Abstract
Background
Epstein-Barr virus (EBV) and human cytomegalovirus (CMV) infections are environmental risk factors affecting the outcome of cancer due to an impairment in the cell-mediated immunity. Therefore, this study aimed to detect the frequency of EBV and CMV DNA and their association with clinical characteristics and outcome of pediatric leukemic patients.
Methods
Samples of 50 immunocompromised pediatric leukemic patients and 30 apparently healthy children were subjected to the amplification of EBV DNA by one version of PCR targeting the Bam H1 W region of the genomic region of EBV, and the amplification of CMV DNA by targeting the CMV UL97 genomic region by a second round PCR. All investigations were performed on WBCs and sera. Results were correlated with the clinical and laboratory characteristics of the disease, and with overall survival.
Results
EBV and CMV DNA were detected in 20 and 54% of leukemic patients, respectively. Nine out of ten patients with EBV DNA (90%) were positive for CMV DNA in their sera. The presence of EBV DNA or CMV DNA was associated with neutropenia and a low total leukocyte count (TLC) (p = 0.02, 0.03, respectively). The presence of severe CMV disease, longer duration of febrile neutropenia, neutropenia, lymphopenia, thrombocytopenia and the presence of EBV DNA in patients’ sera were significantly associated with worse overall survival.
Conclusion
The detection of CMV disease and EBV DNA is relatively common in leukemic children and is significantly associated with a decline in the overall survival.
Keywords: EBV, CMV, Pediatric leukemia, PCR, Survival
Background
It is now known that Cytomegalovirus (CMV) and Epstein-Barr virus (EBV) infect about 90% of the population globally, and that infection continues in a latent phase [1]. Viral infections, particularly caused by herpesviruses, were documented as important cause of morbidity and mortality in immunocompromised patients with a hematological malignancy [2, 3]. Immunosuppression conditions –like those faced in cancer patients- can lead to reactivation years later [1, 4]. Recent evidence shows that the activity of viral proteins interferes with cellular pathways controlling growth and survival, which may lead to crucial cellular transformation [5]. EBV was first identified in cultured Burkitt’s lymphoma cells 40 years ago [6], and coinfection with EBV has been linked with Hodgkin’s disease, large cell lymphoma, and chronic lymphocytic leukemia in adults [7, 8]. Therefore, the detection and quantification of these viruses help in clinical management; and prompt early treatment in order to prevent further progression in disease course [9, 10].
CMV and EBV are the most common infections in pediatric leukemic patients as a result of leukemia-associated immunosuppression status [11, 12]. There are a few reports about the role of EBV and CMV in pediatric leukemic patients. A higher exposure to herpes simplex virus types 1 and 2 (HSV-1 and 2) among acute lymphoblastic leukemia (ALL) children in Egypt has been previously reported [2]. Moreover, Lehtinen and colleagues reported a possible association between maternal EBV infection and childhood ALL [13].
Direct detection for herpesvirus infections in immunocompromised patients allows simplifying preemptive therapy through early detection of EBV replication and is considered a high positive predictive value for the related infections [14]. The current study aimed, therefore, to detect the frequency of EBV and CMV DNA and their association with the clinical characteristics and the outcome of pediatric leukemic patients.
Methods
Patients
This prospective study included 50 pediatric patients with leukemia diagnosed and treated at the pediatric oncology department, National Cancer Institute (NCI), Cairo University between January 2013 and December 2014. Thirty apparently healthy normal individuals with comparable age and sex were included as a control group. The Institutional Review board (IRB) of the NCI approved the protocol (IRP NO. IRP00004025). Informed consent was obtained from all participants enrolled in the study. The inclusion criteria were children till 18 years old, both sexes, suffering from acute lymphocytic leukemia (ALL) or acute myeloid leukemia (AML). All patients were subjected to detailed history, physical examination, as well as routine clinical and laboratory investigations. As for clinical assessment, the patients were checked for fever, organomegaly, mucositis, lymphadenopathy, chest infection and duration of febrile neutropenia. Main laboratory tests included; complete blood count, lactate dehydrogenase level (LDH), erythrocyte sedimentation rate (ESR), cerebrospinal fluid examination (CSF), bone marrow aspiration (BMA) and liver and renal function tests. Echocardiographic examination, immunophenotyping, karyotyping and abdominal ultrasound were also performed. Accordingly; diagnosis, disease extent, risk stratification [15], and comorbidities were determined, and correlated with the patients’ treatment schedule. The patients were followed up during the course of treatment for at least 18 months [16].
Response and survival
Assessment of response of acute leukemia was done after induction treatment using WHO criteria. Scoring of CMV was calculated according to Plotkin scoring system [17] (Table 1). Total score of seven or more was considered severe CMV infection. Overall survival was calculated from the date of diagnosis till the date of death or last follow-up.
Table 1.
System for scoring severity of CMV disease [17]
| Manifestation | Points |
|---|---|
| Fever | 1–3 |
| Leucopenia (<4000) | 1 |
| Thrombocytopenia (<100,000) | 1 |
| Hepatitis | 1–3 |
| Pneumonia | 1–3 |
| CNS changes | 1–3 |
| Glomerulonephritis | 1–3 |
| Arthritis | 2 |
| Muscle wasting | 2 |
| Super infection | 3 |
| GI bleed | 3 |
| Death | 4 |
Specimen collection
Five ml of whole blood specimen was obtained from each patient and control. Serum and leukocyte samples were extracted according to Vander Bij et al.[18] and our previously published protocols [19, 20], and stored in aliquots of 100 μl saline at −20 °C until being processed for nucleic acid extraction.
