Abstract
Background:
At present, a diagnostic tool with high specificity for impaired endometrial receptivity, which may lead to implantation failure, remains to be developed. We aimed to assess the different endometrial microRNA (miRNA) signatures for impaired endometrial receptivity by microarray analysis.
Methods:
A total of 12 repeated implantation failure (RIF) patients and 10 infertile patients, who conceived and delivered after one embryo transfer attempt, were recruited as RIF and control groups, respectively. Endometrial specimens from the window of implantation (WOI) were collected from these two groups. MiRNA microarray was conducted on seven and five samples from the RIF and control groups, respectively. Comparative, functional, and network analyses were performed for the microarray results. Quantitative real-time polymerase chain reaction (PCR) was performed on other samples to validate the expression of specific miRNAs.
Results:
Compared with those in the control group, the expression levels of 105 miRNAs in the RIF group were found to be significantly up- or down-regulated (at least 2-fold) by microarray analysis. The most relevant miRNA functional sets of these dysregulated miRNAs were miR-30 family, human embryonic stem cell regulation, epithelial-mesenchymal transition, and miRNA tumor suppressors by tool for annotations of microRNA analysis. Network regulatory analysis found 176 miRNA-mRNA interactions, and the top 3 core miRNAs were has-miR-4668-5p, has-miR-429, and has-miR-5088. Expression levels of the 18 selected miRNAs in new samples by real-time PCR were found to be regulated with the same trend, as the result of microarray analysis.
Conclusions:
There is a significant different expression of certain miRNAs in the WOI endometrium for RIF patients. These miRNAs may contribute to impaired endometrial receptivity.
Keywords: Embryo Implantation, Endometrial Receptivity, MicroRNA Microarray, Repeated Implantation Failure, Window of Implantation
Introduction
In the past three decades, since the first “test tube baby”, Louise Brown, was born in 1978, in vitro fertilization-embryo transfer (IVF-ET) has experienced rapid and momentous development. However, the pregnancy rate of IVF-ET remains relatively low up to now.[1] Only approximately 30% of the embryos transferred into the uterus lead to a successful pregnancy.[2] Successful implantation depends on the embryo's quality, embryo-endometrium interaction, and endometrial receptivity, of which inadequate endometrial receptivity is responsible for approximately two-thirds of implantation failures.[3,4,5]
The term, “endometrial receptivity”, is introduced to define the state of the endometrium during the window of implantation (WOI), which onsets 4–5 days after the endogenous/exogenous progesterone stimulation and ends 9–10 days afterward.[6] During this period, the endometrium acquires new adhesive properties allowing embryo adhesion and subsequent invasion.[7] Given its key role in successful implantation, predicting and improving endometrial receptivity is critical and may ultimately improve the pregnancy success rate of IVF-ET.[8] Unfortunately, no effective diagnostic tools are yet available to precisely predict endometrial receptivity.[9]
MicroRNAs (miRNAs) are small RNA fragments (18–25 nucleotides) that act as posttranscriptional regulators of various gene targets (either negatively or positively) rather than encoding proteins themselves.[10] miRNAs play a role in some biological processes, such as cellular differentiation, proliferation, and apoptosis, which are involved in implantation.[11,12,13] Therefore, several studies have been conducted to explore their role in endometrial receptivity. The miRNA expression profiles in human endometrium at different phases have been previously investigated. Kuokkanen et al.[14] studied the mRNA and miRNA profiles of fertile women's endometrial epithelial cells in the late proliferative and mid-secretory phases, respectively. They found that miRNA played a role in influencing endometrial receptivity through regulating the relevant genes’ expression. Altmäe et al.[15] compared the miRNA profile of prereceptive (LH+2) and receptive endometrium (LH+7) from fertile, nonstimulated women and revealed miR-30b, miR-30d, and miR-494's roles in regulating endometrium receptivity. Revel's study[16] showed the different miRNA profiles of the secretory endometrium between patients with repeated implantation failure (RIF) and fertile women. These data have clearly demonstrated that miRNA expression profiles of different populations/stages may differ and therefore should be applied in the diagnosis of endometrial receptivity, but further investigation is required due to study limitations.
Despite its diverse definitions, RIF is generally defined as failure to achieve a clinical pregnancy after transferring at least four good-quality embryos in at least three fresh or frozen cycles.[17] We hypothesized that the endometrial receptivity of RIF patients is low, while that of infertile women, who conceived after only one embryo transfer attempt, is high. The aim of this study was to identify the different miRNA expression profiles between these two populations, which may further provide a good predictor for helping to differentiate the discrepant endometrial receptivity.
Methods
Patients
A total of 22 female infertile patients were enrolled in this study. Twelve patients (numbered RIF1–RIF12), who all had a history of RIF, participated in the study group (RIF group). These participants had previously received in vitro fertilization/intracytoplasmic sperm injection (IVF/ICSI) treatment and had suffered at least three embryo transfer failures, in which at least four morphologically high-grade embryos were transferred in total. Further, in this group, there were no other obvious explanations for their RIFs, such as polycystic ovary syndrome, ovarian tumors, polyps, fibroids, endometriosis, hydrosalpinx, adenomyosis, and uterine malformation. Ten infertile patients (due to male infertility, tubal factors, or unexplained infertility; numbered C1–C10), who conceived and delivered after the first attempt of embryo transfer, were recruited as the control group.
Inclusion criteria for all participants were age <40 years; regular menstrual cycles; normal uterine cavity confirmed by hysteroscopy, and more specifically, without intrauterine adhesions or inflammation; endometrial thickness in the late follicular phase of ≥7 mm in ultrasonography; normal ovarian reserve (follicle-stimulating hormone <9.6 mU/ml);[18] a normal ovarian response to the stimulation protocol (>8 oocytes retrieved in a controlled ovary hyperstimulation cycle); and no hormone (estradiol/progesterone) applied during the endometrial biopsy cycle.
The study was approved by the Institutional Review Board at Peking University People's Hospital (No. 2011-87) and all participants signed written informed consent.
Endometrial biopsy specimens
Endometrial biopsies were performed by dilation and curettage during hysteroscopy, 5–7 days after ovulation. Ovulation was determined according to ultrasound combined with morning urine LH detection. Endometrial tissue was immediately sent to the laboratory to make sure it was processed within 1 h after the biopsy. Each sample was divided into two portions: one of which was fixed in 10% formalin and processed for histological evaluation (hematoxylin-eosin [H-E]); the second portion was frozen at −80°C for subsequent RNA extraction.
