Abstract
GATA-1 is essential for the development of erythroid and megakaryocytic lineages. We found that GATA-1 gene knockdown female (GATA-1.05/X) mice frequently develop a hematopoietic disorder resembling myelodysplastic syndrome that is characterized by the accumulation of progenitors expressing low levels of GATA-1. In this study, we demonstrate that GATA-1.05/X mice suffer from two distinct types of acute leukemia, an early-onset c-Kit-positive nonlymphoid leukemia and a late-onset B-lymphocytic leukemia. Since GATA-1 is an X chromosome gene, two types of hematopoietic cells reside within heterozygous GATA-1 knockdown mice, bearing either an active wild-type GATA-1 allele or an active mutant GATA-1.05 allele. In the hematopoietic progenitors with the latter allele, low-level GATA-1 expression is sufficient to support survival and proliferation but not differentiation, leading to the accumulation of progenitors that are easily targeted by oncogenic stimuli. Since such leukemia has not been observed in GATA-1-null/X mutant mice, we conclude that the residual GATA-1 activity in the knockdown mice contributes to the development of the malignancy. This de novo model recapitulates the acute crisis found in preleukemic conditions in humans.
GATA-1 belongs to a small family of transcription factors, members of which bind to the consensus WGATAR sequence utilizing two phylogenetically conserved zinc fingers (44). Of the six members of the family, GATA-1, GATA-2, and GATA-3 comprise the hematopoietic GATA factor subfamily (3, 24, 29). Expression profiles of the hematopoietic subfamily members are lineage and differentiation stage restricted; in the hematopoietic lineages, the expression of GATA-1 is confined to erythroid cells, megakaryocytes, eosinophils, and mast cells, whereas GATA-2 is expressed mainly in hematopoietic stem cells and immature progenitors (8, 18, 34). GATA-3 is predominantly expressed in T lymphocytes. Although GATA-1 and GATA-2 show overlapping expression profiles, gene knockout experiments revealed that both factors play distinct roles, probably resulting from their differential spatiotemporal expression patterns (8, 35, 37, 38).
The GATA-1 gene has two first exons (11). The distal (IT) and proximal (IE) first exons are utilized mainly for testis- and hematopoiesis-specific transcription of the GATA-1 gene, respectively (11, 27). GATA-1 gene transcription in hematopoietic cells requires several cis-acting regions within the GATA-1 gene hematopoietic regulatory domain (G1-HRD) (23, 25). In primitive erythroid cells, gene transcription requires the GATA-1 gene hematopoietic enhancer, double-GATA motif, and CACCC motif. In definitive erythroid cells, GATA-1 gene transcription requires another element in the first intron, in addition to the three regions in the G1-HRD (23, 28, 45).
Analyses of loss-of-function and gain-of-function mutant animals and cell lines have revealed several critical roles for GATA-1. Dysfunction or overproduction of GATA-1 affects the differentiation, survival, and proliferation of the erythroid and megakaryocytic progenitors (5, 42). Thus, the expression level of GATA-1 is a critical determinant of the function of GATA-1 in vivo. For instance, we have generated a GATA-1 gene knockdown allele using a promoter interference approach, and this allele shows attenuated GATA-1 gene expression at approximately 5% of the normal level. We therefore refer to this allele as GATA-1.05 (35). This level of GATA-1 expression is insufficient to support erythropoiesis during embryonic development. In contrast, approximately 20% of the normal level of expression of GATA-1 is sufficient to support erythroid cell maturation (16).
Because the GATA-1 gene is located on the X chromosome, male embryos hemizygous for the knockdown mutation (GATA-1.05/Y) are defective in primitive erythropoiesis and do not survive beyond 12.5 embryonic days (E12.5). Female mice heterozygous for the mutation (GATA-1.05/X) can survive, owing to random inactivation of the X chromosome bearing the GATA-1.05 mutant allele. Indeed, various degrees of anemia and thrombocytopenia are observed in heterozygous female mice during the neonatal period, most likely depending on the relative proportion of cells with an active GATA-1.05 allele. These defects usually improve before adolescence, and the fertility of GATA-1.05/X mice is normal (35). However, we found that GATA-1.05/X mice suffer from a hematopoietic disorder which has similarity to human myelodysplastic syndrome (MDS) (36). Previously, we could not delineate whether the disorder develops into overt leukemia or not. Furthermore, mechanisms linking GATA-1 deficiency to MDS also remain to be clarified.
Children with Down's syndrome (DS) show a high incidence of acute megakaryocytic leukemia (AMKL) and transient myeloproliferative disorder (TMD). TMD is a leukemoid reaction occurring in newborn infants with DS which is characterized by the rapid growth of abnormal blast cells expressing megakaryocytic markers (13, 46). Although the majority of TMD cases resolve spontaneously, DS-AMKL develops in approximately 30% of TMD patients in the first four years of life, often preceded by a myelodysplastic phase. Recently, a high incidence of acquired mutations in the GATA-1 gene was found in individuals with DS-AMKL and TMD (9, 10, 40, 43). In each, the mutation results in the introduction of a premature stop codon in the gene sequence encoding the N-terminal activation domain and a lack of expression of the 50-kDa full-length GATA-1 protein. Instead, an alternative 40-kDa translation product is expressed from a downstream initiation site, which retains the intact zinc fingers and binds appropriately to the GATA consensus sequence. The N-terminal domain of GATA-1 appears to be indispensable for erythroid cell differentiation in vivo (32). These results suggest that the GATA-1 mutant protein may contribute to the expansion of TMD blast cells, and along with other genetic changes, the mutant GATA-1 protein may also contribute to the development of DS-AMKL.
In this study, we have characterized the nature of the hematopoietic disorder in GATA-1.05/X mice in detail. We report results with two germ line GATA-1 mutants, the GATA-1.05 (knockdown) and GATA-1-null (knockout) mouse models. An important observation is that these two GATA-1 mutant lines bear different sensitivities to leukemogenesis. A cohort study with more than 500 GATA-1.05/X mice unequivocally demonstrated that the average life span of these mice is markedly shortened compared to that of control animals. This is due to a very high incidence of two types of leukemias, c-Kit+ nonlymphoid leukemia and CD19+ B-lymphoid leukemia. We also aimed to elucidate how the impairment of erythropoiesis and megakaryopoiesis are related to the development of leukemia in the GATA-1 knockdown mice. The results indicate that the leukemias are most likely caused by decreased expression of GATA-1. Remarkably, however, the presence of a small amount of GATA-1 is critical for the process. Taken together, these results demonstrate that GATA-1 orchestrates differentiation, proliferation, and survival of the erythroid cells in vivo. As each function requires a different amount of GATA-1, the GATA-1.05 knockdown provokes an imbalance of GATA-1 function and establishes the grounds for leukemogenesis. Thus, the GATA-1.05 line of mice is a useful animal model for the analysis of leukemias arising from MDS.
MATERIALS AND METHODS
Mice.
GATA-1.05/X mice were prepared at the University of Tsukuba (35). GATA-1.05/X mice (n = 574) and reporter transgenic mice (n = 225) utilized in this study were born from 1 December 1996 to 31 July 2002. GATA-1-null mouse lines were prepared at the Erasmus Medical Center, Rotterdam, The Netherlands (14). GATA-1-null/X mice were born from 14 December 2001 to 29 April 2003. Survival time was measured from time of birth to the end of follow-up. Mice were bred in a clean animal room at the Center for Tsukuba Advanced Research Alliance, University of Tsukuba. The GATA-1.05 allele and GATA-1-null allele were genotyped by PCR using a pair of primers corresponding to the neomycin resistance gene in the original targeting vector (14, 35). All mice were treated according to the regulations of the Standards for Humane Care and Use of Laboratory Animals (University of Tsukuba).
Histological analysis.
Tissue samples were fixed in 10% buffered formalin and embedded in paraffin. Thin sections were stained with hematoxylin and eosin for histological examination. Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining was performed by using an in situ apoptosis detection kit (TaKaRa). The fluorescein isothiocyanate-labeled DNA fragments were visualized using anti-fluorescein isothiocyanate peroxidase antibody solution, followed by staining with diaminobenzidine (WAKO). Cells were counterstained with 0.5% hematoxylin.
Immunophenotypic analysis.
For fluorescence-activated cell sorting (FACS) analysis, mononuclear cells of hematopoietic tissues of GATA-1.05/X mice and recipient nude mice (CLEA Japan) were stained with phycoerythrin-conjugated anti-CD71, -Ter119, and -CD19 antibodies (BD PharMingen), and allophycoerythrin-conjugated anti-c-Kit antibody (BD PharMingen). The cells were analyzed with a FACSCalibur (Becton Dickinson). c-Kit+ cells were sorted by the Vantage FACS (Becton Dickinson). Dead cells were excluded by propidium iodide.
Immunoglobulin locus rearrangement.
