Abstract
Transcriptional activation of gene expression by Wnt signaling is driven by the association of β-catenin with TCF/LEF factors and the recruitment of transcriptional coactivators. It has been shown that the LIM protein FHL2 and the acetyltransferase CBP/p300 individually stimulate β-catenin transactivating activity and that β-catenin is acetylated by p300. Here, we report that FHL2 and CBP/p300 synergistically enhanced β-catenin/TCF-mediated transcription from Wnt-responsive promoters and that the acetyltransferase activity of CBP/p300 was involved in the cooperation. CBP/p300 interacted directly with FHL2, predominantly through the CH3 domain but not the histone acetyltransferase domain, and different regions of CBP/p300 were involved in FHL2 and β-catenin binding. We provided evidence for the formation of a ternary complex by FHL2, CBP/p300, and β-catenin and for colocalization of the three proteins in the nucleus. In murine FHL2−/− embryo fibroblasts, the transactivation activity of β-catenin/TCF was markedly reduced, and this defect could be restored by exogenous expression of FHL2. However, CBP/p300 were still able to coactivate the β-catenin/TCF complex in FHL2−/− cells, suggesting that FHL2 is dispensable for the coactivator function of CBP/p300 on β-catenin. Furthermore, we found that FHL2 significantly increased acetylation of β-catenin by p300 in vivo. Finally, we showed that FHL2, CBP/p300, and β-catenin could synergistically activate androgen receptor-mediated transcription, indicating that the synergistic coactivator function is not restricted to TCF/LEF.
Tight regulation of gene expression is in large part achieved at the transcription level by DNA-binding factors and coactivators-corepressors. The notion that transcription cofactors-adapters lacking DNA-binding affinity are important components of transcriptional regulation is clearly illustrated by the role of β-catenin and its cofactors in controlling the expression of genes targeted by Wnt signaling.
β-Catenin is a key effector of the canonical Wnt pathway, which plays a crucial role in development and carcinogenesis (15, 33). Wnt signaling promotes expression of a large spectrum of genes involved in various developmental and oncogenic processes, such as siamois, c-myc, cyclin D1, ephrin receptors, and interleukin-8 (IL-8) (2, 4, 16, 25, 36, 40). Transcriptional activation of Wnt-responsive genes is achieved through the formation of a transcriptional complex including β-catenin and TCF/LEF family factors, in which TCF/LEF provide the DNA-binding domain and β-catenin provides the transcription activation domain. In the absence of Wnt signaling, β-catenin is degraded in the cytoplasm, and TCF/LEF negatively regulate Wnt target genes by recruiting transcriptional corepressors such as Groucho/TLE, CtBP, and HDAC (5).
Several mechanisms by which β-catenin stimulates transcription have been proposed. They are based mainly on the properties of recently identified cellular cofactors of the protein. It has been shown that the transcriptional activity of β-catenin is controlled by its association with Legless/BCL9 and the PHD finger protein Pygopus, which target β-catenin to the nucleus (21, 41). There, β-catenin is able to interact directly with the TATA-binding protein, thus linking the β-catenin/TCF complex to the basal transcription machinery (17). In addition, β-catenin can recruit nuclear factors involved in chromatin remodeling and unwinding, such as Brg-1, a core component of the Swi/Snf complex (1), and the coactivators CBP/p300, which play a crucial role in Wnt target gene activation during development (18, 29, 38, 39). CBP/p300 are multifunctional transcriptional coactivators that serve as a protein scaffold for the assembly of multicomponent transcriptional complexes, linking various factors such as the androgen receptor (AR) and the cAMP-responsive element-binding protein (CREB) to the general transcription machinery (6). In addition, CBP/p300 possess intrinsic acetyltransferase (AT) activity, which is responsible for acetylation of a variety of substrates, including histones and transcription factors (24). β-Catenin is one of the substrates of p300 (26, 45). In contrast to a number of β-catenin partners which interact with the central armadillo repeat (arm) domain of β-catenin, CBP/p300 are more likely to bind to the region extending from β-catenin arm 10 to the C terminus (18, 39) or to both the N- and C-terminal regions of β-catenin (29).
We and others have recently demonstrated physical and functional interactions between β-catenin and the LIM protein FHL2 (for four-and-a-half LIM-only protein 2) (27, 44). The LIM domain consists of double zinc finger motif, and LIM family proteins play multiple roles as adapters capable of mediating protein-protein interactions (10). Containing exclusively LIM domains, FHL2 has no DNA-binding activity but possesses an intrinsic transactivation function (14, 31). It has been shown that FHL2 associates with several DNA-binding factors, including AR, CREB, WT1, and AP1, to activate gene expression (11, 13, 30, 31). In a recent report, we demonstrated that FHL2 strongly potentiates β-catenin transactivating activity on TCF/LEF target genes (44). Furthermore, recent data suggest that signaling events might control the cellular localization of FHL2. Rho signaling and stimulation with serum and UV light have been shown to induce nuclear translocation of FHL2 (30, 32). Furthermore, nuclear expression of FHL2 in prostate cancer cells correlates with tumor dedifferentiation and progression to a more malignant carcinoma phenotype (32).
Because LIM proteins are known to mediate protein-protein interactions, we searched for functional interactions between FHL2 and known coactivators of β-catenin. In the present study, we show that FHL2 and CBP/p300 synergistically enhance β-catenin transcriptional activity. The acetyltransferase activity of CBP/p300 is implicated in this synergistic effect, and FHL2 increases acetylation of β-catenin by p300, which may strengthen the binding of β-catenin to TCF4. Moreover, we provide evidence of physical interactions between FHL2, CBP/p300, and β-catenin in vivo. By immunofluorescence, we show colocalization of the three proteins in the nucleus. Finally, we demonstrate that the synergistic interaction of FHL2, CBP/p300, and β-catenin is independent of TCF/LEF, because it also functions on AR-responsive promoters.
MATERIALS AND METHODS
Plasmids.
Expression vectors for β-catenin T41A and FHL2 have been described previously (43, 44). To construct hemagglutinin (HA)-FHL2, full-length FHL2 cDNA carrying a N-terminal HA tag was inserted into pcDNA3. Glutathione S-transferase (GST)-LIM1-4 was constructed by insertion of the fragment coding for FHL2 LIM 1 to LIM4 (amino acids [aa] 36 to 279) into pGEX5X-1 (Pharmacia). FHL2 deletion mutants (L1/2-2, L1/2-3, L1-4, L2-4, L2-3, and L3-4) were cloned into the pCS2 expression vector (a gift of R.T. Moon). Gal4-FHL2 was constructed by insertion of the FHL2-coding region in frame with the Gal4 DNA-binding domain into the pM vector (Clontech Laboratories). Standard recombinant DNA techniques, including PCR followed by sequencing, were used. To construct pCRCBP(1-495), used for in vitro transcription and translation, the fragment encoding amino acids 1 to 495 of CBP was amplified by PCR with primer 1 (5′-ACTAGGATCCCTATGGCCGAGAACTTGCT-3′) and primer 2 (5′-TCACTCGAGTCAAGGCTGGTTCATGTAGGG-3′), followed by insertion into pCRII-TOPO (Invitrogen). Reporter plasmids carrying luciferase under control of the cyclin D1 promoter (−163CD1LUC and −163mtLefCD1LUC) were provided by R. Pestell; pTOPFLASH, pFOPFLASH, pTCF4, and pΔNTCF4 by were provided by H. Clevers, and pSiamois-luciferase was provided by R. T. Moon. The IL-8 promoter-luciferase construct was as described previously (25). pCRE-Luc, which carries four consensus cellular cAMP-responsive element (CRE) sites, was purchased from Stratagene. GST-CBP fusion constructs were kindly provided by T. Kouzarides and A. Harel-Bellan; CBP was provided by R. H. Goodman; p300, p300ΔHAT (with amino acids 1413 to 1721 deleted), p300(1-670), and p300(1135-2414) were provided by Y. Nakatani; wild-type (wt) E1A and the E1AmRb and E1AmCBP mutants were provided by A. Hecht and R. Kemler; pMMTV-Luc was provided by P. Chambon; pSG5-hAR was provided by G. Castoria; G5-E1b-Luc was provided by J.-G. Judde; protein kinase A (PKA) was provided by R. Gaynor; human T-cell lymphotropic virus 1 long terminal repeat luciferase reporter (HTLV1-LTR-LUC) was provided by J. Brady, Tax was provided by K. T. Jeang, and FLAG-p65 was provided by M. Benkirane.
Cell culture, transient transfections, and luciferase assays.