MicroRNA extraction and purifying
Total RNA was isolated from endometrial specimens using Trizol reagent (Invitrogen, USA) following the suppliers’ protocol, and miRNA was then purified using the mirVan miRNA Isolation Kit (AM1561, Ambion, USA) according to the manufacturer's instructions. The purity and concentration of RNA was determined by OD260/280 from a spectrophotometer (NanoDrop, ND-1000). The RNA integrity was examined by 1% formaldehyde denaturing gel electrophoresis. RNA with an OD260/280 between 1.8 and 2.0 and no degradation by electrophoresis was considered of good-quality and was included in further experiments.
MicroRNA array and microarray experiments
The transcription analysis of miRNA was performed using an miRNA Array (ID: 046064, Agilent, USA), which contains probes interrogating 2006 human mature miRNAs from miRBase R19.0 and 2164 Agilent control probes.
The miRNA microarray experiments were conducted according to the manufacturer's instructions for the miRNA Complete Labeling and Hyb Kit (Agilent). Then, 200 ng isolated RNA per sample was dephosphorylated and ligated with Cyanine3-pCp, and the labeled RNA was purified and hybridized to miRNA arrays. Images were scanned using the Agilent microarray scanner (G2565CA, Agilent). The arrays were then gridded and analyzed using Agilent Feature Extraction software version 10.10 (Agilent).
Microarray data analysis
The miRNA array data were analyzed for data summarization, normalization, and quality control using GeneSpring software version 13.0 (Agilent). The significance (P value) of the normalized value for raw data from each sample of the RIF and control group was calculated by an unpaired t-test and then corrected by the Benjamini-Hochberg method. The fold change was also calculated using the normalized value of the raw data. Two criteria were used to select the differentially expressed genes: a fold change ≥2 and a P < 0.05. To reduce the false discovery rate of genes, we excluded from our analysis miRNAs whose expression was detected in less than three samples in either the RIF or control groups. Furthermore, we adjusted the threshold to 5- and 10-fold changes to disclose miRNAs whose expression levels were more significantly different between the two groups.
Supervised hierarchical clustering with average linkage clustering analysis was further carried out on these differentially expressed miRNAs using Cluster version 3.0 software and Java Treeview (Stanford University School of Medicine, Stanford, CA, USA) to visually assess the differentially expressed miRNA profiles of the RIF and control groups.
Functional analysis of differentially expressed microRNAs
To discover the patterns and rules of the differentially expressed miRNAs, functional enrichment analysis was performed using tool for annotations of microRNAs (TAM) software (http://www.cuilab.cn/tam).
TAM, the tool for annotations of human miRNAs, is a web-accessible program that integrates miRNAs into different sets according to various rules and provides us with functions of interested miRNAs. Currently, TAM collects 238 miRNA sets, which include 413 distinct miRNAs.[19]
Regulatory network analysis of differentially expressed microRNAs and mRNAs
Based on the idea that miRNAs reduce, at least partially, the expression of targeted mRNAs, we constructed the miRNA-mRNA regulatory network of these differentially expressed miRNAs and those differentially expressed mRNAs we found from mRNA microarray study on the same samples. To improve the quality of prediction, the regulatory relationships were predicted by combining four existing algorithms: TargetScan, miRanda, Pictar, and DIANA, which were implemented with a Bioconductor package (http://bioconductor.org/), miRNAtap, in the R software environment (http://www.r-project.org). The diagram of the network was generated by Cytoscape.
Validation of the microarray data by quantitative real-time polymerase chain reaction
To validate our microarray findings, 10 new samples consisting of 5 from the RIF group (RIF8, RIF9, RIF10, RIF11, and RIF12) and 5 from the control group (C6, C7, C8, C9, and C10) were used to assess the expression of some miRNAs by quantitative real-time polymerase chain reaction (PCR). We selected miRNAs with a high-fold change and/or miRNAs reported in other similar literature before performing the validation. The names of the selected miRNAs and the corresponding primer sequences are listed in Supplementary Table S1.
Supplementary Table S1.
Sequences of miRNAs primers used for real-time PCR amplification
| Primers | miRNAs |
|---|---|
| F: ATAATACAACCTGATAAGTG | hsa-miR-374a-5p |
| F: GTCCAGTTTTCCCAGGAATCCC | hsa-miR-145-5p |
| F: GTAAACATCCTACACTCAGC | hsa-miR-30b-5p |
| F: TAGGTAGTTTCCTGTTGTTGGG | hsa-miR-196b-5p |
| F: CCCAGTGTTCAGACTACCTGTTC | hsa-miR-199a-5p |
| F: CCCAGTGTTTAGACTATCTGTTC | hsa-miR-199b-5p |
| F: TGGCAGTGTATTGTTAGCTGGT | hsa-miR-449a |
| F: CAGCAGCAATTCATGTTTT | hsa-miR-424-5p |
| F: TCCCTGAGACCCTTTAACCTGTG | hsa-miR-125b-5p |
| F: TAGCTTATCAGACTGATGTTG | hsa-miR-21-5p |
| F: TGGCAGGGAGGCTGGGAG | hsa-miR-1207-5p |
| F: TGGAGAGAAAGGCAGTAA | hsa-miR-4306 |
| F: CGCTCGGCGGTGGC | hsa-miR-572 |
| F: GCGGAGAGAGAATGGGGAGC | hsa-miR-5739 |
| F: AGAGATGAAGCGGGGGGG | hsa-miR-6088 |
| F: AGGGAAAAAAAAAAGGATTTGTC | hsa-miR-4668-5p |
| F: TAATACTGTCTGGTAAAACCGT | hsa-miR-429 |
| F: CAGGGCTCAGGGATTGGATG | hsa-miR-5088-5p |
| F: CTCGCTTCGGCAGCACA R: AACGCTTCACGAATTTGCGT |
U6 |
miRNAs: MicroRNAs; PCR: Polymerase chain reaction.
We applied the poly(A) method to confirm the expression of miRNAs. After being purified with the mirVanaTM miRNA Isolation Kit (Applied Biosystems, USA), total RNA was used for the RT reaction to generate the first strand cDNA using the miRcute miRNA cDNA First-Strand reverse transcription mixture (KR201). Quantitative real-time PCR was then performed according to the miRcute miRNA reverse transcription PCR (RT-PCR) protocol, using U6 as the housekeeping gene. The relative expression was calculated using 2−ΔΔCt method and analyzed with an unpaired t-test.
Results
Patients
The clinical characteristics of the two groups are listed in Table 1. There were no significant differences between the two groups in mean age, body mass index, length of menstrual cycle, menstrual duration, or endometrial thickness on the day of LH surge. Participants’ additional detailed clinical information is presented in Supplementary Table S2. The histological evaluation results for each sample reported normal mid-secretory endometrium. The micrograph of H-E staining for each sample was similar to that of RIF10 [Supplementary Figure S1 (108.5KB, tif) ].