High-molecular-weight DNA was isolated from spleen cells of GATA-1.05/X and recipient nude mice. Fifteen micrograms of DNA was digested with EcoRI, size separated in 0.8% agarose gels, and transferred to Zeta-Probe nylon membranes (Bio-Rad). Immunoglobulin gene configuration was analyzed with the mJH4 probe.
Cell culture.
A total of 500 fluorescence-activated cell-sorted c-Kit+ cells were plated on OP9 cells (20) in the presence of erythropoietin (Epo; 2 U/ml) and stem cell factor (SCF; 100 ng/ml). Cells were trypsinized, and stromal cells were eliminated after passing the suspensions through a G10 column (Amersham Pharmacia Biotech). For the differentiation of c-Kit+ cells, a total of 500 cells were also plated on OP9 cells or FLS-5 cells (26) in the presence of Epo (2 U/ml) or on YSCL-71 cells (7) in the presence of Epo (2 U/ml) and SCF (100 ng/ml). Nonadherent cells were obtained after passing the suspensions through a G10 column. For morphological examination, cytospin samples were stained with Wright-Giemsa solution. E14 embryonic stem (ES) cells were maintained with embryonic fibroblast cells and kept undifferentiated in the presence of recombinant leukemia inhibitory factor (1,000 U/ml; ESGRO; Chemicon International). ES cells were differentiated in vitro by following the method reported previously (33). Floating cells observed on day 11, which represent definitive-stage erythroid cells, were analyzed in this study.
RT-PCR analysis.
Total RNA was prepared with the RNA-sol extraction system (Tel-Test) from sorted c-Kit+ cells. cDNAs were synthesized with Superscript reverse transcriptase (RT) (Life Technologies). For semiquantitative RT-PCR analysis, the amount of cDNA was adjusted by dilution, giving rise to an equivalent amount of the endogenous hypoxanthine guanine phosphoribosyltransferase amplicon. Sequences of the primers used were 5′-GCTGAATCCTCTGCATCAAC and 5′-TAGGCCTCAGCTTCTCTGTA for erythroid cell-specific GATA-1, 5′-GCAACACACCACCCGATACC and 5′-CAATTTGCACAACAGGTGCCC for GATA-2, and 5′-GCTGGTGAAAAGGACCTCT and 5′-CACAGGACTAGAACACCTGC for hypoxanthine guanine phosphoribosyltransferase.
RESULTS
Survival of GATA-1.05/X mice.
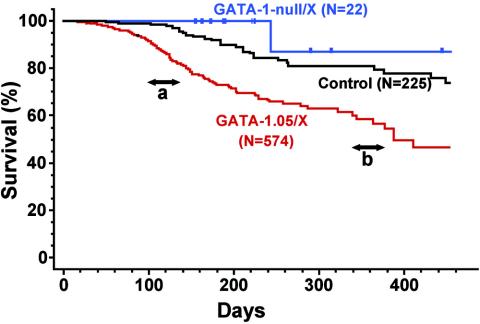
We previously found that female mice with the GATA-1.05/X genotype develop a hematological disorder closely resembling MDS that includes anemia, thrombocytopenia, and accumulation of immature cells in their hematopoietic organs (36). To investigate whether these mice eventually suffered from overt leukemia, we propagated a large number of GATA-1.05/X mice and conducted a cohort study exploiting a line of reporter gene transgenic mice as a control. We examined clinical courses of 574 GATA-1.05/X mice, 225 control mice, and 21 GATA-1-null/X mice. The results of Kaplan-Meier analyses clearly show that the control group of mice lived longer than the GATA-1.05/X mice (Fig. 1). The average survival times of control mice and GATA-1-null/X mice (see below) are comparable to those of other mice in our facility (data not shown). In contrast, the GATA-1.05/X group of mice was prone to early mortality and the mean survival time of this group of mice was 388 days. This result shows that the presence of the GATA-1.05 allele is a strong predictor of overall survival time (P < 0.0001).
FIG. 1.
Statistical evaluation of GATA-1.05/X mice. Kaplan-Meier curves of the overall lengths of survival of 574 GATA-1.05/X (red line), 21 GATA-1-null/X (blue line), and 225 control (black line) mice are shown. The median overall length of survival of GATA-1.05/X mice is 388 days. a and b show the two peaks of mortality of GATA-1.05/X mice.
The age distribution of mortality appeared to be bimodal, with a distinct peak at around 100 days and a second peak slightly after 350 days (Fig. 1). Full autopsy data were available for 98 dead GATA-1.05/X mice. Of the 98 mice, we found that 62 (63.3%) mice had severely enlarged spleens of more than 0.5 g and that 36 (36.7%) mice had moderately enlarged spleens with an average mass of 0.25 ± 0.08 g. The average spleen weight of control mice was 0.12 ± 0.03 g (n = 10), demonstrating that the GATA-1.05/X mice display a high incidence of splenomegaly (P < 0.0001).
Hematological and physiological analysis of GATA-1.05/X mice.
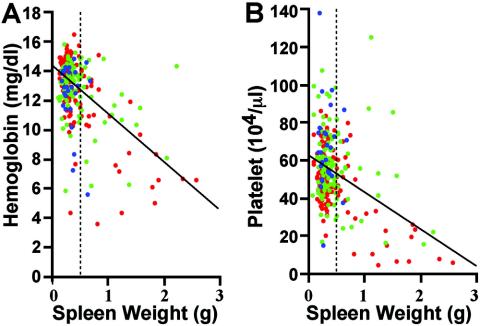
We then randomly selected 265 live adult GATA-1.05/X mice (49 to 449 days old) and analyzed their physical appearance and hematological indices. We found a highly significant negative correlation between the spleen weight and hemoglobin content (Fig. 2A) (P < 0.0001) and platelet count (Fig. 2B) (P < 0.0001). In contrast, the spleen weight showed no significant correlation with the age of the mice. The red, green, and blue spots in Fig. 2 represent 131 young (49 to 149 days old), 107 middle-aged (150 to 299 days old), and 27 senescent (more than 300 days old) mice, respectively. Grossly enlarged spleens (more than 0.5 g) (Fig. 2) were found in 65 mice (24.5%). Of the 65 mice, 48 mice had increased white blood cell numbers and/or the appearance of abnormal cells in their blood smears, in addition to severe anemia and thrombocytopenia (data not shown).
FIG. 2.
Correlation between spleen weight and hemoglobin content or platelet counts in GATA-1.05/X mice. After we screened both hemoglobin and platelet counts from 265 live GATA-1.05/X mice, the spleens were removed and their weights were measured. Red spots indicate 131 mice that were 49 to 149 days old, green spots indicate 107 mice that were 150 to 299 days old, and blue spots indicate 27 mice that were more than 300 days old. Simple regression equations demonstrate linkages between spleen weight (Sp) and hemoglobin content (Hb) as follows: Hb = −3.1Sp + 14.2 (P < 0.0001) (A) and spleen weight and platelet counts (Plt) (Plt = −16.7Sp + 63.2 (P < 0.0001) (B). The regression lines are superimposed in the figures. Note that spleen weight was not related to the age of the mice. Dashed line, grossly enlarged spleens.
The remaining 200 mice had various degrees of spleen enlargement, but spleen weights did not exceed 0.5 g. These mice developed thrombocytopenia. The average hemoglobin concentration and platelet count (± standard deviation) were 13.3 ± 1.7 g/dl and (56.9 ± 16.7) × 104/μl (13.6 ± 0.5 g/dl and 97.9 ± 20.5/μl in wild-type mice), respectively. Histological examination of the enlarged spleens revealed an increase of both proerythroblasts and megakaryocytes within the extended red pulp area (data not shown). These features were completely different from the appearance of the severely enlarged spleens (see below). Based on these observations, we conclude that live GATA-1.05/X mice may be categorized into two groups. The first group with mild splenomegaly displays a myelodysplastic condition with erythroid and megakaryocytic hyperplasia, while the second group with severe splenomegaly suffers from more-advanced hematopoietic dysfunction.
GATA-1.05/X mice are predisposed to leukemias of two different lineages.
