ABSTRACT
The bipolar kinesin-5 Cin8 switches from minus- to plus-end-directed motility under various conditions in vitro. The mechanism and physiological significance of this switch remain unknown. Here, we show that under high ionic strength conditions, Cin8 moves towards and concentrates in clusters at the minus ends of stable and dynamic microtubules. Clustering of Cin8 induces a switch from fast minus- to slow plus-end-directed motility and forms sites that capture antiparallel microtubules (MTs) and induces their sliding apart through plus-end-directed motility. In early mitotic cells with monopolar spindles, Cin8 localizes near the spindle poles at microtubule minus ends. This localization is dependent on the minus-end-directed motility of Cin8. In cells with assembled bipolar spindles, Cin8 is distributed along the spindle microtubules. We propose that minus-end-directed motility is required for Cin8 clustering near the spindle poles before spindle assembly. Cin8 clusters promote the capture of microtubules emanating from the neighboring spindle poles and mediate their antiparallel sliding. This activity is essential to maximize microtubule crosslinking before bipolar spindle assembly and to induce the initial separation of the spindle poles.
KEY WORDS: Cin8, Kinesin-5, Microtubules, Mitosis, Mitotic spindle
Highlighted Article: We show that clustering of the mitotic kinesin-5 Cin8 on microtubules controls its motile properties and directionality, and we provide a refined model of how Cin8 performs its essential functions in mitosis.
INTRODUCTION
The conserved kinesin-5 mitotic motors perform essential functions in bipolar mitotic spindle assembly and elongation. These motors are organized as homo-tetrameric bipolar complexes with two pairs of motor domains located at opposite sides of a 60 nm long mini-filament (Gordon and Roof, 1999; Kashina et al., 1996; Scholey et al., 2014). This organization enables kinesin-5 motors to crosslink and slide apart antiparallel microtubules (MTs) (Gheber et al., 1999; Kapitein et al., 2005), and to perform essential functions in the assembly and maintenance of the bipolar mitotic spindle (Asraf et al., 2015; Blangy et al., 1997; Mayer et al., 1999; Saunders and Hoyt, 1992), as well as anaphase spindle elongation (Fridman et al., 2009; Gerson-Gurwitz et al., 2009; Movshovich et al., 2008; Sharp et al., 1999; Straight et al., 1998).
In contrast to the plus-end-directed motility expected for kinesins with N-terminal motor domains, the Saccharomyces cerevisiae kinesin-5 Cin8 undergoes minus-end-directed motility along single MTs (Gerson-Gurwitz et al., 2011; Roostalu et al., 2011; Thiede et al., 2012) and switches motility direction towards the plus ends of the MTs as a function of ionic strength, in a multi-motor gliding assay or upon binding two antiparallel MTs (Gerson-Gurwitz et al., 2011; Roostalu et al., 2011). A large insert in loop 8 (L8) of the motor domain of Cin8, its C-terminal tail and phosphorylation at the catalytic domain of Cin8 have been found to influence Cin8 directionality (Duselder et al., 2015; Gerson-Gurwitz et al., 2011; Shapira and Gheber, 2016). However, the mechanism of the minus-end-directed motility of Cin8 and its ability to switch directionality remain poorly understood. Recently, two additional yeast kinesin-5 motors have been reported to be bi-directional, the S. cerevisiae kinesin-5 Kip1 (Fridman et al., 2013) and the Schizosaccharomyces pombe Cut7 (Edamatsu, 2014). These findings indicate that minus-end-directed motility in kinesins with N-terminal motor domains may be more common than previously believed.
Plus-end-directed motility of kinesin-5 motors is required for the sliding apart of interpolar spindle MTs (iMTs) and to provide the outward-directed force to separate the spindle pole bodies (SPBs) during spindle assembly and anaphase spindle elongation (Kapitein et al., 2005; Movshovich et al., 2008; Saunders and Hoyt, 1992; Saunders et al., 1995). Thus far, a possible physiological role for the minus-end-directed motility of Cin8 and other kinesin-5 motors has not yet been proposed. Here, we studied the motile properties of single Cin8 motors and of Cin8 clusters, and examined Cin8-induced capturing and sliding apart of antiparallel stabilized and dynamic MTs. We also examined the intracellular localization of Cin8 before and following spindle assembly. Based on our results, we propose a revised model for the physiological function of Cin8 in S. cerevisiae spindle assembly. In this model, before spindle assembly, Cin8 undergoes minus-end-directed motility along nuclear MTs and clusters near the SPBs. This clustering promotes capturing and plus-end-directed sliding apart of spindle MTs emanating from different SPB poles, thus providing the initial SPB separation required for spindle assembly.
RESULTS
Cin8 clustering slows motility and induces a switch of directionality
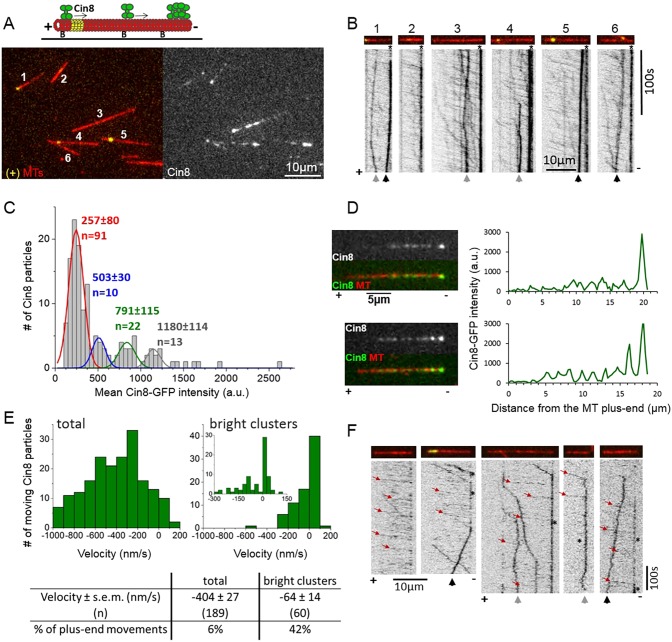
We studied the single-motor motility of full-length Cin8 tagged with three GFP molecules (Cin8–3GFP) along fluorescently-labeled polarity-marked MTs, stabilized by a non-hydrolyzable GTP analog, GMPCPP. In these polarity-marked MTs, plus ends were marked by polymerizing αβ-tubulin labeled with two different fluorophores (Fig. 1A, MTs 1, 4 and 5). In accordance with previous reports (Duselder et al., 2015; Gerson-Gurwitz et al., 2011; Roostalu et al., 2011; Shapira and Gheber, 2016), the majority of Cin8 motors exhibited fast and processive motility towards the minus ends of the MTs under high ionic strength conditions (Fig. 1B; Movie 1). We found that the rapid minus-end-directed motility of Cin8 motors led to their accumulation in bright clusters at or near MT minus ends (Fig. 1A,B). To study the clustering of Cin8 motors, we first examined their intensity distribution (Fig. 1C). Because in our experiments each Cin8 tetramer was labeled with twelve GFP molecules (three per Cin8 monomer), single imaging-induced GFP-photobleaching steps could not be observed (Gerson-Gurwitz et al., 2011). We used the TrackMate (Jaqaman et al., 2008) plugin of ImageJ to detect Cin8 particles (Fig. S1) and to determine their fluorescence intensities. The distribution of Cin8 fluorescence intensity exhibited several peaks, with the first peak (lowest intensity) containing ∼65% of the Cin8 particle population (Fig. 1C). The average Cin8 particle intensities of the second, third and fourth peaks were approximately twice, three and four times that of the first peak, respectively (Fig. 1C). The Cin8 molecule population in the first peak consisted of mainly fast minus-end-directed molecules (see below). We have previously demonstrated, by photobleaching of a mixture of kinesin-5 dimers tagged with a single GFP molecule and Cin8 tagged with three GFP molecules, that single Cin8–3GFP tetramers exhibit fast minus-end-directed motility under high ionic strength conditions (Gerson-Gurwitz et al., 2011). Therefore, we conclude that in the present case, the intensities of Cin8 represented by the first peak reflect single Cin8 motors, while those in the second, third and fourth peaks are likely to represent clusters containing two, three and four Cin8 tetramers, respectively (Fig. 1C).
