Abstract
Membrane-anchored receptors are essential cellular signaling elements for stimulus sensing, propagation, and transmission inside cells. However, the contributions of lipid interactions to the function and dynamics of embedded receptor kinases have not been described in detail. In this study, we used amide hydrogen/deuterium exchange mass spectrometry, a sensitive biophysical approach, to probe the dynamics of a membrane-embedded receptor kinase, EnvZ, together with functional assays to describe the role of lipids in receptor kinase function. Our results reveal that lipids play an important role in regulating receptor function through interactions with transmembrane segments, as well as through peripheral interactions with nonembedded domains. Specifically, the lipid membrane allosterically modulates the activity of the embedded kinase by altering the dynamics of a glycine-rich motif that is critical for phosphotransfer from ATP. This allostery in EnvZ is independent of membrane composition and involves direct interactions with transmembrane and periplasmic segments, as well as peripheral interactions with nonembedded domains of the protein. In the absence of the membrane-spanning regions, lipid allostery is propagated entirely through peripheral interactions. Whereas lipid allostery impacts the phosphotransferase function of the kinase, extracellular stimulus recognition is mediated via a four-helix bundle subdomain located in the cytoplasm, which functions as the osmosensing core through osmolality-dependent helical stabilization. Our findings emphasize the functional modularity in a membrane-embedded kinase, separated into membrane association, phosphotransferase function, and stimulus recognition. These components are integrated through long-range communication relays, with lipids playing an essential role in regulation.
Introduction
Membrane-anchored receptor proteins in cells function as the first responders to extracellular stimuli. One of the fundamental questions in the field of membrane proteins revolves around how lipid-protein contacts in the lipid bilayer alter embedded receptor function (1). Lipids are significant effectors of receptor function and their regulatory roles are increasingly being recognized, although the full impact of lipids on diverse classes of membrane proteins has yet to be uncovered (2, 3). Our model membrane receptor is EnvZ, a dimeric inner membrane anchored kinase and a member of a two-component signal transduction system (TCS) that is widespread in bacteria and lower eukaryotes (4, 5). TCSs recognize and transduce stimuli associated with various stresses to mediate cellular responses. TCSs typically consist of a sensor component, a membrane-anchored histidine kinase that undergoes altered autophosphorylation upon sensing extracellular stimuli. This phosphate group is subsequently transferred to a conserved aspartate on a cognate response regulator, which mediates the cellular response. The EnvZ/OmpR system is a prototypical TCS in which EnvZ senses changes in environmental osmolality and transmits the signal to its response regulator, OmpR. OmpR is a transcription factor that regulates the transcription of the outer-membrane porin genes ompF and ompC (6, 7, 8, 9, 10, 11, 12, 13, 14, 15). Phosphorylation of OmpR changes its affinity for promoters of outer membrane porin genes, resulting in altered porin expression (16).
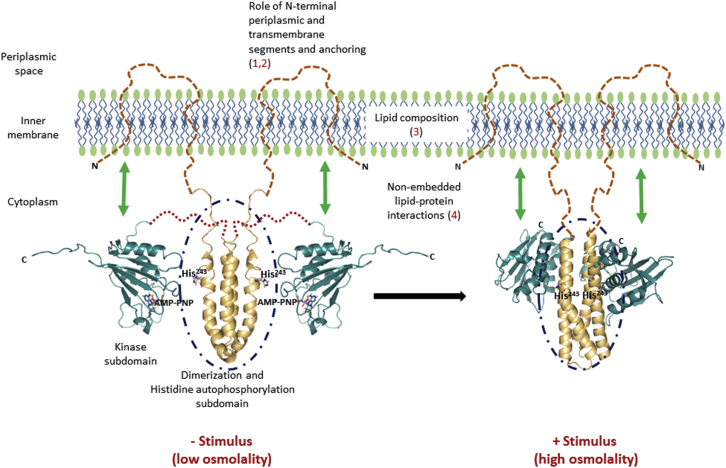
EnvZ has five putative domains (17): a short N-terminus, a periplasmic domain, two transmembrane (TM) regions, an intracellular HAMP (histidine kinase, adenylate cyclase, methyl-accepting proteins and phosphatases) domain (18), and a large C-terminal domain. The C-terminal cytoplasmic domain is organized into a four-helix bundle subdomain, with two helices being contributed by each monomer and linked to the ATP-binding kinase subdomain through a flexible linker. Structures of the four-helix bundle (PDB: 1JOY) (19) and the kinase subdomains (PDB: 1BXD) (20) solved by NMR offered structural insights into the isolated subdomains at low osmolality. Crystallographic structures of the two subdomains tethered together were solved recently in a chimeric construct of EnvZ formed from Thermotoga maritima and Escherichia coli EnvZ (PDB: 4KP4) (21), representing the structure of EnvZ at high osmolality (Fig. 1). The four-helix bundle subdomain forms the dimeric core of the protein and also contains the site of autophosphorylation (His243). No high-resolution structural information is available for the periplasmic or TM domains.
