Abstract
La is a RNA-binding protein implicated in multiple pathways related to the production of tRNAs, ribosomal proteins, and other components of the translational machinery (D. J. Kenan and J. D. Keene, Nat. Struct. Mol. Biol. 11:303-305, 2004). While most La is phosphorylated and resides in the nucleoplasm, a fraction is in the nucleolus, the site of ribosome production, although the determinants of this localization are incompletely known. In addition to its conserved N-terminal domain, human La harbors a C-terminal domain that contains an atypical RNA recognition motif and a short basic motif (SBM) adjacent to phosphoserine-366. We report that nonphosphorylated La (npLa) is concentrated in nucleolar sites that correspond to the dense fibrillar component that harbors nascent pol I transcripts as well as fibrillarin and nucleolin, which function in early phases of rRNA maturation. Affinity purification and native immunoprecipitation of La and fluorescence resonance energy transfer in the nucleolus reveal close association with nucleolin. Moreover, La lacking the SBM does not localize to nucleoli. Lastly, La exhibits SBM-dependent, phosphorylation-sensitive interaction with nucleolin in a yeast two-hybrid assay. The data suggest that interaction with nucleolin is, at least in part, responsible for nucleolar accumulation of La and that npLa may be involved in ribosome biogenesis.
La is an abundant protein, found in the nucleoplasm, nucleolus, and cytoplasm, that interacts with a wide variety of RNAs. Most La (∼85%) is phosphorylated on S366 and resides in the nucleoplasm, where it binds the UUU-OH 3′ ends of RNA polymerase III (pol III) transcripts, which include precursors to tRNAs, 5S rRNA, and other snRNAs, and chaperones them during their maturation (39, 47-49, 75). La has also been found associated with certain small nucleolar RNAs (snoRNAs) whose precursor intermediates contain 3′ oligo(U) (37; reviewed in references 48, 49, and 75). The importance of proper nuclear localization of La is revealed by disruption of its nuclear retention, which causes dysfunctional processing of associated pre-tRNAs (38).
The conserved N-terminal domain of human La (hLa) is composed of two different RNA-binding motifs, a La motif and a canonical RNA recognition motif (RRM) that cooperate to recognize UUU-OH, while its C-terminal domain (CTD) is composed of an atypical RRM with an integral helix that serves as a nuclear retention element, as well as a short basic motif (SBM), a nuclear localization signal (NLS), and phospho-S366 (see below and Fig. 3P) (1, 8, 14, 42; reviewed in reference 45). S366-phosphorylated La (pLa) is active for tRNA maturation, while nonphosphorylated La (npLa) is not (40).
FIG. 3.
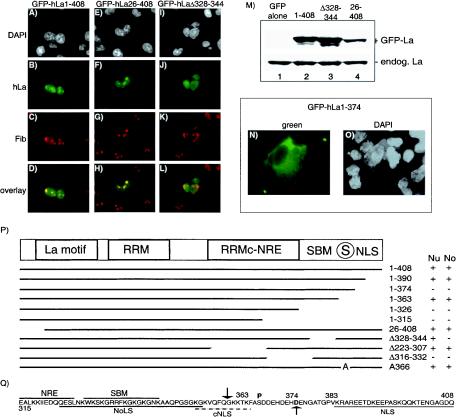
Localization of GFP-hLa fusion proteins in primate cells. CMT3 cells were transfected with GFP-hLa1-408 (A to D), GFP-hLa26-408 (E to H), or GFP-hLaΔ328-344 (I to L), as indicated above the columns, fixed, incubated with antifibrillarin, and processed for immunofluorescence by using Texas red. Cells were visualized for DAPI (white), hLa (green), and fibrillarin (Fib; red), as indicated on the left, and were overlaid to visualize colocalization. (M) Western blot of extracts prepared from transfected cells by using polyclonal anti-La for detection. 1-408, GFP-hLa1-408; Δ328-344, GFP-hLaΔ328-344; 26-408, GFP-hLa26-408; endog. La, endogenous La. (N and O) Localization of GFP-hLa1-374. A field of cells was visualized for hLa (green) and nuclei (DAPI; white). (P) Schematic representation of hLa protein, under which is a summary of the GFP-hLa mutants examined for nuclear (Nu) and nucleolar (No) accumulation. Note that some of these have been documented previously (40). GFP-hLaA366 exhibits only somewhat more nucleolar accumulation than GFP-hLa1-408, consistent with prior data (4), suggesting that cells contain nearly saturating amounts of npLa in their nucleoli. RRMc-NRE, atypical RRM and associated nuclear retention element (42). The encircled S indicates the phosphorylation site at 366; the numbers in the column indicate GFP-hLa fusion proteins. + and −, localization and no localization, respectively, of the indicated protein to the nucleus or nucleolus. (Q) Summary of trafficking elements of hLa spanning residues 315 through the terminus at 408. NoLs, nucleolar localization signal (35); cNLS, cryptic NLS. Downward- and upward-pointing arrows indicate poliovirus protease 3C and caspase 3 cleavage sites, respectively (3, 65).
pLa is almost exclusively nucleoplasmic and associated with pre-tRNAs and other pol III transcripts, while npLa is also cytoplasmic and is bound to a set of specific mRNAs that bear 5′-terminal oligopyrimidine, a common motif found on mRNAs that produce ribosomal proteins, translation elongation factors, and snoRNAs involved in rRNA biogenesis (39, 51, 68). La also associates with poliovirus and other viral and cellular mRNAs that bear internal ribosome entry site elements and promotes their translation, as it does for Mdm2 mRNA (13, 34, 49, 50, 57, 71, 75).
Although La is mainly nuclear, its redistribution to the cytoplasm is directed by poliovirus 3C protease, which cleaves La at Q358, removing its C-terminal NLS (50, 65). As another example of regulated localization, La is dephosphorylated and cleaved at D374 by caspase 3 or a related protease and redistributed to the cytoplasm during apoptosis (3, 63), which is potentially related to translation of the internal ribosome entry site mRNA for X-linked inhibitor of apoptosis (34).
Although the profile of La-associated pre-tRNAs, pre-5S rRNA, pre-snoRNAs, and 5′-terminal oligopyrimidine mRNAs suggests a role for La in the coordinate production of the translation machinery (45), the molecular determinants of involvement of La in the nucleolus, the site of ribosome biogenesis, are only beginning to be characterized (35). Here, we report that npLa is most concentrated in distinct nucleolar sites colocalized with nascent pol I transcripts and with fibrillarin and nucleolin, which bind to snoRNAs and pre-rRNA, respectively, during the early stages of rRNA maturation (26). The data show that the SBM is required for efficient interaction with nucleolin and that this interaction is sensitive to phosphorylation. The data suggest that interaction with nucleolin is responsible, at least partly, for nucleolar accumulation of npLa and that npLa may participate in ribosome biogenesis.
MATERIALS AND METHODS
Immunofluorescence.
Coimmunofluorescence was performed as described previously (18) by using HSC172 cells (cultured human fibroblasts; gift of B. Howard, Bethesda, Md.) (29). Ab1, AbP, and AbNP were described previously, as was polyclonal anti-La (Go), which was a gift of J. D. Keene and D. J. Kenan (Duke Medical Center) (39). Fibrillarin was detected with autoimmune serum S4 (Deitz) (provided by R. L. Ochs, Precision Therapeutics, Pittsburgh, Pa.) at 1:150. B23 was detected by using anti-B23 monoclonal antibody (mAb) (provided by P. K. Chan, Baylor College of Medicine, Houston, Tex.) at 1:20. Primary Ab was detected with secondary Ab conjugated to Texas red for La or fluorescein isothiocyanate for fibrillarin and B23. Western blotting and immunoprecipitation were done by following standard procedures (40, 46, 69). Monoclonal antinucleolin (3G4B2) was from Upstate Biotechnology.
Sequential affinity purification of double epitope-tagged La-associated protein.
