Abstract
Background:
DNA hydroxymethylation refers to a chemical modification process in which 5-methylcytosine (5mC) is catalyzed to 5- hydroxymethylcytosine (5hmC) by ten-eleven translocation (TET) family proteins. Recent studies have revealed that aberrant TETs expression or 5hmC level may play important roles in the occurrence and development of various pathological and physiological processes including cancer and aging. This study aimed to explore the relation between aberrant DNA hydroxymethylation with skin photoaging and to investigate the levels of TETs, 5mC, and 5hmC expression 24 h after 40 mJ/cm2 and 80 mJ/cm2 doses of ultraviolet B (UVB) irradiation to HaCaT cells.
Methods:
To explore whether aberrant DNA hydroxymethylation is also related to skin photoaging, 40 mJ/cm2 and 80 mJ/cm2 doses of UVB were chosen to treat keratinocytes (HaCaT cells). After 24 h of UVB irradiation, 5mC and 5hmC levels were determined by immunohistochemistry (IHC) and immunofluorescence (IF), and at the same time, the expression levels of matrix metalloproteinase 1 (MMP-1) and TETs were assessed by reverse transcription-polymerase chain reaction or Western blot analysis.
Results:
After 40 mJ/cm2 and 80 mJ/cm2 doses of UVB exposure, both IHC and IF results showed that 5hmC levels increased significantly, while the 5mC levels did not exhibit significant changes in HaCaT cells, compared with HaCat cells without UVB exposure. Moreover, compared with HaCat cells without UVB exposure, the levels of TET1, TET2, and TET3 mRNA and protein expression were significantly upregulated (mRNA: P = 0.0022 and 0.0043 for TET1; all P < 0.0001 for TET2; all P = 0.0006 for TET3; protein: P = 0.0012 and 0.0006 for TET1; all P = 0.0022 for TET2; and all P = 0.0002 for TET3), and the levels of MMP-1 mRNA expression increased dose dependently in 40 mJ/cm2 and 80 mJ/cm2 UVB-irradiated groups.
Conclusion:
UVB radiation could cause increased 5hmC and TET expression, which might become a novel biomarker in UVB-related skin aging.
Keywords: 5-hydroxymethylcytosine, 5-methylcytosine, DNA Hydroxymethylation, Ten-eleven Translocation, Ultraviolet B
Introduction
Both intrinsic and extrinsic factors could lead to skin aging. Extrinsic skin aging, also named as skin photoaging, is mainly caused by sun exposure, which could eventually result in photocarcinogenesis including malignant melanoma. Ultraviolet B (UVB), covering a wavelength range of 290–320 nm, makes up only 1–10% of solar UV radiation. Nevertheless, UVB has been proved to be the biggest factor to induce skin disorders partly through affecting the stability of DNA.[1,2]
In 2009, Tahiliani et al.[3] found that ten-eleven translocation (TET) family proteins could catalyze the conversion of 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC), which made a huge contribution to the understanding of active DNA demethylation. Human TET proteins are composed of TET1, TET2, and TET3, which belong to the α-ketoglutarate-dependent and Fe (II)-dependent dioxygenase superfamily.[3,4] Recent studies have found that TET gene mutation and aberrant TET expression and 5hmC levels might play vital roles in various pathological and physiological processes, such as hematological malignancies,[5,6] melanoma,[7] neurodegeneration,[8] and aging.[9] Nevertheless, whether aberrant DNA hydroxymethylation is also involved in the occurrence and development of skin photoaging is not confirmed yet.
The molecular mechanism of skin photoaging still remains to be studied. In the current study, to explore the relation between aberrant DNA hydroxymethylation and skin photoaging, we chose 40 mJ/cm2 and 80 mJ/cm2 doses of UVB irradiation to treat HaCaT cells and investigated the levels of TETs, 5mC, and 5hmC expression, 24 h after irradiation.
