantages it possesses over high-order nonhuman primates for experiments, the tree shrew has received wide attention in biomedical research1,3,4. However, the lack of gene editing methods applicable to the species has limited the acceptance of the tree shrew as an important laboratory animal for genetic studies. Here, we report for the first time the development of culture conditions for the long-term expansion of tree shrew SSCs and its application in generating transgenic tree shrews. On average, 5.7% of offspring generated from recipient males carried the transgene. Moreover, CRISPR/Cas9-mediated gene editing can work in tree shrew SSCs. Thus, SSC-based approach is both feasible and practical for sophisticated gene editing in tree shrew. The key to using SSCs in making transgenic animals is to expand the SSCs in vitro without losing their stem cell abilities. In our research we found two critical factors, the addition of exogenous Wnt3a, and the use of Sertoli cells as feeder, are both required to successfully expand tree shrew SSCs in vitro. Wnt/β-catenin signaling is implicated as a stem cell self-renewal mechanism in pluripotent stem cells53 as well as in several tissue stem cells of the skin, intestine, mammary gland, and stomach54. The functions of Wnt/β-catenin signaling on SSCs have also been reported in the mouse46,55. Interestingly, the beneficial role of exogenous Wnt3a on tree shrew undifferentiated spermatogonia cells is restricted to the initial culture stage and is dispensable once SSCs start to proliferate. In our study, SSCs at P2 and thereafter were cultured on the feeder of tree shrew Sertoli cells, which have been shown to express Wnt ligands in the mouse46. Therefore, it is possible that Sertoli cells secret Wnt ligands and the paracrine Wnt ligands from Sertoli cells are sufficient to support SSC survival and proliferation. Surprisingly, higher concentration of Wnt3a (20 ng/mL) did not show a beneficial effect and the underlying mechanism warrants further investigation. Previous studies reported the involvement of non-canonical Wnt signaling in mouse SSC self-renewal56, and the inhibitory relations between the canonical and non-canonical Wnt pathways56,57. For instance, overexpression of Wnt5a, a non-canonical Wnt pathway ligand, could inhibit canonical Wnt signaling57. In addition, one ligand is able to engage multiple receptors to affect different signaling outcomes57. Thus, it is possible that higher concentration of Wnt3a compromised the non-canonical Wnt signaling in tree shrew SSCs, or engaged different receptors to elicit distinct signaling outcomes.
SSC activities are regulated by the surrounding microenvironment (niche) composed of Sertoli cells, extracellular matrix, and the vascular network58,59. Sertoli cells express glial cell line-derived neurotrophic factor (GDNF) and fibroblast growth factor 2 (FGF2), both of which are critical regulators of SSC self-renewal29,41,60. Sertoli cells also secret other factors including Wnt ligands46. Therefore, as in the rat31, tree shrew Sertoli cells were better than MEFs in supporting SSC survival and expansion in vitro.
In short, for the first time, we characterized the signaling pathways necessary for tree shrew SSCs self-renewal. By using tree shrew Sertoli cells and exogenous Wnt3a, we successfully achieved long-term expansion of tree shrew SSCs and generated transgenic tree shrews following SSC transplantation. Furthermore, we demonstrate that the widely utilized gene editing tool CRISPR/Cas9 is workable in tree shrew SSCs. Thus, in the future, tree shrews with precise gene modification can be obtained by applying CRISPR/Cas9 on SSCs. Our current study paves the way for the generation of various human disease models using the tree shrew and facilitates the wider application of this animal in biomedical research.
Materials and Methods
Animal use and care
Wild-type tree shrews were obtained from the Kunming Primate Research Center in Kunming Institute of Zoology, Chinese Academy of Sciences. All experimental procedures and animal care were performed according to the protocols approved by the Institutional Animal Care and Use Committee of the Kunming Institute of Zoology, Chinese Academy of Sciences. In total, 13 3-month-old tree shrews and about 60 1-year-old tree shrews were used in this study.
Preparation of testicular tissue single-cell suspensions
A pair of tree shrew testes was used in each experiment. Single-cell suspensions were prepared using a two-step enzymatic digestion. Briefly, the testes were washed in PBS solution and cut into pieces after stripping off the albuginea. The tissue pieces were then incubated in 5% collagenase type IV (Lifetech Gibco) at 37 °C for 20 min with gentle agitation every 5 min. After washing in PBS twice, they were incubated in 0.25% trypsin (Lifetech Gibco) at 37 °C for < 10 min until most of the cells were dispersed. Digestion was terminated by adding 10% fetal bovine serum (FBS, Hyclone) and the cell suspensions were centrifuged at 400 g for 5 min to remove the supernatant. The cell pellets were re-suspended in 1% BSA and the clumps were removed through a 100 μm nylon cell strainer (B&D).
