Abstract
Wnt signaling plays an important role in breast carcinogenesis. DAPPER2 (DACT2) functions as an inhibitor of canonical Wnt signaling and plays distinct roles in different cell contexts, with its role in breast tumorigenesis unclear. We investigated DACT2 expression in breast cancer cell lines and primary tumors, as well as its functions and molecular mechanisms. Results showed that DACT2 expression was silenced in 9/9 of cell lines. Promoter CpG methylation of DACT2 was detected in 89% (8/9) of cell lines, as well as in 73% (107/147) of primary tumors, but only in 20% (1/5) of surgical margin tissues and in none of normal breast tissues. Demethylation of BT549 and T47D cell lines with 5-aza-2'-deoxycytidine restored DACT2 expression along with promoter demethylation, suggesting that its downregulation in breast cancer is dependent on promoter methylation. Furthermore, ectopic expression of DACT2 induced breast cell apoptosis in vitro, and further inhibited breast tumor cell proliferation, migration and EMT, through antagonizing Wnt/β-catenin and Akt/GSK-3 signaling. Thus, these results demonstrate that DACT2 functions as a tumor suppressor for breast cancer but was frequently disrupted epigenetically in this cancer.
Keywords: DACT2, tumor suppressor, methylation, EMT, breast cancer
INTRODUCTION
Breast cancer is a highly heterogeneous cancer associated with alterations in multiple signal pathways. Wnt signaling plays a key role during breast carcinogenesis and progression [1–2]. Wnt signaling pathway is involved in cell proliferation, apoptosis, differentiation, motility, and survival in a context-dependent manner [3].
Wnt signaling mainly includes the Wnt/β-catenin pathway, the noncanonical planar cell polarity (PCP) pathway, and the Wnt/calcium (Ca2+) pathway. In Wnt/β-catenin pathway, in the absence of Wnt signaling, β-catenin is phosphorylated in a destruction complex and ubiquitinated, leading to its degradation. Binding of Wnt to frizzled (Fz) and low density lipoprotein receptor related protein (LRP)-5/6 results in the activation of dishevelled (Dvl), which promotes β-catenin stabilization and nuclear translocation to act as a transcriptional coactivator of lymphoid enhancing factor-1 and T-cell factor-1 (LEF/TCF) transcription factors to activate the transcription of multiple target genes [4]. The Wnt/Ca2+-PCP pathway also appears to involve a Wnt ligand and Fz, but does not involve LRP-5/6 co-receptors and β-catenin. Activation of this pathway leads to the association of Dvl with Rac, Rho and Rho-associated kinase (ROCK), resulting in the restructuring of cytoskeleton.
Dapper, an antagonist of β-catenin, homolog 2 (DACT2) belongs to the DACT (Dpr/Frodo) gene family, located at 6q27, a region frequently associated with loss of heterozygosity in human cancers [5]. The DACT family consists of DACT1, 2 and 3 [6]. DACT family member and Dvl form a complex with Axin, GSK-3, CKI, and β-catenin, resulting in a decrease of activated β-catenin and suppression of target genes. These members also inhibit the activation of c-Jun N-terminal kinase (JNK) by Dvl. Thus, DACT genes are predicted to be potent cancer-associated genes [7].
In addition to genetic mutations in Wnt/β-catenin pathway components, epigenetic events also contribute to the abnormal activation of this signaling pathway in tumor cells. Epigenetic silencing of negative regulators of Wnt signaling is crucial for the aberrant activation of Wnt/β-catenin signaling in tumor pathogenesis [8–9]. For examples, promoter methylation of Secreted Frizzled-Related Proteins (SFRPs), Wnt Inhibitory Factor-1 (WIF-1), and DICKKOPFs (DKK-1, 2, 3) have been reported in multiple cancers [10–12].
DACT1 expression has been shown to be significantly downregulated in hepatocellular carcinoma (HCC) [13], gastrointestinal stromal tumors [14], non-small cell lung cancer (NSCLC) [15] and breast cancer [16]. Dysregulated DACT1 was associated with poor prognosis in non-small cell lung cancer patients [15]. DACT2 and DACT1 show 28.8% total-amino-acid identity. The expression level of DACT2 is reduced in some colorectal tumors [17]. However, little is known about the signaling function of DACT2 and its relevance to breast oncogenesis.
We previously identified DACT2 as a methylated target in our breast cancer methylome study. Here, we further examined DACT2 as a negative regulator of Wnt signaling and found that its transcription is repressed in breast cancer cell lines and primary tumors, which is associated with its promoter CpG methylation. The biological functions of DACT2 in breast cancer cells were assessed in vitro in the context of Wnt/β-catenin signaling.
RESULTS
Downregulation of DACT2 in breast cell lines by promoter CpG methylation
Promoter sequence analysis of the DACT2 gene identified a typical CpG island spanning the proximal promoter and exon 1 regions (http://cpgislands.usc.edu/) (Figure 1A). We next performed RT-PCR analysis to examine DACT2 expression in nine breast cancer cell lines. Semi-quantitative RT-PCR showed that DACT2 was abundantly expressed in normal human tissues including breast, while silenced or downregulated in all breast cell lines analyzed (Figure 1B, 1C). Thus, the methylation status of DACT2 promoter was examined. MSP showed that DACT2 was methylated in 7 cell lines (BT549, MB231, MB468, MCF7, T47D, ZR-75-1 and YCCB1), with weak methylation in SK-BR-3 and no methylation in YCCB3 detected (Figure 1C). Pharmacological demethylation was used to assess whether promoter CpG methylation directly regulates DACT2 expression. BT549 and T47D cells with methylated and silenced DACT2 were treated with Aza with or without the histone deacetylase inhibitor TSA. Both treatments resulted in the upregulation of DACT2 expression accompanied by a decrease in the methylated alleles of DACT2 (Figure 1D). The results indicated that promoter methylation is a major mechanism of DACT2 silencing in breast cancer cells. BGS of MB231 cells confirmed the results of MSP analysis, showing heavily methylated DACT2 promoter alleles, while Aza treatment decreased its methylation in MB231 cells, leading to DACT2 upregulation (Figure 1D).