293, COS-7, SW480, and CV1 cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Primary mouse embryo fibroblasts (MEFs) were prepared from wild-type C57BL/6 or FHL2−/− embryos at embryonic day 16.5 as described previously (46) and grown in DMEM supplemented with 10% FBS and nonessential amino acids. For luciferase assays, CV1, COS-7, SW480, or 293 cells (3 × 105 to 5 × 105 cells per well) were seeded in six-well plates. MEFs at the third passage (105 cells per well) were seeded in 12-well plates. Transient transfections were performed by the calcium phosphate precipitation method with 293 cells and by use of the Lipofectamine Plus reagent (Invitrogen) with other cell lines. Cells were harvested 24 h later (48 h for CV1), lysed, and assayed for luciferase activity as described previously (44). For AR transcriptional activity assays, CV1 cells were washed 3 h after transfection and the medium was replaced with DMEM supplemented with 10% hormone-depleted FBS (charcoal-dextran-stripped FBS), in the presence or absence of 10 nM dihydrotestosterone (Sigma). The total amount of transfected DNA was kept constant by adding pcDNA3. Internal controls were performed by cotransfection of the thymidine kinase-β-galactosidase expression vector. Transfections were performed in duplicate, and each experiment was repeated at least four times.
Production of full-length FHL2 protein from insect cells.
Full-length human FHL2 cDNA was cloned in the pAcGP67-A vector (PharMingen) containing the secretion signal sequence of GP67. Recombinant baculoviruses were generated in Sf-9 cells, FHL2 was produced in insect cells with the baculovirus system described previously (28), and the protein was secreted into the culture medium after cleavage of the N-terminal signal peptide. The culture medium was harvested 3 days after infection, cleared by centrifugation at 2,000 × g for 5 min, and stored at −20°C until used.
GST pull-down assays.
35S-labeled proteins were generated by in vitro transcription-translation with the TNT-coupled reticulocyte lysate system (Promega). The GST fusion proteins were purified from Escherichia coli and linked to glutathione-Sepharose beads. Incubation of 2 μg of GST or GST fusion proteins with in vitro-translated proteins was carried out for 1 to 4 h at 4°C in binding buffer (20 mM HEPES-KOH [pH 7.5], 100 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 0.02% bovine serum albumin, 0.8% NP-40, 1 mM dithiothreitol [DTT], and protease inhibitor cocktail [Roche]). The beads were washed three times in binding buffer, and bound proteins were eluted with Laemmli buffer at 95°C and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gels were dried and analyzed by autoradiography.
Immunofluorescence.
Immunofluorescence analysis was performed as described previously (44). 293 cells (2 × 105 per well) were grown on coverslips in six-well plates and transfected with expression vectors for HA-FHL2, FLAG-p300, and β-catenin T41A (0.5 μg each) with calcium phosphate. Forty-eight hours later, cells were washed, fixed with 3.7% paraformaldehyde, and permeabilized with 0.5% Triton X-100. The cells were then incubated with polyclonal anti-FLAG antibody (Sigma), monoclonal anti-HA (Babco), monoclonal anti-β-catenin antibody (Transduction Laboratories), or polyclonal anti-promyelocytic leukemia (anti-PML) antibody (a gift of J. Seeler), either alone or in different combinations. This was followed by incubation with the corresponding Cy3- and fluorescein isothiocyanate-coupled secondary antibodies (Jackson ImmunoResearch Laboratories). For fluorescence microscopy, images were recorded with a Leica DMRB microscope equipped with a Princeton CoolSnapFX charge-coupled device camera controlled by MetaVue software. Confocal microscopy was performed with the Leica TCS4D laser-scanning microscope, using excitation wavelengths of the argon-krypton laser at 488 nm for fluorescein isothiocyanate and 568 nm for Cy3. Acquisitions were made in sequential mode to prevent false colocalization.
Immunoprecipitation, immunoblotting, and in vivo acetylation assays.
COS-7 cells (2 × 106) were seeded in 10-cm-diameter dishes and transfected with expression vectors for CBP (10 μg) and HA-FHL2 (2 μg). Cells were harvested 48 h later and lysed in 500 μl of lysis buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM MgCl2, 0.5% NP-40, 1 mM DTT, and protease inhibitor cocktail [Roche]). The lysate was cleared by centrifugation, followed by incubation with either anti-CBP polyclonal antibodies (A22; Santa Cruz) or the control anti-FLAG antibody (polyclonal; Sigma) overnight at 4°C in the presence of protein A/G-Sepharose beads (Pierce). The beads were washed five times with lysis buffer, and bound proteins were eluted with Laemmli buffer at 95°C and subjected to SDS-PAGE. Proteins were transferred to nitrocellulose and analyzed by immunoblotting with anti-CBP and anti-HA antibodies (Babco) by using the Western-Star chemiluminescent detection system (Tropix).
293 cells were transfected with vectors expressing HA-FHL2 (2 μg), FLAG-p300(1-670), or FLAG-p300(1135-2414) (5 μg) and β-catenin T41A (2 μg). The lysate was immunoprecipitated with either anti-FLAG polyclonal antibody or anti-GAL4DBD antibody (Santa Cruz). Precipitated proteins were detected by immunoblotting with anti-HA, anti-FLAG, or anti-β-catenin antibodies.
For in vivo acetylation assays, COS-7 or 293 cells were transfected with FHL2, p300, and either β-catenin T41A or FLAG-p65 (5 μg each) and were lysed 48 h later in Triton lysis buffer (50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 0.5% Triton X-100, 1 mM DTT, 0.2 μM trichostatin A [Sigma], and protease inhibitor cocktail). For detection of acetylated β-catenin, cell lysates were immunoprecipitated with anti-β-catenin monoclonal antibodies, and immunoprecipitated proteins were analyzed by Western blotting with a mixture of two anti-acetyl-lysine monoclonal antibodies (Cell Signaling Technology and Upstate Biotechnology). For detection of acetylated FLAG-p65, cell lysates were immunoprecipitated with anti-acetyl-lysine monoclonal antibodies (Cell Signaling), followed by immunoblotting with anti-FLAG polyclonal antibody. The antiactin monoclonal antibody used for a loading control was from Sigma. Immunoblots were quantified by using Gene Snap/Gene Tools from SynGene.
RNA analysis.
Total RNA was prepared from MEFs by a guanidinium isothiocyanate-phenol method (RNA PLUS; Bioprobe) and analyzed by Northern blotting as previously described (34). The blot was hybridized sequentially with the following probes: a 1.1-kb cDNA fragment of the human cyclin D1 gene (a gift of P. Hinds), full-length mouse FHL2 cDNA (a gift of J. Chen), and a 0.2-kb PstI cDNA fragment of the mouse 18S rRNA gene (Valbiotech), which served as control for RNA loading. The relative intensities of cyclin D1 and 18S RNAs were quantified with a PhosphorImager (Storm Imaging System; Molecular Dynamics).
RESULTS
FHL2 and CBP/p300 synergistically stimulate β-catenin transcriptional activity on TCF/LEF target genes.
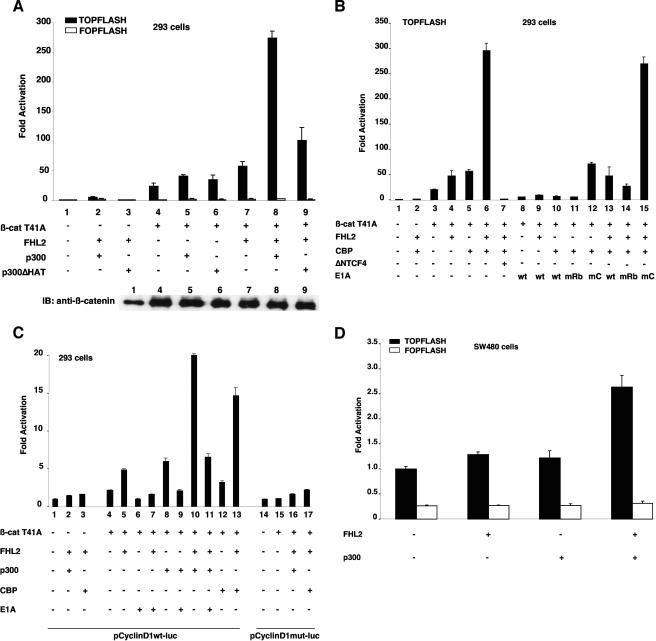
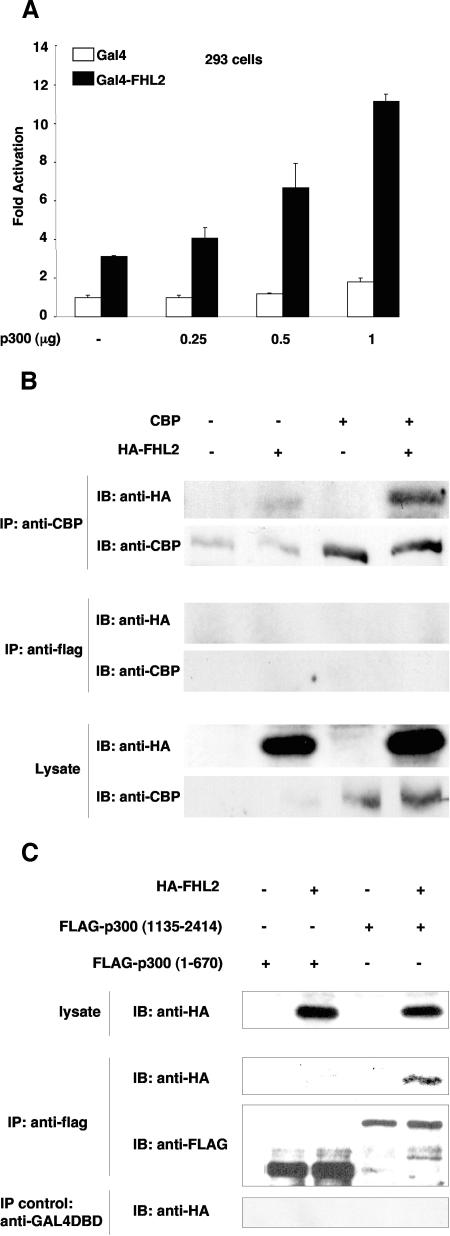
Previous studies have shown that FHL2 and CBP/p300 interact with different domains of β-catenin (arm repeats 1 to 9 for FHL2 and arm 10 to the C terminus for CBP/p300), suggesting that these coactivators might bind β-catenin simultaneously (18, 38, 39, 44). The effect of FHL2 and CBP/p300 on the β-catenin transactivating function was assessed in 293 cells by using the TOPFLASH luciferase reporter, which contains consensus TCF/LEF-binding sites, and the constitutively active β-catenin mutant T41A. In agreement with previous reports, FHL2 and p300 individually activated β-catenin activity on reporter gene expression by about twofold (Fig. 1A) (18, 39, 44). Importantly, coexpression of FHL2 and p300 with β-catenin further enhanced TOPFLASH reporter activity by at least 10-fold (Fig. 1A). β-Catenin expression remained unchanged in the presence of the coactivators (Fig. 1A, lower panel), indicating that this effect was not due to transcriptional activation of the β-catenin expression vector by FHL2 and p300. In addition, synergistic activation was dependent on TCF/LEF family factors, as the reporter gene under the control of mutant TCF/LEF-binding elements (FOPFLASH) showed minimal changes in the presence of FHL2, p300, and β-catenin (Fig. 1A). Interestingly, deletion of the p300 histone acetyltransferase domain (p300ΔHAT) severely reduced the synergy with FHL2 (Fig. 1A), while mutant and wild-type p300 were expressed at similar levels (data not shown).
