Abstract
The antimetabolite 5-fluorouracil (5FU) is a widely used chemotherapeutic for the treatment of solid tumors. Although 5FU slows DNA synthesis by inhibiting the ability of thymidylate synthetase to produce dTMP, the drug also has significant effects on RNA metabolism. Recent genome-wide assays for 5FU-induced haploinsufficiency in Saccharomyces cerevisiae identified genes encoding components of the RNA processing exosome as potential targets of the drug. In this report, we used DNA microarrays to analyze the effect of 5FU on the yeast transcriptome and found that the drug causes the accumulation of polyadenylated fragments of the 27S rRNA precursor and that defects in the nuclear exoribonuclease Rrp6p enhance this effect. The size distribution of these RNAs and their sensitivity to Rrp6p suggest that they are normally degraded by the nuclear exosome and a 5′-3′ exoribonuclease. Consistent with this hypothesis, 5FU inhibits the growth of RRP6 mutants with defects in the degradation function of the enzyme and it interferes with the degradation of an rRNA precursor. The detection of poly(A)+ pre-RNAs in strains defective in various steps in ribosome biogenesis suggests that the production of poly(A)+ pre-rRNAs may be a general result of defects in rRNA processing. These findings suggest that 5FU inhibits an exosome-dependent surveillance pathway that degrades polyadenylated precursor rRNAs.
5-Fluorouracil (5FU) is one of the most successful and widely used chemotherapeutics for the treatment of solid tumors in cancer patients (27). The drug was developed in a rational strategy to inhibit DNA synthesis by interfering with the activity of thymidylate synthetase (TS). Three mechanisms have been proposed to explain the cytotoxic effects of 5FU, and each relies on the efficient import of the base into cells and its rapid conversion into 5-fluoro-UMP (5F-UMP) and 5F-dUMP (27, 33). 5F-UMP is ultimately converted into 5F-UTP, which serves as a substrate for RNA synthesis. Normally, dUMP is converted into dTMP by TS and ultimately into dTTP, which serves as a substrate for DNA synthesis. The first proposed mechanism of action for 5FU relies on the fact that the 5-fluoro linkage in 5F-dUMP inhibits the TS-catalyzed, 5-position methylation that normally converts dUMP into dTMP. The ultimate decrease in dTTP levels should inhibit DNA synthesis. Indeed, inactivation of TS, encoded by CDC21 in Saccharomyces cerevisiae, or thymidine starvation, results in S-phase cell cycle arrest (7). The second proposed mechanism follows from TS inhibition that results in elevation of dUTP levels. The decrease in dTTP and the increase in dUTP levels enhance the incorporation of dUTP into DNA. Repair of uracil-containing DNA begins with uracil removal by uracil-DNA glycosylase to produce apyrimidinic sites in the DNA and proceeds with a DNA chain break and subsequent repair by DNA polymerase and DNA ligase. Thus, inhibition of DNA synthesis and/or the requirement for DNA repair may result in the cell cycle arrest associated with 5FU treatment. However, assessment of the potential for DNA fragmentation in 5FU-treated cells indicated that the drug levels required for the effect are 10 to 100 times those needed for cytotoxicity (33). The third possible mechanism of 5FU cytotoxicity features the incorporation of 5FU into RNA. The strongest argument for the role of RNA in 5FU action comes from experiments showing that cotreatment of cells with uridine, but not thymidine, relieves the cytotoxic and apoptotic effects of 5FU (6, 13, 26, 38). Since 5FU is a competitive inhibitor of TS, thymidine abolishes the decrease in TTP levels caused by treatment with the drug. Thus, relief of 5FU's cytotoxic effects by uridine, but not thymidine, is inconsistent with mechanisms in which defects in DNA metabolism feature in the antiproliferative effects of the drug.
Research into the effect of 5FU on RNA metabolism has focused largely on rRNA synthesis since rRNA is the most abundant class of RNA produced by proliferating cells (reviewed in references 27 and 33). Eukaryotic rRNAs are synthesized in the nucleus by RNA polymerase I (25S, 18S, and 5.8S rRNAs) and RNA polymerase III (5S rRNA). The 28S, 18S, and 5.8S rRNAs are transcribed into a single precursor RNA (pre-rRNA) that undergoes a series of endo- and exonucleotytic processing steps that produce the mature rRNAs found in ribosomes. The nuclear RNA processing exosome, a complex of 10 exoribonucleases, plays an important role in rRNA processing as it is required for proper 3′-end processing of 5.8S rRNA and for the degradation of abnormal pre-rRNA processing intermediates (2, 4, 30). rRNA processing is best understood in yeast, but evidence suggests that the same major features are used to produce the 28S, 18S, and 5.8S rRNAs in humans (18). 5FU is rapidly incorporated into pre-rRNA in treated cells, and some experiments suggest that it may cause inhibition of processing at early steps that normally lead to the production of 20S pre-rRNAs in human cells (10, 16, 17). Experiments with an rRNA processing system in vitro suggest a role for 5FU inhibition of a primary cleavage site of pre-rRNA, but the physiological relevance of this site has not been addressed (14). The importance of these findings is difficult to assess since the experiments were carried out prior to the availability of information on the specific pathway of rRNA processing and because of the lack of reagents for measuring effects on specific steps in the process. Moreover, the interpretation of the experiments in vivo is hampered by the possibility that the observed effects are secondary to some other target of 5FU. In some sense, the choice of rRNA synthesis as a potential target was ad hoc and inhibition alone cannot be used to argue for specificity since 5FU is incorporated into all types of RNA and has been shown to alter pre-mRNA metabolism as well (25).
Recent evidence has refocused attention on rRNA processing as a target of 5FU since genome-wide screens for gene products sensitive to 5FU identified components of the rRNA processing pathway (15, 28). Specifically, yeast strains heterozygous for deletions of genes encoding proteins required for pre-rRNA processing and ribosome biogenesis grew more slowly in the presence of 5FU than all other heterozygotes, including CDC21+/− strains. Heterozygosity of some components of the exosome, in particular RRP6+/− heterozygosity, caused the highest observed sensitivity, suggesting that 5FU may inhibit the activity of this RNA processing complex. Indeed, analysis of the effects of 5FU on the processing of pre-rRNAs in normal strains and the sensitive heterozygotes showed defects in rRNA maturation steps consistent with inhibition of the exosome (28). The exosome functions in mRNA decay pathways in the cytoplasm and in RNA processing and degradation pathways in the nucleus, where its activity requires Rrp6p (8, 30). Defects in exosome components including Rrp6p lead to the accumulation of polyadenylated noncoding RNAs including snRNAs, snoRNAs, rRNAs, and hypomodified initiator tRNA (1, 21, 23, 44). Here we report that a genome-wide comparison of yeast strains treated with 5FU revealed the accumulation of polyadenylated pre-rRNAs in a strain deficient in the nuclear exosome component Rrp6p. Analysis of the structure of these products and the effect of 5FU on specific RRP6 mutants suggests that the drug may inhibit a general pathway for the degradation of rRNA precursors.
MATERIALS AND METHODS
Yeast strains, oligonucleotides, and reagents.