Full-length wild-type and mutant human La open reading frames were ligated into the retroviral vector pOZ for the purpose of immunoaffinity purification of associated proteins as described previously (52). The multicloning site in pOZ was altered to the sequence gcggccgctgccggaggaGACTACAAGGACGACGATGACAAGtcggccgctggaggaTACCCCTACGACGTGCCCGACTACGCCTAG in order to express La with FLAG and hemagglutinin epitopes (uppercase) separated by linkers (lowercase) fused to the C-terminal end of La. HeLa cells were transduced with pOZ recombinant viruses by using the Clontech protocol described for the pRetro-Off vector. Transduced cells expressing IL2Ra on their surface were collected by affinity cell sorting by using anti-IL2Ra antibodies (Upstate Biotechnology) and Dynabeads (Dynal) according to Dynal's protocol. Cells were grown to 5 × 105 cells/ml, collected, and extracted for 30 min with 2 ml of B100 buffer (20 mM Tris-HCl [pH 7.9], 100 mM KCl, 5 mM MgCl2, 10% glycerol, 0.1% Nonidet P-40, 10 μM leupeptin, 1 μM pepstatin, 1 μM phenylmethylsulfonyl fluoride, 0.5 μg of aprotinin/ml, 10 mM NaF, 1 mM sodium orthovanadate, and 1.15 mM sodium molybdate). All steps were carried out on ice or at 4°C. The extract was cleared by centrifugation at 14,000 × g for 30 min and incubated with 400 μl of M2 anti-FLAG agarose beads (Sigma) for 4 h with rotation. The beads were washed in B100, and the bound proteins were eluted by incubation for 1 h with 0.2 mg of FLAG peptide (Sigma)/ml in B100 with rotation. The eluate was incubated for 2 h with 400 μl of protein A-Sepharose (Pharmacia) that was coupled to 12CA5 anti-HA antibody (Roche) as described previously (31). The beads were washed with B100 and eluted for 1 h with 0.5 mg of HA peptide (Roche)/ml in B100 buffer. The yield from 109 cells was 4 μg of La-FLAG-HA protein.
Protein identification by mass spectrometry.
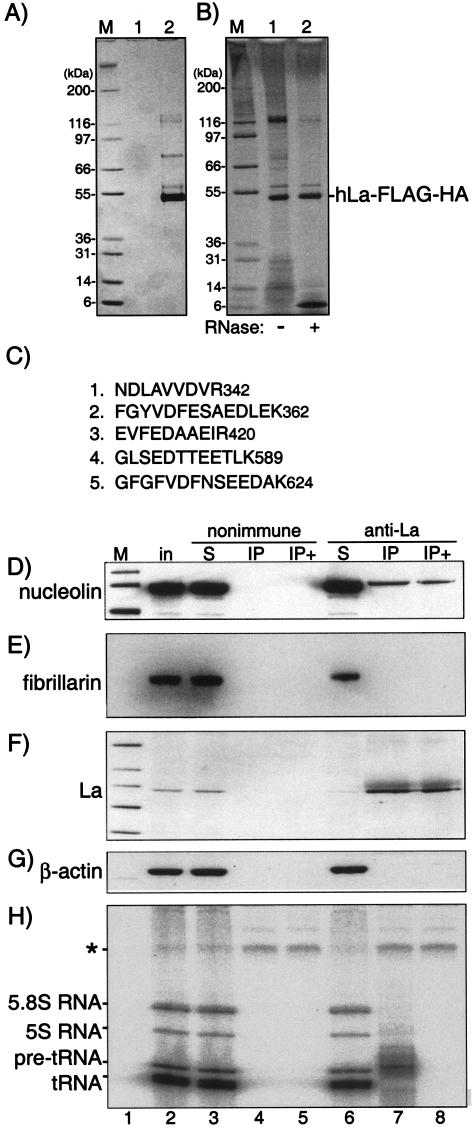
A band migrating at ∼116 kDa after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)-Coomassie blue staining was excised and digested with trypsin, and the spectra of the resulting peptides were acquired by nano-high-pressure liquid chromatography LCQ mass spectrometry (77). Protein sequence database searches of the mass spectrometry data with MASCOT software led to the identification of five peptides whose masses matched tryptic fragments derived from human nucleolin (22).
In situ labeling of sites of transcription.
HSC172 cells growing on 22- by 22-mm coverslips were pretreated with α-amanitin (Roche) at concentrations of 0, 2, or 100 μg/ml for 30 min (or longer; data not shown). Cells were washed, permeabilized, and incubated for 10 min at 37°C in transcription buffer containing 0.5 mM 5-bromouridine 5′-triphosphate (BrUTP; Sigma) containing the appropriate concentration of α-amanitin and processed as described previously with primary monoclonal Ab against BrUTP (1:200; Caltag) and AbNP (1:50) in phosphate-buffered saline for 1 h, washed with phosphate-buffered saline, and incubated with secondary Ab conjugated with fluorescein or Texas red (Vector Laboratories) (16, 19).
Fluorescence resonance energy transfer (FRET) experiments were performed in transfected HeLa cells on a Zeiss 510 confocal microscope with a ×100 and 1.3 numerical aperture planapochromat oil objective and ×3 zoom. FRET was measured by using the acceptor photobleaching method (15, 43). In the presence of FRET, bleaching of the acceptor (yellow fluorescent protein [YFP]) results in a significant increase in fluorescence of the donor (cyan fluorescent protein [CFP]). A single nucleolus was bleached for 500 ms in the YFP channel by using the 514 argon laser line at 100% intensity. Before and after the bleaching, CFP images were collected to assess changes in donor fluorescence. To minimize the effect of photobleaching due to imaging, images were collected at 0.1% laser intensity. To ensure that bleaching due to imaging was minimal, the level of bleaching in each experiment was monitored by collecting five CFP and YFP prebleach and postbleach image pairs. Each image was collected first in the CFP channel and then in the YFP channel. The gain of the photomultiplier tubes in the microscope was adjusted to obtain the best possible dynamic range. The FRET energy transfer efficiency expressed as a percentage, EF, was calculated as EF = (Ipost − Ipre) × 100/Ipost, where Ipre is the prebleach CFP intensity in the last prebleach image and Ipost is the postbleach CFP intensity in the bleached region of the first postbleach image (15). A minimum of 30 measurements (i.e., ≥30 separate cells) were collected and quantitated for each construct to produce the data shown in Fig. 5B.
FIG. 5.
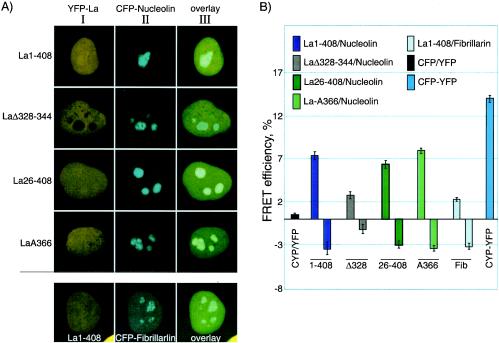
FRET analysis reveals SBM-dependent interaction of La with nucleolin in the nucleolus. (A) Constructs used for the interaction analysis below were first examined for expression in HeLa cells prior to FRET analysis. Column I shows the yellow channel fluorescence of the YFP-hLa constructs, column II shows the cyan channel fluorescence of the CFP-nucleolin constructs, and column III shows the overlaid views for the indicated constructs. Note that the last row shows CFP-fibrillarin. (B) Acceptor photobleaching was used to monitor FRET between the YFP-hLa (acceptor) constructs and the CFP-nucleolin or CFP-fibrillarin (donor) after specific bleaching of the acceptor with YFP-wavelength light. FRET efficiencies reflective of positive interactions were plotted as bars above the horizontal line (see Materials and Methods). Bars below the horizontal line reflect control measurements in the same cells in nucleolar areas in which there was no prebleaching of the acceptor. At least 30 measurements were collected and quantitated for each bar in the graph. CFP/YFP, CFP alone plus YFP alone; CFP-YFP, CFP-YFP fusion protein.
Yeast two-hybrid assay.
The ProQuest Two-Hybrid system (Invitrogen 10835) was used according to the manufacturer's instructions. The hLa constructs were cloned into the SalI and SpeI sites of pDBleu, nucleolin was cloned into the MluI and EcoRI sites of pPC86, and the constructs were verified by sequencing. Yeast transformation was performed with an EasyComp kit and an X-Gal (5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside) beta-galactosidase assay was performed as described in the manufacturer's manual (Invitrogen K5050-01).
RESULTS
Nonphosphorylated La is found at discrete nucleolar sites.