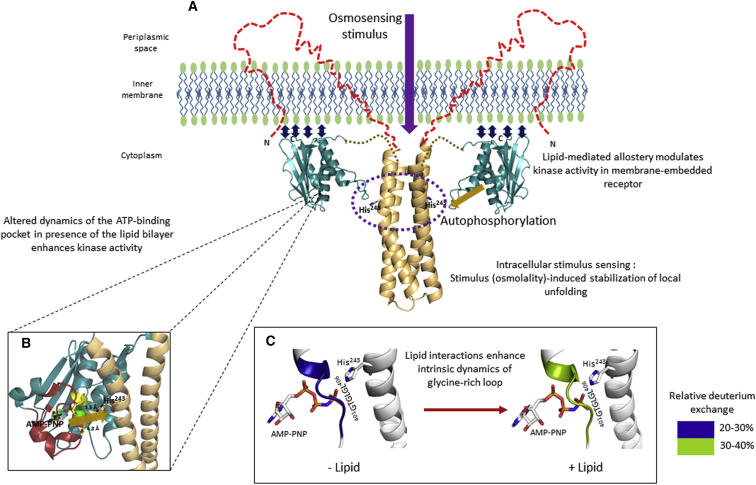
Figure 1.
Membrane lipids alter receptor dynamics and function. The cytoplasmic domain of EnvZ (EnvZc; 180–450) lacking N-terminal TM and periplasmic segments is capable of osmosensing, achieved through stabilization of a locally disordered locus (dark blue dashed circles) spanning the autophosphorylation site His243 (shown as sticks) within the four-helix dimerization subdomain. This establishes this His243-containing locally disordered region as the osmosensing core in EnvZ. However, this raises a question regarding the contribution of 1) the N-terminal 179 residues (the TM and periplasmic domains), 2) membrane anchoring, 3) membrane composition, and 4) potential interactions between the cytoplasmic domain and the membrane to EnvZ dynamics and function. Left: solution NMR structures of the four-helix bundle subdomain (gold, PDB: 1JOY) and the kinase subdomain (teal, PDB: 1BXD) solved separately, denoting the low-osmolality conformation. Large-scale conformational changes are observed in the presence of the osmolality stimulus. Right: x-ray crystal structure of a chimeric construct of the entire EnvZ cytoplasmic domain encompassing both the four-helix bundle (gold) and the kinase subdomains (teal, PDB: 4KP4), denoting the high-osmolality conformation. Structures of the TM and periplasmic regions are currently not available and are depicted as brown dashed lines.