Fig. 1.
Clustering of Cin8 at the minus end of MTs reverses its directionality. (A) Top: schematic presentation of a single-molecule Cin8 motility assay on plus-end-labeled MTs; Alexa-Fluor-633-labeled MTs (red); biotinylated MTs were marked at their plus ends with Alexa-Fluor-543 (yellow) and immobilized to the surface using an anti-biotin antibody. Arrows: direction of single molecules of Cin8 (green). ‘B’, biotin. Bottom: representative snapshot of Cin8–3GFP motility under high ionic strength conditions (right) on plus-end-labeled fluorescent MTs (left). MTs are numbered for presentation purposes (Movie 1). (B) Kymographs of Cin8 movement along MTs 1–6, numbered as in A. For representation purposes, the black and white colors in the kymographs have been inverted. The minus-ends of the MTs are always on the right. Directionality of the MTs is assigned based on plus-end labeling and/or the direction of fast-moving Cin8 molecules. (C) Intensity distribution of 141 Cin8 motors, collected from the first frame of the experiment shown in A and B. The red, blue, green and the gray lines represent the Gaussian fit of the first, second, third and fourth peaks, respectively. The mean intensities of Cin8 particles ±s.d. of the four peaks are indicated in matching colors. (D) Line-scan analysis of Cin8 distribution on the MTs. Left: images of Cin8 (top), and Cin8 and MT merge (bottom) of two representative MTs. Plus- and minus-ends are indicated along the bottom, based on the direction of fast-moving Cin8 particles. Right: line-scan analysis of Cin8 distribution along the length of the MT, shown on the left. Note: (i) the increased intensity of bright clusters at the minus ends of the MTs in the top and bottom panels, and (ii) an additional bright cluster of Cin8 on the bottom MT. (E) Velocity distribution of the total population of Cin8 particles (left) and bright clusters only, whose intensity is more than four times higher than the average intensity of single Cin8 molecules (∼1200 a.u.; right), the insert in the right-hand panel presents the velocity distribution of bright clusters within a smaller velocity range, divided into smaller bins. Mean velocity values, s.e.m., and the percentage of plus-end-directed movements are indicated at the bottom. (F) Representative kymographs of Cin8–3GFP movement on MTs under high ionic strength conditions, in controlled-start experiments, in which the imaging began 30–70 s following addition of Cin8. Red arrows point to the fast minus-end-directed movements, asterisks indicate the accumulation of Cin8 at the minus ends of the MTs. (B,F) Black arrows point to bright clusters moving in a plus-end direction. Gray arrows point to clusters moving in both plus- and minus-end directions. Asterisks indicate the accumulation of Cin8 at the minus-ends of the MTs.
We next examined the distribution of clusters along the MTs using line-scan analysis along the MT length (Fig. 1D). We found that 5–15 min following the addition of Cin8 to the MTs, ∼96% of the MTs (53/55) exhibited a bright Cin8 cluster at the minus-end of the MTs, whose intensity was four times higher than that of the single Cin8 motors (Fig. 1D, top). These data indicate that Cin8 clusters exhibit high affinity to the MT minus ends (Fig. 1A,B,D). Interestingly, high affinity to the plus ends of the MTs has been demonstrated previously for plus-end-directed kinesin-5 motors (Chen and Hancock, 2015; Kapitein et al., 2005). Similar minus-end-directed clustering was also observed on Taxol-stabilized MTs (Fig. S2), although with a lower frequency of occurrence, where only ∼40% of the MTs exhibited clusters at minus ends. The difference may be due to fraying or instability at the minus ends of MTs stabilized with Taxol in contrast to MTs stabilized with GMPCPP (Arnal and Wade, 1995). We also found that 44% of the MTs (24/55) exhibited at least one additional bright cluster along the GMPCPP-stabilized MT lattice (Fig. 1D, bottom). Only ∼4% of the MTs (2/55) exhibited Cin8 clusters at both plus and minus ends of the MTs.
We found that about 60% of the bright Cin8 clusters observed on the MTs were motile, with 75% of the MTs (41/55) containing at least one moving Cin8 cluster. Surprisingly, in contrast to the rapid minus-end-directed motility observed for the majority of Cin8 motors (Fig. 1B; Movie 1), the velocity distribution of the bright clusters revealed a considerably slower average velocity as well as a considerably higher percentage of plus-end-directed motility events, compared to those aspects of the total population of motile Cin8 particles (Fig. 1E). Slow minus- and plus-end-directed movements of bright Cin8 clusters were also observed on Taxol-stabilized MTs (Fig. S2, green and white arrows), although these events were less frequent compared to those on MTs stabilized with GMPCPP (Fig. 1). These results demonstrate that while single Cin8 motors show fast motility in the minus-end direction, accumulation of Cin8 into clusters slows the motility and induces plus-end directionality. To observe Cin8 clustering formation kinetics, we performed an assay in which Cin8 motors were injected into flow-chambers containing MTs immobilized to the glass surface, while being mounted for total internal reflection fluorescence (TIRF) imaging. Using this approach, we could monitor Cin8 motor motility as soon as 30–70 s after their interaction with the immobilized MTs on the glass surface (Fig. 1F). We found that along some MTs, Cin8 clusters formed at MT minus ends as early as 30 s following Cin8 interaction with MTs. After 5 min, 80% of all MTs exhibited Cin8 clusters at their minus ends (Fig. 1F). The distribution of the plus- and minus-end-directed motility of the clusters in these experiments was similar to that in the steady-state experiments (Fig. 1E).
