Abstract
Two ubiquitously expressed isoforms of c-Jun N-terminal protein kinase (JNK), JNK1 and JNK2, have shared functions and different functions. However, the molecular mechanism is unknown. Here we report that JNK1, but not JNK2, is essential for tumor necrosis factor alpha (TNF-α)-induced c-Jun kinase activation, c-Jun expression, and apoptosis. Using mouse fibroblasts deficient in either Jnk1 or Jnk2, we found that JNK1 was activated by TNF-α, whereas JNK2 activation was negligible. In addition, JNK2 interfered with JNK1 activation via its “futile” phosphorylation by upstream kinases. Consequently, expression and activation of c-Jun, which depends on JNK activity, were impaired in Jnk1 null cells but enhanced in Jnk2 null cells. TNF-α-induced apoptosis was also suppressed in Jnk1 null fibroblasts but increased in Jnk2 null cells. Thus, our results provide a molecular mechanism underlying the different biological functions of JNK isoforms.
c-Jun N-terminal protein kinase (JNK) (also known as stress-activated protein kinase [SAPK]) (8) is a subfamily of the mitogen-activated protein kinase (MAPK) superfamily. JNK has two ubiquitously expressed isoforms, JNK1 and JNK2, and a tissue-specific isoform, JNK3, all of which have two different splicing forms (p54 and p46) (1, 3, 16). JNK is activated by sequential protein phosphorylation through a MAP kinase module (i.e., mitogen-activated protein kinase kinase kinase [MAP3K] → MAP2K → MAPK [12, 16]) in response to a variety of extracellular stimuli, including tumor necrosis factor alpha (TNF-α) and UV. Two MAP2Ks (JNKK1/MKK4/SEK1 and JNKK2/MKK7) for JNK have been identified (5, 17, 18, 21). Several MAP3Ks, such as MEKK1, ASK1, MLK, TAK1, and TPL-2, have been reported to act as MAP3Ks for JNK (16). Activation of JNK is also regulated by scaffold proteins, such as JIP, β-arrestin, and JSAP1, protein phosphatases, and NF-κB-mediated inhibition (13, 16, 19, 24, 25).
JNK has been implicated in regulation of many cellular activities from gene expression to programmed cell death (apoptosis) (1, 3, 16). JNK regulates the activity of several transcription factors, including c-Jun, ATF-2, Elk-1, p53, and c-Myc, as well as other factors, such as members of the Bcl-2 family (12, 30). Of the transcription factors, c-Jun autoregulates its expression via binding to its own promoter as a c-Jun/ATF-2 dimer (12). Phosphorylation of c-Jun at Ser63 and Ser73 by JNK increases the transcriptional activity of c-Jun (12), leading to increased expression of c-Jun (12).
The evidence showing that JNK plays a critical role in both cell survival and apoptosis is overwhelming. In the absence of NF-κB activation, prolonged JNK activation promotes TNF-α-induced apoptosis (24, 25), probably via its contribution to the proteolysis of the proapoptotic molecule BID (4). JNK activation is also required for UV-induced apoptosis (27). However, there is evidence that JNK also contributes to cell survival. Genetic evidence reveals that JNK1 and JNK2 are involved in survival of neuronal cells in mouse hindbrain and forebrain regions during development (14, 20). Recently, it has been shown that JNK activation is required for interleukin 3 (IL-3)-mediated survival via phosphorylation and inactivation of the proapoptotic Bcl-2 family protein BAD (30). Thus, the function of JNK in apoptosis depends on the cell type, nature of the apoptotic stimulus, duration of its activation, and activity of other signaling pathways (16).
Different JNK isoforms have shared functions and different functions. JNK1 and JNK2 have been considered redundant isoforms, both of which contribute to JNK activity (3, 8, 11). This assumption is reinforced by the finding that while mouse embryos deficient in Jnk1 or Jnk2 survive, genetic disruption of both Jnk1 and Jnk2 alleles in mouse embryos is lethal (14, 20). Recent studies suggest that JNK1 and JNK2 also have different functions. For example, activation of CD8+ T cells is impaired in Jnk1−/− mice but enhanced in Jnk2−/− mice (2). Skin tumorigenesis is enhanced in Jnk1−/− mice but suppressed in Jnk2−/− mice (23). Genetic disruption of the Jnk1 allele in mice is sufficient to suppress obesity (9). However, the molecular mechanism underlying the differences between the JNK1 and JNK2 isoforms is unknown. Here we show that JNK1, but not JNK2, is activated by TNF-α and UV and only JNK1 is required for c-Jun expression and apoptosis. Thus, our results provide a molecular mechanism underlying the different biological functions of JNK isoforms.
MATERIALS AND METHODS
Materials.
Antibodies against JNK (antibody 333 for immunoprecipitation and antibody 666 for immunoblotting) and poly(ADP-ribose) polymerase (PARP) were from PharMingen. Antibodies against c-Jun, phospho-c-Jun(Ser63) (phosphorylated c-Jun [phosphorylated at Ser63]), and phospho-c-Jun(Ser73), phospho-JNK, p38, phospho-p38, extracellular signal-regulated kinase (ERK), and phospho-ERK were from Cell Signaling. Cycloheximide (CHX), anisomycin, Hoechst 33258, and antibodies against M2 tag and β-actin were from Sigma. Antibody against hemagglutinin (HA) was from Santa Cruz. Mouse TNF-α was from R & D Systems. [γ-32P]ATP (3,000 mCi/nmol) was from Dupont NEN.
Plasmids.
Expression vectors encoding HA-tagged JNK1 (HA-JNK1), M2-tagged JNK1 (M2-JNK2), HA-JNK2, HA-JNK2(KM) mutant, c-Jun, GAL4-c-Jun, GAL4-luciferase (GAL4-Luc), AP-1 Luc, and enhanced green fluorescent protein (EGFP) have been described previously (18, 25, 30). An adenoviral vector encoding the superrepressor IκBα(AA) has also been described previously (24).
Cell culture, transfection, and transcription assays.
Wild-type (WT), Jnk1−/−, and Jnk2−/− mouse fibroblasts were prepared from WT, Jnk1−/−, and Jnk2−/− mice. Jnk1−/− and Jnk2−/− mice were a gift from Michael Karin (20) (generation of Jnk1 and Jnk2 null mice was supported by National Institutes of Health grant ES04151 to Michael Karin). The mouse fibroblasts were immortalized by the 3T3 protocol. Cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Transfection was performed using Exgene 500 (MBI Fermentas), according to the manufacturer's protocol. GAL4-Luc and AP-1 Luc activities were determined as described previously (17).
Protein kinase assays and immunoblotting.
Immune complex kinase assays and glutathione S-transferase (GST)-c-Jun pulldown kinase assays were performed and quantitated as described previously (8, 17). Immunoblotting was performed as described previously (25).
Apoptosis and caspase 3 assays.
Apoptosis assays and caspase 3 assays were performed as previously described (26).
RESULTS
Differential activation of JNK1 and JNK2 by TNF-α.
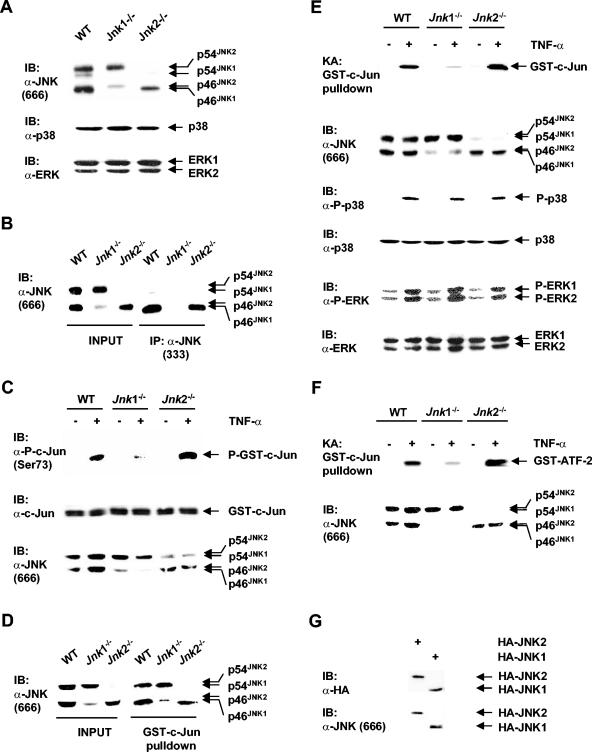
Two closely related JNK isoforms, JNK1 and JNK2, have shared functions (14, 20); however, they also have distinct biological activities (2, 9, 23). To understand the molecular mechanism underlying the difference between JNK1 and JNK2, we employed immortalized mouse embryonic fibroblasts (MEFs) that are deficient in either the Jnk1 or Jnk2 allele. Immunoblotting analysis using an anti-JNK antibody (antibody 666) (PharMingen), which can detect JNK isoforms only in immunoblots, showed that WT cells had both JNK1 (p46JNK1, which is the prominent splicing form of JNK1, and p54JNK1) and JNK2 (p54JNK2, which is the prominent splicing form of JNK2, and p46JNK2) (Fig. 1A). As predicted, Jnk1−/− cells had two different splicing forms of JNK2, whereas Jnk2−/− cells had two different splicing forms of JNK1 (Fig. 1A). Expression of two other MAPKs, p38 and ERK, was comparable in WT, Jnk1−/−, and Jnk2−/− cells (Fig. 1A).