Enrichment of Thy1+ cell population
A single-cell suspension of tree shrew testicular tissue was subject to FACS sorting or immunomagnetic isolation to obtain a Thy1+ cell population according to the manufacturer's instructions. For FACS sorting, cell samples were incubated with Thy1 antibody (BioLegend, 328110, 1:50 dilution) for 30 min before sorting with Influx flow cytometer (Becton Dickinson). For immunomagnetic isolation, cell suspensions were incubated with Thy1 antibody (BioLegend, 328110, 1:50 dilution) for 30 min and the Thy1+ cells were isolated by microbeads (Miltenyi Biotech).
RNA-seq library preparation, sequencing and data analyses
For RNA sequencing, Thy1+ cells and Thy1− cells were sorted by Influx flow cytometer (Becton Dickinson). Two independent biological sample repeats were prepared. Total RNA was extracted using TRIZOL (Invitrogen) and a RNA-Seq library was constructed following the instructions of the TruSeq RNA Sample Preparation kit (Illumina). The libraries were sequenced using an Illumina HiSeq 2000 platform. Clean reads were mapped to the tree shrew genome using Tophat2 software (Version 2.0.8). The genome and annotation files for the tree shrew were downloaded from the tree shrew database (http://www.treeshewdb.org). FPKM (fragments per kilobase million) values of the transcripts were calculated using Cufflinks (Version 2.1.1)61. Differential expression genes were determined by Cuffdiff (Version 2.1.1) with default parameters. The heat maps were generated by the “gplots” R package with the default hierarchical clustering method. PCA was performed using the “prcomp” function of R package. The enrichment of the GO and KEGG pathways was analyzed using DAVID (http://david.abcc.ncifcrf.gov/)62,63,64. The P-value was adjusted using a hypergeometric test with the default parameters. The RNA-Seq data can be retrieved at www.treeshrewdb.org.
Feeder cells preparation
MEFs were prepared according to the standard protocol. Tree shrew Sertoli cells were prepared according to the previous report48. In brief, testicular tissue single-cell suspensions were plated on a dish coated by 0.1% (w/v) gelatin (AMRESCO). After overnight incubation, the medium and floating germ cells were removed and the adherent cells were collected. A Sertoli cell preparation was considered as enriched and not pure as a small number of other somatic cells remain in the culture. The preparation was then cultured in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies) with 10% FBS (HyClone) for propagation. The Sertoli cell identity was verified by RT-PCR examination of several marker gene expressions. To prepare feeder cells, MEFs (∼50 000 cells/cm2) or Sertoli cells (∼50 000 cells/cm2) were treated with 10 μg/mL Mitomycin C (Sigma) for 3 h to inactive the mitotic activity.
Tree shrew SSCs culture
The basic SSCs culture medium is composed of StemPro-34 SFM (Invitrogen) supplemented with StemPro supplement (Invitrogen), 10% fetal calf serum (FBS, HyClone), 1 mM penicillin (Lifetech Gibco), 2 mM ℒ-Glutamine (Sigma), 0.1 mM β-mercaptoethanol (Sigma), 1 mM sodium pyruvate (Lifetech Gibco), 1 mM NEAA (nonessential amino acid, Lifetech Gibco), 60 μM putrescine, 40 ng/mL human GDNF (recombinant human glial cell line-derived neurotrophic factor, R&D), 10 ng/mL human EGF (epidermal growth factor, Invitrogen), 1 000 U mouse LIF (leukemia inhibitory factor, Millipore), 2 ng/mL human bFGF (basic fibroblast growth factor, Millipore). Recombinant Wnt3a (R&D) was dissolved in 1% BSA. For the initial culture of SSC-enriched cells (passages 0-4), 500 μL SSC culture medium were used per well and the medium was changed every other day. SSCs were manually collected and passaged every 7 days at a 1 : 3-5 dilution. Culture was maintained in a 37 °C and 5% CO2 atmosphere.