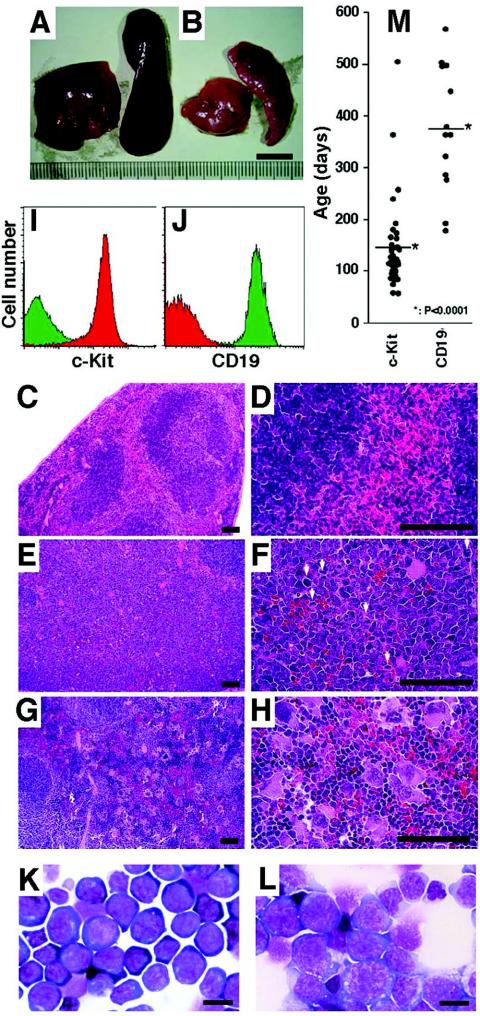
In the necropsy analysis, we noticed that GATA-1.05/X mice in the second group could be subdivided further into two types. Type 1 mice had uniformly enlarged spleens and livers of a dark-red color (Fig. 3A), whereas type 2 mice had pale spleens and livers, with multiple nodules (Fig. 3B) and systemic lymphoadenopathy. Histological examination of the type 1 mouse spleens revealed that these mice contain markedly enlarged red pulp areas, occupied by mononuclear cells. The architecture of the spleens was substantially destroyed (Fig. 3E; Fig. 3C shows results for the wild-type control). An expansion of heterochromatic immature cells and an increase in the number of mitotic figures were observed at higher magnifications (Fig. 3F). In contrast, the architecture of the enlarged type 2 mouse spleens was relatively preserved (Fig. 3G). We found an increase in the number of large megakaryocytes in the red pulp of these spleens (Fig. 3G and H).
FIG. 3.
Differences between type 1 and type 2 GATA-1.05/X splenomegaly. (A and B) Enlarged spleens and livers removed from type 1 (A) or type 2 (B) GATA-1.05/X mice. (C to H) Histological analyses of the type 1 and type 2 mouse spleens with hematoxylin-eosin staining. Both red and white pulps are observed in the normal control spleen (C and D), but these structures are destroyed in both type 1 (E and F) and type 2 (G and H) enlarged spleens. Whereas proliferating cells with a blast-like appearance are seen in the enlarged type 1 spleen (E and F), megakaryocytes are increased in number with lymphocyte infiltration in the red pulp of the type 2 enlarged spleen (G and H). Mitotic cells are observed in type 1 splenomegaly (white arrows in panel F). Scale bars, 50 μm. (I and J) Typical flow cytometry analysis of mononuclear cells from the enlarged spleens of type 1 (I) and type 2 (J) leukemic mice. Red and green histograms show mononuclear cells positive for c-Kit and CD19 antibody, respectively. (K and L) Wright-Giemsa staining for a cytospin specimen of mononuclear cells from type 1 (K) or type 2 (L) mouse spleens. Blast-like cells are increased in number in both types of spleens. The morphologies of these blast-like cells are completely different between type 1 and type 2 spleens. Scale bars, 10 μm. (M) Ages of onset of type 1 and type 2 leukemias. The average age of onset of type 1 leukemia was 143 ± 82 days, and that of type 2 leukemia was 387 ± 128 days (*, P < 0.0001).
To examine characteristics of the hematopoietic cells expanded in the spleen, we analyzed the expression profiles of surface markers on splenic mononuclear cells. For this purpose, we euthanized a number of aged GATA-1.05/X mice. Among those with severely enlarged spleens, we found 40 spleens with a type 1 appearance and 14 spleens with a type 2 appearance. To our surprise, while c-Kit+/CD19− cells were the predominant cell type expanded in the type 1 spleens (Fig. 3I, red fraction), c-Kit−/CD19+ cells were predominant in the type 2 spleens (Fig. 3J, green fraction), suggesting that progenitors of the B-cell lineage were expanded in the spleens of type 2 mice. The correlation between surface marker profiles and the gross morphological appearance of the spleens was highly consistent throughout the analysis.
Hematopoietic indices of type 1 mice and type 2 mice are shown in Table 1. All mice exhibited anemia and thrombocytopenia compared to control mice. Especially, type 1 mice suffered from macrocytic anemia. Morphological analysis showed that c-Kit+ cells resembled proerythroblasts (Fig. 3K) and that CD19+ cells showed anaplastic morphology with monomorphic round nuclei and prominent nucleoli (Fig. 3L). Thus, these analyses of primary expanded spleen cells demonstrate that GATA-1.05/X mice are predisposed to leukemias of c-Kit+ nonlymphoid and CD19+ B-lymphoid lineages.
TABLE 1.
Hematopoietic indices of GATA-1.05/X mutant micea
| Mouse | No. of mice analyzed | Red blood cell count (104/μl) | Hematocrit (%) | Hemoglobin (g/dl) | MCV (fl) | MCH (pg) | MCHC (g/dl) | Platelet count (104/μl) | White blood cell count (102/μl) |
|---|---|---|---|---|---|---|---|---|---|
| Control | 7 | 925 ± 35 | 45.0 ± 2.2 | 13.6 ± 0.5 | 48.7 ± 2.2 | 14.7 ± 0.6 | 30.2 ± 1.0 | 97.9 ± 20.5 | 71 ± 28 |
| c-Kit+ type | 19 | 424 ± 201*** | 25.5 ± 8.6*** | 8.1 ± 2.4*** | 65.3 ± 14.1††† | 21.0 ± 4.6†† | 32.4 ± 3.8 | 26.7 ± 15.8*** | 397 ± 318† |
| B-cell type | 4 | 535 ± 220†† | 29.5 ± 10.0†† | 9.0 ± 2.8†† | 56.3 ± 4.0 | 17.3 ± 2.1† | 30.6 ± 2.9 | 46.5 ± 30.2* | 298 ± 417 |
†, P < 0.05; ††, P < 0.005; †††, P < 0.0005; *, P < 0.01; **, P < 0.001; ***, P < 0.0001. MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular concentration.
Ages of onset of c-Kit+ and CD19+ leukemias.
It is noteworthy that the ages of onset of leukemia are markedly different between the mice with nonlymphoid leukemia and those with B-cell leukemia. The average age of onset of mice with c-Kit+/CD19− leukemia is 143 days after birth, whereas that of mice with c-Kit−/CD19+ leukemia is 387 days (Fig. 3M). This difference is statistically significant (P < 0.0001). Importantly, these onset times correlate very well with the bimodal mortality peaks shown in Fig. 1. We surmise that GATA-1.05/X mice may contain hematopoietic cells in the myelodysplastic stage, with proliferating cells of the erythroid and megakaryocytic lineages. A number of mice may progress to an advanced stage with clonal proliferation of c-Kit+ cells during their early adulthood age (100 to 200 days after birth), and these mice die due to leukemia. However, some mice escape from this crisis but develop a monoclonal proliferation of CD19+ cells after 300 days or later, and these mice die due to leukemia with systemic lymphoadenopathy. In this respect, the leukemic phenotype of GATA-1.05/X mice appears to mirror the leukemic transformation process observed in human patients suffering from MDS.
Autonomous proliferation of GATA-1.05/X spleen cells in nude mice.
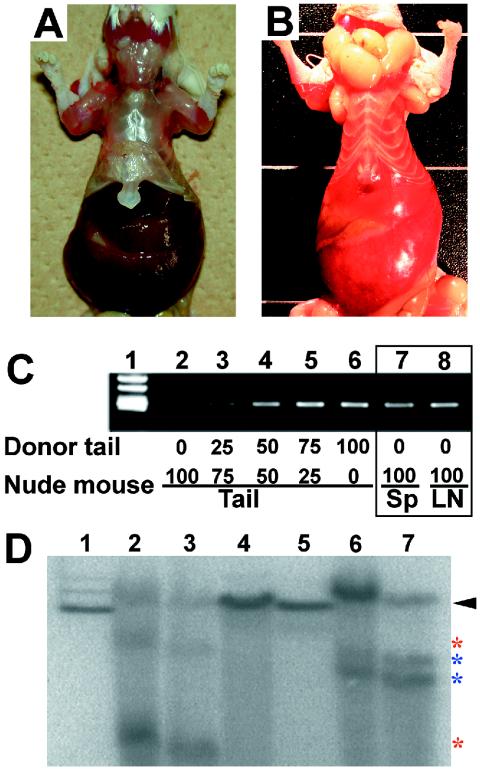
To ascertain whether the c-Kit+ and CD19+ spleen cells proliferate autonomously, we isolated mononuclear cells from the spleens of nine type 1 and six type 2 mice and injected these cells into three different nude mice via the tail vein (106 cells/mouse). All recipient nude mice developed giant splenomegaly within 2 months after transplantation; typical examples are shown in Fig. 4A and B. The recipient mouse reproducibly showed a phenotype closely resembling that of the donor mice; nude mice transplanted with type 1 cells contained enlarged dark-red spleens and livers (Fig. 4A), whereas nude mice transplanted with type 2 cells showed hepatosplenomegaly with multiple nodules and systemic lymphoadenopathy (Fig. 4B). Recipient mice died due to the burden of leukemia, irrespective of the transplanted cell types.