FIG. 1.
Differential activation of JNK1 and JNK2 by TNF-α. (A) Expression of JNK, p38, and ERK were analyzed by immunoblotting (IB) using anti-JNK antibody (antibody 666; PharMingen) (α-JNK), anti-p38 antibody, and anti-ERK antibody. (B) Immunoprecipitation (IP) of JNK isoforms from cell extracts (500 μg each) by a second anti-JNK antibody (α-JNK) (antibody 333; PharMingen) were analyzed by immunoblotting (IB) with the anti-JNK antibody (antibody 666; PharMingen). Whole-cell extracts were used as a control (INPUT gel). (C) Extracts from WT, Jnk1−/−, and Jnk2−/− cells treated with TNF-α (5 ng/ml) for 15 min (+) or not treated with TNF-α (−) were incubated with purified GST-c-Jun proteins (50 ng) in a kinase reaction buffer containing nonradioactive ATP (100 μM). Phosphorylation and amount of GST-c-Jun proteins were measured by immunoblotting analysis with anti-phospho-c-Jun(Ser73) (α-P-c-Jun) and anti-c-Jun antibodies, respectively. P-GST-c-Jun, phosphorylated GST-c-Jun. (D) Purified GST-c-Jun proteins (8 μg) preloaded on glutathione beads were mixed with cell extracts (500 μg each) for 3 h and washed extensively. Isolation of JNK by a GST-c-Jun pulldown kinase assay was detected by immunoblotting analysis as described for panelB. (E) Cells were stimulated with TNF-α (5 ng/ml) for 15 min (+) or not stimulated by TNF-α (−), and the activity of JNK isolated by GST-c-Jun pulldown was measured by in vitro kinase assays (KA) (17). The amount of JNK isoforms being isolated by GST-c-Jun pulldown was measured by immunoblotting (IB) analysis using an anti-JNK antibody (antibody 666; PharMingen). The levels of expressed protein and activation of p38 and ERK were measured by the appropriate antibodies, as indicated. (F) Activation of JNK in WT, Jnk1−/−, and Jnk2−/− cells by TNF-α was measured by GST-c-Jun pulldown kinase assays as described above for panel E, except that GST-ATF-2 was used as a substrate. (G) Jnk2−/− cells were transfected with an expression vector encoding HA-JNK1 or HA-JNK2 (2 μg each). The levels of HA-JNK1 and HA-JNK2 expressed were measured by immunoblotting using the anti-HA antibody and anti-JNK antibody (antibody 666; PharMingen).
It is possible that JNK1 and JNK2 have different biological activities because their enzymatic activities are differentially regulated. To test this scenario, activation of endogenous JNK1 or JNK2 by TNF-α was examined. Immunoprecipitation using a second anti-JNK antibody (antibody 333) (PharMingen) (which has commonly been used for immunoprecipitation of JNK), in combination with immunoblotting using an anti-JNK antibody (antibody 666) (PharMingen) revealed that only JNK1, not JNK2, was immunoprecipitated by the anti-JNK antibody (antibody 333; PharMingen) (Fig. 1B, compare the INPUT gel to the IP gel). Thus, the routine immune complex kinase assays using the anti-JNK antibody (antibody 333; PharMingen) were unable to measure JNK2 activity in Jnk1−/− cells. Similar results were obtained when another commonly used anti-JNK antibody (sc-571; Santa Cruz) was tested (data not shown).
To overcome the limitations of anti-JNK antibodies (antibody 333 [PharMingen] or sc-571 [Santa Cruz]), whole-cell extracts prepared from cells stimulated with TNF-α or left alone were used to phosphorylate purified GST-c-Jun fusion proteins in the presence of nonradioactive ATP. Immunoblotting analysis of GST-c-Jun phosphorylation using anti-phospho-c-Jun(Ser73) antibody revealed that JNK activation by TNF-α was robust in both WT and Jnk2−/− fibroblasts but was very weak in Jnk1−/− fibroblasts (Fig. 1C). Note that Jnk2−/− cell extracts appeared to have even higher JNK activity than WT cell extracts, although the amount of JNK1 in Jnk2−/− cells was slightly less than the amount of JNK1 and JNK2 in WT cells (Fig. 1C). These results suggest that while JNK1 was significantly activated by TNF-α, JNK2 was poorly activated by TNF-α.
To precisely compare the activation of JNK1 and JNK2, we employed the approach of GST-c-Jun pulldown kinase assays, which have been used successfully to analyze the activity of both JNK1 and JNK2 (8, 11). Indeed, GST-c-Jun pulldown performed in combination with immunoblotting analysis using an anti-JNK antibody (antibody 666; PharMingen) demonstrated that both JNK1 and JNK2 were isolated from WT, Jnk1−/−, and Jnk2−/− fibroblasts (Fig. 1D). In vitro kinase assays showed that activation of JNK by TNF-α was robust in both WT and Jnk2−/− fibroblasts (Fig. 1E). Note that JNK activation was even higher in Jnk2−/− cells than in WT cells (Fig. 1E). In contrast, there was negligible activation of JNK2 in Jnk1−/− cells (Fig. 1E). This was not the result of different amounts of JNK1 and JNK2 isolated, as demonstrated by GST-c-Jun pulldown in combination with immunoblotting analysis (Fig. 1E). There was not a general defect in TNF-α signaling in Jnk1−/− cells either, since activation of p38 and ERK by TNF-α was comparable in WT, Jnk1−/−, and Jnk2−/− cells, as measured by anti-phospho-p38 and anti-phospho-ERK antibodies (Fig. 1E). However, it is possible that endogenous JNK1 and JNK2 have different substrate specificities, resulting in the apparent differences in their activation when GST-c-Jun was used as the substrate. This was unlikely. Under the same conditions, JNK activation by TNF-α in Jnk1−/− cells was also greatly reduced when GST-ATF-2 was used as the substrate (Fig. 1F). Similar results were obtained with multiple established cell lines prepared from individual embryos (data not shown).
Another possibility is that the anti-JNK antibody (antibody 666; PharMingen) may have a higher affinity for JNK2. In that case, the amount of JNK2 isolated by GST-c-Jun pulldown could be overestimated by immunoblotting using this antibody, and consequently, the inability of TNF-α to activate JNK2 in Jnk1−/− cells might be due to less JNK2 being isolated by GST-c-Jun pulldown. To exclude this possibility, Jnk2 null cells were transfected with expression vectors encoding HA-JNK1 or HA-JNK2. Immunoblotting analysis using anti-HA antibody revealed that the levels of HA-JNK1 and HA-JNK2 expressed were similar (Fig. 1G, top gel). In a parallel experiment, anti-JNK antibody 666 (PharMingen) detected similar amounts of HA-JNK1 and HA-JNK2 (Fig. 1G, bottom gel), suggesting that the anti-JNK antibody 666 (PharMingen) had similar affinities for JNK1 and JNK2. Taken together, activation of JNK2 by TNF-α was impaired in Jnk1 null fibroblasts.
