Abstract
Nucleolar function and the cellular response to DNA damage have long been studied as distinct disciplines. New research and a new appreciation for proteins holding multiple functional roles, however, is beginning to change the way we think about the crosstalk among distinct cellular processes. Here, we focus on the crosstalk between the DNA damage response and the nucleolus, including a comprehensive review of the literature that reveals a role for conventional DNA repair proteins in ribosome biogenesis, and conversely, ribosome biogenesis proteins in DNA repair. Furthermore, with recent advances in nucleolar proteomics and a growing list of proteins that localize to the nucleolus, it is likely that we will continue to identify new DNA repair proteins with a nucleolar-specific role. Given the importance of ribosome biogenesis and DNA repair in essential cellular processes and the role that they play in diverse pathologies, continued elucidation of the overlap between these two disciplines will be essential to the advancement of both fields and to the development of novel therapeutics.
Graphical abstract
We review the role for conventional DNA repair proteins in ribosome biogenesis and ribosome biogenesis factors in DNA repair.
Introduction
The nucleolus is non-membranous nuclear organelle found in eukaryotic organisms. Its primary function is in the making of ribosomes, and it forms in response to the transcription of the ribosomal DNA (rDNA) and to the recruitment of ribosome biogenesis factors1–3. The pre-ribosomal RNA (pre-rRNA) is transcribed by RNA polymerase I (RNAPI) from the rDNA that is arranged in tandem arrays on chromosomes 13, 14, 15, 21, and 22 in human diploid cells4–6. A series of endo- and exo- nucleolytic cleavages removes the transcribed spacer sequences from the pre-rRNA to release the mature 18S, 5.8S, and 28S rRNAs7, 8. In addition to these RNA processing steps, post-transcriptional modifications by small nucleolar ribonucleoprotein complexes (snoRNPs) and assembly with ribosomal proteins and the RNA polymerase III-transcribed 5S rRNA are required for the maturation of a fully functional ribosome3, 9–11 (Figure 1). The process is complex and defects at any step can be embryonic lethal, or can lead to rare congenital disorders known as ribosomopathies12–20, or even cancer21–24. Much of what we know about ribosome biogenesis in eukaryotes stems from extensive work in budding yeast25–27; as a consequence, however, many aspects of the process in humans, including its multifaceted regulation, remain to be elucidated8, 28.
Figure 1. Ribosome biogenesis at a glance.
The making of a mature ribosome begins with the transcription of the pre-rRNA from an rDNA locus. The pre-rRNA is then processed to remove internal- and external- transcribed spacer sequences (5’ETS; ITS1; ITS2; 3’ETS) and modified by snoRNPs. In addition, the pre-ribosomal subunits are assembled with ribosomal proteins and the RNAPIII transcribed 5S rRNA, and then exported to the cytoplasm where they can join to form a translationally competent ribosome. The black sticks with a ball on top ( ) indicate rRNA modifications, and the blue (
) indicate rRNA modifications, and the blue ( ) and orange (
) and orange ( ) circles represent ribosomal proteins.
) circles represent ribosomal proteins.
The cellular response to DNA damage, like the efficient and accurate production of ribosomes, is an essential cellular function and is critical for the maintenance of genomic fidelity and cell viability. Each cell in the human body has the potential to experience tens of thousands of DNA lesions daily from both internal (e.g. oxidation; alkylation) and external (e.g. ultraviolet light; γ-irradiation) sources that if left alone can lead to genetic mutations, chromosomal aberrations, and disease29, 30. As a result, an extensive network has evolved to sense and respond to these lesions, varying from cell-cycle arrest and DNA repair to apoptosis31. A common player in the DNA damage response is the tumor suppressor protein, p53, implicated in the regulation of cell cycle arrest and proapoptotic functions of the DNA damage response32.
Repair of genotoxic damage, on the other hand, occurs by a number of pathways depending on the type of DNA lesion (Table 1). The base excision repair (BER) pathway, for instance, repairs small lesions catalyzing the removal and replacement of damaged bases; whereas, the nucleotide excision repair (NER) pathway addresses more bulky lesions like cyclobutane pyrimidine dimers typically caused by UV exposure33. Interstrand crosslinks are repaired by the Fanconi anemia pathway, mismatched bases are corrected by mismatch repair, and multiple mechanisms have evolved to repair DNA double strand breaks (DSBs), including the non-homologous end joining (NHEJ) pathway and homology-directed recombination (HDR). In HDR, DNA ends are resected and a sister chromatid is used as a template, thus occurring commonly in S and G2 phases of the cell cycle. NHEJ, on the other hand, is commonly considered error-prone with DNA ends simply recognized and ligated30.
Table 1.
DNA damage response pathways and their protein components.
| DNA repair pathway | Abbreviation | Targeted DNA lesions | Common proteins involved |
|---|---|---|---|
| Base excision repair | BER | Oxidation; Alkylation | UNG; APEX1; POLB; LIG3 |
| Nucleotide excision repair | NER | Cyclobutane pyrimidine dimers |
XPA- XPG; XPV; ERCC1; POLD; LIG1 |
| Mismatch repair | MMR | Mismatched nucleotide incorporation |
MLH1; PMS2; PCNA; RFC1-5; EXO1; POLD |
| Fanconi anemia pathway | FA | Interstrand crosslinks | FANC proteins |
| Non-homologous end joining | NHEJ | Double-strand breaks | 53BP1; XRCC4; XRCC5 (Ku80); XRCC6 (Ku70); PRKDC (DNA- PK) |
| Homology-directed repair | HDR | Double-strand breaks | 53BP1; BRCA1; BRCA2; RPA2; RAD51; RAD52; SMC5; SMC6 |
Common pathways involved in DNA repair, the lesions each pathway targets, and the proteins commonly associated with each pathway.
Despite the treatment of DNA repair and ribosome biogenesis as distinct disciplines, new research is beginning to recognize a clear crosstalk between the nucleolus and the DNA damage response. Here, we review the literature on the current state of nucleolar proteomics, the nucleolar response to DNA damage, and DNA repair in the nucleolus itself. In addition, we highlight nucleolar proteins with roles in DNA repair, and conversely, DNA repair proteins with roles in ribosome biogenesis. These interconnections suggest that our current way of labeling proteins with a single functional role is in the process of being upended. Furthermore, as the number of proteins localized to the nucleolus increases, it is conceivable that more proteins will be assigned unexpected novel roles in the making of ribosomes. Given the importance of these processes and their role in a multitude of diseases, continued probing of the overlap of these disciplines will enrich the way we think about human disease pathogenesis and treatment.
Nucleolar proteomics reveals proteins involved in DNA repair
The proteomics field has advanced significantly over the past decade due in large part to substantial improvements in the sensitivity and accuracy of mass spectrometry platforms34. Coupled with these advancements is progress in the biochemical or biological fractionation of cells using methods such as chromatography and centrifugation34. The field of nucleolar proteomics has benefited largely from these developments, which has allowed not only the determination of proteins that comprise the human nucleolus, but also how the nucleolar proteome changes after exposure to different cellular stressors. Pioneering work in this field, for instance, evaluated the changes in the nucleolar proteome in cells treated with the transcriptional inhibitor actinomycin D (ActD)35–37. More recent work, on the other hand, examined the nucleolar proteome of the human T-cell38, and how it changes in response to infection by the Human Immunodeficiency Virus (HIV)39. The significance of these advancements lies in providing the foundation for better understanding the role of the nucleolus in human cells by analysis of its protein constituents.
The nucleolar proteome, according to the most recent update of the Nucleolar Proteome Database (NOPdb3.0), includes 4,500 proteins from purified nucleoli that localize to the human nucleolus40. These data not only represent a significant increase when compared to the 150 proteins so far identified in the yeast nucleolar proteome41, but also represent the relatively novel concept of an expanded role for the human nucleolus in diverse cellular processes. Proteins that localize to the nucleolus represent functional classes including not only ribosome biogenesis factors (ribosomal proteins; RNA helicases; RNA-binding proteins; RNA-modifying enzymes), but also DNA-binding proteins, and DNA repair proteins36, 42, 43 (Figure 2). In some respects, these functional classes are not surprising given that the “plurifunctional nucleolus” was first hypothesized based on evidence for a role for the nucleolus in the synthesis and processing of some mRNAs, the signal recognition particle, telomerase RNA and tRNAs44. Subsequent research has also supported a role for the nucleolus in the regulation of mitosis, the cell cycle, the stress response, and apoptosis42. What is surprising, however, is the large number of nucleolar proteins that are involved in DNA-associated processes, like DNA repair, of which we will discuss in further detail here in this review.
Figure 2. Classification of nucleolar proteins by functional category.