Table 1.
Characteristics of the women undergoing endometrial biopsy sampling
| Variables | RIF group (n = 12) | Control group (n = 10) | t | P |
|---|---|---|---|---|
| Age (years) | 31.6 ± 4.1 | 32.1 ± 2.9 | −0.33 | 0.74 |
| BMI (kg/m2) | 22.77 ± 2.63 | 21.70 ± 2.22 | 1.01 | 0.32 |
| Cycle length (days) | 30.83 ± 3.10 | 30.40 ± 4.34 | 0.27 | 0.79 |
| Menses duration (days) | 5.08 ± 0.90 | 5.05 ± 0.98 | 0.08 | 0.94 |
| Endometrial thickness* (cm) | 0.95 ± 0.23 | 0.97 ± 0.26 | −0.19 | 0.85 |
Data were presented as mean ± SD. *Endometrial thickness: The thickness of the endometrium on the day when then biopsy was taken. BMI: Body mass index; RIF: Repeated implantation failure; SD: Standard deviation.
Supplementary Table S2.
More clinical information of the women undergoing endometrial biopsy sampling
| Case number | Age | Cause of infertility | Number of failed cycles | IVF/ICSI | Number of transferred embryos | Number of high quality embryos | Endometrial thickness on the day of LH surge | Endometrial type on the day of LH surge | The day of sample (post the day of LH surge) |
|---|---|---|---|---|---|---|---|---|---|
| RIF1 | 34 | Tubal | 11 | ICSI | 22 | 8 | 1 | A | +6 |
| RIF2 | 33 | Tubal | 4 | IVF | 11 | 7 | 0.7 | A | +7 |
| RIF3 | 38 | Tubal | 8 | IVF | 20 | 10 | 0.9 | A | +6 |
| RIF4 | 23 | Male | 4 | ICSI | 11 | 9 | 0.9 | A | +8 |
| RIF5 | 34 | Unexplained | 3 | IVF | 9 | 6 | 0.9 | A | +6 |
| RIF6 | 31 | Tubal | 3 | IVF | 7 | 7 | 1.2 | B | +7 |
| RIF7 | 28 | Male | 3 | ICSI | 7 | 6 | 1 | A | +6 |
| RIF8 | 35 | Male | 3 | ICSI | 6 | 4 | 0.8 | A | +6 |
| RIF9 | 32 | Tubal | 3 | IVF | 7 | 6 | 0.7 | A | +7 |
| RIF10 | 32 | Tubal | 3 | IVF | 6 | 4 | 1.2 | A | +6 |
| RIF11 | 33 | Tubal | 4 | ICSI | 8 | 4 | 1.0 | A | +7 |
| RIF12 | 26 | Male | 4 | IVF | 9 | 4 | 0.7 | A | +8 |
| C1 | 32 | Unexplained | 0 | IVF | 2 | 2 | 1.1 | A | +7 |
| C2 | 35 | Tubal | 0 | IVF | 2 | 1 | 0.9 | A | +7 |
| C3 | 29 | Male | 0 | ICSI | 2 | 2 | 1.1 | A | +8 |
| C4 | 33 | Unexplained | 0 | IVF | 2 | 2 | 1.1 | B | +7 |
| C5 | 26 | Tubal | 0 | ICSI | 2 | 1 | 1.2 | A | +7 |
| C6 | 31 | Male | 0 | IVF | 2 | 2 | 0.9 | A | +7 |
| C7 | 35 | Male | 0 | ICSI | 2 | 2 | 0.8 | A | +8 |
| C8 | 35 | Tubal | 0 | IVF | 3 | 2 | 0.7 | A | +8 |
| C9 | 33 | Tubal | 0 | IVF | 2 | 1 | 1.4 | B | +7 |
| C10 | 32 | Male | 0 | ICSI | 2 | 2 | 1.2 | A | +7 |
The samples with case number marked by underline were used for real-time PCR and the other samples were used for microarray. IVF: In vitro fertilization; ICSI: Intracytoplasmic sperm injection; LH: Luteinizing hormone; PCR: Polymerase chain reaction.
The micrograph of H and E dying for the sample of RIF10. The micrographs for the other 21 samples were similar to this, and all the reports were: normal mid-secretory endometrium. (a) Extreme glandular coiling secretory glands set within a spindled edematous stroma. Luminal secretion is most prominent (H and E, original magnification ×100). (b) The coiled spiral arteries are seen within an edematous stroma. Perivascular predecidual reaction has not occurred (H and E, original magnification ×200). RIF: Repeated implantation failure.
Results of microarray analysis
The miRNA array identified 105 microarray probes with expression levels in RIF patients that were 2-fold greater compared with those in the control group (93 upregulated and 12 downregulated). With a threshold of 5-fold and 10-fold changes, 70 (67 upregulated and 3 downregulated) and 49 (46 upregulated and 3 downregulated) miRNAs were identified, respectively [Table 2]. However, after the raw signal value correction (>50 for each sample), only 15 miRNAs were found to express in a significantly different way using 2-fold change as the threshold [Table 3]. All the differentially expressed genes are listed in Supplementary Table S3, and the raw data have been uploaded into the Gene Expression Omnibus database (number: GSE71332).
Table 2.
The number of differentially expressed miRNAs with different FCs*
| RIF versus control | Total dysregulated | Upregulated | Downregulated |
|---|---|---|---|
| FCs | |||
| >2 | 105 | 93 | 12 |
| >5 | 70 | 67 | 3 |
| >10 | 49 | 46 | 3 |
*The criteria for differentially expressed genes were: a greater than 2-FC with a P<0.05 by an unpaired t-test. FCs: Fold changes; RIF: Repeated implantation failure; miRNA: MicroRNA.
Table 3.
List of the differentially expressed miRNAs between the RIF and control group with the microarray raw signal value of all samples >50
| Systematic name | FC | Mirbase accession number |
|---|---|---|
| Upregulated miRNAs | ||
| hsa-miR-374a-5p | 7.74524 | MIMAT0000727 |
| hsa-miR-145-5p | 3.2018807 | MIMAT0000437 |
| hsa-miR-30b-5p | 2.9336023 | MIMAT0000420 |
| hsa-miR-196b-5p | 2.5407631 | MIMAT0001080 |
| hsa-miR-199a-5p | 2.5355365 | MIMAT0000231 |
| hsa-miR-199b-5p | 2.4879646 | MIMAT0000263 |
| hsa-miR-449a | 2.3427818 | MIMAT0001541 |
| hsa-miR-424-5p | 2.190957 | MIMAT0001341 |
| hsa-miR-125b-5p | 2.1353264 | MIMAT0000423 |
| hsa-miR-21-5p | 2.0441828 | MIMAT0000076 |
| Downregulated miRNAs | ||
| hsa-miR-1207-5p | 2.6758146 | MIMAT0005871 |
| hsa-miR-4306 | 2.2878602 | MIMAT0016858 |
| hsa-miR-572 | 2.0804768 | MIMAT0003237 |
| hsa-miR-5739 | 2.1607096 | MIMAT0023116 |
| hsa-miR-6088 | 2.1698172 | MIMAT0023713 |
RIF: Repeated implantation failure; miRNAs: MicroRNAs; FC: Fold change.