Nucleic acid extraction
Viral DNA was extracted using QIAamp viral RNA extraction kit (Qiagen, Valencia, USA). The extraction was done according to the manufacturer’s instructions. The amount of viral DNA was measured by spectrophotometry using a Nano-Drop 2000 spectrophotometer (Thermo Scientific/US, Canada) and 100 ng of DNA template was used in the PCR assays. DNA extracts were placed on ice and used immediately for PCR, then stored at −80 °C until further subsequence analysis.
Molecular detection
Molecular detection of EBV and CMV DNA: All samples were subjected to one version PCR assay targeting nucleotide position from 32104 to 32256 bp of BamH1 W genomic region of EBV (Accession AB850660) and nucleotide position from 2692 to 2933 bp of UL 97 genomic region of CMV (Accession FJ616285.1). The amplification was conducted in a final volume of 25 μl of AmpliTaq PCR master mix (Biosystems, Barcelona, Spain) containing 0.2 μM of outer primers using the thermal cycler (Perkin-Elmer Cetus), according to previously published protocols [21, 22]. Nucleic acids of EBV VR-1492 (ATCC-USA) and CMV VR-538 (ATCC-USA) were extracted using the same extraction procedure for the samples. EBV and CMV-positive and negative (water) controls were run in each PCR assay.
Detection of amplified product by agarose gel electrophoresis: 17 μl of the PCR product was subjected to electrophoresis on a 2% agarose gel (Sigma) in Tris-acetate buffer (TAE 1X) pH 8.2, stained with 0.5 μg/ml ethidium bromide and examined under UV transillumination and photographed. The product sizes were estimated by comparing with 100 bp DNA ladder (Gendirex, Taiwan). The amplified fragments were 153 bp for EBV [21] and 240 bp for CMV [22].
Sensitivity of qualitative PCR for detection of EBV and CMV DNA in clinical specimens: Serial dilutions were prepared from CMV and EBV-positive controls and PCR assays were performed according to the previously published protocols [21, 22].
Specificity of qualitative PCR for detection of EBV and CMV DNA in clinical specimens: It was performed by testing samples positive for HSV-1 and 2, herpesvirus 6, Varicella Zoster Virus, adenovirus and CMV or EBV according to the tested virus. Then, the PCR assay was performed as mentioned before.
Statistical analysis
Statistical analysis was done using IBM© SPSS© Statistics version 22 (IBM© Corp., Armonk, NY, USA). Numerical data were expressed as a mean and standard deviation or median and range as appropriate. Qualitative data were expressed as frequency and percentage. Chi-square test or Fisher’s exact test was used to examine the relation between qualitative variables. Survival analysis was done using Kaplan-Meier method and comparison between two survival curves was done using log-rank test. All tests were two-tailed. A p-value < 0.05 was considered significant.
Results
The median age of the 50 pediatric leukemic patients was 7 years (range: 9.5 months–18 years) while that of the 30 healthy controls was 11 years (range: 4–18 years). Male to female ratio was 1.08:1 in patients, and 2:1 in controls. The two groups were comparable in age (p = 0.267) and sex (p = 0.199). The demographic, clinical, and laboratory characteristics of patients are summarized in Table 2. All patients had fever and signs of chest infections, and 70% of them had organomegaly. Laboratory (hematological and biochemical) data of the leukemia group are illustrated in Table 3. The most observed abnormal biochemical parameters are LDH and SGOT levels observed in about 94 and 30% of patients, respectively.
Table 2.
Demographics and clinical characteristics of pediatric leukemia patients (n = 50)
| Characteristics | N (%) |
|---|---|
| Age, median (Range) | 7 years (9.5 months–18 years) |
| Sex | |
| Males | 26 (52%) |
| Females | 24 (48%) |
| Clinical characteristics | |
| Fever | 50 (100%) |
| Organomegaly | 35 (70%) |
| Mucositis | 22 (44%) |
| Lymphadenopathy | 7 (14%) |
| Chest infection | 50 (100%) |
| Duration of Febrile neutropenia, median (range) | 26.5 days (9–60) |
| Severe CMV disease | 32 (64%) |
Table 3.
Some laboratory parameters of leukemia patients
| Laboratory parameter | Leukemia group N = 50 | ||
|---|---|---|---|
| Normal value | High risk N (%) |
High risk Median (range) |
|
| 1- Liver function test (LFT): | |||
| Serum glutamic-pyruvic transaminase (SGPT) | 0-41 IU/L | 7 (14) | 20.1 (1–317) |
| High risk: Level more than 41 IU/L | |||
| Serum glutamic oxaloacetic transaminase (SGOT) | 0-32 IU/L | 15 (30) | 23.50 (9–273) |
| High risk: Level more than 32 IU/L | |||
| Total Bilirubin | 0-1.2 mg/dl | 1 (2) | 0.45 (0.1–1.7) |
| High risk: Level more than 1.2 mg/dl | |||
| 2- Kidney function test (KFT): | |||
| Creatinine | 0.8-1.3 mg/dl | 3 (6) | 0.4 (0.1–2.2) |
| High risk: Level more than 1.3 mg/dl | |||
| Uric Acid | 3.4-7 mg/dl | 3 (6) | 3.4 (1–24) |
| High risk: Level more than >7 mg/dl | |||
| Lactate dehydrogenase (LDH) | 125-220 IU/L | 47 (94) | 907 (11–9635) |
| High risk: Level more than 220 IU/L | |||
| 3- Complete blood count (CBC): | |||
| Hemoglobin concentration (Hb) | 11.5-15.5 g/dl | 40 (80) | 7.4 (3.9–14.1) |
| High risk: Level less than <11.5 g/dl | |||
| Total leukocyte count (TLC) | 4500-11000 cell/cmm | 11 (22) | 11950 (1080–731500) |
| High risk: Leucopenia <4500cell/cmm | |||
| Platelet count (Plt) | 150-440 × 109/l | 40 (80) | 39.5 (7–694) |
| High risk: Thrombocytopenia <150 × 109/l | |||
| Absolute neutrophile count (ANC) | 2-7 × 109/l | 42(84) | 0.13 (0–3.83) |
| High risk: Neutropenia <2 × 109/l | |||
| Absolute Monocyte count (AMC) | 0.2-1.2 × 109/l | 29 (58) | 0.11 (0–9.66) |
| High risk: Monocytopenia <0.2 × 109/l | |||
| Absolute Lymphocyte count (ALC) | 1-3.5 × 109/l | 23 (46) | 1.2 (0–143.3) |
| High risk: Lymphopenia <1 × 109/l | |||
IU/L International unit per liter, mg/dl milligram per deciliter, g/dl gram per deciliter
Disease characteristics and outcome of patients
Ten patients had AML and 40 had ALL; 10 of them were T-ALL. Eighty-eight percent of patients received consolidation treatment, 6% were under maintenance therapy and 6% were under salvage therapy. At the end of the study, the mortality rate was 32/50 (64%).