FIG. 4.
Autonomous proliferation of GATA-1.05 spleen cells in recipient nude mice. (A and B) Phenotype of nude mice transplanted with type 1 (A) or type 2 (B) spleen cells. The gross morphological appearance of livers and spleens of the recipient nude mice is similar to that of the donors (Fig. 2). (C) neo gene products obtained by PCR amplification of DNA extracted from nude mice. The amount of donor tail DNA was adjusted by dilution of recipient tail DNA to give a standard curve (lanes 1 to 5). The neo gene was detected in both spleen (Sp) and lymph node (LN) DNAs of recipient nude mice (lanes 6 and 7). (D) Southern blot analyses of spleen cells of donor and recipient mice with the JH4 probe after digestion with EcoRI. Lane 1 contains ES cell DNA showing a germ line band (arrowhead). Lanes 2, 4, and 6 are independent samples from a type 2 enlarged spleen. Lanes 3, 5, and 7 are samples from recipient nude mice transplanted with type 2 spleen cells corresponding to those shown in lanes 2, 4, and 6. Red and blue asterisks show rearranged bands in the immunoglobulin heavy-chain gene.
Flow cytometry analysis further revealed that the mononuclear cells in the spleens of recipient nude mice expressed the same surface markers as the transplanted donor cells. Type 1 cells gave rise to a c-Kit+ CD71+ Ter119−/dull CD19− profile, while type 2 cells gave rise to a c-Kit− Sca-1+ CD43+ CD19+ profile (data not shown). Moreover, the spleen mononuclear cells from the recipient nude mice were successfully transplanted into the next generation of nude mice, maintaining their primary phenotype (data not shown). We therefore conclude that GATA-1.05/X mice develop autonomously proliferating cells of two distinct lineages in their hematopoietic tissues.
To verify that the proliferating cells in the recipient mice are truly derived from the donor GATA-1.05/X mice, we examined the presence of the neomycin resistance (neo) gene in the cells using semiquantitative PCR. Since the neo gene exists only in the genomic DNA of donor cells and their progeny, detection of the neo gene in recipient mouse tissues should verify the expansion of the donor-derived cells. Indeed, a neo gene amplicon was found exclusively in the tail DNA of donor mice but not in the recipient mouse DNA (Fig. 4C, lane 2). In contrast, the neo gene was detected in DNA isolated from the spleens and lymph nodes of recipient nude mice (lanes 7 and 8, respectively), indicating that the leukemic cells are derived from donor cells. More than 75% of the cells in the spleens and lymph nodes of recipient nude mice were composed of GATA-1.05/X mouse-derived cells (Fig. 4C, compare lanes 4 and 5 to lanes 7 and 8).
To assess the clonality of leukemic cells, we utilized rearrangements in the immunoglobulin heavy-chain locus within the CD19+ cell population. A Southern blot hybridization analysis was carried out using the mJH4 probe (Fig. 4D). We extracted DNA from three type 2 donor mouse spleens and three recipient nude mouse spleens. Lane 1 of Fig. 4D shows a control germ line band. Immunoglobulin rearrangement was found in two out of three CD19+ type 2 spleens (lanes 2 and 6) but not in the third spleen (lane 4). The same configurations were found in the corresponding nude mouse spleens (lanes 3 and 7; compare to lanes 2 and 6, respectively). These results support our contention that leukemic cells committed to the B-lymphoid lineage autonomously proliferate in type 2 mice. We could not detect rearranged immunoglobulin bands in both donor and recipient spleen cells from one type 2 mouse, suggesting that the cells had been transformed in an immature stage before the rearrangement of the immunoglobulin locus. Collectively, we conclude that GATA-1.05/X mice develop either c-Kit+ nonlymphoid leukemia or pro-B-cell leukemia.
c-Kit+ leukemia cells have the potential to differentiate into multilineages.
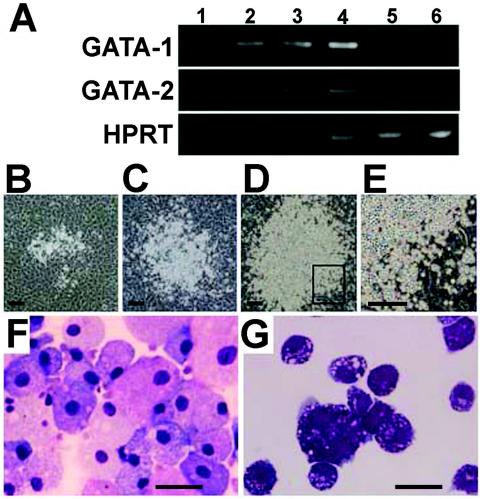
To examine the mechanisms of leukemogenesis in the GATA-1.05/X mice, we focused on the c-Kit+ leukemic cells. We collected c-Kit+ cells from spleens of GATA-1.05/X mice in either the preleukemic stage or the c-Kit+ leukemic stage by FACS. The mononuclear cells accumulating in the severely enlarged spleens contain an active GATA-1.05 allele and an inactive wild-type GATA-1 allele, indicating that the leukemic cells are derived from GATA-1-deficient cells. To verify this notion, we analyzed the expression levels of mRNAs for GATA-1 and GATA-2 in the c-Kit+ cells by RT-PCR (Fig. 5A). The expression of GATA-2 was upregulated in immature cells of GATA-1.05/X mice in both the preleukemic and leukemic stages. In contrast, the expression of GATA-1 was severely repressed in the c-Kit+ leukemia cells (lanes 5 and 6). In the preleukemic stage, however, GATA-1 was expressed at rather higher levels than in the wild-type mice (lanes 3 and 4). This finding suggests that immature erythroid cells with a wild-type GATA-1 allele were expanded in these mice as part of the compensatory response to the anemia. Since GATA-1 expression is known to peak in proerythroblasts (1, 34), this would result in increased GATA-1 mRNA levels in the preleukemic spleens.
FIG. 5.
Properties of c-Kit-positive leukemic cells. (A) Expression patterns of GATA-1 and GATA-2 in c-Kit+ cells of wild-type mice (lanes 1 and 2) and GATA-1.05/X mice in either the stable stage (lanes 3 and 4) or leukemia stage (lanes 5 and 6) are shown. Semiquantitative RT-PCR was performed by means of changing the amplification cycles. HPRT, hypoxanthine phosphoribosyltransferase. (B to E). Appearance of fast expansion of c-Kit+ leukemia cells on the OP9 stromal cell layer. Typical colonies derived from single c-Kit+ leukemia cells at day 3 (B), day 5 (C), and day 6 (D) of coculture with OP9 are shown. (E) Colony grown adhering to OP9 stromal cells. Scale bar, 250 μm. (F and G) Morphological changes of c-Kit+ leukemia cells by the induction of differentiation. c-Kit+ leukemia cells differentiate into macrophages (F) or mast cells (G) on FLS-5 and YSCL-71 stroma cells, respectively. Scale bar, 20 μm.
To further characterize the c-Kit+ leukemia cells, the cells were induced to differentiate in vitro by using three stromal cell lines, OP9, FLS-5, and YSCL-71, which were established from macrophage colony-stimulating factor-deficient cells, fetal liver stromal cells, and yolk sac endothelial cells, respectively. These stromal cell lines provide hematopoietic environments for the proliferation and/or differentiation of hematopoietic progenitors (7, 20, 26). We sorted c-Kit+ cells from type 1 mouse spleens and cocultured them with OP9 or YSCL-71 cells in the presence of Epo and SCF or with FLS-5 cells in the presence of Epo. Colonies from single c-Kit+ leukemia cells appeared 3 days after the start of incubation with OP9 cells (Fig. 5B), and the cells in individual colonies proliferated rapidly (Fig. 5C and D) with a Lin− c-Kit+ phenotype (data not shown). The high-magnification photograph of the boxed area in Fig. 5D shows the adherence of immature cells to the stromal cells (Fig. 5E). This adherence is consistent with the previous finding that c-Kit is important for the proliferation of hematopoietic stem cells through their adhesion to stromal cells (12). Furthermore, c-Kit+ leukemia cells differentiated into macrophages on the FLS-5 cells and into mast cells on YSCL-71 cells (Fig. 5F and G). These findings demonstrate that c-Kit+ leukemia cells retain characteristic features of hematopoietic progenitors.
Low rate of leukemogenesis in GATA-1-null/X mice.
An important question in GATA-1.05 leukemogenesis is whether the residual GATA-1 activity in the GATA-1.05/X mice contributes to the leukemic process. To answer this question, we monitored the survival of GATA-1-null/X mice (n = 22) in the same facility that we used for the GATA-1.05/X mice. We have monitored the lives and health of this cohort of mice for approximately 500 days. To our surprise, the GATA-1-null/X mice show a survival curve similar to that of the control mice (Fig. 1). One mouse died during the early phase of the analysis (less than 250 days), but this mouse did not show any signs of leukemia. These results indicate that the residual GATA-1 activity in GATA-1.05/X mice is necessary for the propensity to develop leukemias.