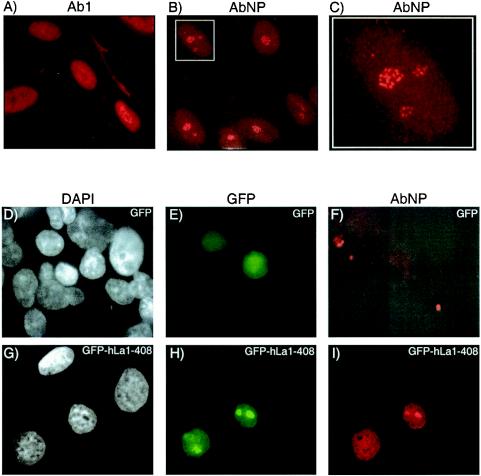
We previously characterized an affinity-purified antibody, referred to as AbNP, that is highly specific for npLa (39). Since only ∼15% of total cellular La is npLa, AbNP recognizes a small fraction with high specificity (39). Ab1 is a control affinity-purified anti-La Ab that recognizes pLa and npLa equally and produces nucleoplasmic staining (Fig. 1A) similar to standard polyclonal anti-La (39). By contrast, AbNP stained discrete sites within nuclei, with less intense staining of the nucleoplasm and cytoplasm (Fig. 1B). Figure 1C is an enlargement of a nucleus in Fig. 1B that reveals discrete sites reminiscent of the dense fibrillar components known to harbor rRNA in the early stages of maturation (17, 54, 64).
FIG. 1.
Immunofluorescence reveals that npLa localizes to discrete nucleolar sites reminiscent of dense fibrillar centers. Cultured human fibroblasts (HSC172) were incubated with Ab1 (A) or AbNP (B and C) and processed for immunofluorescence by using Texas red-conjugated secondary antibody. Panel C is an enlargement of the inset in panel B. CMT3 cells were transfected with GFP (D to F) or GFP-hLa1-408 (G to I), fixed, and incubated with AbNP for immunofluorescence. Cells were visualized with DAPI (D and G), green fluorescence (E and H), and Texas red fluorescence (F and I).
To ensure that AbNP detected hLa and not another antigen, we tested for specificity by transfecting nonhuman cells that are nonreactive with AbNP. AbNP does not recognize endogenous La in these cells by Western blotting, consistent with the high variability of mammalian La sequences corresponding to the sequence of the peptide antigen (data not shown). Cells were transfected with constructs expressing green fluorescent protein (GFP) alone (Fig. 1D to F) or GFP-hLa1-408 fusion protein (Fig. 1G to I) and counterstained with AbNP for red immunofluorescence. Transfected cells were also stained with DAPI (4′,6′-diamidino-2-phenylindole) to visualize nuclei (panels D and G) and visualized for green (panels E and H) and red (panels F and I) fluorescence to examine GFP and AbNP, respectively. Transient transfection led to expression in a fraction of the cells as expected (compare Fig. 1D and E and Fig. 1G and H). Although GFP alone could be detected (Fig. 1E), as expected there was no appreciable nuclear staining by AbNP in either the GFP-expressing or nonexpressing cells (panel F). GFP-hLa1-408 exhibited nucleoplasmic as well as more intense localized staining suggestive of nucleoli (panel H). Only the expressing cells exhibited significant staining by AbNP (panel I), and this staining occurred in a pattern similar to that by GFP-hLa1-408 (panel H), with the most intense staining suggestive of nucleoli. The data confirm that AbNP reacts with hLa specifically and suggest that npLa accumulates in nucleoli.
NpLa colocalizes with fibrillarin and nascent RNA polymerase I transcripts.
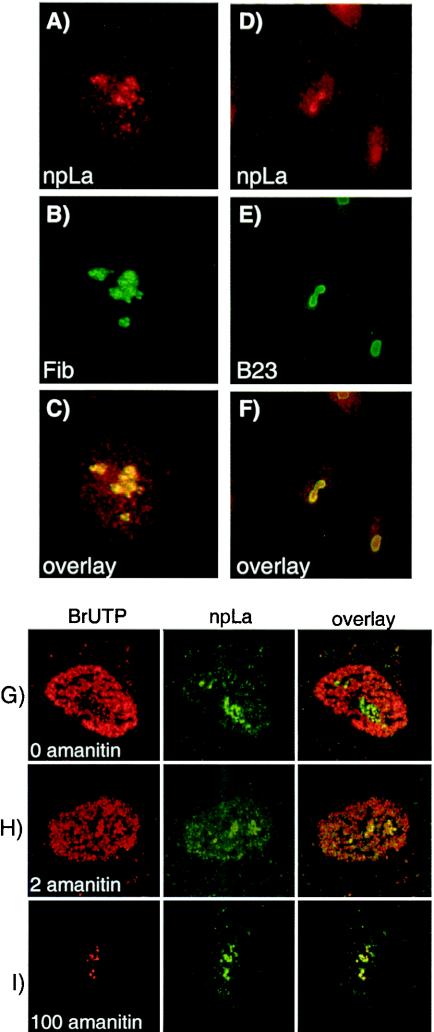
We examined native endogenous La in human cells by using AbNP together with Abs that recognize B23 and fibrillarin (Fig. 2), the latter two of which function in rRNA biogenesis (54, 64). Fibrillarin localizes to sites of nascent pol I transcripts in the dense fibrillar component, whereas B23 is mostly in the granular component, reflective of later stages of ribosome maturation (54, 64). The most intense AbNP staining colocalized with fibrillarin (Fig. 2A to C). The most intense B23 staining appeared as a peripheral ring surrounding the area of AbNP staining (Fig. 2D to F).
FIG. 2.
npLa accumulates in nucleoli and colocalizes with fibrillarin and newly synthesized RNA polymerase I transcripts. Normal human fibroblasts (HSC172) were incubated with AbNP and with either antifibrillarin (A to C) or anti-B23 (D to F). AbNP is represented in red (A and D), and antifibrillarin and anti-B23 (fluorescein isothiocyanate) are represented in green (B and E). Although these colocalizations were done in parallel, the images in panels A to C are intentionally shown at higher magnification than those in panels D to F. Panels A and B were overlaid to produce panel C, and panels D and E were overlaid to produce panel F. (G to I) Anti-BrUTP and npLa are visualized in the red and green channels, respectively, by confocal microscopy, and the red and green images were overlaid to visualize colocalization. BrUTP incorporation occurred in the presence of 0 (G), 2 (H), and 100 (I) μg of α-amanitin/ml.
We next used permeabilized cells to label nascent transcripts with BrUTP, employing α-amanitin to distinguish pol I from pol II and pol III transcripts (16, 19). Anti-BrUTP was observed in the red channel and AbNP was observed in the green channel by confocal microscopy, and the images were overlaid to visualize colocalization. At 0 μg of α-amanitin/ml, most transcripts were observed in the nucleoplasm (Fig. 2G). Because 2 μg of amanitin/ml selectively inhibits pol II, the BrUTP signal observed in its presence reflects nascent pol I and pol III transcripts (Fig. 2H). pol II and pol III are inhibited by 100 μg of α-amanitin/ml, while pol I remains active, as reflected by the most intense BrUTP labeling in discrete sites in the nucleolus with relatively less labeling elsewhere (Fig. 2I). At 100 μg of α-amanitin/ml, the most concentrated npLa overlaps with the BrUTP-labeled nascent products of pol I (Fig. 2I). We conclude that npLa resides in discrete nucleolar sites, colocalized with nascent pol I transcripts and the rRNA processing factor, fibrillarin.
Evidence that the SBM of La is required for nucleolar accumulation.
Mutant hLa proteins were previously characterized for the ability to (i) stimulate pol III transcription in vitro, (ii) stabilize the 5′ and 3′ ends of pre-tRNAs, and (iii) promote tRNA maturation in vivo (20, 21, 30, 40). Here, we examined these proteins as GFP-hLa fusions for colocalization with fibrillarin (Fig. 3A to L). The ability of anti-GFP Ab to immunoprecipitate (IP) pre-tRNA from cells expressing GFP-hLa1-408 indicated that the fusion was functional for pre-tRNA binding (data not shown). We visualized nuclei by DAPI (white), fibrillarin (red), and GFP-hLa (green) and produced overlaid views of the red and green images. The most germane findings are represented by the GFP-hLa constructs shown in Fig. 3A to L. Full-length GFP-hLa1-408 exhibited the expected nucleoplasmic and nucleolar pattern (Fig. 3A to D).