Methods
HaCaT cell culture and experiment groups
HaCaT cells, a spontaneously immortalized human keratinocyte cell line, were brought from Xiangya Cell Bank, Central South University, China. HaCaT cells were grown in Roswell Park Memorial Institute 1640 medium (GIBCO, USA), supplemented with 10% fetal bovine serum (Biological Industries, Israel), 100 U/ml penicillin, and 0.1 mg/ml streptomycin (GIBCO). They were cultured at 37°C in a humidified atmosphere of 5% CO2. According to our aim of this experiment, we divided our cells into three groups, one group treated with 40 mJ/cm2 of UVB irradiation, one with 80 mJ/cm2, and the rest one, as a control group, with the same culture except UVB irradiation.
Ultraviolet B irradiation
HaCaT cells were plated on culture dishes. When cells reached 70–80% confluence, the medium was discarded, and the cells were covered with a thin layer of phosphate-buffered saline (PBS; GIBCO) after being washed with PBS three times and exposed to different doses of UVB (280–320 nm) using Ultraviolet Irradiator SS-03B (Shanghai Sigma High-tech Co., Ltd., China). And then, the cells were covered with new complete medium for further trials.
RNA extraction and reverse transcription-polymerase chain reaction
Total RNA was isolated using E.Z.N.A.™ Total RNA Kit II (Omega, USA) according to the manufacturer's instructions. Total RNA was converted into first-strand cDNA using the ReverTra Ace qPCR-RT Master Mix (Toyobo Co., Ltd., Osaka, Japan) according to the manufacturer's guidelines. Reverse transcription-polymerase chain reaction (RT-PCR) was performed with THUNDERBIRD SYBR qPCR Mix (Toyobo Co., Ltd.,). The β-actin, an endogenous housekeeping gene, was used to normalize the results. RT-PCR reactions were performed in 96-well plates in triplicate, under the following thermo-cycling conditions: 60 s at 95°C, 40 cycles of 15 s at 95°C, 20 s at 55°C, and 30 s at 72°C. The primers for RT-PCR are listed in Table 1. The point at which the PCR product was first detected above a fixed threshold (the cycle threshold [Ct]) was determined for each sample. Changes in the expression of target genes were calculated using 2−ΔΔCt, where ΔΔCt = (Cttarget − Ctβ-actin) sample − (Cttarget − Ctβ-actin) control, taking the mean of Ct in the HaCat cells without UVB exposure as the control.
Table 1.
The primer sequences for quantitative real-time PCR
| Genes | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| β-actin | 5’-TGGCACCCAGCACAATGAA-3’ | 5’-CTAAGTCATAGTCCGCCTAGAAGCA-3’ |
| TET1 | 5’-ATACCACTTTGCTACCGACTT-3’ | 5’-ATGGCTGTTCTTTCTGTTCTT-3’ |
| TET2 | 5’-TCAGCAGCAGCCAATAGGACA-3’ | 5’-TGGGAGGTGATGGTATCAGGAAT-3’ |
| TET3 | 5’-CCTCCTTCTCCTTTGGTTGTT-3’ | 5’-TTCTTCCTCTTTGGGATTGTC-3’ |
| MMP-1 | 5’-ATGCTGAAATGAAGGTG-3’ | 5’-CTGCTTGACCCTCAGAGACC-3’ |
TET1: Ten-eleven translocation 1; TET2: Ten-eleven translocation 2; TET3: Ten-eleven translocation 3; MMP-1: Matrix metalloproteinase 1; PCR: Polymerase chain reaction.