Yeast strains are described in Table 1. Yeast strains were grown in yeast extract-peptone-dextrose (YPD) or synthetic complete dextrose (SCD) medium. Oligonucleotides used for reverse transcription (RT)-PCR and Northern blot analysis are listed in Table 2. Enzymes were purchased from Invitrogen (Carlsbad, Calif.) or Promega (Madison, Wis.). Radioisotopes for random-primed hexamer probe labeling (5′-[α-32P]dCTP; 3,000 Ci/mmol) and oligonucleotide labeling (5′-[γ-32P]dATP; 3,000 Ci/mmol) were purchased from NEN Life Science Products (Boston, Mass.).
TABLE 1.
Yeast strains used in this study
| Strain | Designation | Genotypea | Reference |
|---|---|---|---|
| YSB2001 | RRP6+/+ | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ/MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ | 45 |
| YSB2002 | RRP6+/− | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ RRP6/MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ rrp6::Kan | 45 |
| YSB2003 | RRP4+/− | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ RRP4/MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ rrp4::Kan | 45 |
| YSB2004 | RRP41+/− | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ RRP41/MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ rrp41::Kan | 45 |
| YSB2005 | RRP42+/− | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ RRP42/MATα his3-Δleu2-Δ lys2-Δ ura3-Δ rrp42:Kan | 45 |
| YSB2006 | RRP43+/− | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ RRP43/MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ rrp43::Kan | 45 |
| YSB2007 | RRP44+/− | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ RRP44/MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ rrp44::Kan | 45 |
| YSB2008 | RRP45+/− | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ RRP45/MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ rrp45::Kan | 45 |
| YSB2009 | RRP46+/− | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ RRP46/MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ rrp46::Kan | 45 |
| YSB2010 | LRP1+/− | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ LRP1/MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ lrp1::Kan | 45 |
| YSB2012 | CSL4+/− | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ CSL4/MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ csl4::Kan | 45 |
| YSB2013 | RAT1+/− | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ RAT1/MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ rat1::Kan | 45 |
| YSB2014 | CBF5+/− | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ CBF5/MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ cbf5::Kan | 45 |
| YSB2015 | MAK21+/− | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ MAK21/MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ mak21::Kan | 45 |
| YSB2016 | NOP4+/− | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ NOP4/MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ nop4::Kan | 45 |
| YSB2018 | rrp6−/− | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ rrp6::Kan/MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ rrp6::Kan | 45 |
| YSB1002 | RRP6 | MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ | 45 |
| YSB1005 | rrp6-Δ | MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ rrp6::Kan | 45 |
| YSB1007 | lrp1-Δ | MATα his3-Δ leu2-Δ lys2-Δ ura3-Δ lrp::Kan | 45 |
| R1158 | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ URA3::CMV-tTA | 22 | |
| YSB3002 | tetO7-RRP43 | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ URA3::CMV-tTA tetO7-RRP43 | 22 |
| YSB3003 | tetO7-RRP46 | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ URA3::CMV-tTA tetO7-RRP46 | 22 |
| YSB3007 | tetO7-CBF5 | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ URA3::CMV-tTA tetO7-CBF5 | 22 |
| YSB3008 | tetO7-MAK21 | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ URA3::CMV-tTA tetO7-MAK21 | 22 |
| YSB3009 | tetO7-NOP4 | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ URA3::CMV-tTA tetO7-NOP4 | 22 |
| YSB3010 | tetO7-UTP20 | MATahis3-Δ leu2-Δ met15-Δ ura3-Δ URA3::CMV-tTA tetO7-UTP20 | 22 |
| YSB151 | rrp6-Δ | MATα ade1 ade2 lys2 gal1 ura3-52 rrp6::Kan YCpGFP | 37 |
| YSB152 | RRP6 | MATα ade1 ade2 lys2 gal1 ura3-52 rrp6::Kan YCpGFPH (RRP6) | 37 |
| YSB153 | rrp6-3 | MATα ade1 ade2 lys2 gal1 ura3-52 rrp6::Kan YCpGFPH1 (rrp6-3) | 37 |
| YSB154 | rrp6-13 | MATα ade1 ade2 lys2 gal1 ura3-52 rrp6::Kan YCpGFPH10 (rrp6-13) | 37 |
CMV-tTA, tetracycline-responsive transcriptional activator under control of the cytomegalovirus promoter.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea | Source or reference |
|---|---|---|
| OSB151 | 5′-GTCTAGCCGCGAGGAAGG-3′ | 28 |
| OSB155 | 5′-TCCAGTTACGAAAATTCTTGTTTTTGACAA-3′ | 28 |
| OSB157 | 5′-GGCCAGCAATTTCAAGTTA-3′ | 28 |
| OSB377 | 5′-CTCCTACCTGATTTGAGGTCAAAC-3′ | This study |
| OSB379 | 5′-ACAAATCAGACAACAAAGGCTTAATC-3′ | This study |
| OSB380 | 5′-T21ACAAATCAGACAACAAAGGCTTAATCTCAGCAGAG-3′ | This study |
| OSB402 | 5′-GGCGGAAAGGCCTTGGGTGCTTGCTGGCG-3′ | This study |
| OSB403 | 5′-CACTCGAGTT30-3′ | This study |
| OSB451 | 5′-TGGGTTTGGAATCTGCCGGTATTG-3′ | This study |
| OSB452 | 5′-TTAGAAACACTTGTGGTGAACGATAG-3′ | This study |
T21 and T30 indicate sequences of 21 and 30 T's, respectively.
Northern blot analysis.
Total RNA or poly(A)+ RNA was isolated from yeast strains grown to an A600 of 0.6 to 0.9 (in SCD medium) or 1.0 to 1.2 (in YPD medium) as previously described (34), and Northern blot analysis was carried out as described by Briggs et al. (4).
RT-PCR.
One microgram of DNase-treated total RNA and 1 μl of oligo(dT) (OSB403; 50 pmol/μl) were used for cDNA synthesis in a 20-μl reaction mixture using Moloney murine leukemia virus reverse transcriptase (200 U/μl) according to the instructions of the manufacturer (Invitrogen). Fifty-microliter PCR mixtures contained 0.25 μl of cDNA product, 12.5 pmol of oligonucleotide primer, 0.025 mM (each) deoxynucleoside triphosphates, 2.5 mM MgCl2, and 0.25 μl of GoTaq DNA polymerase (Promega; 5 U/μl). PCR was performed with the following conditions: 94°C for 3 min followed by 20 cycles (94°C for 30 s, 55°C for 30 s, and 72°C for 30 s) followed by 72°C for 7 min. PCR products were stored at 4°C.
Microarray analysis.
Cells were grown in YPD at an A600 of 1.0 to 1.2 and treated with 20 μM 5FU for 1 h. Total RNA was isolated as previously described (34). Affymetrix Yeast S98 arrays were used for expression profiling. Sample labeling, hybridization, and scanning were conducted according to guidelines from Affymetrix (Santa Clara, Calif.). Sample labeling involved oligo(dT)-primed cDNA synthesis followed by transcription to yield single-stranded, biotinylated RNA probes. Duplicate experiments were carried out for each strain and condition, and the data were analyzed with GeneTraffic 3.1 (Iobion, La Jolla, Calif.) and Excel (Microsoft Corp., Redmond, Wash.). Data were normalized to the RRP6+/+ data set for each duplicate group of RNA samples. Within each of the two groups of array hybridizations, the total signal intensity from each chip varied from the others by 3.2% or less, and so no corrections for signal intensity were made. ACT1 mRNA was chosen as a control for RT-PCR experiments used to verify changes in RNA levels because its signal in the microarray analysis changed by 2% or less in response to the rrp6 mutations and/or treatment with 5FU. The insensitivity of ACT1 mRNA to Rrp6p activity and 5FU treatment was verified by Northern blot analysis and will be discussed elsewhere.