Supplementary Table S3.
List of differentially expressed miRNAs
| Systematic name | FC | Mirbase accession number |
|---|---|---|
| Upregulated miRNAs | ||
| hsa-miR-186-5p | 111.32307 | MIMAT0000456 |
| hsa-miR-135b-5p | 87.14255 | MIMAT0000758 |
| hsa-miR-3125 | 76.56718 | MIMAT0014988 |
| hsa-miR-136-5p | 73.41167 | MIMAT0000448 |
| hsa-miR-204-5p | 72.42954 | MIMAT0000265 |
| hsa-miR-3907 | 72.28242 | MIMAT0018179 |
| hsa-miR-30d-3p | 67.86486 | MIMAT0004551 |
| hsa-miR-1288 | 66.41283 | MIMAT0005942 |
| hsa-miR-371b-5p | 59.9805 | MIMAT0019892 |
| hsa-miR-374c-5p | 53.332508 | MIMAT0018443 |
| hsa-miR-32-5p | 42.89187 | MIMAT0000090 |
| hsa-miR-6512-5p | 40.44899 | MIMAT0025480 |
| hsa-miR-1914-3p | 35.808 | MIMAT0007890 |
| hsa-miR-205-5p | 32.111767 | MIMAT0000266 |
| hsa-miR-505-3p | 29.47475 | MIMAT0002876 |
| hsa-miR-7-1-3p | 29.342558 | MIMAT0004553 |
| hsa-miR-449b-5p | 29.015373 | MIMAT0003327 |
| hsa-miR-145-3p | 28.210178 | MIMAT0004601 |
| hsa-miR-4734 | 26.772724 | MIMAT0019859 |
| hsa-miR-144-5p | 25.90218 | MIMAT0004600 |
| hsa-miR-9-5p | 24.925808 | MIMAT0000441 |
| hsa-miR-4690-5p | 24.287247 | MIMAT0019779 |
| hsa-miR-744-5p | 19.679468 | MIMAT0004945 |
| hsa-miR-4486 | 18.95102 | MIMAT0019020 |
| hsa-miR-6132 | 18.29064 | MIMAT0024616 |
| hsa-miR-149-5p | 17.349735 | MIMAT0000450 |
| hsa-miR-1307-5p | 16.641792 | MIMAT0022727 |
| hsa-miR-424-3p | 16.024895 | MIMAT0004749 |
| hsa-miR-4324 | 15.442569 | MIMAT0016876 |
| hsa-miR-154-5p | 14.907551 | MIMAT0000452 |
| hsa-miR-10a-3p | 14.901987 | MIMAT0004555 |
| hsa-miR-141-5p | 14.643423 | MIMAT0004598 |
| hsa-miR-501-3p | 14.513452 | MIMAT0004774 |
| hsa-miR-1290 | 14.159889 | MIMAT0005880 |
| hsa-miR-206 | 14.120981 | MIMAT0000462 |
| hsa-miR-4257 | 13.665979 | MIMAT0016878 |
| hsa-miR-3127-5p | 13.506273 | MIMAT0014990 |
| hsa-miR-375 | 13.398406 | MIMAT0000728 |
| hsa-miR-3156-5p | 13.002842 | MIMAT0015030 |
| hsa-miR-598 | 11.697124 | MIMAT0003266 |
| hsa-miR-5088 | 11.453611 | MIMAT0021080 |
| hsa-miR-5096 | 11.370991 | MIMAT0020603 |
| hsa-miR-4746-3p | 11.153188 | MIMAT0019881 |
| hsa-miR-4726-5p | 11.049062 | MIMAT0019845 |
| hsa-miR-4656 | 10.935905 | MIMAT0019723 |
| hsa-miR-33a-5p | 10.576019 | MIMAT0000091 |
| hsa-let-7i-3p | 9.9141245 | MIMAT0004585 |
| hsa-miR-34c-3p | 9.809227 | MIMAT0004677 |
| hsa-miR-1285-3p | 9.787075 | MIMAT0005876 |
| hsa-miR-29c-5p | 9.758672 | MIMAT0004673 |
| hsa-miR-16-2-3p | 9.330457 | MIMAT0004518 |
| hsa-miR-142-3p | 8.779017 | MIMAT0000434 |
| hsa-miR-34a-3p | 8.748133 | MIMAT0004557 |
| hsa-miR-4695-5p | 8.440075 | MIMAT0019788 |
| hsa-miR-193a-5p | 7.9020715 | MIMAT0004614 |
| hsa-miR-374a-5p | 7.74524 | MIMAT0000727 |
| hsa-miR-182-5p | 7.1148996 | MIMAT0000259 |
| hsa-miR-203a | 6.915573 | MIMAT0000264 |
| hsa-miR-301b | 6.900287 | MIMAT0004958 |
| hsa-miR-450a-5p | 6.619105 | MIMAT0001545 |
| hsa-miR-6131 | 6.455001 | MIMAT0024615 |
| hsa-miR-887 | 6.105473 | MIMAT0004951 |
| hsa-miR-19b-1-5p | 6.0583606 | MIMAT0004491 |
| hsa-miR-590-5p | 5.9550056 | MIMAT0003258 |