Presence of EBV and CMV DNA in studied groups
The distribution of EBV and CMV DNA in serum were significantly different between leukemic and control groups (p = 0.048). Likewise, the distribution of both viruses in leukocytes was significantly different between leukemic and control groups (p < 0.001) (Table 4).
Table 4.
Detection of herpesviruses by PCR in both sera and WBCs of the studied groups expressed as number and percentage
| Group | EBV DNA | CMV DNA | Both EBV/CMV DNA | Negative for herpesviruses |
|---|---|---|---|---|
| Serum | ||||
| Leukemia (n = 50) | 1 (2.0%) | 18 (36.0%) | 9 (18.0%) | 22 (44.0%) |
| Control (n = 30) | 0 (0.0%) | 14 (46.7%) | 0 (0.0%) | 16 (53.3%) |
| p Value | =0.048 | |||
| WBCs | ||||
| Leukemia (n = 50) | 14 (28%) | 10 (20%) | 9 (18%) | 17 (34%) |
| Control (n = 30) | 0 (0%) | 12 (40%) | 0 (0.0%) | 18 (60%) |
| p Value | <0.001 | |||
EBV and/or CMV frequencies were higher in sera of leukemic patients (28/50, 56%). EBV and CMV co-infection was detected in nine leukemia patients (18%) and none of the controls. The number of patients with co-infection was too small to be associated statistically with the development of severe CMV disease (n = 9). There was no significant association of any of the clinical and laboratory parameters with the active co-infection.
Sensitivity and specificity of qualitative PCR assay for detection of EBV and CMV DNA
Such assays were found to be highly specific for EBV BamH1 W region and CMV UL97 as none of the other viruses tested was amplified. The sensitivity level of qualitative PCR assay was approximately 100 copies/μl for both viruses, which represents the lowest standard dilution that could be detected.
Relation between presence of EBV and/or CMV and demographic, clinical and biochemical parameters in leukemia patients
Mucositis was more common in leukemic patients negative for herpesviruses in serum than in positive patients, with borderline statistical significance (p = 0.07). Neutropenia (ANC <0.13 × 109/l) was more observed in patients with EBV and/ or CMV DNA in serum than in those with EBV/CMV co-infection and negative patients, with borderline significance (p = 0.07) (Table 5). No significant association of presence of herpesvirus DNA with other high risk clinical parameters; lymphopenia (<1.2 × 109/l), thrombocytopenia (Plt < 39.5 × 109/l), and low Hb level (<7.4 g/dl) (Tables 5 and 6).
Table 5.
Relation between demographic, clinical, and biochemical findings and CMV and/or EBV as detected in serum by qualitative PCR assay in leukemic patients
| CMV orEBV alone n = 19 |
Both positive n = 9 |
Both negative n = 22 |
p value | |
|---|---|---|---|---|
| Age | 0.61 | |||
| ≤6 years (n = 23) | 7 (30.4%) | 5 (21.7%) | 11 (47.8%) | |
| >6 years (n = 27) | 12 (44.4%) | 4 (14.8%) | 11 (40.7%) | |
| Sex | 0.53 | |||
| Male (n = 26) | 11 (42.3%) | 3 (11.5%) | 12 (46.2%) | |
| Female (n = 24) | 8 (33.3%) | 6 (25%) | 10 (41.7%) | |
| Mucositis | 0.07 | |||
| Yes (n = 22) | 7 (31.8%) | 7 (31.8%) | 8 (36.4%) | |
| No (n = 28) | 12 (42.9%) | 2 (7.1%) | 14 (50.0%) | |
| Organomegaly | 0.72 | |||
| Yes (n = 35) | 14 (40.0%) | 7 (20.0%) | 14 (40.0%) | |
| No (n = 15) | 5 (33.3%) | 2 (13.3%) | 8 (53.4%) | |
| DFN (days) | 0.75 | |||
| <26.5 (n = 25) | 8 (32%) | 5 (20%) | 12 (48%) | |
| >26.5 (n = 25) | 11 (44%) | 4 (16%) | 10 (40%) | |
| ANC (×109/L) | 0.07 | |||
| <0.13 (n = 25) | 13 (52%) | 2 (8%) | 10 (40%) | |
| ≥0.13 (n = 25) | 6 (24%) | 7 (28%) | 12 (48%) | |
| Plt count (×109/L) | 0.27 | |||
| <39.5 (n = 25) | 7 (28%) | 4 (16%) | 14 (56%) | |
| ≥39.5 (n = 25) | 12 (48%) | 5 (20%) | 8 (32%) | |
| Hb (gm/dL) | 0.65 | |||
| <7.4 (n = 25) | 10 (40%) | 3 (12%) | 12 (48%) | |
| ≥7.4 (n = 25) | 9 (36%) | 6 (24%) | 10 (40%) | |
| AMC(×109/L) | 0.43 | |||
| <0.11 (n = 25) | 9 (36%) | 3 (12%) | 13 (52%) | |
| ≥0.11 (n = 25) | 10 (40%) | 6 (24%) | 9 (36%) | |
| TLC(cell/mm3) | 0.57 | |||
| <11950 (n = 25) | 11 (44%) | 5 (20%) | 9 (36%) | |
| ≥11950 (n = 25) | 8 (32%) | 4 (16%) | 13 (52%) | |
| ALC(×109/L) | 0.93 | |||
| <1.2 (n = 24) | 10 (41.7%) | 4 (16.7%) | 10 (41.7%) | |
| ≥1.2 (n = 26) | 9 (34.6%) | 5 (19.2%) | 12 (46.2%) |
Numerical factors were divided according to their median values
DFN Duration of febrile neutropenia, HG hemoglobin, TLC Total leukocytic count, Plt platelets, ANC Absolute neutrophilic count, AMC Absolute monocytic count, ALC Absolute lymphocytic count
Table 6.