Polyribosome analysis.
Yeast polyribosome lysates were prepared from cells grown to an A600 of 1 to 2 at 30°C in YPD, and the lysates were separated by ultracentrifugation through 15 to 50% sucrose gradients and analyzed as previously described (39).
RESULTS
5FU inhibits the growth of strains with exosome mutations.
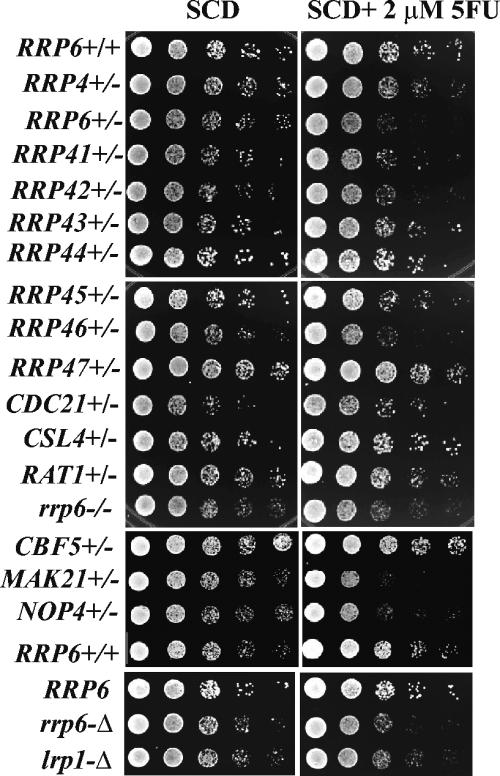
Recent growth competition experiments indicated that yeast strains heterozygous for deletions of genes encoding components of the RNA processing exosome and several other genes whose products play a role in 60S ribosome biogenesis exhibit slowed growth in the presence of the antimetabolite 5FU (15, 28). Specifically, strains heterozygous for deletions of RRP6, RRP41, RRP46, and RRP44 grow more slowly than other heterozygotes in the presence of 2 μM 5FU. We directly tested the effect of 5FU on the growth of strains heterozygous for each of the exosome genes, as well as CDC21, which encodes TS, the predicted target of 5FU (27). The results shown in Fig. 1 and summarized in Table 3 generally agree with the previous findings with the exception that our test indicates modest sensitivity of the RRP42+/− heterozygote to 2 μM 5FU. The CDC21+/− strain did not appear to be sensitive to 5FU under these conditions, in agreement with its previously reported low sensitivity to 5FU (28).
FIG. 1.
5FU-induced growth deficiency in various yeast strains. Yeast strains were grown to an A600 of 0.5 to 1.0, and 10-fold serial dilutions were spotted onto synthetic complete plates containing 2% dextrose (SCD) in the absence or presence of 2 μM 5FU. The plates were incubated for 2 days at 30°C. All diploid strains contained Kanr knockouts of the indicated genes in the background of the normal (+/+) strain YSB2001. All haploid strains contained Kanr knockouts of the indicated genes in the background of the normal (RRP6) strain YSB1002.
TABLE 3.
Relative sensitivities of yeast heterozygotes to 5FU
| Relevant genotype | Growtha on:
|
|
|---|---|---|
| SCD | SCD + 2 μM 5FU | |
| RRP6+/+ | +++++ | +++++ |
| RRP4+/− | +++++ | +++++ |
| RRP6+/− | +++++ | ++ |
| RRP41+/− | +++++ | +++ |
| RRP42+/− | +++++ | ++++ |
| RRP43+/− | +++++ | +++++ |
| RRP44+/− | +++++ | +++++ |
| RRP45+/− | +++++ | +++++ |
| RRP46+/− | +++++ | ++ |
| LRP1+/− | +++++ | +++++ |
| CSL4+/− | +++++ | +++++ |
| RAT1+/− | +++++ | +++++ |
| rrp6−/− | +++++ | +++ |
| CDC21+/− | ++++ | ++++ |
| CBF5+/− | +++++ | +++++ |
| MAK21+/− | +++++ | ++ |
| NOP4+/− | +++++ | ++ |
| RRP6 | +++++ | +++++ |
| rrp6-Δ | +++++ | +++ |
| lrp1-Δ | +++++ | ++++ |
++, least growth; +++++, most growth.
We also tested RAT1+/− and LRP1+/− strains, neither of which appeared to be sensitive to 5FU. RAT1 encodes a nuclear 5′-3′ riboexonuclease involved in rRNA processing (20), and LRP1 (RRP47) encodes a protein involved in some aspects of Rrp6p function (29, 35). Although the LRP1+/− heterozygote showed no sensitivity to 5FU under these conditions, a haploid deletion of the gene rendered the cells slightly sensitive to 5FU. These findings confirm that some, but not all, exosome mutations render heterozygotes hypersensitive to 5FU. These results, as well as the previous findings, indicate that defects in the nuclear exosome component RRP6 cause the greatest sensitivity to 5FU (28).
5FU-treated cells accumulate polyadenylated rRNAs.
To gain a better understanding of the effects of 5FU on RNA metabolism in yeast cells, we compared duplicate experiments that analyzed the abundance of RNAs in treated and untreated strains by using the Affymetrix S98 microchip system. A striking feature of the microarray results is a significant increase in signal from array probes sets corresponding to portions of the rRNA locus (Fig. 2A). Because the oligonucleotide arrays were probed with fluorescent RNAs generated by transcription of cDNAs synthesized by oligo(dT) priming of reverse transcriptase, the original transcripts from the treated strains must carry poly(A) tracts to produce signals on the chips.
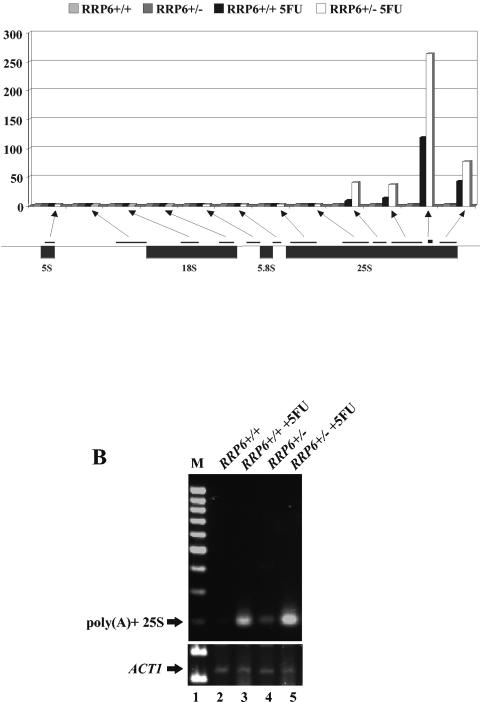
FIG. 2.
5FU causes the accumulation of poly(A)+ rRNAs. (A) Fold change in signal as a function of the microarray oligonucleotide probe set specific for the rRNA locus from each of the indicated strains grown in YPD at 30°C and treated or untreated with 20 μM 5FU for 60 min. The diagram below the chart represents the rRNA locus. The lines above represent the positions and lengths of sequence coverage of individual probe sets on the Affymetrix S98 microarray chips. The arrows point to positions in the chart where the fold changes from the respective probe sets are illustrated. (B) RT-PCR analysis of the levels of 27S rRNA from the indicated strains. Total RNA from each of the strains was reverse transcribed using an oligo(dT) primer, followed by PCR for 20 cycles using oligo(dT) and OSB402 as primers. M, molecular weight markers.