Pie chart from an early study by Andersen and colleagues that depicts the functional categories of proteins that localize to the human HeLa cell nucleolus36. The number of proteins that fall into each category is indicated in parentheses. Notably, a proportion of nucleolar proteins were classified as DNA repair proteins. Now, as many as 4,500 proteins may localize to the human nucleolus40, and in our analysis of NOPdb40, the human T-cell nucleolar proteome38, and the Human Protein Atlas142, 166 unique nucleolar proteins can be classified as DNA repair proteins (Supplemental Table 1). Reprinted by permission from Macmillan Publishers Ltd: Nature Publishing Group. http://www.nature.com/nature/journal/v433/n7021/full/nature03207.html
Analysis of the two published nucleolar proteomes and the Human Protein Atlas database reveal that 166 unique DNA repair proteins localize to the nucleolus based on Gene Ontology (GO) Consortium categorization (GO: 0006281) (Table 2; Supplemental Table 1). One hypothesis to account for the abundance of DNA repair proteins in the nucleolus is that they are sequestered there until required for their functional role in DNA repair, implicating the nucleolus as part of the DNA damage response. Moore and colleagues observed a mobilization of DNA repair proteins associated with the NHEJ pathway from the nucleolus to the nucleoplasm upon UV and ionizing radiation45. Similar observations have been made with the BER enzymes apurinic/apyrmidinic endodeoxyribonuclease 1 (APEX1)46 and Alpha-Ketoglutarate-Dependent Dioxygenase AlkB Homolog 2 (ALKBH2)47, and the DNA helicase Werner Syndrome protein (WRN)48, 49. An alternative hypothesis, however, is that these proteins possess a nucleolar function in the absence of DNA damage. It is the latter hypothesis that we elaborate on in this review.
Table 2.
Human nucleolar proteomes include proteins involved in DNA repair.
| NOPDB*40 | T-cell nucleolar proteome38 |
Human Protein Atlas142 |
|
|---|---|---|---|
| Total proteins | 2717 | 872 | 1153 |
| DNA repair proteins | 136 | 38 | 40 |
| DNA repair proteins with yeast ortholog |
89 | 30 | 23 |
Current database not available; total protein number based on a prior version of the database.
Three databases exist on proteins that localize to the human nucleolus. Within each database, a subset of proteins are classified as DNA repair proteins based on Gene Ontology (GO) Consortium categorization (GO: 0006281).
Functional roles for DNA repair proteins in the nucleolus
A wealth of evidence is beginning to support a dual role for DNA repair proteins in not only DNA repair, but in the nucleolar function of making ribosomes. Antoniali and colleagues, for example, reviewed nicely the role of DNA repair proteins in RNA metabolism50. Here, we focus more narrowly on the role of DNA repair proteins in the synthesis of ribosomes, and highlight a few examples, beginning with a brief summary of their more commonly recognized role in DNA repair, followed by evidence for a role in ribosome biogenesis.
Apurinic/Apyrmidinic endodeoxyribonuclease 1 (APEX1; APE1; REF-1)
Nucleolar localization of APEX1 was identified in two out of the three nucleolar proteome datasets; however, it is more commonly known for its nucleoplasmic role in the BER DNA repair pathway that recognizes and repairs oxidized or otherwise modified DNA bases. Following identification and removal of a damaged base by a DNA glycosylase, APEX1 is the primary endonuclease that then catalyzes the incision of the DNA backbone at the abasic (AP) site allowing for repair by a DNA polymerase and ligase51. In addition to its role in BER however additional roles for the protein are beginning to emerge52, 53, including a nucleolar-specific role in ribosome biogenesis.
Multiple lines of evidence exist in support of a role for APEX1 in the accurate and efficient production of ribosomes. APEX1 co-immunoprecipitates known proteins involved in ribosome biogenesis, including nucleophosmin (NPM1), ribosomal protein lateral stalk subunit P0 (RPLP0), and ribosomal protein SA (uS2; RPSA)54. NPM1 is a multifunctional protein involved in multiple aspects of ribosome biogenesis, RPLP0 is a ribosomal protein associated with the large (60S) subunit of the ribosome, and uS2 is protein required for the maturation of the small (40S) subunit of the ribosome. These proteins thus have diverse functions in ribosome biogenesis, which suggests that APEX1’s role may too be wide-ranging in terms of its effect on the production of ribosomes. Further supporting this claim is that APEX1 also co-immunoprecipitates the 47S primary transcript, and both the 18S and 28S mature rRNA species54. One role APEX1 may be performing is in the role of RNA quality control by removing damaged RNA bases. APEX1-depleted cells treated with hydrogen peroxide to generate oxidative damage revealed that oxidation of rRNA was higher when compared to control cells, and protein synthesis and cell proliferation was decreased54. Additionally, in APEX1-depleted cells, metabolic labeling experiments revealed impaired pre-rRNA transcription55. These data therefore suggest that APEX1 may be important in the surveillance and removal of damaged RNA that directly affects the production of ribosomes.
With data supporting two distinct functional roles for APEX1 in different subcellular locations, the question as to how these roles are regulated remains. What we know is that sequestration of APEX1 in the nucleolus is dependent on rDNA transcription and caused by the direct interaction between APEX1 and NPM154, and specific lysine residues in APEX1’s N-terminus56. Acetylation of these N-terminal lysine residues results in the disruption of the interaction between NPM1 and APEX1 and decreased nucleolar localization46. Interestingly, treatment of cells with methyl methanesulfonate (MMS), which is an alkylating agent causing DNA damage repaired by the BER pathway, also leads to lysine acetylation of APEX1, suggesting that this may be a mechanism by which this protein switches between its nucleolar function and role in DNA repair. Evidence also supports a role for the nucleoplasmic deacetylase sirtuin 1 (SIRT1) in the reversal of this process, presumably when DNA repair has been completed46. The regulation of APEX1 in its roles in both DNA repair and ribosome biogenesis at this time thus appears to be largely governed by protein-protein interactions and post-translational modifications; however, this is likely to be expanded upon with further experimentation.
Werner syndrome RecQ like helicase (WRN)
The Werner syndrome RecQ like helicase (WRN) is the protein implicated in Werner syndrome, a disease characterized by premature aging and a predisposition to cancer57. Most commonly, WRN has been implicated in a number of DNA repair pathways including DNA DSB repair and repair of lesions to DNA bases. WRN has both helicase and exonuclease activity, which are both required for the repair of DSBs in HDR and NHEJ58. In addition, WRN-depleted cells treated with MMS (requiring BER repair) results in decreased cell survival59. It has also been demonstrated that WRN enhances the activity of the BER enzymes, DNA polymerase β (POLβ)59, 60 and the DNA glycosylase NEIL161. These functional roles in DNA repair however are not the only roles identified for WRN; indeed, a significant literature exists in support of a functional role for WRN in ribosome biogenesis as well.
WRN has been identified as a nucleolar protein in only one out of the three human nucleolar proteome datasets (Table 2; Supplemental Table 1). This lack of agreement among the studies however is not concerning given that these datasets were collected by different means and in different cells lines. Additional support for nucleolar localization includes immunofluorescence with polyclonal anti-WRN antibodies in simian and human cell lines that reveals clear enrichment of WRN in nucleoli62. Nucleolar localization is dependent on transcription of the rDNA62, 63, suggesting a putative role for WRN in RNAPI transcriptional regulation. Later work supports this with evidence that WRN co-immunoprecipitates the RNAPI subunit RPA4063. Furthermore, pulse-chase analysis reveals that in the absence of WRN in the nucleolus, levels of the 18S and 28S mature rRNA species are reduced63. Thus data support a nucleolar-specific role for WRN in ribosome biogenesis, in addition to its nucleoplasmic role in DNA repair processes.
The coordination of WRN between its functional roles in DNA repair and in ribosome biogenesis, like APEX1, is regulated by protein-protein interactions and post-translational modifications. WRN, for instance, has been identified in a complex with valosin-containing protein (VCP). The VCP/WRN complex is localized to the nucleolus, however, treatment with a DSB-inducing agent leads to reduced sequestration of WRN in the nucleolus and disruption of its interaction with VCP64. This interaction may be the result of tyrosine phosphorylation. A previous study had identified that cells treated with a transcriptional inhibitor resulted in WRN translocation to the nucleoplasm; treatment with a tyrosine phosphatase inhibitor, however, maintained WRN’s sequestration in the nucleolus62. VCP is known to be phosphorylated on tyrosine residues in its C-terminus 64. Whether this is directly related to its interaction with WRN remains to be tested. Finally, in addition to phosphorylation, acetylation may also be important in the regulation of WRN function. Treatment with a deacetylase inhibitor, Trichostatin A, enhanced recruitment to the nucleoplasm following DNA damage49. In contrast, inhibition of the deacetylase SIRT1 maintained nucleolar sequestration48. These data are in clear contradiction, and it has been proposed that perhaps SIRT1 does not act on WRN, but instead on WRN-interacting proteins.48 This too however remains to be tested. Regardless of these discrepancies, there is a clear role for acetylation, phosphorylation, and protein-protein interactions in the regulation of WRN between its nucleolar function and its role in DNA repair.
