Abstract
Microdialysis (MD) has been shown to be a promising technique for sampling of biomarkers. Implantation of MD probe causes an acute tissue trauma and provokes innate response cascades. In order to normalize tissue a two hours equilibration period for analysis of small molecules has been reported previously. However, how the proteome profile changes due to this acute trauma has yet to be fully understood. To characterize the early proteome events induced by this trauma we compared proteome in muscle dialysate collected during the equilibration period with two hours later in “post-trauma”. Samples were collected from healthy females using a 100 kDa MW cut off membrane and analyzed by high sensitive liquid chromatography tandem mass spectrometry. Proteins involved in stress response, immune system processes, inflammatory responses and nociception from extracellular and intracellular fluid spaces were identified. Sixteen proteins were found to be differentially abundant in samples collected during first two hours in comparison to “post-trauma”. Our data suggests that microdialysis in combination with mass spectrometry may provide potentially new insights into the interstitial proteome of trapezius muscle, yet should be further adjusted for biomarker discovery and diagnostics. Moreover, MD proteome alterations in response to catheter injury may reflect individual innate reactivity.
Microdialysis (MD) is an established technique for the in vivo sampling of various substances from the extracellular space of various tissues giving the possibility to monitor localized molecular events in tissues before changes occurs on the blood level. MD of muscle tissues has been used by our group to study nociceptive and metabolic mechanisms in different chronic musculoskeletal pain states. The exact nociceptive/inflammatory and neuropathic pain mechanisms underlying chronic muscle pain are very complex and not fully clarified1 and identification of myalgia “signatures” requires a detection method that allows sensitive, specific and reproducible identification, and quantification of potential biomarkers within a high dynamic range. During MD, a catheter with a porous membrane is implanted into the muscle, which is aimed to mimic the functions of a capillary blood vessel, where molecules from extracellular space are diffusing to the physiological saline (perfusion) solution along the concentration gradient and can be collected for following analyses. MD has been initially developed for sampling of rather small molecules (metabolites, amino acids, energy substrates, drugs and neurotransmitters). During the last decade the high molecular weight cut-off MD has demonstrated to be a promising technique for the sampling of protein biomarkers; however this application is rather complex in comparison to small molecule MD. To turn this technique into the robust and reliable daily clinical routine method for in situ monitoring of protein biomarkers, and to explore new clinically relevant biomarkers a deeper method understanding and optimization is needed.
MD causes minimal discomfort to patients and due to the small size of the probe is called a minimally-invasive procedure2. Nevertheless, the insertion of MD catheter induces mechanical damage of the surrounding tissue and blood vessels3,4,5, which results in local blood flow interruption and bleeding5,6,7, followed by cascades of fast signaling molecular events6. MD probe implantation itself can lead to inflammatory responses, both acute and chronic6, leading to a variety of substances being released from the damaged tissue, including inflammatory mediators, such as chemokines and cytokines. MD injury can also led to activation of sensory receptors including nociceptors if trauma is localized in the vicinity to them, generating sensation of pain8.
Therefore, there are reasonable concerns about inaccurate interpretation of rather complex stress, wound, wound repair, and disease response events: the disease-related and catheter injury-associated events can involve the same proteins/molecules and thus the sum of events, which can, potentially, compromise study outcomes6.
In order to minimize the influence of needle trauma on experimental outcomes, the “recovery and equilibration” period is often introduced to allow the vascular reaction to return to the normal (or stabilized) state9. Thus an equilibration period of two hours after probe injection is typically used in small molecular MD sampling protocols, and initial fractions under 2 hours often are discarded or not analyzed. The same two hours equilibration period is often used even in case of protein biomarkers sampling10,11,12, thus discarding potentially useful information about the individual innate response, which might be different depending on individual subject condition3. In this respect a better understanding of processes that occur in tissues during microdialysis is needed.
The present study aimed to evaluate changes in the proteome over time following catheter insertion in the trapezius muscle of healthy subjects. We performed proteomic analyses of MD fraction collected immediately after insertion of MD probe (trauma protein fraction (T)) and compared it to the fractions collected 2 hours later (post trauma fraction (PT)).
Results and Discussion
In present study we used advantages of high-resolution mass spectrometry for the analysis of peptides obtained from two different muscle dialysate fractions of individual subjects: equilibration period “trauma” fraction (T), collected immediately after catheter insertion and a “post-trauma” fraction (PT) collected 2 hours later. Proteins from dialysate fractions obtained from trapezius muscle interstitium of six healthy female subjects were subjected to minimal sample preparation: desalted, concentrated, and digested with trypsin. Equal amounts of peptides were analyzed in duplicate by LC-MS/MS. The obtained raw files were analyzed using Sequest HT algorithm in Proteome Discoverer (Thermo). The parsimony principle was applied and proteins with similar sequences sharing the same identified peptides were reported as one group.
This straightforward approach allowed the identification of 2516 proteins, in total, merged in to 579 protein groups. A substantial variance between individual samples was observed. Only about 40% of all proteins were identical between all individual subjects from both T and PT periods. Datasets for T and PT samples for proteins detected with top PSMs and identified with the largest number of peptides, were generally the same for all samples (Fig. 1). Proteins identified with top scores were generally the same for T and PT samples (Tables 1 and 2) and includes structural proteins and proteins abundant in plasma/serum. Actin and myosin isoforms are major proteins in muscle cells known to be released into blood in case of muscle damage13. Ten proteins (serum albumin, hemoglobin, carbonic anhydrase-1, immunoglobulins, α1-antitrypsin, serotransferrin, fibrinogen, complement C3, hemoplexin, and prothrombin) were identified with top scores in all fractions are well known as the most abundant blood/serum/plasma proteins (Fig. 1).
Figure 1. Scatter diagram of the protein changes.
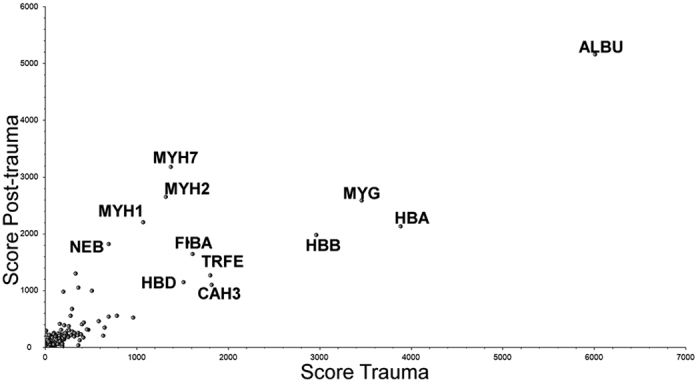
Scatter plot of T versus PT protein scores (sum of the scores of individual peptides) of all confidently identified proteins. Each dot in the figure represents one of the proteins.
