Abstract
The yeast SNM1/PSO2 gene specifically functions in DNA interstrand cross-link (ICL) repair, and its role has been suggested to be separate from other DNA repair pathways. In vertebrates, there are three homologs of SNM1 (SNM1A, SNM1B, and SNM1C/Artemis; SNM1 family proteins) whose functions are largely unknown. We disrupted each of the SNM1 family genes in the chicken B-cell line DT40. Both SNM1A- and SNM1B-deficient cells were sensitive to cisplatin but not to X-rays, whereas SNM1C/Artemis-deficient cells exhibited sensitivity to X-rays but not to cisplatin. SNM1A was nonepistatic with XRCC3 (homologous recombination), RAD18 (translesion synthesis), FANCC (Fanconi anemia), and SNM1B in ICL repair. SNM1A protein formed punctate nuclear foci depending on the conserved SNM1 (metallo-β-lactamase) domain. PIAS1 was found to physically interact with SNM1A, and they colocalized at nuclear foci. Point mutations in the SNM1 domain, which disrupted the interaction with PIAS1, led to mislocalization of SNM1A in the nucleus and loss of complementation of snm1a cells. These results suggest that interaction between SNM1A and PIAS1 is required for ICL repair.
DNA interstrand cross-links (ICLs) covalently bind the two complementary strands of the double helix of DNA. They severely impair fundamental processes of DNA metabolism such as transcription or replication, leading to cell death if they are left unrepaired. Current protocols for cancer chemotherapy often include ICL-inducing agents, i.e., mitomycin C (MMC) or cisplatin, for effective killing of malignant cells (reviewed in references 7 and 21).
The molecular mechanism of ICL repair is still poorly understood. In the yeast Saccharomyces cerevisiae, three distinct pathways, including nucleotide excision repair (NER), homologous recombination (HR), and translesion synthesis (TLS), participate in ICL repair (7, 20). In addition, yeast SNM1 (also known as PSO2) specifically functions in ICL repair without apparently participating directly in any of these pathways (2, 9). During the repair process, NER mediates incisions on both sides of the cross-linked DNA (sometimes termed “unhooking”) (5), and double-strand breaks (DSBs), which are probably associated with the collapse of a replication fork, are formed. The DSBs are then repaired by HR mechanisms. The interdependence between unhooking and formation of DSBs remains unclear (7). Yeast snm1 mutants are proficient in unhooking but are unable to process DSB intermediates (7, 19, 40).
In mammalian cells, the situation is even more complex. Among NER factors, XPF/ERCC1 endonuclease appears to be particularly important for the unhooking of ICLs (5, 21, 26, 32). Hamster mutant cells lacking the RAD51 paralog XRCC2 or XRCC3 display extreme sensitivity to ICLs, indicating an important role of HR in mammalian ICL repair (17). Cells from Fanconi anemia (FA) patients are also highly sensitive to ICL reagents (33), but the role of FA proteins in ICL repair is still unclear (4). Furthermore, there are three homologs of SNM1 (referred to as SNM1A, SNM1B, and SNM1C/Artemis) in vertebrate cells whose functions are largely unknown (6, 23). The SNM1 family proteins share a region of approximately 300 amino acids that is similar to the C-terminal region of yeast SNM1 (we termed this region the “SNM1 domain” for simplicity), which is homologous to metallo-β-lactamase (3, 23). Interestingly, human SNM1A (hSNM1A) has been shown to reside in nuclear dots or foci (29). DNA repair proteins often form such foci in response to DNA damage. Mouse embryonic stem (ES) cells lacking SNM1A exhibited increased sensitivity to MMC but not to other cross-linking agents or to ionizing radiation (IR), leading to the speculation that relatively mild phenotypes of the cells might be due to functional redundancy between the SNM1 homologs (6). Until now, the function of SNM1B has not been analyzed. SNM1C is identical to Artemis, a causative gene for a subset of radiation-sensitive severe combined immunodeficiency syndrome. Since Artemis participates in DSB repair by nonhomologous end joining, patients of this disorder display a defect in V(D)J recombination of antigen receptor genes together with IR sensitivity (23, 25). Artemis has a nuclease activity in its conserved SNM1 domain that cleaves a hairpin structure formed at coding joints during V(D)J recombination (18, 27, 28).
In this study, we disrupted all three SNM1 family genes in the chicken (ch) B-cell line DT40. We found that cells deficient in SNM1A or SNM1B, but not SNM1C/Artemis, were sensitive to ICL treatment, while only SNM1C-deficient cells displayed X-ray sensitivity. By analyzing double knockout cells, we showed nonepistasis in terms of cisplatin sensitivity between SNM1A and SNM1B, or between SNM1A and the genes involved in several DNA repair pathways, including XRCC3 (HR), RAD18 (TLS), and FANCC (FA). We also found physical interaction and nuclear colocalization of SNM1A with PIAS1, a small ubiquitin-like modifier (SUMO) E3 ligase (24, 35), originally identified as a transcriptional repressor of STAT1 (16). Significantly, point mutations in the SNM1 domain abolished the interaction, proper nuclear focus formation, and normal ICL repair function of SNM1A.
MATERIALS AND METHODS
Cells, antibodies, and expression vectors.
Wild-type and various mutant chicken DT40 cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 1% chicken serum, 2 mM l-glutamine, 50 μM 2-mercaptoethanol, penicillin, and streptomycin in 5% CO2 at 39.5°C. HeLa, 293T, and MCF-7 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, and nonessential amino acids in 5% CO2 at 37°C. DT40 cells deficient in the DNA ligase IV gene (LIG4) and RAD18 were described previously (1, 42). FANCC-deficient DT40 cells will be described elsewhere (S. Hirano et al., submitted for publication).