Figure 1. The expression and methylation status of DACT2 in breast cancer cell lines and normal mammary tissues.
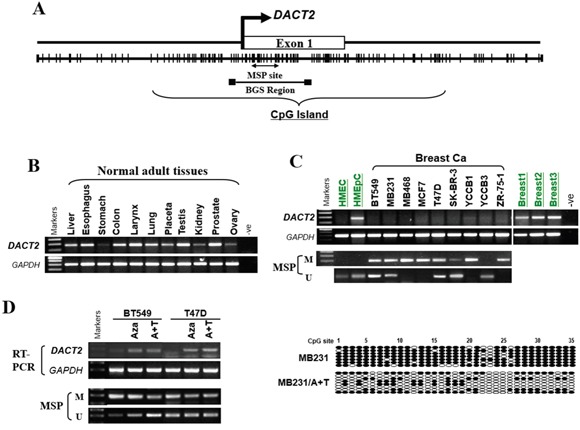
A. Schematic structure of the DACT2 promoter CpG island (CGI). The white rectangle represents exon 1, and the CpG sites in the CGI are indicated with short black lines. B. Robust mRNA expression of DACT2 in human normal adult tissues detected by semiquantitative RT-PCR, GAPDH as a control. C. Expression of DACT2 in breast cancer cell lines, and the methylation status of DACT2 in breast cancer and normal mammary epithelial cells. D. Pharmacological demethylation of the DACT2 CGI by Aza (A) with or without TSA (T) induced its expression. DACT2 expression before and after drug treatment was determined by RT–PCR, and demethylation was confirmed by MSP and BGS.
DACT2 methylation in breast tumors and its correlation with clinical features
DACT2 expression in human breast cancer samples was analyzed using the online database Oncomine and qRT-PCR. We found that the expression of DACT2 mRNA in breast tumor specimens was significantly lower than that in non-tumor breast tissue specimens. The average level of DACT2 mRNA expression in breast cancer tissues was 2.25-fold lower than that in adjacent non-cancerous tissues (p<0.05) (Figure 2A, 2B). TCGA breast studies demonstrated a significant 14.7-fold and 9.3-fold decrease in DACT2 mRNA expression in Invasive Ductal Breast Carcinoma (IDBC) and Invasive Lobular Breast Carcinoma (ILBC), respectively, and an 11.2-fold decrease in Invasive Breast Carcinoma (IBC) compared to normal breast tissues (Figure 2C). Curtis Breast Statistics showed similar results (Figure 2C).
Figure 2. The expression and methylation status of DACT2 in breast tumor tissues.
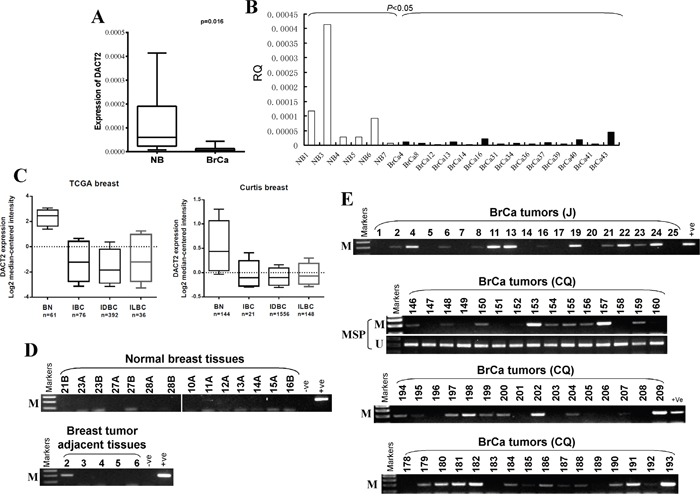
A, B. Expression of DACT2 in human normal and breast tumor tissues detected by qRT-PCR, C. The expression of DACT2 (median of expression intensity) in breast cancer from Oncomine database. D. Methylation of DACT2 in normal breast tissues and breast tumor adjacent tissues. E. Representative images of methylation of the DACT2 promoter in breast tumor tissues. M: methylated; U: unmethylated.
We further investigated DACT2 methylation in primary tumors, surgical margin tissues and normal breast tissues. DACT2 methylation was detected in 73% (107/147) of breast cancer tissues, 20% (1/5) of breast tumor adjacent tissues and none of normal breast tissues (Figure 2D, 2E; Table 1), suggesting the tumor-specific methylation of DACT2 in breast cancer. We next analyzed the correlation between DACT2 methylation and clinicopathological features of breast cancer patients, including age, tumor size, tumor grade, lymph node metastasis, and estrogen receptor (ER), progesterone receptor (PR) and HER2 status. However, no significant correlation between DACT2 methylation and clinicopathological features was observed (Table 2).
Table 1. Methylation status of the DACT2 promoter in primary breast tumors.
| Samples | DACT2 promoter | Frequency of methylation | |
|---|---|---|---|
| methylated | unmethylated | ||
| BrCa (n=147) | 107 | 40 | 107/147 (73%) |
| BA (n=5) | 1 | 4 | 1/5 (20%) |
| BNP (n= 14) | 0 | 14 | 0/14 (0%) |
Note: BrCa, breast cancer; BA, breast cancer adjacent tissues; BNP, breast normal tissues.