Table 1. Top 30 most abundant proteins during the equilibration period.
| Protein ID | Protein name | Sum Xcorr | Sum Coverage, % | Mw, kDa | |
|---|---|---|---|---|---|
| 1 | ALBU_HUMAN | Serum albumin | 6008,93 | 90,31 | 69,3 |
| 2 | HBA_HUMAN | Hemoglobin subunit alpha | 3885,41 | 87,32 | 15,2 |
| 3 | MYG_HUMAN | Myoglobin OS | 3458,04 | 80,52 | 17,2 |
| 4 | HBB_HUMAN | Hemoglobin subunit beta | 2965,12 | 95,92 | 16,0 |
| 5 | CAH3_HUMAN | Carbonic anhydrase 3 | 1818,56 | 88,85 | 29,5 |
| 6 | TRFE_HUMAN | Serotransferrin | 1807,00 | 66,91 | 77,0 |
| 7 | FIBA_HUMAN | Fibrinogen alpha chain | 1611,09 | 48,38 | 94,9 |
| 8 | HBD_HUMAN | Hemoglobin subunit delta | 1515,40 | 93,20 | 16,0 |
| 9 | MYH7_HUMAN | Myosin-7 | 1373,51 | 28,63 | 223,0 |
| 10 | MYH2_HUMAN | Myosin-2 | 1319,36 | 27,41 | 222,9 |
| 11 | MYH1_HUMAN | Myosin-1 | 1074,00 | 23,36 | 223,0 |
| 12 | CAH1_HUMAN | Carbonic anhydrase 1 | 962,34 | 68,20 | 28,9 |
| 13 | A1AT_HUMAN | Alpha-1-antitrypsin | 785,71 | 52,63 | 46,7 |
| 14 | FHL1_HUMAN | Isoform 5 of Four and a half LIM domains protein | 698,88 | 71,96 | 33,6 |
| 15 | F8WCP0_HUMAN | Nebulin | 697,60 | 17,01 | 986,1 |
| 16 | HEMO_HUMAN | Hemopexin | 653,26 | 55,84 | 51,6 |
| 17 | TPIS_HUMAN | Isoform 2 of Triosephosphate isomerase | 638,12 | 81,93 | 26,7 |
| 18 | PEBP1_HUMAN | Phosphatidylethanolamine-binding protein 1 | 589,24 | 75,40 | 21,0 |
| 19 | MYH3_HUMAN | Myosin-3 | 512,92 | 11,55 | 223,8 |
| 20 | FABPH_HUMAN | Fatty acid-binding protein, heart | 475,80 | 63,91 | 14,8 |
| 21 | IGHG1_HUMAN | Ig gamma-1 chain C region | 457,88 | 50,91 | 36,1 |
| 22 | THRB_HUMAN | Prothrombin | 425,90 | 32,96 | 70,0 |
| 23 | HBG1_HUMAN | Hemoglobin subunit gamma-1 | 421,21 | 45,58 | 16,1 |
| 24 | D6RF35_HUMAN | Vitamin D-binding protein | 420,17 | 50,84 | 53,0 |
| 25 | IGKC_HUMAN | Ig kappa chain C region | 403,16 | 82,08 | 11,6 |
| 26 | BLVRB_HUMAN | Flavin reductase (NADPH) | 395,55 | 75,73 | 22,1 |
| 27 | B7Z7A9_HUMAN | Phosphoglycerate kinase | 379,70 | 57,33 | 41,4 |
| 28 | K2C1_HUMAN | Keratin, type II cytoskeletal 1 | 366,51 | 39,44 | 66,0 |
| 29 | A1AG1_HUMAN | Alpha-1-acid glycoprotein 1 | 363,39 | 48,76 | 23,5 |
| 30 | KCRM_HUMAN | Creatine kinase M-type | 363,20 | 49,87 | 43,1 |
The ranking is based on Peptide Spectrum match (PSM) from high to low.
Table 2. Top 30 most abundant proteins during the PT period.
| Protein ID | Protein name | Sum Xcorr | Sum Coverage, % | Mw, kDa | |
|---|---|---|---|---|---|
| 1 | ALBU_HUMAN | Serum albumin | 5153,68 | 87,85 | 69,3 |
| 2 | MYH7_HUMAN | Myosin-7 | 3180,15 | 43,72 | 223,0 |
| 3 | MYH2_HUMAN | Myosin-2 | 2646,74 | 37,82 | 222,9 |
| 4 | MYG_HUMAN | Myoglobin | 2582,55 | 75,32 | 17,2 |
| 5 | MYH1_HUMAN | Myosin-1 | 2200,63 | 36,10 | 223,0 |
| 6 | HBB_HUMAN | Hemoglobin subunit alpha | 2132,33 | 87,32 | 15,2 |
| 7 | HBB_HUMAN | Hemoglobin subunit beta | 1974,66 | 89,80 | 16,0 |
| 8 | F8WCP0_HUMAN | Nebulin | 1815,88 | 30,92 | 986,1 |
| 9 | FIBA_HUMAN | Fibrinogen alpha chain | 1640,35 | 48,27 | 94,9 |
| 10 | K1C10_HUMAN | Keratin, type I cytoskeletal 10 | 1298,61 | 61,64 | 58,8 |
| 11 | TRFE_HUMAN | Serotransferrin | 1264,25 | 72,06 | 77,0 |
| 12 | MYH4_HUMAN | Myosin-4 | 1219,40 | 24,34 | 222,9 |
| 13 | HBD_HUMAN | Hemoglobin subunit delta | 1151,19 | 87,07 | 16,0 |
| 14 | CAH3_HUMAN | Carbonic anhydrase 3 | 1099,53 | 84,23 | 29,5 |
| 15 | K2C1_HUMAN | Keratin, type II cytoskeletal 1 | 1051,04 | 57,92 | 66,0 |
| 16 | MYH3_HUMAN | Myosin-3 | 998,14 | 16,65 | 223,8 |
| 17 | K22E_HUMAN | Keratin, type II cytoskeletal 2 epidermal | 978,82 | 74,65 | 65,4 |
| 18 | ACTS_HUMAN | Actin, alpha skeletal muscle | 675,47 | 62,86 | 42,0 |
| 19 | ACTB_HUMAN | Actin, cytoplasmic 1 | 558,25 | 52,53 | 41,7 |
| 20 | A1AT_HUMAN | Alpha-1-antitrypsin | 558,04 | 58,13 | 46,7 |
| 21 | FHL1_HUMAN | Isoform 5 of Four and a half LIM domains protein 1 | 536,29 | 61,82 | 33,6 |
| 22 | CAH1_HUMAN | Carbonic anhydrase 1 | 524,27 | 68,20 | 28,9 |
| 23 | PEBP1_HUMAN | Phosphatidylethanolamine-binding protein 1 | 463,35 | 75,40 | 21,0 |
| 24 | THRB_HUMAN | Prothrombin | 433,85 | 34,41 | 70,0 |
| 25 | CO3_HUMAN | Complement C3 | 413,23 | 27,30 | 187,0 |
| 26 | IGKC_HUMAN | Ig kappa chain C region | 405,29 | 82,08 | 11,6 |
| 27 | GDIR2_HUMAN | Rho GDP-dissociation inhibitor 2 | 388,45 | 70,15 | 23,0 |
| 28 | K1C9_HUMAN | Keratin, type I cytoskeletal 9 | 373,16 | 52,49 | 62,0 |
| 29 | HEMO_HUMAN | Hemopexin | 350,64 | 56,28 | 51,6 |
| 30 | ALBU_HUMAN | Ig gamma-1 chain C region | 318,41 | 52,42 | 36,1 |
The ranking is based on Peptide Spectrum match (PSM) from high to low.