Anti-green fluorescent protein (GFP) (MBL, Nagoya, Japan), anti-Xpress (detecting Max epitope) (Invitrogen, Carlsbad, Calif.), anti-Myc (Invitrogen), anti-His (MBL), and Alexa fluor 594-conjugated secondary antibody (Molecular Probes, Eugene, Oreg.) were purchased from the indicated manufacturers.
hSNM1C/Artemis cDNA was obtained from normal human peripheral mononuclear cells by use of reverse transcription-PCR (RT-PCR) (12). The plasmid containing hSNM1A/KIAA0086 or hSNM1B/FLJ12810 was obtained from Kazusa DNA Research Institute (Kisarazu, Chiba, Japan) or Helix Research Institute (Kisarazu, Chiba, Japan), respectively. Tagged hSNM1 family genes were constructed by cloning each cDNA into pcDNA4/HisMax (Invitrogen), pcDNA3.1Myc-His (Invitrogen), pEGFP-C1 or pEGFP-N1 (BD Clontech, Palo Alto, Calif.). Mutations in enhanced GFP (EGFP)-hSNM1A were introduced by QuikChange (Stratagene, La Jolla, Calif.). EGFP-hPIAS1 was kindly provided by T. Nishida (11). Transfections into DT40 cells were done as described previously (41). HeLa, 293T, or MCF-7 cells were transiently transfected by use of Lipofectamine reagents (Invitrogen) in accordance with the manufacturer's protocol.
Generation of SNM1 family gene-deficient DT40 cells.
Partial chicken SNM1A, SNM1B, and SNM1C/Artemis cDNAs were identified in the chicken expressed sequence tag (EST) databases (http://swallow.gsf.de/dt40.html or http://www.chick.umist.ac.uk/index.html). Full-length SNM1 family cDNAs were then obtained by screening a DT40 cDNA library (kindly provided by R. Goitsuka) and/or RT-PCR from DT40 cDNA on the basis of the sequences found in the chicken genome database (http://www.genome.ucsc.edu/).
The genomic regions of chSNM1 family genes were obtained by either screening a library (Stratagene) or PCR amplification from DT40 genomic DNA. We designed each targeting vector by replacing a genomic segment within the SNM1 domain with a bsr or hisD resistance gene cassette (41). The SNM1A gene-targeting events were expected to delete an ∼3.5-kb genomic fragment containing three exons that correspond to SNM1A amino acids (aa) 698 to 818. In the case of SNM1B gene targeting, an ∼0.7-kb fragment containing part of one exon that corresponds to SNM1B aa 190 to 410 was deleted. The targeting vector for the SNM1C/Artemis gene replaces an ∼1.5-kb genomic fragment that corresponds to SNM1C aa 180 to 329 with a resistance gene cassette. Strategies for the gene targeting are summarized below (see Fig. 1B to D).
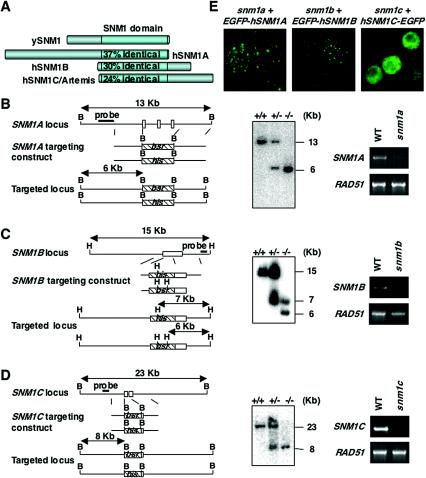
FIG. 1.
SNM1 family genes in vertebrates. (A) Schematic diagrams of SNM1A family. Yeast (y) SNM1 and hSNM1 family proteins are shown. Numbers in the SNM1 domain indicate percentages of identity to the yeast SNM1 domain. (B to D) Targeted disruption of chicken SNM1A (B), SNM1B (C), and SNM1C (D) loci in DT40 cells. Schematic representations of part of each loci, the gene targeting constructs, the configuration of targeted allele, and results of the Southern blot analysis and RT-PCR analysis are shown. White boxes indicate the positions of exons that were disrupted. B, BamHI; H, HindIII. Southern blot analysis was carried out with genomic DNA digested by BamHI (SNM1A and SNM1C) or HindIII (SNM1B) from cells with the indicated genotypes by use of flanking probes. mRNA expression of each disrupted gene or control (RAD51) in wild-type and mutant DT40 cells was analyzed by RT-PCR. (E) Nuclear localization patterns of EGFP-fused SNM1 family proteins in complemented DT40 mutant cells. WT, wild type.
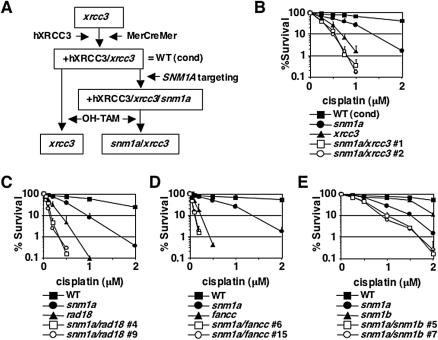
For making doubly deficient DT40 cells, SNM1A loci were disrupted in conditional xrcc3 cells, which was done as described below (see Fig. 3A). Briefly, xrcc3 cells (36) were simultaneously transfected with the human XRCC3 expression vector pCR3-loxP-hXRCC3/IRES-EGFP-loxP (8) and the MerCreMer expression vector (44). The XRCC3 expression vector contains the loxP sequences on both sides of the hXRCC3-internal ribosome entry site (IRES)-EGFP expression cassette. MerCreMer protein consists of Cre recombinase fused to two mutated ligand binding domains of human estrogen receptor and translocates to the nucleus upon addition of 4-hydroxytamoxifen (Sigma, St. Louis, Mo.). After SNM1A gene disruption, the hXRCC3-IRES-EGFP expression cassette (8) was excised by MerCreMer recombinase (44) activated by the addition of 4-hydroxytamoxifen (see Fig. 3A). Removal of the expression cassette was ensured by subcloning and was further verified by the loss of EGFP fluorescence and Southern blotting with a human XRCC3 probe. The SNM1A gene was also targeted in cells lacking RAD18 (42) or SNM1B. The FANCC gene (Hirano et al., submitted) was targeted in SNM1A-deficient cells.
FIG. 3.