Western blot analysis
Cells were lysed on ice with lysis buffer containing 1 mmol/L phenylmethanesulfonyl fluoride (PMSF; KeyGen Biotech Co., Ltd, Nanjing, China) for 0.5 h. The protein concentration was determined with a BCA protein assay kit (KeyGen Biotech Co., Ltd.,). Cell lysates (16 μg protein) were subjected to 6% sodium dodecyl sulfate polyacrylamide gel electrophoresis, and then transferred onto a polyvinylidene-fluoride (PVDF) membrane. The blot was blocked with 5% skim milk in tris-buffered saline tween (TBST) at room temperature for 1 h, and then incubated with the primary rabbit-polyclonal antibodies against TET1 or TET2 or TET3 (1:500 dilution; Abcam, USA), and glyceraldehyde-phosphate dehydrogenase (1:3000 dilution; CWBIO, China) at 4°C overnight. Then, the blot was incubated with secondary anti-rabbit antibody (1:6000 dilution; CWBIO) at room temperature for 1 h. After PVDF membrane being washed with TBST three times, protein bands were developed with the ECL kit (KeyGen Biotech Co., Ltd.,). Protein densities were analyzed with the ImageJ software (NIH, Bethesda, Maryland, USA).
Immunohistochemistry
HaCaT cells were seeded onto coverslips when cells reached the confluence of approximately 60–70% in 24-well plate. Twenty-four hours after UVB irradiation, cells on coverslips were blocked using 5% bovine serum albumin (Sigma, USA) for 45 min and incubated with primary mouse-anti-5mC (1:500 dilution; Auragene Bioscience Corporation, China) and rabbit-anti-5hmC (1:500 dilution; Auragene Bioscience Corporation) for 3 h at 37°C. Then, cells were incubated with secondary anti-mouse/rabbit antibody (1:1000 dilution; Auragene Bioscience Corporation) separately. Moreover, streptavidin/peroxidase (Beijing OriGene Technology Co., Ltd., China) was needed to incubate cells 30 min after washing with PBS for three times. Images were performed by fluorescence microscope (Nikon Corporation, Japan) after the cells were incubated in 3,3’-diaminobenzidine (Beijing OriGene Technology Co., Ltd.,) for 5 min and stained in hemalum, and all images were measured by Image Pro-Plus 6.0 software (Media Cybernetics, Silver Spring, MD, USA).
Immunofluorescence
HaCaT cells were treated and incubated with primary antibodies almost the same with immunohistochemistry (IHC). As secondary antibodies, we used cy3-goat-anti-rabbit IgG diluted at 1:400 (Auragene Bioscience Corporation) and cy3-goat-anti-mouse IgG diluted at 1:400 (Auragene Bioscience Corporation). For nuclear staining, we used 4’,6-diamidin-2-phenylindol (Biosharp, USA). Images were performed by fluorescence microscope (Nikon Corporation) and were measured by Image Pro-Plus 6.0 software (Media Cybernetics).
Statistical analysis