Bloom syndrome RecQ like helicase (BLM)
Bloom Syndrome RecQ like helicase (BLM) is a helicase in the same family as WRN. Defects in BLM result in a syndrome characterized by severe growth defects and a predisposition to cancer and other diseases65, 66. Its commonly recognized functional role in the cell is in DNA replication and repair processes. BLM, for instance, plays an important role in resolving stalled replication forks as well as other forms of replication stress67. Additionally, BLM has been implicated in the DSB response in both the protection against deletion of large segments in the repair of single-strand breaks and in promoting DNA end resection in DSB repair68, 69. Recent studies, however, also support a novel, nucleolar-specific role for BLM distinct from these roles in the maintenance of genome integrity.
BLM, like WRN, has been identified as a nucleolar protein in only one out of the three human nucleolar proteome datasets (Table 2; Supplemental Table 1). BLM, however, has been localized to the nucleolus by immunofluorescence70, 71, and co-immunoprecipitates the RNAPI subunit, RPA19471. Functionally, it has been shown that BLM facilitates the transcription of the pre-rRNA though its helicase activity on the rDNA71. Pulse-chase analysis for instance in BLM-deficient cells reveals a decrease in the production of the 45S pre-rRNA primary transcript71. Defects in pre-rRNA transcription can lead to decreases in mature 18S and 28S rRNA species, and/or the triggering of the cell stress response and apoptosis72. Thus evidence supports a critical role for BLM in the nucleolus and the production of ribosomes.
The regulation of BLM between its roles in DNA repair and in ribosome biogenesis is likely due to interaction with other proteins and/or post-translational modifications, similar to what has been observed for APEX1 and WRN. There is a clear regulation of BLM’s roles in DNA replication and repair by post-translational modifications73; however, a paucity of research exists on what regulates BLM’s nucleolar localization and mobilization to the nucleoplasm. One possible regulatory interaction occurs between BLM and DNA Topoisomerase I (TOPI). This interaction has been shown to occur in the nucleolus and to stimulate BLM helicase activity on RNA:DNA hybrids74. Ubiquitin modification is another possible mechanism by which the localization of BLM may be managed. Tikoo and colleagues suggest that ubiquitylation of BLM is required for recruitment to sites of DNA damage and that depletion of an E3-ubiquitin ligase, RNF8, results in nucleolar sequestration75. Despite these gaps, it is clear that BLM localizes to the nucleolus and maintains a nucleolar-specific function when sequestered there.
Genetic screens reveal roles for DNA repair proteins in human ribosome biogenesis
While there are specific examples of proteins with dual roles in both DNA repair and ribosome biogenesis like APEX1, WRN, and BLM, recent genetic screens of nucleolar proteins as well as unbiased genome-wide approaches have also revealed a role for DNA repair proteins in ribosome biogenesis. An siRNA-based screen of 625 nucleolar proteins in HeLa cervical epithelial cells revealed by northern blot a role for 286 in the normal processing of the pre-rRNA76. Interestingly, 28 of these hits we have classified as DNA repair proteins (GO: 0006281) including BLM and APEX1 (Table 3; Supplemental Table 2). Another siRNA screen in HeLa cells using fluorescently-tagged ribosomal protein reporters as read-outs identified proteins involved in 40S and 60S maturation. In this study, 153 hits out of 464 putative ribosome biogenesis factors were identified,77 with only four classified as DNA repair proteins (HUWE1; POLR2A; RPS27A/eS31; uS3). While the number here is small, the screen was restricted to ribosomal proteins, nuclear pore complex components, nucleo-cytoplasmic trafficking machinery, and homologues of yeast ribosome biogenesis factors, and therefore is not unexpected. In a follow-up to this study, however, the first unbiased genome-wide approach was used to identify proteins involved solely in 40S maturation in HeLa cells that identified 302 hits28. Here the number increased to 19 classified as DNA repair proteins. Thus, intriguingly, if this screen were further expanded to identify proteins required for 60S maturation it is conceivable that the number of DNA repair proteins would also increase.
Table 3.
Genetic screens for ribosome biogenesis proteins reveal proteins involved in DNA repair.
| Badertscher et al.28 |
Tafforeau et al.76 | Wild et al.77 | Neumuller et al.78 (Fly/Yeast) |
|
|---|---|---|---|---|
| Proteins screened |
Genome-wide | 625 | 464 | Genome-wide |
| Hits | 302 | 286 | 153 | 757/388 |
| DNA repair proteins |
19 | 28 | 4 | 36/30 |
Hits in a number of recent screens for proteins involved in human ribosome biogenesis reveal a subset of proteins classified as DNA repair proteins based on Gene Ontology (GO) Consortium categorization (GO: 0006281). A similar screen for proteins regulating nucleolar size in flies and yeast also reveal proteins involved in DNA repair.
While dual roles for DNA repair proteins in the nucleolus is only recently coming to light, this may not solely be a human phenomenon. In an unbiased genome-wide screen for proteins that affect nucleolar size in flies (Drosophila), 757 hits were identified78, where 36 are DNA repair proteins (Table 3; Supplemental Table 2). A similar screen in yeast revealed 388 hits78, with 30 classified as DNA repair proteins. Nucleolar size is known to reflect the rate of pre-rRNA transcription,79 and can be a useful biomarker for cancer malignancy80. Therefore, a nucleolar function for DNA repair proteins may be a highly conserved phenomenon in all eukaryotes.
Functional roles for nucleolar proteins in DNA repair
While evidence supports an increasing likelihood for DNA repair proteins to also perform functional roles in ribosome biogenesis, does evidence also support the converse: are proteins involved in ribosome biogenesis also performing roles in DNA repair? Interestingly, accumulating evidence suggests the answer is yes. Here, we highlight a few examples of ribosome biogenesis factors with clearly identified roles in DNA repair.
Treacle (TCOF1; Treacher Collins syndrome protein)
TCOF1 is a nucleolar protein implicated in Treacher Collins syndrome, a congenital disease characterized by abnormal craniofacial development14. In ribosome biogenesis, TCOF1 has been implicated in not only the regulation of RNAPI, but also the modification of the pre-rRNA. Pulse-chase analysis of TCOF1-depleted HeLa cells revealed a nearly 50% reduction in the 47S pre-rRNA81. Similarly, TCOF1 heterozygous mice showed a 50% decrease in the levels of the 47S primary rRNA transcript81. TCOF1 directly interacts with the RNAPI transcription factor UBTF, which may be the mechanism by which this protein regulates RNAPI transcription81. TCOF1 also interacts with NOP56, which is a core component in the box C/D snoRNP82. And, TCOF1-depletion in Xenopus oocytes and TCOF1 heterozygous mice both reveal a decrease in pre-rRNA 2’-O-methylation on the pre-18S rRNA82. These data thus support a role for TCOF1 in not only RNAPI transcription, but pre-rRNA modification as well. In addition to these roles in ribosome biogenesis, however, a relatively novel role in the DNA damage response is emerging.
The role of TCOF1 in DNA repair includes the recognition of DNA damage and a role in the transient RNAPI silencing that occurs as a result. In the osteosarcoma cell line, U-2 OS, a subset of cellular TCOF1 is recruited to sites of DNA damage in a poly ADP-ribose polymerase (PARP)-dependent manner83. Additionally, the recognition of DNA damage by TCOF1 is required for the Ataxia telangiectasia mutated (ATM)- and TCOF1- dependent recruitment of Nijmegen breakage syndrome protein 1 (NBS1) to nucleoli required for RNAPI transcriptional silencing83, 84. The regulation of this response and the interaction of TCOF1 with NBS1 appears to be dependent on post-translational modifications. Mutation of residue T210 to an alanine, for instance, disrupts TCOF1’s interaction with NBS183. Phosphorylation of S1199 also appears to be required for the localization of TCOF1 and NBS1 to nucleoli following DNA damage84. These data therefore support a role for TCOF1 in the DNA damage response by regulatory post-translational modifications and protein-protein interactions.
Nucleophosmin (NPM1; B23; NO38; numatrin)
NPM1 is a protein in the small nucleoplasmin family of molecular chaperones. It has diverse functional roles in a range of cellular processes, which include regulation of centrosome duplication, histone chaperone activity, tumor suppressor protein regulation, ribosome biogenesis, and DNA repair85. Given these diverse roles in essential cellular processes it is thus not surprising that NPM1 has also been implicated in disease. For example, NPM1 is mutated in approximately 30% of acute myeloid leukemia patients86, 87, is overexpressed in a variety of tumors88, and is essential for proper embryonic development and genome stability89. NPM1 levels are also increased in oncogenic human papilloma virus (HPV) infection90, and has been implicated in aiding viral replication including the replication of HIV91, 92 and T-cell leukemia virus93, among others94–97.