Epistasis analyses between SNM1A and the indicated repair pathways. (A) Generation of an snm1a/xrcc3 double disruptant from conditional xrcc3 cells was done as described in Materials and Methods. (B to E) Survival of SNM1A double mutants with XRCC3 (B), RAD18 (C), FANCC (D), and SNM1B (E) compared to that of the wild type (WT) and corresponding single mutants after continuous exposure to cisplatin. Two clones of each double mutant were included in the analyses. The data shown are means ± standard deviations of results for at least three separate experiments.
Measurement of sensitivity of cells to DNA-damaging agents.
Colony formation was assayed in medium containing methylcellulose as described previously (41). Serially diluted cells were plated and then irradiated with 4-MV X-rays (linear accelerator; Mitsubishi Electric, Tokyo, Japan). UV irradiation was performed with a cell suspension in phosphate-buffered saline. Cells were exposed for 1 h to MMC (Kyowa Hakkou Kogyo, Tokyo, Japan) or continuously to cisplatin (Nihon-Kayaku, Tokyo, Japan), methyl methanesulfonate (Sigma), and bleomycin (Nihon-Kayaku). After the cells were cultured for 10 to 14 days, visible colonies were counted.
Yeast two-hybrid screening.
A yeast two-hybrid assay was performed by use of the Matchmaker LexA two-hybrid system (BD Clontech) in accordance with the manufacturer's protocol. A carboxyl-terminal fragment of chSNM1A containing the SNM1 domain (chSNM1AΔ1-553, equivalent to the hSNM1AΔ1-554 fragment) was cloned in-frame with the LexA DNA binding domain in the bait plasmid pEG202. EGY48 yeast cells containing both the bait plasmid and the reporter plasmid pSH18-34 were transformed with a pJG4.5-DT40 cDNA library (kindly provided by R. Goitsuka). Using two reporter expression genes (lacZ and LEU2), double-positive clones were selected from 8 × 106 transfectants. Plasmid DNAs were recovered, and the cDNA inserts were sequenced. To detect the interaction among human proteins, the hSNM1AΔ1-554 fragment or hPIAS1 cDNA was cloned into the bait or prey plasmid, respectively. Two point mutations were introduced independently using QuikChange reagent (Stratagene). The β-galactosidase activity (Miller units) was measured by a liquid culture assay using o-nitrophenyl-β-d-galactopyranoside (Nacalai tesque, Kyoto, Japan) as substrate, and Miller units were calculated according to the protocol from BD Clontech.
Ni-NTA resin pull-down assay.
Transfected 293T cells were solubilized by a brief sonication in lysis buffer (10 mM Tris-HCl, 50 mM sodium phosphate, 100 mM NaCl [pH 8]) containing 3 M guanidine HCl and protease inhibitors (Roche, Mannheim, Germany). After centrifugation, cleared lysates were diluted with 1 M guanidine and incubated with Ni-nitrilotriacetic acid (Ni-NTA) resin (QIAGEN, Valencia, Calif.) for 3 h at 4°C. Resins were washed four times with wash buffer (50 mM sodium phosphate, 100 mM NaCl, 4 M urea [pH 7]) containing protease inhibitors. Samples were separated by use of sodium dodecyl sulfate-6% polyacrylamide gel electrophoresis gels and analyzed by Western blotting.
Fluorescence microscopy.
Cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 in phosphate-buffered saline, and then stained with an appropriate antibody followed by Alexa fluor 594-conjugated secondary antibody. Images were captured with a TCS-SP2 confocal laser-scanning microscope (Leica Microsystems, Wetzler, Germany).
General techniques for DT40 cell analysis.
RT-PCR analysis, Western blotting, flow cytometric analysis of cell cycle, cell growth determination, measurement of targeted integration frequencies, and analysis of HR-mediated repair of DSBs induced by I-SceI expression were described previously (41).
Nucleotide sequence accession numbers.
The cDNA sequences for chicken SNM1A, SNM1B, SNM1C, and PIAS1 were deposited in GenBank under accession numbers AY376896, AY376897, AY376898, and AY549568.
RESULTS
Generation of SNM1 family gene-deficient DT40 cells.
Previous studies have shown the existence of yeast SNM1 homologs in many organisms, including humans and mice, based on homology with the C-terminal region of yeast SNM1 (SNM1 domain). Mammalian species have at least three SNM1 homologs (SNM1 family), including SNM1A, SNM1B, and SNM1C/Artemis (Fig. 1A) (3, 6, 23). Of these, SNM1A displays the highest homology in the SNM1 domain, as well as overall structural similarity, to yeast SNM1. For example, the SNM1 domain in SNM1A is located at the C terminus, while this domain is located in the N terminus in both SNM1B and SNM1C.
To characterize the function of the SNM1 family genes, we aimed to generate gene disruptants in the chicken B-cell line DT40. We performed database searches and found several EST sequences that have significant homology to SNM1 family genes. chSNM1 family cDNAs and genomic DNAs were cloned using the PCR method and/or library screening. The percentages of amino acid sequence identity of chSNM1A (encoding 972 aa), chSNM1B (457 aa), and chSNM1C (714 aa) with their human counterparts are 46, 53, and 62%, respectively. We created gene targeting vectors for each family member by replacing a genomic segment containing the SNM1 domain with a drug-resistant gene cassette (see Material and Methods) (Fig. 1B to D). The gene targeting was achieved by serial transfections with the vectors.
In the resulting SNM1 family mutants (hereafter designated as snm1a, snm1b, and snm1c), gene disruption was verified by the loss of transcripts based on RT-PCR analysis (Fig. 1B to D). The proliferative properties of these mutant cells, determined by growth curve and cell-cycle analysis, were indistinguishable from those of wild-type cells (data not shown).
Distinct functions of SNM1 family genes in repairing DNA damage.