FIG. 1.
Synergistic transactivation of β-catenin/TCF-responsive promoters by FHL2 and CBP/p300. (A) 293 cells were transfected with the TOPFLASH or FOPFLASH reporter plasmid (0.5 μg) together with different combinations of β-catenin (β-cat) T41A (50 ng), FHL2 (1 μg), p300 or p300ΔHAT (0.5 μg), or the empty vector as indicated. Luciferase activities were determined 48 h later. The basal activity of the TOPFLASH reporter cotransfected with empty vector was arbitrarily set at 1, and data are presented as the mean induction in luciferase activity ± standard deviation from duplicate samples. The results shown are representative of those from more than four independent assays. Bottom: expression levels of transfected β-catenin determined by immunoblotting (IB). (B) 293 cells were transfected with TOPFLASH and CBP (0.5 μg), ΔNTCF4 (0.25 μg), and wild-type (wt) or mutant E1A (0.1 μg) as indicated. E1AmRb: mutant with the Rb-binding region deleted; E1AmC: mutant with the CBP-binding region deleted. (C) 293 cells were transfected with cyclin D1 promoter reporter plasmids harboring either the wt or mutant LEF site (0.5 μg) and with β-cateninT41A (0.25 μg), FHL2 (1 μg), p300 or CBP (0.5 μg), or wt E1A (0.1 μg) as indicated. (D) SW480 cells were transfected with the TOPFLASH and FOPFLASH reporters and with FHL2 (0.5 μg) or p300 (0.5 μg) and assayed for luciferase activity 24 h later.
Similarly, CBP cooperated with FHL2 to synergistically increase β-catenin activity on the TOPFLASH reporter (Fig. 1B). Expression of dominant negative TCF4 (ΔNTCF4), which retains DNA binding activity but fails to interact with β-catenin, completely abolished this effect (Fig. 1B, compare bars 6 and 7). To assess the role of endogenous CBP in FHL2 coactivator function, we used the CBP inhibitor E1A in this assay. Both wild-type E1A and the mutant E1AmRb, which selectively affects interaction with the retinoblastoma protein (Rb), decreased luciferase expression to the level found in the absence of cotransfected CBP (Fig. 1B). In contrast, the mutant E1AmCBP (E1AmC), which lacks the interaction domain with CBP, had little inhibitory effect on the cooperation of CBP with β-catenin (Fig. 1B). Furthermore, coexpression of E1A with β-catenin and FHL2 inhibited the activity of FHL2 on β-catenin (Fig. 1B, compare bars 8 and 9 with bars 3 and 4), suggesting that endogenous CBP/p300 may be involved in the coactivator function of FHL2 in this context.
We then investigated the response of natural TCF/LEF-responsive promoters to the combined action of FHL2 and CBP/p300. As found with the synthetic promoter, the cyclin D1 promoter was synergistically activated by FHL2, CBP/p300, and β-catenin (Fig. 1C), and this effect was strictly dependent on the presence of the TCF/LEF-binding site (Fig. 1C, compare bars 10 and 13 with bars 16 and 17). Similar results were obtained with other TCF/LEF target promoters, including the IL-8 and siamois promoters (data not shown). As expected, E1A inhibited not only the coactivation of β-catenin and p300 but also the stimulating effect of FHL2 and the synergy between p300 and FHL2, indicating that endogenous p300 has impact on FHL2 coactivator function on β-catenin.
To test the effect of FHL2 and CBP/p300 on endogenous β-catenin, we performed transfection assays with the TOPFLASH reporter in SW480 colon carcinoma cells, in which endogenous wild-type β-catenin is constitutively active because of defective adenomatous polyposis coli. Exogenous coexpression of FHL2 and p300 cooperatively stimulated β-catenin activity on transcription from the synthetic promoter in SW480 cells (Fig. 1D). Taken together, these results show that FHL2 and CBP/p300 synergistically activate transcription mediated by β-catenin/TCF and that endogenous CBP/p300 is required for FHL2-induced activation of β-catenin-dependent transcription.
Physical interaction between FHL2 and CBP/p300.
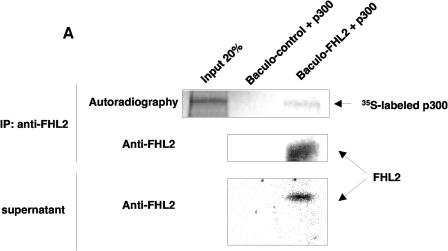
To better understand the mechanisms underlying the cooperation between FHL2 and CBP/p300, we next explored the ability of FHL2 to interact directly with p300 in vitro. To this end, full-length, untagged FHL2 protein was produced in the supernatant of insect cells by a baculovirus system (28). The presence of FHL2 in the culture medium was verified by immunoblotting with anti-FHL2 polyclonal antibodies (44) (Fig. 2A), while the protein could not be detected in culture medium of cells infected with nonrecombinant baculovirus. The supernatant containing FHL2 was then immunoprecipitated with purified anti-FHL2 polyclonal antibodies (32) in the presence of in vitro-translated, 35S-labeled full-length p300 protein. 35S-labeled p300 was readily detected together with FHL2 in the immune complex, but none of the proteins could be seen in precipitated proteins from the supernatant of empty baculovirus-infected cells (Fig. 2A). Similar data were obtained with 35S-labeled full-length CBP (data not shown).
FIG. 2.
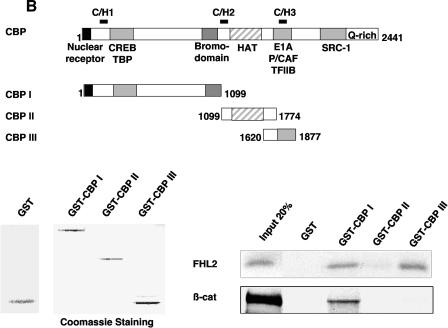
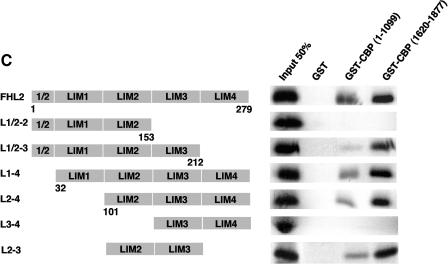
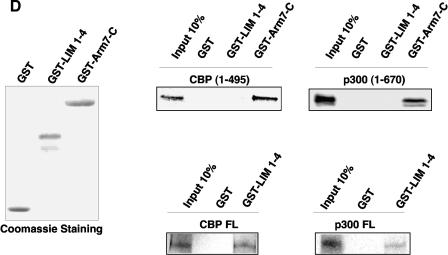
FHL2 interacts with CBP/p300 in vitro. (A) Insect cell culture medium containing or not containing baculovirus-expressed FHL2 as indicated was incubated with [35S]methionine-labeled full-length p300 and A/G-Sepharose beads and subjected to immunoprecipitation (IP) with anti-FHL2 antibodies. The beads were washed four times, and bound proteins were eluted, subjected to SDS-PAGE, and analyzed by immunoblotting with anti-FHL2 antibodies and by autoradiography. Input reflects 20% of the proteins incubated with the beads. Bottom panel: portions of the insect cell culture medium were immunoblotted for FHL2. (B) Mapping of CBP domains that interact with FHL2 by GST pull-down assay. A schematic illustration of full-length CBP with representative domains and binding partners is shown on the top, followed by CBP constructs used as GST fusion proteins. Bottom left: expression of GST and different GST-CBP constructs was analyzed by polyacrylamide gel electrophoresis and staining with Coomassie blue. Bottom right: for GST pull-down assays, [35S]methionine-labeled FHL2 or β-catenin (β-cat) was incubated with GST or GST-CBP fusion proteins as indicated. Input reflects 20% of the protein incubated with the beads. (C) Determination of the CBP-binding region of the FHL2 protein. The FHL2 constructs are depicted on the left. [35S]methionine-labeled FHL2 and different deletion mutants were incubated with GST, GST-CBP(1-1099), or GST-CBP(1620-1877). Input reflects 50% of the protein incubated with the beads. (D) Coomassie blue-stained GST, GST-FHL2 fusion protein carrying the LIM domains 1 to 4 (GST-LIM1-4), and GST-β-catenin spanning the region from arm 7 to the C terminus (GST-Arm7-C) are shown in the left panel. [35S]methionine-labeled CBP(1-495), p300(1-670), and full-length CBP and p300 were incubated with GST or GST-LIM1-4 as indicated in the right panel. Input reflects 10% of the protein incubated with the beads. FL, full length.