Relation between demographic, clinical, and biochemical findings and CMV and/or EBV as detected in leukocytes by qualitative PCR assay in leukemic patients
| EBV alone n = 14 |
CMV alone n = 10 |
Both positive n = 9 |
Both negative n = 17 |
p value | |
|---|---|---|---|---|---|
| Age | 0.45 | ||||
| ≤6 years (n = 23) | 8 (34.8%) | 6 (26.1%) | 3 (13%) | 6 (26.1%) | |
| >6 years (n = 27) | 6 (22.2%) | 4 (14.8%) | 6 (22.2%) | 11 (40.7%) | |
| Sex | 0.94 | ||||
| Male (n = 26) | 7 (26.9%) | 6 (23.1%) | 5 (19.2%) | 8 (30.8%) | |
| Female (n = 24) | 7 (29.2%) | 4 (16.7%) | 4 (16.7%) | 9 (37.5%) | |
| Mucositis | 0.60 | ||||
| Yes (n = 22) | 4 (18.2%) | 5 (22.7%) | 5 (22.7%) | 8 (36.4%) | |
| No (n = 28) | 10 (35.7%) | 5 (17.9%) | 4 (14.3%) | 9 (32.1%) | |
| Organomegaly | 0.61 | ||||
| Yes (n = 35) | 11 (31.4%) | 8 (22.9%) | 6 (17.1%) | 10 (28.6%) | |
| No (n = 15) | 3 (20.0%)) | 2 (13.3%) | 3 (20.0%) | 7 (46.7%) | |
| DFN (days) | 0.79 | ||||
| <26.5 (n = 25) | 7 (28.0%) | 4 (16.0%) | 4 (16.0%) | 10 (40.0%) | |
| >26.5 (n = 25) | 7 (28.0%) | 6 (24.0%) | 5 (20.0%) | 7 (28.0%) | |
| ANC(×109/L) | 0.84 | ||||
| <0.13 (n = 25) | 8 (32%) | 4 (16%) | 5 (20%) | 8 (32%) | |
| ≥0.13 (n = 25) | 6 (24%) | 6 (24%) | 4 (16%) | 9 (36%) | |
| Plt count(×109/L) | 0.90 | ||||
| <39.5 (n = 25) | 7 (28.0%) | 6 (24.0%) | 4 (16.0%) | 8 (32.0%) | |
| ≥39.5 (n = 25) | 7 (28.0%) | 4 (16.0%) | 5 (20.0%) | 9 (36.0%) | |
| Hb(gm/dL) | 0.12 | ||||
| <7.4 (n = 25) | 8 (32%) | 5 (20%) | 7 (28%) | 5 (20%) | |
| ≥7.4 (n = 25) | 6 (24.0%) | 5 (20.0%) | 2 (8.0%) | 12 (48.0%) | |
| AMC(×109/L) | 0.67 | ||||
| <0.11 (n = 25) | 8 (32%) | 6 (24%) | 3 (12%) | 8 (32%) | |
| ≥0.11 (n = 25) | 6 (24%) | 4 (16%) | 6 (24%) | 9 (36%) | |
| TLC(cell/mm3) | 0.59 | ||||
| <11950 (n = 25) | 6 (24%) | 6 (24%) | 6 (24%) | 7 (28%) | |
| ≥11950 (n = 25) | 8 (32%) | 4 (16%) | 3 (12%) | 10 (40%) | |
| ALC(×109/L) | 0.79 | ||||
| <1.2 (n = 24) | 8 (33.3%) | 4 (16.7%) | 5 (20.8%) | 7 (29.2%) | |
| ≥1.2 (n = 26) | 6 (23.1%) | 6 (23.1%) | 4 (15.4%) | 10 (38.5%) |
DFN Duration of febrile neutropenia, HG hemoglobin, TLC Total leukocytic count, Plt platelets, ANC Absolute neutrophilic count, AMC Absolute monocytic count, ALC Absolute lymphocytic count
Relation between presence of EBV and/or CMV and overall survival in leukemia patients
The median follow-up period of leukemia patients was 23.4 months (range: 18–28.7 months). The cumulative overall survival at 24 months was 47.4%. The Survival of pediatric leukemia patients was significantly worse in association with severe CMV disease (p = 0.002), longer duration of febrile neutropenia (p = 0.024), thrombocytopenia (p = 0.024), presence of active EBV infection (p = 0.038), lymphopenia (p = 0.042), and neutropenia (p = 0.044). EBV and CMV co-infection had borderline effect on the overall survival of patients (p = 0.079) (Table 7, Figs. 1, 2, 3, 4, 5 and 6).