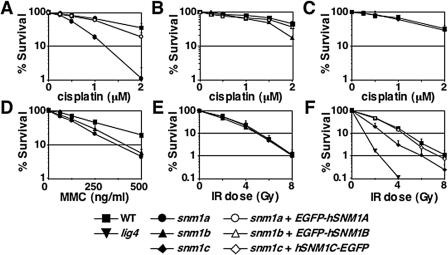
The DNA repair capacity of each mutant cell was assessed in colony survival assays following exposure to DNA-damaging agents. Both snm1a and snm1b DT40 cells were sensitive to cisplatin or MMC, albeit the sensitivity in the snm1b mutant seemed to be lower (Fig. 2A, B, and D). On the other hand, we could not find any significant sensitivity of snm1a and snm1b cells to X-ray, suggesting their specific role in ICL repair (Fig. 2E). Reassuringly, the defects were complemented by the expression of EGFP-fused hSNM1A or hSNM1B cDNAs in the respective mutants (Fig. 1E and 2A and B).
FIG. 2.
Sensitivities of wild-type and SNM1 family-deficient DT40 cells to DNA-damaging agents. The fractions of the surviving colonies after treatment compared to the nontreated control of the same genotype are shown as percent survival. (A to C) Survival of snm1a (A), snm1b (B), and snm1c (C) mutant cells compared to wild-type (WT) and complemented control cells after continuous exposure to cisplatin. (D and E) Survival of snm1a, snm1b, and WT cells after 1 h of exposure to MMC (D) or IR (E). (F) Survival of snm1c, ligase4, and WT cells and SNM1C-complemented control cells after IR. The data shown are means ± standard deviations of results for at least three separate experiments.
Next, we tested cisplatin and MMC sensitivity in snm1c cells and found no difference in the sensitivities of the mutant and wild-type DT40 cells (Fig. 2C and data not shown). In contrast, snm1c cells were sensitive to X-ray (Fig. 2F), as expected from the results of previous studies (23, 25, 30, 31). This defect was restored by the expression of hSNM1C-EGFP cDNA (Fig. 1E and 2F). snm1c cells were less X-ray sensitive than LIG4-deficient cells, in keeping with the report that Artemis-deficient mouse embryo fibroblast and ES cells were less sensitive to gamma rays than were XRCC4-deficient mouse embryo fibroblasts and ES cells, respectively (30, 31). XRCC4 is a cofactor for LIG4, and they function at the same step in the nonhomologous end joining pathway (reviewed in reference 15). Taken together, these results demonstrate that SNM1 family genes can be functionally divided into two groups: both SNM1A and SNM1B function in ICL repair but not in DSB repair, while SNM1C is involved in repair of DSB but not of ICL.
Nonepistatic relationship of SNM1A with other ICL repair pathways.
Multiple pathways, including the HR, TLS, and FA pathways, participate in ICL repair. To further dissect the role of SNM1A in ICL repair, we next sought to investigate any epistatic relationship of SNM1A with these pathways. We chose XRCC3 for the HR pathway, RAD18 for the TLS pathway, and FANCC for the FA pathway. In addition, we included SNM1B in our epistasis analysis since snm1b DT40 cells were also sensitive to cisplatin and MMC (Fig. 2B and D). Since XRCC3 is required for gene targeting (36), we first made conditional XRCC3-deficient cells (Fig. 3A) and disrupted the SNM1A gene in this background.
While all single mutants were more sensitive to cisplatin to various extents than wild-type cells were, the double mutants with SNM1A displayed higher sensitivities than the single mutants did in every combination tested (reflecting an additive phenotype) (Fig. 3A to D). These analyses suggest that SNM1A has a function distinct from the HR, TLS, or FA pathways.
In addition, we also tested whether SNM1A-deficient cells display any defects in HR assays. In assays of target integration frequency and HR repair of I-SceI restriction enzyme-induced DSBs, we observed similar levels of HR activity in SNM1A-deficient and wild-type control cells (data not shown).
SNM1 domain is required for ICL repair.
To examine whether the SNM1 domain is involved in ICL repair, we introduced a point mutation (D838N or H994A) into the SNM1 domain of the EGFP-hSNM1A construct (Fig. 4A and B). The equivalent amino acid changes in hSNM1C/Artemis were reported to abolish the nuclease activity of Artemis protein (18, 27), and alignment of chicken, human, mouse, and yeast SNM1 domains revealed that these were among the residues that were particularly well conserved (Fig. 4B). These mutants were stably transfected into snm1a DT40 cells, and clones expressing similar levels of EGFP were selected by fluorescence-activated cell sorting analysis (Fig. 4C). The function of each mutant protein was assayed by determining colony survival in the presence of cisplatin (Fig. 4C). As stated above, expression of EGFP-hSNM1A (wild type) could reverse the cisplatin sensitivity of snm1a cells to wild-type levels, while the mutant proteins were clearly defective in this ability. Although the D838N mutant was capable of complementation to some extent, the H994A mutant showed no complementation at all, indicating an essential role of the domain in ICL repair.
FIG. 4.
Mutational analysis of SNM1A. (A) Schematic representations of SNM1A point mutants. Cellular distributions of each construct determined by transient transfections of HeLa cells (E) and complementation data (C) are summarized. (B) Alignment of conserved regions in the SNM1 domain of SNM1 family proteins. The amino acid sequences of the SNM1 domain of human (Hs), mouse (Mm), chicken (Gd), and yeast (Sc) SNM1 family gene products were aligned. Amino acids that were identical across 10 proteins or 6 to 9 proteins are indicated by black or grey shading, respectively. Arrowheads indicate the mutated amino acid residues (D838 and H994 in hSNM1A) in this study. (C) Survival of snm1a cells stably expressing EGFP-hSNM1A mutants in the presence of cisplatin. Fluorescence-activated cell sorting analyses of wild-type and mutant EGFP-SNM1A expression are shown in the lower panels. Nontransfected snm1a cells were used as a negative control, and their fluorescence levels are shown as grey lines. (D) Localization of EGFP-SNM1A mutants transiently expressed in HeLa cells. Cells were examined with a confocal laser microscope.
Focus formation of hSNM1A depends on SNM1 domain.
Next, we examined localization of EGFP-fused hSNM1 proteins expressed in the mutant cells. As shown in Fig. 1E, both EGFP-hSNM1A and EGFP-hSNM1B localized as distinct subnuclear foci, while hSNM1C/Artemis-EGFP was diffusely distributed in the nucleus. These SNM1A or SNM1B foci existed in every cell, and MMC treatment did not significantly change their numbers, intensities, or distributions (data not shown).