We next investigated the CBP/p300 domains involved in the interaction with FHL2 in pull-down assays, using different fragments of CBP in fusion with GST and 35S-labeled full-length FHL2. As shown in Fig. 2B, the N-terminal domain in CBP (aa 1 to 1099) and the CH3 domains in CBP (aa 1620 to 1877), but not the HAT domain in CBP (aa 1099 to 1774), were able to pull down FHL2. In parallel experiments, 35S-labeled β-catenin was able to bind to CBP(1-1099) in GST pull-down assays, but it failed to interact with the CH3 domain (Fig. 2B). We next mapped the CBP-interacting domain in FHL2, using CBP(1-1099) and CBP(1620-1877) recombinant proteins and a series of truncated 35S-labeled FHL2 proteins. In this assay, FHL2 fragments were capable of interacting with both CBP constructs as long as they contained at least the LIM domains 2 and 3 (Fig. 2C). Thus, LIM2 and LIM3 are required and sufficient for the interaction with CBP, predominantly with the CH3 domain.
The interaction between FHL2 and CBP/p300 was further evaluated with the 35S-labeled mutants CBP(1-495) and p300(1-670), which retain only the N-terminal part of each protein. In this assay, we used the LIM domains 1 to 4 of FHL2 in fusion with GST, because full-length FHL2 fused to GST was found to be insoluble, as previously reported (12), and the in vitro-translated LIM1-4 segment interacted with GST-CBP as well as full-length FHL2 (Fig. 2C). While GST-LIM 1-4 was able to bind full-length CBP and p300, it failed to interact with the CBP 1-495) and p300(1-670) mutants (Fig. 2D). The corresponding region of CBP/p300 (aa 302 to 530 of CBP and aa 451 to 682 of p300) has been reported to bind β-catenin (38, 39). As expected, we observed that the C-terminal region of β-catenin (arm 7 to the C terminus) was able to pull down both CBP and p300 mutant proteins (Fig. 2D). Taken together, our data show that FHL2 directly interacts with CBP and p300 in vitro and that CBP/p300 domains involved in the interaction with FHL2 are distinct from the β-catenin-binding region.
To extend these observations, functional interaction between FHL2 and p300 was assessed in a luciferase reporter assay with 293 cells, using full-length FHL2 fused with the Gal4 DNA-binding domain and different amounts of p300. As previously reported, FHL2-Gal4 was able to activate the Gal4-dependent reporter gene by threefold (Fig. 3A) (31). More importantly, luciferase activity was further enhanced by p300 in a dose-dependent manner, while p300 had limited effect on Gal4-mediated activation of the reporter (Fig. 3A). These results suggest functional interaction between FHL2 and p300.
FIG. 3.
FHL2 interacts with CBP/p300 in vivo. (A) 293 cells were transfected with a luciferase reporter plasmid carrying five Gal4-binding sites followed by the E1b minimal promoter (0.5 μg) together with expression vectors for Gal4-FHL2 or Gal4 (1 μg each) and increasing amounts of p300 as indicated. Conditions were as for Fig. 1. Error bars indicate standard deviations. (B) Coimmunoprecipitation of CBP andFHL2. Lysates from COS-7 cells transfected with HA-tagged FHL2 and CBP were immunoprecipitated (IP) with CBP antibody or the irrelevant anti-FLAG antibody, and coprecipitated FHL2 was detected by immunoblotting (IB) with anti-HA antibodies. Control immunoprecipitation was performed with an anti-FLAG antibody. Bottom: portions of the original cell lysates were immunoblotted for HA-FHL2 and CBP. (C) Coimmunoprecipitation of FHL2 and different fragments of p300. 293 cells were transfected with expression plasmids for HA-tagged FHL2, FLAG-tagged p300(1-670), and FLAG-tagged p300(1135-2414). Cell lysates were immunoprecipitated with anti-FLAG antibodies and the control GAL4DBD antibody, and immune complexes were analyzed by immunoblotting with anti-HA and anti-FLAG antibodies as indicated.
To confirm that FHL2 can interact with CBP/p300 in cells, coimmunoprecipitation experiments were carried out with COS-7 cells. At 48 h after transfection of CBP and HA-FHL2, cells lysates were precipitated with anti-CBP antibodies, followed by immunoblotting analysis with anti-HA antibody. FHL2 was revealed in the immune complexes containing overexpressed or endogenous CBP (Fig. 3B). The presence of FHL2 in the CBP-containing immune complexes was specific, as anti-FLAG antibody failed to immunoprecipitate FHL2 (Fig. 3B). Similar data were obtained with full-length FLAG-p300 and HA-FHL2 in 293 cells (data not shown). For mapping the CBP/p300-interacting region in FHL2 in vivo, we analyzed the ability of FHL2 to interact with different fragments of p300 by coimmunoprecipitation. HA-FHL2 was cotransfected with FLAG-p300(1-670) or FLAG-p300(1135-2414) in 293 cells, and cell lysates were precipitated with anti-FLAG antibodies, followed by immunoblotting with anti-HA antibody. Consistent with our in vitro data, FHL2 was coprecipitated with p300(1135-2414) (Fig. 3C), whereas the irrelevant anti-GAL4DBD antibody failed to precipitate FHL2. In contrast, we could not detect any interaction of FHL2 with p300(1-670) (Fig. 3C), suggesting that this region of p300 is not capable of binding to FHL2 in vivo, as also shown in vitro (see Fig. 2D).
FHL2, p300, and β-catenin form a ternary complex.
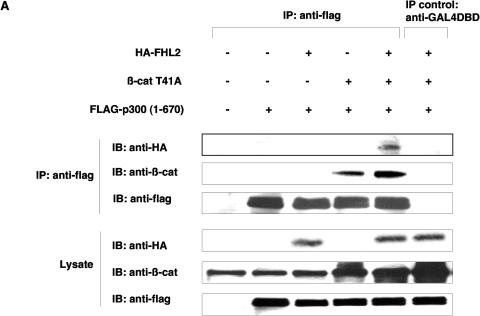
The finding of direct physical interaction between FHL2 and CBP/p300 suggested that FHL2, CBP/p300, and β-catenin might form a ternary complex. To test this hypothesis, we used the p300(1-670) fragment (which is able to interact with β-catenin but not with FHL2) in coimmunoprecipitation assays, with the view that FHL2 could be coprecipitated with p300(1-670) through its association with β-catenin. After transfection of different combinations of FLAG-tagged p300(1-670), HA-FHL2, and β-catenin T41A in 293 cells, cell lysates were precipitated by anti-FLAG antibody, followed by immunoblotting analysis with anti-HA or anti-β-catenin antibodies. When cotransfected with p300, β-catenin was found in the immune complex, confirming previous reports (38, 39). Remarkably, in the presence of transfected β-catenin, FHL2 was readily detected in the immune complex precipitated by anti-FLAG antibody, while it was barely detectable in the absence of transfected β-catenin (Fig. 4A). The irrelevant anti-GAL4DBD antibody failed to immunoprecipitate any of the three factors (Fig. 4A). These data strongly suggest that β-catenin is able to bind simultaneously to the N-terminal domain of p300 and FHL2 and that β-catenin, p300, and FHL2 might form a ternary complex.
FIG. 4.