Table 7.
Overall survival of the leukemic patients and its relation to different prognostic factors
| Cumulative survival at 24 month (%) | Median Survival (months) | p value | |
|---|---|---|---|
| Diagnosis | |||
| ALL | 47.4 | 23.5 | 0.518 |
| AML | 33.3 | 15.6 | |
| DFN (days) | |||
| <26.5 | 57.9 | 32.2 | |
| >26.5 | 35.4 | 16.5 | 0.024 |
| ANC (×109/L) | |||
| <0.13 | 15.6 | 16.5 | |
| ≥0.13 | 56.6 | 32.3 | 0.044 |
| Plt count (×109/L) | |||
| <39.5 | 14.7 | 16.5 | |
| ≥39.5 | 68.3 | 44.3 | 0.024 |
| ALC (×109/L) | |||
| <1.2 | 12.2 | 16.5 | 0.042 |
| ≥1.2 | 62.6 | 32.3 | |
| CMV score | |||
| <7 (n = 8) | 80.0 | 15.1 | 0.002 |
| ≥7 (n = 20) | 26.1% | 23.5 | |
| EBV PCR Serum | |||
| Positive (n = 9) | 68.6 | 91.6 | |
| Negative (n = 29) | 33.7 | 21.5 | 0.038 |
| CMV&EBV in serum | |||
| Both negative(n = 15) | 30.8 | 21.6 | 0.079 |
| CMV alone/EBV alone(n = 15) | 25.9 | 32.3 | |
| Both positive(n = 8) | 62.5 | 91.6 | |
Numerical factors were divided according to their median values
DFN Duration of febrile neutropenia, Plt platelets, ANC Absolute neutrophilic count, ALC Absolute lymphocytic count
Fig. 1.
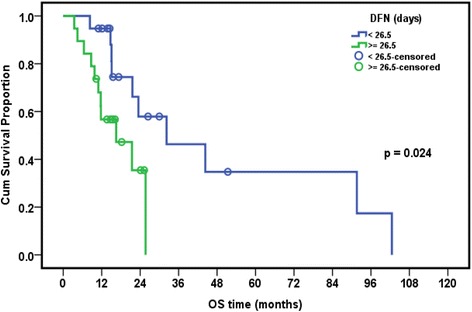
Relation of overall survival (OS) with duration of febrile neutropenia (DFN) in pediatric ALL patients
Fig. 2.
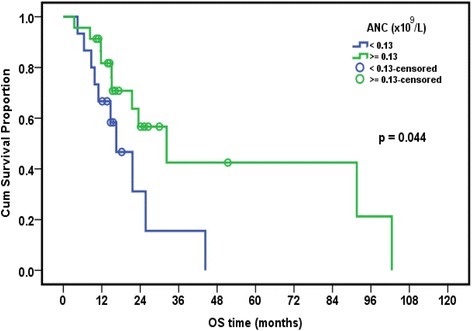
Relation of overall survival with absolute neutrophilic count (ANC) in pediatric ALL patients
Fig. 3.
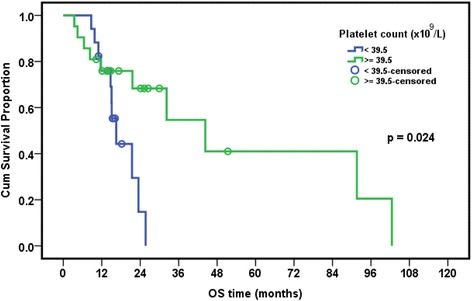
Relation of overall survival with platelet count in pediatric ALL patients
Fig. 4.
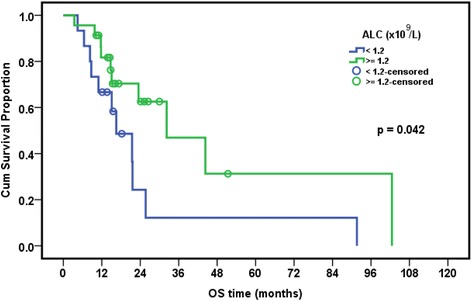
Relation of overall survival with absolute lymphocytic count in pediatric ALL patients
Fig. 5.
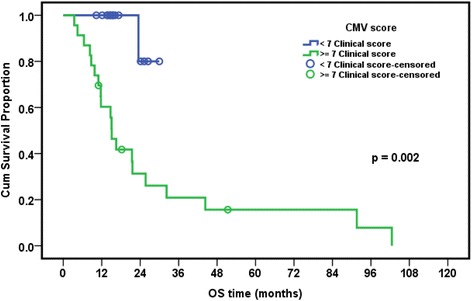
Relation of overall survival with severe CMV disease (score ≥7) in pediatric ALL patients
Fig. 6.