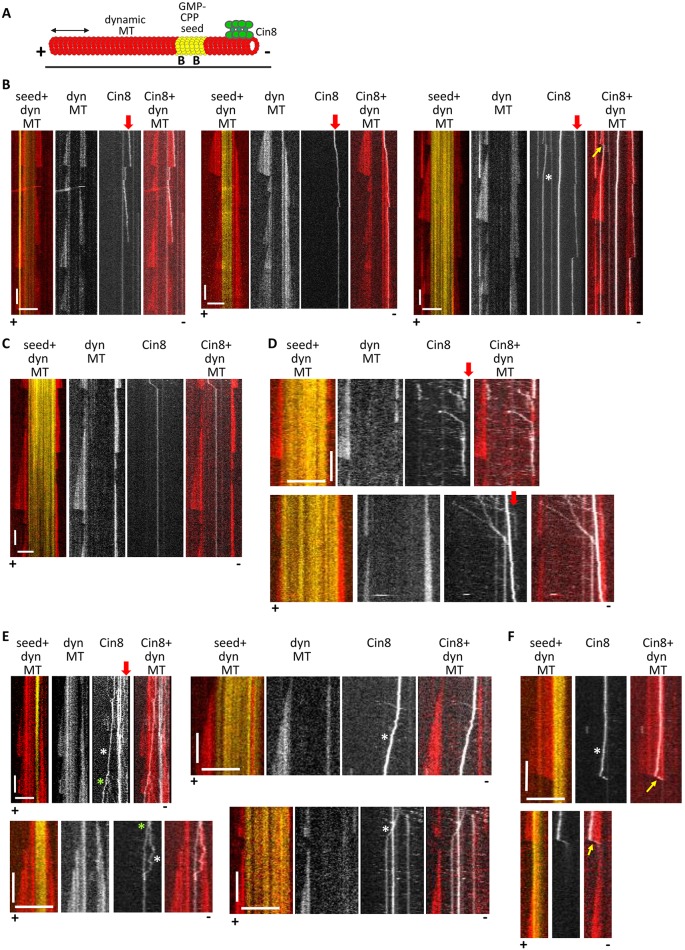
We next examined Cin8 motility along dynamic MTs to compare that with its behavior along GMPCPP-stabilized MTs, as the dynamic MTs represent the native tracks for Cin8 motors during mitosis. Dynamic MTs were polymerized in the presence of 1 mM GTP and 6 µM free tubulin (Fig. 2; Movies 2–4). We found that, similar to its behavior on stabilized MTs, Cin8 exhibited attachment and clustering at the minus ends of dynamic MTs (Fig. 2B,D and E; Movies 2, 3). Furthermore, we observed Cin8 tracking along the MT minus ends (Fig. 2B; Movies 2, 3). This clustering and tracking did not significantly affect the dynamic polymerization properties of the MT minus ends (Fig. 2B and C). Along dynamic MTs, Cin8 maintained rapid minus-end-directed motility (Fig. 2D) as well as slower and bi-directional motility when assembled into clusters (Fig. 2, white asterisks; Movie 4), with no significant binding to or tracking along the plus ends, except for a few sporadic events (Fig. 2F). Our results indicate that Cin8 accumulates at the minus ends of stabilized and dynamic MTs, and that in both cases, clustering of Cin8 slows its motility and induces plus-end directionality, suggesting that this behavior may be relevant to the physiological functions of Cin8.
Fig. 2.
Cin8 accumulates at and tracks along the minus ends of dynamic MTs. (A) Schematic presentation of a MT dynamics assay. Yellow, GMPCPP-stabilized seed adsorbed onto the surface via an anti-biotin antibody; red, dynamic MTs; green, Cin8. ‘B’, biotin. (B–F) Representative kymographs of Cin8 movements on dynamic (dyn) MTs. In the merged images: red, dynamic MTs; yellow, GMPCPP-stabilized seeds; white, Cin8. The plus and minus ends of the MTs are indicated at the bottom, based on the dynamics characteristics at the two ends. Scale bars: 10 µm (horizontal); 100 s (vertical). (B,C) Long-term and (D) short-term imaging. Vertical red arrows point to Cin8 attachment and tracking along the minus-end of dynamic MTs. MT 1 in Movie 2 is shown in B, left panels. (C) Example of lack of Cin8 attachment to the MT minus end. Yellow arrows indicate the tracking of Cin8 along MT plus ends. (E) Examples of bi-directional and plus-end-directed movements of Cin8 clusters on dynamic MTs and GMPCPP-stabilized seeds. White asterisks indicate slow bi-directional and plus-end-directed bright Cin8 particles. Green asterisks indicate Cin8 cluster-splitting events. The splitting of the Cin8 cluster shown in the upper-left panel is shown in Movie 4. (F) Two sporadic events of Cin8 tracking on a depolymerizing plus end. Minus ends of the MTs were not observed in these examples. The plus end was assigned based on the dynamics of this end. Note: Cin8 tracking on growing and shrinking MT plus ends was extremely rare and was only observed on one occasion (Fig. 2B, right MT).
Cin8 clustering at the minus ends promotes MT capturing and plus-end-directed antiparallel sliding of stable and dynamic MTs
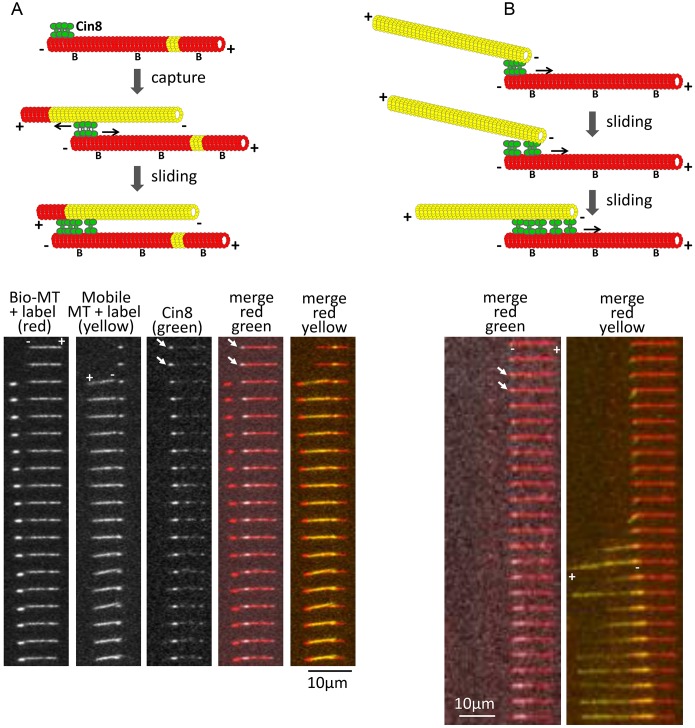
Kinesin-5 motors perform their essential mitotic functions by crosslinking and sliding apart antiparallel MTs. Thus, we next examined how the minus-end clustering of Cin8 is linked to its ability to crosslink and slide apart antiparallel MTs. We applied a three-color fluorescence TIRF microscopy strategy (Figs 3 and 4; Movies 5–9) in which one set of MTs was immobilized to the glass surface while the second set was free in solution. The addition of Cin8 motors promoted antiparallel sliding of the free MTs onto the immobilized MTs. We first confirmed that the Cin8 motors could slide apart MTs in a plus-end-directed manner, using polarity-marked GMPCPP-stabilized MTs (Fig. 3A; Fig. S3A–C, Movies 5, 6). Cin8-induced MT sliding was slow, 17±2 nm/s (mean±s.e.m., n=27), consistent with previous data (Gerson-Gurwitz et al., 2011) and with the rates of Cin8-induced spindle elongation in early anaphase in S. cerevisiae cells (Movshovich et al., 2008; Straight et al., 1998).
Fig. 3.
Cin8, clustered at the minus ends of MTs, promotes MT capture and antiparallel sliding. Two examples of capture events are shown. Biotinylated (‘B’) Alexa-Fluor-633-labeled (red) MTs were immobilized on the surface via an anti-biotin antibody. Alexa-Fluor-543-labeled free MTs (yellow) were then added in the presence of Cin8. Top: schematic representation of capturing and sliding. Bottom: time-lapse images. Cin8 clustered at the minus end of the immobilized MT is marked by a white arrow. Bio-MT, biotinylated MT. (A) Plus ends of the MTs are labeled with a different fluorophore. The plus and minus ends of the MTs are indicated based on the polarity mark of the MTs. Cin8 promoted antiparallel plus-end-directed sliding with the minus ends of the MTs leading. Time difference between frames: 11.1 s; Movie 5. In the schematic in A, yellow in the immobilized MT represents a GMPCPP-stabilized seed. (B) Cin8 promotes initial attachment of the minus ends of two MTs. Time difference between frames: 29 s. The minus and plus ends of the MTs were assigned based on the location of Cin8 clusters and the direction of antiparallel sliding, with the minus ends leading; Movie 6, MT 1.