Apparently, all these highly abundant proteins are present in extracellular space of the muscle as a result of catheter injury and can potentially obscure low-abundant species for proteomic analysis: this problem is well known for many proteomic studies for biomarkers14. The excess of serum proteins compared to other proteins was identified as a big technical challenge of MD15,16,17 and proteomic analysis of muscle interstitium can be compromised by the presence of high abundant proteins from blood/serum/plasma. Indeed, a depletion step would decrease sample complexity and improve detection and quantification of extremely low abundant proteins, however there is always the risk to remove some proteins of interest associated with high-abundance proteins. At the same time, many of skeletal muscle enzymes are known as the useful markers of muscle injury. For example, levels of creatine kinase, myoglobin, troponin, and carbonic anhydrase (CAIII) vary widely in both pathological and physiological conditions18.
Data files obtained in Proteome Discoverer were exported and further validated in Scaffold software with 90% peptide identification probability, 99% protein identification probability parameters and with minimum 2 peptides required for protein identification, resulting in a 0.0% peptide decoy false discovery rate. Proteins were merged into 212 clusters according to Scaffold’s protein cluster analysis algorithm. (Supplementary data S1).
Scaffold software was used to perform a comparative differential analysis of T and PT samples, employing Student’s t-test using normalized spectral abundance factor method (NSAF) to normalize run-to-run variations19. Sixteen proteins were found to have statistically significant differential abundance in T and PT samples (Table 3). Most of these 16 proteins are known to leak out into the blood in case of tissue damage and represent markers of cellular damage. Four proteins related to muscle energy metabolism: isoform 2 of triosephosphate isomerase; phosphoglycerate kinase; creatine kinase and beta-enolase M-type were significantly higher at initial T and decreased in PT fractions.
Table 3. Proteins significantly (p < 0.05) altered between T and PT samples.
| Protein | Protein ID | Mw, kDa | Fold change | |
|---|---|---|---|---|
| Protein S100-A9 | S10A9_HUMAN | 13 kDa | post-trauma high, trauma low | 5.5 |
| Protein S100-A8 | S10A8_HUMAN | 11 kDa | 6.3 | |
| Profilin-1 | PROF1_HUMAN | 15 kDa | 2.2 | |
| Lysozyme C | LYSC_HUMAN | 17 kDa | 2.5 | |
| Rho GDP-dissociation inhibitor 2 | GDIR2_HUMAN | 23 kDa | 3.1 | |
| Histone H1.5 | H15_HUMAN | 23 kDa | 3.1 | |
| Troponin T, fast skeletal muscle | TNNT3_HUMAN | 30 kDa | 4.7 | |
| Isoform 2 of Troponin T, slow skeletal muscle | TNNT1_HUMAN | 30 kDa | 7.4 | |
| Troponin I, slow skeletal muscle | TNNI1_HUMAN | 22 kDa | 3.1 | |
| Myozenin-1 | MYOZ1_HUMAN | 32 kDa | 1.7 | |
| Vimentin | VIME_HUMAN | 54 kDa | 8.7 | |
| Alpha-crystallin B chain | CRYAB_HUMAN | 20 kDa | 17 | |
| Isoform 2 of Triosephosphate isomerase | TPIS_HUMAN | 27 kDa | post-trauma low, trauma high | 0.3 |
| Cluster of Phosphoglycerate kinase | PGK1_HUMAN | 41 kDa | 0.2 | |
| Creatine kinase M-type | KCRM_HUMAN | 43 kDa | 0.06 | |
| Cluster of Beta-enolase | ENOB_HUMAN | 47 kDa | 0.1 |
Creatine kinase plays a central role in energy transduction especially in skeletal muscle tissues, where rapid energy consumption is needed, and creatine kinase levels/activity in plasma are known to vary between healthy subjects depending on age, gender, race, and physical activity20. In conditions that damage skeletal muscles or brain (heart attacks, myositis, strenuous exercises, muscular dystrophy, cerebral diseases, any mechanical muscle damage), creatine kinase is released from muscle into the blood. The routine test for elevated levels of creatine kinase in blood is traditionally used to detect inflammation of muscles or serious muscle damage21. Phosphoglycerate kinase, triosephosphate isomerase, and muscle specific glycolytic enzyme beta-enolase play important roles in glycolysis and glucogenesis and are essential for efficient energy production. Beta- enolase is also known as a serum marker for muscle damage22.
The levels of 12 proteins (myozenin-1, troponins, vimentin, profilin, RhoGDI2, alpha-crystallin B, histone H1.5, S100-A8 and S100-A9) were increased in PT samples in comparison to initial T samples. Four of them were muscle contraction and calcium signal transduction -related proteins: myozenin-1 and troponins. In skeletal muscle, calcium plays a pivotal role in signal transduction and is essential for cellular processes such as excitation-contraction coupling23. Troponins (Troponin T, fast skeletal muscle, Troponin I, slow skeletal muscle, Isoform 2 of Troponin T, slow skeletal muscle) are regulatory proteins in skeletal and cardiac muscle. Raised troponin level is used as plasma marker of skeletal muscle damage and particularly as an indication of cardiac muscle cell death.
Proteins involved in actin cytoskeleton regulation (vimentin and profilin): Profilin-1 is one of the most important regulators of F-actin dynamics, regulating many intracellular functions implicated to play a role in many pathological conditions24. Vimentin is known to be implicated in the regulation of cell migration and proliferation during the wound healing process25,26,27; desmin and vimentin are markers of regeneration in muscle diseases28,29,30, and up-regulated vimentin level is a marker of skeletal muscle injury31.