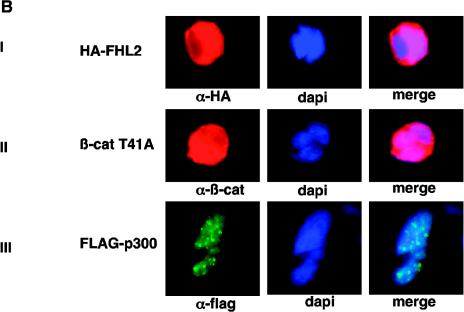
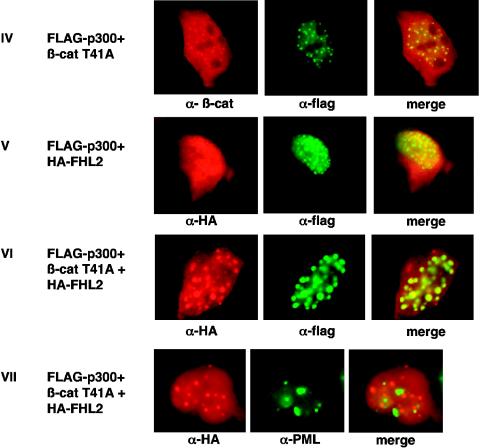
Formation of a ternary protein complex by FHL2, p300, and β-catenin in vivo. (A) Coimmunoprecipitation of FHL2, p300, and β-catenin (β-cat). 293 cells were transfected with vectors expressing HA-tagged FHL2, FLAG-tagged p300(1-670), and β-catenin T41A. Immunoprecipitation (IP) was performed as described for Fig. 3A. Immune complexes were analyzed by immunoblotting (IB) with anti-HA, anti-β-catenin, and anti-FLAG antibodies as indicated. The bottom panel shows the protein levels of FHL2, β-catenin, and p300 in the initial cell lysates. (B) Colocalization of FHL2, p300, and β-catenin T41A in the nucleus. 293 cells were transiently transfected with HA-FHL2, FLAG-p300, or β-catenin, either alone (panels I to III) or in different combinations (panels IV to VII). Cells were fixed and doubly labeled with either specific antibodies or DAPI (4′,6′-diamidino-2-phenylindole). To localize PML nuclear bodies, endogenous PML was immunostained with a specific PML polyclonal antibody (panel VII). Immunostained proteins were detected by fluorescence microscopy. Colocalization of p300 with FHL2 and β-catenin (panels V and VI) and absence of colocalization of FHL2 with PML (panel VII) were verified by laser-scanning confocal microscopy (data not shown). Merged images are shown in the right panels.
We next examined the cellular localizations of FHL2, β-catenins and p300 by immunofluorescence analysis. In a first step, each expression vector was individually transfected into 293 cells. When exogenously expressed alone, FHL2 was localized in both the cytoplasm and the nucleus (Fig. 4B, panel I), as was also observed for β-catenin (panel II). In contrast, overexpression of p300 resulted in nuclear aggregates (Fig. 4B, panel III), as previously reported (3). p300 was then cotransfected with FHL2, β-catenin, or both. In agreement with previous reports (29, 35), we observed that β-catenin localized mostly in p300-containing nuclear dots (Fig. 4B, panel IV). Interestingly, two-color immunofluorescence staining showed a clear colocalization of FHL2 and p300 in nuclear bodies (panel V). The shuttling of FHL2 to these bodies was more prominent when β-catenin was exogenously coexpressed (Fig. 4B, panel VI), suggesting that β-catenin may induce the formation of a ternary complex. Colocalization of FHL2, β-catenin, and p300 in the nucleus was also observed in HeLa cells (data not shown). Since it has been suggested that β-catenin and p300 colocalized in PML bodies (29, 35), we next sought to investigate whether FHL2 may be associated with this nuclear structure. Double-labeling experiments were carried out with HA-FHL2-, p300-, and β-catenin-transfected 293 cells by using polyclonal anti-PML antibodies and monoclonal anti-HA antibodies, and images were analyzed by fluorescence and confocal microscopy. As shown in Fig. 4B, panel VII, the PML nuclear bodies were clearly distinct from the sites of FHL2 localization, and they were adjacent to FHL2 bright dots. Taken together, these results indicate that FHL2, p300, and β-catenin colocalize in the nucleus but not in PML bodies.
Effects of endogenous FHL2 on transcription mediated by β-catenin/TCF.
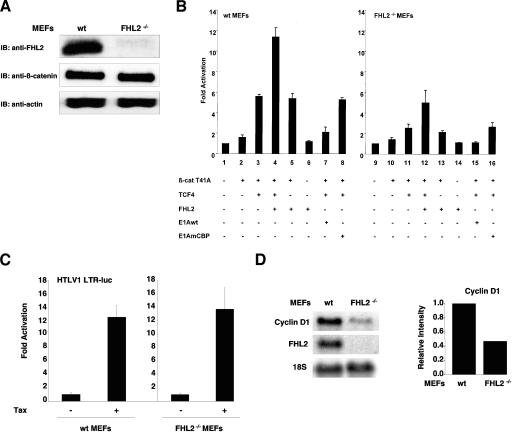
To explore the role of endogenous FHL2 in regulating transcription mediated by β-catenin/TCF, we examined TOPFLASH reporter activity in MEFs prepared from normal and FHL2-null mice (9). Endogenous β-catenin was expressed at similar levels in wild-type and FHL2−/− MEFs (Fig. 5A), and it was localized mainly at the cell membrane in both cell types (data not shown). In transient-transfection assays, exogenous expression of β-catenin T41A weakly increased TOPFLASH luciferase activity in normal MEFs, while coexpression of β-catenin with TCF4 activated the reporter gene by sixfold (Fig. 5B, bars 2 and 3), suggesting that TCF4 is a limiting factor in this condition. In contrast, β-catenin and TCF4 increased luciferase activity only by 2.5-fold in FHL2-null cells (Fig. 5B, bar 11), showing significantly lower activity than in normal cells. To determine whether this defect might be specifically attributed to the absence of FHL2, we expressed FHL2 by transient transfection in FHL2-null cells. β-Catenin/TCF4-mediated activation of the TOPFLASH reporter was restored in FHL2−/− cells after FHL2 transfection (Fig. 5B, compare bars 3 and 12). In wild-type MEFs, coexpression of FHL2, β-catenin, and TCF-4 resulted in a strong enhancement of TOPFLASH activity (bar 4). Activation of the reporter by β-catenin and TCF4 depended on the presence of TCF-binding sites in the promoter, since the FOPFLASH reporter activity was not modified by β-catenin and TCF4 in MEFs (data not shown). Besides, the basal activity of the FOPFLASH reporter was similar in normal and FHL2−/− cells (data not shown).
FIG. 5.
Effect of endogenous FHL2 on β-catenin/TCF target gene expression. (A) Lysates from MEFs at the third passage were analyzed by immunoblotting (IB) with anti-FHL2 or anti-β-catenin antibodies. Actin was used as a loading control. wt, wild type. (B) MEFs from wt or FHL2−/− mice were transfected with the TOPFLASH reporter plasmid (0.25 μg) and expression vectors for TCF4 (0.125 μg), β-catenin (β-cat) T41A (0.125 μg), wt or mutant E1A (0.125 μg), and FHL2 (0.5 μg). Error bars indicate standard deviations. (C) wt or FHL2-null MEFs were transfected with the HTLV1-LTR-LUC reporter (0.5 μg) and expression vector for Tax (0.5 μg). Luciferase activities were measured 24 h after transfection. (D) Analysis of cyclin D1 transcript in MEF cells. Total RNA was prepared from FHL2+/+ and FHL2−/− cells and analyzed by Northern blotting with cyclin D1 and FHL2 probes. Equal loading in each lane was verified by hybridization with a probe of the 18S rRNA gene. The relative cyclin D1/18S intensities were quantified with a PhosphorImager and are graphically depicted in the right panel. The intensity of cyclin D1/18S in FHL2+/+ cells was arbitrarily set to 1.
To assess the effect of CBP/p300 in the absence of FHL2, we coexpressed either CBP or p300 with β-catenin and TCF4 in MEFs and assayed for TOPFLASH reporter activity. CBP/p300 was fully able to induce β-catenin/TCF4-mediated activation in FHL2−/− cells, suggesting that the cooperation of CBP/p300 with β-catenin can bypass FHL2 (data not shown). It is known that expression of CBP/p300 starts at early stages of embryogenesis (22). We then examined the contribution of endogenous CBP/p300 in activating the TOP promoter in MEFs. When E1A was coexpressed with β-catenin and TCF4 in normal or FHL2−/− cells, luciferase activity was drastically decreased by wild-type E1A but not by E1AmCBP (Fig. 5B, compare bars 7 and 8 with bar 3 and bars 15 and 16 with bar 11), strongly suggesting that endogenous CBP/p300 is required for β-catenin/TCF-mediated activation. Thus, although FHL2 plays a significant role in β-catenin transcriptional activity, it is not required for the coactivation function of p300 on β-catenin.
The specificity of FHL2 in transcription activation was evaluated by testing the transactivating activity of the Tax gene of HTLV-1 on the viral LTR. As shown in Fig. 5C, the luciferase reporter gene was strongly activated by Tax in both wild-type and FHL2−/− cells. Thus, FHL2-independent transactivation was not impaired in FHL2-null cells.
To determine the role of FHL2 in regulation of β-catenin target gene transcription in vivo, we analyzed the expression of the cyclin D1 gene in normal and FHL2−/− MEFs by Northern blotting. As shown in Fig. 5D, the production of endogenous cyclin D1 transcripts was reduced by twofold in FHL2−/− cells compared to normal cells. This result further confirms that FHL2 participates in the regulation of β-catenin/TCF target genes in vivo.