Preparation of EGFP-expressing lentivirus and infection of tree shrew SSCs
Simian immunodeficiency virus (SIV)-based vector GAE-CAG-eGFP/WPRE65 was utilized to transfer the EGFP gene into the genome of tree shrew SSCs. To produce virus, 293T cells were transfected with DNA mixtures, which contains the SIV lentiviral expression vector GAE-CAG-EGFP/WPRE and packaging vectors (SIV : SIV3+ : G-REV = 1.3 : 1 : 1) by lipo2000 transfection method (Invitrogen 11668-019). Supernatant of the transfected 293T cells was collected and filtered by 0.45 μm filter (Millipore), followed by ultra-centrifugation at 25 000 rpm for 2.5 h. The SSCs were suspended and infected at a density of ∼100 viruses per cell for 24 h. The infected cells were purified by FACS sorting.
Preparation of sterilized recipient tree shrews
Busulfan (Sigma) was dissolved in corn oil. Adult male tree shrews were weighed and certain amount of busulfan (40 mg/kg body weight) was administrated by intraperitoneal injection. At 4-5 weeks following busulfan treatment, the elimination of pre-existing germ cells was confirmed by hematoxylin and eosin stain of testicular sections and recipients were prepared for cell transplantation.
SSC transplantation into recipient tree shrew testis
Tree shrew SSC transplantation into testis was performed according to the methods described in the mouse52. Briefly, a glass pipette with an inner diameter of ∼50-80 μm at the tip was used. About 0.04% Trypan blue dye was added into the cell suspension (3-5 × 104 cells/L) to monitor the injection. Recipient tree shrews were anesthetized with ketamine (0.2 mg/g body weight, intramuscular injection). The testicle skin wall was then lifted by using small forceps to make a ∼0.3 cm small incision. Carefully cut the bursa to open the peritoneal cavity and use one pair of forceps to hold the skin wall and the other pair to search the fat pads attached to the testis in the cavity. Gently pull the fat out until the testis is exteriorized. Identify the efferent ducts that connect the testis to the epididymis. Cells were transplanted into the seminiferous tubules of the testis through the efferent ducts. In general, total 0.5-0.7 million SSCs were injected into each recipient testis. The injection was stopped when almost all the surface tubules had been filled as indicated by the Trypan blue dye.
Construction of the CRISPR/Cas9 plasmids and electroporation of tree shrew SSCs
The sgRNA targeting sequence for the App gene was designed by using the CRISPR Design Tool (http://tools.genome-engineering.org)66. The sgRNA targeting sequence pair sgRNA-App-Forward (5′-CACCGGTTTCTTCTTCAGCATCACCA-3′) and sgRNA-App-Reverse (5′-AAACTGGTGATGCTGAAGAAGAAACC-3′) were cloned into the pX330-T7 vector expressing mcherry66.
About 105 SSCs were used for electroporation. SSCs were detached from feeder cells through gentle pipetting. After collection and wash in PBS for three times, cells were then suspended in 100 μL OPTI-MEM (Gibco, 31985-062) with 1 μg targeting plasmids. Electroporation was conducted with Super Electroporator NEPA21 (NEPA GENE), using the parameters of 125 V voltage and 5 ms pulse length. Cells were transferred to culture medium for further culture following electroporation. After 20-24 h of culture, cells were examined for the electroporation efficiency under the fluorescence microscope and the individual SSCs, which had normal morphology and expressed mcherry, were manually picked with mouth pipette for downstream examination of gene targeting.
Genomic DNA preparation and DNA sequencing of target loci
Genomic DNA was extracted from single cell by alkaline lysis method67. Lysis buffer is composed of 25 mM NaOH and 0.2 mM EDTA. Individual SSC was transferred into 5 μL of lysis buffer and heated at 95 °C for 10 min. After heating, cell lysis was cooled down on ice for 1 min and neutralized by 5 μL neutralization buffer (40 mM Tris HCl). All lysis solution was then used for PCR amplification. The gene region spanning the sgRNA targeting site was PCR amplified by primer pair (App forward: 5′-GCCCGAGTACCATGGAGGAA-3′ App reverse: 5′-TTCAAGGTGTTCTTTGCGGA-3′). The PCR products were first sequenced using the PCR primer to screen which sample contains potential mutation. Subsequently, the PCR products containing potential mutation were cloned into T-Vector pMD19 (TAKARA BIO Inc.) for transformation. About 10 clones were randomly picked and sequenced for each transformation. The sequence was analyzed by using DNASTAR Lasergene 7.1 (DNAS Inc., Madison, WI, USA).
Other Materials and Methods including chromosome metaphase spread preparation and G-band staining, tree shrew SSCs cryopreservation and thawing, preparation of frozen tissue sections and immunofluorescence staining, genotyping of transgenic tree shrews and sperm, reverse transcription, regular RT-PCR, quantitative RT-PCR, antibodies and primers information were provided in Supplementary information, Data S1.