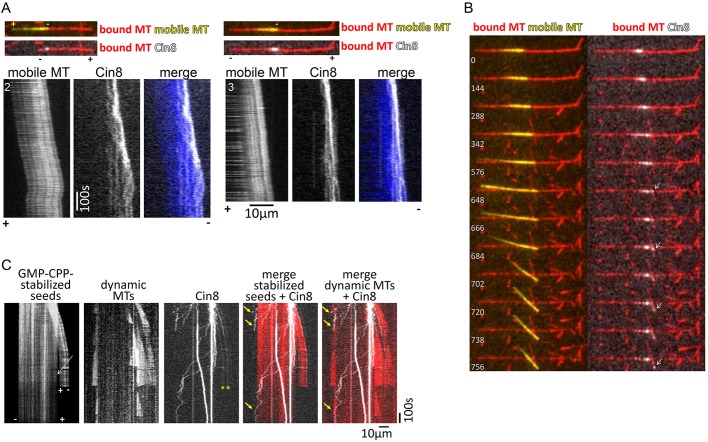
Fig. 4.
Cin8-induced antiparallel MT sliding of stable and dynamic MTs. (A,B) Cin8, clustered at the minus ends of the MTs, drives plus-end-directed antiparallel MT sliding (Movie 6). (A) Kymographs of the mobile MTs and of Cin8 motility (MTs 2 and 3 in Movie 6). Merged images of the MTs and Cin8 are shown at the top. Minus and plus ends of the MTs are indicated based on the directionality of the mobile (yellow) MT. In all cases, part of the Cin8 cluster moved along the leading minus end of the mobile MT, and some of the cluster became dispersed in the overlap region. (B) MT 3 (Movie 6) switched between two MTs while sliding. A montage of images at selected time points is shown, indicated at the bottom left of each frame (seconds). Cin8 leads the switch between the two red MTs. Arrows indicate the cluster of Cin8 that leads the switch of the yellow MT on top of the two red MTs. (C) Cin8 promotes crosslink and sliding apart of dynamic MTs (Movie 9). Kymographs of Cin8-mediated sliding of MTs in a dynamics-permitting experiment, as described in Fig. 2. In the left-hand kymographs, sliding of one GMPCPP-stabilized seed on top of the other is observed. Both seeds promote MT dynamics, second kymograph from the left. White arrow points to a gap between the two stable seeds, indicating that they are held together by Cin8-mediated crosslinking of at least one dynamic MT. Yellow arrows point to Cin8 tracking along the minus ends of the dynamic MTs. Yellow asterisks indicate the Cin8 particles that are likely to crosslink the dynamic MTs (Movie 9).
We found that capture of a second MT and the subsequent MT plus-end-directed sliding was mediated by Cin8 clusters at the minus ends on at least one of the two crosslinked MTs (Fig. 3A,B, arrows; Movies 5, 6, MTs 1, 3, and Movie 7). In our experiments, the concentration of the free (yellow) MTs was kept low, to prevent excessive bundling by Cin8 motors. Therefore, using this approach, MT capturing and crosslinking events were fairly rare. Nonetheless, we observed a total of seven MT capture and subsequent crosslinking–sliding events, and in six of these cases, at least one of the two MTs was interacting via a Cin8 cluster located at its minus end. The directionality of the MTs was determined either by plus-end labeling (Fig. 3A; Movie 5) or by the presence of Cin8 clusters (Movies 6 and 7), which, as we have demonstrated (Fig. 1), are located at the MT minus end in ∼96% of cases. Finally, we also observed ten pairs of MTs interacting at their ends, without sliding, whose interaction was mediated by a Cin8 cluster located at the ends of these MTs (Fig. S3D,E and Movie 8), further supporting the notion that clustering of Cin8 at the minus-end of the MTs promotes MT capturing.
During antiparallel MT sliding, Cin8 motors were often localized to and co-transported with the leading minus end of the MTs, while some Cin8 clusters remained stationary in the overlapping zone (Fig. 4A and B; Movie 6, MTs 2, 3), suggesting that clusters of Cin8 mediate MT sliding. Indeed, in the examples shown in Fig. 3A (Movie 5) and Movie 7, during sliding, a free MT was observed pivoting around a single Cin8 motor cluster located at the minus end of the surface-immobilized MT, indicating that these clusters of Cin8 drive the antiparallel sliding of the free MT. Moreover, on a single occasion, we observed a sliding MT that switched between two stationary MTs on the surface that were nearly perpendicular to one another. The switching between the two stationary MTs was mediated through translocation of a Cin8 cluster located near to the minus end of the sliding MT (Fig. 4B; Movie 6, MT 3). Overall, we observed seven examples of Cin8 cluster-mediated MT sliding, arising through either MT pivoting (Fig. 3A; Movies 5 and 7) or through a change in the direction of a mobile MT within a stationary MT array (Fig. 4B; Movie 6, MT 3), supporting the idea that Cin8 clusters promote the antiparallel sliding of MTs.
In addition to sliding apart stabilized MTs, Cin8 was also able to slide apart dynamic MTs by forming clusters near to dynamic MT minus ends (Fig. 4C; Movie 9). Taken together, our results demonstrate that in contrast to the view that the kinesin-5 Cin8 is equally distributed in the overlapping zone between two antiparallel MTs, clusters of Cin8 motors mediate the initial antiparallel MT capturing and sliding apart, driven by plus-end-directed motility near to the MT minus ends. Our data also demonstrate that as the overlap between the two antiparallel MTs increases, the clusters of Cin8 become dispersed and Cin8 motors become more equally distributed between the two sliding MTs (Figs 3 and 4A). Thus, it is likely that as the overlap between the antiparallel MTs increases, the contribution to sliding of Cin8 in the overlapping region increases, consistent with previous findings relating to kinesin-5 in higher eukaryotes (Kapitein et al., 2005).