The N-terminal region of hLa that is absent in hLa26-408 is required for high-affinity recognition of 3′ UUU-OH-containing RNA (9, 30, 44; reviewed in reference 49; see also reference 53). Accordingly, this protein does not stabilize pre-tRNA 3′ ends in vitro or in vivo and fails to support tRNA maturation in vivo (20, 40). The most concentrated areas of GFP-hLa26-408 colocalized with fibrillarin (Fig. 3E to H). This result suggested that nucleolar localization is an activity of hLa that is distinct from 3′ end recognition of UUU-OH-containing RNAs. GFP-hLaΔ223-307, which lacks the C-terminal RRM, also exhibited no defect in nucleolar accumulation (Fig. 3P).
GFP-hLaΔ328-344, which lacks the SBM, localized diffusely throughout nuclei but, unlike the proteins above, did not concentrate in regions characteristic of nucleoli (Fig. 3J). GFP-hLaΔ328-344 exhibited less colocalization with fibrillarin than with the other GFP-hLa constructs. Thus, while GFP-hLa1-408 and GFP-hLa26-408 produced a yellow color when overlaid with fibrillarin (Fig. 3D and 4H), GFP-hLaΔ328-344 produced a more orange-red color in the region stained for fibrillarin (Fig. 3L). Immunoblotting showed that the relative paucity of nucleolar accumulation of GFP-hLaΔ328-344 was not due to low expression (Fig. 3M). These data indicate that GFP-hLaΔ328-344, which lacks the SBM, is less concentrated in nucleoli than all other GFP-hLa mutant proteins examined that contain the SBM.
FIG. 4.
Nucleolin interacts with epitope-tagged La-FLAG-HA or native endogenous La in HeLa cells. (A) SDS-PAGE followed by Coomassie blue staining after two epitope-directed affinity purifications of extract from cells transfected with empty plasmid (lane 1) or cells expressing hLa-FLAG-HA (lane 2). (B) The affinity-purified material derived from cells transfected with hLa-FLAG-HA was untreated (lane 1) or treated with RNase A (lane 2) and then subjected to SDS-PAGE analysis followed by silver staining. (C) Identity of the five peptide sequences identified from the La-FLAG-HA-associated band with an apparent molecular mass of ∼116 kDa (see text). (D to G) Immunoprecipitation of HeLa cell extract by anti-La and nonimmune antibodies and analysis by Western blotting to detect nucleolin(D), fibrillarin (E), La (F), and β-actin (G). Samples are numbered below the lanes. in, input; S, supernatant; IP+, immunoprecipitate after treatment with RNase A. For panel H, material was collected and labeled with 32P-Cp and RNA ligase and analyzed by denaturing PAGE and autoradiography. The asterisk represents RNase A-resistant, protease-sensitive material that results from the labeling reaction (data not shown), which serves as a recovery-loading control for labeling. Molecular mass markers (lanes M) are expressed in kilodaltons.
Cryptic nuclear localization activity in the C-terminal domain of hLa.
As reported previously (38) and summarized in Fig. 3P, GFP-hLa1-363 exhibited localization indistinguishable from that of GFP-hLa1-408, indicating that the NLS mapped to residues 383 to 408 (66) is not required for nuclear accumulation of GFP-hLa1-363. Because others had demonstrated that GFP-hLa1-374, which represents truncation by caspase 3, was cytoplasmic (3), we obtained and examined their mutant and also constructed the mutant ourselves and confirmed that it was mostly cytoplasmic (Fig. 3N and O). The cumulative data suggested that GFP-hLa1-363 may harbor a context-specific NLS (62). Since GFP-hLa1-328 is cytoplasmic (Fig. 3P), the region between residues 328 and 363 may therefore function as an NLS in the context of GFP-hLa1-363. Indeed, sequence examination reveals a tract of basic-rich residues, KGKVQFQGKKTK, at the terminus of La1-363 (Fig. 3Q), which may be recognized as an NLS in this context as described for other truncated proteins (62), while cleavage at 374 leaves acidic residues DDEHDEHD at the terminus (see Discussion).
La interacts with nucleolin in HeLa cells.
Affinity chromatography can be a powerful way to examine protein-protein associations that occur in vivo (52, 73, 74). We expressed double-epitope-tagged hLa-FLAG-HA in HeLa cells and subjected it to two sequential affinity chromatography purifications. Figure 4A shows a Coomassie blue-stained gel after purification from cells transduced with empty vector (lane 1) and cells expressing hLa-FLAG-HA (lane 2). The major band just below the 55-kDa marker was isolated and subjected to tryptic digestion followed by mass spectrometric analyses combined with database searching, and it was identified as hLa (data not shown). The band just above the 55-kDa marker was identified by the same approach as for Ro60, a known La-associated protein (data not shown) (33). The affinity-purified material was also examined by silver staining (Fig. 4B, lane 1) and by treatment with RNase A (Fig. 4B, lane 2). As expected, some of the bands visualized by silver staining were decreased after treatment with RNase A, indicating that they were associated RNAs. An RNase-resistant band that copurified with La whose electrophoretic mobility suggested a mass of ∼116 kDa was isolated from the gel and subjected to mass spectrometry to reveal a mass of 76,298 Da. Tryptic digestion of this band followed by mass spectrometric analyses and database searching revealed five peptide fragments that matched human nucleolin (Fig. 4C). The disparity between the apparent and actual mass has been noted previously and presumably reflects the multiple highly charged tracts within nucleolin (28). Other proteins that copurified with La-FLAG-HA will be characterized elsewhere.
To test whether native endogenous La was associated with nucleolin, we performed immunoprecipitation by anti-La, and examined the input, the IP, and the supernatants by immunoblotting with antinucleolin, antifibrillarin, antiactin, and standard polyclonal anti-La (Fig. 4D to H). This examination revealed that a fraction of nucleolin was coimmunoprecipitated with La, while fibrillarin and actin were not (Fig. 4D to G, lanes 7). Specificity was revealed by the failure to detect nucleolin by using nonimmune immunoglobulin G (lane 4). To test whether co-IP of nucleolin might be dependent on RNA, we subjected the IP samples to RNase A (lanes 5 and 8) prior to elution from the anti-La immunoglobulin G-protein A beads. We assessed the efficacy of RNase treatment by examining the associated RNAs by [32P]pCp ([32P]cytidine-3′,5′-diphosphate) labeling with RNA ligase (Fig. 4H). La-associated RNAs migrate between 5.8S rRNA and pre-tRNAs (Fig. 4H, lane 7) (33). Although all associated RNAs were efficiently removed by RNase as expected (Fig. 4H, lane 8 and data not shown), nucleolin remained associated with La in the same samples (Fig. 4D, lane 8). Thus, nucleolin was specifically associated with endogenous La.
FRET between La and nucleolin in the nucleolus.
We used acceptor photobleaching as a means to monitor FRET to determine whether La and nucleolin are in close proximity in the nucleolus (15, 43). This required the creation of CFP-nucleolin and YFP-La fusion proteins. Four different La-YFP constructs were each cotransfected with nucleolin-CFP into HeLa cells and visualized for either YFP or CFP or both (Fig. 5A, columns I, II, and III, respectively). Nucleolar intensity appeared to be only slightly higher than the surrounding nucleoplasm for some of the YFP-La constructs and not as intense as for the same GFP-hLa fusions in the single transfectants (see Discussion). Nonetheless, YFP-LaΔ328-344 was specifically deficient for nucleolar accumulation (Fig. 5A), in agreement with the GFP-La results. These analyses indicated that the CFP-nucleolin and YFP-La fusion constructs can be used for FRET analysis as described below.
Acceptor photobleaching by laser scanning confocal microscopy with single channel collection allows accurate and specific assessment of FRET between YFP and CFP; increase in donor fluorescence after acceptor photobleaching reflects the degree of FRET between the CFP and YFP (15, 43). By photobleaching in the yellow wavelength in the nucleolus followed by monitoring cyan fluorescence in the same space, nucleolar FRET between La-YFP and nucleolin-CFP was assessed. Cotransfection of CFP alone plus YFP alone revealed a low level of background (Fig. 5B, CFP/YFP bar), while the fusion protein CFP-YFP presumably represents a maximal obtainable signal from a covalent connection between CFP and YFP (Fig. 5B, CFP-YFP bar) (15, 43). As noted above, although npLa and fibrillarin were found colocalized by immunofluorescence, fibrillarin was not stably associated with La (Fig. 4E). We observed significantly more nucleolar FRET between La1-408 and nucleolin than between La1-408 and fibrillarin (Fig. 5B). Thus, FRET analysis indicated close proximity of La and nucleolin in nucleoli.