FHL2 enhances β-catenin acetylation by p300.
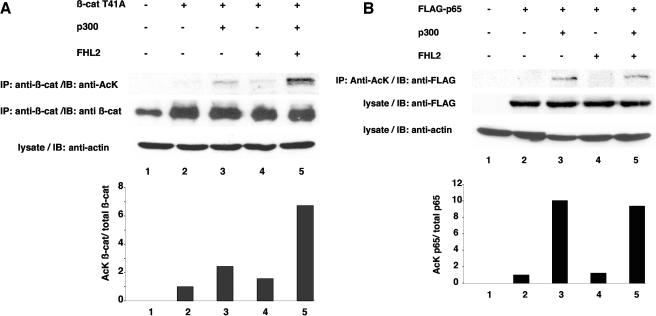
It has been shown that β-catenin can be acetylated by CBP/p300 (26, 45) and that acetylation at residue K345 might enhance the affinity of β-catenin for TCF4. In this study, we have shown that the synergistic cooperation of FHL2 with p300 in β-catenin-mediated transactivation was largely compromised by deletion of the HAT domain of p300 (Fig. 1A). This observation prompted us to investigate whether FHL2 might play a role in β-catenin acetylation by p300. β-Catenin was transiently transfected in COS-7 cells, either alone or in combination with FHL2 and p300. After immunoprecipitation with anti-β-catenin monoclonal antibodies, the immune complex was analyzed by immunoblotting with anti-acetyl-lysine antibodies. The acetylated form of β-catenin was detected in cells when β-catenin was exogenously overexpressed (Fig. 6A). Cotransfection of β-catenin with p300 resulted in a twofold increase of the acetylated pool of β-catenin. Importantly, exogenous expression of FHL2 increased the acetylated pool of β-catenin to the same extent. In this condition, β-catenin acetylation is likely achieved by endogenous CBP/p300. Coexpression of p300 and FHL2 significantly enforced acetylation of β-catenin by at least sixfold (Fig. 6A). Similar data were obtained with 293 cells (not shown).
FIG. 6.
FHL2 enhances acetylation of β-catenin by p300. (A) COS-7 cells were transfected with expression vectors for β-catenin (β-cat) T41A, p300, or FHL2 (5 μg each) as indicated. After immunoprecipitation (IP) with anti-β-catenin antibodies, the samples were resolved by SDS-PAGE. Acetylated β-catenin was revealed by immunoblotting with anti-acetyl-lysine antibodies (anti-AcK). Input levels of immunoprecipitated β-catenin were determined with anti-β-catenin antibodies. (B) FLAG-p65 was transfected in COS-7 cells, either alone or in combination with FHL2 and p300. The lysates were immunoprecipitated with anti-acetylated-lysine monoclonal antibodies, followed by immunoblotting (IB) with anti-FLAG polyclonal antibodies. Expression levels of FLAG-p65 in the lysates were determined with anti-FLAG polyclonal antibodies. Similar protein amounts in the lysates were verified by actin immunoblotting. For each lane, the ratio of acetylated β-catenin or FLAG-p65 on total β-catenin or FLAG-p65 was quantified and is graphically depicted in the bottom panel. The ratio was arbitrarily set to 1 for cells transfected with β-catenin T41A or FLAG-p65 alone.
In order to examine whether FHL2 might also enhance p300-mediated acetylation of other protein substrates, we next investigated the effect of FHL2 on the acetylation of the p65 subunit of NF-κB, which is known to be achieved by p300 (19). Moreover, FHL2 has been shown to enhance NF-κB activation (37). FLAG-p65 was transfected in COS-7 cells, either alone or in combination with FHL2 and p300. The lysates were immunoprecipitated with anti-acetylated-lysine antibodies, and the precipitate was analyzed by SDS-PAGE and immunoblotting with anti-FLAG antibodies. Cotransfection of p65 with p300 resulted in a 10-fold increase of the acetylated form of p65 (Fig. 6B). However, coexpression of FHL2 did not further increase the acetylated pool of p65. These results indicate that FHL2 does not modulate the acetylation of p65 by p300. Thus, FHL2 might play a specific role in enhancing β-catenin acetylation, presumably by stabilizing the interaction between β-catenin and p300.
Synergistic induction of the AR-responsive promoter by FHL2, CBP/p300, and β-catenin.
Since FHL2, CBP/p300, and β-catenin are involved in multiple transcriptional regulations, we asked whether their cooperation might function in a TCF/LEF-independent promoter context. To this end, we performed reporter assays using AR- and CREB-mediated transcription as model systems.
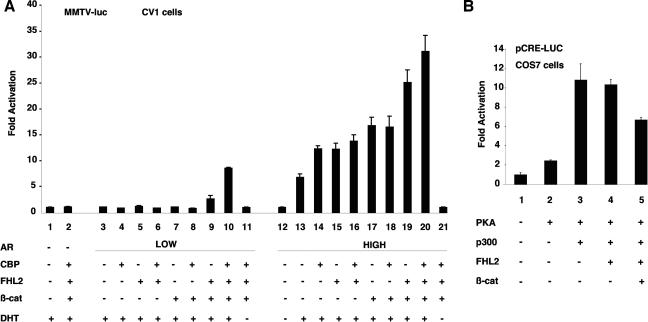
It has been known that FHL2, CBP/p300, and β-catenin individually participate in enhancement of AR transcription activity by direct binding with this nuclear receptor (31, 42). To study the effects of the three coactivators, we employed a transfection protocol recently devised by Lee et al. that allows comparison of coactivator activities on AR expressed at different levels (23). Either 5 or 50 ng of AR expression vector was transiently cotransfected with a mouse mammary tumor virus (MMTV) luciferase reporter in 3 × 105 CV-1 cells in the presence of FHL2, CBP/p300, and β-catenin. The three coactivators functioned synergistically on AR in a hormone-dependent manner only under conditions where AR was expressed at a low level (Fig. 7A, bar 10). When AR was expressed at a high level, each coactivator efficiently stimulated the reporter gene, but their coexpression resulted in only a minor additive effect (Fig. 7A, bars 14 to 20). These results are in agreement with a previous study showing that the synergy of the protein methyltransferase CARM1, p160, and CBP/p300 depends on low-level expression of AR in cells (23). Although transcriptional activity of AR alone was detectable only when AR was transfected at the higher dose (Fig. 7A, compare bars 13 and 3), both AR and the hormone were indispensable for synergistic action of the coactivators (Fig. 7A, compare bars 10 and 11), showing that the synergy operated specifically on AR.
FIG. 7.
Coactivator function of FHL2, CBP/p300, and β-catenin in an AR-dependent promoter context. (A) Synergistic activation of the MMTV promoter by FHL2, CBP/p300, and β-catenin (β-cat) at low levels of AR. CV1 cells were transfected with the MMTV-Luc reporter plasmid (0.5 μg) and expression vectors for FHL2, CBP, or wild-type β-catenin (0.5 μg each) or AR (low, 5 ng; high, 50 ng), as indicated. The medium was supplemented with 10 nM dihydrotestosterone (DHT) after transfection, and luciferase activities were determined 2 days later. Values are expressed as fold activation compared with cells transfected with the reporter and the empty vector. Error bars indicate standard deviations. (B) COS-7 cells were transfected with a luciferase reporter plasmid harboring four CREs followed by a basic promoter element (0.5 μg) and expression vectors for FHL2 (0.5 μg), p300 (0.5 μg), PKA (0.25 μg), or wild-type β-catenin (0.5 μg) as indicated. Conditions were as described for Fig. 1.
FHL2 and CBP/p300 have been shown to interact with and coactivate CREB (8, 13). To test whether FHL2 cooperates with CBP/p300 in CREB-mediated transcription, we carried out reporter assays in COS-7 cells with a luciferase reporter gene under control of CREs. Although FHL2 and p300 individually increased endogenous CREB activity, together they had little additional effect on reporter gene expression (data not shown). Phosphorylation of CREB by PKA enhanced the overall level of transactivation but failed to reveal cooperation between the coactivators (Fig. 7B). In addition, β-catenin, either alone or in combination with FHL2 and p300, decreased CREB activity (data not shown and Fig. 7B), suggesting that β-catenin may titrate p300 and FHL2 from the CREB complex. Collectively, our data show that while FHL2, CBP/p300, and β-catenin can synergistically activate AR-dependent transcription, this synergy is no longer observed for CREB-dependent transcription, in which β-catenin is not directly involved.
DISCUSSION
Coactivation of β-catenin by FHL2 and CBP/p300.