Statistical analysis was performed with SPSS version 19.0 (SPSS Inc., Chicago, IL, USA). All data were expressed as mean ± standard derivation (SD). Statistical significance was determined by one-way analysis of variance (ANOVA), followed by Dunnett's multiple comparison test, and a P < 0.05 was considered statistically significant.
Results
The levels of 5-methylcytosine and 5-hydroxymethylcytosine in ultraviolet B-irradiated HaCaT cells
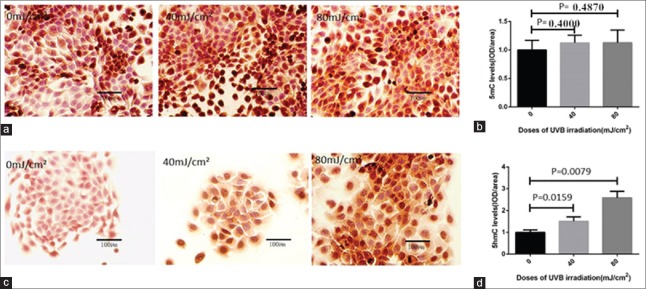
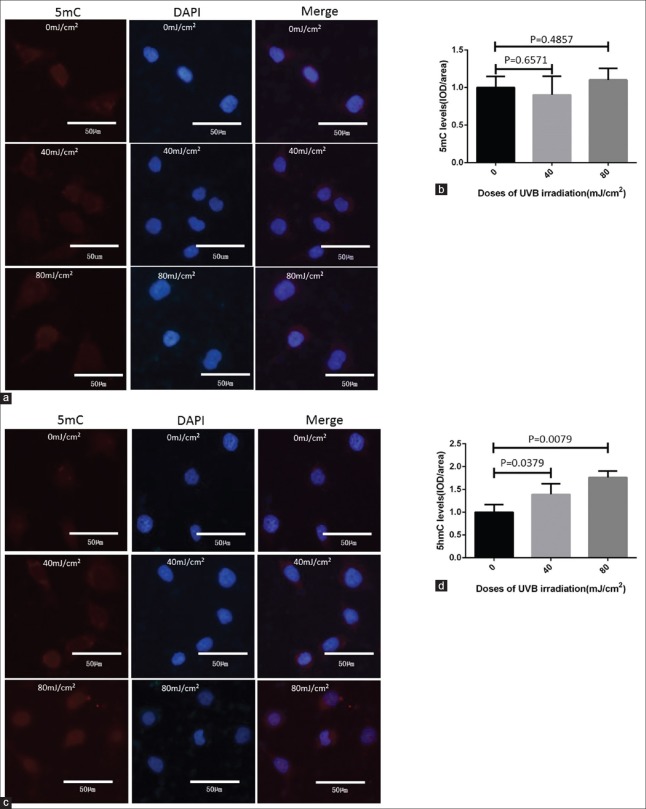
To explore the effect of UVB radiation on HaCaT cells, the 40 mJ/cm2 and 80 mJ/cm2 doses of UVB were chosen to treat HaCaT cells. Initially, to identify the DNA hydroxymethylation levels of UVB-irradiated HaCaT cells, we used IHC and immunofluorescence (IF) to determine whether the levels of 5mC and 5hmC changed after UVB irradiation. The results of IHC showed that compared to HaCat cells without UVB exposure, 5hmC levels significantly increased by 1.5- and 2.6-folds (P = 0.0159 and 0.0079, respectively), while 5mC levels did not exhibit significant change (P = 0.4000 and 0.4870, respectively) in 40 mJ/cm2 and 80 mJ/cm2 doses of UVB-irradiated HaCat cells [Figure 1]. Interestingly, IF examination showed almost the same results of IHC. IF showed 5hmC levels increased by 1.4- and 1.8-folds (P = 0.0379 and 0.0079, respectively) after 40 mJ/cm2 and 80 mJ/cm2 UVB irradiation, but 5mC levels did not show significant change [P = 0.4857 and 0.6571, respectively; Figure 2].
Figure 1.
IHC results of 5mC and 5hmC levels in HaCaT cells with and without UVB irradiation. IHC was used to detect 5mC and 5hmC levels in HaCaT cells treated by 40 mJ/cm2 and 80 mJ/cm2 doses of UVB. (a and b) IHC results showed that 5mC levels in UVB-irradiated HaCaT cells treated by 40 mJ/cm2 and 80 mJ/cm2 doses did not appear to have significant changes compared to the HaCat cells without UVB exposure. (c and d) IHC results showed that 5hmC levels increased significantly in both 40 mJ/cm2 and 80 mJ/cm2 doses of UVB-irradiated HaCaT cells compared to the HaCat cells without UVB exposure. The data were shown as mean ± standard deviation. IHC: Immunohistochemistry; UVB: Ultraviolet B; 5mC: 5-methylcytosine; 5hmC: 5-hydroxymethylcytosine.
Figure 2.
IF results of 5mC and 5hmC levels in HaCaT cells with and without UVB irradiation. (a and b) Compared to the HaCat cells without UVB exposure, the 5mC levels did not change significantly in treated group. (c and d) The 5hmC levels increased significantly in UVB-treated group compared to the HaCat cells without UVB exposure. The data were shown as mean ± standard deviation. DAPI: 4’,6-diamidino-2-phenylindole; IF: Immunofluorescence; UVB: Ultraviolet B; 5mC: 5-methylcytosine; 5hmC: 5-hydroxymethylcytosine.