Discomposure of GATA-1 function predisposes hematopoietic cells to preleukemia.
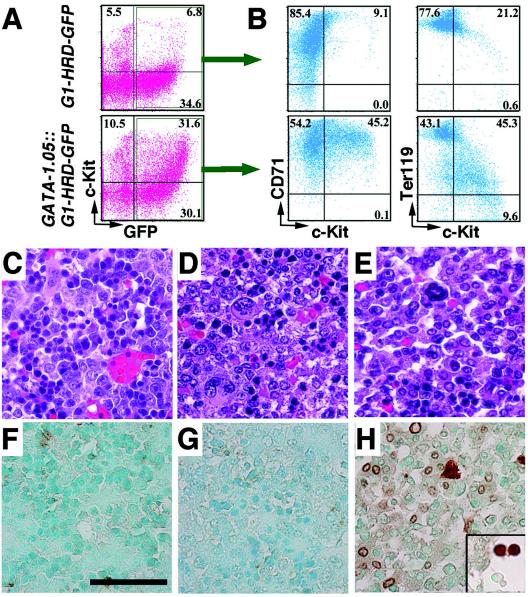
We have established G1-HRD-green fluorescent protein (GFP) transgenic mouse lines to obtain cells that express a GFP reporter under the regulatory influence of the GATA-1 gene (37). We crossed GATA-1.05/X mice with G1-HRD-GFP mice and analyzed the hematopoietic cells in the compound mutant embryos. Although at E17.5, GATA-1.05/X::G1-HRD-GFP embryos were slightly paler than G1-HRD-GFP transgenic embryos (data not shown), flow cytometry analysis of E17.5 fetal liver cells revealed that definitive erythropoiesis was robustly activated and the number of GFP+ cells in the GATA-1.05/X::G1-HRD-GFP embryos was increased relative to that in X/X::G1-HRD-GFP embryos (Fig. 6A).
FIG. 6.
Phenotypes of GATA-1.05/X embryos. (A, B) Flow cytometric analyses of the livers of E17.5 GATA-1.05/X::G1-HRD-GFP compound mutant embryos. GFP+ c-Kit+ cells increase in number in the lower grid in panel A compared with that in a normal littermate (upper grid). These GFP+ c-Kit+ cells show CD71+ and Ter119−/dull marker profiles (B). (C to H) Immunohistochemical analysis of the livers of E15.5 GATA-1.05/X and GATA-1-null/X embryos. Hematoxylin-eosin staining shows that mature erythroid cells decrease in number in both GATA-1.05/X (D) and GATA-1-null/X (E) fetal livers compared with the number in normal control cells (C). (F to H) Expression of TUNEL-positive cells in the livers of E15.5 wild-type (F), GATA-1.05/X (G), and GATA-1-null/X (H) embryos. A large number of immature cells positive for TUNEL staining (brown) were seen in the livers of GATA-1-null/X embryos, whereas a small number of TUNEL-positive cells were found in the livers of GATA-1.05/X embryos and normal control embryos. TUNEL-positive cells in circulating blood are shown in the column inserted in H. These sections were counterstained by methyl green. Scale bar, 50 μm.
The GFP+ fetal liver cells were also stained with transferrin receptor (CD71) and Ter119 antibodies. CD71 is expressed predominantly in the proerythroblast stage, while Ter119 is a marker of mature erythroid progenitors. We recently defined a late-stage erythroid progenitor (LEP) fraction that contains CFU-erythroid (CFU-E) cells and proerythroblasts (34). Two-color staining demonstrated that the GFP+ c-Kit+ cells coincided with the CD71+ and Ter119−/dull cells (Fig. 6B, upper panels) and that these GFP+ c-Kit+ CD71+ Ter119−/dull cells correspond to the LEP fraction. The number of GFP+ cells was markedly increased in the GATA-1.05/X::G1-HRD-GFP embryos (lower panels), and this marked increase of GFP+ c-Kit+ CD71+ Ter119−/dull cells was also observed in 14-day-old GATA-1.05/X::G1-HRD-GFP mouse spleens, the sizes of which were within the normal range (data not shown). These results thus suggest that the immature erythroid cells or LEP cells that express GATA-1 are already expanding in the hematopoietic organs of GATA-1.05/X mice at the late-embryonic and early-juvenile stages.
Histological examination of E15.5 GATA-1.05/X fetal livers showed that the number of mature erythroid cells with condensed chromatin was decreased but that the number of immature cells with multiple nucleoli was increased (Fig. 6D), relative to the numbers found in the wild-type fetal liver (Fig. 6C). These observations are consistent with those of previous analyses, demonstrating that the residual 5% expression of GATA-1 is not sufficient to drive terminal differentiation of erythroid progenitors (35). It is of interest to examine whether the residual level of GATA-1 in GATA-1.05 proerythroblasts is capable of preventing apoptosis, as GATA-1-null proerythroblasts are known to undergo apoptosis.
To evaluate the extent of apoptosis in the fetal livers of mutant embryos (Fig. 6E to G), we carried out a TUNEL analysis. We found only a small number of TUNEL-positive cells in the fetal livers of GATA-1.05/X and wild-type mice. In sharp contrast, we observed a significant increase in the number of TUNEL-positive cells, corresponding to immature cells with multiple nucleoli, in the GATA-1-null/X fetal livers. These findings suggest that many erythroid progenitors succumb to apoptosis in GATA-1-null/X fetal livers. Our present data indicate that maturation-arrested proerythroblasts in the GATA-1.05/X fetal livers are not susceptible to apoptosis.
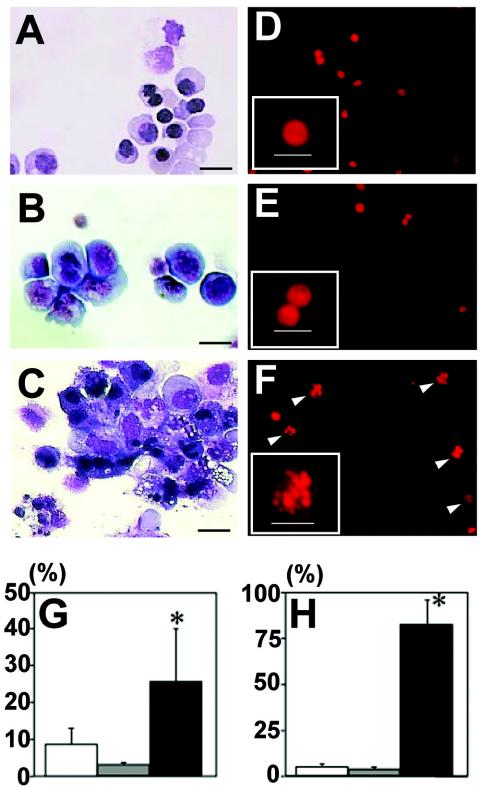
To further verify this possibility, we carried out in vitro differentiation analyses of ES cells. We exploited the OP9/ES cell differentiation method, which has been developed for the detection of primitive and definitive erythropoiesis (33). Cytospin samples of the wild-type, GATA-1.05/Y, and GATA-1-null/Y ES cells at the definitive erythroid cell stage (i.e., 11 days after the differentiation induction) clearly showed that both GATA-1.05/Y and GATA-1-null/Y ES cells are arrested at the progenitor stage but that wild-type ES cells differentiate into various stages of erythroid cells, including enucleated erythrocytes (Fig. 7A to C).
FIG. 7.
In vitro ES cell differentiation analysis. (A to C) Wright-Giemsa staining of cytospin specimens of ES cells 11 days after the induction of differentiation. Culture of wild-type ES cells in this system generates erythroid cells at various differentiation stages, including that of terminally enucleated red blood cells. In contrast, monotonous proerythroblast-like cells were observed in the culture of both GATA-1.05/Y (B) and GATA-1-null/Y (C) ES cells. Note that a large amount of cellular debris was observed in the latter cell culture. (D to F) Morphology of PI stain-positive cells. Apoptotic cells bearing fragmented nuclei (arrowhead) were observed in the GATA-1-null/Y ES cell culture (F) but not in the wild-type (D) and GATA-1.05/Y (E) ES cell cultures. PI stain-positive cells are shown at a higher magnification in the insets of panels. Scale bars, 10 μm. (G) Mortality evaluated by the trypan blue dye exclusion test. A GATA-1-null/Y ES cell culture shows higher mortality of the hematopoietic cells than that of a GATA-1.05/Y ES cell culture (*, P < 0.01). (H) Frequencies of apoptotic cells. The percentage of apoptotic cells with nuclear fragmentation was much higher in the GATA-1-null/Y ES cell culture than in the wild-type and GATA-1.05/Y ES cell cultures. In contrast, GATA-1.05/Y and wild-type ES-derived hematopoietic cells do not show a significant difference in apoptosis (*, P < 0.01).