Before spindle assembly, Cin8 clusters near to the SPBs through its minus-end-directed motility
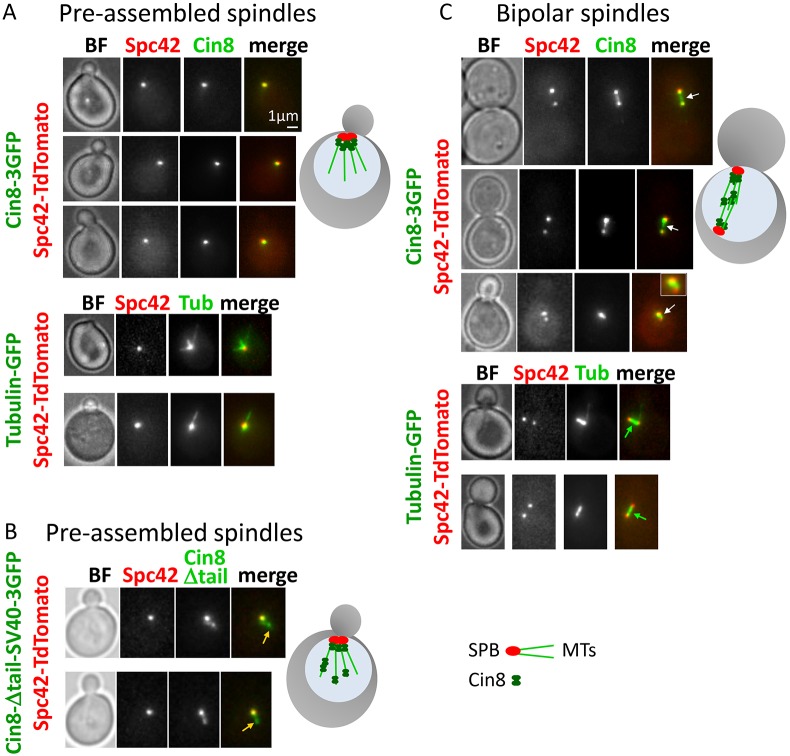
Our in vitro data indicate that Cin8 exhibits fast minus-end-directed motility and clustering at the minus end of single MTs (Fig. 1), and that it distributes in the MT overlap region during antiparallel MT sliding (Figs 3 and 4A). Based on these data, we predicted that in cells, before spindle assembly, Cin8 should accumulate near the spindle poles, at the minus end of the nuclear MTs, while in the assembled spindles, Cin8 should be distributed between the poles, on the overlapping spindle MTs. To test this prediction, we examined the in vivo localization of Cin8 motors before and following the assembly of mitotic spindles (Fig. 5). Indeed, we found that in small-budded early mitotic yeast cells with monopolar spindles, Cin8 localized at the SPBs and not along the MTs, consistent with its minus-end-directed motility on the nuclear MTs. These data are also consistent with previous reports showing that when Cin8 is expressed in the cytoplasm due to a mutated nuclear localization sequence, Cin8 concentrates near the SPBs, likely as a result of its minus-end-directed motility along cytoplasmic MTs (Duselder et al., 2015; Roostalu et al., 2011). Furthermore, this localization of wild-type Cin8 is similar to that of the bi-directional (Edamatsu, 2014) S. pombe kinesin-5 Cut7 in pre-assembled spindles (Hagan and Yanagida, 1992). In contrast to wild-type Cin8, a tail-less mutant of Cin8 that contains the SV40 nuclear localization sequence did not concentrate solely at the SPBs in the pre-assembled spindles. Instead, it was found in different locations within the nucleus (Fig. 5B). Under high ionic strength conditions, the tail-less Cin8 mutant moves bi-directionally while the wild-type Cin8 is exclusively minus-end directed (Duselder et al., 2015). Thus, SPB localization of wild-type Cin8 but not of the tail-less variant strongly indicates that before spindle assembly, the minus-end-directed motility of Cin8 is required for its localization near to the SPBs.
Fig. 5.
Spindle localization of Cin8 before and following the formation of a bipolar spindle. Images of cells co-expressing the SPB-localizing Spc42–tdTomato with 3GFP-tagged Cin8 variants or tubulin–GFP (Tub), indicated on the left of each panel. Schematic presentations of yeast cells, MTs, SPBs and Cin8 are displayed on the right of each group. (A) Cells with pre-assembled spindles expressing wild-type Cin8–3GFP (top) or tubulin–GFP (bottom). When SPBs were not separated, Cin8 localized as a single cluster overlapping with SPBs (top), in spite of the fact that the MTs were present and emanating from SPBs (bottom). BF, bright-field image. (B) Cells with pre-assembled spindles expressing a GFP-tagged tail-less mutant of Cin8 (Cin8-Δtail-SV40–3GFP). In contrast to the wild-type Cin8, this mutant has been previously shown to move bi-directionally under high ionic strength conditions (Duselder et al., 2015). Orange arrows point to Cin8 localization away from the SPBs. (C) Cells with bipolar spindles expressing Cin8–3GFP (top) or tubulin–GFP (bottom). Cin8 localized near to the SPBs, as well as between the SPBs (white arrows), probably along the spindle MTs (green arrows).
In contrast to the pre-assembled spindles, when the SPBs separated, Cin8 localized in the region between the SPBs, along the spindle MTs (Fig. 5C). This localization is consistent with Cin8 cross-linking activity on antiparallel interpolar MTs. At this stage, some Cin8 was also located near to the SPBs (Fig. 5C), which can result from Cin8 clustering at the minus end of the interpolar MTs, similarly to its localization at the pre-assembled spindles (Fig. 5A). Alternatively, the localization of Cin8 in the vicinity of the SPBs in the bipolar spindles can result from Cin8 binding to the kinetochores, which at this stage are congressed near to the SPBs (Tytell and Sorger, 2006), and/or at the short kinetochore MTs (Gardner et al., 2008). Thus, we find that although before SPB separation, Cin8 is localized exclusively at the minus end on nuclear MTs near to the SPBs, part of Cin8 relocates to the antiparallel spindle MTs following SPB separation. We propose that before spindle assembly, the localization of Cin8 near to the SPBs through its minus-end-directed motility is essential for the spindle assembly function of Cin8.
DISCUSSION
Since the discovery of the bi-directional motility of Cin8, the mechanism of this bi-directionality has been examined in several studies. On the one hand, it has been suggested that in multi-motor and antiparallel sliding assays, the switch to the plus-end directionality is induced by the Cin8 motors coupled through the MT that they interact with (Roostalu et al., 2011). This model was mainly based on the finding that the switch from minus- to plus-end-directed motility in the multi-motor gliding assay was dependent on MT length – the longer the MTs, the greater the likelihood of plus-end-directed motility – indicating that the number of motors interacting with the same MT determines the directionality (Roostalu et al., 2011). On the other hand, it has also been shown that the ability of Cin8 to move in both directions is an intrinsic property of Cin8 motors since the large insert in loop 8 of the catalytic domain affects the directionality of Cin8 in the single-molecule motility assay (Gerson-Gurwitz et al., 2011) and a dimeric chimera comprising the motor domain of Cin8 and dimerization domain of kinesin-1 moves in both plus- and minus-end directions (Duselder et al., 2015). Here, we describe a different, new mode of directionality switch of Cin8, induced by its accumulation in clusters on single MTs (Fig. 1). This new directionality-switch behavior suggests that it is controlled by allosteric intermolecular interactions between multiple tetrameric Cin8 motors in a cluster. These interactions are likely to be labile since we occasionally observed the splitting of clusters on MTs, resulting in sub-clusters with different directionalities (Fig. 2E, green asterisk; Movie 4). Possible pathways of cluster formation in vitro are proposed in Fig. S4; more work is required to elucidate the mechanism of motor-cluster formation and how clustering of motors controls their motile properties. Intermolecular interactions among Cin8 motors in a cluster may involve the conserved kinesin-5 tail domain, which emerges in proximity to the motor domain in the kinesin-5 tetramer (Scholey et al., 2014) and has been shown to influence the directionality of Cin8 (Duselder et al., 2015). Cin8 motor clustering may also involve additional structural elements such as the uniquely long loop 8 in the Cin8 motor (Gerson-Gurwitz et al., 2011), or phosphorylation of Cdk1-specific sites in the motor domain of Cin8, which have been shown to affect the spindle localization of Cin8 during anaphase (Avunie-Masala et al., 2011) as well as its motile properties and directionality in vitro (Shapira and Gheber, 2016).
