Main Text
Membraneless organelles are micron or sub-micron-sized bodies that consist of multiple proteins and, in many cases, RNA molecules (1, 2). Unlike typical intracellular organelles such as mitochondria and nuclei, membraneless organelles lack a surrounding phospholipid membrane. A variety of nuclear (1, 2, 3) and cytoplasmic functions (1, 2, 4, 5, 6) are associated with membraneless organelles and these include ribosomal biogenesis, RNA processing, and stress response. How do membraneless organelles form, and how are the hundreds of distinct protein and RNA molecules organized within these organelles? In 2009, Brangwynne et al. (6) made the startling discovery that Caenorhabditis elegans germline P granules are liquid droplets that flow, fuse, and drip like classical liquids. Subsequent studies established that the formation of P-granules bodies is driven by phase separation whereby specific proteins separate from the milieu to make dense liquidlike droplets (5). This process is also known as liquid-liquid demixing and is analogous to the demixing observed in a binary mixture of oil and vinegar. Past a critical concentration, referred to more precisely as a saturation concentration, specific types of proteins drive phase separation leading to the formation of a protein-rich liquidlike phase that coexists with a protein-deficient dispersed phase. The internal environments and material properties of protein-rich liquid droplets will be determined by their constituent components (7). A recurring theme is internal fluidity with the possibility of gelation either due to mutations or aging of droplets. The internal fluidity of membraneless organelles is postulated to mimic the features of micro-reactors that enable efficient biochemical reactions and information transduction through the advantages of proximal spatial localization.
A typical eukaryotic cell comprises of ∼104 proteins and RNA molecules. Distinct membraneless organelles simultaneously coexist and each organelle comprises of ∼102 types of macromolecules. Furthermore, specific proteins or combinations of protein and RNA molecules drive the formation of specific membraneless organelles whereas other macromolecules preferentially partition into these organelles. These observations raise two key questions, i.e., 1) how many distinct phases are conceivable if there are on average n distinct copies of each of the N protein and RNA molecules? 2) Why do we observe only a small number of types organelles with specific compositional biases as opposed to either a unitary droplet that encompasses all macromolecules or an entire continuum of options with different compositional biases? The answer to the first question comes from the Gibbs phase rule. Given N distinct protein and RNA components, the phase rule states that, for fixed temperature and pressure, there can be at most N + 2 coexisting phases. And yet, cells are not characterized by ∼104 types of droplets of homogeneous compositions. This implies that there might be certain generic organizing principles that determine the number of coexisting phases within cells.
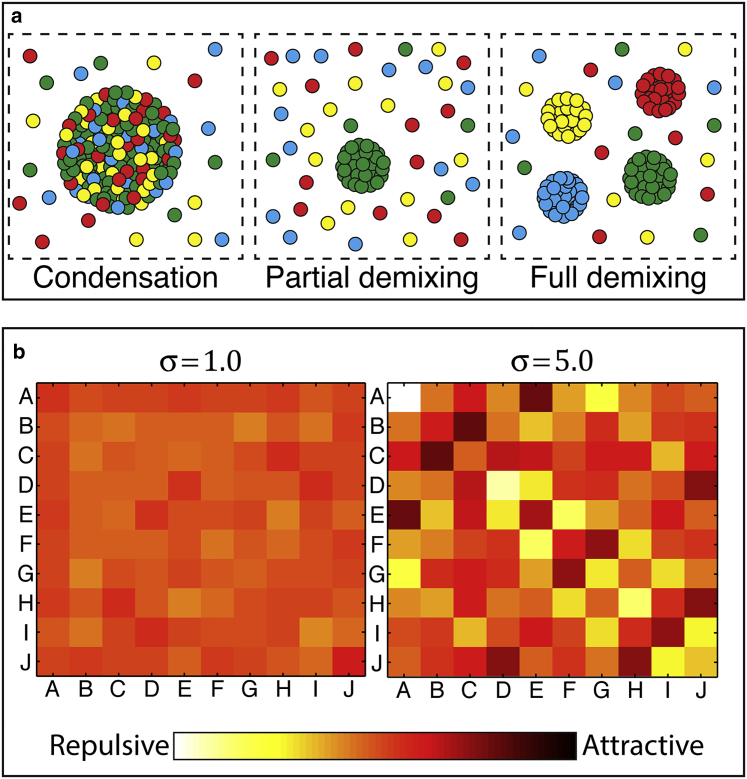
In this issue, Jacobs and Frenkel (8) present a key insight to set physical expectations regarding the number of distinct phases that can coexist in multicomponent mixtures. Although the Gibbs phase rule places an upper limit on the number of conceivable coexisting phases, it does not provide insights regarding the number of phases that one might realistically expect for a system with N distinct macromolecular components. This will be governed by the balance of homo- and heterotypic intermolecular and solvent-macromolecule interactions for all the distinct components in the system. Jacobs and Frenkel (8) developed a simple model to model the effects of these competing interactions. They do so by defining effective pairwise interaction strengths between every pair of components. These interaction strengths are extracted from a Gaussian distribution. Schematics of two interaction matrices generated in this manner are shown in Fig. 1 b. Here, each element in the matrix is defined by selecting a value from a Gaussian distribution with a mean of 0 and a variance (σ) of either 1.0 or 5.0. Effectively, this approach assigns an arbitrary number of different types of sticky particles (in the case of Fig. 1 b, 10 particle types designated A–J), where the stickiness between each pairwise combination of particles is defined using the Gaussian random variable. The phase behavior of a large collection of particles is then simulated using a Grand Canonical lattice based Monte Carlo approach.