In the course of this analysis, we noticed a large amount of cellular debris in the culture of GATA-1-null/Y ES cells (Fig. 7C). When assessed with the trypan blue dye exclusion test, the GATA-1-null/Y ES cell culture showed a significantly higher number of dead cells than did the wild-type and GATA-1.05/Y ES cell cultures (Fig. 7G). We also performed propidium iodide (PI) staining of the ES cells. Showing very good agreement with results of the dye exclusion analysis, the number of apoptotic cells bearing fragmented nuclei was markedly increased in the population derived from the GATA-1-null/Y ES cells compared to that of the populations derived from wild-type and GATA-1.05/Y ES cells (Fig. 7D to F and H). Thus, we conclude that differentiation-defective erythroid progenitors accumulating in the GATA-1.05/X animals are protected from apoptosis due to the residual activity of GATA-1.
DISCUSSION
In this analysis, we conducted a cohort study of more than 500 female mice with a heterozygous GATA-1 gene knockdown allele (GATA-1.05/X). We found that these GATA-1.05/X mice suffer from leukemia with a very high incidence, but such a high incidence of leukemia was not observed in GATA-1-null/X and wild-type mice. The genetic enfeeblement of GATA-1 function was found to elicit two distinct types of leukemia. One is c-Kit+ nonlymphoid leukemia, and the other is CD19+ B-cell leukemia. Closer examination of the GATA-1.05/X leukemic mice revealed that almost all of the mutant mice first developed thrombocytopenia and dyserythropoiesis in their hematopoietic tissues. Some of the mutant mice then proceeded to the accelerating stage with a grossly enlarged livers and spleens, unregulated proliferation of monoclonal leukemic cells, and suppression of normal hematopoietic cells. This course of phenotypic changes shares high similarity to the transformation process of preleukemic myelodysplastic cells to overt leukemia in humans.
The cohort analysis further demonstrates unequivocally that GATA-1 expression at 5% of the normal level is insufficient to sustain normal erythroid differentiation, as GATA-1.05 homozygous and GATA-1.05/Y pups have never been born (references 32, 35, and 37 and this study). Another interesting feature of GATA-1.05/X mice is that they contain two types of hematopoietic cells, owing to the process of X inactivation (15). In one cell type, the X chromosome bearing the GATA-1.05 allele is inactivated, but the one bearing the wild-type GATA-1 allele is active. These hematopoietic progenitors express normal amounts of GATA-1 and are able to differentiate into their appropriate cell types, including enucleated erythrocytes and platelets. In the other cell type, the X chromosome bearing the GATA-1.05 allele is active, but the one bearing the wild-type GATA-1 allele is inactive. In the latter case, the expression level of GATA-1 in erythroid and megakaryocytic progenitors is very low. This reduction appears to cause both arrest of differentiation and stimulation of proliferation (31). Therefore, this type of progenitor remains in an immature stage.
Whereas the GATA-1.05/X embryos showed various degrees of anemia depending on the extent of inactivation of the X chromosome with the wild-type GATA-1 allele, most of the GATA-1.05/X mice acquired close to normal erythroid indices after birth. The expression of GATA-1 mRNA was severely repressed in the c-Kit+ leukemic cells. In contrast, GATA-1 mRNA was expressed at higher levels in hematopoietic tissues at the preleukemic stage of GATA-1.05/X mice than in those of wild-type mice. Thus, a compensatory expansion of hematopoietic progenitors with an active wild-type GATA-1 allele appears to take place in response to the anemia. These results further support the notion that the mouse hematopoietic system has the capacity to compensate for the substantial lack of erythroid and megakaryocytic progenitors caused by the heterozygous GATA-1.05 knockdown mutation.
In this regard, it is noteworthy that GFP+ c-Kit+ CD71+ Ter119−/dull cells, corresponding to late-stage erythroid progenitors (34), have already accumulated in the livers of GATA-1.05/X embryos. Histological analyses of the GATA-1.05/X fetal livers suggest that these cells arise from the immature progenitors with the inactivated wild-type GATA-1 allele. In addition, the neo gene is actively expressed in the GFP+ c-Kit+ CD71+ Ter119−/dull cells in type 1 leukemia mice. These results indicate that GFP+ c-Kit+ CD71+ Ter119−/dull cells are differentiation-arrested erythroid progenitors with an inactivated wild-type GATA-1 allele.
GATA-1-null proerythroblasts are known to undergo apoptosis (42). Upon examination of apoptosis, we found that there was a significant increase of TUNEL+ cells in the GATA-1-null/X embryos. Consistent with the previous observations on the apoptosis of GATA-1-null proerythroblasts in culture (42), this result indicates that maturation-defective erythroid progenitors succumb to apoptosis in GATA-1-null/X embryos. Importantly, this process is prevented effectively in the GATA-1.05/X fetal livers, which display numbers of apoptotic cells similar to those observed in wild-type fetal livers.
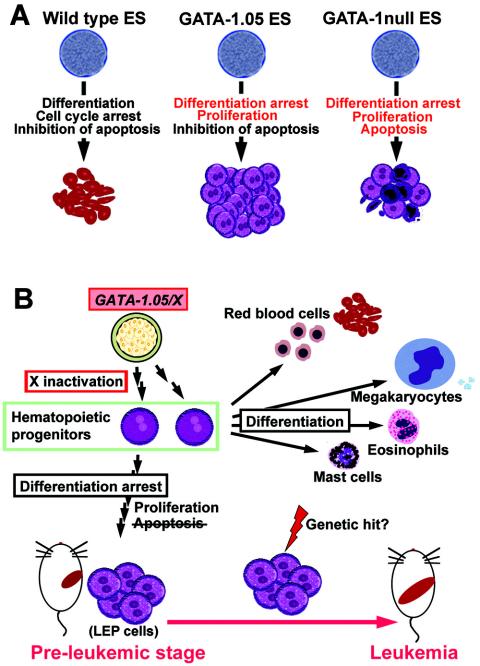
We carried out in vitro ES cell differentiation analyses and found that the differentiation of hematopoietic cells derived from GATA-1.05/Y and GATA-1-null/Y mutant ES cells was severely arrested at the LEP stage. The analyses also delineated that erythroid progenitors derived from GATA-1.05/Y and GATA-1-null/Y mutant ES cells proliferate much more vigorously than those derived from the wild-type ES cells (data not shown). In contrast, these two mutant ES cell lines differ sharply in terms of their sensitivities to apoptosis. While hematopoietic cells derived from GATA-1-null/Y ES cells suffer extensively from apoptosis, GATA-1.05/Y ES cells do not suffer from apoptosis, suggesting that the low level of GATA-1 expression is sufficient to prevent hematopoietic cells undergoing apoptosis. These mechanisms are summarized in Fig. 8A.
FIG. 8.
Model for the development of leukemia in GATA-1.05/X mice. (A) GATA-1 function during erythroid-cell differentiation of ES cells in vitro. Wild-type ES cells develop into mature erythroid cells depending on the three distinct but cooperative functions of GATA-1. Whereas GATA-1-null/Y ES cells remain in the immature proerythroblast stage, the cells are finally removed through apoptosis. In contrast, erythroblasts derived from GATA-1.05/Y ES cells increase vigorously because of the inhibition of apoptosis. (B) Model for the development of leukemia in GATA-1.05/X mice. GATA-1.05/X mice possess two types of hematopoietic cells, in which either the wild-type GATA-1 allele or the mutant GATA-1.05 allele is activated owing to the random inactivation of the X chromosome. While hematopoietic progenitor cells with the wild-type GATA-1 allele are able to differentiate normally, differentiation of the cells with the active GATA-1.05 allele is blocked, but the mutant progenitor cells proliferate actively. In addition, the presence of a residual amount of GATA-1 effectively prevents the progenitor cells from apoptosis. Thus, the progenitor cells with the active GATA-1.05 allele accumulate in the hematopoietic tissues of the GATA-1.05/X mice. These hematopoietic progenitor cells are eventually transformed into leukemic cells most likely through the multistep hit mechanism.
Importantly, this conclusion shows excellent agreement with the in vivo analyses that we carried out in this study and strongly argues that the differentiation-arrested LEP erythroblasts are eliminated through active pressure toward apoptosis in the hematopoietic tissues of GATA-1-null/X embryos. This determination is also in very good agreement with the previous observation that caspase-mediated down-regulation of GATA-1 prevents differentiation of normal proerythroblasts without inducing apoptosis (4). Thus, the prevention of apoptosis, in conjunction with the differentiation arrest and stimulation of cell proliferation at the LEP stage, provokes the accumulation of erythroid progenitors in the livers of GATA-1.05/X embryos. These accumulating progenitors provide a pool of cells that may progress into overt leukemia. Based on these observations, we conclude that GATA-1.05/X mice are in a preleukemic condition from the embryonic stage of development (Fig. 8B).