We observed significant FRET between nucleolin and La1-408, La26-408, and LaS366A, but significantly less for LaΔ328-344 (Fig. 5B). Although the paucity of LaΔ328-344 in the nucleolus (Fig. 5A) makes it difficult to directly compare this mutant for nucleolar FRET with the other constructs, its lower FRET is consistent with its failure to accumulate in nucleoli. Fibrillarin, however, does accumulate in nucleoli but exhibits significantly less FRET with La (Fig. 5B). These data indicate that while La, fibrillarin, and nucleolin co-occupy the nucleolus, La maintains closer physical contact with nucleolin than with fibrillarin, consistent with their coimmunoprecipitation characteristics (Fig. 4).
Yeast two-hybrid analysis reveals that interaction of La with nucleolin requires the SBM and is sensitive to phosphorylation of S366.
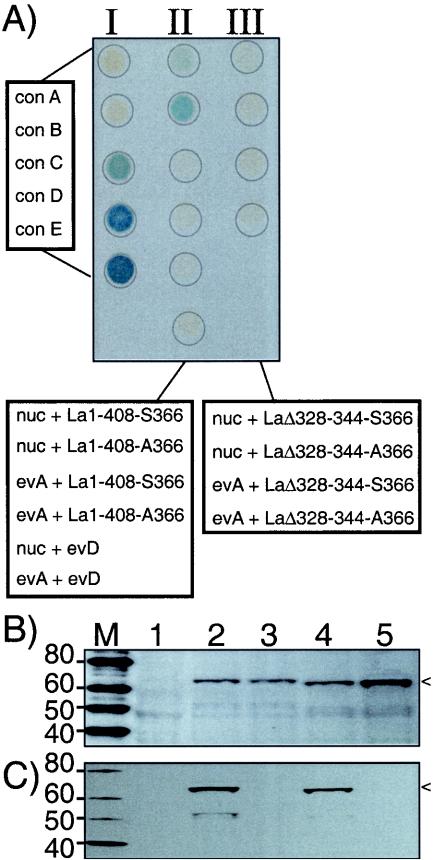
We next used yeast two-hybrid analysis as an indicator of interaction between La and nucleolin, employing a beta-galactosidase activity assay (23). In this approach, La is fused to the DNA-binding domain of Gal4 and nucleolin is fused to the activation domain of Gal4. Beta-galactosidase is produced only if both fusion proteins interact with each other at the promoter of the beta-galactosidase gene in the yeast cells (23). As an indicator of the range of activity of our assay, five controls (provided by the supplier; see Materials and Methods) that produce no interaction to the strongest interaction, reflected by no color to dark blue (Fig. 6A, column I), were used. The bottom four samples in column II reflect our negative controls, in which the nucleolin activation domain plasmid, the La-DNA-binding domain plasmid, or both plasmids were empty, as indicated. It is therefore significant that coexpression of La1-408-S366 and nucleolin fusion proteins activated beta-galactosidase (column II, top sample) and that La1-408-A366 was significantly more active (column II, second from top). Column III shows that neither LaΔ328-344-S366 nor LaΔ328-344-A366, which both lack the SBM, interacted with nucleolin. The data indicate that there is La SBM-dependent interaction with nucleolin and that the nonphosphorylatable mutant La1-408-A366 interacts more strongly than does hLa1-408-S366.
FIG. 6.
SBM-dependent, S366 phosphorylation-sensitive interaction of hLa with nucleolin as assayed by yeast two-hybrid analysis. (A) Beta-galactosidase assay performed on immobilized, lysed, and transformed yeast cells. Column I contains control cells transformed with plasmids engineered to indicate no interaction to strong interaction from top to bottom (con A to con E, respectively) (see text). nuc, nucleolin fusion construct; evD, empty vector carrying the Gal4 DNA-binding domain; evA, empty vector carrying the Gal4 activation domain. (B and C) Western blots of extracts from cells from panel A. Lane 2, nuc + La1-408-S366; lane 3, nuc + La1-408-A366; lane 4, nuc + LaΔ328-344-S366; lane 5, nuc + LaΔ328-344-A366. The La fusion proteins are indicated by arrowheads. The membrane shown in panel B was developed by using polyclonal anti-La that recognizes npLa and pLa; that shown in panel C was developed by using AbP specific for pLa (39). M, molecular mass in kilodaltons.
Although it was established that hLa is efficiently phosphorylated on S366 in fission yeast (40), this phosphorylation had not been examined with Saccharomyces cerevisiae, the yeast used for two-hybrid analysis (23). S. cerevisiae extracts made from the same cells as shown in the top two samples of columns II and III, as well as control cells from Fig. 6A, were examined by Western blotting for their La-S366 phosphorylation status (Fig. 6B and C). For the membrane shown in Fig. 6B, we used polyclonal anti-La that recognizes both pLa and npLa; for the membrane shown in Fig. 6C, we used AbP, which is specific for pLa (39). The La fusion proteins were recognized by polyclonal anti-La, indicating their expression (Fig. 6A, lanes 2 to 5), whereas this band was not detected in the control extract (lane 1). The hLa fusion was recognized by AbP, indicating that the expressed protein was phosphorylated on S366, while the hLaA366 fusion protein was not, as expected (Fig. 6C, lanes 2 and 3) (39). The LaΔ328-344-S366 fusion was recognized by AbP, indicating S366-phosphorylation, while its nonphosphorylatable derivative, LaΔ328-344-A366, was not recognized by AbP (Fig. 6C, lanes 4 and 5). Probing of the membrane in C with antinucleolin revealed the nucleolin fusion protein in lanes 2 to 5, but not in lane 1, indicating coexpression of the nucleolin fusion protein in the same cells as expected (data not shown). The data indicate that the SBM of La directs interaction with nucleolin and that phosphorylation of nearby S366 decreases this interaction. Since S366 phosphorylation is only 80 to 90% complete in HeLa and fission yeast cells (39, 40), it seems likely that a fraction of La1-408-S366 will be nonphosphorylated in the two-hybrid yeast. Therefore, the slight positive effect of hLa1-408-S366 (Fig. 6A) may result from the small fraction of npLa in these yeast cells.
DISCUSSION
Novel data reported here indicate that the isoform of hLa that is nonphosphorylated on S366 resides in discrete sites within nucleoli that correspond to the dense fibrillar component where rRNA processing occurs. While npLa, nascent pol I transcripts and fibrillarin all appeared to be largely limited to these discrete sites, nucleolin is present at but not limited to these sites, as its localization is more complex (28). Nucleolin is a most abundant nucleolar protein that has been implicated in multiple steps in ribosome biogenesis (reviewed in reference 2). Thus, while immunolocalization shows overlap of npLa and nucleolin (data not shown), the most intense nucleolin staining is at the periphery of the nucleolus as reported previously (18), presumably reflecting its role in later stages of rRNA biogenesis (28). Nonetheless, close association with nucleolin was indicated by FRET between CFP-nucleolin and YFP-La in the nucleolus and by coimmunoprecipitation with native endogenous La as well as with epitope-tagged La expressed in HeLa cells. FRET between LaA366 and nucleolin provides evidence that nucleolin interacts with npLa in the nucleolus. Localization of fluorescent La mutants revealed that the SBM is required for nucleolar localization (Fig. 3J and 5A). These data are consistent with that obtained with two different antibodies specific for S366-phosphorylated La, which did not detect concentration of pLa in nucleoli (39, 61). Finally, yeast two-hybrid assays indicate that the interaction with nucleolin is dependent on the SBM of La, comprising amino acids 328 to 344, and that the nonphosphorylatable mutant, hLaA366, interacts more strongly than does S366-phosphorylated La.