Among proteins which were up-regulated in all PT samples we identified a two proteins with chaperon activities: Rho GDP-dissociation inhibitor 2 (RhoGDI2) and alpha-crystallin B. RhoGDI2 (a member of a small family of chaperone proteins, which controls Rho GTPases32) is linked to apoptosis-induced cytoskeletal reorganization and is potentially associated with oxidative stress, apoptosis, and wound healing33. Alpha-crystallin B (Heat shock protein beta-5, HspB5) is part of the small heat shock protein family and is able to interact with misfolded proteins to prevent protein aggregation and a wide range of cell stress conditions, as well as inhibit apoptosis and contribute to intracellular architecture34. During cell stress, induction and secretion of HSPs leads to pro-inflammatory cytokine and chemokine release activating immune responses35. Alpha-crystallin B was found to be upregulated in trapezius muscle of chronic widespread pain subjects in comparison to healthy control group in our previous MD-study12.
The level of Histone H1.5 was also increased in PT samples. Beside the classical gene-regulating role, extracellular histones bind to receptors and trigger activation of multiple signaling pathways. Histone levels are known to be significantly elevated in response to injury and involved in the regulation of inflammation36. Furthermore, the linker Histone H1.5 was suggested to play a role in the development of muscle37,38 and may stimulate the proliferation of satellite myoblasts during skeletal muscle regeneration39.
Calcium binding proteins S100-A8 and its binding partner S100-A9 (calgranulin A and B respectively) are involved in the regulation of inflammatory, anti-inflammatory, and immune responses. As well as histone H1.5 and HSPs, these proteins have both intracellular (calcium- binding; regulation of microtubule) and extracellular functions. S100A8 and A9 are known to be released from damaged cells in response to stress and are classified as damage associated molecular pattern (DAMP) molecules40. Serum protein levels of S100A8/A9 were found to be elevated in many inflammatory diseases and these proteins are known to be involved in the wound healing process41.
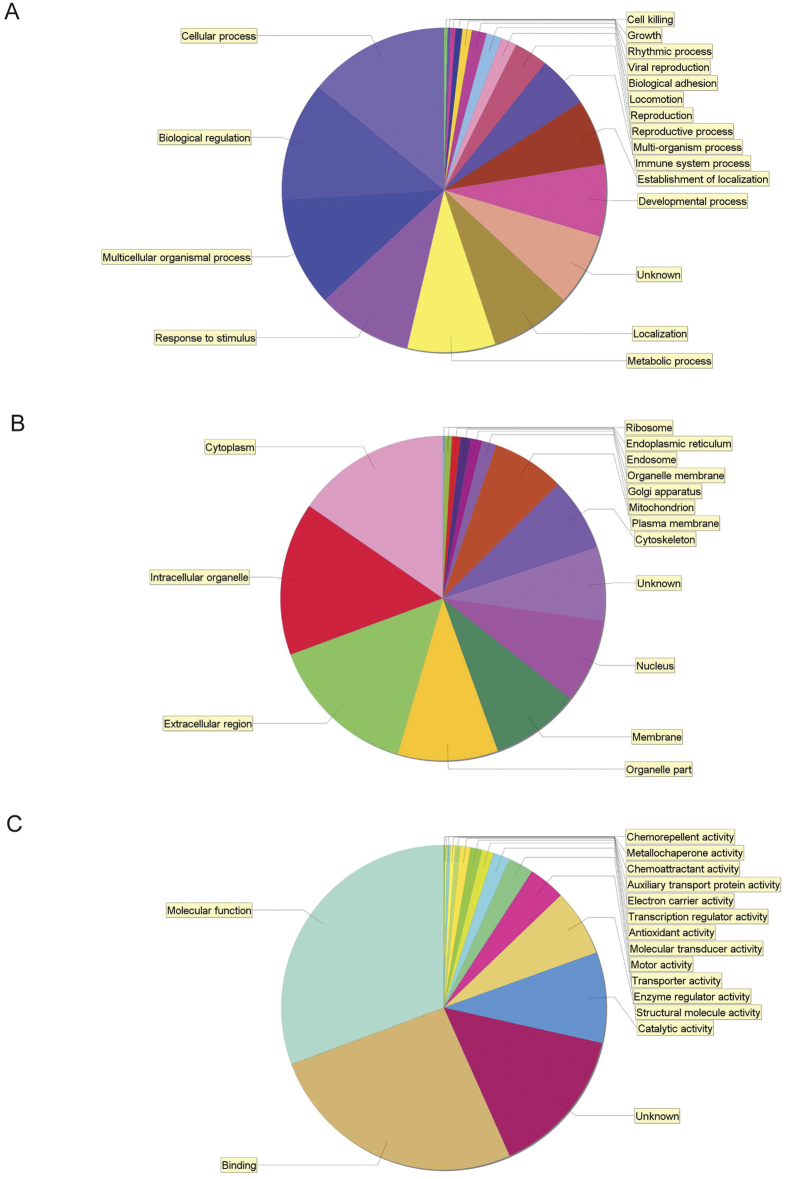
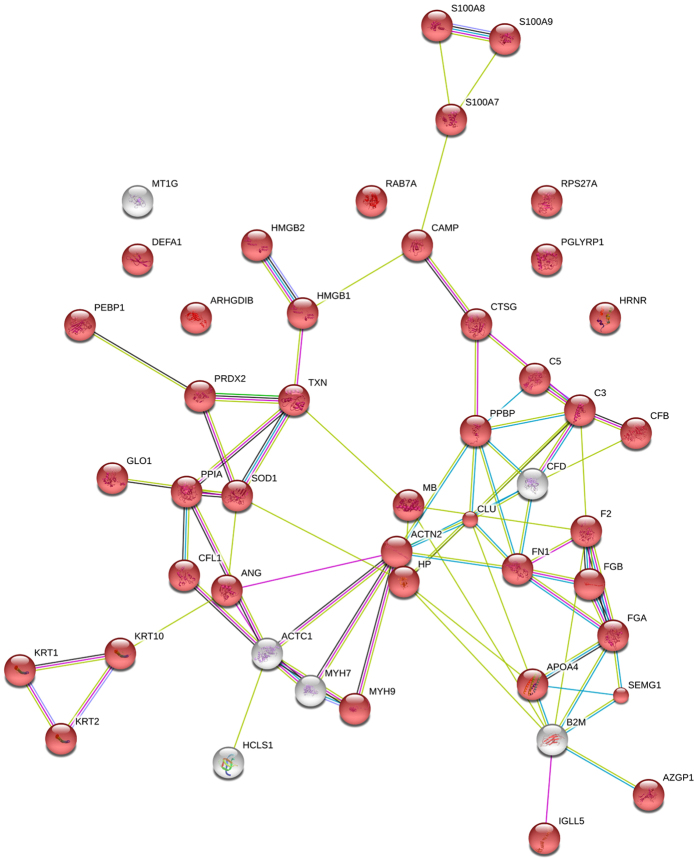
To spot possible protein candidates of potential diagnostic value we performed the Scaffold Gene ontology analysis of all identified proteins, which revealed large groups correlated to stimuli response, metabolic process, localization and developmental process (Fig. 2). Fifty-four protein clusters were identified to be involved in immune system processes (Table 4). An interaction map of these proteins was build using String (Search Tool for the Retrieval of Interacting Genes/Proteins, version 10.) (Fig. 3) and according to bioinformatics analysis 41 of 54 inflammatory proteins can be classifies as extracellular region proteins (Fig. 3, red color).