| hsa-miR-200c-5p | 5.630886 | MIMAT0004657 |
| hsa-miR-214-5p | 5.396898 | MIMAT0004564 |
| hsa-miR-30e-3p | 5.378995 | MIMAT0000693 |
| hsa-miR-218-5p | 4.7786775 | MIMAT0000275 |
| hsa-miR-423-3p | 4.703984 | MIMAT0001340 |
| hsa-miR-455-5p | 4.035282 | MIMAT0003150 |
| hsa-miR-30c-5p | 3.5054796 | MIMAT0000244 |
| hsa-miR-1260b | 3.3699887 | MIMAT0015041 |
| hsa-miR-145-5p | 3.2018807 | MIMAT0000437 |
| hsa-miR-362-3p | 3.0494413 | MIMAT0004683 |
| hsa-miR-374b-5p | 3.0005975 | MIMAT0004955 |
| hsa-miR-30b-5p | 2.9336023 | MIMAT0000420 |
| hsa-miR-429 | 2.7092197 | MIMAT0001536 |
| hsa-miR-4428 | 2.6921449 | MIMAT0018943 |
| hsa-miR-196b-5p | 2.5407631 | MIMAT0001080 |
| hsa-miR-199a-5p | 2.5355365 | MIMAT0000231 |
| hsa-miR-199b-5p | 2.4879646 | MIMAT0000263 |
| hsa-miR-143-3p | 2.4430275 | MIMAT0000435 |
| hsa-miR-449a | 2.3427818 | MIMAT0001541 |
| hsa-miR-6717-5p | 2.3069673 | MIMAT0025846 |
| hsa-miR-301a-3p | 2.30314 | MIMAT0000688 |
| hsa-miR-424-5p | 2.190957 | MIMAT0001341 |
| hsa-miR-3653 | 2.1542947 | MIMAT0018073 |
| hsa-miR-335-5p | 2.1516027 | MIMAT0000765 |
| hsa-miR-125b-5p | 2.1353264 | MIMAT0000423 |
| hsa-miR-1305 | 2.1121273 | MIMAT0005893 |
| hsa-miR-365a-3p | 2.0961003 | MIMAT0000710 |
| hsa-miR-146b-5p | 2.0629325 | MIMAT0002809 |
| hsa-miR-21-5p | 2.0441828 | MIMAT0000076 |
| Downregulated miRNAs | ||
| hsa-miR-4668-5p | 103.51767 | MIMAT0019745 |
| hsa-miR-4254 | 14.588222 | MIMAT0016884 |
| hsa-miR-4701-5p | 13.288958 | MIMAT0019798 |
| hsa-miR-134 | 2.7737036 | MIMAT0000447 |
| hsa-miR-1207-5p | 2.6758146 | MIMAT0005871 |
| hsa-miR-4306 | 2.2878602 | MIMAT0016858 |
| hsa-miR-3162-3p | 2.2871423 | MIMAT0019213 |
| hsa-miR-4788 | 2.2722929 | MIMAT0019958 |
| hsa-miR-6088 | 2.1698172 | MIMAT0023713 |
| hsa-miR-5739 | 2.1607096 | MIMAT0023116 |
| hsa-miR-6165 | 2.133992 | MIMAT0024782 |
| hsa-miR-572 | 2.0804768 | MIMAT0003237 |
miRNAs: MicroRNAs; FC: Fold change.
In terms of the supervised hierarchical clustering analysis, the dendrograms showed satisfying segregation of the gene expression levels for samples from the two groups, based on the differentially expressed miRNAs [Figure 1]. The first branch in the miRNA heat maps was able to differentiate samples from the RIF group and the control group. This finding suggested a diverse miRNA expression profile for WOI endometrium between RIF patients and those who conceived after their first attempt of IVF/ICSI.
Figure 1.
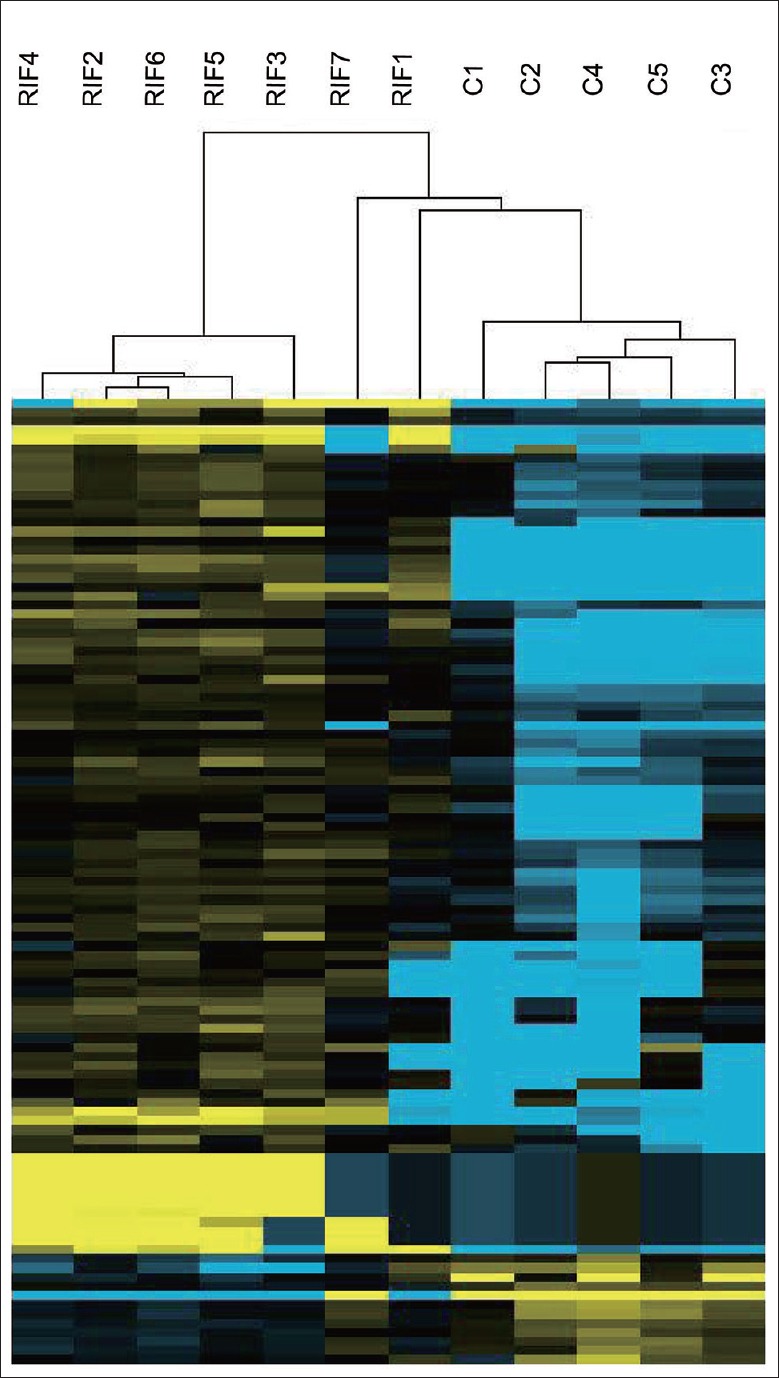
Dendrogram and hierarchical clustering. Expression data from all the differentially expressed miRNAs are analyzed. Each row presents one gene and each column represents an endometrial sample. Column RIF1, RIF2, RIF3, RIF4, RIF5, RIF6, and RIF7 are RIF samples and column C1, C2, C3, C4, and C5 are control samples. Up- and down-regulated miRNAs are, respectively, indicated by yellow and blue, and miRNAs that are lack of significant change are indicated by black. miRNA: MicroRNA; RIF: Repeated implantation failure.