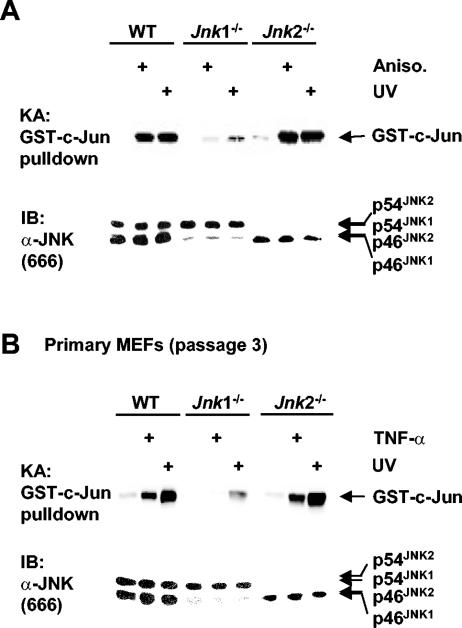
To determine whether other JNK inducers also differentially activate JNK1 and JNK2, cells were stimulated by UV and anisomycin, both of which strongly activate JNK. Like TNF-α, JNK activation by UV and anisomycin was robust in WT and Jnk2−/− fibroblasts but was greatly reduced in Jnk1−/− cells as measured by GST-c-Jun pulldown kinase assays (Fig. 2A). Again, JNK activation was enhanced in the absence of JNK2 (Fig. 2A). These results suggest that JNK1 is the main JNK isoform that is activated by other JNK inducers, such as UV and anisomycin.
FIG. 2.
Differential activation of JNK1 and JNK2 is not limited to TNF-α and immortalized fibroblasts. (A) Activation (+) of JNK isoforms by UV (60 J/m2, 30 min) or anisomycin (Aniso.) (50 ng/ml, 30 min) was measured by GST-c-Jun pulldown kinase assays as described in the legend to Fig. 1E. (B) Primary MEFs of WT, Jnk1−/−, and Jnk2−/− (passage 3) were stimulated by TNF-α (5 ng/ml, 15 min) or UV (60 J/m2, 30 min) (+), and activation of JNK isoforms was measured by GST-c-Jun pulldown kinase assays as described in the legend to Fig. 1E.
The immortalization process may change the response of cells to extracellular stimuli and account for the poor activation of JNK2 by TNF-α in Jnk1−/− cells. To exclude this possibility, primary MEFs (passage 3) were used to examine activation of JNK isoforms by TNF-α or UV. GST-c-Jun pulldown kinase assays showed that JNK activation by TNF-α or UV was robust in WT and Jnk2−/− primary MEFs (Fig. 2B). Like immortalized Jnk1−/− fibroblasts, primary Jnk1−/− MEFs had negligible JNK activation, whereas primary Jnk2−/− MEFs had enhanced JNK activation compared to primary WT MEFs (Fig. 2B). Similar results were obtained when multiple preparations of primary MEFs from different embryos were tested (data not shown). Thus, JNK1 is also the main functional JNK isoform in primary MEFs.
JNK2 interferes with JNK1 activation.
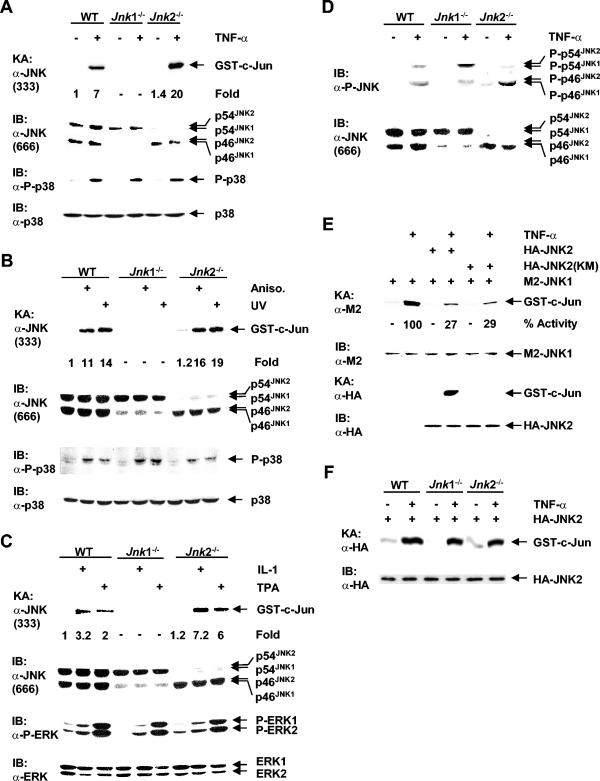
The increase in JNK activation in Jnk2−/− cells prompted us to test the hypothesis that JNK2 may negatively regulate JNK1 activity. Since the anti-JNK antibody (antibody 333; PharMingen) was able to immunoprecipitate only JNK1, not JNK2 (Fig. 1B), we used it to analyze the effect of JNK2 on JNK1 activation. Immune complex kinase assays using anti-JNK antibody 333 (PharMingen) showed that JNK1 activation by TNF-α was twofold higher in Jnk2−/− cells than in WT cells, even though there was slightly less JNK1 in Jnk2−/− cells than in WT cells as measured by immunoblotting analysis (Fig. 3A). This was not the result of upregulation of TNF-α signaling in Jnk2−/− cells, since activation of p38 by TNF-α was comparable in WT, Jnk1−/−, and Jnk2−/− cells (Fig. 3A). As predicted, JNK activation was not detected in Jnk1−/− cells (Fig. 3A). Activation of JNK1 by UV and anisomycin was also modestly enhanced in Jnk2−/− cells (Fig. 3B). Again, p38 activation was comparable in WT, Jnk1−/−, and Jnk2−/− cells (Fig. 3B). The increase in JNK1 activation in Jnk2−/− cells was much more profound in response to IL-1 and 12-O-tetradecanoylphorbol-13-acetate (TPA), which induced JNK weakly in fibroblasts (Fig. 3C). In contrast, ERK activation by IL-1 and TPA was comparable in WT, Jnk1−/−, and Jnk2−/− cells (Fig. 3C). These results suggest that JNK2 does indeed negatively regulate JNK1 activation.
FIG. 3.
JNK2 interferes with JNK1 activation. Cells were stimulated (+) with TNF-α (5 ng/ml, 15 min) (A), UV (60 J/m2, 30 min) and anisomycin (Aniso.) (50 ng/ml, 30 min) (B), and IL-1β (2 ng/ml, 15 min) and TPA (100 ng/ml, 30 min) (C). Endogenous JNK1 was isolated by using the anti-JNK antibody (α-JNK) (antibody 333; PharMingen), and its activity was measured by immune complex kinase assays (KA). The levels of JNK isoforms expressed were measured by immunoblotting (IB) with an anti-JNK antibody (antibody 666; PharMingen). The levels of protein expressed and activation of p38 and ERK were analyzed by immunoblotting with the appropriate antibodies. P-p38, phosphorylated p38. (D) Phosphorylation of endogenous JNK isoforms was analyzed by immunoblotting with anti-phospho-JNK antibody (α-P-JNK). The levels of JNK expressed were analyzed by immunoblotting with the anti-JNK antibody (antibody 666; PharMingen). (E) Jnk2−/− cells were transfected (+) with expression vectors encoding M2-JNK1, HA-JNK2, the kinase-deficient HA-JNK2(KM) mutant, or empty vector (2 μg each) and stimulated with TNF-α (5 ng/ml, 15 min) (+) or left alone. The activities of M2-JNK1 and HA-JNK2, as well as expression of M2-JNK1 and HA-JNK2, were analyzed by immune complex kinase assays (KA) and immunoblotting (IB), respectively. (F) Cells were transfected with an expression vector encoding HA-JNK2 (2 μg) (+) and stimulated with TNF-α (5 ng/ml, 15 min) (+) or not stimulated with TNF-α (−). The activity and expression of HA-JNK2 were measured above as described for panel E.