The evidence for NPM1’s role in ribosome biogenesis is substantial and supports a multifaceted role for it in making ribosomes. NPM1 localizes to the granular component of the nucleolus98–100, and is found in RNA precursors to both the small and large ribosomal subunits99. NPM1 promotes transcription through its histone chaperone activity by facilitating the remodeling of the nucleolar chromatin. Chromatin immunoprecipitation (ChIP) experiments reveal a direct interaction between NPM1 and the chromatin around the rDNA locus101. Furthermore, NPM1-depletion decreases the rate of pre-rRNA transcription and cell proliferation101. In addition to its effect on transcription, NPM1 has also been implicated in the processing of the pre-rRNA through its intrinsic ribonuclease activity102, 103, and more specifically as a phosphodiesterase that cleaves in the ITS2 region of the pre-rRNA103. Finally, several studies have evaluated a role for NPM1 in ribosomal subunit assembly, a natural follow-up to the early work by Borer and colleagues that identified NPM1 as a protein that “shuttles” between the cytoplasm and the nucleus104. Ribosomal protein S9 (uS4; RPS9) interacts with NPM1 in an RNA-independent interaction105. In addition, over-expression of NPM1 led to an increase of uS4 in the nucleolus, whereas NPM1-depletion resulted in a decrease in uS4 levels105. NPM1 is also required for the export of ribosomal protein L5 (uL18; RPL5), and is required for its colocalization with pre-60S particles106. Additionally, a recent genetic screen in HeLa cells for proteins that interact with NPM1 found that it interacts with 60 different ribosomal proteins107, suggesting that the role for NPM1 in ribosomal assembly is likely much larger. Therefore, NPM1 has roles in multiple aspects of ribosome biogenesis, however a role in DNA repair has only recently emerged.
Early work had suggested that NPM1 likely played a role in the DNA damage response. For example, NPM1 protein expression is increased following UV irradiation108, 109. Additionally, NPM1 is mobilized from the nucleolus to the nucleoplasm upon induction of DSBs110, and a subset is recruited specifically to sites of damage111. More recently however, specific roles for NPM1 in both the BER and NER repair pathways, as well as a DNA damage bypass mechanism, have emerged. For instance, in addition to its interaction with the BER protein APEX1 as discussed previously, NPM1 has also been implicated in the regulation of additional BER proteins (POLD1; POLβ; LIG1; LIG3; XRCC1) and their sequestration in the nucleolus55. Furthermore, in NPM1-depleted cells, BER activity is impaired, suggesting a role for NPM1 too in promoting efficient repair by this pathway55. NPM1 has also been implicated in the transcriptional regulation of NER proteins through its interaction with the retinoblastoma protein (pRB)112. Finally, although not a repair mechanism, NPM1 has also been implicated in translesion synthesis (TLS). TLS is a mechanism by which damaged DNA is bypassed by low fidelity polymerases. NPM1 stabilizes one of these polymerases, polymerase ?, protecting it from degradation until it is needed for TLS113. Thus NPM1 maintains functional roles in multiple DNA damage response pathways, including BER, NER, and TLS. A recent review, in fact, also highlights the many roles of NPM1 (and nucleolin) in DNA repair, and in much greater depth than presented here114.
Evidence for the regulation of NPM1 in response to DNA damage is limited but suggests a number of mechanisms. First, as described previously, NPM1’s interaction with the BER protein APEX1 is likely due to acetylation. Second, phosphorylation on T199 is required for the recruitment of NPM1 to sites of DNA damage111. In addition, dephosphorylation of NPM1 is required for NPM1’s interaction with pRB and the increased expression of genes associated with NER112. Finally, NPM1, like BLM, is regulated by the RNF8 E3-ubiquitin ligase in response to DNA damage111. Thus, it is likely that both protein-protein interactions and varied post-translational modifications regulate NPM1s switch between its role in DNA repair and ribosome biogenesis.
Nucleolin (NCL, C23)
Nucleolin (NCL), like NPM1, is a multifunctional protein in the nucleoplasmin family of molecular chaperones. It is localized to the nucleolus115, and directly interacts with both the rDNA116 and pre-rRNA117. Its nucleolar function in ribosome biogenesis is thus wide-ranging and includes roles in RNAPI transcription, pre-rRNA processing, and ribosomal subunit assembly. NCL-depleted cells for instance show a decrease in histone modifications associated with active rDNA chromatin116. NCL depletion also leads to a decrease in UBTF at the rDNA, and an increase in the binding of the RNAPI termination factor TTF-1 to the promoter-proximal terminator T0116. These results thus provide strong evidence for the role of NCL in promoting pre-rRNA transcription. Furthermore, NCL has also been implicated in the first step of pre-rRNA processing in the 5’ ETS, with data supporting a direct interaction with the pre-rRNA and U3 snoRNP, both of which are required for its activity117. Finally, NCL has been implicated in ribosomal subunit assembly through its shuttling activity104, and interaction with 18 different ribosomal proteins divided among both large- and small- subunit proteins118. In addition to these roles in ribosome biogenesis, however, there also lies evidence for NCL in the DNA damage response.
NCL’s role in the DNA damage response is likely in the cellular response to DNA DSBs. NCL is not only recruited to DSB-induced foci, but interacts with proteins involved in the DNA DSB response. For instance, NCL was identified as a protein that interacts with phosphorylated histone H2AX (λH2AX), a known marker for DNA DSBs, as well as both HDR (RPA34; NBS1) and NHEJ (XRCC6) factors119. In NCL-depleted cells, for instance, access of repair factors to DSBs is decreased due to decreased nucleosome disruption by NCL’s histone chaperone activity and its interaction with the MRE11-NBS1-RAD50 DSB repair complex120. This, at least in part, is a likely reason for observed decreases in the HDR and NHEJ activities119.
The regulation of NCL between its roles in DNA repair and ribosome biogenesis may be governed by p53. First, p53 is known to interact with NCL’s C-terminal domain121. NCL is also mobilized from the nucleolus to the nucleoplasm following treatment by ionizing radiation and other stressors; however, in p53-deficient cells, localization to the nucleoplasm is impaired121. Finally, ribosomal proteins also interact with a region in NCL’s C-terminal domain118; thus perhaps regulation is through post-translational modifications here, acting as a switch between interactions with ribosomal proteins and interaction with p53, although this remains to be tested. Despite lacking a comprehensive understanding of the regulation between NCL’s roles, there is clear evidence in support of NCL playing roles in both ribosome biogenesis and DNA repair.
Ribosomal protein S3 (uS3; RPS3)
uS3 is a ribosomal protein required for the maturation of the small ribosomal subunit and likely the mRNA helicase activity of the ribosome122. Interestingly, and somewhat surprising, however, roles in DNA repair have also been described for this protein. uS3 has also been found to localize to sites of oxidative DNA damage123, and interact with the BER proteins, APEX1 and 8-oxoguanine DNA glycosylase (OGG1)124. uS3 also has a high affinity for the oxidized base, 8-oxoguanine, and increases the glycosylase activity of OGG1 in the removal of these DNA lesions124. The regulation of this activity appears to be dependent on extracellular signal-regulated kinase 1/2 (ERK1/2) phosphorylation of residue T42, where mutations at this residue failed to recruit uS3 to nucleoplasmic loci upon DNA damage123. Thus, even proteins with more restricted roles pertaining to ribosome biogenesis may also have functional roles in the DNA damage response.
The nucleolar response to DNA damage
The crosstalk between the nucleolus and the DNA damage response is not limited to proteins functional in both DNA repair and ribosome biogenesis. In addition, there is the well-known p53 tumor suppressor response to nucleolar stress125, 126, as well as an emerging ATM-dependent mechanism by which RNAPI transcription is transiently silenced in response to DNA damage. In the first study to identify this response, mouse embryonic fibroblasts (MEFs) were exposed to ionizing radiation, and RNAPI transcriptional silencing was observed in nucleoli proximal to the DNA DSBs127 (Figure 3A). ATM kinase, NBS1 and mediator of DNA damage checkpoint 1 (MDC1) were required for RNAPI silencing, and ChIP experiments suggest that the NBS1/MDC1-dependent displacement of RNAPI at the rDNA locus is responsible127. This finding is supported by work that was published the same year that found NBS1 immunoprecipitates the rDNA in MCF7 cells upon DSB-induction by the 28S site-specific endonuclease, I-Ppol128.
Figure 3. Nucleolar response to DNA damage.
There are currently two mechanisms proposed for the transient RNAPI transcriptional silencing caused by DSBs in nuclear chromatin. A. RNAPI inhibition requires ATM, NBS1 and MDC1. Kruhlak and colleagues identified the ATM-dependent inhibition of RNAPI in nucleoli proximal to DSBs127. Inhibition required ATM, NBS1 and MDC1. B. RNAPI inhibition requires ATM, PARP, NBS1, and TCOF1 (not MDC1). Larsen and colleagues on the other hand identified ATM-dependent RNAPI transcriptional silencing in all nucleoli83. This silencing was not only dependent on (1) PARP recruitment of TCOF1 to sites of DSBs, but also on (2) the interaction between TCOF1 and NBS1 and their recruitment to nucleoli.