The rRNA locus of yeast is composed of the 5S rRNA gene, transcribed by RNA polymerase III, and the 18S, 5.8S, and 25S genes, transcribed as a single pre-rRNA by RNA polymerase I. The Affymetrix S98 microarray chip uses a series of probe sets to distinguish between transcripts produced from different regions of this locus (Fig. 2A). Analysis of the signals from these probe sets revealed a dramatic increase in rRNA levels in response to 5FU treatment that was significantly more pronounced in RRP6+/− cells than in RRP6+/+ cells. As illustrated in Fig. 2A, the results show a distinct polarity of the signals coming from these probe sets. The chart shows that the most significant increases in rRNA signal in response to 5FU treatment came from the 3′ half of the 25S rRNA region. This polarity most likely reflects the inability of oligo(dT)-primed reverse transcriptase to reach the end of the long poly(A)+ 27S rRNA template but may also partly result from the polarity of degradation of these molecules (see below).
As an initial test of whether 5FU induced the accumulation of polyadenylated forms of 25S rRNA, we amplified such transcripts by RT-PCR with an oligo(dT) primer and a primer internal to 25S under nonsaturating PCR conditions and compared these with ACT1 mRNA, which is unaffected by rrp6 mutations and 5FU treatment. The result, shown in Fig. 2B, supports the idea that 5FU treatment enhances the levels of rRNAs polyadenylated at the 25S rRNA 3′ end. DNA sequence analysis of 10 independent clones of these PCR products confirmed the presence of a poly(A) tail and showed that, in 9 of 10 cases, polyadenylation occurred at the mature 3′ end of 25S rRNA while in the remaining case it occurred one nucleotide before the mature 3′ end. Since the sequence just 3′ of the mature end of 25S rRNA does not contain a stretch of adjacent adenosines, it is unlikely that these products resulted from reverse transcriptase priming at template-encoded adenosines. Similar oligo(dT)-dependent RT-PCR experiments with primers internal to 5S, 18S, and 5.8S rRNAs did not show significant enhancement of the products by 5FU (data not shown).
5FU causes accumulation of polyadenylated forms of 27S rRNA in RRP6 mutants.
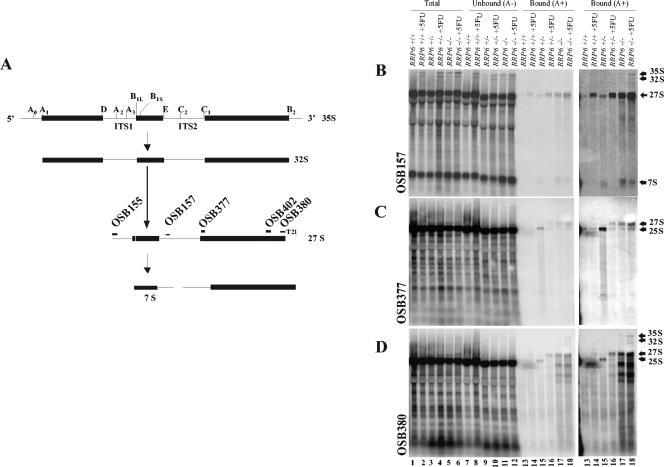
We addressed directly the issue of whether polyadenylated forms of 25S rRNA accumulate in RRP6 mutants treated with 5FU by analyzing rRNAs that bind to oligo(dT)-cellulose with the use of Northern blots probed with radiolabeled oligonucleotides spanning the 5.8S and 25S regions of the rRNA locus (Fig. 3A). In this experiment, we compared RRP6, RRP6+/−, and rrp6−/− diploid strains treated and untreated with 5FU. Detection with an oligonucleotide (OSB157) that hybridized to the internal transcribed spacer region 2 (ITS2) just 3′ of the mature 3′ end of 5.8S rRNA revealed 27S, 7S, 35S, and 32S pre-rRNAs that bind to oligo(dT) (Fig. 3). Consistent with the microarray results, the levels of polyadenylated 27S pre-rRNA increased significantly upon addition of 5FU to the RRP6+/+ and RRP6+/− cells (Fig. 3B, lanes 13 to 16). The homozygous deletion strain (rrp6−/−) accumulated polyadenylated 27S rRNA in the absence of 5FU, and addition of the drug enhanced this effect (Fig. 3B, lanes 17 and 18). Small amounts of polyadenylated RNAs also appeared in the 35S-32S region of the blots, suggesting that these precursors may also accumulate as polyadenylated forms as a result of deletion of RRP6 and addition of 5FU. Polyadenylated 7S pre-rRNA showed a 5FU-dependent pattern opposite to that of 27S rRNA. Blots probed with OSB157 revealed that the level of polyadenylated 7S RNA decreased in the presence of 5FU. This is consistent with previous results that showed that 5FU inhibits the accumulation of 7S and 5.8S rRNAs in yeast, possibly by inhibiting early cleavage steps in rRNA processing (28).
FIG. 3.
Ribosomal RNAs from 5FU-treated strains bind to oligo(dT)-cellulose. (A) Simplified diagram of the processing of 35S pre-rRNA into 32S, 27S, and 7S pre-rRNAs. The diagram indicates the position to which the indicated oligonucleotide probes bind 27S pre-rRNA. T21, sequence of 21 T's following the 3′ end of OSB380. (B to D) Northern blot analysis of oligo(dT)-selected RNAs. Total RNA from the indicated strains untreated or treated with 20 μM 5FU for 60 min was separated into unbound [poly(A)− (A−)] and bound [poly(A)+ (A+)] fractions and analyzed by Northern blotting with the indicated radiolabeled oligonucleotide probes. Lanes 13 to 18 are shown once at the same exposure as that of lanes 1 to 12 (left) and again at a longer exposure (right panel).
Hybridization of the same blot with an oligonucleotide that hybridized near the 5′ end of mature 25S rRNA (OSB377) (Fig. 3A) detected a similar pattern of accumulation of poly(A)+ 27S rRNA (Fig. 3C, lanes 12 to 18). Due to the large amounts of mature 25S rRNA in total RNA samples, various amounts of this RNA carried over and were detected in the bound fractions of the oligo(dT) separations (Fig. 3C and D, lanes 13 and 15). Northern blot analysis with a probe carrying 21 T's following the mature 3′ end of 25S rRNA (OSB380) (Fig. 3A) revealed 27S rRNA, as well as numerous shorter RNAs, in the poly(A)+ fractions. This probe was expected to hybridize strongly to polyadenylated 27S rRNAs based on the findings that such molecules accumulate in rrp6−/− mutants and that poly(A) is added to the mature 3′ end of 25S rRNA. The fact that the oligo(dT)-containing probe OSB380 hybridized more efficiently to oligo(dT)-selected 27S rRNA than to 25S rRNA, in contrast to OSB377, supports the idea that the 27S transcripts are poly(A)+ and the 25S transcripts are not. Comparison of the blots analyzed with the OSB157 and OSB377 probes that hybridized near the 5′ ends of 27S rRNA and 25S rRNA, respectively, with the hybridization pattern revealed by OSB380 suggests the presence of rRNAs polyadenylated at the end of 25S and truncated at various positions upstream of the 3′ end. This pattern is consistent with the polarity of hybridization seen in the microarray experiment (Fig. 2A) and suggests that the polyadenylated 27S rRNAs may undergo 5′-3′ degradation.
rRNA processing defects cause accumulation of poly(A)+ 27S rRNAs.