When EGFP-hSNM1A was transiently transfected into HeLa, 293T, MCF-7, and DT40 cells, we observed similar distributions in the nucleus: the vast majority of cells expressing EGFP-hSNM1A displayed punctate and distinct foci (Fig. 4D and see Fig. 6A; data not shown). Transiently introduced N-terminal His/Max-tagged and C-terminal His/Myc-tagged hSNM1A were also detected as similar nuclear “dots” by use of appropriate antibodies (see Fig. 6B; data not shown).
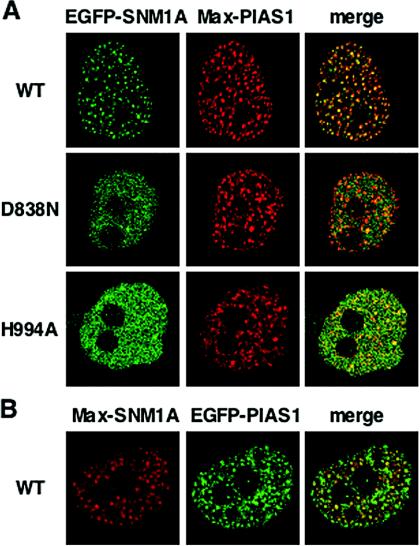
FIG. 6.
Colocalization of SNM1A and PIAS1 in HeLa cells. (A) Localization of transiently transfected EGFP-hSNM1A or its mutants (with the D838N or H994A mutation) and His/Max-hPIAS1. PIAS1 was detected by use of anti-Xpress and Alexa fluor 594-conjugated secondary antibody. Images were captured by confocal microscopy. (B) Localization of His/Max-SNM1A and EGFP-PIAS1 were analyzed as described for panel A.
To test whether SNM1 domain mutations affect focus formation of SNM1A, we transiently introduced the D383N or H994A expression plasmids into HeLa cells and examined the EGFP distribution. We found that the SNM1 domain point mutants were expressed in the nucleus diffusely, with barely recognizable (H994A mutant) or slight (D838N mutant) focus formation (Fig. 4D), indicating an important role of the SNM1 domain in focus formation. Localization of the mutant proteins in DT40 stable transformants was essentially the same as that observed in HeLa cells (data not shown). The degree of focus formation and the ability to complement cisplatin sensitivity seemed to be correlated (Fig. 4C and D, compare the D838N and H994A mutants). Taken together, these experiments suggest that the SNM1 domain of SNM1A, along with focus formation, is required for cisplatin tolerance. Consistently, the ICL repair function of yeast SNM1 also depends on the SNM1 domain (14).
Two-hybrid screening for SNM1A-interacting factors.
In order to find clues for the molecular mechanisms of SNM1A in ICL repair, we employed the yeast two-hybrid system to search for proteins that bind to SNM1A, by use of the C-terminal half of chSNM1A, which contains the entire SNM1 domain, as a bait (chSNM1AΔ1-531). In a screen of 8 × 106 DT40 cDNA library clones, 14 clones specifically interacted with chSNM1AΔ1-531. DNA sequencing of the interacting clones revealed that three of them were identical clones encoding chPIAS1 protein (lacking the N-terminal 12 residues) fused in-frame to the transcription activation domain in the prey plasmid.
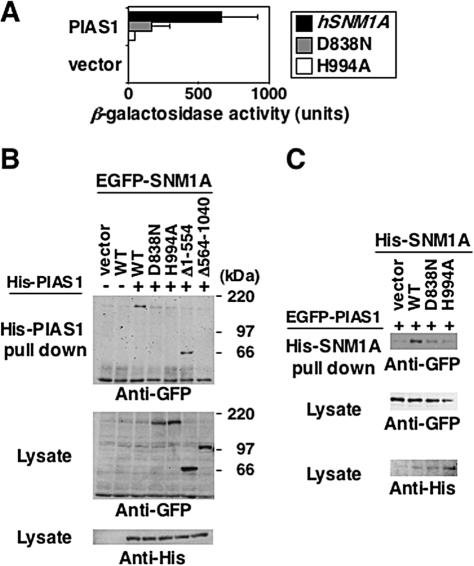
We confirmed this result by two-hybrid analysis of hSNM1A and hPIAS1. Full-length hPIAS1 or hSNM1AΔ1-554 (equivalent to chSNM1AΔ1-531) were cloned in-frame to the prey or bait plasmid, respectively. The hSNM1AΔ1-554 bait strongly interacted with hPIAS1, whereas mutations in the SNM1 domain (D838N and H994A) markedly reduced their interaction (Fig. 5A). The interaction with the H994A mutant was more severely affected than that of the D838N mutant, a finding which seemed to correlate with their ICL repair functions as well as their abilities to form foci.
FIG. 5.
Interaction of hSNM1A and hPIAS1. (A)Yeast two-hybrid assay. Yeast cells were cotransformed with the bait plasmid containing either hSNM1AΔ1-554 or its derivatives (with the D838N or H994A mutation) and the prey plasmid containing hPIAS1. Empty bait plasmid was used for control. β-Galactosidase activity was determined by a liquid culture assay and calculated in Miller units. (B and C) 293T cells were cotransfected with the indicated expression plasmids. Ni-NTA resin pull-down analysis was carried out as described in Materials and Methods. Samples were analyzed by Western blotting with anti-GFP or anti-His antibodies.
The SNM1A and PIAS1 interaction was further confirmed by pull-down analysis of His- or EGFP-tagged proteins expressed in 293T cells. Although the solubility of wild-type SNM1A was rather low compared to those of its mutants (Fig. 5B and C; see wild-type SNM1A bands in lysate), we could detect binding of full-length hSNM1A protein to full-length hPIAS1 (Fig. 6B and C). This interaction was strongly reduced by the D838N or H994A mutation, in good agreement with the two-hybrid analysis. Furthermore, hSNM1AΔ1-554 was able to bind to hPIAS1, while a fragment representing the N-terminal half of SNM1A (SNM1AΔ564-1040) could not (Fig. 5B). Collectively, these observations demonstrated that PIAS1 binds to the C-terminal region of SNM1A containing the SNM1 domain.