Table 2. Clinicopathological features of DACT2 methylation in breast cancer.
| Clinicopathological features | Number(n=158) | DACT2 promoter methylation status | P value | |
|---|---|---|---|---|
| methylated | unmethylated | |||
| Age | 0.602 | |||
| ≤40 | 13 | 11(85%) | 2(15%) | |
| >40 | 103 | 74(72%) | 29(28%) | |
| unknown | 31 | 22(71%) | 9(29%) | |
| Grade | 0.431 | |||
| I | 7 | 5(71%) | 2(29%) | |
| II | 80 | 57(71%) | 23(29%) | |
| III | 7 | 7(100%) | 0 | |
| unknown | 53 | 38(72%) | 15(28%) | |
| Tumor size | 0.289 | |||
| <2.0 cm | 43 | 36(84%) | 7(16%) | |
| ≥2.0 cm≤5.0cm | 62 | 43(69%) | 19(31%) | |
| >5.0cm | 9 | 6(67%) | 3(33%) | |
| unknown | 33 | 22(67%) | 11(33%) | |
| Lymph node metastasis | 0.9 | |||
| Positive | 50 | 37(74%) | 13(26%) | |
| Negative | 64 | 47(73%) | 17(27%) | |
| unknown | 33 | 23(70%) | 10(30%) | |
| ER status | 0.479 | |||
| Positive | 52 | 36(69%) | 16(31%) | |
| Negative | 44 | 35(80%) | 9(20%) | |
| unknown | 51 | 36(71%) | 15(29%) | |
| PR status | 0.048 | |||
| Positive | 40 | 25(63%) | 15(27%) | |
| Negative | 56 | 47(84%) | 9(16%) | |
| unknown | 51 | 35(69%) | 16(31%) | |
| HER2 status | 0.606 | |||
| >+++ | 9 | 6(67%) | 3(33%) | |
| ++ | 45 | 36(80%) | 9(20%) | |
| <+ | 41 | 28(68%) | 13(32%) | |
| unknown | 52 | 37(71%) | 15(29%) | |
| p53 expression | 0.662 | |||
| Positive | 35 | 27(77%) | 8(23%) | |
| Negative | 44 | 30(68%) | 14(32%) | |
| unknown | 68 | 50(74%) | 18(26%) | |
Re-expression of DACT2 inhibits cell growth and induces apoptosis in breast cancer cells
To determine whether DACT2 suppresses cell growth in breast cancer, colony formation assays were performed using MB231 and MCF7 cells transfected with a DACT2-expressing plasmid or the empty vector. Cells transfected with DACT2 showed a dramatic decrease in colony numbers compared to control cells (Figure 3A). Moreover, MB231 cells ectopically expressing DACT2 inhibited cell proliferation by inducing a G1-phase cell cycle arrest and displayed condensed nuclei typical of apoptosis (Figure 3B, 3C). These results further demonstrated that DACT2 functions as a potential tumor suppressor in breast cancer.
Figure 3. Growth inhibitory effect of DACT2 on breast cancer cell lines.
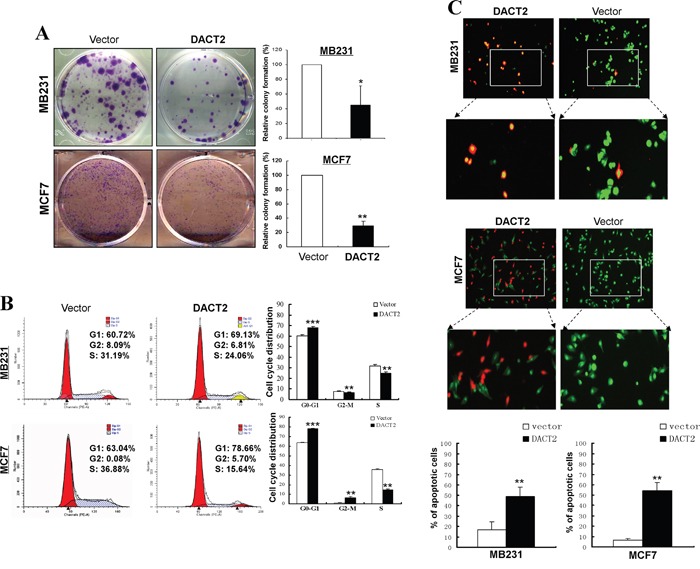
A. Representative colony formation assay in vector- and DACT2-expressing MB231 and MCF7 cells. The values are shown as the mean ± S.E. from three independent experiments (*p< 0.05, **p< 0.01). B. Effect of cell cycle distribution of vector- and DACT2-transfected MB231 and MCF7 cells was detected by flow cytometry analysis. Representative flow cytometry plots (left) and histograms of cell cycle alterations (right). C. Induction of apoptosis was detected by AO/EB assay.
DACT2 reverses EMT through antagonizing β-catenin activity and inhibiting Akt/GSK-3β pathway in breast cells
We next examined whether epithelial-to-mesenchymal transition (EMT) in breast cancer cells is affected by DACT2 using the aggressive triple-negative breast cancer cell line MB231. The results showed that forced transfection of DACT2 effectively reversed EMT to mesenchymal-to- epithelial transition in MB231 cells, resulting in the upregulation of the epithelial marker E-cadherin and downregulation of the mesenchymal marker Vimentin (Figure 4A–4D). Activation of Wnt/β-catenin pathway and Akt/GSK-3 pathway play a central role in the maintenance of EMT. DACT family members in various species have previously been shown to interact with Dvl proteins through a highly conserved C-terminal motif and to negatively regulate β-catenin function. Therefore, we investigated whether DACT2 functions as a tumor suppressor by antagonizing Wnt/β-catenin signaling. Expression of DACT2 led to the downregulation of active β-catenin and Dvl2 in MB231 cells. Furthermore, overexpression of DACT2 in MB231 breast cancer cell lines resulted in a profound reduction of both total and phosphorylated forms of Akt and GSK-3β (Figure 5D).