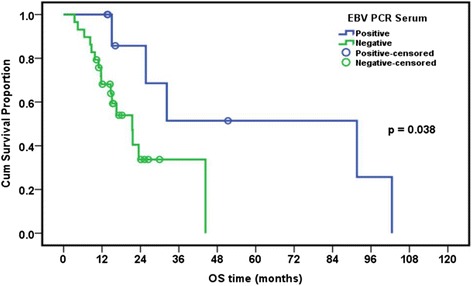
Relation of overall survival with presence of EBV DNA in serum of pediatric ALL patients
Discussion
In recent years, a causal relationship between EBV and a variety of childhood leukemia has been demonstrated. Our previous report showed the synergistic effect of HHV6 on CMV-associated infection resulting in severe outcome among pediatric lymphoma patients [19]. This encouraged us to investigate the frequency of EBV and CMV infections and their impact when existed together on the clinical course of leukemia disease. EBV DNA whether alone or with CMV was detected in the sera of 10/50 (20%) of our leukemia patients but not in control subjects. There are few reports in literature on the role of EBV in childhood leukemia. Sehgal and coworkers have detected EBV DNA in 8/25 (32%) of pediatric ALL patients by PCR amplification of BamH1-W EBV nuclear antigen (EBNA) promoter [23]. They have also observed the presence of EBV LMP1 gene transcripts in 29/80 (36.3%) Sudanese patients with leukemia disease, but not in healthy controls (p = 0.0001) [24]. Moreover, It has been reported that the presence of EBV DNA in 22.8% of pediatric ALL patients, but not in the control group [25].
In the present study, CMV DNA alone or combined with EBV DNA was detected in 27/50 (54%) of the sera of leukemic patients. Debaugnies and coworkers have detected EBV and CMV DNA in 19 and 27% in the whole blood samples from immunocompromised adult and pediatric patients respectively [26].
Regarding the detection of both herpesviruses in WBCs in the present study, EBV DNA was found in 23/50 (46%) while CMV DNA in 19/50 (38%) among leukemic patients. This was higher than that reported by Lu and coworkers who have detected EBV DNA in 8/35 (22.86%) of pediatric leukemia patients using fluorescent quantitative PCR (FQ-PCR) of peripheral blood mononuclear cells [25], whereas Bonon and coworkers have reported CMV DNA in 69.5% of WBCs samples taken from adult and pediatric immunocompromised patients using nested PCR assay [27].
In the current study, CMV/EBV co-infection was found in nine patients (18%), while only one patient had a single EBV DNA detected in his serum. Zawilińska and team work have reported infections with two or three viruses dominated particularly by CMV and EBV in 65% of adult ALL-HSCT recipients’ peripheral leukocytes using nested PCR method [28]. Earlier, Drouct and colleagues had briefly reported signs of CMV and EBV co-infections in graft recipients and suggested that CMV might encourage EBV replication and dissemination [29]. Moreover, Aalto and team work have suggested that CMV may induce EBV immune reactivation following stimulation of EBV-specific memory B-cells due to iterated EBV replication [30]. EBV may require another virus like CMV to increase immunosuppressive status and mediate a suitable environment for EBV reactivation as previously reported [30, 31].
Few studies have demonstrated the impact of both EBV and/or CMV infections on the clinical course of leukemia disease. Our results revealed that no significant association of age and sex with the presence of EBV and CMV DNA Conversely, Terrazzini and his colleagues observed that effect of CMV infection depends on age and sex [32]. In the current study, the presence of EBV DNA and or CMV DNA in serum was mostly associated with mucositis and low absolute neutrophil count (ANC <1.3 × 109/l) with borderline significance (p = 0.07). This apparently may suggest that the suppressive effect of EBV on some hematological parameters could be attributed to the presence of CMV DNA infection. However, we cannot rule out the role of leukemia treatment but could be a confounder factor that aggravated the suppressive effect of EBV or CMV. Moreover, the presence of herpesvirus-6 (HHV6) needs to be evaluated in the pediatric leukemic patients as a reactivation of HHV6 has been previously reported following treatment of Tunisian acute leukemia patients [33]. Furthermore, we have previously reported that the presence of HHV6 and CMV DNA in pediatric lymphoma patients using PCR was highly associated with more frequent episodes of febrile neutropenia, absolute neutrophil count (<0.8), lymphocytes (<0.5) and low hemoglobin level (<9.1)18 amongst pediatric lymphoma patients.
Severe CMV infection and active EBV infection were significantly associated with worse overall survival of leukemia patients (p = 0.002 and 0.038, respectively). EBV/CMV co-infection showed a borderline association with worse overall survival of patients (p = 0.079). These findings suggest that severe CMV infection and possibly EBV infection might play a role in the progression of leukemia and might be a useful indicator of worsening the clinical course of the disease. Similar findings were reported by Yoo and his colleagues on Korean pediatric umbilical cord blood transplant patients where the presence of CMV disease was among factors that adversely affected the survival rates of all patients [34].
Conclusion
CMV and EBV infections are rather common in pediatric leukemic patients. Qualitative PCR assay is a hallmark for detection of both EBV and CMV DNA among leukemic patients. These infections, especially severe CMV infection, were associated with worse overall survival. Further analysis is still required to evaluate the role of HHV6 when present with EBV and CMV on woresing the immune status of pediatric leukemic patients.
Acknowledgement
We are indebted to the virology unit, school of medicine in Kuwait for providing us with ATCC of CMV and EBV. We are also thankful to Wassim Chehadeh, Associate Professor in the Faculty of Medicine, Kuwait University, for his critical review of the manuscript.
Funding
The current research is funded by National Cancer Institute, Cairo University.