Later studies however, in contrast, identified TCOF1, and not MDC1, in the RNAPI transcriptional silencing response to ionizing radiation83, 84. As discussed previously, one study found in U-2 OS cells that a subset of cellular TCOF1 is recruited to sites of DNA damage in a PARP-dependent manner that is required for the ATM- and TCOF1- dependent recruitment of NBS1 to nucleoli83 (Figure 3B). Interestingly, this study also found, in contrast to previous work, that this interaction resulted not only in RNAPI silencing near DSBs, but also the propagation of silencing to all nucleoli83.
The crosstalk between the DNA damage response and the nucleolus however may not be universal for all forms of DNA damage. For example, in skin WS1 fibroblasts and U-2 OS cells, RNAPI transcriptional silencing was only observed following damage by UV radiation, and not by ionizing radiation45. Thus, while the differences among the results are hard to reconcile, they could be attributed to differences in the cell lines used. U-2 OS cells are derived from a human osteosarcoma and MCF7 cells are derived from a metastatic breast carcinoma, and both maintain a highly abnormal chromosome ploidy129, 130; whereas WS1 cells, on the other hand, are non-tumorigenic in origin, propagated through amino-acid restriction131, and MEFs are the fibroblasts of a different species altogether (mouse). Additionally, we know that different cell lines have different average numbers of nucleoli per cell132. While the meaning of more or fewer nucleoli is not well defined132, its affects could confound results dealing with a nucleolar response to stress by DNA damage. Despite these discrepancies, overall the evidence does support a crosstalk between DNA repair and the nucleolus through RNAPI transcriptional silencing in response to DNA damage.
DNA repair in the nucleolus itself
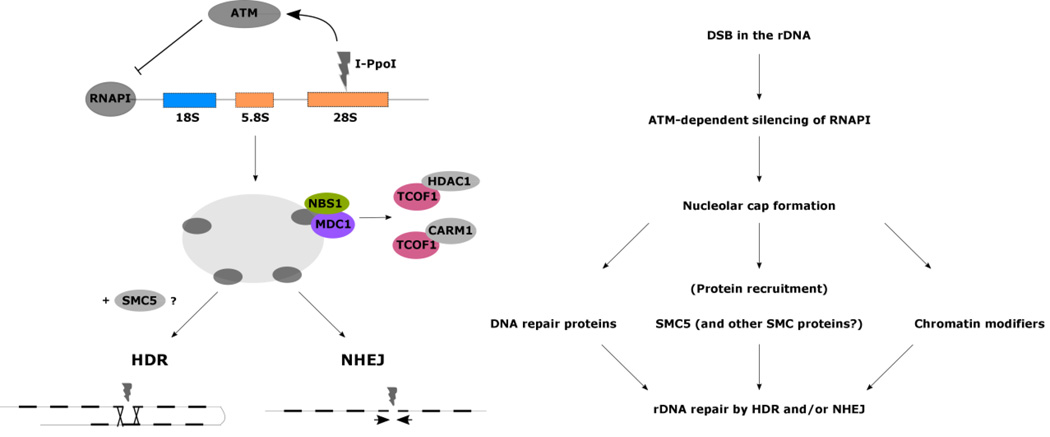
Crosstalk between the DNA damage response and the nucleolus is also evident when DNA damage occurs specifically in the rDNA. The rDNA is unique among transcribed regions in the genome. In humans, there are approximately 300 repeats of the rDNA transcriptional unit, arranged in tandem, and spread across 5 different chromosomes (10 total in a diploid cell); these regions of rDNA clusters are also commonly referred to as nucleolar organizing regions2. Because of the repetitive nature of the rDNA loci, loss of repeats by HDR can lead to genomic instability and cancer133. Repair at the rDNA loci is thus critically important, yet can be challenging. Recent studies have elucidated the response to DSBs by the I-Ppol endonuclease in the rDNA, and have arrived at different conclusions as to how repair occurs in the nucleolus itself (Figure 4).
Figure 4. DNA repair in the nucleolus.
DSBs in the 28S rRNA result in the ATM-dependent silencing of RNAPI transcription, which results in nucleolar cap formation and recruitment of proteins involved in the DNA damage response. Repair may be primarily through the HDR response supported by unscheduled DNA synthesis in G1 cells134 and recruitment of HDR proteins to nucleolar caps134, 136, or by NHEJ supported by prolonged RNAPI silencing upon DNA-PK inhibition and greater DNA damage in cells depleted of NHEJ proteins (DNA-PK; XRCC4)138. The DNA damage response may be mediated by SMC5136, or NBS1 and MDC1139, as well as chromatin modifications by HDAC1 and CARM1 interactions with TCOF1139.
Repair by HDR or NHEJ?
In one study, cleavage of the rDNA resulted in nucleolar cap formation and ATM-dependent inhibition of RNAPI in retinal pigment epithelial (RPE1) cells134. Caps were positive for λH2AX and p53 binding protein 1 (53BP1), and about half of all caps were enriched in proteins involved in HDR (BRCA1; RPA2; RAD51; RAD52), whereas NHEJ proteins were found localized to the nucleoplasm. Unscheduled DNA synthesis in G1 caps was also observed, suggesting cis-templated HDR in the repair of nucleolar DSBs134. These results are not without precedent; in budding yeast, it was found that the presence of multiple rDNA copies, many of which are not transcribed, is important for the maintenance of genome integrity through repair by HDR135.
Warmerdam and colleagues also support a role for HDR in the response to nucleolar DNA damage. In this study, I-Ppol endonuclease induction in U2-OS and RPE1 cells led to transient ATM-dependent inhibition of RNAPI and nucleolar cap formation136. Caps were positive for damage markers 53BP1 and λH2AX, UBTF, and HDR proteins (RAD51; RPA; BRCA1; BRCA2), suggesting a role for HDR in the repair of damage to the rDNA. By quantifying levels of UBTF, the authors also suggest that rDNA repeats are lost following repair136; however, it may not be appropriate to evaluate rDNA copy number based on UBTF levels – reduced UBTF levels may indicate a loss of repeats or could also simply represent fewer active repeats. Additionally, through siRNA-mediated inhibition of either HDR or NHEJ, the authors find that inhibition of HDR led to more efficient repair of breaks in the rDNA suggesting perhaps a role for NHEJ too in nucleolar DNA repair. Finally, inhibition of the HDR-associated protein structural maintenance of chromosome 5 (SMC5) led to more efficient repair of rDNA breaks136. SMC5 has been implicated before in HDR repair of the rDNA locus in yeast; however, the complex in yeast was required for “controlled H[D]R,” and in the absence of functional SMC complex proteins, rDNA hyper-recombination and loss of repeats was observed, rather than the converse seen here in this study in human cells137.
In contrast, a study in MCF10A breast epithelial cells implicates NHEJ in the repair of the rDNA. Harding and colleagues similarly found that I-Ppol cleavage of the rDNA induced ATM-dependent RNAPI silencing and localization of 53BP1, as well as HDR proteins BRCA1 and RAD51 to peripheral nucleolar caps138. Further experimentation however revealed pharmacological inhibition of the NHEJ protein DNA-PK led to prolonged RNAPI transcriptional silencing suggesting inefficient repair. Additionally, siRNA-depletion of NHEJ proteins, and not HDR proteins, increased DNA damage revealed by comet assays. Thus, altogether these data implicate NHEJ as the predominant form of DNA repair in nucleolar chromatin. These data are also supported by previous work from the Kastan group in MCF7 cells that found ATM-dependent recruitment of the NHEJ protein, XRCC4, at induced DSBs in the rDNA by I-Ppol128.
Other factors involved in rDNA repair
Lastly, a recent study in ES-D3 mouse embryonic stem cells validated transient RNAPI silencing and nucleolar cap formation upon damage to the rDNA by I-Ppol, but did not address the question of which pathway is implicated in repair. In this study, damage by I-Ppol led to nucleolar caps positive for 53BP1 and BRCA1, but also NBS1 and MDC1139. While the latter two proteins have not previously been observed to play a role in the response to nucleolar DNA damage, they have been suggested to play a role in RNAPI silencing in response to damage in extra-nucleolar chromatin as discussed previously. In addition, the authors here addressed the contribution of chromatin modifies to the repair of the rDNA. They found by co-immunoprecipitation that damage in the rDNA led to the interaction of nucleolar protein TCOF1 with histone deacetylase I (HDAC1) and coactivator-associated arginine methyltransferase I (CARM1)139. They also found, as expected, an increase in H3R17me2a histone marks mediated by CARM1 in the caps as well as the absence of histone acetylation presumably mediated by HDAC1.