Figure 4.
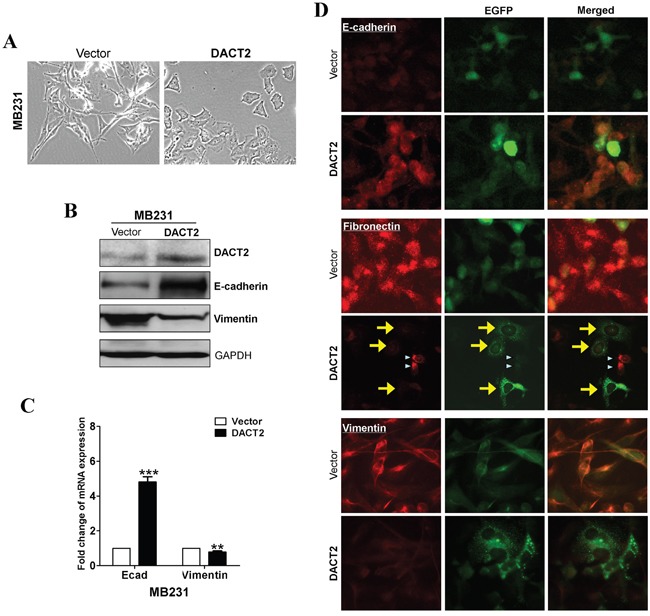
A. Morphology changes of MB231 cells transfected with DACT2 or empty vector by phase-contrast microscopy. B. Western blot analysis of E-cadherin and Vimentin. GAPDH was used as an internal control. C. qRT-PCR analysis of E-cadherin and Vimentin. β-actin was used as an internal control. D. Subcellular localization of EMT markers was detected by immunofluorescence staining.
Figure 5. Ectopic expression of DACT2 inhibited the migration and invasion of breast tumor cells.
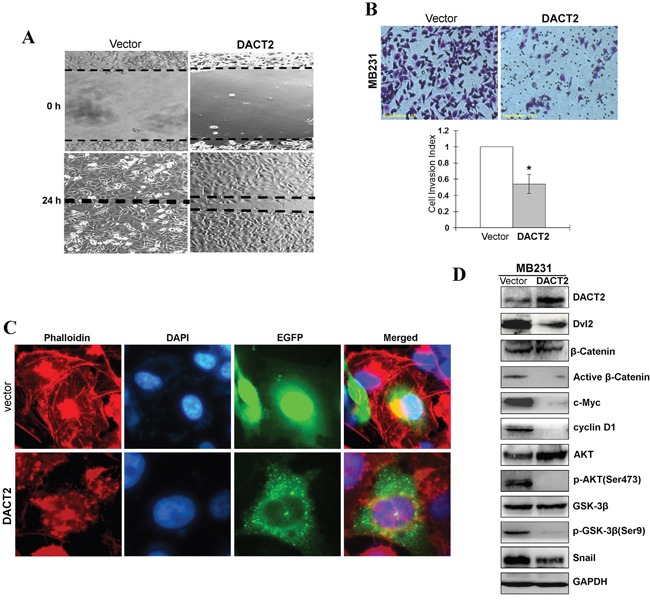
A. Representative images of wound healing assay. Photographs were taken at 0 and 24 h to determine the different mobility between Vector- and DACT2-transfected MB231 cells. B. Representative images of the transwell invasion assay. The pictures were taken at 24 h after seeding (magnification, ×200). The numbers of invaded cells were counted in seven representative high power fields per transwell. The values are shown as mean±SD. Three independent experiments were carried out in triplicate, *p<0.05. C. Cells transfected with DACT2 or empty vector were stained with FITC-phalloidin to observe the organization of actin fibers. Pictures were taken under 400× magnification. D. Ectopic expression of DACT2 in MB231 cells disrupted Wnt/β-catenin signaling and AKT/GSK-3β pathway. Western blot analysis of β-catenin and AKT, as well as their downstream targets in MB231 cells. GAPDH was used as an internal control.
DACT2 suppresses breast cancer cell migration and invasion and induces actin cytoskeleton reorganization
EMT is a phenomenon that is accompanied by increased cell motility and invasion and involves loss of epithelial cell-cell junctions and actin cytoskeleton reorganization. The effect of DACT2 on the migration and invasiveness of breast cancer cells was analyzed using a wound-healing assay and Matrigel invasion chambers. The results of wound-healing assays showed that MB231 cells migrated into the scraped areas within 24 h, whereas DACT2 expression reduced wound closure by approximately 55% after 24 h (Figure 5A). Furthermore, DACT2-transfected cells had lower invasion rates than control cells (Figure 5B), suggesting that DACT2 attenuates wound-induced cell migration and inhibits the invasiveness of breast cancer cells. Cells transfected with DACT2 or empty vector were stained with FITC-phalloidin to observe the organization of actin fibers. Cells transfected with DACT2 displayed diffuse cytoplasmic actin, arranged irregularly, and much fewer thin stress fibers, whereas control group of cells present formation of actin stress fibers, supporting a role for DACT2 regulating actin cytoskeleton reorganization. (Figure 5C).
DISCUSSION
Epigenetic disruption of tumor suppressor genes (TSGs) by promoter CpG methylation and histone modification is a key mechanism of gene deregulation in cancer [18–19]. DACT2 is frequently downregulated by promoter methylation in HCC [20] and colorectal tumors [17]. High DACT2 protein levels were correlated with better differentiation and better survival rates in HCC and esophageal squamous cell carcinoma patients [20–21]. Here, we showed that DACT2 is expressed in normal breast tissues but frequently downregulated/silenced by promoter methylation in breast cancer cell lines and primary tumors, but not in normal breast tissues. No methylation was detected in the YCCB3 cell line with silenced DACT2, suggesting that histone modifications or other mechanisms are also involved in DACT2 downregulation in this cell line. We showed the tumor-specific methylation of DACT2 in breast cancer, which suggests its potential as a tumor marker. However, no obvious correlation between DACT2 methylation and clinicopathological features was observed in breat cancer, although this needs to be further confirmed in a larger cohort study.