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Authors’ contributions
SA put plan and design of the study, supervised and edited manuscript. MAS supervised the work, edited and revised the manuscript. MF participated in the design of the study, supervised clinical part, evaluated the results and participated in editing clinical part of the study. MEW, carried out full clinical investigations, drafted the data in tables, edited clinical part and participated in revising the important intellectual contents. ShA, carried out the practical part, collected clinical and hematological data from patient’s sheets SK carried out the practical part. NF, carried out practical part and collected samples, collected clinical and hematological data from patient’s sheets, drafted the data in tables. MM and AE has performed the statistical analysis and explicitly reviewed and edited the whole manuscript including tables and graphs. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The research has been approved by the National Cancer Institute-Cairo University, IRB. IRB00004025. Organization No. IORG0003381.
Contributor Information
Samah Aly Loutfy, Phone: +201222840964, Email: samaly183@yahoo.com.
Maha A. Abo-Shadi, Email: m_a_shadi@hotmail.com
Mohamed Fawzy, Email: Amfawzyonc@hotmail.com.
Mohamed El-Wakil, Email: wakilmohamed@yahoo.com.
Shimaa A. Metwally, Email: shemoo88elawady@yahoo.com
Manar M. Moneer, Email: manar_moneer@yahoo.com
Nasra F. Abdel Fattah, Email: nasra_fathy87@yahoo.com.
Sara Kassem, Email: Sara_kaseem@yahoo.com.
Ahmed Elgebaly, Email: Ahmedelgebaly94@gmail.com.
References
- 1.Cohen J. Eptein-barr virus infection. N Engl J Med. 2000;343:481–92. doi: 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- 2.Loutfy SA, Alam El-Din HM, Ibrahim MF, Hafez MM. Seroprevalence of herpes simplex virus types 1 and 2, Epstein-Barr virus, and cytomegalovirus in children with acute lymphoblastic leukemia in Egypt. Saudi Med J. 2006;27:1139–45. [PubMed] [Google Scholar]
- 3.Wade JC. Viral infections in patients with hematological malignancies. Hematology Am Soc Hematol Educ Program. 2006;368–74. doi:10.1182/asheducation-2006.1.368. [DOI] [PubMed]
- 4.Mocarski ES, Shenk T. Cytomegalovirus. In: Knipe DMHP, editor. Fields virol. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2701–72. [Google Scholar]
- 5.Dolcetti R, Masucci MG. Epstein-Barr virus: Induction and control of cell transformation. J Cell Physiol. 2003:207–18. doi:10.1002/jcp.10263. [DOI] [PubMed]
- 6.Epstein MA, Henle G, Achong BG, Barr YM. Morphological and biological studies on a virus in cultured lymphoblasts from Burkitt’S lymphoma. J Exp Med [Internet] 1965;121:761–70. doi: 10.1084/jem.121.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keegan THM, Glaser SL, Clarke CA, Gulley ML, Craig FE, DiGiuseppe JA, et al. Epstein-Barr virus as a marker of survival after Hodgkin’s lymphoma: a population-based study. J Clin Oncol. 2005;23:7604–13. doi: 10.1200/JCO.2005.02.6310. [DOI] [PubMed] [Google Scholar]
- 8.Laytragoon-Lewin N, Chen F, Avila-Cariño J, Zou JZ, Mellstedt H, Ernberg I, et al. Epstein barr virus (EBV)-carrying cells of a chronic lymphocytic leukemia (CLL) subpopulation express EBNA1 and LMPS but not EBNA2 in vivo. Int J Cancer. 1995;63:486–90. doi: 10.1002/ijc.2910630404. [DOI] [PubMed] [Google Scholar]
- 9.Bonon SH, Menoni SM, Rossi CL, De Souza CA, Vigorito AC, Costa DB, et al. Surveillance of cytomegalovirus infection in haematopoietic stem cell transplantation patients. J Infect. 2005;50:130–7. doi: 10.1016/j.jinf.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Meerbach A, Wutzler P, Häfer R, Zintl F, Gruhn B. Monitoring of Epstein-Barr virus load after hematopoietic stem cell transplantation for early intervention in post-transplant lymphoproliferative disease. J Med Virol. 2008;80:441–54. doi: 10.1002/jmv.21096. [DOI] [PubMed] [Google Scholar]
- 11.Parasuraman R, Samarapungavan D, Venkat KK. Updated principles and clinical caveats in the management of infection in renal transplant recipients. Transplant Rev. 2010;24:43–51. doi: 10.1016/j.trre.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Fishman JA. Infection in solid-organ transplant recipients. N Engl J Med. 2007;357:2601–14. doi: 10.1056/NEJMra064928. [DOI] [PubMed] [Google Scholar]
- 13.Lehtinen M. Maternal Herpesvirus Infections and Risk of Acute Lymphoblastic Leukemia in the Offspring [Internet]. Am J Epidemiol. 2003. Available from: http://aje.oupjournals.org/cgi/doi/10.1093/aje/kwg137. [DOI] [PubMed]
- 14.Hess RD. Routine Epstein-Barr virus diagnostics from the laboratory perspective: still challenging after 35 years. J Clin Microbiol. 2004;42(8):3381–7. doi: 10.1128/JCM.42.8.3381-3387.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pizzo PA, Poplack DG. Principles and practice of pediatric oncology. Wolters Kluwer Health/Lippincott Williams & Wilkins; 2011.