Functional analysis of differentially expressed microRNAs
TAM analysis was used to gain an in-depth understanding of the biological functions of the differentially expressed miRNAs. According to the TAM analysis results, mir-30 family, human embryonic stem cell regulation, epithelial-mesenchymal transition, and miRNA tumor suppressors were the most relevant miRNA functional sets [Figure 2].
Figure 2.
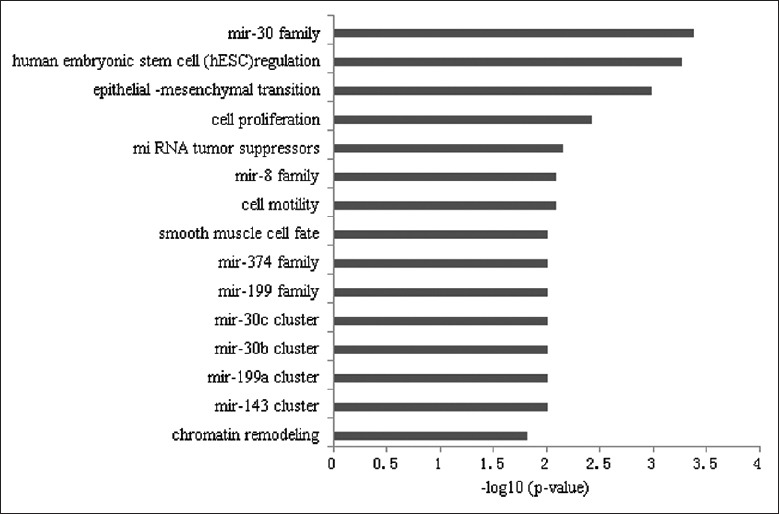
Results of the tool for annotations of microRNA analysis for the deregulated miRNAs between the RIF and control endometrial samples. Mir-30 family, human embryonic stem cell regulation and epithelial-mesenchymal transition were the top 3 relevant miRNA functional sets. miRNA: MicroRNA; RIF: Repeated implantation failure.
Construction of a regulatory network of differentially expressed microRNAs and mRNAs
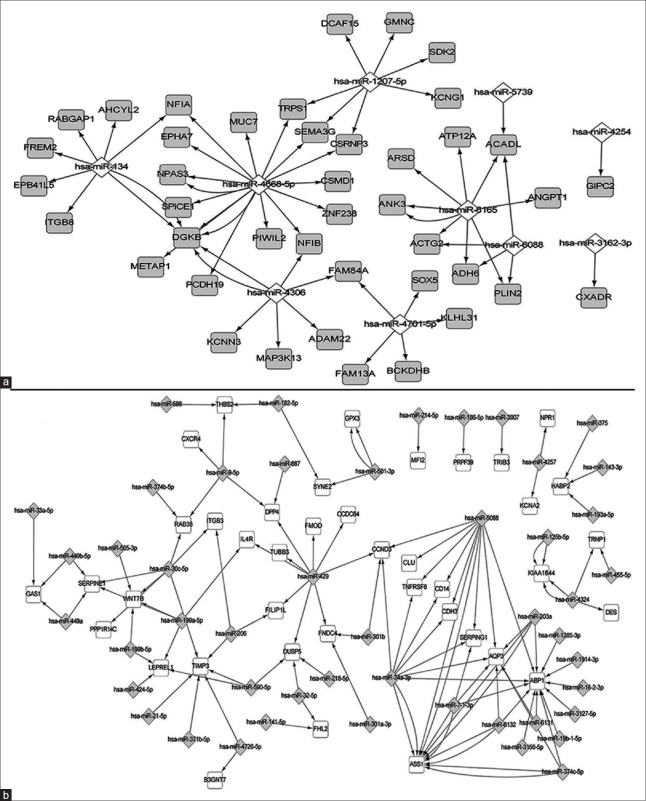
The relationships between the dysregulated miRNAs and mRNAs were predicted by network regulatory analysis software. A total of 176 interactions between miRNAs and mRNAs were found, of which 122 were for upregulated miRNAs and downregulated mRNAs and 54 were for downregulated miRNAs and upregulated mRNAs. The top core mRNA was ABP1, which was regulated by 13 miRNAs, followed by AQP3, ASS1, and TIMP3 (regulated by 6 miRNAs). The top core miRNA was has-miR-4668-5p, which regulated 14 mRNAs, followed by has-miR-429 and has-miR-5088 (which regulated 9 mRNAs) [Figure 3].
Figure 3.
The layout of the miRNA-mRNA regulatory network. The network consists of 54 regulations (a) between downregulated miRNAs to upregulated mRNAs and 122 regulations (b) between upregulated miRNAs to downregulated mRNAs. A diamond marks miRNA and a rectangle marks the mRNA. An edge represents a regulation from miRNA to one of its targets. The miRNAs and mRNAs are colored based on their dysregulation pattern. If the miRNAs (or mRNAs) are upregulated in the RIF group, the nodes are marked by gray, otherwise they are marked by white. miRNA: MicroRNA; RIF: Repeated implantation failure.
Validation of microRNA expression using quantitative reverse transcription polymerase chain reaction
To validate the differences in transcript levels found in the microarrays, a selected set of miRNAs was chosen for quantitative RT-PCR. New endometrial samples from the RIF group (n = 5; RIF8, RIF9, RIF10, RIF11, and RIF12) and control group (n = 5; C6, C7, C8, C9, and C10) were used for this validation.
Selection for validated miRNAs was done according to the following criteria: (i) miRNAs, the raw signal for each sample in the miRNA microarray analysis was >50 and was differentially up- or down-regulated in samples from the RIF group compared with the control group; and (ii) miRNAs that were in the core mRNA-miRNA network results. The RT-PCR results were in agreement with that of the microarray for all miRNAs: hsa-miR-374a-5p, hsa-miR-145-5p, hsa-miR-30b-5p, hsa-miR-196b-5p, hsa-miR-199a-5p, hsa-miR-199b-5p, hsa-miR-449a, hsa-miR-424-5p, hsa-miR-125b-5p, and hsa-miR-21-5p were elevated and hsa-miR-1207-5p, hsa-miR-4306, hsa-miR-572, hsa-miR-5739, hsa-miR-6088, hsa-miR-4668-5p, hsa-miR-429, and hsa-miR-5088 were reduced in the RIF group compared with the control group [Figure 4].
Figure 4.