Our findings suggest that 5FU interferes with the ability of the nuclear exosome to degrade poly(A)+ 27S rRNAs. However, since mutations in RRP6 lead to defects in degradation of aberrant RNAs, as well as defects in rRNA processing, it remained unclear whether poly(A)+ rRNAs arise from defects in rRNA processing alone. Accordingly, we asked whether inhibition of rRNA processing independent of 5FU treatment leads to the accumulation of poly(A)+ 27S rRNAs. We employed an RT-PCR assay in which the level of poly(A)+ 27S rRNA detected was directly proportional to the amount of DNase-treated total RNA added at the RT step of the assay (Fig. 4A). Amplification of ACT1 mRNA served as an internal control for each sample.
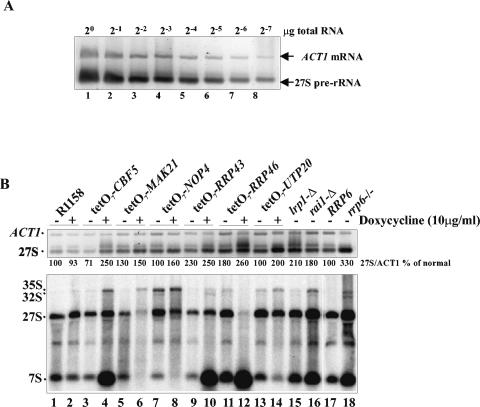
FIG. 4.
Ribosomal processing mutations cause accumulation of poly(A)+ 27S rRNAs. (A) RNA titration of the RT-PCR assay mixture. Reverse transcription with an oligo(dT) primer was carried out with twofold dilutions of DNase-treated total RNA from an rrp6−/− strain, followed by PCR using primers OSB451 and OSB452 for ACT1 mRNA and OSB402 and OSB379 for 27S rRNA. (B) RT-PCR analysis of DNase-treated total RNA from the indicated strains. The top panel shows 2% agarose gel electrophoretic analysis of the RT-PCR products. The bottom panel shows a Northern blot of the same RNA samples. The blot was hybridized with OSB157. Strains producing RNA in lanes 1 to 14 were grown in SCD at 30°C and treated with 10 μg of doxycycline/ml for 24 h before RNA isolation.
First, we asked whether depletion of proteins required for rRNA processing causes an increase in poly(A)+ 27S rRNA levels. Strains with rRNA processing factors under control of the tetO7 promoter were treated with doxycycline for 24 h to repress transcription and allow depletion of the proteins. RNA was extracted from these cells before and after depletion and analyzed by Northern blotting for rRNA processing defects and by RT-PCR for poly(A)+ 27S rRNA (22). MAK21, NOP4, RRP43, and RRP46 each encode proteins required for 60S ribosomal subunit biogenesis, and diploids heterozygous for deletions of these genes, with the exception of RRP43, exhibit 5FU hypersensitivity (12, 28, 30, 43) (Fig. 1). CBF5 and UTP20 encode factors required for efficient processing of pre-rRNAs that form the 40S and 60S ribosomal subunits (22, 24). However, strains heterozygous for deletion of CBF5 (Fig. 1) or UTP20 (15, 28) do not exhibit hypersensitivity to 5FU. Depletion of these gene products caused the expected defects in rRNA processing as judged by Northern blot analysis with an oligonucleotide probe that hybridized just downstream of the mature 3′ end of 5.8S rRNA (OSB157) (Fig. 3A). In the cases of NOP4, RRP46, and UTP20, a clear defect in rRNA processing occurred in the absence of doxycycline, suggesting that replacement of the native promoters with tetO7 results in inadequate expression of the respective gene products. All of the depletions tested resulted in the accumulation of polyadenylated 27S rRNA, which was enhanced by repression of expression by the addition of doxycycline (Fig. 4B).
Finally, we analyzed the effects of deletions of the nonessential RAI1 and LRP1 genes, whose products play roles in 5′- and 3′-end processing of 5.8S rRNA, respectively. Both deletions caused the expected accumulation of 7S rRNA, and both resulted in increased levels of poly(A)+ 27S rRNA (Fig. 4B). These findings suggest that the production of poly(A)+ 27S rRNA may be a general result of the inhibition of rRNA processing.
Evidence that 5FU treatment inhibits the degradation activity of Rrp6p.
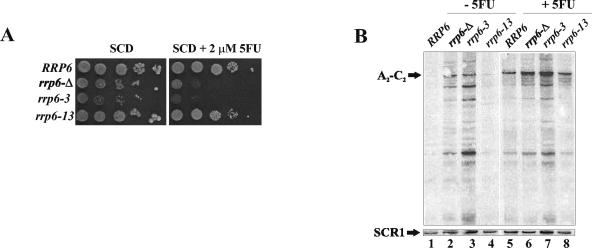
Rrp6p functions in the formation of the mature 3′ ends of 5.8S rRNAs and various snRNAs and snoRNAs and in the degradation of aberrant rRNAs and mRNAs (3, 8, 44). The rrp6-3 and rrp6-13 mutations partially separate the 3′-end formation and degradation functions of Rrp6p (37). For example, the rrp6-Δ, rrp6-3, and rrp6-13 mutations all cause the accumulation of a 5.8S pre-rRNA extended by 27 nucleotides at its 3′ end, indicating defects in 5.8S 3′-end processing. However, while the rrp6-Δ and rrp6-3 mutations abolish the degradation of the rRNA 5′ external transcribed spacer fragment, the rrp6-13 mutation has no effect and does not cause accumulation of poly(A)+ snRNAs and snoRNAs (37). These findings suggest that rrp6-13 strains are defective for the processing, but not the degradation, functions of Rrp6p. Analysis of the 5FU growth sensitivities of strains carrying these mutations revealed 5FU sensitivity of the rrp6-3 strain comparable to that of a strain with a deletion of RRP6 (Fig. 5A). In contrast, the rrp6-13 mutant showed no growth defect under these conditions. These results suggest that sensitivity to 5FU requires a defect in the degradation activity of Rrp6p.
FIG. 5.
5FU inhibits degradation of rRNA. (A) Tenfold serial dilutions of strains with the indicated genotypes were incubated at 30°C on SCD-URA-MET plates with or without the indicated amount of 5FU. (B) Northern blot analysis of RNA from the indicated strains grown as described for panel A with (+) or without (−) 20 μM 5FU. The blot in the top panel was probed with OSB155 and the blot in the bottom panel was probed with OSB151 for SCR1 RNA. The positions of A2-C2 RNA and SCR1 RNA are indicated to the left of the panels.