DACT family members have N-terminal leucine zipper domains and C-terminal PDZ binding motifs and can bind to many factors in both cytoplasmic and nuclear compartments [6, 22]. DACT2 interacts with PKC, Dvl3, β-catenin, PITX2 and Lef-1 [23]. Several studies have shown that DACT2 has distinct roles in different cells, indicating tissue type- or species- specific roles of DACT2. DACT2 in zebrafish regulates the noncanonical Wnt/Ca2+−PCP pathway [4], but also inhibits TGF-β/Nodal pathway [24] by targeting the TGF-β receptors ALK5 and ALK4, accelerating their lysosomal degradation [25], thus preventing the activation of Smad2/3. Mouse Dact2 antagonizes TGF-β signaling [26], as well as Wnt/β-catenin and Wnt/JNK signaling without directly altering β-catenin level [22, 27–28]. DACT2 remarkably suppressed TGF-β signaling via both proteasomal and lysosomal pathways in esophageal squamous cell carcinoma [21]. Recent studies further indicated that DACT2 could suppress Wnt signaling by inhibiting TCF/LEF in lung cancer [29]. However, in esophageal squamous cell carcinoma, DACT2 significantly decreases pSmad2/3 binding to their response elements, but has no effect on regulatory elements of canonical Wnt or other pathways [21].
Wnt/β-catenin and Akt/GSK-3 pathways have been well characterized to induce EMT in epithelial cell models [30]. Indeed, DACT2 re-expression significantly increased E-cadherin and decreased Vimentin levels in our study. EMT is associated with actin reorganization, tubular basement membrane disruption, and cell migration. DACT2 inhibited breast cancer cell proliferation by inducing apoptosis, and further suppressed tumor cell migration. We further showed that Dvl2 and activated β-catenin were inhibited by DACT2 re-expression in breast tumor cells. As expected, expression of p-AKT and p-GSK-3β dramatically decreased in MB231 cells upon DACT2 re-expression. Snail, which is tightly regulated by GSK-3β at the protein level, was also significantly downregulated.
In summary, DACT2 was found to be an antagonist to Wnt/β-catenin signaling and was frequently downregulated/silenced in breast cancer. Our results indicate that DACT2 acts as an important TSG for breast cancer, which may serve as a potential biomarker or therapeutic stargeting strategy for breast cancer.
MATERIALS AND METHODS
Cell lines, tumor samples and normal tissues
Breast cancer cell lines (BT549, MB231, MB468, MCF7, T47D, SK-BR-3, YCCB1, YCCB3 and ZR-75-1) were used. Normal breast tissue RNA samples were purchased commercially (Stratagene, La Jolla, CA; Millipore Chemicon, Billerica, MA; BioChain Institute, Hayward, CA). Primary tumor samples, paired surgical margin tissues and normal breast tissues were obtained from the First Affiliated Hospital of Chongqing Medical University [31–33]. All samples were reviewed and subjected to histological diagnosis by pathologists to ensure the percentage of tumor cells was ~70%. Clinical information was collected for most tumor samples. The study was approved by the ethics committee of the First Affiliated Hospital of Chongqing Medical University (Approval notice: 2010/2012(23)).
Semiquantitative reverse transcription PCR (RT-PCR) and quantitative RT-PCR (qRT-PCR)
Total RNA was isolated using the TRIzol® Reagent (Invitrogen, Carlsbad, CA). Semiquantitative RT-PCR was performed in a final volume of 10 μl of reaction mixture containing 2 μl cDNA using Go-Taq (Promega, Madison, WI) [34]. GAPDH was amplified as the control. Primer sequences are listed in Table 1. Real-time PCR was performed using Thermo Fisher Maxima SYBR Green qPCR Master Mixes (Thermo-Fisher Scientific, former Fermentas, Schwerte, Germany). Thermal cycling reactions were performed using the 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA). Relative expression levels of DACT2 in breast tissues were standardized to β-actin levels. Online cancer database Oncomine (http://www.oncomine.org) was used to examine the mRNA expression of DACT2 in normal versus breast tumor tissues. p-values for each group were calculated using Student's t-test. Standardized normalization techniques and statistical calculations are provided on the Oncomine website and published.
5-aza-2'-deoxycytidine (Aza) and trichostatin a treatment
Cell lines were treated with 10 mmol/l Aza (Sigma-Aldrich, St Louis, MO) for 3 days and further treated with 100 nmol/l trichostatin A (TSA, Cayman Chemical Co., Ann Arbor, MI) for 24 h.
Methylation-specific PCR (MSP) and bisulfite genomic sequencing (BGS)
Genomic DNA was extracted from cell pellets, normal tissues and tumors using the DNAzol® Reagent (Invitrogen, Carlsbad, CA). Bisulfite modification of DNA and MSP were performed as described previously [35–36]. Bisulfite-treated DNA was amplified by MSP with DACT2 methylation-specific primers (Table 3) using AmpliTaq-Gold DNA Polymerase (Applied Biosystems, Foster City, CA). Methylated and unmethylated MSP primer sets target the same CpG sites in the DACT2 promoter, and do not amplify genomic DNA with no bisulfite treatment. For BGS, bisulfite-treated DNA was amplified using a BGS primer (Table 3). The PCR products were then cloned into a pCR4-Top vector (Invitrogen, Carlsbad, CA). Eight to twelve colonies were randomly chosen and sequenced by Beijing Genomics Institute.