While the differences among these studies, again, could be attributed to cell type differences, it has also been suggested that the level or persistence of DSBs induced by I-Ppol may cause the different responses observed140. Despite these discrepancies, RNAPI silencing mediated by ATM signaling followed by cap formation appears central to the DNA damage response in the nucleolus. While we are beginning to investigate how DNA repair occurs in the nucleolus, however, why inhibition of RNAPI transcription remains an outstanding question. One hypothesis relates to the observed cap formation, suggesting that RNAPI inhibition and consequent movement of damaged nucleolar chromatin to the periphery is necessary for DNA damage recognition and access by DNA repair proteins134, 138. Antoniali and colleagues, on the other hand, suggest that RNAPI transcriptional silencing may represent a coupling mechanism that allows for the efficient repair of damaged DNA by synchronizing the DNA damage response with cellular growth and homeostasis50. Similarly, it is possible that the silencing of RNAPI is protective against further genome instability that might be caused by collision between transcription and DNA repair machinery. For example, it has been shown that the Fanconi anemia pathway resolves R-loops (DNA:RNA hybrids) at sites of transcription, that if left unresolved can lead to stalled replication forks and DNA damage141. Although the latter wouldn’t adequately explain RNAPI silencing from damage in extra-nucleolar chromatin. Despite lacking a clear understanding for why transient RNAPI transcriptional silencing occurs in response to DNA damage, what is clear is that DNA repair and ribosome biogenesis do not operate in isolation and it seems likely that we will continue to see novel roles for proteins traditionally considered DNA repair proteins in steps of ribosome biogenesis, and vice versa.
Conclusions
The crosstalk between the DNA damage response and the nucleolar function of making ribosomes is not without precedent. We have reviewed the literature supporting an increasing number of proteins that localize to the human nucleolus, including proteins involved in DNA repair. We have highlighted a few examples of DNA repair proteins with defined roles in ribosome biogenesis (APEX1; WRN; BLM), as well as ribosome biogenesis proteins with newly identified roles in DNA repair (TCOF1; NPM1; NCL; uS3), largely regulated by protein-protein interactions and post-translational modifications (phosphorylation; acetylation; ubiquitylation). A summary of these findings are illustrated in Figure 5. Furthermore, given recent genetic screens that revealed DNA repair proteins in aspects of human ribosome biogenesis, it is likely that the number of examples of proteins with defined roles in both DNA repair and ribosome biogenesis will increase. We also reviewed the amassing literature on the crosstalk between DNA repair, both in extra-nucleolar chromatin and the nucleolus itself, and RNAPI transcriptional silencing. While some disparities among studies remain, a greater move towards work in primary cells and model organisms will likely lead to a more comprehensive understanding of the nucleolar response to DNA damage. It is an exciting time for the fields of ribosome biogenesis and DNA repair, and a greater appreciation for the overlap in these disciplines will surely aid in gaining a better understanding of diseases, including congenital disorders and cancer, and lead to novel therapeutic options.
Figure 5. Crosstalk between the nucleolus and the DNA damage response.
Diagram of the role of DNA repair proteins in ribosome biogenesis, and conversely the role of ribosome biogenesis proteins in the DNA damage response. A. Roles for DNA repair proteins in ribosome biogenesis. A number of DNA repair proteins have been localized to the nucleolus. Based on studies of APEX1, WRN, and BLM, these proteins may play important roles in pre-rRNA transcription and pre-rRNA quality control. B. Roles for ribosome biogenesis factors in the DNA damage response. Evidence also supports a role for proteins involved in ribosome biogenesis to have a dual role in DNA repair. Based on studies of TCOF1, NPM1, NCL, and uS3, these proteins may play important roles in both the recognition of DNA damage and chromatin remodeling at sites of DNA damage, as well as both the functional and transcriptional regulation of DNA repair proteins.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health [grant numbers R01GM115710 (to S.J.B.) and CMB TG T32GM007223 (to S.J.B. and L.M.O.)]. We would also like to acknowledge and thank Sam Sondalle, Vincent Yip, Nick Vincent, Sarah Ludwin-Peery, and Katie Farley for helpful discussions during the preparation of this manuscript. Finally, thanks again to Sam Sondalle for his assistance with figure formatting.
References
- 1.Kressler D, Hurt E, Bassler J. Biochim Biophys Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Pederson T. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomson E, Ferreira-Cerca S, Hurt E. J Cell Sci. 2013;126:4815–4821. doi: 10.1242/jcs.111948. [DOI] [PubMed] [Google Scholar]
- 4.McStay B, Grummt I. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 5.Grummt I, Langst G. Biochim Biophys Acta. 2013;1829:393–404. doi: 10.1016/j.bbagrm.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Smirnov E, Cmarko D, Mazel T, Hornacek M, Raska I. Histochem Cell Biol. 2016;145:359–372. doi: 10.1007/s00418-016-1407-x. [DOI] [PubMed] [Google Scholar]
- 7.Henras AK, Plisson-Chastang C, O’Donohue MF, Chakraborty A, Gleizes PE. Wiley Interdiscip Rev RNA. 2015;6:225–242. doi: 10.1002/wrna.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mullineux ST, Lafontaine DL. Biochimie. 2012;94:1521–1532. doi: 10.1016/j.biochi.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 9.de la Cruz J, Karbstein K, Woolford JL., Jr Annu Rev Biochem. 2015;84:93–129. doi: 10.1146/annurev-biochem-060614-033917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lafontaine DL. Nat Struct Mol Biol. 2015;22:11–19. doi: 10.1038/nsmb.2939. [DOI] [PubMed] [Google Scholar]
- 11.Nerurkar P, Altvater M, Gerhardy S, Schutz S, Fischer U, Weirich C, Panse VG. Int Rev Cell Mol Biol. 2015;319:107–140. doi: 10.1016/bs.ircmb.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Sondalle SB, Baserga SJ. Biochim Biophys Acta. 2014;1842:758–764. doi: 10.1016/j.bbadis.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCann KL, Teramoto T, Zhang J, Tanaka Hall TM, Baserga SJ. Elife. 2016;5 doi: 10.7554/eLife.16381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCann KL, Baserga SJ. Science. 2013;341:849–850. doi: 10.1126/science.1244156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narla A, Ebert BL. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farley KI, Baserga SJ. Biochem Soc Trans. 2016;44:1035–1044. doi: 10.1042/BST20160064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fumagalli S, Thomas G. Semin Hematol. 2011;48:97–105. doi: 10.1053/j.seminhematol.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Yelick PC, Trainor PA. Rare Dis. 2015;3:e1025185. doi: 10.1080/21675511.2015.1025185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruggero D, Shimamura A. Blood. 2014;124:2784–2792. doi: 10.1182/blood-2014-04-526301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danilova N, Gazda HT. Dis Model Mech. 2015;8:1013–1026. doi: 10.1242/dmm.020529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruggero D, Pandolfi PP. Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 22.Brighenti E, Trere D, Derenzini M. Oncotarget. 2015;6:38617–38627. doi: 10.18632/oncotarget.5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woods SJ, Hannan KM, Pearson RB, Hannan RD. Biochim Biophys Acta. 2015;1849:821–829. doi: 10.1016/j.bbagrm.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 24.Orsolic I, Jurada D, Pullen N, Oren M, Eliopoulos AG, Volarevic S. Semin Cancer Biol. 2016;37–38:36–50. doi: 10.1016/j.semcancer.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Granneman S, Baserga SJ. Exp Cell Res. 2004;296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 26.Woolford JL, Jr, Baserga SJ. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turowski TW, Tollervey D. Wiley Interdiscip Rev RNA. 2015;6:129–139. doi: 10.1002/wrna.1263. [DOI] [PubMed] [Google Scholar]
- 28.Badertscher L, Wild T, Montellese C, Alexander LT, Bammert L, Sarazova M, Stebler M, Csucs G, Mayer TU, Zamboni N, Zemp I, Horvath P, Kutay U. Cell Rep. 2015;13:2879–2891. doi: 10.1016/j.celrep.2015.11.061. [DOI] [PubMed] [Google Scholar]