Figure 2.
Pie charts classifying the identified proteins according to their biological processes (A), cellular components (B) and molecular functions (C). The identified proteins were grouped according to GO annotations.
Table 4. Proteins involved in immune system processes.
| Identified Proteins and Clusters | Gene name | Protein ID |
|---|---|---|
| Myoglobin | MB | MYG_HUMAN |
| Phosphatidylethanolamine-binding protein 1 | PEBP1 | PEBP1_HUMAN |
| Protein S100-A9 | S100A9 | S10A9_HUMAN |
| Isoform 2 of Fibrinogen alpha chain | FGA | FIBA_HUMAN |
| Protein S100-A8 | S100A8 | S10A8_HUMAN |
| Superoxide dismutase [Cu-Zn] | SOD1 | SODC_HUMAN |
| Keratin, type II cytoskeletal 1 | KRT1 | K2C1_HUMAN |
| Neutrophil defensin 1 | DEFA1 | DEF1_HUMAN |
| Rho GDP-dissociation inhibitor 2 | ARHGDIB | GDIR2_HUMAN |
| Keratin, type I cytoskeletal 10 | KRT10 | K1C10_HUMAN |
| Peptidoglycan recognition protein 1 | PGLYRP1 | PGRP1_HUMAN |
| High mobility group protein B2 | HMGB2 | HMGB2_HUMAN |
| Keratin, type II cytoskeletal 2 epidermal | KRT2 | K22E_HUMAN |
| Peptidyl-prolyl cis-trans isomerase A | PPIA | PPIA_HUMAN |
| Cofilin-1 OS = Homo sapiens | CFL1 | COF1_HUMAN |
| Fibrinogen beta chain | FGB | FIBB_HUMAN |
| Prothrombin | F2 | THRB_HUMAN |
| Ubiquitin-40S ribosomal protein S27a | RPS27A | RS27A_HUMAN |
| Actin, alpha cardiac muscle 1 | ACTC1 | ACTC_HUMAN |
| Complement C3 | C3 | CO3_HUMAN |
| Myosin-9 | MYH9 | MYH9_HUMAN |
| Myosin-7 | MYH7 | MYH7_HUMAN |
| Ig lambda-2 chain C regions | IGLC2 | LAC2_HUMAN |
| Complement component C4B (Childo blood group | C4B-1 | A2BHY4_HUMAN |
| Metallothionein-1G | MT1G | MT1G_HUMAN |
| Ig gamma-1 chain C region | IGHG1 | IGHG1_HUMAN |
| Alpha-actinin-2 | ACTN2 | ACTN2_HUMAN |
| Haptoglobin | HP | HPT_HUMAN |
| Ig alpha-1 chain C region | IGHA1 | IGHA1_HUMAN |
| Ig heavy chain V-III region BUT | N/A | HV306_HUMAN |
| Complement factor B | CFB | B4E1Z4_HUMAN |
| Ig kappa chain C region | IGKC | IGKC_HUMAN |
| Zinc-alpha-2-glycoprotein | AZGP1 | ZA2G_HUMAN |
| Ig gamma-2 chain C region | IGHG2 | IGHG2_HUMAN |
| Beta-2-microglobulin | B2M | B2MG_HUMAN |
| Ig gamma-4 chain C region | IGHG4 | IGHG4_HUMAN |
| Thioredoxin | TXN | THIO_HUMAN |
| Isoform 2 of Semenogelin-1 | SEMG1 | SEMG1_HUMAN |
| Isoform 2 of Clusterin | CLU | CLUS_HUMAN |
| Apolipoprotein A-IV | APOA4 | APOA4_HUMAN |
| Angiogenin | ANG | ANGI_HUMAN |
| Cathelicidin antimicrobial peptide | CAMP | CAMP_HUMAN |
| Cathepsin G | CTSG | CATG_HUMAN |
| Complement C5 | C5 | CO5_HUMAN |
| Complement factor D | CFD | CFAD_HUMAN |
| Hematopoietic lineage cell-specific protein SV = 3 | HCLS1 | HCLS1_HUMAN |
| High mobility group protein B1 | HMGB1 | HMGB1_HUMAN |
| Hornerin | HRNR | HORN_HUMAN |
| Isoform 11 of Fibronectin | FN1 | FINC_HUMAN |
| Lactoylglutathione lyase | GLO1 | LGUL_HUMAN |
| Peroxiredoxin-2 | PRDX2 | PRDX2_HUMAN |
| Platelet basic protein | PPBP | CXCL7_HUMAN |
| Protein S100-A7 | S100A7 | S10A7_HUMAN |
| Ras-related protein Rab-7a | RAB7A | RAB7A_HUMAN |
Figure 3. STRING Bioinformatic analysis of the proteins involved in immune system response shown in Table 4.
Colored network nodes represent query proteins. Edges represent protein-protein interactions and include different type of actions depicted by the colored lines. For known interactions: pink, experimentally determined; turquoise, from curated databases. For predicted interactions: green, gene neighborhood; blue, gene co-occurrence. For others interactions: olive green, textmining; black, co-expression; purple, protein homology. Extracellular proteins are marked with red color.