Ultraviolet B irradiation upregulated ten-eleven translocations and matrix metalloproteinase-1 expression
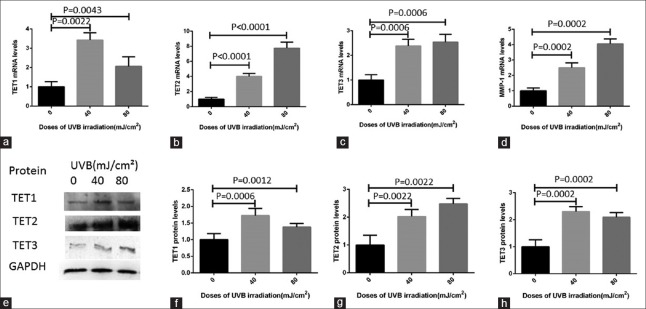
Then, to further explore the molecular mechanism of UVB-induced increase of 5hmC levels, we examined whether UVB exposure of HaCaT cells could stimulate the expression levels of both TET mRNA and protein. The results of RT-PCR revealed that compared to HaCat cells without UVB exposure, the mRNA levels of TET1, TET2, and TET3 increased significantly after being exposed to 40 mJ/cm2 and 80 mJ/cm2 doses of UVB irradiation for 24 h [Figure 3a−3c]. The TET1 mRNA levels increased by 3.4- and 2.1-folds after 40 mJ/cm2 and 80 mJ/cm2 UVB irradiation, respectively (P = 0.0022 and 0.0043, respectively). TET2 mRNA levels were upregulated by 4.0- and 7.8-folds after being treated with 40 mJ/cm2 and 80 mJ/cm2 doses of UVB, respectively (all P < 0.0001). Moreover, TET3 mRNA levels increased by 2.4- and 2.5-folds in 40 mJ/cm2 and 80 mJ/cm2 UVB-irradiated cells in contrast to the HaCat cells without UVB exposure (all P = 0.0006). Besides, in this study, the results of RT-PCR showed that compared to HaCat cells without UVB exposure, matrix metalloproteinase 1 (MMP-1) mRNA levels increased by 2.4- and 4.1-folds at the 40 mJ/cm2 and 80 mJ/cm2 doses of UVB irradiation in HaCaT cells, respectively [all P = 0.0002; Figure 3d].
Figure 3.
The expression levels of TET1, TET2, TET3, and MMP-1 in HaCaT cells with and without UVB irradiation. (a–d) Compared to the HaCat cells without UVB exposure, TET1, TET2, TET3, and MMP-1 mRNA levels upregulated after UVB irradiation, respectively. (e) Western blot analyses of TET1, TET2, TET3, and GAPDH proteins. (f–h) Bar graphs showing quantitative evaluation of the relative gray values between target bands related to GAPDH. All assays were triplicated and demonstrated similar results. The data were shown as mean ± standard deviation. TET1: Ten-eleven translocation 1; TET2: Ten-eleven translocation 2; TET3: Ten-eleven translocation 3; MMP-1: Matrix metalloproteinase 1; GAPDH: Glyceraldehyde-phosphate dehydrogenase.
Interestingly, the results of Western blot analysis showed that the expression levels of TET1, TET2, and TET3 protein were also significantly upregulated compared to the HaCat cells without UVB exposure, which were almost the same as the changes of mRNA levels [Figure 3e−3h]. TET1 proteins were upregulated by 1.7- and 1.4-folds in 40 mJ/cm2 and 80 mJ/cm2 UVB-irradiated cells (P = 0.0006 and 0.0012, respectively). TET2 protein levels were 2.0- and 2.4-folds of the original levels after 40 mJ/cm2 and 80 mJ/cm2 doses of UVB treatment compared to the HaCat cells without UVB exposure (all P = 0.0022). And, TET3 protein expression was increased by 2.3- and 2.1-folds in the above dose of UVB irradiation cells in contrast to the HaCat cells without UVB exposure (all P = 0.0002).