La has been documented to interact with multiple RNAs and their associated proteins, including Ro60, a factor involved in response to UV radiation (10, 11, 33). It is noteworthy that nucleolin was found to be associated with a fraction of cytoplasmic Ro RNPs in an RNase-sensitive manner (24). Because Ro60 is stably associated with the Y RNAs, which are pol III transcripts that largely retain their 3′ oligo(U) tracts, and La and Ro co-occupy a fraction of these heterogenous RNPs, the Y RNAs are found in our anti-La immunoprecipitates and those of others (60, 67, 76). However, this finding does not indicate that the Y RNAs are involved in the La-nucleolin interaction characterized here, since examination for hY1 and hY3 RNAs by Northern blotting after treatment of our anti-La IPs with RNase A revealed no trace of the otherwise abundant hY1 and hY3 RNAs, whereas the La-nucleolin interaction remained in the same samples (data not shown). Our data should be further distinguished from those of the previous study since we performed sequential affinity purifications and immunoprecipitation from low-speed supernatants of detergent-mediated whole-cell extracts as opposed to the detergent-free extract of a cytoplasmic high-speed supernatant (24). Moreover, our FRET data indicate the La-nucleolin interaction in the nucleolus. Thus, it would seem that the nucleolin associated with the cytoplasmic Ro RNPs characterized previously would represent a fraction quite distinct from that which we have characterized here. Our data demonstrate that the RNase-resistant La-nucleolin interaction characterized here occurs in the absence of Y and other La-associated RNAs, consistent with the two-hybrid data for yeast.
We observed more nucleolar accumulation of GFP-hLa in CMT3 cells than YFP-hLa in HeLa cells. This finding appeared not to be due to intrinsic difference in the accumulation of GFP-hLa and YFP-hLa per se, since direct comparisons revealed similar levels in nucleoli in the cell types examined (data not shown). Rather, the data suggest that HeLa and CMT3 cells differ in the amounts of GFP-hLa and YFP-hLa that accumulate in their nucleoli. It is noteworthy that while GFP-hLa26-408 exhibits somewhat more nucleolar accumulation than hLa1-408 in CMT3 cells (Fig. 3B and F), this was not the case in HeLa cells (Fig. 5A). This difference does not affect our major conclusion, since in each cell type, the hLaΔ328-344 fusion exhibited less nucleolar accumulation than did the hLa1-408 fusion.
Reports of specific proteins that interact with La in the absence of RNA are few. Among the limited number of these are hepatitis C virus NS5A protein and DDX15/hPrp43, a putative DEAH-box RNA helicase (25, 36). Because La is a highly abundant protein found in multiple subcellular compartments, it should be expected that only a fraction may be associated with any particular binding protein. Our data suggest that only a fraction of La is associated with nucleolin and, likewise, that only a fraction of nucleolin is stably associated with La. However, since npLa represents only ∼15% of total La (39), the amount associated with nucleolin may be a substantial fraction of npLa. These data indicate that some La-interacting proteins may associate preferentially with npLa and may therefore appear to interact with only a minute fraction of total La.
Nucleolar localization.
In addition to establishing an isoform identity of nucleolar La, we identified the SBM (residues 328 to 344) as required for nucleolar localization, in agreement with recent work that showed that La amino acids 323 to 354 are a nucleolar localization signal (35). The present data extend the recent report because they provide evidence that nucleolar localization is specific for npLa and, additionally, because nucleolin was identified as a specific La-interacting protein. The cumulative data which include RNase-resistant interaction between La and nucleolin, together with the yeast two-hybrid data that indicate SBM-dependent, phosphorylation-sensitive interaction with nucleolin, support the idea that npLa accumulates in the nucleolus as a result of its interaction with nucleolin.
The SBM of hLa resides within a larger basic tract, spanning residues 325 to 365, with an isoelectric point (pI) of 12.5, that is followed by an abrupt transition to an acidic tract, 367DDEHDEHDE375, with a pI of 3.7. Phospho-S366 accentuates the transition with a high-density acidic group at the boundary that presumably leads to structural alterations. These data are reminiscent of a nucleolar retention signal that can be negatively controlled by phosphorylation (5) and a nucleolar localization sequence that is unmasked by deletion of a nearby acidic tract or substitutions that block phosphorylation (32). Thus, hLa may be a member of a class of proteins whose basic determinants of nucleolar accumulation (72) can be neutralized by adjacent acidic regions and phosphorylation.
The present data are entirely consistent with prior characterizations of the interaction potential of the SBM, which had been shown to be involved in RNA binding, and stimulation of pol III activity, in each case negated by S366 phosphorylation (20, 21, 30, 40). Moreover, the data support the idea that the hLa CTD contains a phosphorylation-sensitive interaction domain that may interact with nucleic acid or protein.
A multitude of localization signals in the CTD of La.
The multiple localization signals identified in La include an NLS that is positioned at the extreme terminus (383 to 408), a nuclear retention element (NRE, 316 to 326) whose integrity is important for proper pre-tRNA processing, and a nucleolar localization signal (323 to 354) (35, 38, 66). In addition, functional analyses have indicated the presence of a cryptic signal that can direct the nuclear export of yeast and human La proteins, although the sequence elements responsible for export have yet to be identified (38).
Since GFP-hLa1-328 was mainly cytoplasmic (Fig. 3P), and GFP-hLa1-363 was mainly nuclear, the region between 328 and 363 would appear to harbor an NLS. Examination indeed revealed that truncation at 363 leaves a tract of basic-rich residues, GKGKVQFQGKKTK-363 (Fig. 3Q), at the C terminus which may be recognized as an NLS in this context (62). As mentioned in the introduction, two different proteolytic cleavages of hLa, at positions 358 and 374, provide evidence that cytoplasmic accumulation can be controlled by C-terminal truncation (3, 63, 65). Poliovirus protease 3C cleaves immediately upstream of the terminal basic residues of GFP-hLa1-363 (Fig. 3Q), apparently inactivating the potential NLS activity of the truncated protein. Likewise, caspase 3 cleavage (Fig. 3Q) produces a truncation whose acidic terminus apparently masks the activity of the cryptic NLS. These observations suggest that the activities of these proteases must be precise in order to direct cytoplasmic accumulation (Fig. 3Q). In summary, it would appear that the localization activities in hLa's CTD are comprised of an intricate array of closely associated and sometimes overlapping signals that remain to be fully understood.
Colocalization with nascent pol I transcripts and fibrillarin and interaction with nucleolin provide evidence for a potential role for npLa in rRNA biogenesis.
Recent results indicate that pLa, rather than npLa, is associated with nascent pol III transcripts in the nucleoplasm and therefore does not indicate a role for npLa in association with a pol III transcript in the nucleolus (39). Although an intriguing aspect of the nucleolus is its occupation by proteins involved in diverse activities with no readily apparent direct connection to ribosome biogenesis (6, 12, 41, 54, 70), its best-established function is the production of ribosomes and other ribonucleoprotein complexes (26, 55, 56). In the case of the signal recognition particle, a ribosome-associated RNP composed of multiple subunits that controls production of membrane-bound polypeptides in the cytoplasm, its RNA, and protein components localizes to the nucleolus, and the RNA was found concentrated in a nucleolar subcompartment different from where ribosomes are assembled, suggesting that there are nucleolar regions that are specialized for functions other than ribosome biogenesis (56, 58, 59). Thus, the close proximity of npLa to nascent pol I transcripts, fibrillarin, and nucleolin observed here suggests potential involvement in an early phase of rRNA biogenesis. Although we have been unable to detect the 5′ external transcribed spacer of the nascent pol I transcript in association with La, we cannot rule out the possibility that La and nucleolin interact in the context of a nascent pol I transcript.
Among the best-characterized activities of nucleolin is its ability to bind to specific sequence elements found in the 5′ external transcribed spacer and promote an early processing step (2, 27). Indeed, the mode of nucleolin binding to pre-rRNA, the locations of its binding sites in the 5′ external spacer of the precursor, and the timing of processing have led to the proposal that it serves as a chaperone to promote correct folding of pre-rRNA (2; also see reference 26). Given the recent demonstration that yeast La protein can promote the proper folding of a structured RNA (7) and the interaction between npLa and nucleolin documented here, it is tempting to speculate that npLa may assist nucleolin in its pre-rRNA chaperone activity. In summary, the data presented here suggest that npLa may be involved in rRNA biogenesis.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Child Health and Human Development. R.J.M. was supported by The Commissioned Corp of the United States Public Health Service.
We thank Tom Misteli for access to imaging equipment and reagents and for advice and critical reading, B. Vogler for assistance with fluorescence, and M. Cashel, R. Crouch, B. Howard, and the Friday Seminar and SMCB lab members for discussion and comments, and the following for reagents: J. Aris for mAb D77, R. Ochs for serum S4, P. Chan for anti-B23 mAb, J. Sagara for the fusion construct, and B. Howard for HSC172 cells.