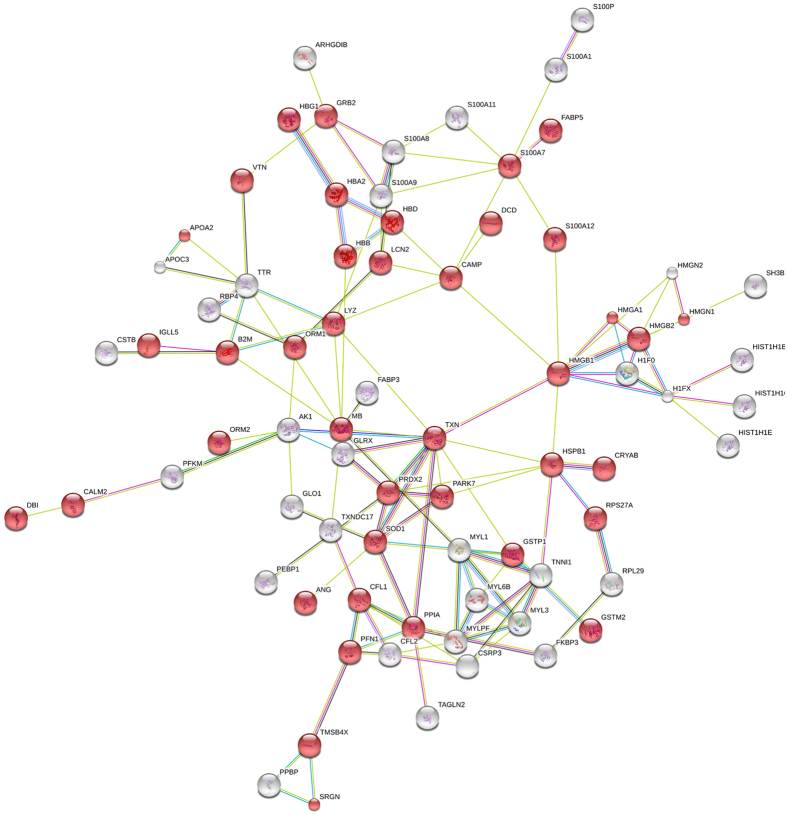
Many proteins were annotated to be functionally involved in the regulation of nociception, inflammation and many other essential major biological processes and reported to be potential predictive biomarkers are low-molecular weight proteins (<25 kDa). According to Scaffold analysis, 106 protein groups identified with at least one peptide were corresponding to proteins with molecular weight of less or equal 25 kDa (Supplementary S2). The list of the identified protein was subjected to STRING analysis to reveal functional interactions between the low mass proteins, most of them (as much as 74) belonging to extracellular region proteins. Interestingly, the function of 46 of these proteins was identified as “response to stress” (Fig. 4, red color).
Figure 4. The interaction map of proteins ≤25 kDa (Supplementary S2) obtained by STRING Bioinformatic analysis.
Colored network nodes represent query proteins, only the connected nodes are shown, red color indicates extracellular region proteins. Edges represent protein-protein interactions and include different type of actions depicted by the colored lines. For known interactions: pink, experimentally determined; turquoise, from curated databases. For predicted interactions: green, gene neighborhood; blue, gene co-occurrence. For others interactions: olive green, text mining; black, co-expression; purple, protein homology.
In our previous study we employed the MD technique in combination with 2-D electrophoresis and in-gel digestion to characterize changes in trapezius muscle interstitial proteome in women with chronic myalgia12. However, proteins with low-molecular weight are often underrepresented in in-gel digestion based proteomic studies or their coverage and, by that; validity of their identification is small. The limited number tryptic cleavage sites and by that limited number of generated tryptic peptides in the small proteins should be taken into consideration as well.
From the collected fractions we were able to identify 13 proteins involved in nociception (Table 5) with rather low levels and low reproducibility with some exceptions: high molecular weight kininogen, neutrophyl cytosolic factor 2 and calmodulin 5 were identified with 3 or more peptides. Consistent with previous publications3,15 no significant levels of interleukins were detected in these sampling conditions.
Table 5. Proteins potentially involved in regulation of nociception pathways.
| Gene name | Protein name | No of peptides sequenced | Mw |
|---|---|---|---|
| KNG1 | High molecular weight kininogen | 16 | 71.9 |
| NCF2 | Neutrophil cytosolic factor 2 | 4 | 50.3 |
| CALML5 | Calmodulin 5 | 3 | 15.9 |
| PKIA | protein kinase inhibitor alpha | 1 | 8.0 |
| RIMS1 | Regulating synaptic membrane exocytosis protein 1 | 1 | 91.5 |
| PRKX | cAMP-dependent protein kinase catalytic subunit PRKX | 1 | 40.9 |
| PRKD1 | Serine/threonine-protein kinase D1 | 1 | 101.6 |
| IRS1 | Insulin receptor substrate 1 | 1 | 131.5 |
| GFRA1 | GDNF family receptor alpha-1 | 1 | 37.7 |
| NTF3 | Neurotrophin-3 | 1 | |
| PRKD1 | Protein kinase D1 | 1 | 29.3 |
| KCNC1 | Potassium voltage-gated channel subfamily C member 1 | 1 | 69.5 |
| CCL14 | Chemokine (C-C motif) ligand 14 | 1 | 10.7 |
Conclusions
Proteins involved virtually in stress responses, immune system processes, inflammatory responses and nociception were identified in the interstitial fluid MD proteome in healthy pain free subjects. Moreover, the comparison proteins in “trauma” dialysate collected immediately after catheter insertion with proteins from “post-trauma” MD-fraction revealed 16 differentially abundant proteins (p < 0.05). Our results demonstrate that MD-LC-MS/MS is a promising approach to provide new insights into the interstitial proteome of muscle, and potentially can be further adjusted for biomarker discovery and diagnostics. The presence of highly abundant serum and muscle structural proteins, sample complexity, multiple protein isoforms, protein modifications and low abundance of many proteins of interest are the key challenges. Moreover, careful validation of biological relevance of biomarkers is of great importance for personalized medicine.
The damage caused by the MD probe should be taken into consideration when analyzing disease biomarkers using MD.
Materials and Methods
Subjects
Six healthy women (age: 35.1 ± 8.3, BMI: 23.8 ± 2.3) were included in this study. Subjects were instructed not to drink any beverages with caffeine on the day of the study, not to smoke and to avoid NSAID-medication the week before the study. The participants arrived at the clinic in the morning after having eaten breakfast. A brief interview was then made by one of the physicians checking that the instructions had been followed. All subjects reported that they had followed the instructions. During the study, they were not allowed to eat, but they could drink water. All participants gave their informed written consent before the start of the study. The study was approved by the Ethical Committee of Linköping University (Dnr: M10-08) and the methods were carried out in accordance with the approved guidelines.
Microdialysis
Microdialysis was performed generally as described in ref. 12. Samples were collected during 20–120 minutes after the catheter insertion (trauma period samples, T) and during 140–200 minutes after the catheter insertion (post-trauma samples, PT). All vials were weighed before the experiment started and after each 20 minutes interval in order to confirm that sampling and fluid recovery (FR) was working according to the perfusion rate set. Vials with visible sign of hemolysis were discarded. The samples were stored on ice to prevent protease activation. The samples were then stored as aliquots in −70 °C until analysis.