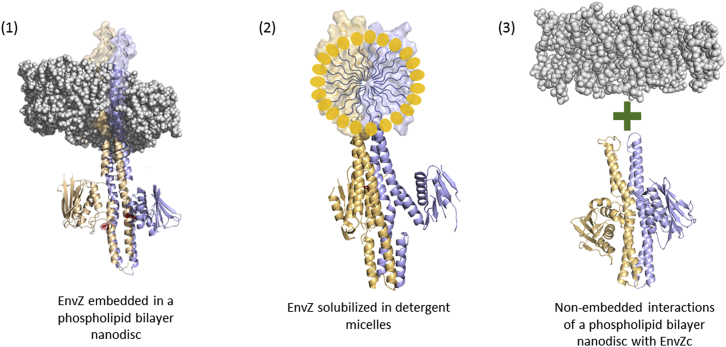
Recent studies have shown that the cytoplasmic C-terminal domain of EnvZ (residues 180–450, EnvZc) is sufficient to restore osmosensing function in strains of E. coli containing a deletion of envZ (22). These results, together with a comparative dynamics analysis performed using amide hydrogen/deuterium exchange mass spectrometry (HDXMS) at low and high osmolality, revealed that the four-helix bundle subdomain was the central osmosensing locus in EnvZ. This showed that the membrane-spanning N-terminal residues of EnvZ are nonessential for osmosensing, and that any roles of the membrane and associated regions are uncoupled from osmosensing in EnvZc. This intriguing and unexpected result raised two obvious questions: what function do the N-terminal 179 residues of EnvZ serve, and what role, if any, does membrane anchoring play in the osmosensing function of EnvZ. To address these questions, we compared the function and dynamics of intact, detergent-solubilized EnvZ in solution and intact EnvZ in phospholipid bilayer nanodiscs (Fig. 2). Membrane proteins have typically been examined in detergent-solubilized conditions, which do not represent the native lipid-protein interface. As an alternative, proteoliposomes offer a more physiologically relevant membrane environment, but their application is limited by their inherent instability and polydispersity (23). Lipid bilayer nanodiscs, on the other hand, are characterized by their monodisperse nature, long-term stability, and application to a wide range of membrane protein targets and biophysical tools (24).
Figure 2.
Use of membrane mimetics to probe receptor dynamics and function. Different artificial membrane environments have been used to investigate the multiple mechanisms through which the cellular membrane may impact receptor dynamics and function in vitro. 1) The effect of membrane anchoring and lipid composition can be studied using EnvZ embedded in phospholipid bilayer nanodiscs. 2) EnvZ solubilized in detergent micelles allows the role of the N-terminal TM and periplasmic domains to be described independently of phospholipid bilayer anchoring. 3) Nonembedded peripheral interactions of the phospholipid bilayer membrane with the soluble cytoplasmic domain can be examined with EnvZc in the presence of a nonembedded phospholipid bilayer nanodisc.
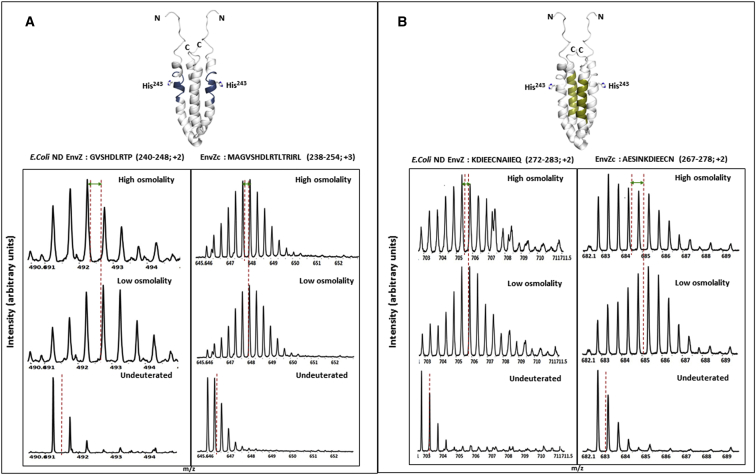
HDXMS is a newly emerging approach for probing the dynamics of membrane-embedded proteins (25, 26, 27). It is particularly suited for this purpose because it causes minimal perturbation of the sample, since the method uses backbone amide hydrogens as conformational reporters and does not necessitate modification of the protein sample. By measuring the average increase in deuterons exchanged at backbone amide positions, HDXMS reports on the solvent accessibility (28) and hydrogen-bonding propensities (29) of different regions in proteins, at peptide resolution. Changes in deuterium exchange correlate with protein dynamics on millisecond and longer timescales, and further allow the effects of diverse perturbations to be mapped (30). Prior studies focusing on EnvZc showed that osmosensing is achieved through intrahelical stabilization of a locally unfolded region containing His243 and a flanking helix that is a putative OmpR-binding site (22). The His243-containing peptide showed a decreased exchange of ∼2 deuterons in the presence of osmolytes, indicative of at least two hydrogen bonds undergoing stabilization at high osmolality. The flanking helix exhibited characteristic bimodal profiles indicative of ensemble behavior, with osmolytes increasing the relative proportion of the lower-exchanging (more ordered) conformation. Importantly, these were the only two regions across EnvZc that showed any significant differences in deuterium exchange with osmolality, and represented the osmosensing core of EnvZ. These peptides additionally offered two reporters to track the dynamics of the four-helix bundle in the full-length protein and map the effects of osmolytes on EnvZ (Fig. 3).