5FU treatment caused the accumulation of an aberrant 5′- and 3′-extended 5.8S rRNA that ended at the A2 and C2 sites within ITS1 and ITS2, respectively (Fig. 3A). This product also accumulated in rrp6-Δ strains, in strains depleted of other exosome components, and after 5FU treatment in strains heterozygous for deletions of some exosome components (2, 28). Strains heterozygous for deletions of RRP6, RRP41, RRP44, and RRP46 all showed enhanced accumulation of A2-C2 rRNA in the presence of 5FU compared to a normal strain (28). A similar pattern resulted from 5FU treatment of haploid rrp6 mutants (Fig. 5B). Northern blot analysis of RNA from rrp6-Δ and rrp6-3 cells revealed the accumulation of A2-C2 RNA and a series of shorter RNA products (Fig. 5B, lanes 1 to 4). These products must differ in the sites of their 3′ ends since they were detected by Northern blot analysis with an oligonucleotide (OSB155) (Fig. 3A) complementary to the region just 3′ of the A2 site in ITS1. Addition of 5FU caused the appearance of these products in the normal and rrp6-13 strains (Fig. 5A, fifth and eighth lanes). The drug also increased the level of the A2-C2 RNA in the rrp6-Δ and rrp6-3 cells but caused a relative decrease in the levels of the shorter products (Fig. 5A, sixth and seventh lanes). Alternatively, the short RNAs we ascribe to A2-C2 may result from processing of other longer precursors. Nevertheless, the observed increase in levels of A2-C2 RNA upon 5FU treatment suggests its stabilization by the drug. Neither the mutations nor 5FU treatment affected the levels of SCR1 RNA, as shown previously (28, 37). These findings suggest that 5FU treatment inhibits the 3′-5′ degradation of the A2-C2 RNA.
5FU causes ribosome biogenesis and translation defects.
Previous experiments showed that 5FU treatment of an RRP6+/− strain causes the accumulation of rRNA processing intermediates to a significantly greater degree than that seen in a normal strain (28). Specifically, these strains accumulate 35S rRNA, 27S rRNA, and A2-C2 RNA. We analyzed polyribosome profiles from these strains to determine the extent to which these changes affect the number of ribosomes and the efficiency of translation in the presence of 5FU. The RRP6+/+ and RRP6+/− strains in the absence of 5FU yielded typical polyribosome profiles showing 40S, 60S, and 80S ribosomes and translating polyribosome peaks (Fig. 6A and C). Growth of these cells in the presence of 5FU dramatically altered these profiles. Specifically, the sizes of the 60S and 80S peaks decreased significantly but the size of the 40S peaks remained nearly the same as those for untreated cells (Fig. 6B and D). This pattern indicates a decrease in the number of 60S ribosomes, suggests a defect in 60S biogenesis, and is consistent with the observation that 5FU causes defects in 25S and 5.8S, but not 18S, rRNA production (28). While the effects of 5FU on the numbers of 80S and 60S ribosomes appear to be similar in RRP6+/+ and RRP6+/− cells, the appearances of the polyribosome portions of the profiles differ in a potentially significant manner. The heights of the largest RRP6+/− strain polyribosome peaks decreased more in relation to those of the smaller polyribosomes upon 5FU treatment than did the largest RRP6+/+ strain polyribosome peaks. This difference in translation between the two strains may represent the basis for the slower growth of RRP6+/− cells in the presence of 5FU.
FIG. 6.
5FU inhibits 60S ribosome formation. Strains with the indicated genotypes were grown in the presence or absence (−) of 20 μM 5FU at 30°C, and cell extracts were prepared in the presence of cycloheximide. Extracts were separated by ultracentrifugation through 15 to 50% sucrose gradients, and the positions of ribosomes were determined by continuous analysis of the absorbance at 254 nM and are drawn in the figure using the same scale. The positions of the polyribosomes, the 80S, 60S, and 40S ribosome subunits, and the 60S/40S ratio are indicated in each panel.
DISCUSSION
In this report we present evidence that the antiproliferative antimetabolite 5FU inhibits ribosome biogenesis and the ability of the nuclear exosome to degrade polyadenylated noncoding RNAs. It was previously reported that 5FU treatment causes the accumulation of aberrant rRNA processing intermediates and that heterozygosity for a deletion of the nuclear exosome subunit Rrp6p enhances this defect (28). Several observations indicate that the accumulation of such products results from inhibition of the degradation function of Rrp6p. First, sensitivity of rrp6− mutants to 5FU requires a defect in the degradation function of the exoribonuclease. A strain with a mutation (rrp6-13) causing a partial defect in 3′-end processing of 5.8S rRNA and snoRNAs, but no defect in degradation of aberrant pre-RNAs such as A2-C2, grows as well as a normal strain in the presence of 5FU levels that inhibit the growth of strains with rrp6-Δ and rrp6-3 mutations, which cause defects in processing and degradation (Fig. 5A). Moreover, in the rrp6-Δ and rrp6-3 mutants, the pattern of A2-C2 RNAs changes upon addition of 5FU such that the relative level of smaller degradation products decreases and the level of full-length A2-C2 increases (Fig. 5B). These observations indicate that 5FU treatment exacerbates the defect in the degradation of the aberrant A2-C2 RNA caused by the rrp6-Δ and rrp6-3 mutations.
Finally, the microarray results revealed a significant increase in the level of 27S rRNA in 5FU-treated cells, indicating that they accumulate polyadenylated forms of this rRNA. Our findings suggest that these RNAs represent polyadenylated forms of 27S pre-rRNA degradation intermediates. This conclusion is supported by the detection of poly(A)+ 27S precursors by Northern blot analysis and by the fact that shorter versions of these RNAs are detected when analyzed with probes complementary to 3′ distal portions of 27S rRNA. The appearance of these RNAs in untreated rrp6−/− homozygotes and in heterozygotes treated with 5FU implies that Rrp6p normally degrades these transcripts, presumably in concert with the rest of the exosome. The truncated 27S pre-rRNAs all carry poly(A) tails and hybridize to a probe complementary to the 3′ end of 25S rRNA. This indicates that their length heterogeneity results from hydrolysis upstream of the 3′ end. Thus, the role of the exosome in limiting the levels of these aberrant pre-rRNAs, as well as their 5′-end heterogeneity, implies that the transcripts are degraded in both the 5′-3′ and 3′-5′ directions. Whether the 5′ heterogeneity of these transcripts reflects exo- or endonucleolytic cleavage remains unclear.
What then is the effect of 5FU on rRNA processing in yeast? The drug appears to cause a defect in ribosome biogenesis as shown by the inability of treated cells to produce normal amounts of 60S ribosomal subunits (Fig. 6). This effect is less pronounced than that seen for an rrp6-Δ strain, which shows a dramatic decrease in the number of 60S ribosomes and a clear formation of half-mer polyribosomes (4). Nevertheless, RRP6+/+ and RRP6+/− cells show a 35 and 33% decrease in the 60S/40S subunit ratio, respectively, upon treatment with 5FU, consistent with a decrease in the rate of 60S formation. The similarity of the effects in RRP6+/+ and RRP6+/− cells suggests that inhibition of protein synthesis may not cause the enhanced sensitivity of the heterozygote to 5FU. The inhibition of 60S ribosome biogenesis caused by 5FU treatment may result from the previously reported 5FU-dependent defects in 25S and 5.8S, but not 18S, rRNA processing and is similar to that seen for other strains with mutations that affect 60S biogenesis (4, 28). We suggest that the imbalance in ribosome production caused by 5FU results from direct defects in the processing steps leading to maturation of 27S rRNA and/or imbalances in ribosome assembly that shunt rRNA precursors into a degradation pathway. Consistent with this model, mutations causing a variety of defects in rRNA processing elevate the levels of poly(A)+ rRNAs (Fig. 5B). One step in such a degradation pathway would catalyze polyadenylation of the excess, or aberrant, pre-rRNAs, which may accelerate their degradation. The major poly(A) polymerase Pap1p is the most likely candidate for polyadenylation of rRNAs, since inactivation of the enzyme in a pap1-1 mutant inhibits polyadenylation of such rRNAs in rrp6-Δ strains (23). These polyadenylated rRNAs would then be degraded by the exosome, possibly in concert with the 5′-3′ exoribonuclease Rat1p. This model reflects the mechanism of RNA degradation by the bacterial degradasome, which is activated to degrade transcripts in a 3′-5′ direction by polyadenylation of the RNAs (9). Interestingly, experiments have shown that high-level expression of poly(A) polymerase in Escherichia coli leads to polyadenylation of rRNAs (31). Alternatively, polyadenylation of noncoding RNAs may be a normal, very low efficiency process resulting from lack of Pap1p specificity. In this case, the nuclear exosome would routinely degrade aberrant molecules before they accumulate to readily detectible levels.