Discussion
The role of modified cytosines, especially DNA methylation of cytosine residues in promoter regions, has been well studied and is associated with gene expression. There are increasing evidences that the imbalance of DNA methylation/demethylation plays a vital role in UVB-associated diseases in skin.[10] Nandakumar et al.[11] found that UVB-induced DNA hypermethylation and enhanced DNA methyltransferase (DNMT, enzymes that could catalyze cytosines to 5mC) activity occurred in UVB-exposed skin and UVB-induced skin tumors. In patients with systemic lupus erythematosus (SLE), UVB exposure could inhibit DNA methylation and DNMT1 expression, which was subsequently related to the epigenetic mechanism of SLE.[12] However, another research showed that the changes of DNA methylation could not be detected in human keratinocytes after UVB irradiation.[13] Nevertheless, whether UVB results in aberrant DNA hydroxymethylation in skin and the involved mechanism remains to be researched so far.
To discover the relationship between UVB and DNA hydroxymethylation in skin keratinocytes, we chose 40 mJ/cm2 and 80 mJ/cm2 doses of UVB to treat HaCaT cells. Upregulated expression of MMP-1 was an important biomarker in skin photoaging according to a recent research.[14] In our study, we found that MMP-1 expression was significantly increased at 40 mJ/cm2 and 80 mJ/cm2 doses of UVB irradiation, which suggested a photoaging state of the irradiated cells.
For a long time, 5mC was considered as a static and stable epigenetic marker of DNA. However, it was not until 2009 that two research groups reported that 5hmC was derived from 5mC, and this phenomenon was proved to be ubiquitous in mammalian DNA, indicating that DNA methylation is not static but dynamic and that 5hmC may serve as an intermediate of active DNA demethylation.[15] Moreover, recent studies revealed that the aberration of 5hmC levels occurred in various diseases. To explore whether UVB induces aberrant DNA hydroxymethylation, we initially studied the 5mC and 5hmC levels in UVB-irradiated cells through IHC and IF. Our results showed that 5hmC levels increased significantly after UVB irradiation, while 5mC levels did not exhibit significant changes, indicating that increased 5hmC in HaCaT cells might become a novel epigenetic marker in UVB-related skin aging.
TET family proteins, consisting of TET1, TET2, and TET3, have been known as the most important regulator for the formation of 5hmC.[16] To further explore the molecular mechanism of UVB-induced increase of 5hmC levels, we examined TET1, TET2, and TET3 levels in both mRNA and protein expression by RT-PCR and Western blot analysis. In this study, we found that the expression levels of TET1, TET2, and TET3 were significantly upregulated in UVB-irradiated HaCaT cells, compared to the HaCat cells without UVB exposure. Moreover, the expression levels of TET2 increased most significantly among the three members. These results indicated that the upregulation of TET family proteins might be the potential molecular mechanism for the increase of 5hmC levels after UVB irradiation. And in the following study, we would explore the changes of 5hmC under UVB irradiation in TET-inhibited cells or TETs knockout animal models to further demonstrate the role of TET1-3 and 5hmC in skin photoaging.
Besides, it is worth mentioning that our findings showed that 5mC did not display significant postirradiation changes, which conformed to the research of Lahtz et al.[13] Although the expression levels of TET proteins were upregulated in UVB-irradiated HaCaT cells, the genome-wide DNA methylation levels did not change significantly, indicating that there were many factors regulating 5mC levels including DNMT in UVB-induced skin aging.