SNM1A subnuclear foci colocalized with PIAS1.
To directly test whether SNM1A and PIAS1 interact in vivo, we looked at HeLa cells expressing these proteins by use of confocal laser-scanning microscopy. When expressed alone, His/Max-tagged PIAS1 was present as punctuate nuclear foci (data not shown) (13, 22). As expected from two-hybrid and pull-down analyses, coexpression of EGFP-SNM1A and His/Max-PIAS1 resulted in colocalization in multiple small foci (Fig. 6A). His/Max-SNM1A and EGFP-PIAS1 proteins also colocalized (Fig. 6B). In contrast, the primarily diffuse distribution of EGFP-SNM1A D838N or H994A mutants was not altered by coexpression with His/Max-PIAS1 (Fig. 6A). We repeated these experiments with 293T cells, and the results were essentially the same (data not shown). Together, these data indicated that SNM1A and PIAS1 interact in vivo, leading to formation of the colocalizing nuclear foci. We conclude that PIAS1 interacts with SNM1A through the SNM1 domain, thereby mediating focus formation and promoting ICL repair.
DISCUSSION
SNM1 family genes and their roles in DNA repair.
In this study, we have established DT40 mutant cells lacking each of three SNM1 family genes and revealed that SNM1 families are classified into two subgroups by their function: SNM1A and SNM1B are involved only in ICL repair, whereas SNM1C/Artemis is involved in repairing DSB, not ICL (Fig. 2). The functions of SNM1A and SNM1B seem to be independent of each other, given the nonepistasis shown by the cisplatin sensitivity of the double knockout mutants.
In agreement with reports on yeast SNM1/PSO2 mutants (reviewed in references 2 and 7), snm1a DT40 cells showed significant sensitivity to cisplatin and MMC but not to IR, UV light at 254 nm, methyl methanesulfonate, or bleomycin (Fig. 2 and data not shown). Our results were also generally in agreement with genetic studies of mice regarding SNM1A (6) and SNM1C/Artemis (30, 31). However, mouse snm1a ES cells were reported to be sensitive to MMC but not to cisplatin (6). This discrepancy might be due to differences in species, design of the gene disruption, or the possible contribution of SNM1B.
The classical epistasis analysis of S. cerevisiae determined that SNM1 belongs to the NER epistasis group with respect to monoadducts and ICL (10). More recent reports have suggested that yeast SNM1 acts in a different epistasis group than do REV3 and RAD51 for ICL repair (9, 14). We carried out similar epistasis analysis using DT40 cells and found that SNM1A was not epistatic with XRCC3 (HR), RAD18 (TLS), or FANCC (FA). We also confirmed that SNM1A did not play a significant role in HR by various assays, consistent with yeast SNM1 (14). Together, these data suggest that SNM1A constitutes a distinct ICL repair pathway, which is independent of the HR, TLS, or FA pathway. However, a more recent work has suggested that yeast SNM1, EXO1, and MUTS mismatch repair factors have a redundant and highly overlapping role in HR, making the defects due to single SNM1 deletion less clear (P. McHugh, personal communication). Thus, it still seems possible that SNM1A has a role in HR but that it is concealed by other redundant factors. More work is needed to clarify this point in the vertebrate DNA repair pathway.
PIAS1 interacts and colocalizes with SNM1A in nuclear foci.
During the course of this study, another group reported that SNM1A forms nuclear foci by use of ectopically expressed tagged hSNM1A as well as antibodies detecting endogenous SNM1A protein (29), although we had noticed similar findings ourselves.
Among SNM1 family genes, an enzymatic function has been identified only for SNM1C/Artemis. Artemis has a nuclease activity that depends on its SNM1 domain and is regulated by phosphorylation by DNA-dependent protein kinase (18, 28). Mutations in the SNM1 domain in SNM1C/Artemis abolished its function in V(D)J recombination (18, 27, 28). The equivalent SNM1A mutants could not effectively restore the ICL repair deficiency in snm1a DT40 cells, suggesting that the putative nuclease function of SNM1A is essential for ICL repair. An obvious scenario is that the nuclease function of SNM1A processes DNA intermediates during ICL repair. Because yeast snm1 mutants incise ICLs and normally form replication-associated DSBs (7, 19, 40), it is unlikely that vertebrate SNM1A is involved in these steps.
Interestingly, the same mutations in hSNM1A also abolished the interaction with PIAS1, leading to mislocalization of SNM1A. The PIAS1-SNM1 domain interaction likely mediates the SNM1A foci, but another region of SNM1A (aa 394 to 616) was also reported to play a role (14, 29), which might be independent of PIAS1. Since we observed a good correlation between the degree of focus formation and ICL repair function, we suggest that PIAS1-dependent focus formation and the putative nuclease function of the SNM1 domain are crucial for ICL repair through SNM1A. Given the SUMO E3 ligase function of PIAS1 and role of SUMO in DNA repair (24, 35), it might be interesting to test whether SNM1A is sumoylated during storage in the foci of ICL repair.
Role of SNM1A focus in ICL repair.
In our study using EGFP-hSNM1A, we could not detect any significant change in the number or intensity of foci following DNA damage in transient (HeLa or 293T) or stable (DT40) transformants (data not shown). The same was true for the SNM1B foci. Indeed, EGFP-SNM1A foci were highly stable, as determined by an assay of fluorescence recovery after photobleaching (data not shown). These data are more consistent with the idea that EGFP-hSNM1A foci act as a storage depot or possibly as sites for protein modification rather than as active DNA repair sites.
It was reported that the majority of unstressed cells contain a few endogenous SNM1A “bodies” and that, in response to IR or ICL reagents, the cells form a large number of SNM1A foci (29), which colocalize with MRE11 or 53BP1 (37, 38). This finding is consistent with the idea that the foci are the sites for DNA repair, although the biological meaning of the SNM1A interaction with 53BP1 is unclear, given the specific role of SNM1A in ICL repair (34, 39). We were not able to detect such a dynamic change of localization of SNM1A; however, this inability could be due to the use of overexpressed EGFP-hSNM1A. In any case, our data suggest that hSNM1A has to be properly stored to exert its function. Focus formation might be crucial for mobilization or recruitment of SNM1A to DNA repair sites following DNA damage. Such a regulated protein storage-mobilization function has been described for the cell cycle checkpoint protein CHK2 stored in the promyelocytic leukemia protein (PML) bodies (43).