At present the mechanism of inhibition of Rrp6p by 5FU remains unclear. Analysis of Rrp6p hydrolysis of synthetic RNAs containing 5FU in vitro indicates that the rate is not decreased compared to that of normal RNAs and the addition of 5FU or 5F-UTP to the assays has no effect (data not shown). The fact that defects in other exosome components cause 5FU sensitivity suggests that detection of the effect of the modified nucleotide may require the presence of the entire exosome. Another possibility follows from the fact that the 5-fluoro substitutions in RNA can inhibit the conversion of uridine to pseudouridine, a modification necessary for efficient rRNA processing (24, 46). In this view, pre-rRNAs containing 5FU would fail to be correctly processed, leading to their polyadenylation and degradation by the nuclear exosome. This model predicts that heterozygosity of CBF5, the major rRNA pseudouridylase, would lead to 5FU sensitivity and that depletion of Cbf5p would cause polyadenylation of rRNAs. However, although depletion of Cbf5p does cause accumulation of poly(A)+ 27S rRNAs, 5FU treatment does not inhibit growth of a CBF5+/− strain, as previously shown by Lum et al. (28) (Fig. 1). Moreover, we have not observed 5FU sensitivity of a tetO7-CBF5 strain treated with a range of doxycyline levels that inhibit growth and rRNA processing (1 to 10 μg/ml) (Fig. 5B), nor have we observed 5FU sensitivity of a cbf5 D95A mutant known to be deficient in pseudouridylation of rRNAs (data not shown) (24, 46).
The inhibition of rRNA processing by 5FU may have significant implications for the drug's role in cancer therapy. Several experiments indicate that cotreatment with uridine, but not thymidine, abolishes the cytotoxic effects of 5FU, suggesting that the drug's effects reflect inhibition of RNA metabolism (5, 13, 26, 38). These effects appear to be mediated by p53 since (i) administration of the drug causes apoptosis in p53+/+, but not p53−/−, intestinal epithelial cells (38) and (ii) drug-induced depletion of ribonucleotides causes cell cycle arrest in a p53-dependent manner in the absence of observable DNA damage, suggesting that p53 may act as a sensor of alterations in ribonucleotide metabolism (26). Interestingly, a dominant-negative version of the murine nucleolar Bop1 gene inhibits rRNA processing and induces G1/S arrest in a p53-dependent manner (36, 41). Erb1p, the yeast homologue of Bop1, interacts with Yph1p, a yeast protein that binds the DNA replication origin recognition complex, as well as with 60S preribosomal particles (11). Mutations in Yph1p, as well as in Noc3p, another origin recognition complex protein, cause defects in rRNA processing, DNA replication, and cell cycle checkpoint control (47). These and other findings gave rise to the hypothesis that nutrient deprivation, cellular insults, or DNA replication blocks may activate a ribosome biogenesis checkpoint that inhibits or slows the production of ribosomes until conditions improve (11, 19, 40, 42). Likewise, conditions that block ribosome biogenesis may result in activation of checkpoints governing cellular proliferation, thereby coordinating cell division and the production of ribosomes (11, 19, 32, 40, 42). In this regard, the 5FU sensitivity of cells deficient in the exosome and other rRNA processing components supports the idea that some of the drug's antiproliferative effects may reflect the inhibition of rRNA processing.
In summary, this report provides evidence for an rRNA degradation pathway that features polyadenylation of rRNA precursors followed by degradation by the nuclear exosome. Activation of this pathway may occur due to defects in rRNA processing or treatment of cells with the anticancer, antimetabolite 5FU, which appears to antagonize the ability of the exosome to degrade polyadenylated rRNAs.
Acknowledgments
We are grateful for many helpful discussions with Fred Sherman, Eric Phizicky, Beth Grayhack, and Letian Kuai. We also thank Letian Kuai for invaluable help with microarray data analysis. The members of the Butler lab provided many spirited discussions and helpful comments on the manuscript.
This work was supported by a grant (CA-95913) to J.S.B. from the National Institutes of Health. J.H. was supported in part by a University of Rochester DeKiewiet Summer Fellowship. Affymetrix gene chip and processing costs were supported in part by the Nathan Shock Center for Excellence in Aging Research Microarray Core and National Institutes of Health grant P30 AG18254.