Table 3. List of primers used in this study.
| PCR | Primer | Sequence (5'-3') | Product size (bp) | PCR Cycles | Annealing temperature (°C) |
|---|---|---|---|---|---|
| MSP | DACT2 M1 | CGTGTAGATTTCGTTTTTCGC | 200 | 40 | 60 |
| DACT2 M2 | CCGAAAATCCGCCCGACG | ||||
| DACT2 U1 | TGTGTGTAGATTTTGTTTTTTGT | 203 | 40 | 58 | |
| DACT2 U2 | CCCCAAAAATCCACCCAACA | ||||
| RT-PCR | DACT2F | AGCCGTGGGGCACATTCTG | 173 | 35 | 55 |
| DACT2R | CCAGGTCCTGCCGATACTTG | ||||
| BGS | DACT2-BGS1 | GGTTATAGATTTTAGTTTATTTTGG | 249 | 40 | 60 |
| DACT2-BGS2 | CTACAACTCCTACAACCCC |
Immunofluorescence staining
Cells transfected with pcDNA3.1-DACT2 or pcDNA3.1 plasmid were seeded in six-well tissue culture plates containing glass coverslips. The cells were washed with PBS, fixed with precooled methanol at −20°C for 10 min, permeabilized with 0.5% Triton X-100 for 5 min and blocked with 5% normal goat serum at room temperature for 30-40 min. Cells were incubated with primary antibodies diluted in TBST at 4°C overnight, washed twice with PBS, and then incubated with Alexa Fluor 594- or 488-conjugated goat anti-rabbit or anti-mouse secondary antibody (Invitrogen Molecular Probes, Carlsbad, Ca) for an additional 30 min. Nuclei were counterstained with 4,6-diamidino- 2- phenylindole (DAPI) (Roche, Palo Alto, CA). To analyze the effects of DACT2 on F-actin, the cells were fixed with 4% paraformaldehyde for 20 min at room temperature and stained by rhodamine-labeled phalloidin (Invitrogen Molecular Probes). The slides were visualized with an Olympus BX51 microscope (Olympus Corporation, Tokyo, Japan) under 400× magnification and the images were captured with a camera.
Colony formation assay
Colony formation assays were performed as described previously [16]. MB231 and MCF7 cells were plated in six-well plates and transfected with 4 μg pcDNA3.1-DACT2 or pcDNA3.1 plasmid using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). At 48 h after transfection, cells were collected, re-plated in six-well plates and selected for 14 days in the presence of G418 (1.2 mg/ml for MB231, 0.8 mg/ml for MCF7). Surviving colonies (≥50 cells/colony) were stained with Gentian Violet and counted. All experiments were performed three times.
Wound healing assay and cell invasion assay
Stably transfected cells were selected using G418, and then cultured in six-well plates until confluent. After scratching the monolayer, cells were photographed at 0, 12 and 24 h under a 10× objective (Olympus Corporation, Tokyo, Japan). Cell invasion assay was performed using 24-well culture plates (Millipore, Billerica, MA) with inserts of 8-μm pore membranes pre-coated with Matrigel (BD Bioscience, San Jose, CA). Briefly, 1×104 cells were seeded in each well. The lower compartment was filled with cell culture medium supplemented with 15% fetal bovine serum. After 24 h, invaded cells on the bottom surface were fixed with methanol and stained with 0.1% Crystal Violet. The invaded cells were counted and photographed under a microscope. Seven fields per membrane were counted in each group. Each experiment was performed twice.
Flow cytometry analysis of cell cycle status and apoptosis assay
To assess cell cycle status, MB231 cells and MCF7 cells were seeded in six-well plates and transfected with 4 μg of DACT2 or empty vector using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) following the manufacturer's protocol. After 48 h, cells were harvested using 0.1% trypsin and washed with PBS, then fixed in ice-cold 70% ethanol for 1 h, and treated with 100 μl of 50 mg/l propidium iodide for 30 min at 4°C in the dark. Data were analyzed with the CELL Quest software (BD Biosciences, San Jose, CA). Acridine orange/ethidium bromide (AO/EB) staining was used for apoptosis analysis. Briefly, the transfected cells were replated in six-well plates containing glass coverslips. After 24 h, cells were washed with phosphate-buffered saline (PBS) then stained with AO/EB for 5 min and visualized immediately under a fluorescence microscope (LEICA CTR4000B). The percentage of apoptotic cells was then calculated.
Western blotting
Transfected cells were washed with ice cold PBS and lysed with lysis buffer (Pierce, Thermo Scientific, Cramlington, UK) containing a protease inhibitor cocktail (Sigma Aldrich, St. Louis, MO). The lysate was centrifuged at 4°C for 10 min at 10,000 g and the supernatant was collected. A total of 50 μg of protein lysate for each sample was separated using sodium dodecyl sulfate/polyacrylamide gel electrophoresis. The lysates were then transferred to polyvinylidene fluoride membranes for antibody incubation. After blocking with 5% nonfat milk and 0.1% Tween 20 in TBS, the membranes were incubated with the following primary antibody overnight at 4°C: β-catenin (1:1000, Cell Signaling, #2677), active β-catenin (1:1000, Cell Signaling, #4270), cyclinD1 (1:1000, Epitomics, #2261), c-Myc (1:10000, Epitomics, #1472-1), p-AKT (Ser473) (1:1000, Cell Signaling #4060), p-GSK-3β (Ser9) (1:1000, Cell Signaling #9323) and GAPDH (1:2000, Epitomics, #2261) was used as control. Next day the membranes were washed and incubated with secondary antibodies. Bands were visualized using the enhanced chemiluminescence detection system.
Statistical analysis
Statistical analyses were performed using the χ2 test and Fisher's exact test to determine the p value. p<0.05 was considered statistically significant.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
GRANT SUPPORT
This study was supported by National Natural Science Foundation of China (#31420103915, #81372238, #81572327) and VC special fund from The Chinese University of Hong Kong.