In this study, we have demonstrated that the coactivator FHL2 directly interacts and functionally cooperates with CBP/p300 in stimulating β-catenin/TCF-mediated transcription. Furthermore, we present evidence showing that FHL2, CBP/p300, and β-catenin form a ternary complex in vivo and that these proteins, when exogenously overexpressed, colocalize in nuclear structures. The ability of β-catenin to bind simultaneously FHL2 and CBP/p300 is strongly supported by previous studies showing that the CBP/p300 interaction domain in β-catenin covers the arm repeats 10 to 12 and the C-terminal region, while the FHL2-binding domain extends through either arm repeats 1 to 9 or the N-terminal region of β-catenin (18, 27, 39, 44). Because LIM domains are known to function as adapters and modifiers in protein interactions (10), FHL2 might act as a cofactor of CBP/p300, by stabilizing the interaction between CBP/p300 and β-catenin. This notion is strongly supported by the finding that β-catenin/TCF transcriptional activity was significantly downregulated in FHL2-null MEFs, while functional inactivation of endogenous CBP/p300 by the adenovirus protein E1A fully inhibited the coactivator function of FHL2 on β-catenin/TCF-dependent transcription.
Although FHL2-null mice do not show any developmental defect (9), we found that β-catenin/TCF-dependent transcription was decreased by twofold in FHL2−/− MEFs and that endogenous expression of the Wnt target gene cyclin D1 was also significantly reduced. It is unlikely that the cellular systems controlling β-catenin turnover or nuclear translocation might be affected in FHL2−/− cells, since they are indispensable for animal development and survival. Moreover, the total amounts and membranous localization of β-catenin were not modified in FHL2−/− MEFs, and nuclear translocation of β-catenin by LiCl treatment was induced to similar extents in FHL2−/− and wild-type cells (unpublished data). Therefore, reduced β-catenin/TCF activity may rather be attributed to the absence of FHL2 coactivator function in these cells. Despite the absence of a grossly detectable phenotype (9), FHL2-null mice show an altered cardiac hypertrophic response to β-adrenergic stimulation, suggesting that FHL2 is capable of modifying cardiac responses to environmental stress (20). It will be important to determine whether FHL2 deficiency might affect tumorigenic processes associated with abnormal reactivation of the Wnt/β-catenin pathway.
Several studies have suggested that cooperation between different coactivators of β-catenin might depend on cellular or promoter context (7, 35). For instance, the oncoprotein SKI was shown to interact with FHL2. These two factors synergistically activated β-catenin-regulated promoters specifically in human melanoma cells, while they repressed β-catenin activity in other cell types (7). Interestingly, CBP/p300 additionally stimulated the SKI/FHL2/β-catenin complex in melanoma cells. In another study, the PML protein was shown to interact with β-catenin and to cooperate with CBP/p300 in enhancing transcription of the siamois and ARF promoters but not the cyclin D1 promoter (35). In the present study, however, FHL2 and CBP/p300 were found to coactivate β-catenin/TCF-dependent transcription from a variety of Wnt-responsive promoters and in different cell types. Because FHL2 has been shown to inhibit β-catenin-induced transcriptional activity specifically in myoblasts (27), it will be interesting to investigate whether CBP/p300 plays a role in this process.
The cooperation between FHL2 and CBP/p300 is dependent on their association with β-catenin.
Our observation that the CH3 domain of CBP can associate with FHL2 but not with β-catenin in GST pull-down assays supports a direct interaction between FHL2 and CBP. Thus, it is possible that FHL2 and CBP/p300 form a regulatory complex that operates independently of β-catenin. This hypothesis was ruled out by our findings that FHL2 and CBP failed to cooperate on the AR-responsive MMTV promoter in the absence of nuclear β-catenin and on a CREB-dependent promoter on which β-catenin is inactive. These results favor the notion that β-catenin may be required for synergistic cooperation between FHL2 and CBP/p300, although further studies in different experimental settings are still needed.
The androgen receptor recruits multiple transcriptional cofactors and represents an interesting model for studying synergistic cooperation among coactivators. It has been shown recently that the functional synergy between the methyltransferase CARM1 and the coactivators p160 and CBP/p300 could be detected only when AR was weakly expressed (23). Those authors argued that highly expressed AR might fulfill the activation function by itself, through maximal occupancy of the cognate promoter elements. Our results, showing effective coactivation of the MMTV promoter by FHL2, CBP/p300, and β-catenin only when AR was expressed at low levels, are consistent with that report. Thus, the relative expression levels of transcription factors and coactivators may be critical in achieving transcriptional activation. Similar considerations might account for discordant results on the ability of p300 to stimulate β-catenin activity on the cyclin D1 promoter (references 18 and 35 and this study).
FHL2 enhances acetylation of β-catenin by p300.
We have shown that deletion of the HAT domain of p300 strongly reduces its coactivator synergy with FHL2 (Fig. 1A), suggesting that the acetyltransferase activity of p300 plays an important role in the cooperation between these coactivators. However, confirming previous reports (38, 39), we found that the HAT domain was not necessary for p300 binding to FHL2 or β-catenin (Fig. 2B). Thus, reduced synergy might be due to the lack of AT activity rather than to defects in interaction between p300ΔHAT, FHL2, and β-catenin. On the other hand, p300ΔHAT displayed residual synergy with FHL2 in stimulating β-catenin activity on reporter gene expression. This suggests that, besides its AT activity, p300 may cooperate with FHL2 and β-catenin through its capacity to associate other regulatory factors with the complex or to link the β-catenin/TCF complex to the general transcription machinery.
A growing list of transcription factors has been shown to be acetylated (for a review, see reference 6). In most cases, acetylation is responsible for transcription stimulation by enhancing DNA-binding activity or regulating protein-protein interaction. We have previously observed that acetylation of β-catenin by p300 increases its binding affinity for TCF4 (26). Furthermore, we have shown in the present study that FHL2 expression enhanced β-catenin acetylation by p300 (Fig. 6). By contrast, FHL2 was not able to stimulate p300-dependent acetylation of p65 NF-κB, although the LIM protein might behave as a coactivator of p65 (37). Thus, increased β-catenin acetylation might be a specific mechanism by which FHL2 contributes to activated transcription of Wnt target genes. Whether acetylation affects β-catenin interaction with other nuclear partners such as TATA-binding protein and whether CBP/p300 also acetylates different known coactivators of β-catenin remain to be determined.
Acknowledgments
We thank J. Chen for providing FHL2 knockout mice and R. Schüle for providing the polyclonal anti-FHL2 antibody. We are grateful to S. Pêtres for producing FHL2 by a baculovirus system and to E. Perret for help with confocal microscopy. We thank M. Benkirane, J. Brady, G. Castoria, P. Chambon, H. Clevers, R. Gaynor, A. Harel-Bellan, A. Hecht, P. Hinds, K. T. Jeang, J.-G. Judde, R. Kemler, T. Kouzarides, R. T. Moon, Y. Nakatani, R. Pestell, and J. Seeler for kindly providing constructs and antibodies and C. Perret for helpful discussions.
This work was supported in part by grant R03/75-31 of the Comité de Paris of La Ligue contre le Cancer. C.L. was supported by the Ecole Normale Supérieure de Lyon.