REFERENCES
- 1.Allmang, C., J. Kufel, G. Chanfreau, P. Mitchell, E. Petfalski, and D. Tollervey. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18:5399-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmang, C., P. Mitchell, E. Petfalski, and D. Tollervey. 2000. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 28:1684-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allmang, C., E. Petfalski, A. Podtelejnikov, M. Mann, D. Tollervey, and P. Mitchell. 1999. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev. 13:2148-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briggs, M. W., K. T. Burkard, and J. S. Butler. 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 273:13255-13263. [DOI] [PubMed] [Google Scholar]
- 5.Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497-1501. [DOI] [PubMed] [Google Scholar]
- 6.Bunz, F., P. M. Hwang, C. Torrance, T. Waldman, Y. Zhang, L. Dillehay, J. Williams, C. Lengauer, K. W. Kinzler, and B. Vogelstein. 1999. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Investig. 104:263-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke, D. J., and D. Church. 1991. Protein synthesis requirements for nuclear division, cytokinesis, and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:3691-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler, J. S. 2002. The yin and yang of the exosome. Trends Cell Biol. 12:90-96. [DOI] [PubMed] [Google Scholar]
- 9.Carpousis, A. J., N. F. Vanzo, and L. C. Raynal. 1999. mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet. 15:24-28. [DOI] [PubMed] [Google Scholar]
- 10.Cory, J. G., J. C. Breland, and G. L. Carter. 1979. Effect of 5-fluorouracil on RNA metabolism in Novikoff hepatoma cells. Cancer Res. 39:4905-4913. [PubMed] [Google Scholar]
- 11.Du, Y. C., and B. Stillman. 2002. Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell 109:835-848. [DOI] [PubMed] [Google Scholar]
- 12.Edskes, H. K., Y. Ohtake, and R. B. Wickner. 1998. Mak21p of Saccharomyces cerevisiae, a homolog of human CAATT-binding protein, is essential for 60 S ribosomal subunit biogenesis. J. Biol. Chem. 273:28912-28920. [DOI] [PubMed] [Google Scholar]
- 13.Engelbrecht, C., I. Ljungquist, L. Lewan, and T. Yngner. 1984. Modulation of 5-fluorouracil metabolism by thymidine. In vivo and in vitro studies on RNA-directed effects in rat liver and hepatoma. Biochem. Pharmacol. 33:745-750. [DOI] [PubMed] [Google Scholar]
- 14.Ghoshal, K., and S. T. Jacob. 1997. An alternative molecular mechanism of action of 5-fluorouracil, a potent anticancer drug. Biochem. Pharmacol. 53:1569-1575. [DOI] [PubMed] [Google Scholar]
- 15.Giaever, G., P. Flaherty, J. Kumm, M. Proctor, C. Nislow, D. F. Jaramillo, A. M. Chu, M. I. Jordan, A. P. Arkin, and R. W. Davis. 2004. Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc. Natl. Acad. Sci. USA 101:793-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenhalgh, D. A., and J. H. Parish. 1990. Effect of 5-fluorouracil combination therapy on RNA processing in human colonic carcinoma cells. Br. J. Cancer 61:415-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenhalgh, D. A., and J. H. Parish. 1989. Effects of 5-fluorouracil on cytotoxicity and RNA metabolism in human colonic carcinoma cells. Cancer Chemother. Pharmacol. 25:37-44. [DOI] [PubMed] [Google Scholar]
- 18.Hadjiolova, K. V., M. Nicoloso, S. Mazan, A. A. Hadjiolov, and J. P. Bachellerie. 1993. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur. J. Biochem. 212:211-215. [DOI] [PubMed] [Google Scholar]
- 19.Itahana, K., K. P. Bhat, A. Jin, Y. Itahana, D. Hawke, R. Kobayashi, and Y. Zhang. 2003. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell 12:1151-1164. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, A. W. 1997. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 17:6122-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadaba, S., A. Krueger, T. Trice, A. M. Krecic, A. G. Hinnebusch, and J. Anderson. 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 18:1227-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D. P. Richards, B. K. Beattie, A. Lalev, W. Zhang, A. P. Davierwala, S. Mnaimneh, A. Starostine, A. P. Tikuisis, J. Grigull, N. Datta, J. E. Bray, T. R. Hughes, A. Emili, and J. F. Greenblatt. 2004. High-definition macromolecular composition of yeast RNA-processing complexes: a panoramic view of yeast noncoding RNA processing. Mol. Cell 13:225-239. [DOI] [PubMed] [Google Scholar]
- 23.Kuai, L., F. Fang, J. S. Butler, and F. Sherman. 2004. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101:8581-8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lafontaine, D. L., C. Bousquet-Antonelli, Y. Henry, M. Caizergues-Ferrer, and D. Tollervey. 1998. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 12:527-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenz, H. J., D. J. Manno, K. D. Danenberg, and P. V. Danenberg. 1994. Incorporation of 5-fluorouracil into U2 and U6 snRNA inhibits mRNA precursor splicing. J. Biol. Chem. 269:31962-31968. [PubMed] [Google Scholar]
- 26.Linke, S. P., K. C. Clarkin, A. Di Leonardo, A. Tsou, and G. M. Wahl. 1996. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 10:934-947. [DOI] [PubMed] [Google Scholar]
- 27.Longley, D. B., D. P. Harkin, and P. G. Johnston. 2003. 5-Fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer 3:330-338. [DOI] [PubMed] [Google Scholar]
- 28.Lum, P. Y., C. D. Armour, S. B. Stepaniants, G. Cavet, M. K. Wolf, J. S. Butler, J. C. Hinshaw, P. Garnier, G. D. Prestwich, A. Leonardson, P. Garrett-Engele, C. M. Rush, M. Bard, G. Schimmack, J. W. Phillips, C. J. Roberts, and D. D. Shoemaker. 2004. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell 116:121-137. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell, P., E. Petfalski, R. Houalla, A. Podtelejnikov, M. Mann, and D. Tollervey. 2003. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell. Biol. 23:6982-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell, P., E. Petfalski, A. Shevchenko, M. Mann, and D. Tollervey. 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91:457-466. [DOI] [PubMed] [Google Scholar]
- 31.Mohanty, B. K., and S. R. Kushner. 1999. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol. 34:1094-1108. [DOI] [PubMed] [Google Scholar]
- 32.Olson, M. O. 16 March 2004, posting date. Sensing cellular stress: another new function for the nucleolus? Sci. STKE 2004:pe10. [Online.] http://stke.sciencemag.org. [DOI] [PubMed]
- 33.Parker, W. B., and Y. C. Cheng. 1990. Metabolism and mechanism of action of 5-fluorouracil. Pharmacol. Ther. 48:381-395. [DOI] [PubMed] [Google Scholar]
- 34.Patel, D., and J. S. Butler. 1992. Conditional defect in mRNA 3′ end processing caused by a mutation in the gene for poly(A) polymerase. Mol. Cell. Biol. 12:3297-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng, W. T., M. D. Robinson, S. Mnaimneh, N. J. Krogan, G. Cagney, Q. Morris, A. P. Davierwala, J. Grigull, X. Yang, W. Zhang, N. Mitsakakis, O. W. Ryan, N. Datta, V. Jojic, C. Pal, V. Canadien, D. Richards, B. Beattie, L. F. Wu, S. J. Altschuler, S. Roweis, B. J. Frey, A. Emili, J. F. Greenblatt, and T. R. Hughes. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113:919-933. [DOI] [PubMed] [Google Scholar]
- 36.Pestov, D. G., Z. Strezoska, and L. F. Lau. 2001. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G1/S transition. Mol. Cell. Biol. 21:4246-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips, S., and J. S. Butler. 2003. Contribution of domain structure to the RNA processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA 9:1098-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pritchard, D. M., A. J. Watson, C. S. Potten, A. L. Jackman, and J. A. Hickman. 1997. Inhibition by uridine but not thymidine of p53-dependent intestinal apoptosis initiated by 5-fluorouracil: evidence for the involvement of RNA perturbation. Proc. Natl. Acad. Sci. USA 94:1795-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Proweller, A., and J. S. Butler. 1997. Ribosome concentration contributes to discrimination against poly(A)− mRNA during translation initiation in Saccharomyces cerevisiae. J. Biol. Chem. 272:6004-6010. [DOI] [PubMed] [Google Scholar]
- 40.Ruggero, D., and P. P. Pandolfi. 2003. Does the ribosome translate cancer? Nat. Rev. Cancer 3:179-192. [DOI] [PubMed] [Google Scholar]
- 41.Strezoska, Z., D. G. Pestov, and L. F. Lau. 2000. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S rRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol. 20:5516-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugimoto, M., M. L. Kuo, M. F. Roussel, and C. J. Sherr. 2003. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol. Cell 11:415-424. [DOI] [PubMed] [Google Scholar]
- 43.Sun, C., and J. L. Woolford, Jr. 1994. The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 13:3127-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Hoof, A., P. Lennertz, and R. Parker. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 46.Zebarjadian, Y., T. King, M. J. Fournier, L. Clarke, and J. Carbon. 1999. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol. 19:7461-7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Y., Z. Yu, X. Fu, and C. Liang. 2002. Noc3p, a bHLH protein, plays an integral role in the initiation of DNA replication in budding yeast. Cell 109:849-860. [DOI] [PubMed] [Google Scholar]