Protein extraction and digestion
Samples were subjected to 3 kDa Amicon spin-filter (Merck Millipore) to desalt and concentrate the protein contents. The desalted proteins were dried by speed vacuum concentrator, redissolved in 6 M urea in 25 mM ammonium bicarbonate and incubated at room temperature for at least 30 minutes. The proteins were reduced by incubating in 25 mM DTT for 15 minutes and alkylated with 75 mM iodacetamide for an additional 15 minutes. The samples were diluted 8 times with 25 mM ammonium bicarbonate and filtered by 3 kDa Amicon spin-filter before digestion with trypsin (1:25, w/w trypsin/protein). The digested peptides were dried in speed vacuum concentrator, reconstituted in 0.1% of formic acid in MilliQ water and approximately 0.25 μg was subjected to LC-MS/MS analysis.
LC-MS/MS analysis
Peptides were separated by reverse phase chromatography on a 20 mm × 100 μm C18 pre column followed by a 100 mm × 75 μm C18 column with particle size 5 μm (NanoSeparatoons, Nieuwkoop, Netherlands) at a fow rate 300 nL/min. EASY-nLC II (Thermo Scientific) by linear gradient of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) (0–100% B in 90 min). Automated online analyses were performed with a LTQ Orbitrap Velos Pro hybrid mass spectrometer (Thermo Scientific) with a nano-electrospray source.
Database searches
Raw files were searched using Sequest HT in Proteome Discoverer (Thermo Fisher Scientific, San Jose, CS, USA; version 1.4.0.288) against a Uniprot Human database (available at UniProtKB website: http://www.uniprot.org/taxonomy/9606) with the following parameters: semi trypsin was used as digestion enzyme; maximum number of missed cleavages 2; fragment ion mass tolerance 0.60 Da; parent ion mass tolerance 10.0 ppm; fixed modification- carbamidomethylation of cysteine; variable modifications - N-terminal acetylation. Data were filtered at 1% false discovery rate, high peptide confidence; rank 1 peptides in top scored proteins.
Data evaluation
Identified proteins were filtered using SCAFFOLD (version 1.4.0.288; Proteome Software Inc., Portland, OR, USA). Identifications were based on a minimum of 2 unique peptides, 90% peptide identification probability (using the Scaffold Local FDR algorithm), and 99% protein identification probability (using the Protein Prophet algorithm), resulting in a 0.0% decoy FDR. Proteins that contained similar peptides and which could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. The label-free quantitative analysis of peptides was performed by spectral counting analysis, using normalized spectral abundance factor (NSAF), calculated for each protein to normalize run-to-run variations19, and quantitative differences were statistically analyzed by a t-test. Differences with p values lower than 0.05 were considered statistically significant. Identified proteins were categorized according to gene ontology terms.
String (Search Tool for the Retrieval of Interacting Genes/Proteins, version 10) and Pathway Studio (Elsevier) were used for bioinformatics analysis.
Additional Information
How to cite this article: Turkina, M. V. et al. Evaluation of dynamic changes in interstitial fluid proteome following microdialysis probe insertion trauma in trapezius muscle of healthy women. Sci. Rep. 7, 43512; doi: 10.1038/srep43512 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors would like to thank Eva-Britt Lind for her technical expertise in conducting MD procedures. The present study was supported by grants from The Swedish research council (K2015-99X-21874-05-04) and AFA Insurance (Dnr: 140341).
Footnotes
The authors declare no competing financial interests.
Author Contributions Conceived and designed the experiments: B.Gh., M.V.T., N.G. & B.G. Performed the experiments: B.Gh., M.V.T. Analyzed the data: B.Gh., M.V.T. Contributed reagents/materials/analysis tools: B.Gh., M.V.T. Wrote the paper: B.Gh., M.V.T. B.G., N.G. Reviewed manuscript: B.Gh., M.V.T., B.G., N.G.
References
- Gerdle B., Ghafouri B., Ernberg M. & Larsson B. Chronic musculoskeletal pain: review of mechanisms and biochemical biomarkers as assessed by the microdialysis technique. J Pain Res 7, 313–326, 10.2147/JPR.S59144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange E. C., de Boer A. G. & Breimer D. D. Methodological issues in microdialysis sampling for pharmacokinetic studies. Adv Drug Deliv Rev 45, 125–148 (2000). [DOI] [PubMed] [Google Scholar]
- Sjogren F. & Anderson C. D. Are cutaneous microdialysis cytokine findings supported by end point biopsy immunohistochemistry findings? Aaps J 12, 741–749, 10.1208/s12248-010-9235-8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S. et al. Novel microdialysis method to assess neuropeptides and large molecules in free-moving mouse. Neuroscience 186, 110–119, 10.1016/j.neuroscience.2011.04.035 (2011). [DOI] [PubMed] [Google Scholar]
- Jaquins-Gerstl A. et al. Effect of dexamethasone on gliosis, ischemia, and dopamine extraction during microdialysis sampling in brain tissue. Anal Chem 83, 7662–7667, 10.1021/ac200782h (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenken J. A., Church M. K., Gill C. A. & Clough G. F. How minimally invasive is microdialysis sampling? A cautionary note for cytokine collection in human skin and other clinical studies. Aaps J 12, 73–78, 10.1208/s12248-009-9163-7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashina M. et al. No release of interstitial glutamate in experimental human model of muscle pain. Eur J Pain 9, 337–343, 10.1016/j.ejpain.2004.09.004 (2005). [DOI] [PubMed] [Google Scholar]
- Egekvist H., Bjerring P. & Arendt-Nielsen L. Pain and mechanical injury of human skin following needle insertions. European journal of pain 3, 41–49, 10.1053/eujp.1998.0099 (1999). [DOI] [PubMed] [Google Scholar]
- Groth L. & Serup J. Cutaneous microdialysis in man: effects of needle insertion trauma and anaesthesia on skin perfusion, erythema and skin thickness. Acta Derm Venereol 78, 5–9 (1998). [DOI] [PubMed] [Google Scholar]
- Mellergard P., Aneman O., Sjogren F., Pettersson P. & Hillman J. Changes in extracellular concentrations of some cytokines, chemokines, and neurotrophic factors after insertion of intracerebral microdialysis catheters in neurosurgical patients. Neurosurgery 62, 151–157, 10.1227/01.NEU.0000311072.33615.3A (2008). [DOI] [PubMed] [Google Scholar]