To determine how JNK2 negatively regulates JNK1 activation, we analyzed phosphorylation of JNK1 and JNK2 at Thr183 and Tyr185, which are required for JNK activation (17), in WT, Jnk1−/−, and Jnk2−/− cells. Immunoblotting analysis using anti-phospho-JNK antibody (Cell Signaling) revealed that phosphorylation of JNK1 was significantly higher in Jnk2−/− cells than in WT cells (Fig. 3D). This was not the result of changes in the levels of JNK1 expressed in WT and Jnk2−/− cells as measured by immunoblotting using anti-JNK antibody 666 (PharMingen) (Fig. 3D). Interestingly, JNK2 was still phosphorylated by upstream kinases of JNK (Fig. 3D). The phosphorylation of JNK2 by upstream kinases was futile, since its activation was negligible (Fig. 1E). This suggests that JNK2 may interfere with JNK1 activation by competing upstream kinases. In support of this notion, expression of HA-JNK2 inhibited activation of cotransfected M2-JNK1 by TNF-α in Jnk2−/− cells (Fig. 3E). The inhibitory effect of HA-JNK2 did not require its enzymatic activity, since the kinase-deficient HA-JNK2(KM) mutant also inhibited M2-JNK1 activation (Fig. 3E). This inhibition was not the result of changes in M2-JNK1 expression (Fig. 3E). Taken together, JNK2 may interfere with JNK1 activation via occupation of JNK upstream kinases.
In contrast to endogenous JNK2, transfected HA-JNK2 was significantly activated by TNF-α in Jnk1−/− cells (Fig. 3F). GST-c-Jun pulldown kinase assays revealed that the c-Jun kinase activity of transfected HA-JNK2 was similar to that of transfected HA-JNK1 (data not shown). These data suggest that activation of endogenous JNK2 may be suppressed by an endogenous inhibitor.
c-Jun expression and activation are impaired in Jnk1 null fibroblasts but are upregulated in Jnk2 null cells.
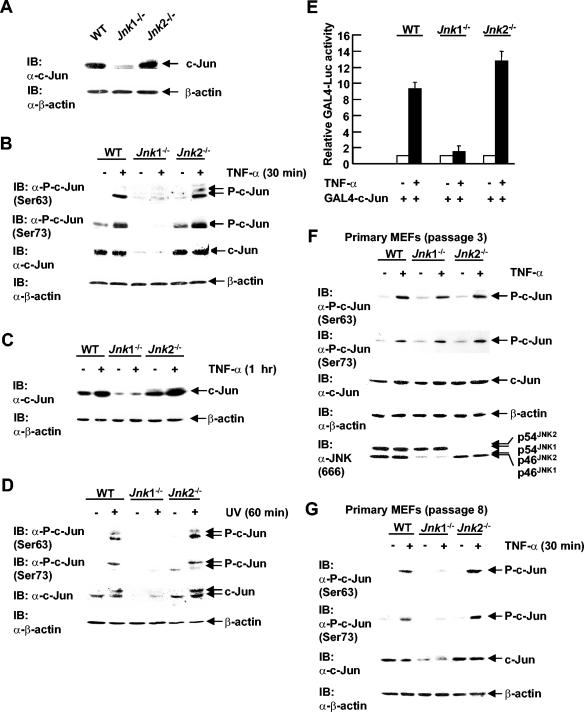
The transcription factor c-Jun autoregulates its expression via binding to its own promoter as a c-Jun/ATF-2 dimer (12). Phosphorylation of c-Jun by JNK at Ser63 and Ser73 increases c-Jun transcription activity, resulting in enhanced expression of c-Jun proteins (12). If JNK1 is the main JNK isoform activated by extracellular stimuli, c-Jun activation and expression should be impaired in Jnk1−/− cells. To test this hypothesis, expression and activation of c-Jun were examined in WT, Jnk1−/−, and Jnk2−/− fibroblasts. Under resting conditions, c-Jun expression was much lower in Jnk1−/− cells than in WT cells, whereas c-Jun expression in Jnk2−/− cells was similar to that in WT cells as measured by immunoblotting using anti-c-Jun antibody (Fig. 4A). As predicted, c-Jun phosphorylation at Ser63 and Ser73 was rapidly induced by TNF-α (30 min) in WT cells as measured by immunoblotting using antibodies against phospho-c-Jun(Ser63) and phospho-c-Jun(Ser73), respectively (Fig. 4B). However, phosphorylation of c-Jun at Ser63 and Ser73 was nearly eliminated in Jnk1−/− cells but was modestly enhanced in Jnk2−/− cells (Fig. 4B). When cells were treated with TNF-α for a longer period of time (1 h), the levels of c-Jun protein expressed also increased in both WT and Jnk2−/− fibroblasts but remained unchanged in Jnk1−/− cells (Fig. 4C). Note that c-Jun expression was even higher in Jnk2−/− cells than in WT cells (Fig. 4C). Similar results were obtained in cells treated with UV (Fig. 4D).
FIG. 4.
Expression and activation of c-Jun are impaired in Jnk1 null fibroblasts. (A) The levels of c-Jun expressed were measured by immunoblotting (IB) using anti-c-Jun antibody (α-c-Jun). (B) Cells were treated with TNF-α (5 ng/ml) for 30 min (+) or not treated with TNF-α (−). c-Jun phosphorylation at Ser63 and Ser73, as well as the levels of protein expressed, were measured by immunoblotting using anti-phospho-c-Jun(Ser63), anti-phospho-c-Jun(Ser73), and anti-c-Jun antibodies. (C) As in panel B, except that the cells were treated with TNF-α for 1 h. (D) Cells were treated with UV (60 J/m2) for 1 h (+) or not treated with UV (−). c-Jun expression and phosphorylation were measured as described for panel B. (E) Cells were transfected with expression vectors encoding GAL4-Luc (0.5 μg), along with GAL4-c-Jun or empty vector (10 ng each), and treated with TNF-α (5 ng/ml) for 10 h (+) or not treated with TNF-α (−). GAL4-Luc activity was measured as described previously (31). (F) Activation and levels of c-Jun expressed in early passage WT, Jnk1−/−, and Jnk2−/− primary MEFs (passage 3) treated by TNF-α as described for panel B. (G) The levels of c-Jun protein expressed and activation of c-Jun by treatment with TNF-α (5 ng/ml) for 30 min in late passage WT, Jnk1−/−, and Jnk2−/− primary MEFs (passage 8) were analyzed as described for panel B.
To determine whether c-Jun transcription activity was also altered in Jnk1 null fibroblasts, cells were transfected with GAL4-c-Jun along with GAL4-Luc. GAL4-c-Jun activity was stimulated by TNF-α in WT fibroblasts as measured by luciferase assays (Fig. 4E). However, TNF-α-induced GAL4-c-Jun activation was significantly lower in Jnk1−/− cells but enhanced in Jnk2−/− cells compared to that in WT cells (Fig. 4E). Taken together, JNK1, but not JNK2, is required for c-Jun expression and activation by TNF-α and UV.
Next, we tested whether c-Jun expression also depends on JNK1 in primary MEFs. Intriguingly, immunoblotting analysis revealed that in early passage primary MEFs (passage 3), the levels of c-Jun expressed were comparable in WT, Jnk1−/−, and Jnk2−/− cells. The levels of c-Jun phosphorylation in response to TNF-α were also comparable in WT, Jnk1−/−, and Jnk2−/− cells (Fig. 4F). However, in late passage MEFs (passage 8), the levels of c-Jun expressed and phosphorylation of c-Jun were much lower in Jnk1−/− MEFs than in WT or Jnk2−/− MEFs (Fig. 4G), similar to that seen in immortalized fibroblasts (Fig. 4A). This suggests that regulation of c-Jun expression and activation may be different in early embryonic development and after exposure to the environmental stresses associated with cell culture.
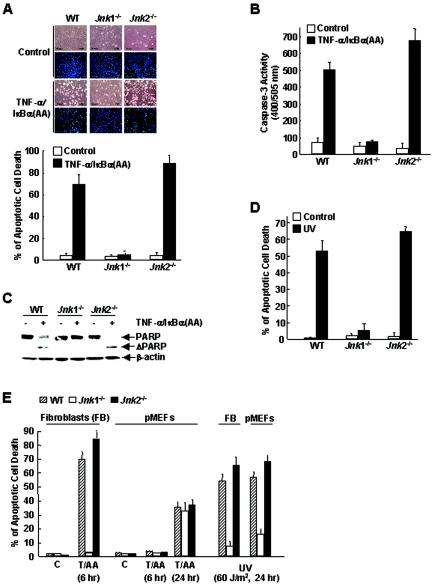
Jnk1-deficient fibroblasts are resistant to apoptosis induced by TNF-α.