In summary, we have shown differential roles of vertebrate SNM1 family genes in DSB and ICL repair. Epistasis analyses suggest a separate and distinct function of SNM1A relative to those of other pathways. We also found that SNM1A function depends on the integrity of the SNM1 domain that interacts with PIAS1. The interaction probably mediates SNM1A storage in nuclear foci, which might be crucial for proper mobilization to DNA damage sites.
Acknowledgments
We thank Ryo Goitsuka (Science University of Tokyo) for λZAPII- and pJG4.5-DT40 cDNA libraries used in cDNA cloning and two-hybrid screening, respectively; Hideki Koyama (Yokohama City University) for LIG4-deficient DT40 cells; Tamotsu Nishida (Tokyo University of Pharmacy and Life Science) for the EGFP-hPIAS1 expression plasmid and helpful comments; Yoshinari Imajo and Jyuichi Kubota for irradiating cells with the linear accelerator; Taichi Shirao (Leica Microsystems), Kuniko Asahara, and Hiroyuki Kitao for advice with immunofluorescence and confocal microscopies; Sohsuke Seki for technical help; Mayu Fujii and Kazuko Hikasa for secretarial assistance; and Peter J. McHugh and Ian D. Hickson for critically reading the manuscript.
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology (M.I. and M.T.); by grants from the Uehara Memorial Foundation, the Okayama Medical Foundation, and the Kawasaki Medical and Medical Welfare Foundation (M.I.); and by grants from the Japan Space Forum (M.T.). Financial support also came from the Kawasaki Medical School (Project Research Grant 14-409).
REFERENCES
- 1.Adachi, N., T. Ishino, Y. Ishii, S. Takeda, and H. Koyama. 2001. DNA ligase IV-deficient cells are more resistant to ionizing radiation in the absence of Ku70: implications for DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 98:12109-12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brendel, M., D. Bonatto, M. Strauss, L. F. Revers, C. Pungartnik, J. Saffi, and J. A. P. Henriques. 2003. Role of PSO genes in repair of DNA damage of Saccharomyces cerevisiae. Mutat. Res. 544:179-193. [DOI] [PubMed] [Google Scholar]
- 3.Callebaut, I., D. Moshous, J.-P. Mornon, and J.-P. de Villartay. 2002. Metallo-β-lactamase fold within nucleic acids processing enzymes: the β-CASP family. Nucleic Acids Res. 30:3592-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Andrea, A. D., and M. Grompe. 2003. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 3:23-34. [DOI] [PubMed] [Google Scholar]
- 5.De Silva, I. U., P. J. McHugh, P. H. Clingen, and J. A. Hartley. 2000. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell. Biol. 20:7980-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dronkert, M. L. G., J. de Wit, M. Boeve, M. L. Vasconcelos, H. van Steeg, T. L. Tan, J. H. J. Hoeijmakers, and R. Kanaar. 2000. Disruption of mouse SNM1 causes increased sensitivity to the DNA interstrand cross-linking agent mitomycin C. Mol. Cell. Biol. 20:4553-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dronkert, M. L. G., and R. Kanaar. 2001. Repair of DNA interstrand cross-links. Mutat. Res. 486:217-247. [DOI] [PubMed] [Google Scholar]
- 8.Fujimori, A., S. Tachiiri, E. Sonoda, L. H. Thompson, P. K. Dhar, M. Hiraoka, S. Takeda, Y. Zhang, M. Reth, and M. Takata. 2001. Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J. 20:5513-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grossmann, K. F., A. M. Ward, M. E. Matkovic, A. E. Folias, and R. E. Moses. 2001. S. cerevisiae has three pathways for DNA interstrand crosslink repair. Mutat. Res. 487:73-83. [DOI] [PubMed] [Google Scholar]
- 10.Henriques, J. A. P., J. Brozmanova, and M. Brendel. 1997. Role of PSO genes in the repair of photoinduced interstrand cross-links and photooxidative damage in the DNA of the yeast Saccharomyces cerevisiae. J. Photochem. Photobiol. B 39:185-196. [DOI] [PubMed] [Google Scholar]
- 11.Kahyo, T., T. Nishida, and H. Yasuda. 2001. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8:713-718. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi, N., K. Agematsu, K. Sugita, M. Sako, S. Nonoyama, A. Yachie, S. Kumaki, S. Tsuchiya, H. D. Ochs, Y. Fukushima, and A. Komiyama. 2003. Novel Artemis gene mutations of radiosensitive severe combined immunodeficiency in Japanese families. Hum. Genet. 112:348-352. [DOI] [PubMed] [Google Scholar]
- 13.Kotaja, N., U. Karvonen, O. A. J.änne, and J. J. Palvimo. 2002. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 22:5222-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, X., and R. E. Moses. 2003. The β-lactamase motif in Snm1 is required for repair of DNA double-strand breaks caused by interstrand crosslinks in S. cerevisiae. DNA Repair (Amsterdam) 2:121-129. [DOI] [PubMed] [Google Scholar]
- 15.Lieber, M. R., Y. Ma, U. Pannicke, and K. Schwarz. 2003. Mechanism and regulation of human nonhomologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 4:712-720. [DOI] [PubMed] [Google Scholar]
- 16.Liu, B., J. Liao, X. Rao, S. A. Kushner, C. D. Chung, D. D. Chang, and K. Shuai. 1998. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. USA 95:10626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, N., J. E. Lamerdin, R. S. Tebbs, D. Schild, J. D. Tucker, M. R. Shen, K. W. Brookman, M. J. Siciliano, C. A. Walter, W. Fan, L. S. Narayana, Z.-Q. Zhou, A. W. Adamson, K. J. Sorensen, D. J. Chen, N. J. Jones, and L. H. Thompson. 1998. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell 1:783-793. [DOI] [PubMed] [Google Scholar]
- 18.Ma, Y., U. Pannicke, K. Schwarz, and M. R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J. recombination. Cell 108:781-794. [DOI] [PubMed] [Google Scholar]