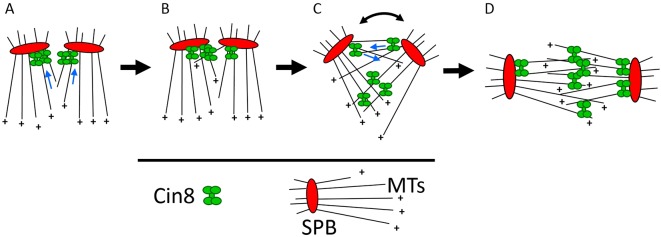
We show that clustering of Cin8 on single MTs promotes capturing, crosslinking and sliding apart of antiparallel MTs (Figs 3 and 4), and that prior to spindle assembly, Cin8 forms clusters near to the SPBs as a result of its minus-end-directed motility (Fig. 5). Based on these biochemical and in vivo data, we propose a revised model for tetrameric Cin8 motor function in mitotic spindle assembly and propose a physiological role for its bi-directional motility (Fig. 6). Before spindle assembly, Cin8 exhibits minus-end-directed motility and concentrates into clusters at the minus ends of nuclear MTs, near to the SPBs (Fig. 6A). At this location, clusters of Cin8 capture MTs emanating from the neighboring SPB (Fig. 6B) and promote their plus-end-directed antiparallel sliding. The antiparallel MT overlap due to Cin8-mediated crosslinking and/or sliding apart induces the initial separation of the two SPBs (Fig. 6C) to form the bipolar mitotic spindle (Fig. 6D). In this model, the accumulation of Cin8 near to the SPBs and capture of antiparallel MTs at this location are the key steps for Cin8-mediated SPB separation and spindle formation. It has previously been shown that before bipolar spindle assembly in S. cerevisiae cells, duplicated SPBs are located in close vicinity with limited overlap between the nuclear MTs (Lim et al., 1996; Peterson and Ris, 1976). We propose that clustering of Cin8 near to the SPBs at this initial state is required to maximize the crosslinking of nuclear MTs emanating from the neighboring SPBs. Moreover, before the initial SPB separation, the nuclear MTs emanating from the neighboring SPBs interdigitate in a near-parallel manner, thus, clusters of Cin8 may provide the required flexibility and force to slide apart iMTs that are not oriented in an antiparallel manner, inducing the initial separation of the SPBs.
Fig. 6.
Model for the physiological role of minus-end-directed motility of Cin8. Schematic presentation of the suggested model. Before spindle assembly, when the two SPBs are located in close vicinity, there is a limited overlap between interpolar MTs. (A) The minus-end motility of Cin8 and its accumulation at the minus ends of the MTs result in the accumulation of Cin8 near to the SPBs, which maximizes MT crosslinking at the location at which the overlap between spindle MTs occurs. (B) Cin8 clusters at the minus end induce capturing of MTs from the neighboring SPB. (C) Capturing and antiparallel sliding apart of MTs from this location promotes the initial separation of the SPBs. This increases the overlap between interpolar MTs, adds sites for Cin8-induced crosslinking and promotes SBP separation until the bipolar spindle is formed (D). The curved double-headed arrow indicates SPB separation. Blue arrows indicate the direction of Cin8 motility.
The role of Cin8 in the assembly and maintenance of the bipolar spindle is well established (Hoyt et al., 1992; Saunders and Hoyt, 1992). It has previously been demonstrated that a motor domain mutant, Cin8-R196K, that is partially impaired in the ability to promote surface MT gliding in a multi-motor motility assay (Gheber et al., 1999) can facilitate spindle assembly in the absence of Kip1, which has overlapping functions with Cin8 (Crasta et al., 2006; Gheber et al., 1999), suggesting that the motility function of Cin8 is not required for spindle assembly. Since the antiparallel MT sliding activity of the Cin8-R196K mutant has not yet been examined, it is to be determined whether force generation by Cin8 is required for spindle assembly. Nevertheless, we propose that localization of Cin8 near to the SPBs is essential before spindle assembly to mediate the initial SPB separation either by capturing or by capturing and sliding apart antiparallel MTs emanating from the neighboring SPBs.
Thus far, three kinesin-5 motors have been shown to be bi-directional: S. cerevisiae Cin8 and Kip1 (Fridman et al., 2013; Gerson-Gurwitz et al., 2011), and S. pombe Cut7 (Edamatsu, 2014). The three homologs are expressed in yeast cells that divide using ‘closed mitosis’ without nuclear envelope disassembly. In higher eukaryotes, which divide using ‘open mitosis’ due to nuclear envelope breakdown, the cytoplasmic dynein has been shown to contribute to the initial spindle-pole separation and cooperate with kinesin-5 function in spindle assembly (Clift and Schuh, 2015; Sommi et al., 2011). In contrast, in S. cerevisiae and S. pombe cells, it has been shown that dynein does not serve a role in mitotic spindle assembly (Courtheoux et al., 2007; Eshel et al., 1993). We propose that with no external pulling force provided by dynein, the minus-end-directed motility of yeast kinesin-5 motors is required to cluster the protein at the MT minus-ends and to provide the initial SPB separation needed for spindle assembly. The minus-end-directed motility of Cin8 and its ability to capture and slide antiparallel MTs by clustering at the MT minus end may therefore be a demonstration of the evolutionary adaptation of this protein to perform its essential function in spindle assembly.
MATERIALS AND METHODS
Overexpression and purification of Cin8 for in vitro assays
Cin8 overexpression was performed in a protease-deficient S. cerevisiae strain containing a 2 μm plasmid for Cin8-3GFP–6His expressed from the pGAL1 promoter [LGY 4054 (MATα, leu2-3,112, reg-1-501, ura3-52, pep4-3, prb1-1122, gal1, (pOS2: 2μ, LEU2, PGAL1-CIN8-3GFP-6HIS)]. Cin8 purification was performed as previously described (Gerson-Gurwitz et al., 2011). Cultures were grown in 1–4 l of minimal medium supplemented with 2% raffinose to mid-log phase. For induction of Cin8 expression, 2% galactose was added and the culture was grown for an additional 5 h. Cells were pelleted, washed once with purified water and twice with lysis–binding buffer [500 mM NaCl, 10% glycerol, 2 mM β-mercaptoethanol, 1 mM PMSF, 1 mM MgCl2, 0.1 mM ATP, 0.2% Triton X-100, Complete protease inhibitor (Roche), protease inhibitor cocktail for histidine-tagged proteins (Sigma), 0.1 µg/ml leupeptin, 0.2 µg/ml aprotinin, 0.1 mM benzamidine, 50 mM Tris-HCl, pH 8] and ground using a cryo-beads mill. The ground cells were thawed and centrifuged at 15,000 g for 20 min at 4°C. Ni-NTA beads (Invitrogen) were added to the cell extract, and binding was allowed to occur for 1.5 h on a shaker at 4°C. The mix was loaded onto a column and washed with 3–5 column volumes of wash buffer (500 mM NaCl or KCl, 30 mM imidazole, 10% glycerol, 1.5 mM β-mercaptoethanol, 1 mM MgCl2, 0.1 mM ATP, 0.2% Triton X-100, 50 mM KH2PO4, pH 8). The protein was eluted with 2–5 ml of elution buffer (500 mM NaCl or KCl, 350 mM imidazole, 10% glycerol, 1.5 mM β-mercaptoethanol, 1 mM MgCl2, 0.1 mM ATP, 0.2% Triton X-100, 50 mM KH2PO4, pH 8). The eluent was collected in 200–500 µl fractions, and aliquots were analyzed by SDS-PAGE and western blotting analysis for detection of Cin8. The selected fractions were aliquoted, snap frozen and stored until use at −80°C.