- 29.Jackson SP, Bartek J. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ciccia A, Elledge SJ. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou BB, Elledge SJ. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Kulesz-Martin M. Carcinogenesis. 2001;22:851–860. doi: 10.1093/carcin/22.6.851. [DOI] [PubMed] [Google Scholar]
- 33.Reardon JT, Sancar A. Genes Dev. 2003;17:2539–2551. doi: 10.1101/gad.1131003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larance M, Lamond AI. Nat Rev Mol Cell Biol. 2015;16:269–280. doi: 10.1038/nrm3970. [DOI] [PubMed] [Google Scholar]
- 35.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 36.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 37.Leung AK, Trinkle-Mulcahy L, Lam YW, Andersen JS, Mann M, Lamond AI. Nucleic Acids Res. 2006;34:D218–D220. doi: 10.1093/nar/gkj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jarboui MA, Wynne K, Elia G, Hall WW, Gautier VW. Mol Immunol. 2011;49:441–452. doi: 10.1016/j.molimm.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Jarboui MA, Bidoia C, Woods E, Roe B, Wynne K, Elia G, Hall WW, Gautier VW. PLoS One. 2012;7:e48702. doi: 10.1371/journal.pone.0048702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmad Y, Boisvert FM, Gregor P, Cobley A, Lamond AI. Nucleic Acids Res. 2009;37:D181–D184. doi: 10.1093/nar/gkn804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 42.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 43.Scherl A, Coute Y, Deon C, Calle A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ. Mol Biol Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pederson T. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore HM, Bai B, Boisvert FM, Latonen L, Rantanen V, Simpson JC, Pepperkok R, Lamond AI, Laiho M. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.009241. M111 009241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lirussi L, Antoniali G, Vascotto C, D’Ambrosio C, Poletto M, Romanello M, Marasco D, Leone M, Quadrifoglio F, Bhakat KK, Scaloni A, Tell G. Mol Biol Cell. 2012;23:4079–4096. doi: 10.1091/mbc.E12-04-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li P, Gao S, Wang L, Yu F, Li J, Wang C, Li J, Wong J. Cell Rep. 2013;4:817–829. doi: 10.1016/j.celrep.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 48.Lee SY, Lee H, Kim ES, Park S, Lee J, Ahn B. Mutat Res. 2015;774:40–48. doi: 10.1016/j.mrfmmm.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 49.Blander G, Zalle N, Daniely Y, Taplick J, Gray MD, Oren M. J Biol Chem. 2002;277:50934–50940. doi: 10.1074/jbc.M210479200. [DOI] [PubMed] [Google Scholar]
- 50.Antoniali G, Lirussi L, Poletto M, Tell G. Antioxid Redox Signal. 2014;20:621–639. doi: 10.1089/ars.2013.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robertson AB, Klungland A, Rognes T, Leiros I. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. Antioxid Redox Signal. 2009;11:601–620. doi: 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tell G, Wilson DM, 3rd, Lee CH. Mol Cell Biol. 2010;30:366–371. doi: 10.1128/MCB.01174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vascotto C, Fantini D, Romanello M, Cesaratto L, Deganuto M, Leonardi A, Radicella JP, Kelley MR, D’Ambrosio C, Scaloni A, Quadrifoglio F, Tell G. Mol Cell Biol. 2009;29:1834–1854. doi: 10.1128/MCB.01337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poletto M, Lirussi L, Wilson DM, 3rd, Tell G. Mol Biol Cell. 2014;25:1641–1652. doi: 10.1091/mbc.E13-12-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fantini D, Vascotto C, Marasco D, D’Ambrosio C, Romanello M, Vitagliano L, Pedone C, Poletto M, Cesaratto L, Quadrifoglio F, Scaloni A, Radicella JP, Tell G. Nucleic Acids Res. 2010;38:8239–8256. doi: 10.1093/nar/gkq691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen JC, Loeb LA. Trends Genet. 2000;16:213–220. doi: 10.1016/s0168-9525(99)01970-8. [DOI] [PubMed] [Google Scholar]
- 58.Chen L, Huang S, Lee L, Davalos A, Schiestl RH, Campisi J, Oshima J. Aging Cell. 2003;2:191–199. doi: 10.1046/j.1474-9728.2003.00052.x. [DOI] [PubMed] [Google Scholar]
- 59.Harrigan JA, Wilson DM, 3rd, Prasad R, Opresko PL, Beck G, May A, Wilson SH, Bohr VA. Nucleic Acids Res. 2006;34:745–754. doi: 10.1093/nar/gkj475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harrigan JA, Opresko PL, von Kobbe C, Kedar PS, Prasad R, Wilson SH, Bohr VA. J Biol Chem. 2003;278:22686–22695. doi: 10.1074/jbc.M213103200. [DOI] [PubMed] [Google Scholar]
- 61.Das A, Boldogh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, de Souza Pinto N, Ramos W, Greenberg MM, Hazra TK, Mitra S, Bohr VA. J Biol Chem. 2007;282:26591–26602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- 62.Gray MD, Wang L, Youssoufian H, Martin GM, Oshima J. Exp Cell Res. 1998;242:487–494. doi: 10.1006/excr.1998.4124. [DOI] [PubMed] [Google Scholar]
- 63.Shiratori M, Suzuki T, Itoh C, Goto M, Furuichi Y, Matsumoto T. Oncogene. 2002;21:2447–2454. doi: 10.1038/sj.onc.1205334. [DOI] [PubMed] [Google Scholar]
- 64.Partridge JJ, Lopreiato JO, Jr, Latterich M, Indig FE. Mol Biol Cell. 2003;14:4221–4229. doi: 10.1091/mbc.E03-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ellis NA, German J. Hum Mol Genet. 1996;(5 Spec No):1457–1463. doi: 10.1093/hmg/5.supplement_1.1457. [DOI] [PubMed] [Google Scholar]
- 66.German J. Dermatol Clin. 1995;13:7–18. [PubMed] [Google Scholar]
- 67.Brosh RM., Jr Nat Rev Cancer. 2013;13:542–558. doi: 10.1038/nrc3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grabarz A, Guirouilh-Barbat J, Barascu A, Pennarun G, Genet D, Rass E, Germann SM, Bertrand P, Hickson ID, Lopez BS. Cell Rep. 2013;5:21–28. doi: 10.1016/j.celrep.2013.08.034. [DOI] [PubMed] [Google Scholar]
- 69.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Proc Natl Acad Sci U S A. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yankiwski V, Marciniak RA, Guarente L, Neff NF. Proc Natl Acad Sci U S A. 2000;97:5214–5219. doi: 10.1073/pnas.090525897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grierson PM, Lillard K, Behbehani GK, Combs KA, Bhattacharyya S, Acharya S, Groden J. Hum Mol Genet. 2012;21:1172–1183. doi: 10.1093/hmg/ddr545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bohm S, Bernstein KA. DNA Repair (Amst) 2014;22:123–132. doi: 10.1016/j.dnarep.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grierson PM, Acharya S, Groden J. Mutat Res. 2013;743–744:89–96. doi: 10.1016/j.mrfmmm.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tikoo S, Madhavan V, Hussain M, Miller ES, Arora P, Zlatanou A, Modi P, Townsend K, Stewart GS, Sengupta S. EMBO J. 2013;32:1778–1792. doi: 10.1038/emboj.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL. Mol Cell. 2013;51:539–551. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 77.Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, Kozak K, Csucs G, Lund E, Kutay U. PLoS Biol. 2010;8:e1000522. doi: 10.1371/journal.pbio.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Neumuller RA, Gross T, Samsonova AA, Vinayagam A, Buckner M, Founk K, Hu Y, Sharifpoor S, Rosebrock AP, Andrews B, Winston F, Perrimon N. Sci Signal. 2013;6 doi: 10.1126/scisignal.2004145. ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Derenzini M, Trere D, Pession A, Montanaro L, Sirri V, Ochs RL. Am J Pathol. 1998;152:1291–1297. [PMC free article] [PubMed] [Google Scholar]
- 80.Derenzini M, Montanaro L, Trere D. Histopathology. 2009;54:753–762. doi: 10.1111/j.1365-2559.2008.03168.x. [DOI] [PubMed] [Google Scholar]
- 81.Valdez BC, Henning D, So RB, Dixon J, Dixon MJ. Proc Natl Acad Sci U S A. 2004;101:10709–10714. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gonzales B, Henning D, So RB, Dixon J, Dixon MJ, Valdez BC. Hum Mol Genet. 2005;14:2035–2043. doi: 10.1093/hmg/ddi208. [DOI] [PubMed] [Google Scholar]
- 83.Larsen DH, Hari F, Clapperton JA, Gwerder M, Gutsche K, Altmeyer M, Jungmichel S, Toledo LI, Fink D, Rask MB, Grofte M, Lukas C, Nielsen ML, Smerdon SJ, Lukas J, Stucki M. Nat Cell Biol. 2014;16:792–803. doi: 10.1038/ncb3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ciccia A, Huang JW, Izhar L, Sowa ME, Harper JW, Elledge SJ. Proc Natl Acad Sci U S A. 2014;111:18631–18636. doi: 10.1073/pnas.1422488112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lindstrom MS. Biochem Res Int. 2011;2011:195209. doi: 10.1155/2011/195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Falini B. Cancer Treat Res. 2010;145:149–168. doi: 10.1007/978-0-387-69259-3_9. [DOI] [PubMed] [Google Scholar]