Despite the limited number of mice in the GATA-1-null/X cohort, our findings provide important insight into the contribution of the residual amount of GATA-1 in GATA-1.05 cells to the transformation of LEP cells into overt leukemic cells. In GATA-1-null/X mice, this population of cells is rapidly eliminated through apoptosis, resulting in efficient prevention of leukemogenesis. Indeed, a large number of GATA-1-null/X mice have been maintained in one of the authors' institutes on average for 25 weeks; neither leukemia cases nor enlarged spleen cases have ever been noticed (S. Philipsen, unpublished observation). Taken together, these results suggest a scenario for the leukemogenesis in GATA-1 gene knockdown mice in which immature erythroid progenitors at the late stage accumulate because they are arrested in differentiation but still capable of proliferation. The accumulation of progenitors, which are resistant to apoptosis by virtue of low-level GATA-1 expression, is critical for the predisposition to leukemogenesis of GATA-1.05/X mice.
Whereas both GATA-1 and GATA-2 are known to play important roles in hematopoiesis, the GATA-1 and GATA-2 genes show distinct expression profiles that are strictly regulated. The expression of GATA-1 increases as the differentiation of erythroid and megakaryocytic lineages continues (19, 33, 41). In contrast, GATA-2 is expressed in hematopoietic stem and progenitor cells, and GATA-2 is essential for cell proliferation (33, 38). It has been reported that persistent expression of GATA-2 stimulates the production of immature progenitor cells that have the potential to differentiate into multiple lineages (2). The expression of GATA-2 is elevated in the bone marrow of MDS patients, and the increase in the GATA-2/GATA-1 ratio correlates with the severity of the disease (6). These data are consistent with our hypothesis that the expanded population of LEP cells in GATA-1.05/X mice, in which GATA-2 expression is elevated, may be easily transformed through a “multistep hit” mechanism (39). In fact, we observed in the course of this study a type 1 case in which p53 mRNA expression was dramatically decreased in leukemia cells compared to that in c-Kit+ cells in control adult spleens (data not shown). Interestingly, it has been reported that mutation of the p53 gene is required for the acute crisis in essential thrombocytemia (21). Thus, these results suggest that the accumulation of GATA-1-knockdown progenitor cells, in combination with secondary genetic events, forms the molecular basis for the leukemic transformation in GATA-1.05/X mice.
Finally, there remains the intriguing question of how defective GATA-1 function provokes leukemia in the B-cell lineage. We do not have any definitive answers to this question yet, but we propose three hypotheses that are not mutually exclusive. The first proposal is based on the observation that B-lineage cells substantially lack the expression of hematopoietic GATA factors (17). The expression of GATA-2 in hematopoietic progenitors is downregulated after the commitment of cells to the lymphoid lineage, and the expression of GATA-3 becomes specific to T-lineage cells. Therefore, it is plausible that the targeted knockdown of GATA-1 creates a situation where the differentiation potential of the precursor cells is shifted from the erythroid to the B-lymphoid lineage. The second proposal is based on reports that GATA-1 represses PU.1 activity. Since PU.1 activity is important for B-cell development (22), an increase of PU.1 activity in progenitor cells with aberrantly low GATA-1 activity may lead the cells to adopt a B-cell fate. Although the expression of PU.1 mRNA in type 2 leukemia cells is comparable with that of normal spleen cells (data not shown), we think that this possibility still exists since the GATA-1/PU.1 interaction is a posttranscriptional event. The third possibility is that the accumulation of erythroid precursor cells may create an environment that is permissive for the oncogenic transformation of normal B cells.
Many chromosomal mutations are implicated in human MDS and various leukemias (30). The present study demonstrates that a simple genetic modification of a lineage-specific transcription factor can confer an unstable condition on progenitor cells, from which leukemias of two distinct lineages are provoked. In GATA-1.05/X mice, the overall activity of GATA-1 is significantly decreased, but there is no mutation in the GATA-1 protein, unlike the situation with DS-AMKL and TMD. Thus, the GATA-1.05 knockdown mouse provides a prime example of a model system for the systematic analysis of the ontogeny of MDS and the transformation from MDS into overt leukemia.
Acknowledgments
We thank Melin Khandekar for the discussion and critical reading of the manuscript. We also thank Yuko Kikuchi, Naomi Kaneko, and Reiko Kawai for help.
This work was supported in part by grants from the Ministry of Education, Science, Sports and Culture (Advanced Research for Cancer and General Area) to R.S. and M.Y., from JST ERATO and the Naito Foundation to M.Y., and from the Dutch Organization for Scientific Research NWO and the Dutch Cancer Society KWF to S.P.
REFERENCES
- 1.Baron, M. H., and S. M. Farrington. 1994. Positive regulators of the lineage-specific transcription factor GATA-1 in differentiating erythroid cells. Mol. Cell. Biol. 14:3108-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briegel, K., K. C. Lim, C. Plank, H. Beug, J. D. Engel, and M. Zenke. 1993. Ectopic expression of a conditional GATA-2/estrogen receptor chimera arrests erythroid differentiation in a hormone-dependent manner. Genes Dev. 7:1097-1109. [DOI] [PubMed] [Google Scholar]
- 3.Cantor, A. B., and S. H. Orkin. 2002. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21:3368-3376. [DOI] [PubMed] [Google Scholar]
- 4.De Maria, R., A. Zeuner, A. Eramo, C. Domenichelli, D. Bonci, F. Grignani, S. M. Srinivasula, E. S. Alnemri, U. Testa, and C. Peschle. 1999. Negative regulation of erythropoiesis by caspase-mediated cleavage of GATA-1. Nature 401:489-493. [DOI] [PubMed] [Google Scholar]
- 5.Dubart, A., P. H. Romeo, W. Vainchenker, and D. Dumenil. 1996. Constitutive expression of GATA-1 interferes with the cell-cycle regulation. Blood 87:3711-3721. [PubMed] [Google Scholar]
- 6.Fadilah, S. A., S. K. Cheong, H. Roslan, M. Rozie-Hanisa, and G. K. Yen. 2002. GATA-1 and GATA-2 gene expression is related to the severity of dysplasia in myelodysplastic syndrome. Leukemia 16:1563-1565. [DOI] [PubMed] [Google Scholar]
- 7.Fennie, C., J. Cheng, D. Dowbenko, P. Young, and L. A Lasky. 1995. CD34+ endothelial cell lines derived from murine yolk sac induce the proliferation and differentiation of yolk sac CD34+ hematopoietic progenitors. Blood 86:4454-4467. [PubMed] [Google Scholar]
- 8.Fujiwara, Y., A. N. Chang, A. M. Williams, and S. H. Orkin. 2004. Functional overlap of GATA-1 and GATA-2 in primitive hematopoietic development. Blood 103:583-585. [DOI] [PubMed] [Google Scholar]
- 9.Greene, M. E., G. Mundschau, J. Wechsler, M. McDevitt, A. Gamis, J. Karp, S. Gurbuxani, R. Arceci, and J. D. Crispino. 2003. Mutations in GATA1 in both transient myeloproliferative disorder and acute megakaryoblastic leukemia of Down syndrome. Blood Cells Mol. Dis. 31:351-356. [DOI] [PubMed] [Google Scholar]
- 10.Gurbuxani, S., P. Vyas, and J. D. Crispino. 2004. Recent insights into the mechanisms of myeloid leukemogenesis in Down syndrome. Blood 103:399-406. [DOI] [PubMed] [Google Scholar]
- 11.Ito, E., T. Toki, H. Ishihara, H. Ohtani, L. Gu, M. Yokoyama, J. D. Engel, and M. Yamamoto. 1993. Erythroid transcription factor GATA-1 is abundantly transcribed in mouse testis. Nature 362:466-468. [DOI] [PubMed] [Google Scholar]
- 12.Kodama, H., M. Nose, S. Niida, S. Nishikawa, and S. Nishikawa. 1994. Involvement of the c-kit receptor in the adhesion of hematopoietic stem cells to stromal cells. Exp. Hematol. 22:979-984. [PubMed] [Google Scholar]
- 13.Lange, B. 2000. The management of neoplastic disorders of haematopoiesis in children with Down's syndrome. Br. J. Haematol. 110:512-524. [DOI] [PubMed] [Google Scholar]