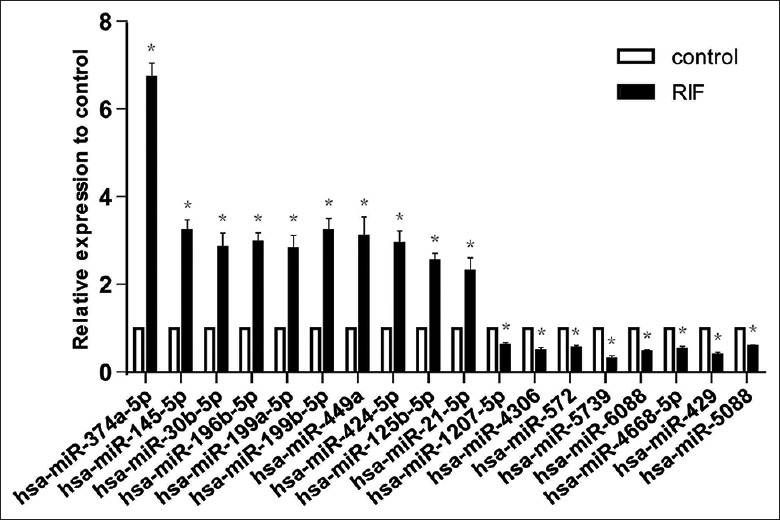
Validation of miRNAs by real-time PCR in new samples (RIF, n = 5; control, n = 5). Relative levels of the transcripts for the selected 18 miRNAs in the RIF group as compared to the control group are shown. The dysregulation pattern of all the selected miRNAs by real-time PCR is coincident with that by microarray. *P < 0.05, vs. control group. miRNA: MicroRNA; RIF: Repeated implantation failure; PCR: Polymerase chain reaction.
Discussion
Until now, an objective diagnosis of endometrial receptivity remained neglected, which limited the improvement of clinical IVF/ICSI success from the endometrial perspective. Therefore, we used a microarray technique to investigate the miRNA profile of women with RIF compared to women who conceived after their first attempt of embryo transfer. We found that 105 differentially expressed miRNAs could result in two distinct groups by hierarchical clustering: RIF endometrium and the control group endometrium.
Previous research using a miRNA microarray to study endometrial receptivity can be generally grouped into two categories: (i) to compare the dynamic genomic expression profiles of endometrium from the proliferative phase to the WOI in fertile women; and (ii) to investigate the differential genomic expression profiles between fertile and infertile women.
In the first category, 4 studies have been reported. Has-miR-30b, has-miR-30d, and has-miR-494 were considered to play important roles in regulating endometrial receptivity. Compared with the prereceptive endometrium, hsa-miR-30b and hsa-miR-30d were found to be significantly upregulated and hsa-miR-494 was found to be downregulated in receptive endometrium.[15] In our study, hsa-miR-30b was also found to be upregulated in the RIF group. It is indicated that the destroyed endometrial receptivity of RIF patients was related to miRNAs other than hsa-miR-30b.
For the second category, only one study by Revel et al.[16] was reported, which found 13 deregulated miRNAs (1 were upregulated and 12 were downregulated). Different microarray platforms contributed mostly to the coincidence of the numbers of dysregulated miRNAs between our results and results of Ariel Revel's study. The miRNA Array card we used contained 2006 mature human miRNAs while the card Revel's group used only contained 381 mature human miRNAs. However, we also obtained two shared deregulated miRNAs: hsa-miR-145 and has-miR-374, which were both upregulated in the RIF patients in our study. ERα, mucin1 and RTKN, which play important roles in the acquisition of endometrial receptivity, have been validated to be the target genes of has-miR-145. In Revel's study, they thought that upregulated hsa-miR-145 might destroy endometrial receptivity in RIF patients by reducing endometrial ERα and mucin1 expression, which was also validated by Western-blot as downregulated.[16] In our study, we also detected the expression of mucin1, ERα, and RTKN by RT-PCR in the WOI endometrium from the two groups. Unexpectedly, ERα and RTKN were found to be upregulated in our RIF group, while mucin1 presented with a similar expression levels in both of our groups. Such a result may due to our small sample size (i.e., bias) or by has-miR-145 impairing the endometrial receptivity via regulating the expression of other target genes. Hsa-miR-374, located on chromosome Xq13.2, has been previously shown to constitutively activate Wnt/b-catenin signaling,[20] which has been reported to participate in the implantation process in several studies.[21,22]
Since miRNAs act as the post-transcriptional regulators of mRNA, usually negatively, we created a regulatory network of differentially expressed mRNAs and miRNAs and found 176 regulated pairs. The top 3 core miRNAs were has-miR-4668-5p, has-miR-429, and has-miR-5088, of which has-miR-4668-5p was downregulated, while has-miR-429 and has-miR-5088 were upregulated in the RIF group. The targeted mRNAs of has-miR-429 and has-miR-5088, DPP4, SERPING1, and AQP3 were validated to be downregulated in the new RIF samples in our previous report. These results indicated that the endometrial receptivity of RIF patients may be impacted by the expression of these mRNAs, which were regulated by specific miRNAs. Hence, we should pay more attention to miRNAs in future studies, which may shed some light on potential treatment for RIF.
In conclusion, we performed miRNA microarray on the samples from the RIF and control groups. Differentially expressed miRNAs were found and analyzed for their role in the establishment of endometrial receptivity. We found that has-miR-145, hsa-miR-374, hsa-miR-4668-5p, hsa-miR-429, and hsa-miR-5088 may be relevant to the low endometrial receptivity of RIF patients. We hypothesize that an array including miRNAs may increase the specificity for diagnosing the endometrial receptivity of patients with RIF, and our report provides clues to this diagnostic tool.
Financial support and sponsorship
This work was supported by grants from the Peking University People's Hospital Research and Development Funds (No. RDU2011-04 and No. RDC2014-07).
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
The authors would like to thank CapitalBio Corp., (Beijing, China) for the microarray service and also would like to thank Dr. Li Jiang for language editing.
Supplementary information is linked to the online version of the paper on the Chinese Medical Journal website.