Author Contributions
CHL established the tree shrew SSC lines, performed the immunostaining, genotyping of transgenic tree shrews, CRISPR/Cas9 gene targeting, and prepared the figures. LZY performed SSC transplantation. WZB constructed the EGFP viral-expressing vectors, performed the genotyping, and RT-PCR examination. QT prepared the recipient males. YW and LW conducted the RNA-seq data analyses. RB constructed the CRISPR/Cas9 targeting vector. SJ tested the Thy1 antibody. YHM and LBL took care of tree shrews. WHN performed karyotyping and G-band staining. PZ designed and supervised the study, and wrote the manuscript. XDZ and YGY supervised the study.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Wei-Dao Zhang for help with the immunoblotting analysis, Guo-Lan Ma for help in FACS sorting, Da-Jian He for help in picking single transfected SSC, and Ian Logan for language editing. This work was supported by the Breakthrough Project of the Strategic Priority Program of the Chinese Academy of Sciences (Grant Nos XDB13000000 and XDA01010203) and the project from Yunnan Province to Ping Zheng (2015HA038).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
Overall study design.
Examination of the Sertoli cell marker gene expressions.
Different passages of the tree shrew SSCs in culture.
Hematoxylin-eosin stain of testicular samples from normal and busulfan-treated tree shrews.
Morphology of SSCs at 20-24 h post transfection of CRISPR/Cas9 targeting plasmids.
Information of EGFP+ offspring derived from therecipient tree shrews.
Supplementary Materials and Methods
References
- Fan Y, Huang ZY, Cao CC, et al. Genome of the Chinese tree shrew. Nat Commun 2013; 4:1426. [DOI] [PubMed] [Google Scholar]
- Rockland KS, Lund JS. Widespread periodic intrinsic connections in the tree shrew visual cortex. Science 1982; 215:1532–1534. [DOI] [PubMed] [Google Scholar]
- Zheng YT, Yao YG, Xu L, eds. Basic biology and disease models of Chinese tree shrew. Kunming: Yunnan Science and Technology Press, 2014.
- Cao J, Yang EB, Su JJ, Li Y, Chow P. The tree shrews: adjuncts and alternatives to primates as models for biomedical research. J Med Primatol 2003; 32:123–130. [DOI] [PubMed] [Google Scholar]
- Vercauteren K, de Jong YP, Meuleman P. HCV animal models and liver disease. J Hepatol 2014; 61:S26–S33. [DOI] [PubMed] [Google Scholar]
- Sun H, Zhang A, Yan G, et al. Metabolomic analysis of key regulatory metabolites in hepatitis C virus-infected tree shrews. Mol Cell Proteomics 2013; 12:710–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Tang ZY, Klumpp B, et al. Primary hepatocytes of Tupaia belangeri as a potential model for hepatitis C virus infection. J Clin Invest 2002; 109:221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Ruan P, Ou C, et al. Chronic hepatitis B virus infection and occurrence of hepatocellular carcinoma in tree shrews (Tupaia belangeri chinensis). Virol J 2015; 12:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Zhong G, Xu G, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 2012; 1:e00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JP, Liao Y, Zhang Y, et al. Experimental infection of tree shrews (Tupaia belangeri) with Coxsackie virus A16. Zool Res 2014; 35:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L, Ding H, Lin S, et al. Experimental Mycobacterium tuberculosis infection in the Chinese tree shrew. FEMS Microbiol Lett 2014; 360:23–32. [DOI] [PubMed] [Google Scholar]
- Familtsev D, Quiggins R, Masterson SP, et al. Ultrastructure of geniculocortical synaptic connections in the tree shrew striate cortex. J Comp Neurol 2016; 524:1292–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanni MP, Thomas S, Petry HM, Bickford ME, Casanova C. Spatiotemporal profile of voltage-sensitive dye responses in the visual cortex of tree shrews evoked by electric microstimulation of the dorsal lateral geniculate and pulvinar nuclei. J Neurosci 2015; 35:11891–11896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Huang X, Fitzpatrick D. Topology of ON and OFF inputs in visual cortex enables an invariant columnar architecture. Nature 2016; 533:90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Frost MR, Siegwart JT Jr, Norton TT. Scleral gene expression during recovery from myopia compared with expression during myopia development in tree shrew. Mol Vis 2014; 20:1643–1659. [PMC free article] [PubMed] [Google Scholar]