- 14.Lindeboom, F., N. Gillemans, A. Karis, M. Jaegle, D. Meijer, F. Grosveld, and S. Philipsen. 2003. A tissue-specific knockout reveals that Gata1 is not essential for Sertoli cell function in the mouse. Nucleic Acids Res. 31:5405-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyon, M. F. 1961. Gene action in the X chromosome of the mouse (mus musculus L.). Nature 190:372-373. [DOI] [PubMed] [Google Scholar]
- 16.McDevitt, M. A., R. A. Shivdasani, Y. Fujiwara, H. Yang, and S. H. Orkin. 1997. A “knockdown” mutation created by cis-element gene targeting reveals the dependence of erythroid cell maturation on the level of transcription factor GATA-1. Proc. Natl. Acad. Sci. USA 94:6781-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minegishi, N., S. Morita, M. Minegishi, S. Tsuchiya, T. Konno, N. Hayashi, and M. Yamamoto. 1997. Expression of GATA transcription factors in myelogenous and lymphoblastic leukemia cells. Int. J. Hematol. 65:239-249. [DOI] [PubMed] [Google Scholar]
- 18.Minegishi, N., N. Suzuki, Y. Yokomizo, X. Pan, T. Fujimoto, S. Takahashi, T. Hara, A. Miyajima, S. Nishikawa, and M. Yamamoto. 2003. Expression and domain-specific function of GATA-2 during differentiation of the hematopoietic precursor cells in midgestation mouse embryos. Blood 102:896-905. [DOI] [PubMed] [Google Scholar]
- 19.Nagai, T., H. Harigae, H. Ishihara, H. Motohashi, N. Minegishi, S. Tsuchiya, N. Hayashi, L. Gu, B. Andres, J. D. Engel, and M. Yamamoto. 1994. Transcription factor GATA-2 is expressed in erythroid, early myeloid, and CD34+ human leukemia-derived cell lines. Blood 84:1074-1084. [PubMed] [Google Scholar]
- 20.Nakano, T., H. Kodama, and T. Honjo. 1994. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science 265:1098-1101. [DOI] [PubMed] [Google Scholar]
- 21.Neri, A., N. S. Fracchiolla, F. Radaelli, A. Boletini, S. Ribera, C. Migliorini, D. Trecca, and A. T. Maiolo. 1996. p53 tumor suppressor gene and RAS oncogenes: molecular analysis in the chronic and leukaemic phases of essential thrombocythaemia. Br. J. Haematol. 93:670-673. [DOI] [PubMed] [Google Scholar]
- 22.Nerlov, C., E. Querfurth, H. Kulessa, and T. Graf. 2000. GATA-1 interacts with the myeloid PU. 1 transcription factor and represses PU. 1-dependent transcription. Blood 95:2543-2551. [PubMed] [Google Scholar]
- 23.Nishimura, S., S. Takahashi, T. Kuroha, N. Suwabe, T. Nagasawa, C. Trainor, and M. Yamamoto. 2000. A GATA box in the GATA-1 gene hematopoietic enhancer is a critical element in the network of GATA factors and sites that regulate this gene. Mol. Cell. Biol. 20:713-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohneda, K., and M. Yamamoto. 2002. Roles of hematopoietic transcription factors GATA-1 and GATA-2 in the development of red blood cell lineage. Acta Haematol. 108:237-245. [DOI] [PubMed] [Google Scholar]
- 25.Ohneda, K., R. Shimizu, S. Nishimura, Y. Muraosa, S. Takahashi, J. D. Engel, and M. Yamamoto. 2002. A minigene containing four discrete cis elements recapitulates GATA-1 gene expression in vivo. Genes Cells 7:1243-1254. [DOI] [PubMed] [Google Scholar]
- 26.Ohneda, O., N. Yanai, and M. Obinata. 1990. Microenvironment created by stromal cells is essential for a rapid expansion of erythroid cells in mouse fetal liver. Development 110:379-384. [DOI] [PubMed] [Google Scholar]
- 27.Onodera, K., K. Yomogida, N. Suwabe, S. Takahashi, Y. Muraosa, N. Hayashi, E. Ito, L. Gu, M. Rassoulzadegan, J. D. Engel, and M. Yamamoto. 1997. Conserved structure, regulatory elements, and transcriptional regulation from the GATA-1 gene testis promoter. J. Biochem. 121:251-263. [DOI] [PubMed] [Google Scholar]
- 28.Onodera, K., S. Takahashi, S. Nishimura, J. Ohta, H. Motohashi, K. Yomogida, N. Hayashi, J. D. Engel, and M. Yamamoto. 1997. GATA-1 transcription is controlled by distinct regulatory mechanisms during primitive and definitive erythropoiesis. Proc. Natl. Acad. Sci. USA 94:4487-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry, C., and H. Soreq. 2002. Transcriptional regulation of erythropoiesis. Eur. J. Biochem. 269:3607-3618. [DOI] [PubMed] [Google Scholar]
- 30.Rabbitts, T. H. 1994. Chromosomal translocations in human cancer. Nature 372:143-149. [DOI] [PubMed] [Google Scholar]
- 31.Rylski, M., J. J. Welch, Y. Y. Chen, D. L. Letting, J. A. Diehl, L. A. Chodosh, G. A. Blobel, and M. J. Weiss. 2003. GATA-1-mediated proliferation arrest during erythroid maturation. Mol. Cell. Biol. 23:5031-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimizu, R., S. Takahashi, K. Ohneda, J. D. Engel, and M. Yamamoto. 2001. In vivo requirements for functional GATA-1 domains during primitive and definitive erythropoiesis. EMBO J. 20:5250-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suwabe, N., S. Takahashi, T. Nakano, and M. Yamamoto. 1998. GATA-1 regulates growth and differentiation of definitive erythroid lineage cells during in vitro ES cell differentiation. Blood 92:4108-4118. [PubMed] [Google Scholar]
- 34.Suzuki, N., N. Suwabe, O. Ohneda, N. Obara, S. Imagawa, X. Pan, H. Motohashi, and M. Yamamoto. 2003. Identification and characterization of 2 types of erythroid progenitors that express GATA-1 at distinct levels. Blood 102:3575-3583. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi, S., K. Onodera, H. Motohashi, N. Suwabe, N. Hayashi, N. Yanai, Y. Nabeshima, and M. Yamamoto. 1997. Arrest in primitive erythroid cell development caused by promoter-specific disruption of the GATA-1 gene. J. Biol. Chem. 272:12611-12615. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi, S., T. Komeno, N. Suwabe, K. Yoh, O. Nakajima, S. Nishimura, T. Kuroha, T. Nagasawa, and M. Yamamoto. 1998. Role of GATA-1 in proliferation and differentiation of definitive erythroid and megakaryocytic cells in vivo. Blood 92:434-442. [PubMed] [Google Scholar]
- 37.Takahashi, S., R. Shimizu, N. Suwabe, T. Kuroha, K. Yoh, J. Ohta, S. Nishimura, K. C. Lim, J. D. Engel, and M. Yamamoto. 2000. GATA factor transgenes under GATA-1 locus control rescue germline GATA-1 mutant deficiencies. Blood 96:910-916. [PubMed] [Google Scholar]
- 38.Tsai, F. Y., G. Keller, F. C. Kuo, M. Weiss, J. Chen, M. Rosenblatt, F. W Alt, and S. H. Orkin. 1994. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371:221-226. [DOI] [PubMed] [Google Scholar]
- 39.Tucker, T., and J. M. Friedman. 2002. Pathogenesis of hereditary tumors: beyond the “two-hit” hypothesis. Clin. Genet. 62:345-357. [DOI] [PubMed] [Google Scholar]
- 40.Wechsler, J., M. Greene, M. A. McDevitt, J. Anastasi, J. E. Karp, M. M. Le Beau, and J. D. Crispino. 2002. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat. Genet. 32:148-152. [DOI] [PubMed] [Google Scholar]
- 41.Weiss, M. J., G. Keller, and S. H. Orkin. 1994. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev. 8:1184-1197. [DOI] [PubMed] [Google Scholar]
- 42.Weiss, M. J., and S. H. Orkin. 1995. Transcription factor GATA-1 permits survival and maturation of erythroid precursors by preventing apoptosis. Proc. Natl. Acad. Sci. USA 92:9623-9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, G., M. Nagano, R. Kanezaki. T. Toki, Y. Hayashi, T. Taketani, T. Taki, T. Mitui, K. Koike, K. Kato, M. Imaizumi, I. Sekine, Y. Ikeda, R. Hanada, M. Sako, K. Kudo, S. Kojima, O. Ohneda, M. Yamamoto, and E. Ito. 2003. Frequent mutations in the GATA-1 gene in the transient myeloproliferative disorder of Down's syndrome. Blood 102:2960-2968. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto, M., L. J. Ko, M. W. Leonard, H. Beug, S. H Orkin, and J. D. Engel. 1990. Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes Dev. 4:1650-1662. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto, M., S. Takahashi, K. Onodera, Y. Muraosa, and J. D. Engel. 1997. Upstream and downstream of erythroid transcription factor GATA-1. Genes Cells 2:107-115. [DOI] [PubMed] [Google Scholar]
- 46.Zipursky, A., A. Poon, and J. Doyle. 1992. Leukemia in Down syndrome. A review. Pediatr. Hematol. Oncol. 9:139-149. [DOI] [PubMed] [Google Scholar]