- 19.Magana-Schwencke, N., J. P. Henriques, R. Chanet, and E. Moustacchi. 1982. The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc. Natl. Acad. Sci. USA 79:1722-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McHugh, P. J., W. R. Sones, and J. A. Hartley. 2000. Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:3425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McHugh, P. J., V. J. Spanswick, and J. A. Hartley. 2001. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2:483-490. [DOI] [PubMed] [Google Scholar]
- 22.Miyauchi, Y., S. Yogosawa, R. Honda, T. Nishida, and H. Yasuda. 2002. Sumoylation of Mdm2 by protein inhibitor of activated STAT (PIAS) and RanBP2 enzymes. J. Biol. Chem. 277:50131-50136. [DOI] [PubMed] [Google Scholar]
- 23.Moshous, D., I. Callebaut, R. de Chasseval, B. Corneo, M. Cavazzana-Calvo, F. Le Deist, I. Tezcan, O. Sanal, Y. Bertrand, N. Philippe, A. Fischer, and J. P. de Villartay. 2001. Artemis, a novel DNA double-strand break repair/V(D)J. recombination protein, is mutated in human severe combined immune deficiency. Cell 105:177-186. [DOI] [PubMed] [Google Scholar]
- 24.Müller, S., A. Ledl, and D. Schmidt. 2004. SUMO: a regulator of gene expression and genome integrity. Oncogene 23:1998-2008. [DOI] [PubMed] [Google Scholar]
- 25.Nicolas, N., D. Moshous, M. Cavazzana-Calvo, D. Papadopoulo, R. de Chasseval, F. Le Deist, A. Fischer, and J.-P. de Villartay. 1998. A human severe combined immunodeficiency (SCID) condition with increased sensitivity to ionizing radiations and impaired V(D)J rearrangements defines a new DNA recombination/repair deficiency. J. Exp. Med. 188:627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niedernhofer, L. J., H. Odijk, M. Budzowska, E. van Drunen, A. Maas, A. F. Theil, J. de Wit, N. G. J. Jaspers, H. B. Beverloo, J. H. J. Hoeijmakers, and R. Kanaar. 2004. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 24:5776-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pannicke, U., Y. Ma, K.-P. Hopfner, D. Niewolik, M. R. Lieber, and K. Schwarz. 2004. Functional and biochemical dissection of the structure-specific nuclease ARTEMIS. EMBO J. 23:1987-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poinsignon, C., D. Moshous, I. Callebaut, R. de Chasseval, I. Villey, and J.-P. de Villartay. 2004. The metallo-β-lactamase/β-CASP domain of Artemis constitutes the catalytic core for V(D)J recombination. J. Exp. Med. 199:315-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richie, C. T., C. Peterson, T. Lu, W. N. Hittelman, P. B. Carpenter, and R. J. Legerski. 2002. hSnm1 colocalizes and physically associates with 53BP1 before and after DNA damage. Mol. Cell. Biol. 22:8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rooney, S., F. W. Alt, D. Lombard, S. Whitlow, M. Eckersdorff, J. Fleming, S. Fugmann, D. O. Ferguson, D. G. Schatz, and J. Sekiguchi. 2003. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J. Exp. Med. 197:553-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rooney, S., J. Sekiguchi, C. Zhu, H.-L. Cheng, J. Manis, S. Whitlow, J. DeVido, D. Foy, J. Chaudhuri, D. Lombard, and F. W. Alt. 2002. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol. Cell 10:1379-1390. [DOI] [PubMed] [Google Scholar]
- 32.Rothfuss, A., and M. Grompe. 2004. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol. Cell. Biol. 24:123-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki, M. S., and A. Tonomura. 1973. A high susceptibility of Fanconi's anemia to chromosome breakage by DNA cross-linking agents. Cancer Res. 33:1829-1836. [PubMed] [Google Scholar]
- 34.Schultz, L. B., N. H. Chehab, A. Malikzay, and T. D. Halazonetis. 2000. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151:1381-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seeler, J.-S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4:690-699. [DOI] [PubMed] [Google Scholar]
- 36.Takata, M., M. S. Sasaki, S. Tachiiri, T. Fukushima, E. Sonoda, D. Schild, L. H. Thompson, and S. Takeda. 2001. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 21:2858-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson, L. H., and D. Schild. 2002. Recombinational DNA repair and human disease. Mutat. Res. 509:49-78. [DOI] [PubMed] [Google Scholar]
- 38.van den Bosch, M., R. T. Bree, and N. F. Lowndes. 2003. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 4:844-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward, I. M., K. Minn, J. van Deursen, and J. Chen. 2003. p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23:2556-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilborn, F., and M. Brendel. 1989. Formation and stability of interstrand cross-links induced by cis- and trans-diamminedichloroplatinum (II) in the DNA of Saccharomyces cerevisiae strains differing in repair capacity. Curr. Genet. 16:331-338. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto, K., M. Ishiai, N. Matsushita, H. Arakawa, J. E. Lamerdin, J.-M. Buerstedde, M. Tanimoto, M. Harada, L. H. Thompson, and M. Takata. 2003. Fanconi anemia FANCG protein in mitigating radiation- and enzyme-induced DNA double-strand breaks by homologous recombination in vertebrate cells. Mol. Cell. Biol. 23:5421-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita, Y. M., T. Okada, T. Matsusaka, E. Sonoda, G. Y. Zhao, K. Araki, S. Tateishi, M. Yamaizumi, and S. Takeda. 2002. RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. EMBO J. 21:5558-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, S., C. Kuo, J. E. Bisi, and M. K. Kim. 2002. PML-dependent apoptosis after DNA damage is regulated by the checkpoint kinase hCds1/Chk2. Nat. Cell Biol. 4:865-870. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, Y., J. Wienands, C. Zürn, and M. Reth. 1998. Induction of the antigen receptor expression on B lymphocytes results in rapid competence for signaling of SLP-65 and Syk. EMBO J. 17:7304-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]