REFERENCES
- 1.Najafov A, Seker T, Even I, Hoxhaj G, Selvi O, Ozel DE, Koman A, Birgul-Iyison N. MENA is a transcriptional target of the Wnt/beta-catenin pathway. PLoS One. 2012;7:e37013. doi: 10.1371/journal.pone.0037013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, Varmus HE. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell. 1988;55:619–625. doi: 10.1016/0092-8674(88)90220-6. [DOI] [PubMed] [Google Scholar]
- 3.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waxman JS, Hocking AM, Stoick CL, Moon RT. Zebrafish Dapper1 and Dapper2 play distinct roles in Wnt-mediated developmental processes. Development. 2004;131:5909–5921. doi: 10.1242/dev.01520. [DOI] [PubMed] [Google Scholar]
- 5.Noviello C, Courjal F, Theillet C. Loss of heterozygosity on the long arm of chromosome 6 in breast cancer: possibly four regions of deletion. Clin Cancer Res. 1996;2:1601–1606. [PubMed] [Google Scholar]
- 6.Fisher DA, Kivimae S, Hoshino J, Suriben R, Martin PM, Baxter N, Cheyette BN. Three Dact gene family members are expressed during embryonic development and in the adult brains of mice. Dev Dyn. 2006;235:2620–2630. doi: 10.1002/dvdy.20917. [DOI] [PubMed] [Google Scholar]
- 7.Katoh M. Identification and characterization of human DAPPER1 and DAPPER2 genes in silico. Int J Oncol. 2003;22:907–913. [PubMed] [Google Scholar]
- 8.Ying Y, Tao Q. Epigenetic disruption of the WNT/beta-catenin signaling pathway in human cancers. Epigenetics. 2009;4:307–312. doi: 10.4161/epi.4.5.9371. [DOI] [PubMed] [Google Scholar]
- 9.Xiang TX, Yuan Y, Li LL, Wang ZH, Dan LY, Chen Y, Ren GS, Tao Q. Aberrant promoter CpG methylation and its translational applications in breast cancer. Chin J Cancer. 2011 doi: 10.5732/cjc.011.10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiang T, Li L, Yin X, Zhong L, Peng W, Qiu Z, Ren G, Tao Q. Epigenetic silencing of the WNT antagonist Dickkopf 3 disrupts normal Wnt/beta-catenin signalling and apoptosis regulation in breast cancer cells. Journal of cellular and molecular medicine. 2013;17:1236–1246. doi: 10.1111/jcmm.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng YY, Yu J, Wong YP, Man EP, To KF, Jin VX, Li J, Tao Q, Sung JJ, Chan FK, Leung WK. Frequent epigenetic inactivation of secreted frizzled-related protein 2 (SFRP2) by promoter methylation in human gastric cancer. Br J Cancer. 2007;97:895–901. doi: 10.1038/sj.bjc.6603968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki M, Shigematsu H, Nakajima T, Kubo R, Motohashi S, Sekine Y, Shibuya K, Iizasa T, Hiroshima K, Nakatani Y, Gazdar AF, Fujisawa T. Synchronous alterations of Wnt and epidermal growth factor receptor signaling pathways through aberrant methylation and mutation in non small cell lung cancer. Clin Cancer Res. 2007;13:6087–6092. doi: 10.1158/1078-0432.CCR-07-0591. [DOI] [PubMed] [Google Scholar]
- 13.Yau TO, Chan CY, Chan KL, Lee MF, Wong CM, Fan ST, Ng IO. HDPR1, a novel inhibitor of the WNT/beta-catenin signaling, is frequently downregulated in hepatocellular carcinoma: involvement of methylation-mediated gene silencing. Oncogene. 2005;24:1607–1614. doi: 10.1038/sj.onc.1208340. [DOI] [PubMed] [Google Scholar]
- 14.Astolfi A, Nannini M, Pantaleo MA, Di Battista M, Heinrich MC, Santini D, Catena F, Corless CL, Maleddu A, Saponara M, Lolli C, Di Scioscio V, Formica S, Biasco G. A molecular portrait of gastrointestinal stromal tumors: an integrative analysis of gene expression profiling and high-resolution genomic copy number. Lab Invest. 2010;90:1285–1294. doi: 10.1038/labinvest.2010.110. [DOI] [PubMed] [Google Scholar]
- 15.Yang ZQ, Zhao Y, Liu Y, Zhang JY, Zhang S, Jiang GY, Zhang PX, Yang LH, Liu D, Li QC, Wang EH. Downregulation of HDPR1 is associated with poor prognosis and affects expression levels of p120-catenin and beta-catenin in nonsmall cell lung cancer. Mol Carcinog. 2010;49:508–519. doi: 10.1002/mc.20622. [DOI] [PubMed] [Google Scholar]
- 16.Yin X, Xiang T, Li L, Su X, Shu X, Luo X, Huang J, Yuan Y, Peng W, Oberst M, Kelly K, Ren G, Tao Q. DACT1, an antagonist to Wnt/beta-catenin signaling, suppresses tumor cell growth and is frequently silenced in breast cancer. Breast cancer research: BCR. 2013;15:R23. doi: 10.1186/bcr3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang X, Tan J, Li J, Kivimae S, Yang X, Zhuang L, Lee PL, Chan MT, Stanton LW, Liu ET, Cheyette BN, Yu Q. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell. 2008;13:529–541. doi: 10.1016/j.ccr.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nature reviews Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 20.Gao S, Yang Z, Zheng ZY, Yao J, Zhang F, Wu LM, Xie HY, Zhou L, Zheng SS. Reduced expression of DACT2 promotes hepatocellular carcinoma progression: involvement of methylation-mediated gene silencing. World J Surg Oncol. 2013;11:57. doi: 10.1186/1477-7819-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou J, Liao LD, Xie YM, Zeng FM, Ji X, Chen B, Li LY, Zhu MX, Yang CX, Qing Z, Chen T, Xu XE, Shen J, Guo MZ, Li EM, Xu LY. DACT2 is a candidate tumor suppressor and prognostic marker in esophageal squamous cell carcinoma. Cancer Prev Res (Phila) 2013;6:791–800. doi: 10.1158/1940-6207.CAPR-12-0352. [DOI] [PubMed] [Google Scholar]