- 16.Pui CH, Relling MV, Sandlund JT, Downing JR, Campana D, Evans WE. Rationale and design of total therapy study XV for newly diagnosed childhood acute lymphoblastic leukemia. Ann Hematol. 2004;83(Suppl 1):S124–6. doi: 10.1007/s00277-004-0850-2. [DOI] [PubMed] [Google Scholar]
- 17.Plotkin SA, Starr SE, Friedman HM, Brayman K, Harris S, Jackson S, et al. Effect of Towne live virus vaccine on cytomegalovirus disease after renal transplant: A controlled trial. Ann Intern Med. 1991;114:525–31. doi: 10.7326/0003-4819-114-7-525. [DOI] [PubMed] [Google Scholar]
- 18.Van Der Bij W, Torensma R, Van Son WJ, Anema J, Schirm J, Tegzess AM, et al. Rapid immunodiagnosis of active cytomegalovirus infection by monoclonal antibody staining of blood leucocytes. J Med Virol. 1988;25:179–88. doi: 10.1002/jmv.1890250208. [DOI] [PubMed] [Google Scholar]
- 19.Loutfy SA, Fawzy M, El-Wakil M, Moneer MM. Presence of human herpes virus 6 (HHV6) in pediatric lymphomas: impact on clinical course and association with cytomegalovirus infection. Virol J. 2010;7:287. doi: 10.1186/1743-422X-7-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohamed WS, Loutfy SA, Rashed RA, ElAzhary NM, Amer MM. Prospective study for CMV detection in cancer patients: comparison between PCR, antigenemia, and serological assays. Life Sci J. 2012;9:4536–41. [Google Scholar]
- 21.Hashimoto YE. Investigation of EB VIRUS and cytomegalovirus in rapidly progressive interstetial pneumonitis in polymyositis/dermatomyositis by in situ hyberidization and polymerase chain reaction. Clin Immunol immuno Pathol. 1995;77:298–306. doi: 10.1006/clin.1995.1156. [DOI] [PubMed] [Google Scholar]
- 22.Madi N, Al-Nakib W, Pacsa A. Does cytomegalovirus develop resistance following antiviral prophylaxis and treatment in renal transplant patients in Kuwait? Adv Virol. 2011;2011:1–6. doi: 10.1155/2011/260561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sehgal S, Mujtaba S, Gupta D, Aggarwal R, Marwaha RK. High incidence of Epstein Barr virus infection in childhood acute lymphocytic leukemia: a preliminary study. Indian J Pathol Microbiol [Internet] 2010;53:63–7. doi: 10.4103/0377-4929.59186. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed HG, Osman SI, Ashankyty IM. Incidence of Epstein-Barr virus in pediatric leukemia in the Sudan. Clin Lymphoma Myeloma Leuk. 2012;12:127–31. doi: 10.1016/j.clml.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Lu Y, Sun LR, Pang XY, Lu ZH, Sui AH. Infection status and clinical significance of Epstein-Barr virus in pediatric leukemia---a report of 35 cases. Chin J Cancer. 2007;26:54–7. [PubMed] [Google Scholar]
- 26.Debaugnies F, Busson L, Ferster A, Lewalle P, Azzi N, Aoun M, et al. Detection of Herpesviridae in whole blood by multiplex PCR DNA-based microarray analysis after hematopoietic stem cell transplantation. J Clin Microbiol. 2014;52:2552–6. doi: 10.1128/JCM.00061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonon CSHA, Ross CL, de Souza CA, Vigorito AC, Costa SCB. Comparison of serology, antigenemia assay and the polymerase chain reaction for monitoring active cytomegalovirus infections in hematopoietic stem cell transplantation patients. Virology. 2006;48. [DOI] [PubMed]
- 28.Zawilińska B, Kosz-Vnenchak M, Piatkowska-Jakubas B, Kopeć J, Daszkiewicz E, Skotnicki AB. Herpesviruses mixed infections in allogeneic steam cell recipients (allo-HSCT) Przegl Epidemiol. 2008;62:39–46. [PubMed] [Google Scholar]
- 29.Drouct E, Garnier JL, Chapuis-Cellier C, Touraine JL. Double CMV and EBV infections are associated with monoclonal immunoglobulins in patients after renal transplantation. Proc. Sixth Int. Cytomegalovirus Work. Alabama; 1997.
- 30.Aalto SM, Linnavuori K, Peltola H, Vuori E, Weissbrich B, Schubert J, et al. Immunoreactivation of Epstein-Barr virus due to cytomegalovirus primary infection. J Med Virol. 1998;56:186–91. doi: 10.1002/(SICI)1096-9071(199811)56:3<186::AID-JMV2>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 31.Tselis A. Epstein–Barr Virus and Cytomegalovirus Infections. Viral Infect. Hum. Nerv. Syst. [Internet]. Basel: Springer Basel; 2013 [cited 2017 Jan 19]. p. 23–46. Available from: http://link.springer.com/10.1007/978-3-0348-0425-7_2
- 32.Terrazzini N, Bajwa M, Vita S, Cheek E, Thomas D, Seddiki N, et al. A novel cytomegalovirus-induced regulatory- type T-cell subset increases in size during older life and links virus-speci fi c immunity to vascular pathology. J Infect Dis. 2014;209:382–92. doi: 10.1093/infdis/jit576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nefzi F, Ben Salem NA, Khelif A, Feki S, Aouni M, Gautheret-Dejean A. Quantitative analysis of human herpesvirus-6 and human cytomegalovirus in blood and saliva from patients with acute leukemia. J Med Virol [Internet] 2015;87:451–60. doi: 10.1002/jmv.24059. [DOI] [PubMed] [Google Scholar]
- 34.Yoo KH, Lee SH, Sung KW, Koo HH, Chung NG, Cho B, et al. Current status of pediatric umbilical cord blood transplantation in Korea: a multicenter retrospective analysis of 236 cases. Am J Hematol. 2011;86:12–7. doi: 10.1002/ajh.21886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.