- Norton TT, Amedo AO, Siegwart JT Jr. Darkness causes myopia in visually experienced tree shrews. Invest Ophthalmol Vis Sci 2006; 47:4700–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Sun YJ, Lv YH, et al. High activity of the stress promoter contributes to susceptibility to stress in the tree shrew. Sci Rep 2016; 6:24905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. Social stress in tree shrews as an animal model of depression: an example of a behavioral model of a CNS disorder. CNS Spectr 2005; 10:182–190. [DOI] [PubMed] [Google Scholar]
- Van Kampen M, Kramer M, Hiemke C, Flugge G, Fuchs E. The chronic psychosocial stress paradigm in male tree shrews: evaluation of a novel animal model for depressive disorders. Stress 2002; 5:37–46. [DOI] [PubMed] [Google Scholar]
- Shen F, Duan Y, Jin S, Sui N. Varied behavioral responses induced by morphine in the tree shrew: a possible model for human opiate addiction. Front Behav Neurosci 2014; 8:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YM, Yang JZ, Sun HY, Ma YY, Wang JH. Establishment of tree shrew chronic morphine dependent model. Zool Res 2012; 33:14–18. [DOI] [PubMed] [Google Scholar]
- Khani A, Rainer G. Recognition memory in tree shrew (Tupaia belangeri) after repeated familiarization sessions. Behav Processes 2012; 90:364–371. [DOI] [PubMed] [Google Scholar]
- Bartolomucci A, de Biurrun G, Czeh B, van Kampen M, Fuchs E. Selective enhancement of spatial learning under chronic psychosocial stress. Eur J Neurosci 2002; 15:1863–1866. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Fuchs E, Taira M, Yamamoto T, Hayashi M. Somatostatin-immunoreactive senile plaque-like structures in the frontal cortex and nucleus accumbens of aged tree shrews and Japanese macaques. J Med Primatol 2012; 41:147–157. [DOI] [PubMed] [Google Scholar]
- Ge GZ, Xia HJ, He BL, et al. Generation and characterization of a breast carcinoma model by PyMT overexpression in mammary epithelial cells of tree shrew, an animal close to primates in evolution. Int J Cancer 2016; 138:642–651. [DOI] [PubMed] [Google Scholar]
- Wu X, Xu H, Zhang Z, et al. Transcriptome profiles using next-generation sequencing reveal liver changes in the early stage of diabetes in tree shrew (Tupaia belangeri chinensis). J Diabetes Res 2016; 2016:6238526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang Z, Li Y, et al. Cholesterol induces lipoprotein lipase expression in a tree shrew (Tupaia belangeri chinensis) model of non-alcoholic fatty liver disease. Sci Rep 2015; 5:15970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan LZ, Sun B, Lyu LB, et al. Early embryonic development and transplantation in tree shrews. Zool Res 2016; 37:252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL. Male germline stem cells: from mice to men. Science 2007; 316:404–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci USA 2004; 101:16489–16494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamra FK, Chapman KM, Nguyen DM, Williams-Stephens AA, Hammer RE, Garbers DL. Self renewal, expansion, and transfection of rat spermatogonial stem cells in culture. Proc Natl Acad Sci USA 2005; 102:17430–17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadri-Ardekani H, Mizrak SC, van Daalen SK, et al. Propagation of human spermatogonial stem cells in vitro. JAMA 2009; 302:2127–2134. [DOI] [PubMed] [Google Scholar]
- Chapman KM, Medrano GA, Jaichander P, et al. Targeted germline modifications in rats using CRISPR/Cas9 and spermatogonial stem cells. Cell Rep 2015; 10:1828–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhou H, Fan X, et al. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res 2015; 25:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehashi M, Kanatsu-Shinohara M, Inoue K, et al. Adenovirus-mediated gene delivery into mouse spermatogonial stem cells. Proc Natl Acad Sci USA 2007; 104:2596–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izsvak Z, Frohlich J, Grabundzija I, et al. Generating knockout rats by transposon mutagenesis in spermatogonial stem cells. Nat Methods 2010; 7:443–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh JR, Nagano MC. Spermatogonial stem cell biomarkers: improved outcomes of spermatogonial transplantation in male fertility restoration? Expert Rev Mol Diagn 2009; 9:109–114. [DOI] [PubMed] [Google Scholar]
- Dym M, Kokkinaki M, He Z. Spermatogonial stem cells: mouse and human comparisons. Birth Defects Res C Embryo Today 2009; 87:27–34. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Ogonuki N, Inoue K, et al. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod 2003; 69:612–616. [DOI] [PubMed] [Google Scholar]