REFERENCES
- 1.Alfano, C., D. Sanfelice, J. Babon, G. Kelly, A. Jacks, S. Curry, and M. R. Conte. 2004. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 11:323-329. [DOI] [PubMed] [Google Scholar]
- 2.Allain, F. H., P. Bouvet, T. Dieckmann, and J. Feigon. 2000. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 19:6870-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayukawa, K., S. Taniguchi, J. Masumoto, S. Hashimoto, H. Sarvotham, A. Hara, T. Aoyama, and J. Sagara. 2000. La autoantigen is cleaved in the COOH terminus and loses the nuclear localization signal during apoptosis. J. Biol. Chem. 275:34465-34470. [DOI] [PubMed] [Google Scholar]
- 4.Broekhuis, C. H., G. Neubauer, A. van der Heijden, M. Mann, C. G. Proud, W. J. van Venrooij, and G. J. Pruijn. 2000. Detailed analysis of the phosphorylation of human La (SS-B) autoantigen. (De)phosphorylation does not affect subcellular distribution. Biochemistry 39:3023-3033. [DOI] [PubMed] [Google Scholar]
- 5.Catez, F., M. Erard, N. Schaerer-Uthurralt, K. Kindbeiter, J.-J. Madjar, and J.-J. Diaz. 2002. Unique motif for nucleolar retention and nuclear export regulated by phosphorylation. Mol. Cell. Biol. 22:1126-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerutti, L., and V. Simanis. 2000. Controlling the end of the cell cycle. Curr. Opin. Genet. Dev. 10:65-69. [DOI] [PubMed] [Google Scholar]
- 7.Chakshusmathi, G., S. D. Kim, D. A. Rubinson, and S. L. Wolin. 2003. A La protein requirement for efficient pre-tRNA folding. EMBO J. 22:6562-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers, J. C., D. Kenan, B. J. Martin, and J. D. Keene. 1988. Genomic structure and amino acid sequence domains of the human La autoantigen. J. Biol. Chem. 263:18043-18051. [PubMed] [Google Scholar]
- 9.Chang, Y.-N., D. J. Kenan, J. D. Keene, A. Gatignol, and K.-T. Jeang. 1994. Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J. Virol. 68:7008-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, X., A. M. Quinn, and S. L. Wolin. 2000. Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Genes Dev. 14:777-782. [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, X., and S. L. Wolin. 2004. The Ro 60 kDa autoantigen: insights into cellular function and role in autoimmunity. J. Mol. Med. 82:232-239. [DOI] [PubMed] [Google Scholar]
- 12.Cockell, M. M., and S. M. Gasser. 1999. The nucleolus: nucleolar space for RENT. Curr. Biol. 9:R575-R576. [DOI] [PubMed] [Google Scholar]
- 13.Costa-Mattioli, M., Y. Svitkin, and N. Sonenberg. 2004. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 24:6861-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong, G., G. Chakshusmathi, S. L. Wolin, and K. M. Reinisch. 2004. Structure of the La motif: a winged helix domain mediates RNA binding via a conserved aromatic patch. EMBO J. 23:1000-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dundr, M., M. D. Hebert, T. S. Karpova, D. Stanek, H. Xu, K. B. Shpargel, U. T. Meier, K. M. Neugebauer, A. G. Matera, and T. Misteli. 2004. In vivo kinetics of Cajal body components. J. Cell Biol. 164:831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dundr, M., U. Hoffmann-Rohrer, Q. Hu, I. Grummt, L. I. Rothblum, R. D. Phair, and T. Misteli. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science 298:1623-1626. [DOI] [PubMed] [Google Scholar]
- 17.Dundr, M., and T. Misteli. 2001. Functional architecture in the cell nucleus. Biochem. J. 356:297-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dundr, M., T. Misteli, and M. O. Olson. 2000. The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol. 150:433-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elbi, C., T. Misteli, and G. L. Hager. 2002. Recruitment of dioxin receptor to active transcription sites. Mol. Biol. Cell 13:2001-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan, H., J. L. Goodier, J. Chamberlain, D. R. Engelke, and R. J. Maraia. 1998. 5′ processing of tRNA precursors can be modulated by the human La antigen phosphoprotein. Mol. Cell. Biol. 18:3201-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan, H., A. L. Sakulich, J. L. Goodier, X. Zhang, J. Qin, and R. J. Maraia. 1997. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell 88:707-715. [DOI] [PubMed] [Google Scholar]
- 22.Fenyo, D., J. Qin, and B. T. Chait. 1998. Protein identification using mass spectrometric information. Electrophoresis 19:998-1005. [DOI] [PubMed] [Google Scholar]
- 23.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interaction. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 24.Fouraux, M. A., P. Bouvet, S. Verkaart, W. J. van Venrooij, and G. J. Pruijn. 2002. Nucleolin associates with a subset of the human Ro ribonucleoprotein complexes. J. Mol. Biol. 320:475-488. [DOI] [PubMed] [Google Scholar]
- 25.Fouraux, M. A., M. J. Kolkman, A. Van der Heijden, A. S. De Jong, W. J. Van Venrooij, and G. J. Pruijn. 2002. The human La (SS-B) autoantigen interacts with DDX15/hPrp43, a putative DEAH-box RNA helicase. RNA 8:1428-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerbi, S. A., A. V. Borovjagin, and T. S. Lange. 2003. The nucleolus: a site of ribonucleoprotein maturation. Curr. Opin. Cell Biol. 15:318-325. [DOI] [PubMed] [Google Scholar]
- 27.Ginisty, H., F. Amalric, and P. Bouvet. 1998. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 17:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ginisty, H., H. Sicard, B. Roger, and P. Bouvet. 1999. Structure and functions of nucleolin. J. Cell Sci. 112:761-772. [DOI] [PubMed] [Google Scholar]
- 29.Goldstein, S., C. M. Fordis, and B. H. Howard. 1989. Enhanced transfection efficiency and improved cell survival after electroporation of G2/M-synchronized cells and treatment with sodium butyrate. Nucleic Acids Res. 17:3959-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodier, J. L., H. Fan, and R. J. Maraia. 1997. A carboxy-terminal basic region controls RNA polymerase III transcription factor activity of human La protein. Mol. Cell. Biol. 17:5823-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Hebert, M. D., and A. G. Matera. 2000. Self-association of coilin reveals a common theme in nuclear body localization. Mol. Biol. Cell 11:4159-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendrick, J. P., S. L. Wolin, J. Rinke, M. R. Lerner, and J. A. Steitz. 1981. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol. Cell. Biol. 1:1138-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holcik, M., and R. G. Korneluk. 2000. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell. Biol. 20:4648-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horke, S., K. Reumann, M. Schweizer, H. Will, and T. Heise. 2004. Nuclear trafficking of La protein depends on a newly identified NoLS and the ability to bind RNA. J. Biol. Chem. 279:26563-26570. [DOI] [PubMed] [Google Scholar]
- 36.Houshmand, H., and A. Bergqvist. 2003. Interaction of hepatitis C virus NS5A with La protein revealed by T7 phage display. Biochem. Biophys. Res. Commun. 309:695-701. [DOI] [PubMed] [Google Scholar]
- 37.Inada, M., and C. Guthrie. 2004. Identification of Lhp1p-associated RNAs by microarray analysis in Saccharomyces cerevisiae reveals association with coding and noncoding RNAs. Proc. Natl. Acad. Sci. USA 101:434-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Intine, R. V., M. Dundr, T. Misteli, and R. J. Maraia. 2002. Aberrant nuclear trafficking of La protein leads to disordered processing of associated precursor tRNAs. Mol. Cell 9:1113-1123. [DOI] [PubMed] [Google Scholar]
- 39.Intine, R. V., S. A. Tenenbaum, A. S. Sakulich, J. D. Keene, and R. J. Maraia. 2003. Differential phosphorylation and subcellular localization of La RNPs associated with precursor tRNAs and translation-related mRNAs. Mol. Cell 12:1301-1307. [DOI] [PubMed] [Google Scholar]
- 40.Intine, R. V. A., A. L. Sakulich, S. B. Koduru, Y. Huang, E. Pierstorrf, J. L. Goodier, L. Phan, and R. J. Maraia. 2000. Transfer RNA maturation is controlled by phosphorylation of the human La antigen on serine 366. Mol. Cell 6:339-348. [DOI] [PubMed] [Google Scholar]
- 41.Isaac, C., K. L. Marsh, W. A. Paznekas, J. Dixon, M. J. Dixon, E. W. Jabs, and U. T. Meier. 2000. Characterization of the nucleolar gene product, treacle, in Treacher Collins syndrome. Mol. Biol. Cell 11:3061-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacks, A., J. Babon, G. Kelly, I. Manolaridis, P. D. Cary, S. Curry, and M. R. Conte. 2003. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure (Cambridge) 11:833-843. [DOI] [PubMed] [Google Scholar]
- 43.Karpova, T. S., C. T. Baumann, L. He, X. Wu, A. Grammer, P. Lipsky, G. L. Hager, and J. G. McNally. 2003. Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J. Microsc. 209:56-70. [DOI] [PubMed] [Google Scholar]
- 44.Kenan, D. J. 1995. RNA recognition by the human La protein and its relevance to transcription, translation and viral infectivity. Ph.D. thesis. Duke University, Durham, N.C.