In the absence of NF-κB activation, prolonged JNK activation contributes to TNF-α-induced apoptosis (13, 16, 19, 24, 25). If JNK1 were the main JNK isoform activated by TNF-α (Fig. 1), TNF-α-induced apoptosis should be impaired in Jnk1−/− cells. To test this hypothesis, cells were infected with an adenoviral vector encoding IκBα(AA) [Ad/IκBα(AA)] (25), which is a superrepressor of NF-κB, or with a control adenoviral vector encoding LacZ (Ad/LacZ) and then treated with TNF-α for 6 h. Under these conditions, WT cells underwent apoptosis as measured by apoptosis assays, caspase 3 activation, and proteolysis of PARP (Fig. 5A, B, and C). However, Jnk1−/− cells were resistant to TNF-α-induced apoptosis (Fig. 5A, B, and C). In contrast, Jnk2−/− cells were more sensitive to TNF-α killing (Fig. 5A, B, and C). Similar results were obtained when the cells were irradiated by UV (Fig. 5D). Thus, JNK1 is required for apoptosis induced by TNF-α and UV, whereas JNK2 negatively regulates the apoptotic process, consistent with JNK activity in these cells (Fig. 1 to 3).
FIG. 5.
JNK1, but not JNK2, is required for apoptosis induced by TNF-α and UV. (A) WT, Jnk1−/−, and Jnk2−/− cells were infected with Ad/IκBα(AA) or control Ad/LacZ (multiplicity of infection of 200 for each) for 16 h and then treated with TNF-α (5 ng/ml) for 6 h or left alone. Apoptosis was measured with Hoechst 33258 staining. (B and C) Caspase 3 activity and PARP proteolysis were measured as described previously (26). (D) As in panel B, except that the cells were treated with UV (60 J/m2) for 24 h or left alone. (E) Primary WT, Jnk1−/−, and Jnk2−/− fibroblasts (FB) or primary MEFs (pMEFs) (passage 3) were stimulated by TNF-α (5 ng/ml) for 6 or 24 h plus Ad/IκBα(AA) (T/AA) or UV (60 J/m2) for 24 h. Immortalized fibroblasts were used as controls (C). Apoptosis was measured with Hoechst 33258 staining.
To determine whether JNK1 and JNK2 also differentially regulate apoptosis in primary MEFs, WT, Jnk1−/−, and Jnk2−/− primary MEFs (passage 3) were infected with Ad/IκBα(AA) or Ad/LacZ and then treated with TNF-α. Immortalized mouse fibroblasts were used as controls. Surprisingly, TNF-α did not induce apoptosis in all these primary MEFs 6 h poststimulation, when TNF-α had already induced significant apoptosis in immortalized WT and Jnk2−/− fibroblasts (Fig. 5E, 70 and 80% apoptosis, respectively; also see Fig. 5B). After 24 h, TNF-α induced modest apoptosis in all three types of primary MEFs (Fig. 5E, ∼30% apoptosis). In contrast, the rate of UV-induced apoptosis in primary MEFs was similar to that in immortalized fibroblasts (Fig. 5E), suggesting that there were no general defects in the apoptosis machinery in primary MEFs. Importantly, UV-induced apoptosis was also impaired in Jnk1−/− primary MEFs, as seen in immortalized fibroblasts (Fig. 5E). Similar results were obtained with cells prepared from several individual embryos (data not shown). These data suggest that primary MEFs were less sensitive to TNF-α-induced apoptosis and that JNK activity was not critical for the delayed apoptosis in response to TNF-α.
Distinct roles of JNK1 and c-Jun in apoptosis induced by TNF-α.
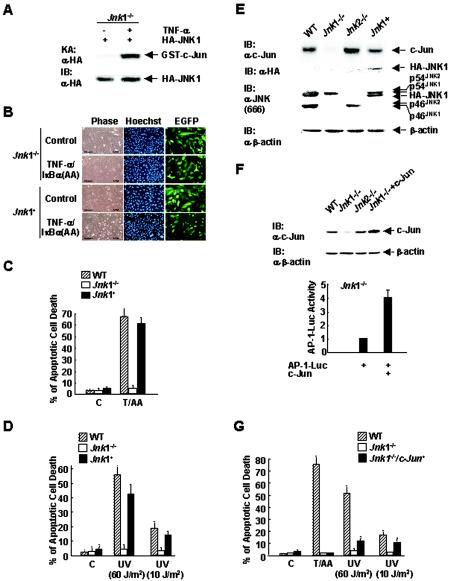
The observations that Jnk1−/− fibroblasts were resistant to apoptosis and had greatly reduced c-Jun expression and activation in response to TNF-α prompted us to determine whether c-Jun mediates the proapoptotic function of JNK1. Jnk1−/− cells were cotransfected with expression vectors encoding EGFP and HA-JNK1 (at a ratio of 1:4) or empty vector, infected with Ad/IκBα(AA), and treated with TNF-α or left alone. Transfected HA-JNK1 was activated by TNF-α (Fig. 6A). Apoptosis assays revealed that TNF-α-induced death of EGFP-positive Jnk1−/− cells transfected with HA-JNK1 was significantly higher than that of Jnk1−/− cells transfected with empty vector (Fig. 6B and C, 60 versus 7%). Thus, the resistance of Jnk1−/− cells to TNF-α-induced apoptosis was due to the absence of JNK1 itself, rather than possible secondary effects caused by disruption of the Jnk1 allele. Similar results were obtained when cells were treated with either high (60 J/m2) or low (10 J/m2) doses of UV (Fig. 6D). Similar results were also obtained when multiple cell lines prepared from several individual embryos were tested (data not shown).
FIG. 6.
Distinct roles of JNK1 and c-Jun in apoptosis. (A) Jnk1−/− cells were transfected with an expression vector encoding HA-JNK1 or empty vector (3 μg each). The activity and expression of HA-JNK1 were measured as previously described (18). An in vitro kinase assay (KA) and immunoblotting (IB) were performed with anti-HA antibody (α-HA). (B) Jnk1 null cells were transfected with an expression vector encoding EGFP (0.8 μg) and HA-JNK1 or empty vector (3.2 μg each) for 12 h, and then the cells were infected with Ad/IκBα(AA) for 16 h. Cells were treated with TNF-α (5 ng/ml) for 6 h or left alone (control). Apoptosis of EGFP-positive cells was measured as described previously (26). The cells were examined by phase-contrast microscopy or by microscopy with Hoechst 33258 staining. (C) Schematic presentation of the results in panel B. C, control; T/AA, TNF-α (5 ng/ml) plus Ad/IκBα(AA). (D) As in panel B, except the cells were irradiated with UV (60 or 10 J/m2) for 24 h. (E) Jnk1 null cells were transfected with expression vectors encoding HA-JNK1 or empty vector (3 μg each), and c-Jun expression was measuredby immunoblotting analysis using anti-c-Jun antibody. Expression of HA-JNK1 and different isoforms of JNK was measured by immunoblotting analysis using anti-HA antibody and anti-JNK antibody (antibody 666; PharMingen), respectively. (F) Jnk1 null cells were transfected with expression vectors encoding the AP-1 Luc reporter gene (0.5 μg), along with c-Jun or empty vector (3 μg each). The levels of transfected c-Jun protein expressed in Jnk1−/− cells were analyzed by immunoblotting using anti-c-Jun antibody. WT, Jnk1−/−, and Jnk2−/− cells were used as controls. AP-1 Luc activity was measured as described previously (17). (G) Jnk1 null cells were transfected with expression vectors encoding EGFP (0.8 μg) and c-Jun or empty vector (3.2 μg each). The cells were then treated with TNF-α (5 ng/ml) for 6 h plus Ad/IκBα(AA) (T/AA) or with UV (60 or 10 J/m2) for 24 h or left alone (control [C]). Apoptosis was analyzed as described in the legend to Fig. 5A.