In vitro single molecule MT–Cin8 motor motility assays
Single-motor motility assays and dynamic MT polymerization assays were performed at 28°C and 37°C, respectively, in flow chambers assembled using a clean and freshly salinized coverslip using a double-sided tape, as previously described (Al-Bassam, 2014). Surfaces were coated with biotinylated bovine serum albumin (Sigma-Aldrich, A8549) and then treated with NeutraAvidin (Life Technologies, A2666) to attach biotinylated MTs along surfaces. Alternatively, surfaces were treated with a mouse monoclonal anti-biotin antibody (Invitrogen), then pacified using 1% pluronic acid F-127, following which biotinylated MTs were attached. Flow chambers were treated with MB1 (10% glycerol, 175 mM NaCl-KCl, 2 mM MgSO2, 1 mM EGTA, 30 µg/ml casein, 1 mM DTT, 3 mM ATP, 50 mM Tris-HCl, pH 7.2, 30 mM PIPES). For Cin8 motility (Fig. 1), 20 µl of 0.1–4 µg/ml Cin8 in MB1 buffer was added to the immobilized MTs, and the MTs were immediately imaged using a TIRF microscope. For the dynamic MT assay (Fig. 2), 0.6–1 mg/ml tubulin, 0.1 mg/ml tubulin–Alexa-Fluor-543 and 3 µg/ml Cin8 in PCB (10% glycerol, 135 mM KCl, 1 mM MgCl2, 1 mM EGTA, 30 µg/ml casein, 1 mM DTT, 1–2 mM GTP, 3 mM ATP, 80 mM PIPES, pH 7.2) were added to immobilized Alexa-Fluor-633-labeled GMPCPP-stabilized seeds. For antiparallel MT sliding assays (Figs 3 and 4), Alexa-Fluor-543-labeled MTs diluted 1:10 in MB1 and 2 µg/ml Cin8 were added to the immobilized Alexa-Fluor-633-labeled MTs. Imaging was performed using a Nikon TI-microscope-equipped TIRF objective and optical launch operating three solid-state lasers at 488 nm, 561 nm and 640 nm wavelengths, with 2–4 s intervals for each of the three wavelengths using optical excitation and emission filters for each wavelength. Kymographs, line-scans and montages were created using the freely available ImageJ software (https://imagej.nih.gov/ij/). Velocities were obtained from kymographs by measuring the slopes of directional movements, as previously described (Shapira and Gheber, 2016). Directionality of the MTs was assigned based on plus-end labeling and/or the direction of fast-moving Cin8 molecules, or based on the direction of movement of a MT during antiparallel sliding. Fluorescence intensity distributions of Cin8 particles were determined with the use of the TrackMate (Jaqaman et al., 2008) ImageJ plugin (http://imagej.net/TrackMate). Statistical significance was determined using a two-tailed Student's t-test.
Live-cell imaging
Live-cell imaging was performed as previously described (Duselder et al., 2015; Fridman et al., 2013) using cells co-expressing the SPB-localizing Spc42–tdTomato with Cin8–3GFP [LGY-4391: a, ura3, leu2, his3, lys2, cin8::LEU2, spc42::KanMX-SPC42-TdTomato (pVF68: CEN, URA, CIN8-3GFP)] or tubulin–GFP (LGY-4425: α, ura3, leu2, his3, lys2, GAL+, ADE2, his3::TUB1-GFP-HIS3, spc42::KanMX-SPC42-TdTomato). Cells were grown overnight in medium lacking tryptophan; 2 h before imaging, the cells were diluted 10-fold. A small sample of cells was placed on a low-fluorescence agarose gel on a slide. Images were acquired at room temperature using a Zeiss Axiovert 200M-based microscope setup equipped with a cooled CCD Andor Neo sCMOS camera, and MetaMorph (MDS Analytical Technologies) software. Images of z-stacks of 11 slices were obtained with a microscope in three channels, with a separation of 0.5 μm between planes.
Note added in proof
During the revisions of this manuscript, it has been reported that the S. pombe kinesin-5 Cut7 switches directionality as a result of MT crowding by motile or non-motile proteins (Britto et al., 2016). This property of Cut7, and the ability of Cin8 to switch directionality by motor-clustering reported here, may result from a similar control mechanism, pointing to generality of the reported phenomena.
Acknowledgements
We thank Levi Gheber and Yael Lavi (Biotechnology Engineering, Ben-Gurion University of the Negev, Israel) for help with TrackMate analysis of Cin8 particle intensities. We thank Ken Kaplan (Molecular Cellular Biology, UC Davis, CA) for introducing L.G. to J.A.-B., and for supporting work in purifying Cin8 from S. cerevisiae. J.A.-B. is grateful for support from Jonathan Scholey and Jodi Nunnari (Molecular Cellular Biology, UC Davis, CA).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
L.G. and J.A.-B. conceived and supervised the study; O.S., A.G. and L.G. performed experiments; O.S. and L.G. analyzed data; L.G. and J.A.-B. wrote the manuscript; O.S., A.G., J.A.-B. and L.G. revised the manuscript.
Funding
This work was supported in part by the Israel Science Foundation (ISF) (grant 165/13 awarded to L.G.), the National Science Foundation (NSF-1615991), United States - Israel Binational Science Foundation (BSF-2015851 awarded to L.G. and J.A.-B.) and the National Institutes of Health (grant NIH-R01-GM11283 awarded to J.A.-B.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.195040.supplemental
References
- Al-Bassam J. (2014). Reconstituting dynamic microtubule polymerization regulation by TOG domain proteins. Methods Enzymol. 540, 131-148. 10.1016/B978-0-12-397924-7.00008-X [DOI] [PubMed] [Google Scholar]
- Arnal I. and Wade R. H. (1995). How does taxol stabilize microtubules? Curr. Biol. 5, 900-908. 10.1016/S0960-9822(95)00180-1 [DOI] [PubMed] [Google Scholar]
- Asraf H., Avunie-Masala R., Hershfinkel M. and Gheber L. (2015). Mitotic slippage and expression of survivin are linked to differential sensitivity of human cancer cell-lines to the Kinesin-5 inhibitor monastrol. PLoS ONE 10, e0129255 10.1371/journal.pone.0129255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avunie-Masala R., Movshovich N., Nissenkorn Y., Gerson-Gurwitz A., Fridman V., Koivomagi M., Loog M., Hoyt M. A., Zaritsky A. and Gheber L. (2011). Phospho-regulation of kinesin-5 during anaphase spindle elongation. J. Cell Sci. 124, 873-878. 10.1242/jcs.077396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A., Arnaud L. and Nigg E. A. (1997). Phosphorylation by p34cdc2 protein kinase regulates binding of the kinesin-related motor HsEg5 to the dynactin subunit p150 Glued. J. Biol. Chem. 272, 19418-19424. 10.1074/jbc.272.31.19418 [DOI] [PubMed] [Google Scholar]
- Britto M., Goulet A., Rizvi S., von Loeffelholz O., Moores C. A. and Cross R. A. (2016). Schizosaccharomyces pombe kinesin-5 switches direction using a steric blocking mechanism. Proc. Natl. Acad. Sci. USA 113, E7483-E7489. 10.1073/pnas.1611581113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. and Hancock W. O. (2015). Kinesin-5 is a microtubule polymerase. Nat. Commun. 6, 8160 10.1038/ncomms9160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift D. and Schuh M. (2015). A three-step MTOC fragmentation mechanism facilitates bipolar spindle assembly in mouse oocytes. Nat. Commun. 6, 7217 10.1038/ncomms8217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtheoux T., Gay G., Reyes C., Goldstone S., Gachet Y. and Tournier S. (2007). Dynein participates in chromosome segregation in fission yeast. Biol. Cell 99, 627-637. 10.1042/BC20070047 [DOI] [PubMed] [Google Scholar]