Figure 1.
(a) Three limiting cases in a simple four-component system. “Condensation” refers to the coexistence of a single dense and single dilute phase. The relative populations of any two components are the same in both phases, although the absolute numbers vary considerably. “Partial demixing” refers to a subset of the components demixing to form homogenous droplets, while the other components remain soluble. “Full demixing” refers to all components undergoing demixing such that there are as many distinct homogenous droplets as there are components. (b) Two examples of random interaction matrices for 10-component systems (A–J) generated by selecting pairwise interactions from a Gaussian distribution with low variance (σ = 1) and high variance (σ = 5). The strength of the interaction between any two of the components is given at the corresponding intersection element on the matrix. For example, in the high variance table (σ = 5.0) (A) and (E) are strongly attractive, while (A) and (D) are strongly repulsive.
Jacobs and Frenkel (8) find that complex, multicomponent systems show two limiting behaviors, and that these are defined by the variance associated with the Gaussian distribution used to construct the interaction matrix. When the variance is small, relative to the number of components, a condensation transition is observed, whereby all the components condense into a single well-mixed dense phase or a single mixed dilute phase (e.g., Fig. 1 a, condensation). The proportion of different components relative to one another in each of those two phases is identical, although the absolute concentrations will be different. In effect, there are two phases with identical composition but different concentrations. In contrast, when the variance is large relative to the number of components, a small number of components will undergo demixing while the rest remain soluble (Fig. 1 a, partial demixing). As the variance changes across different simulations, the transition between these two limiting cases is sharp, suggesting that the formation of many demixed phases of distinct compositions or several distinct-condensed phases is unlikely.
The variance (σ) defines the extent of similarity among different components. When the variance is small vis-à-vis thermal energy, then all the different components have equivalent effective affinities for another and the result of phase separation is condensation (see left panel in Fig. 1 a). When the variance is large, a small number of components interact preferentially with one another. Accordingly, these components demix to form droplets whereas the weakly interacting components remain part of the dispersed bulk phase. The organizing principle is astonishingly simple: If the effective homo- and heterotypic interactions are of equivalent strength, then condensation results. In contrast, if the interactions are sufficiently different, vis-à-vis thermal energy, then a small number of demixed droplets with distinct compositional biases will result. The transition between the two scenarios is sharp and suggests that intermediate scenarios that lie between condensation and a limited number of heterogeneous demixed droplets are not readily realized. Therefore, the observation of distinct liquidlike intracellular membraneless organelles suggests that cells are poised whereby small changes in expression levels of key driver proteins is sufficient to induce the formation of droplets with specific compositional biases that can then be dissolved either through post-translational modifications, through dilution or through other effects (9, 10).
Although the work of Jacobs and Frenkel (8) sets default expectations for the number of coexisting phases, their work does not take into account the role of interfacial tensions. The formation of distinct phases does not say anything about the spatial organization within droplets. However, layered droplets are also consistent with multiple coexisting phases and this has been reported for the nucleolus (3). It remains to be seen if spatially organized droplets are a unique feature of nucleoli or a general phenomenon associated with membraneless organelles that are driven by the interplay between solute-solute and solvent-solvent interactions.
The use of random interaction models has a long, rich history in physical chemistry and protein biophysics, ranging from theories of ideal solutions to the understanding of protein folding and gene expression. Deviations from random interaction models or agreement between experimental data and simplifying models help uncover insights into the functionally important components. The work of Jacobs and Frenkel (8) represents an important step forward toward achieving a quantitative understanding of phase separation in biological systems. Their random interaction model sets up well-defined expectations for in depth exploration both in vitro and in vivo.
Editor: Leslie Loew.
References
- 1.Mitrea D.M., Kriwacki R.W. Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 2016;14:1. doi: 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergeron-Sandoval L.-P., Safaee N., Michnick S.W. Mechanisms and consequences of macromolecular phase separation. Cell. 2016;165:1067–1079. doi: 10.1016/j.cell.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 3.Feric M., Vaidya N., Brangwynne C.P. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su X., Ditle J.A., Vale R.D. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016;352:595–599. doi: 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brangwynne C.P., Tompa P., Pappu R.V. Polymer physics of intracellular phase transitions. Nat. Phys. 2015;11:899–904. [Google Scholar]
- 6.Brangwynne C.P., Eckmann C.R., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 7.Nott T.J., Craggs T.D., Baldwin A.J. Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nat. Chem. 2016;8:569–575. doi: 10.1038/nchem.2519. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs W.M., Frenkel D. Phase transitions in biological systems with many components. Biophys. J. 2017;112:683–691. doi: 10.1016/j.bpj.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elbaum-Garfinkle S., Kim Y., Brangwynne C.P. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pak C.W., Kosno M., Rosen M.K. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell. 2016;63:72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]