- Noce T, Okamoto-Ito S, Tsunekawa N. Vasa homolog genes in mammalian germ cell development. Cell Struct Funct 2001; 26:131–136. [DOI] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvonen ME, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 2000; 287:1489–1493. [DOI] [PubMed] [Google Scholar]
- Lustig B, Jerchow B, Sachs M, et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol 2002; 22:1184–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/β-catenin-responsive stem cells in the mammary gland. Cell Stem Cell 2012; 11:387–400. [DOI] [PubMed] [Google Scholar]
- Katoh M. Network of WNT and other regulatory signaling cascades in pluripotent stem cells and cancer stem cells. Curr Pharm Biotechnol 2011; 12:160–170. [DOI] [PubMed] [Google Scholar]
- Hao J, Li TG, Qi X, Zhao DF, Zhao GQ. WNT/β-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Dev Biol 2006; 290:81–91. [DOI] [PubMed] [Google Scholar]
- Takase HM, Nusse R. Paracrine Wnt/β-catenin signaling mediates proliferation of undifferentiated spermatogonia in the adult mouse testis. Proc Natl Acad Sci USA 2016; 113:E1489–E1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness MP, Linder CC, Morales CR, Heckert LL, Pikus J, Griswold MD. Relationship of a mouse Sertoli cell line (MSC-1) to normal Sertoli cells. Biol Reprod 1994; 51:116–124. [DOI] [PubMed] [Google Scholar]
- Karl AF, Griswold MD. Sertoli cells of the testis: preparation of cell cultures and effects of retinoids. Methods Enzymol 1990; 190:71–75. [DOI] [PubMed] [Google Scholar]
- Paranko J, Kallajoki M, Pelliniemi LJ, Lehto VP, Virtanen I. Transient coexpression of cytokeratin and vimentin in differentiating rat Sertoli cells. Dev Biol 1986; 117:35–44. [DOI] [PubMed] [Google Scholar]
- Muller S, Stanyon R, O'Brien PC, Ferguson-Smith MA, Plesker R, Wienberg J. Defining the ancestral karyotype of all primates by multidirectional chromosome painting between tree shrews, lemurs and humans. Chromosoma 1999; 108:393–400. [DOI] [PubMed] [Google Scholar]
- Niu Y, Yu Y, Bernat A, et al. Transgenic rhesus monkeys produced by gene transfer into early-cleavage-stage embryos using a simian immunodeficiency virus-based vector. Proc Natl Acad Sci USA 2010; 107:17663–17667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci USA 1994; 91:11298–11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D, Kurek D, Blauwkamp T, et al. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol 2011; 13:1070–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 2014; 346:1248012. [DOI] [PubMed] [Google Scholar]
- Golestaneh N, Beauchamp E, Fallen S, Kokkinaki M, Uren A, Dym M. Wnt signaling promotes proliferation and stemness regulation of spermatogonial stem/progenitor cells. Reproduction 2009; 138:151–162. [DOI] [PubMed] [Google Scholar]
- Yeh JR, Zhang X, Nagano MC. Wnt5a is a cell-extrinsic factor that supports self-renewal of mouse spermatogonial stem cells. J Cell Sci 2011; 124:2357–2366. [DOI] [PubMed] [Google Scholar]
- Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits β-catenin-TCF signaling depending on receptor context. PLoS Biol 2006; 4:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Sukeno M, Nabeshima Y. A vasculature-associated niche for undifferentiated spermatogonia in the mouse testis. Science 2007; 317:1722–1726. [DOI] [PubMed] [Google Scholar]
- de Rooij DG. The spermatogonial stem cell niche. Microsc Res Tech 2009; 72:580–585. [DOI] [PubMed] [Google Scholar]
- Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Annu Rev Cell Dev Biol 2008; 24:263–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 2007; 35:W169–W175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wianny F, Bernat A, Huissoud C, et al. Derivation and cloning of a novel rhesus embryonic stem cell line stably expressing tau-green fluorescent protein. Stem Cells 2008; 26:1444–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 2000; 29:52, 54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overall study design.
Examination of the Sertoli cell marker gene expressions.
Different passages of the tree shrew SSCs in culture.
Hematoxylin-eosin stain of testicular samples from normal and busulfan-treated tree shrews.
Morphology of SSCs at 20-24 h post transfection of CRISPR/Cas9 targeting plasmids.
Information of EGFP+ offspring derived from therecipient tree shrews.
Supplementary Materials and Methods