- Crasta K., Huang P., Morgan G., Winey M. and Surana U. (2006). Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. EMBO J. 25, 2551-2563. 10.1038/sj.emboj.7601136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duselder A., Fridman V., Thiede C., Wiesbaum A., Goldstein A., Klopfenstein D. R., Zaitseva O., Janson M. E., Gheber L. and Schmidt C. F. (2015). Deletion of the tail domain of the kinesin-5 Cin8 affects its directionality. J. Biol. Chem. 19, 620799 10.1074/jbc.m114.620799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edamatsu M. (2014). Bidirectional motility of the fission yeast kinesin-5, Cut7. Biochem. Biophys. Res. Commun. 446, 231-234. 10.1016/j.bbrc.2014.02.106 [DOI] [PubMed] [Google Scholar]
- Eshel D., Urrestarazu L. A., Vissers S., Jauniaux J. C., van Vliet-Reedijk J. C., Planta R. J. and Gibbons I. R. (1993). Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc. Natl. Acad. Sci. USA 90, 11172-11176. 10.1073/pnas.90.23.11172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman V., Gerson-Gurwitz A., Movshovich N., Kupiec M. and Gheber L. (2009). Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Rep. 10, 387-393. 10.1038/embor.2009.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman V., Gerson-Gurwitz A., Shapira O., Movshovich N., Lakamper S., Schmidt C. F. and Gheber L. (2013). Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. J. Cell Sci. 126, 4147-4159. 10.1242/jcs.125153 [DOI] [PubMed] [Google Scholar]
- Gardner M. K., Bouck D. C., Paliulis L. V., Meehl J. B., O'Toole E. T., Haase J., Soubry A., Joglekar A. P., Winey M., Salmon E. D.. et al. (2008). Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell 135, 894-906. 10.1016/j.cell.2008.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerson-Gurwitz A., Movshovich N., Avunie R., Fridman V., Moyal K., Katz B., Hoyt M. A. and Gheber L. (2009). Mid-anaphase arrest in S. cerevisiae cells eliminated for the function of Cin8 and dynein. Cell. Mol. Life Sci. 66, 301-313. 10.1007/s00018-008-8479-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerson-Gurwitz A., Thiede C., Movshovich N., Fridman V., Podolskaya M., Danieli T., Lakämper S., Klopfenstein D. R., Schmidt C. F. and Gheber L. (2011). Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 30, 4942-4954. 10.1038/emboj.2011.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheber L., Kuo S. C. and Hoyt M. A. (1999). Motile properties of the kinesin-related Cin8p spindle motor extracted from Saccharomyces cerevisiae cells. J. Biol. Chem. 274, 9564-9572. 10.1074/jbc.274.14.9564 [DOI] [PubMed] [Google Scholar]
- Gordon D. M. and Roof D. M. (1999). The kinesin-related protein Kip1p of Saccharomyces cerevisiae is bipolar. J. Biol. Chem. 274, 28779-28786. 10.1074/jbc.274.40.28779 [DOI] [PubMed] [Google Scholar]
- Hagan I. and Yanagida M. (1992). Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature 356, 74-76. 10.1038/356074a0 [DOI] [PubMed] [Google Scholar]
- Hoyt M. A., He L., Loo K. K. and Saunders W. S. (1992). Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J. Cell Biol. 118, 109-120. 10.1083/jcb.118.1.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S. L. and Danuser G. (2008). Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695-702. 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein L. C., Peterman E. J. G., Kwok B. H., Kim J. H., Kapoor T. M. and Schmidt C. F. (2005). The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435, 114-118. 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- Kashina A. S., Baskin R. J., Cole D. G., Wedaman K. P., Saxton W. M. and Scholey J. M. (1996). A bipolar kinesin. Nature 379, 270-272. 10.1038/379270a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H. H., Goh P. Y. and Surana U. (1996). Spindle pole body separation in Saccharomyces cerevisiae requires dephosphorylation of the tyrosine 19 residue of Cdc28. Mol. Cell. Biol. 16, 6385-6397. 10.1128/MCB.16.11.6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer T. U., Kapoor T. M., Haggarty S. J., King R. W., Schreiber S. L. and Mitchison T. J. (1999). Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 286, 971-974. 10.1126/science.286.5441.971 [DOI] [PubMed] [Google Scholar]
- Movshovich N., Fridman V., Gerson-Gurwitz A., Shumacher I., Gertsberg I., Fich A., Hoyt M. A., Katz B. and Gheber L. (2008). Slk19-dependent mid-anaphase pause in kinesin-5-mutated cells. J. Cell Sci. 121, 2529-2539. 10.1242/jcs.022996 [DOI] [PubMed] [Google Scholar]
- Peterson J. B. and Ris H. (1976). Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J. Cell Sci. 22, 219-242. [DOI] [PubMed] [Google Scholar]
- Roostalu J., Hentrich C., Bieling P., Telley I. A., Schiebel E. and Surrey T. (2011). Directional switching of the Kinesin cin8 through motor coupling. Science 332, 94-99. 10.1126/science.1199945 [DOI] [PubMed] [Google Scholar]
- Saunders W. S. and Hoyt M. A. (1992). Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell 70, 451-458. 10.1016/0092-8674(92)90169-D [DOI] [PubMed] [Google Scholar]
- Saunders W. S., Koshland D., Eshel D., Gibbons I. R. and Hoyt M. A. (1995). Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J. Cell Biol. 128, 617-624. 10.1083/jcb.128.4.617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey J. E., Nithianantham S., Scholey J. M. and Al-Bassam J. (2014). Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers. Elife 3, e02217 10.7554/eLife.02217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira O. and Gheber L. (2016). Motile properties of the bi-directional kinesin-5 Cin8 are affected by phosphorylation in its motor domain. Sci. Rep. 6, 25597 10.1038/srep25597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D. J., McDonald K. L., Brown H. M., Matthies H. J., Walczak C., Vale R. D., Mitchison T. J. and Scholey J. M. (1999). The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 144, 125-138. 10.1083/jcb.144.1.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommi P., Cheerambathur D., Brust-Mascher I. and Mogilner A. (2011). Actomyosin-dependent cortical dynamics contributes to the prophase force-balance in the early Drosophila embryo. PLoS ONE 6, e18366 10.1371/journal.pone.0018366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A. F., Sedat J. W. and Murray A. W. (1998). Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 143, 687-694. 10.1083/jcb.143.3.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiede C., Fridman V., Gerson-Gurwitz A., Gheber L. and Schmidt C. F. (2012). Regulation of bi-directional movement of single kinesin-5 Cin8 molecules. BioArchitecture 2, 70-74. 10.4161/bioa.20395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytell J. D. and Sorger P. K. (2006). Analysis of kinesin motor function at budding yeast kinetochores. J. Cell Biol. 172, 861-874. 10.1083/jcb.200509101 [DOI] [PMC free article] [PubMed] [Google Scholar]