- Helmy A., Antoniades C. A., Guilfoyle M. R., Carpenter K. L. & Hutchinson P. J. Principal component analysis of the cytokine and chemokine response to human traumatic brain injury. PLoS One 7, e39677, 10.1371/journal.pone.0039677 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olausson P., Gerdle B., Ghafouri N., Larsson B. & Ghafouri B. Identification of proteins from interstitium of trapezius muscle in women with chronic myalgia using microdialysis in combination with proteomics. PLoS One 7, e52560, 10.1371/journal.pone.0052560 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero M. et al. Fast and slow myosins as markers of muscle injury. Br J Sports Med 42, 581–584, 10.1136/bjsm.2007.037945 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millioni R. et al. High abundance proteins depletion vs low abundance proteins enrichment: comparison of methods to reduce the plasma proteome complexity. PLoS One 6, e19603, 10.1371/journal.pone.0019603 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough G. F. Microdialysis of large molecules. Aaps J 7, E686–692, 10.1208/aapsj070369 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill C. et al. A qualitative and quantitative proteomic study of human microdialysate and the cutaneous response to injury. Aaps J 13, 309–317, 10.1208/s12248-011-9269-6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen L. J., Sorensen M. A., Codrea M. C., Zacho H. D. & Bendixen E. Large pore dermal microdialysis and liquid chromatography-tandem mass spectroscopy shotgun proteomic analysis: a feasibility study. Skin Res Technol 19, 424–431, 10.1111/srt.12063 (2013). [DOI] [PubMed] [Google Scholar]
- Brancaccio P., Lippi G. & Maffulli N. Biochemical markers of muscular damage. Clin Chem Lab Med 48, 757–767; 10.1515/CCLM.2010.179 (2010). [DOI] [PubMed] [Google Scholar]
- Zybailov B. et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res 5, 2339–2347, 10.1021/pr060161n (2006). [DOI] [PubMed] [Google Scholar]
- Brancaccio P., Maffulli N. & Limongelli F. M. Creatine kinase monitoring in sport medicine. Br Med Bull 81–82, 209–230, 10.1093/bmb/ldm014 (2007). [DOI] [PubMed] [Google Scholar]
- Baird M. F., Graham S. M., Baker J. S. & Bickerstaff G. F. Creatine-kinase- and exercise-related muscle damage implications for muscle performance and recovery. J Nutr Metab 2012, 960363, 10.1155/2012/960363 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chosa E. et al. Evaluation of human beta-enolase as a serum marker for exercise-induced muscle damage. Clin J Sport Med 13, 209–212 (2003). [DOI] [PubMed] [Google Scholar]
- Gehlert S., Bloch W. & Suhr F. Ca2+ -dependent regulations and signaling in skeletal muscle: from electro-mechanical coupling to adaptation. Int J Mol Sci 16, 1066–1095, 10.3390/ijms16011066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizwani W. et al. S137 phosphorylation of profilin 1 is an important signaling event in breast cancer progression. PLoS One 9, e103868, 10.1371/journal.pone.0103868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornemann A. & Schmalbruch H. Desmin and vimentin in regenerating muscles. Muscle Nerve 15, 14–20, 10.1002/mus.880150104 (1992). [DOI] [PubMed] [Google Scholar]
- Menko A. S. et al. A central role for vimentin in regulating repair function during healing of the lens epithelium. Mol Biol Cell 25, 776–790, 10.1091/mbc.E12-12-0900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S., Marcal H. & Foster L. J. Towards scarless wound healing: a comparison of protein expression between human, adult and foetal fibroblasts. Biomed Res Int 2014, 676493, 10.1155/2014/676493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallanti A. et al. Desmin and vimentin as markers of regeneration in muscle diseases. Acta Neuropathol 85, 88–92 (1992). [DOI] [PubMed] [Google Scholar]
- Vater R., Cullen M. J. & Harris J. B. The expression of vimentin in satellite cells of regenerating skeletal muscle in vivo. Histochem J 26, 916–928 (1994). [PubMed] [Google Scholar]
- Vaittinen S. et al. The expression of intermediate filament protein nestin as related to vimentin and desmin in regenerating skeletal muscle. J Neuropathol Exp Neurol 60, 588–597 (2001). [DOI] [PubMed] [Google Scholar]
- Hamilton S. M., Bayer C. R., Stevens D. L., Lieber R. L. & Bryant A. E. Muscle injury, vimentin expression, and nonsteroidal anti-inflammatory drugs predispose to cryptic group A streptococcal necrotizing infection. J Infect Dis 198, 1692–1698, 10.1086/593016 (2008). [DOI] [PubMed] [Google Scholar]
- Moissoglu K., McRoberts K. S., Meier J. A., Theodorescu D. & Schwartz M. A. Rho GDP dissociation inhibitor 2 suppresses metastasis via unconventional regulation of RhoGTPases. Cancer Res 69, 2838–2844, 10.1158/0008-5472.CAN-08-1397 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T. L., Wang P. W., Al-Suwayeh S. A., Chen C. C. & Fang J. Y. Skin toxicology of lead species evaluated by their permeability and proteomic profiles: a comparison of organic and inorganic lead. Toxicol Lett 197, 19–28, 10.1016/j.toxlet.2010.04.019 (2010). [DOI] [PubMed] [Google Scholar]
- Boelens W. C. Cell biological roles of alphaB-crystallin. Prog Biophys Mol Biol 115, 3–10, 10.1016/j.pbiomolbio.2014.02.005 (2014). [DOI] [PubMed] [Google Scholar]
- Tsan M. F. & Gao B. Heat shock proteins and immune system. J Leukoc Biol 85, 905–910, 10.1189/jlb.0109005 (2009). [DOI] [PubMed] [Google Scholar]
- Chen R., Kang R., Fan X. G. & Tang D. Release and activity of histone in diseases. Cell Death Dis 5, e1370, 10.1038/cddis.2014.337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Habas R. & Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science 304, 1675–1678, 10.1126/science.1098096 (2004). [DOI] [PubMed] [Google Scholar]
- Harshman S. W., Young N. L., Parthun M. R. & Freitas M. A. H1 histones: current perspectives and challenges. Nucleic Acids Res 41, 9593–9609, 10.1093/nar/gkt700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriquez J. P., Casar J. C., Fuentealba L., Carey D. J. & Brandan E. Extracellular matrix histone H1 binds to perlecan, is present in regenerating skeletal muscle and stimulates myoblast proliferation. J Cell Sci 115, 2041–2051 (2002). [DOI] [PubMed] [Google Scholar]
- Averill M. M., Kerkhoff C. & Bornfeldt K. E. S100A8 and S100A9 in cardiovascular biology and disease. Arterioscler Thromb Vasc Biol 32, 223–229, 10.1161/ATVBAHA.111.236927 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoff C. et al. Novel insights into the role of S100A8/A9 in skin biology. Exp Dermatol 21, 822–826, 10.1111/j.1600-0625.2012.01571.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.