Footnotes
Edited by: Li-Shao Guo
References
- 1.Kupka MS, Ferraretti AP, de Mouzon J, Erb K, D’Hooghe T, Castilla JA, et al. Assisted reproductive technology in Europe 2010: Results generated from European registers by ESHREdagger. Hum Reprod. 2014;29:2099–113. doi: 10.1093/humrep/deu175. doi: 10.1093/humrep/deu175. [DOI] [PubMed] [Google Scholar]
- 2.Stern JE, Cedars MI, Jain T, Klein NA, Beaird CM, Grainger DA, et al. Assisted reproductive technology practice patterns and the impact of embryo transfer guidelines in the United States. Fertil Steril. 2007;88:275–82. doi: 10.1016/j.fertnstert.2006.09.016. doi: 10.1016/j.fertnstert.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 3.Koler M, Achache H, Tsafrir A, Smith Y, Revel A, Reich R. Disrupted gene pattern in patients with repeated in vitro fertilization (IVF) failure. Hum Reprod. 2009;24:2541–8. doi: 10.1093/humrep/dep193. doi: 10.1093/humrep/dep193. [DOI] [PubMed] [Google Scholar]
- 4.Dessolle L, Daraï E, Cornet D, Rouzier R, Coutant C, Mandelbaum J, et al. Determinants of pregnancy rate in the donor oocyte model: A multivariate analysis of 450 frozen-thawed embryo transfers. Hum Reprod. 2009;24:3082–9. doi: 10.1093/humrep/dep303. doi: 10.1093/humrep/dep303. [DOI] [PubMed] [Google Scholar]
- 5.Liu SM, Zhou YZ, Wang HB, Sun ZY, Zhen JR, Shen K, et al. Factors Associated with effectiveness of treatment and reproductive outcomes in patients with thin endometrium undergoing estrogen treatment. Chin Med J. 2015;128:3173–7. doi: 10.4103/0366-6999.170258. doi: 10.4103/0366-6999.170258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rashid NA, Lalitkumar S, Lalitkumar PG, Gemzell-Danielsson K. Endometrial receptivity and human embryo implantation. Am J Reprod Immunol. 2011;66(Suppl 1):23–30. doi: 10.1111/j.1600-0897.2011.01048.x. doi: 10.1111/j.1600-0897. [DOI] [PubMed] [Google Scholar]
- 7.Achache H, Revel A. Endometrial receptivity markers, the journey to successful embryo implantation. Hum Reprod Update. 2006;12:731–46. doi: 10.1093/humupd/dml004. doi: 10.1093/humupd/dml004. [DOI] [PubMed] [Google Scholar]
- 8.Herrler A, von Rango U, Beier HM. Embryo-maternal signalling: How the embryo starts talking to its mother to accomplish implantation. Reprod Biomed Online. 2003;6:244–56. doi: 10.1016/s1472-6483(10)61717-8. [DOI] [PubMed] [Google Scholar]
- 9.Altmäe S, Esteban FJ, Stavreus-Evers A, Simón C, Giudice L, Lessey BA, et al. Guidelines for the design, analysis and interpretation of ’ OMICS’ data: Focus on human endometrium. Hum Reprod Update. 2014;20:12–28. doi: 10.1093/humupd/dmt048. doi: 10.1093/humupd/dmt048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambros V. MicroRNAs: Tiny regulators with great potential. Cell. 2001;107:823–6. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 11.Galliano D, Pellicer A. MicroRNA and implantation. Fertil Steril. 2014;101:1531–44. doi: 10.1016/j.fertnstert.2014.04.023. doi: 10.1016/j.fertnstert. [DOI] [PubMed] [Google Scholar]
- 12.Yang C, Zheng SD, Wu HJ, Chen SJ. Regulatory mechanisms of the molecular pathways in fibrosis induced by microRNAs. Chin Med J. 2016;129:2365–72. doi: 10.4103/0366-6999.190677. doi: 10.4103/0366-6999.190677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu W, Niu Z, Li Q, Pang RT, Chiu PC, Yeung WS. MicroRNA and embryo implantation. Am J Reprod Immunol. 2016;75:263–71. doi: 10.1111/aji.12470. doi: 10.1111/aji.12470. [DOI] [PubMed] [Google Scholar]
- 14.Kuokkanen S, Chen B, Ojalvo L, Benard L, Santoro N, Pollard JW. Genomic profiling of microRNAs and messenger RNAs reveals hormonal regulation in microRNA expression in human endometrium. Biol Reprod. 2010;82:791–801. doi: 10.1095/biolreprod.109.081059. doi: 10.1095/biolreprod.109.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altmäe S, Martinez-Conejero JA, Esteban FJ, Ruiz-Alonso M, Stavreus-Evers A, Horcajadas JA, et al. MicroRNAs miR-30b, miR-30d, and miR-494 regulate human endometrial receptivity. Reprod Sci. 2013;20:308–17. doi: 10.1177/1933719112453507. doi: 10.1177/1933719112453507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Revel A, Achache H, Stevens J, Smith Y, Reich R. MicroRNAs are associated with human embryo implantation defects. Hum Reprod. 2011;26:2830–40. doi: 10.1093/humrep/der255. doi: 10.1093/humrep/der255. [DOI] [PubMed] [Google Scholar]
- 17.Coughlan C, Ledger W, Wang Q, Liu F, Demirol A, Gurgan T, et al. Recurrent implantation failure: Definition and management. Reprod Biomed Online. 2014;28:14–38. doi: 10.1016/j.rbmo.2013.08.011. doi: 10.1016/j.rbmo.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Kdous M, Merdassi G, Zhioua F, Elloumi H, Kacem K, Zhioua A. Basal follicle stimulating hormone level correlated to age is a good prognostic criterion for the outcome of intracytoplasmic sperm microinjection. Tunis Med. 2016;94:181–5. [PubMed] [Google Scholar]
- 19.Lu M, Shi B, Wang J, Cao Q, Cui Q. TAM: A method for enrichment and depletion analysis of a microRNA category in a list of microRNAs. BMC Bioinformatics. 2010;11:419. doi: 10.1186/1471-2105-11-419. doi: 10.1186/1471-2105-11-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Xia H, Zhuang Z, Miao L, Chen X, Cai H. Axl-altered microRNAs regulate tumorigenicity and gefitinib resistance in lung cancer. Cell Death Dis. 2014;5:e1227. doi: 10.1038/cddis.2014.186. doi: 10.1038/cddis.2014.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonderegger S, Pollheimer J, Knöfler M. Wnt signalling in implantation, decidualisation and placental differentiation – Review. Placenta. 2010;31:839–47. doi: 10.1016/j.placenta.2010.07.011. doi: 10.1016/j.placenta.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Horst PH, Wang Y, van der Zee M, Burger CW, Blok LJ. Interaction between sex hormones and WNT/ß-catenin signal transduction in endometrial physiology and disease. Mol Cell Endocrinol. 2012;358:176–84. doi: 10.1016/j.mce.2011.06.010. doi: 10.1016/j.mce.2011.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The micrograph of H and E dying for the sample of RIF10. The micrographs for the other 21 samples were similar to this, and all the reports were: normal mid-secretory endometrium. (a) Extreme glandular coiling secretory glands set within a spindled edematous stroma. Luminal secretion is most prominent (H and E, original magnification ×100). (b) The coiled spiral arteries are seen within an edematous stroma. Perivascular predecidual reaction has not occurred (H and E, original magnification ×200). RIF: Repeated implantation failure.