- 87.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, La Starza R, Diverio D, Colombo E, Santucci A, Bigerna B, Pacini R, Pucciarini A, Liso A, Vignetti M, Fazi P, Meani N, Pettirossi V, Saglio G, Mandelli F, Lo-Coco F, Pelicci PG, Martelli MF, Party GALW. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 88.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 89.Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, Pandolfi PP. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 90.McCloskey R, Menges C, Friedman A, Patel D, McCance DJ. J Virol. 2010;84:5131–5139. doi: 10.1128/JVI.01965-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li YP. J Virol. 1997;71:4098–4102. doi: 10.1128/jvi.71.5.4098-4102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fankhauser C, Izaurralde E, Adachi Y, Wingfield P, Laemmli UK. Mol Cell Biol. 1991;11:2567–2575. doi: 10.1128/mcb.11.5.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Adachi Y, Copeland TD, Hatanaka M, Oroszlan S. J Biol Chem. 1993;268:13930–13934. [PubMed] [Google Scholar]
- 94.Duan Z, Chen J, Xu H, Zhu J, Li Q, He L, Liu H, Hu S, Liu X. Virology. 2014;452–453:212–222. doi: 10.1016/j.virol.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 95.Lee SJ, Shim HY, Hsieh A, Min JY, Jung G. J Microbiol. 2009;47:746–752. doi: 10.1007/s12275-009-2720-z. [DOI] [PubMed] [Google Scholar]
- 96.Tsuda Y, Mori Y, Abe T, Yamashita T, Okamoto T, Ichimura T, Moriishi K, Matsuura Y. Microbiol Immunol. 2006;50:225–234. doi: 10.1111/j.1348-0421.2006.tb03789.x. [DOI] [PubMed] [Google Scholar]
- 97.Okuwaki M, Iwamatsu A, Tsujimoto M, Nagata K. J Mol Biol. 2001;311:41–55. doi: 10.1006/jmbi.2001.4812. [DOI] [PubMed] [Google Scholar]
- 98.Spector DL, Ochs RL, Busch H. Chromosoma. 1984;90:139–148. doi: 10.1007/BF00292451. [DOI] [PubMed] [Google Scholar]
- 99.Schmidt-Zachmann MS, Hugle-Dorr B, Franke WW. EMBO J. 1987;6:1881–1890. doi: 10.1002/j.1460-2075.1987.tb02447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feuerstein N, Chan PK, Mond JJ. J Biol Chem. 1988;263:10608–10612. [PubMed] [Google Scholar]
- 101.Murano K, Okuwaki M, Hisaoka M, Nagata K. Mol Cell Biol. 2008;28:3114–3126. doi: 10.1128/MCB.02078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Herrera JE, Savkur R, Olson MO. Nucleic Acids Res. 1995;23:3974–3979. doi: 10.1093/nar/23.19.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Savkur RS, Olson MO. Nucleic Acids Res. 1998;26:4508–4515. doi: 10.1093/nar/26.19.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 105.Lindstrom MS, Zhang Y. J Biol Chem. 2008;283:15568–15576. doi: 10.1074/jbc.M801151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yu Y, Maggi LB, Jr, Brady SN, Apicelli AJ, Dai MS, Lu H, Weber JD. Mol Cell Biol. 2006;26:3798–3809. doi: 10.1128/MCB.26.10.3798-3809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, Hyman AA, Mann M. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 108.Wu MH, Chang JH, Yung BY. Carcinogenesis. 2002;23:93–100. doi: 10.1093/carcin/23.1.93. [DOI] [PubMed] [Google Scholar]
- 109.Wu MH, Yung BY. J Biol Chem. 2002;277:48234–48240. doi: 10.1074/jbc.M206550200. [DOI] [PubMed] [Google Scholar]
- 110.Lee SY, Park JH, Kim S, Park EJ, Yun Y, Kwon J. Biochem J. 2005;388:7–15. doi: 10.1042/BJ20042033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Koike A, Nishikawa H, Wu W, Okada Y, Venkitaraman AR, Ohta T. Cancer Res. 2010;70:6746–6756. doi: 10.1158/0008-5472.CAN-10-0382. [DOI] [PubMed] [Google Scholar]
- 112.Lin CY, Tan BC, Liu H, Shih CJ, Chien KY, Lin CL, Yung BY. Mol Biol Cell. 2010;21:4409–4417. doi: 10.1091/mbc.E10-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ziv O, Zeisel A, Mirlas-Neisberg N, Swain U, Nevo R, Ben-Chetrit N, Martelli MP, Rossi R, Schiesser S, Canman CE, Carell T, Geacintov NE, Falini B, Domany E, Livneh Z. Nat Commun. 2014;5:5437. doi: 10.1038/ncomms6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Scott DD, Oeffinger M. Biochem Cell Biol. 2016;94:419–432. doi: 10.1139/bcb-2016-0068. [DOI] [PubMed] [Google Scholar]
- 115.Olson MO, Orrick LR, Jones C, Busch H. J Biol Chem. 1974;249:2823–2827. [PubMed] [Google Scholar]
- 116.Cong R, Das S, Ugrinova I, Kumar S, Mongelard F, Wong J, Bouvet P. Nucleic Acids Res. 2012;40:9441–9454. doi: 10.1093/nar/gks720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ginisty H, Amalric F, Bouvet P. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bouvet P, Diaz JJ, Kindbeiter K, Madjar JJ, Amalric F. J Biol Chem. 1998;273:19025–19029. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- 119.Kobayashi J, Fujimoto H, Sato J, Hayashi I, Burma S, Matsuura S, Chen DJ, Komatsu K. PLoS One. 2012;7:e49245. doi: 10.1371/journal.pone.0049245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Goldstein M, Derheimer FA, Tait-Mulder J, Kastan MB. Proc Natl Acad Sci U S A. 2013;110:16874–16879. doi: 10.1073/pnas.1306160110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Daniely Y, Dimitrova DD, Borowiec JA. Mol Cell Biol. 2002;22:6014–6022. doi: 10.1128/MCB.22.16.6014-6022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Takyar S, Hickerson RP, Noller HF. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 123.Yadavilli S, Hegde V, Deutsch WA. DNA Repair (Amst) 2007;6:1453–1462. doi: 10.1016/j.dnarep.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hegde V, Wang M, Deutsch WA. Biochemistry. 2004;43:14211–14217. doi: 10.1021/bi049234b. [DOI] [PubMed] [Google Scholar]
- 125.Golomb L, Volarevic S, Oren M. FEBS Lett. 2014;588:2571–2579. doi: 10.1016/j.febslet.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 126.Burger K, Eick D. Biol Chem. 2013;394:1133–1143. doi: 10.1515/hsz-2013-0153. [DOI] [PubMed] [Google Scholar]
- 127.Kruhlak M, Crouch EE, Orlov M, Montano C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- 128.Berkovich E, Monnat RJ, Jr, Kastan MB. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 129.Ponten J, Saksela E. Int J Cancer. 1967;2:434–447. doi: 10.1002/ijc.2910020505. [DOI] [PubMed] [Google Scholar]
- 130.Soule HD, Vazguez J, Long A, Albert S, Brennan M. J Natl Cancer Inst. 1973;51:1409–1416. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- 131.Corfield VA, Hay RJ. In Vitro. 1978;14:787–794. doi: 10.1007/BF02617973. [DOI] [PubMed] [Google Scholar]
- 132.Farley KI, Surovtseva Y, Merkel J, Baserga SJ. Chromosoma. 2015;124:323–331. doi: 10.1007/s00412-015-0507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stults DM, Killen MW, Williamson EP, Hourigan JS, Vargas HD, Arnold SM, Moscow JA, Pierce AJ. Cancer Res. 2009;69:9096–9104. doi: 10.1158/0008-5472.CAN-09-2680. [DOI] [PubMed] [Google Scholar]
- 134.van Sluis M, McStay B. Genes Dev. 2015;29:1151–1163. doi: 10.1101/gad.260703.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ide S, Miyazaki T, Maki H, Kobayashi T. Science. 2010;327:693–696. doi: 10.1126/science.1179044. [DOI] [PubMed] [Google Scholar]
- 136.Warmerdam DO, van den Berg J, Medema RH. Cell Rep. 2016;14:2519–2527. doi: 10.1016/j.celrep.2016.02.048. [DOI] [PubMed] [Google Scholar]
- 137.Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M. Nat Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- 138.Harding SM, Boiarsky JA, Greenberg RA. Cell Rep. 2015;13:251–259. doi: 10.1016/j.celrep.2015.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Franek M, Kovarikova A, Bartova E, Kozubek S. J Histochem Cytochem. 2016 doi: 10.1369/0022155416668505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.McStay B. Genes Dev. 2016;30:1598–1610. doi: 10.1101/gad.283838.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schwab RA, Nieminuszczy J, Shah F, Langton J, Lopez Martinez D, Liang CC, Cohn MA, Gibbons RJ, Deans AJ, Niedzwiedz W. Mol Cell. 2015;60:351–361. doi: 10.1016/j.molcel.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, Berglund L, Bergstrom K, Brumer H, Cerjan D, Ekstrom M, Elobeid A, Eriksson C, Fagerberg L, Falk R, Fall J, Forsberg M, Bjorklund MG, Gumbel K, Halimi A, Hallin I, Hamsten C, Hansson M, Hedhammar M, Hercules G, Kampf C, Larsson K, Lindskog M, Lodewyckx W, Lund J, Lundeberg J, Magnusson K, Malm E, Nilsson P, Odling J, Oksvold P, Olsson I, Oster E, Ottosson J, Paavilainen L, Persson A, Rimini R, Rockberg J, Runeson M, Sivertsson A, Skollermo A, Steen J, Stenvall M, Sterky F, Stromberg S, Sundberg M, Tegel H, Tourle S, Wahlund E, Walden A, Wan J, Wernerus H, Westberg J, Wester K, Wrethagen U, Xu LL, Hober S, Ponten F. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.