Reintroduction of HA-JNK1 not only restored the apoptotic response of Jnk1−/− cells but also restored expression of c-Jun proteins as measured by immunoblotting analysis (Fig. 6E). To determine the roles of JNK1 and c-Jun in apoptosis induced by TNF-α and UV, we examined whether expression of c-Jun itself, which has transcriptional activity when overexpressed, can restore the apoptotic response of Jnk1−/− cells. Cells were transfected with an expression vector encoding c-Jun or an empty vector, along with the AP-1 Luc reporter gene. Immunoblotting analysis confirmed that the level of transfected c-Jun expressed in Jnk1−/− cells was about twofold higher than that of endogenous c-Jun in WT cells (Fig. 6F). As expected, overexpression of c-Jun stimulated AP-1 transcription activity (Fig. 6F). In parallel, cells were transfected with expression vectors encoding EGFP and c-Jun (at a ratio of 1:4) or empty vector, infected with Ad/IκBα(AA), and treated with TNF-α or left alone. Apoptosis assays showed that Jnk1−/− cells transfected with an expression vector encoding c-Jun remained resistant to apoptosis induced by TNF-α (Fig. 6G). Similar results were obtained when cells were treated with high doses of UV (60 J/m2) (Fig. 6G). Thus, JNK1, but not c-Jun, is required for apoptosis induced by TNF-α or high doses of UV (60 J/m2) in fibroblasts. However, when cells were treated with low doses of UV (10 J/m2), expression of c-Jun partially restored the apoptotic response of Jnk1 null cells to UV (Fig. 6G), suggesting that c-Jun is involved in the proapoptotic function of JNK1 in response to low doses of UV (10 J/m2). Similar results were obtained when c-Jun−/− fibroblasts were used (data not shown).
DISCUSSION
The MAPK JNK regulates many important cellular events from gene expression to apoptosis (1, 3, 16). The JNK family has two ubiquitously expressed isoforms, JNK1 and JNK2, and a tissue-specific isoform, JNK3, all of which have two different splicing forms, p54 and p46 (12). At this time, the paradigm is that both JNK1 and JNK2 contribute to cellular JNK activity in response to extracellular stimuli (3, 8, 11). Indeed, JNK1 and JNK2 have shared biological functions (14, 20). However, there is evidence that JNK1 and JNK2 also have distinct or even opposing biological functions (2, 9, 23). The molecular mechanism underlying the different biological functions of JNK isoforms is unknown. In this report, we demonstrate that only JNK1 is activated by various extracellular stimuli, including TNF-α, UV, anisomycin, IL-1, and TPA, whereas activation of JNK2 is not only negligible but also interferes with JNK1 activation. The differential activation of JNK1 and JNK2 determines the role of these two JNK isoforms in regulation of c-Jun expression and apoptosis. Thus, our results provide a molecular mechanism underlying the different biological functions of JNK isoforms.
The enzymatic activity of JNK1 and JNK2 is differentially regulated by extracellular stimuli. Using immortalized mouse fibroblasts deficient in the Jnk1 or Jnk2 allele, we found that the commercial anti-JNK antibodies (antibody 333 [PharMingen] or sc-571 [Santa Cruz]), which have been used widely in immune complex kinase assays to study JNK activation by extracellular stimuli, were unable to immunoprecipitate endogenous JNK2 (Fig. 1B). Using the GST-c-Jun pulldown approach, which isolates both endogenous JNK1 and JNK2 (Fig. 1D), we found that JNK1 was significantly activated by the extracellular stimuli examined, including TNF-α, UV, and anisomycin, whereas JNK2 activation was negligible (Fig. 1E and 2A). This was not the result of defects in TNF-α signaling in Jnk1−/− cells (Fig. 1E), different substrate specificities (Fig. 1F), different affinities of GST-c-Jun for JNK1 versus JNK2 (Fig. 1D and G), or altered responses of Jnk1−/− and Jnk2−/− cells to TNF-α after immortalization (Fig. 2B). Thus, JNK1 is the main JNK isoform that is activated by extracellular stimuli.
The regulation and function of JNK2 are complicated. Unlike JNK1, JNK2 was poorly activated by various extracellular stimuli, such as TNF-α, UV and anisomycin in Jnk1−/− cells (Fig. 1E and 2A). Unlike TNF-α and anisomycin, UV slightly activated JNK2 (Fig. 2A and B), consistent with a previous report (27). This suggests that endogenous JNK2 is not an inert enzyme. The differential activation of JNK2 cannot be attributed to the differences in the strength of stimulation by UV, anisomycin, and TNF-α, since activation of JNK2 by anisomycin, which activates JNK as strongly as UV does, was also negligible (Fig. 2A). Future studies are needed to elucidate the molecular mechanism underlying this phenomenon.
Not only is JNK2 poorly activated, it also interferes with JNK1 activation. Our results showed that JNK1 activation was higher in Jnk2−/− cells than in WT cells (Fig. 1 to 3). Interestingly, JNK2 was still phosphorylated by upstream kinases of JNK in response to TNF-α (Fig. 3D). Since activation of JNK2 by TNF-α was negligible (Fig. 1), this phosphorylation was futile. These observations suggest that through its futile phosphorylation, JNK2 may compete with JNK1 for upstream kinases, thereby interfering with JNK1 activation. This hypothesis was supported by the observation that phosphorylation of JNK1 was higher in Jnk2−/− cells than in WT cells (Fig. 3D). Furthermore, expression of WT HA-JNK2 or the kinase-deficient HA-JNK2(KM) mutant also inhibited activation of cotransfected M2-JNK1 by TNF-α (Fig. 3E), suggesting that the inhibition was independent of the enzymatic activity of HA-JNK2. In contrast to endogenous JNK2, transfected HA-JNK2 was activated by TNF-α (Fig. 3F). It is possible that endogenous JNK2 associates with a yet-to-be-identified inhibitor, which may suppress the enzymatic activity of JNK2 (Fig. 1 and 2) but not its phosphorylation by upstream kinases (Fig. 3D). Since the amount of endogenous inhibitor is limited, overexpression of exogenous HA-JNK2 should overcome this inhibition (Fig. 3F). This hypothesis also explains why JNK2 was active in in-gel kinase assays, where JNK2 would have already been separated from its inhibitor during sodium dodecyl sulfate-polyacrylamide gel electrophoresis (8). Future studies are needed to test this hypothesis.
Is genetic disruption of the Jnk1 allele alone sufficient to eliminate c-Jun expression and activation? It is known that JNK is required for c-Jun expression and activation (12). Our results showed that c-Jun expression and activation were impaired in Jnk1−/− fibroblasts but enhanced in Jnk2−/− fibroblasts stimulated by TNF-α and UV (Fig. 4). c-Jun expression was also greatly reduced in nonstimulated Jnk1−/− cells (Fig. 4), suggesting that basal JNK1 activity was required for maintaining c-Jun expression. Introduction of exogenous HA-JNK1 into Jnk1−/− fibroblasts restored c-Jun expression (Fig. 6E), suggesting that the greatly reduced c-Jun expression was due to the absence of JNK1, rather than a consequence of immortalization or a secondary effect of genetic disruption of the Jnk1 allele. These data further support the conclusion that JNK1 is the main JNK isoform that is activated by extracellular stimuli, such as TNF-α and UV, whereas JNK2 may interfere with JNK1 activation.
It is intriguing that c-Jun expression and activation in an early passage of Jnk1−/− primary MEFs (passage 3) were similar to those in WT and Jnk2−/− primary MEFs (Fig. 4F). However, expression and phosphorylation of c-Jun in a late passage of Jnk1−/− primary MEFs (passage 8) were much lower than in an early passage of cells (Fig. 4G), as seen in Jnk1−/− fibroblasts (Fig. 4A). One possible explanation is that the molecular mechanism governing c-Jun expression and activation may be different during embryonic development and after exposure to the environmental stress of cell culture. During embryonic development, the weak activity of JNK2 or other kinases may contribute to maintaining c-Jun expression and activity in Jnk1−/− mice. After exposure to the environmental stress of cell culture, JNK1 is essential for maintaining c-Jun expression and activation. This is analogous to the role of JNKK2/MKK7 in the regulation of cdc2 expression (28). JNKK2/MKK7 was required for cdc2 expression in immortalized fibroblasts and late passage primary MEFs but dispensable in early passage primary MEFs (28). It will be interesting to study how c-Jun expression and activation are differentially regulated during development and under environmental stress.
Is JNK1, but not JNK2, required for TNF-α-induced apoptosis? Previous studies have shown that in the absence of NF-κB activation, prolonged JNK activation promotes TNF-α killing in cultured cells and in vivo (13, 16, 19, 24, 25). Our results consistently showed that TNF-α-induced apoptosis was lower in Jnk1−/− fibroblasts but higher in Jnk2−/− fibroblasts (Fig. 5). Introduction of exogenous HA-JNK1 restored the apoptotic response of Jnk1−/− fibroblasts to TNF-α (Fig. 6A, B, and C), suggesting that the resistance of Jnk1−/− fibroblasts to TNF-α-induced apoptosis is due to the absence of JNK1, rather than a possible secondary effect of gene disruption or immortalization. Similar results were obtained when several established cell lines from multiple embryos were tested (data not shown). These data suggest that JNK1, but not JNK2, is required for TNF-α-induced apoptosis.