- 45.Kenan, D. J., and J. D. Keene. 2004. La gets its wings. Nat. Struct. Mol. Biol. 11:303-305. [DOI] [PubMed] [Google Scholar]
- 46.Maraia, R., M. Zasloff, P. Plotz, and S. Adeniyi-Jones. 1988. Pathway of B1-Alu expression in microinjected oocytes: Xenopus laevis proteins associated with nuclear precursor and processed cytoplasmic RNAs. Mol. Cell. Biol. 8:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maraia, R. J. 2001. La protein and the trafficking of nascent RNA polymerase III transcripts. J. Cell Biol. 153:F13-F17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maraia, R. J., and R. V. Intine. 2002. La protein and its associated small nuclear and nucleolar precursor RNAs. Gene Expr. 10:41-57. [PMC free article] [PubMed] [Google Scholar]
- 49.Maraia, R. J., and R. V. Intine. 2001. Recognition of nascent RNA by the human La antigen: conserved and diverged features of structure and function. Mol. Cell. Biol. 21:367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meerovitch, K., Y. V. Svitkin, H. S. Lee, F. Lejbkowicz, D. J. Kenan, E. K. Chan, V. I. Agol, J. D. Keene, and N. Sonenberg. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67:3798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyuhas, O., and E. Hornstein. 2000. Translational control of TOP mRNAs, p. 671-693. In N. Sonenberg, J. W. B. Hershey, and M. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Nakatani, Y., and V. Ogryzko. 2003. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370:430-444. [DOI] [PubMed] [Google Scholar]
- 53.Ohndorf, U. M., C. Steegborn, R. Knijff, and P. Sondermann. 2001. Contributions of the individual domains in human La protein to its RNA 3′-end binding activity. J. Biol. Chem. 276:27188-27196. [DOI] [PubMed] [Google Scholar]
- 54.Olson, M. O., M. Dundr, and A. Szebeni. 2000. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 10:189-196. [DOI] [PubMed] [Google Scholar]
- 55.Pederson, T. 1998. The plurifunctional nucleolus. Nucleic Acids Res. 26:3871-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pederson, T., and J. C. Politz. 2000. The nucleolus and the four ribonucleoproteins of translation. J. Cell Biol. 148:1091-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pellizzoni, L., B. Cardinali, N. Lin-Marq, D. Mercanti, and P. Pierandrei-Amaldi. 1996. A Xenopus laevis homologue of the La autoantigen binds the pyrimidine tract of the 5′ UTR of ribosomal protein mRNAs in vitro: implication of a protein factor in complex formation. J. Mol. Biol. 259:904-915. [DOI] [PubMed] [Google Scholar]
- 58.Politz, J. C., L. B. Lewandowski, and T. Pederson. 2002. Signal recognition particle RNA localization within the nucleolus differs from the classical sites of ribosome synthesis. J. Cell Biol. 159:411-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Politz, J. C., S. Yarovoi, S. M. Kilroy, K. Gowda, C. Zwieb, and T. Pederson. 2000. Signal recognition particle components in the nucleolus. Proc. Natl. Acad. Sci. USA 97:55-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pruijn, G. J., R. L. Slobbe, and W. J. van Venrooij. 1991. Analysis of protein-RNA interactions within Ro ribonucleoprotein complexes. Nucleic Acids Res. 19:5173-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raats, J. M., W. F. Roeffen, S. Litjens, I. Bulduk, G. Mans, W. J. van Venrooij, and G. J. Pruijn. 2003. Human recombinant anti-La (SS-B) autoantibodies demonstrate the accumulation of phosphoserine-366-containing la isoforms in nucleoplasmic speckles. Eur. J. Cell Biol. 82:131-141. [DOI] [PubMed] [Google Scholar]
- 62.Roberts, B. L., W. D. Richardson, and A. E. Smith. 1987. The effect of protein context on nuclear location signal function. Cell 50:465-475. [DOI] [PubMed] [Google Scholar]
- 63.Rutjes, S. A., P. J. Utz, A. der Heijden, C. Broekhuis, W. J. van Venrooij, and G. J. Pruijn. 1999. The La (SS-B) autoantigen, a key protein in RNA biogenesis, is dephosphorylated and cleaved early during apoptosis. Cell Death Differ. 6:976-986. [DOI] [PubMed] [Google Scholar]
- 64.Shaw, P. J., and E. G. Jordan. 1995. The nucleolus. Annu. Rev. Cell Dev. Biol. 11:93-121. [DOI] [PubMed] [Google Scholar]
- 65.Shiroki, K., T. Isoyama, S. Kuge, T. Ishii, S. Ohmi, S. Hata, K. Suzuki, Y. Takasaki, and A. Nomoto. 1999. Intracellular redistribution of truncated La protein produced by poliovirus 3Cpro-mediated cleavage. J. Virol. 73:2193-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simons, F. H., F. J. Broers, W. J. Van Venrooij, and G. J. Pruijn. 1996. Characterization of cis-acting signals for nuclear import and retention of the La (SS-B) autoantigen. Exp. Cell Res. 224:224-236. [DOI] [PubMed] [Google Scholar]
- 67.Slobbe, R. L., W. Pluk, W. J. van Venrooij, and G. J. M. Pruijn. 1992. Ro ribonucleoprotein assembly in vitro. Identification of RNA-protein and protein-protein interactions. J. Mol. Biol. 227:361-366. [DOI] [PubMed] [Google Scholar]
- 68.Smith, C. M., and J. A. Steitz. 1998. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 18:6897-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Steitz, J. A. 1989. Immunoprecipitation of ribonucleoproteins using autoantibodies. Methods Enzymol. 180:468-481. [DOI] [PubMed]
- 70.Szekely, A. M., Y. H. Chen, C. Zhang, J. Oshima, and S. M. Weissman. 2000. Werner protein recruits DNA polymerase-δ to the nucleolus. Proc. Natl. Acad. Sci. USA 97:11365-11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trotta, R., T. Vignudelli, L. Pecorari, R. V. Intine, C. Guerzoni, G. Santilli, O. Candini, M. W. Byrom, S. Goldoni, L. P. Ford, M. A. Caligiuri, R. Maraia, et al. 2003. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell 3:145-160. [DOI] [PubMed] [Google Scholar]
- 72.van Eenennaam, H., A. van der Heijden, R. J. Janssen, W. J. van Venrooij, and G. J. Pruijn. 2001. Basic domains target protein subunits of the RNase MRP complex to the nucleolus independently of complex association. Mol. Biol. Cell 12:3680-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vassilev, A., K. J. Kaneko, H. Shu, Y. Zhao, and M. L. DePamphilis. 2001. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 15:1229-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vassilev, A., J. Yamauchi, T. Kotani, C. Prives, M. L. Avantaggiati, J. Qin, and Y. Nakatani. 1998. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol. Cell 2:869-875. [DOI] [PubMed] [Google Scholar]
- 75.Wolin, S. L., and T. Cedervall. 2002. The La protein. Annu. Rev. Biochem. 71:375-403. [DOI] [PubMed] [Google Scholar]
- 76.Wolin, S. L., and J. A. Steitz. 1984. The Ro small cytoplasmic ribonucleoproteins: identification of the antigenic protein and its binding site on the Ro RNAs. Proc. Natl. Acad. Sci. USA 81:1996-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao, Y., Y. Zhang, Y. Kho, and Y. Zhao. 2004. Proteomic analysis of integral plasma membrane proteins. Anal. Chem. 76:1817-1823. [DOI] [PubMed] [Google Scholar]