In summary, our current findings suggested that the 5hmC levels increased significantly while 5mC did not show detectable changes in UVB-irradiated HaCaT cells, indicating that increased 5hmC levels might become a novel biomarker in UVB-related skin aging. Moreover, TETs, especially TET2 expression, were significantly upregulated in both mRNA and protein levels after UVB irradiation, providing a potential mechanism underlying the 5hmC reduction. Nevertheless, the mechanism of how the abnormal DNA hydroxymethylation affects skin photoaging still requires exploration. Further understanding of the molecular mechanism in skin photoaging could help us to treat UVB-related diseases more effectively, including skin photoaging.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Xin Chen
References
- 1.Kammeyer A, Luiten RM. Oxidation events and skin aging. Ageing Res Rev. 2015;21:16–29. doi: 10.1016/j.arr.2015.01.001. doi: 10.1016/j.arr.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Rinnerthaler M, Bischof J, Streubel MK, Trost A, Richter K. Oxidative stress in aging human skin. Biomolecules. 2015;5:545–89. doi: 10.3390/biom5020545. doi: 10.3390/biom5020545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito S, D’ Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–33. doi: 10.1038/nature09303. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meldi KM, Figueroa ME. Cytosine modifications in myeloid malignancies. Pharmacol Ther. 2015;152:42–53. doi: 10.1016/j.pharmthera.2015.05.002. doi: 10.1016/j.pharmthera.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Pan F, Weeks O, Yang FC, Xu M. The TET2 interactors and their links to hematological malignancies. IUBMB Life. 2015;67:438–45. doi: 10.1002/iub.1389. doi: 10.1002/iub.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–46. doi: 10.1016/j.cell.2012.07.033. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherwani SI, Khan HA. Role of 5-hydroxymethylcytosine in neurodegeneration. Gene. 2015;570:17–24. doi: 10.1016/j.gene.2015.06.052. doi: 10.1016/j.gene.2015.06.052. [DOI] [PubMed] [Google Scholar]
- 9.Zampieri M, Ciccarone F, Calabrese R, Franceschi C, Bürkle A, Caiafa P. Reconfiguration of DNA methylation in aging. Mech Ageing Dev. 2015;151:60–70. doi: 10.1016/j.mad.2015.02.002. doi: 10.1016/j.mad.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Xu QF, Zheng Y, Chen J, Xu XY, Gong ZJ, Huang YF, et al. Ultraviolet A Enhances Cathepsin L Expression and Activity via JNK Pathway in Human Dermal Fibroblasts. Chin Med J. 2016;129:2853–2860. doi: 10.4103/0366-6999.194654. doi: 10.4103/0366-6999.194654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nandakumar V, Vaid M, Tollefsbol TO, Katiyar SK. Aberrant DNA hypermethylation patterns lead to transcriptional silencing of tumor suppressor genes in UVB-exposed skin and UVB-induced skin tumors of mice. Carcinogenesis. 2011;32:597–604. doi: 10.1093/carcin/bgq282. doi: 10.1093/carcin/bgq282. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Zhu X, Li F, Yang B, Liang J, Qin H, Xu J. Effects of ultraviolet B exposure on DNA methylation in patients with systemic lupus erythematosus. Exp Ther Med. 2013;5:1219–25. doi: 10.3892/etm.2013.960. doi: 10.3892/etm.2013.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lahtz C, Kim SI, Bates SE, Li AX, Wu X, Pfeifer GP. UVB irradiation does not directly induce detectable changes of DNA methylation in human keratinocytes. F1000Res. 2013;2:45. doi: 10.12688/f1000research.2-45.v1. doi: 10.12688/f1000research.2-45.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta A, Kaur CD, Jangdey M, Saraf S. Matrix metalloproteinase enzymes and their naturally derived inhibitors: Novel targets in photocarcinoma therapy. Ageing Res Rev. 2014;13:65–74. doi: 10.1016/j.arr.2013.12.001. doi: 10.1016/j.arr.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Tang J, Lai M, Zhang H. 5-Hydroxymethylcytosine and disease. Mutat Res Rev Mutat Res. 2014;762:167–75. doi: 10.1016/j.mrrev.2014.09.003. doi: 10.1016/j.mrrev.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Kroeze LI, van der Reijden BA, Jansen JH. 5-Hydroxymethylcytosine: An epigenetic mark frequently deregulated in cancer. Biochim Biophys Acta. 2015;1855:144–54. doi: 10.1016/j.bbcan.2015.01.001. doi: 10.1016/j.bbcan.2015.01.001. [DOI] [PubMed] [Google Scholar]