REFERENCES
- 1.Barker, N., A. Hurlstone, H. Musisi, A. Miles, M. Bienz, and H. Clevers. 2001. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 20:4935-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batlle, E., J. T. Henderson, H. Beghtel, M. M. van den Born, E. Sancho, G. Huls, J. Meeldijk, J. Robertson, M. van de Wetering, T. Pawson, and H. Clevers. 2002. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 111:251-263. [DOI] [PubMed] [Google Scholar]
- 3.Boisvert, F. M., M. J. Kruhlak, A. K. Box, M. J. Hendzel, and D. P. Bazett-Jones. 2001. The transcription coactivator CBP is a dynamic component of the promyelocytic leukemia nuclear body. J. Cell Biol. 152:1099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brannon, M., M. Gomperts, L. Sumoy, R. T. Moon, and D. Kimelman. 1997. A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 11:2359-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brantjes, H., N. Barker, J. van Es, and H. Clevers. 2002. TCF: Lady Justice casting the final verdict on the outcome of Wnt signalling. Biol. Chem. 383:255-261. [DOI] [PubMed] [Google Scholar]
- 6.Chan, H. M., and N. B. La Thangue. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114:2363-2373. [DOI] [PubMed] [Google Scholar]
- 7.Chen, D., W. Xu, E. Bales, C. Colmenares, M. Conacci-Sorrell, S. Ishii, E. Stavnezer, J. Campisi, D. E. Fisher, A. Ben-Ze'ev, and E. E. Medrano. 2003. SKI activates Wnt/beta-catenin signaling in human melanoma. Cancer Res. 63:6626-6634. [PubMed] [Google Scholar]
- 8.Chrivia, J. C., R. P. Kwok, N. Lamb, M. Hagiwara, M. R. Montminy, and R. H. Goodman. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365:855-859. [DOI] [PubMed] [Google Scholar]
- 9.Chu, P. H., W. M. Bardwell, Y. Gu, J. Ross, Jr., and J. Chen. 2000. FHL2 (SLIM3) is not essential for cardiac development and function. Mol. Cell. Biol. 20:7460-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawid, I. B., J. J. Breen, and R. Toyama. 1998. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 14:156-162. [DOI] [PubMed] [Google Scholar]
- 11.Du, X., P. Hublitz, T. Gunther, D. Wilhelm, C. Englert, and R. Schule. 2002. The LIM-only coactivator FHL2 modulates WT1 transcriptional activity during gonadal differentiation. Biochim. Biophys. Acta 1577:93-101. [DOI] [PubMed] [Google Scholar]
- 12.El Mourabit, H., S. Muller, L. Tunggal, M. Paulsson, and M. Aumailley. 2003. Characterization of recombinant and natural forms of the human LIM domain-containing protein FHL2. Protein Expr. Purif. 32:95-103. [DOI] [PubMed] [Google Scholar]
- 13.Fimia, G. M., D. De Cesare, and P. Sassone-Corsi. 1999. CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature 398:165-169. [DOI] [PubMed] [Google Scholar]
- 14.Fimia, G. M., D. De Cesare, and P. Sassone-Corsi. 2000. A family of LIM-only transcriptional coactivators: tissue-specific expression and selective activation of CREB and CREM. Mol. Cell. Biol. 20:8613-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giles, R. H., J. H. van Es, and H. Clevers. 2003. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys. Acta 1653:1-24. [DOI] [PubMed] [Google Scholar]
- 16.He, T. C., A. B. Sparks, C. Rago, H. Hermeking, L. Zawel, L. T. da Costa, P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Identification of c-MYC as a target of the APC pathway. Science 281:1509-1512. [DOI] [PubMed] [Google Scholar]
- 17.Hecht, A., C. M. Litterst, O. Huber, and R. Kemler. 1999. Functional characterization of multiple transactivating elements in beta-catenin, some of which interact with the TATA-binding protein in vitro. J. Biol. Chem. 274:18017-18025. [DOI] [PubMed] [Google Scholar]
- 18.Hecht, A., K. Vleminckx, M. P. Stemmler, F. van Roy, and R. Kemler. 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 19:1839-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiernan, R., V. Bres, R. W. Ng, M. P. Coudart, S. El Messaoudi, C. Sardet, D. Y. Jin, S. Emiliani, and M. Benkirane. 2003. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J. Biol. Chem. 278:2758-2766. [DOI] [PubMed] [Google Scholar]
- 20.Kong, Y., J. M. Shelton, B. Rothermel, X. Li, J. A. Richardson, R. Bassel-Duby, and R. S. Williams. 2001. Cardiac-specific LIM protein FHL2 modifies the hypertrophic response to beta-adrenergic stimulation. Circulation 103:2731-2738. [DOI] [PubMed] [Google Scholar]
- 21.Kramps, T., O. Peter, E. Brunner, D. Nellen, B. Froesch, S. Chatterjee, M. Murone, S. Zullig, and K. Basler. 2002. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell 109:47-60. [DOI] [PubMed] [Google Scholar]
- 22.Kung, A. L., V. I. Rebel, R. T. Bronson, L. E. Ch'ng, C. A. Sieff, D. M. Livingston, and T. P. Yao. 2000. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 14:272-277. [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, Y. H., S. S. Koh, X. Zhang, X. Cheng, and M. R. Stallcup. 2002. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol. Cell. Biol. 22:3621-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehrmann, H., L. L. Pritchard, and A. Harel-Bellan. 2002. Histone acetyltransferases and deacetylases in the control of cell proliferation and differentiation. Adv. Cancer Res. 86:41-65. [DOI] [PubMed] [Google Scholar]
- 25.Levy, L., C. Neuveut, C. A. Renard, P. Charneau, S. Branchereau, F. Gauthier, J. T. Van Nhieu, D. Cherqui, A. F. Petit-Bertron, D. Mathieu, and M. A. Buendia. 2002. Transcriptional activation of interleukin-8 by beta-catenin-Tcf4. J. Biol. Chem. 277:42386-42393. [DOI] [PubMed] [Google Scholar]
- 26.Levy, L., Y. Wei, C. Labalette, Y. Wu, C. A. Renard, M. A. Buendia, and C. Neuveut. 2004. Acetylation of beta-catenin by p300 regulates beta-catenin-Tcf4 interaction. Mol. Cell. Biol. 24:3404-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, B., R. Schneider, S. Janetzky, Z. Waibler, P. Pandur, M. Kuhl, J. Behrens, K. von der Mark, A. Starzinski-Powitz, and V. Wixler. 2002. The LIM-only protein FHL2 interacts with beta-catenin and promotes differentiation of mouse myoblasts. J. Cell Biol. 159:113-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matos, A. R., A. d'Arcy-Lameta, M. Franca, S. Petres, L. Edelman, J. Kader, Y. Zuily-Fodil, and A. T. Pham-Thi. 2001. A novel patatin-like gene stimulated by drought stress encodes a galactolipid acyl hydrolase. FEBS Lett. 491:188-192. [DOI] [PubMed] [Google Scholar]
- 29.Miyagishi, M., R. Fujii, M. Hatta, E. Yoshida, N. Araya, A. Nagafuchi, S. Ishihara, T. Nakajima, and A. Fukamizu. 2000. Regulation of Lef-mediated transcription and p53-dependent pathway by associating beta-catenin with CBP/p300. J. Biol. Chem. 275:35170-35175. [DOI] [PubMed] [Google Scholar]
- 30.Morlon, A., and P. Sassone-Corsi. 2003. The LIM-only protein FHL2 is a serum-inducible transcriptional coactivator of AP-1. Proc. Natl. Acad. Sci. USA 100:3977-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller, J. M., U. Isele, E. Metzger, A. Rempel, M. Moser, A. Pscherer, T. Breyer, C. Holubarsch, R. Buettner, and R. Schule. 2000. FHL2, a novel tissue-specific coactivator of the androgen receptor. EMBO J. 19:359-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller, J. M., E. Metzger, H. Greschik, A. K. Bosserhoff, L. Mercep, R. Buettner, and R. Schule. 2002. The transcriptional coactivator FHL2 transmits Rho signals from the cell membrane into the nucleus. EMBO J. 21:736-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peifer, M., and P. Polakis. 2000. Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science 287:1606-1609. [DOI] [PubMed] [Google Scholar]
- 34.Renard, C. A., G. Fourel, M. P. Bralet, C. Degott, A. De La Coste, C. Perret, P. Tiollais, and M. A. Buendia. 2000. Hepatocellular carcinoma in WHV/N-myc 2 transgenic mice: oncogenic mutations of beta-catenin and synergistic effect of p53 null alleles. Oncogene 19:2678-2686. [DOI] [PubMed] [Google Scholar]
- 35.Shtutman, M., J. Zhurinsky, M. Oren, E. Levina, and A. Ben-Ze'ev. 2002. PML is a target gene of beta-catenin and plakoglobin, and coactivates beta-catenin-mediated transcription. Cancer Res. 62:5947-5954. [PubMed] [Google Scholar]
- 36.Shtutman, M., J. Zhurinsky, I. Simcha, C. Albanese, M. D'Amico, R. Pestell, and A. Ben-Ze'ev. 1999. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 96:5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stilo, R., A. Leonardi, L. Formisano, B. Di Jeso, P. Vito, and D. Liguoro. 2002. TUCAN/CARDINAL and DRAL participate in a common pathway for modulation of NF-kappaB activation. FEBS Lett. 521:165-169. [DOI] [PubMed] [Google Scholar]
- 38.Sun, Y., F. T. Kolligs, M. O. Hottiger, R. Mosavin, E. R. Fearon, and G. J. Nabel. 2000. Regulation of beta-catenin transformation by the p300 transcriptional coactivator. Proc. Natl. Acad. Sci. USA 97:12613-12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takemaru, K. I., and R. T. Moon. 2000. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J. Cell Biol. 149:249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tetsu, O., and F. McCormick. 1999. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422-426. [DOI] [PubMed] [Google Scholar]
- 41.Townsley, F. M., A. Cliffe, and M. Bienz. 2004. Pygopus and Legless target Armadillo/beta-catenin to the nucleus to enable its transcriptional co-activator function. Nat. Cell Biol. 6:626-633. [DOI] [PubMed] [Google Scholar]
- 42.Truica, C. I., S. Byers, and E. P. Gelmann. 2000. Beta-catenin affects androgen receptor transcriptional activity and ligand specificity. Cancer Res. 60:4709-4713. [PubMed] [Google Scholar]
- 43.Wei, Y., M. Fabre, S. Branchereau, F. Gauthier, G. Perilongo, and M. A. Buendia. 2000. Activation of beta-catenin in epithelial and mesenchymal hepatoblastomas. Oncogene 19:498-504. [DOI] [PubMed] [Google Scholar]
- 44.Wei, Y., C. A. Renard, C. Labalette, Y. Wu, L. Levy, C. Neuveut, X. Prieur, M. Flajolet, S. Prigent, and M. A. Buendia. 2003. Identification of the LIM protein FHL2 as a coactivator of beta-catenin. J. Biol. Chem. 278:5188-5194. [DOI] [PubMed] [Google Scholar]
- 45.Wolf, D., M. Rodova, E. A. Miska, J. P. Calvet, and T. Kouzarides. 2002. Acetylation of beta-catenin by CREB-binding protein (CBP). J. Biol. Chem. 277:25562-25567. [DOI] [PubMed] [Google Scholar]
- 46.Zindy, F., D. E. Quelle, M. F. Roussel, and C. J. Sherr. 1997. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene 15:203-211. [DOI] [PubMed] [Google Scholar]