The requirement of JNK1 for apoptosis in primary MEFs is complicated. Our results showed that when NF-κB activation was blocked, it took 24 h for TNF-α to induce modest apoptosis (35%) in primary MEFs (Fig. 5E). In contrast, TNF-α induced significant apoptosis (70%) in fibroblasts as early as 6 h (Fig. 5A and E), when apoptosis was not detected in primary MEFs (Fig. 5E). Thus, the apoptotic machinery that mediates TNF-α-induced apoptosis may be less efficient in primary MEFs than in fibroblasts. One possibility is that some components in the apoptotic machinery may be underexpressed in primary MEFs. We also found that primary MEFs were insensitive to Fas ligand-induced apoptosis but immortalized fibroblasts were sensitive (data not shown). Another difference between fibroblasts and primary MEFs is that there was no significant difference in apoptotic responses to TNF-α in WT, Jnk1−/−, and Jnk2−/− primary MEFs (Fig. 5E). Considering the fact that JNK activation was impaired in Jnk1−/− primary MEFs (Fig. 2B), it is likely that JNK activation may not be essential for the delayed TNF-α-induced apoptosis in primary MEFs, probably due to the different mode of TNF-α-induced apoptosis in these cells. In contrast, UV-induced apoptosis occurred at the same rate in both fibroblasts and primary MEFs (Fig. 5E). More importantly, like Jnk1−/− fibroblasts, Jnk1−/− primary MEFs were resistant to UV killing (Fig. 5E). These data support the notion that the requirement of JNK (JNK1) for apoptosis depends on cellular contents and death stimuli (16). This is analogous to the function of c-Jun in cell death, which differs in primary and transformed hepatocytes (7). The different roles of JNKK2/MKK7 in the regulation of cdc2 expression in fibroblasts and primary MEFs are another example (28). While JNKK2/MKK7 was required for cdc2 expression in immortalized fibroblasts and late passage primary MEFs, it was dispensable in early passage primary MEFs (28). Future studies will elucidate the mechanism underlying the different apoptotic responses of Jnk1−/− fibroblasts and primary MEFs to TNF-α.
Our results that JNK activation is not essential for TNF-α-induced apoptosis in primary MEFs contradicts the results of some previous reports. In the absence of NF-κB activation, prolonged JNK activation promotes TNF-α-induced apoptosis in cultured cells and in vivo (13, 16, 19, 24, 25). However, it was proposed that JNK inhibits TNF-α-induced apoptosis in primary MEFs, since primary MEFs that are deficient in both Jnk1 and Jnk2 alleles were more sensitive to killing by TNF-α plus CHX (15). The requirement for JNK may be different in apoptosis induced by TNF-α plus CHX and apoptosis induced by TNF-α plus Ad-IκBα(AA) (4). Another report suggested that JNK inhibits TNF-α-induced apoptosis, since Jnk1 or Jnk2 null primary MEFs were more sensitive to TNF-α plus CHX (10). However, it is puzzling that 500 ng of TNF-α per ml, in combination with 10 μg of CHX per ml, was used to induce apoptosis in Jnk1 or Jnk2 null primary MEFs in that study (10), whereas only 5 ng of TNF-α per ml was sufficient to activate JNK and apoptosis in primary MEFs in our study (Fig. 2B and 5E). The proposed antiapoptotic role of JNK in TNF-α-induced apoptosis also contradicted the finding that Jnk2−/− primary MEFs had higher JNK activity but were more sensitive to TNF-α killing than WT MEFs (10). A recent report showed that Jnk2−/− primary MEFs were resistant to apoptosis induced by TNF-α and CHX, whereas Jnk1−/− primary MEFs were more sensitive (6), suggesting that TNF-α killing is inhibited by JNK1 but enhanced by JNK2. However, the levels of JNK1 expressed in Jnk2−/− primary MEFs were very low for unknown reasons (6). In contrast, the levels of JNK1 expressed in WT and Jnk2−/− MEFs were comparable in our experiments (Fig. 2B) or the experiments of other investigators (10, 27). The extremely low levels of JNK1 expressed in Jnk2−/− primary MEFs may account for the apoptotic response of these cells in the previous report (6). It is possible that multiple factors influence the role of JNK in primary MEFs, such as the method used to measure apoptosis, the genetic background of mice, and experimental conditions. Future studies are needed to clarify the apparent controversy regarding the role of JNK in TNF-α-induced apoptosis in primary MEFs.
Is the proapoptotic function of JNK1 mediated by c-Jun in TNF-α killing? It was reported that c-Jun suppressed or did not affect TNF-α-induced apoptosis (10, 29). Our results showed that expression of exogenous JNK1 restored the apoptotic response of Jnk1 null cells to TNF-α (Fig. 6B and C), as well as c-Jun expression (Fig. 6E). However, expression of exogenous c-Jun, which was active in transcription (Fig. 6F), failed to restore the apoptotic response (Fig. 6G). Similar results were obtained in c-jun−/− cells (data not shown). Thus, it is unlikely that the proapoptotic function of JNK1 in TNF-α-induced apoptosis is mediated by c-Jun in fibroblasts.
Whether c-Jun mediates the proapoptotic function of JNK in UV-induced apoptosis is also controversial, as c-Jun has been suggested to be inhibitory, essential, or dispensable (22, 27, 29). Our results showed that expression of exogenous JNK1 in Jnk1 null cells restored c-Jun expression (Fig. 6E) and the apoptotic response to UV (Fig. 6D). In contrast, expression of exogenous c-Jun, which was sufficient to stimulate AP-1 activity (Fig. 6F), did not restore the apoptotic response of Jnk1 null cells to high doses of UV (60 J/m2) (Fig. 6G). However, expression of exogenous c-Jun partially restored the apoptotic response of Jnk1 null cells to low doses of UV (10 J/m2) (Fig. 6G). Similar results were obtained in c-jun−/− cells (data not shown). The different effects of c-Jun on apoptosis induced by high or low doses of UV is likely due to its effect on the cell cycle. While a low dose of UV (10 J/m2) induces p53-dependent cell cycle arrest, which is negatively regulated by c-Jun (22), a high dose of UV (60 J/m2) does not induce cell cycle arrest. Thus, c-Jun is involved in the proapoptotic function of JNK1 in apoptosis induced by low doses of UV (10 J/m2) but is not involved in the proapoptotic function of JNK1 in apoptosis induced by high doses of UV (60 J/m2).
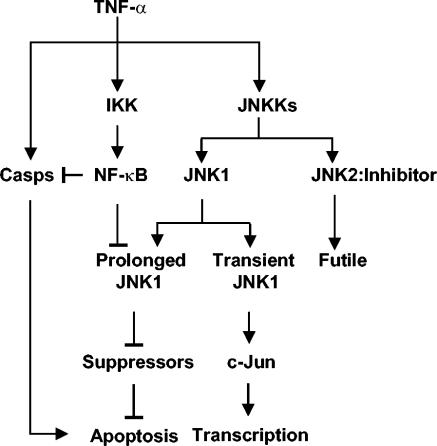
It is a paradox how two closely related JNK isoforms, JNK1 and JNK2, have distinct or even opposing biological functions. For example, activation of CD8+ T cells was impaired in Jnk1−/− mice but enhanced in Jnk2−/− mice (2). Skin tumorigenesis was also found to be enhanced in Jnk1−/− mice but suppressed in Jnk2−/− mice (23). Finally, genetic disruption of the Jnk1 allele alone in mice was sufficient to suppress obesity (9). These observations can be explained by our findings that JNK1 is the main JNK isoform that is activated by extracellular stimuli, whereas JNK2 may interfere with JNK1 activation (Fig. 7). However, JNK1 and JNK2 may have overlapping functions during development, since Jnk null mouse embryos died, but Jnk1 or Jnk2 null mice developed normally (14, 20). Future studies are needed to examine how JNK1 and JNK2 are differentially regulated in vivo.
FIG. 7.
A model of differential activation of JNK1 and JNK2 by TNF-α. Casps, caspases.