- 22.Cheyette BN, Waxman JS, Miller JR, Takemaru K, Sheldahl LC, Khlebtsova N, Fox EP, Earnest T, Moon RT. Dapper, a Dishevelled-associated antagonist of beta-catenin and JNK signaling, is required for notochord formation. Dev Cell. 2002;2:449–461. doi: 10.1016/s1534-5807(02)00140-5. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Florez S, Wang J, Cao H, Amendt BA. Dact2 represses PITX2 transcriptional activation and cell proliferation through Wnt/beta-catenin signaling during odontogenesis. PLoS One. 2013;8:e54868. doi: 10.1371/journal.pone.0054868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Zhou H, Su Y, Sun Z, Zhang H, Zhang Y, Ning Y, Chen YG, Meng A. Zebrafish Dpr2 inhibits mesoderm induction by promoting degradation of nodal receptors. Science. 2004;306:114–117. doi: 10.1126/science.1100569. [DOI] [PubMed] [Google Scholar]
- 25.Su Y, Zhang L, Gao X, Meng F, Wen J, Zhou H, Meng A, Chen YG. The evolutionally conserved activity of Dapper2 in antagonizing TGF-beta signaling. FASEB J. 2007;21:682–690. doi: 10.1096/fj.06-6246com. [DOI] [PubMed] [Google Scholar]
- 26.Meng F, Cheng X, Yang L, Hou N, Yang X, Meng A. Accelerated re-epithelialization in Dpr2-deficient mice is associated with enhanced response to TGFbeta signaling. J Cell Sci. 2008;121:2904–2912. doi: 10.1242/jcs.032417. [DOI] [PubMed] [Google Scholar]
- 27.Lee WC, Hough MT, Liu W, Ekiert R, Lindstrom NO, Hohenstein P, Davies JA. Dact2 is expressed in the developing ureteric bud/collecting duct system of the kidney and controls morphogenetic behavior of collecting duct cells. Am J Physiol Renal Physiol. 2010;299:F740–751. doi: 10.1152/ajprenal.00148.2010. [DOI] [PubMed] [Google Scholar]
- 28.Kettunen P, Kivimae S, Keshari P, Klein OD, Cheyette BN, Luukko K. Dact1-3 mRNAs exhibit distinct expression domains during tooth development. Gene Expr Patterns. 2010;10:140–143. doi: 10.1016/j.gep.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia Y, Yang Y, Brock MV, Zhan Q, Herman JG, Guo M. Epigenetic regulation of DACT2, a key component of the Wnt signalling pathway in human lung cancer. J Pathol. 2013;230:194–204. doi: 10.1002/path.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim EY, Kim A, Kim SK, Kim HJ, Chang J, Ahn CM, Chang YS. Inhibition of mTORC1 induces loss of E-cadherin through AKT/GSK-3beta signaling-mediated upregulation of E-cadherin repressor complexes in non-small cell lung cancer cells. Respir Res. 2014;15:26. doi: 10.1186/1465-9921-15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiang T, Li L, Fan Y, Jiang Y, Ying Y, Putti TC, Tao Q, Ren G. PLCD1 is a functional tumor suppressor inducing G(2)/M arrest and frequently methylated in breast cancer. Cancer biology & therapy. 2010;10:520–527. doi: 10.4161/cbt.10.5.12726. [DOI] [PubMed] [Google Scholar]
- 32.Xiang T, Li L, Yin X, Yuan C, Tan C, Su X, Xiong L, Putti TC, Oberst M, Kelly K, Ren G, Tao Q. The ubiquitin peptidase UCHL1 induces G0/G1 cell cycle arrest and apoptosis through stabilizing p53 and is frequently silenced in breast cancer. PLoS One. 2012;7:e29783. doi: 10.1371/journal.pone.0029783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin H, Wang X, Ying J, Wong AH, Cui Y, Srivastava G, Shen ZY, Li EM, Zhang Q, Jin J, Kupzig S, Chan AT, Cullen PJ, Tao Q. Epigenetic silencing of a Ca(2+)-regulated Ras GTPase-activating protein RASAL defines a new mechanism of Ras activation in human cancers. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12353–12358. doi: 10.1073/pnas.0700153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ying J, Li H, Seng TJ, Langford C, Srivastava G, Tsao SW, Putti T, Murray P, Chan AT, Tao Q. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene. 2006;25:1070–1080. doi: 10.1038/sj.onc.1209154. [DOI] [PubMed] [Google Scholar]
- 35.Tao Q, Swinnen LJ, Yang J, Srivastava G, Robertson KD, Ambinder RF. Methylation status of the Epstein-Barr virus major latent promoter C in iatrogenic B cell lymphoproliferative disease. Application of PCR-based analysis. Am J Pathol. 1999;155:619–625. doi: 10.1016/S0002-9440(10)65157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao Q, Huang H, Geiman TM, Lim CY, Fu L, Qiu GH, Robertson KD. Defective de novo methylation of viral and cellular DNA sequences in ICF syndrome cells. Hum Mol Genet. 2002;11:2091–2102. doi: 10.1093/hmg/11.18.2091. [DOI] [PubMed] [Google Scholar]