Acknowledgments
We thank Michael Karin for providing the Jnk1−/− and Jnk2−/− mice that made this work possible and anonymous reviewers for constructive criticism.
This work was partially supported by National Institutes of Health grants CA92650 and CA100460 (to A.L.).
REFERENCES
- 1.Chang, L., and M. Karin. 2001. Mammalian MAP kinase signaling cascades. Nature 410:37-40. [DOI] [PubMed] [Google Scholar]
- 2.Conze, D., T. Krahl, N. Kennedy, L. Weiss, J. Lumsden, P. Hess, R. A. Flavell, G. Le Gros, R. J. Davis, and M. Rincon. 2002. c-Jun NH2-terminal kinase (JNK)1 and JNK2 have distinct roles in CD8+ T cell activation. J. Exp. Med. 195:811-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis, R. J. 2000. Signal transduction by the JNK group of MAP kinases. Cell 103:239-252. [DOI] [PubMed] [Google Scholar]
- 4.Deng, Y., X. Ren, L. Yang, Y. Lin, and X. Wu. 2003. A JNK-dependent pathway is required for TNFα-induced apoptosis. Cell 115:61-70. [DOI] [PubMed] [Google Scholar]
- 5.Derijard, B., J. Raingeaud, T. Barrett, I. H. Wu, J. Han, R. J. Ulevitch, and R. J. Davis. 1995. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science 267:682-685. [DOI] [PubMed] [Google Scholar]
- 6.Dietrich, N., J. Thastrup, C. Holmberg, M. Gyrd-Hansen, N. Fehrenbacher, U. Lademann, M. Lerdrup, T. Herdegen, M. Jaattela, and T. Kallunki. 2004. JNK2 mediates TNF-induced cell death in mouse embryonic fibroblasts via regulation of both caspase and cathepsin protease pathways. Cell Death Differ. 11:301-313. [DOI] [PubMed] [Google Scholar]
- 7.Eferl, R., R. Ricci, L. Kenner, R. Zenz, J. P. David, M. Rath, and E. F. Wagner. 2003. Liver tumor development: c-Jun antagonizes the proapoptotic activity of p53. Cell 112:181-192. [DOI] [PubMed] [Google Scholar]
- 8.Hibi, M., A. Lin, T. Smeal, A. Minden, and M. Karin. 1993. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 7:2135-2148. [DOI] [PubMed] [Google Scholar]
- 9.Hirosumi, J., G. Tuncman, L. Chang, C. Z. Gorgun, K. T. Uysal, K. Maeda, M. Karin, and G. S. Hotamisligil. 2002. A central role for JNK in obesity and insulin resistance. Nature 420:333-336. [DOI] [PubMed] [Google Scholar]
- 10.Hochedlinger, K., E. F. Wagner, and K. Sabapathy. 2002. Differential effects of JNK1 and JNK2 on signal specific induction of apoptosis. Oncogene 21:2441-2445. [DOI] [PubMed] [Google Scholar]
- 11.Kallunki, T., B. Su, I. Tsigelny, H. K. Sluss, B. Derijard, G. Moore, R. Davis, and M. Karin. 1994. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 8:2996-3007. [DOI] [PubMed] [Google Scholar]
- 12.Karin, M. 1995. The regulation of AP-1 activity by mitogen-activated protein kinases. J. Biol. Chem. 270:16483-16486. [DOI] [PubMed] [Google Scholar]
- 13.Karin, M., and A. Lin. 2002. NF-κB at the crossroads of life and death. Nat. Immunol. 3:221-227. [DOI] [PubMed] [Google Scholar]
- 14.Kuan, C. Y., D. D. Yang, D. R. Samanta Roy, R. J. Davis, P. Rakic, and R. A. Flavell. 1999. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron 22:667-676. [DOI] [PubMed] [Google Scholar]
- 15.Lamb, J. A., J. J. Ventura, P. Hess, R. A. Flavell, and R. J. Davis. 2003. JunD mediates survival signaling by the JNK signal transduction pathway. Mol. Cell 11:1479-1489. [DOI] [PubMed] [Google Scholar]
- 16.Lin, A. 2003. Activation of the JNK signaling pathway: breaking the brake on apoptosis. Bioessays 25:1-8. [DOI] [PubMed] [Google Scholar]
- 17.Lin, A., A. Minden, H. Martinetto, F. X. Claret, C. Lange-Carter, F. Mercurio, G. L. Johnson, and M. Karin. 1995. Identification of a dual specificity kinase that activates the Jun kinases and p38-Mpk2. Science 268:286-290. [DOI] [PubMed] [Google Scholar]
- 18.Lu, X., S. Nemoto, and A. Lin. 1997. Identification of c-Jun NH2-terminal protein kinase (JNK)-activating kinase 2 as an activator of JNK but not p38. J. Biol. Chem. 272:24751-24754. [DOI] [PubMed] [Google Scholar]
- 19.Maeda, S., L. Chang, Z.-W. Li, J.-L. Luo, H. Leffert, and M. Karin. 2003. IKKβ is required for prevention of apoptosis mediated by cell-bound but not by circulating TNFα. Immunity 19:725-737. [DOI] [PubMed] [Google Scholar]
- 20.Sabapathy, K., W. Jochum, K. Hochedlinger, L. Chang, M. Karin, and E. F. Wagner. 1999. Defective neural tube morphogenesis and altered apoptosis in the absence of both JNK1 and JNK2. Mech. Dev. 89:115-124. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez, I., R. T. Hughes, B. J. Mayer, K. Yee, J. R. Woodgett, J. Avruch, J. M. Kyriakis, and L. I. Zon. 1994. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature 372:794-798. [DOI] [PubMed] [Google Scholar]
- 22.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 23.She, Q. B., N. Chen, A. M. Bode, R. A. Flavell, and Z. Dong. 2002. Deficiency of c-Jun-NH2-terminal kinase-1 in mice enhances skin tumor development by 12-O-tetradecanoylphorbol-13-acetate. Cancer Res. 62:1343-1348. [PubMed] [Google Scholar]
- 24.Tang, F., G. Tang, J. Xiang, Q. Dai, M. R. Rosner, and A. Lin. 2002. Absence of NF-κB-mediated inhibition of c-Jun N-terminal kinase activation contributes to tumor necrosis factor alpha-induced apoptosis. Mol. Cell. Biol. 24:8571-8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang, G., Y. Minemoto, B. Dibling, N. H. Purcell, Z. Li, M. Karin, and A. Lin. 2001. Inhibition of JNK activation through NF-κB target genes. Nature 414:313-317. [DOI] [PubMed] [Google Scholar]
- 26.Tang, G., J. Yang, Y. Minemoto, and A. Lin. 2001. Blocking caspase 3-mediated proteolysis of IKKβ suppresses TNF-α-induced apoptosis. Mol. Cell 8:1005-1016. [DOI] [PubMed] [Google Scholar]
- 27.Tournier, C., P. Hess, D. D. Yang, J. Xu, T. K. Turner, A. Nimnual, D. Bar-Sagi, S. N. Jones, R. A. Flavell, and R. J. Davis. 2000. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288:870-874. [DOI] [PubMed] [Google Scholar]
- 28.Wada, T., N. Joza, H. Y. Cheng, T. Sasaki, I. Kozieradzki, K. Bachmaier, T. Katada, M. Schreiber, E. F. Wagner, H. Nishina, and J. M. Penninger. 2004. MKK7 couples stress signalling to G2/M cell-cycle progression and cellular senescence. Nat. Cell Biol. 6:215-226. [DOI] [PubMed] [Google Scholar]
- 29.Wisdom, R., R. S. Johnson, and C. Moore. 1999. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 18:188-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu, C., Y. Minemoto, J. Zhang, J. Liu, F. Tang, T. N. Bui, J. Xiang, and A. Lin. 2004. JNK suppresses apoptosis via phosphorylation of the proapoptotic Bcl-2 family protein BAD. Mol. Cell 13:329-340. [DOI] [PubMed] [Google Scholar]
- 31.Zheng, C., J. Xiang, T. Hunter, and A. Lin. 1999. The JNKK2-JNK1 fusion protein acts as a constitutively active c-Jun kinase that stimulates c-Jun transcription activity. J. Biol. Chem. 274:28966-28971. [DOI] [PubMed] [Google Scholar]