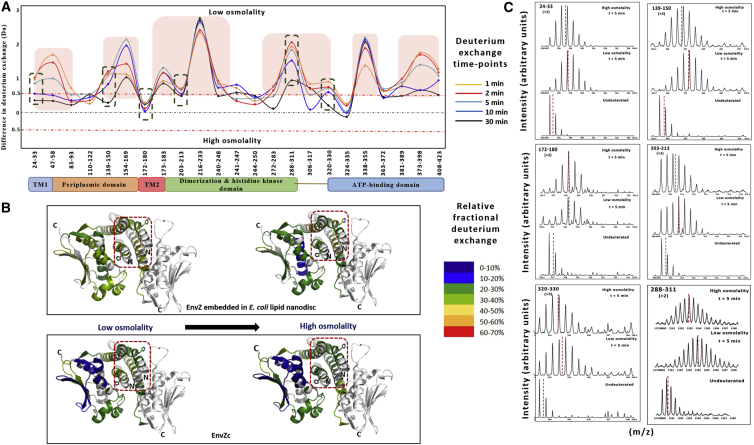
Figure 3.
The four-helix bundle subdomain constitutes the osmosensing core in both EnvZc and EnvZ embedded in E. coli lipid nanodiscs. In EnvZc, two peptides within the four-helix bundle showed decreased deuterium exchange with an increase in osmolality, and were identified as the osmosensing core of EnvZc (22). These represented important peptide reporters to measure the effects of osmolality on EnvZ dynamics. In EnvZ embedded in E. coli lipid nanodiscs, the same regions exhibited significant decreases in deuterium exchange, highlighting the importance of these regions for EnvZ function. The electrospray ionization quadrupole time-of-flight mass spectra shown here demonstrate the decrease in deuterium exchange with increased osmolality, as represented by the shift in centroid mass (red dashed vertical line) of the mass isotopic envelope. (A) The locally unfolded region containing His243 (highlighted in blue; PDB: 1JOY) is stabilized under high osmolality (20 mM Tris-HCl, 20% sucrose w/v; 780 mOsm/kg) in both EnvZ in E. coli nanodiscs (peptide 240–248) and EnvZc (peptide 238–254). (B) The flanking helix (highlighted in gold), a putative OmpR-binding region, also showed decreased deuterium exchange under high osmolality in both EnvZ in E. coli lipid nanodiscs and EnvZc. A shift of the mass spectral envelope to the left denotes stabilization of the peptide, as highlighted by the green double-headed arrows.
We set out to examine any potential effects of lipid anchoring and the N-terminal 179 residues on intrinsic dynamics and basal autophosphorylation. For this purpose, we serially probed the contributions of the following factors to the intrinsic dynamics and osmosensing function of EnvZ: 1) N-terminal residues (1–179), 2) peripheral interactions between EnvZc and the membrane, 3) anchoring of full-length EnvZ, and 4) lipid composition (Fig. 1). We further tested the effects of osmolytes on EnvZ dynamics and function to confirm the uncoupling of osmosensing from membrane associations.
Our results reveal that while the four-helix bundle subdomain remains the osmosensing core, there are broader osmolality-dependent changes apparent within full-length EnvZ, which are not present in EnvZc. These include the HAMP domain and the kinase subdomain, which provide allosteric relays that directly communicate the effects of membrane anchoring and lipid composition to osmosensing. In addition, the TM and periplasmic regions exhibited osmolality-dependent changes in deuterium exchange. This accounts for the higher basal activity in membrane-anchored EnvZ as compared with the cytoplasmic domain alone. Lipid-mediated allostery thus highlights the critical contributions of membrane composition to receptor function. The membrane lipid interface directly regulates EnvZ kinase dynamics and function to amplify the basal activity of the kinase. The extracellular stimulus (osmolality) in turn regulates the kinase substrate, which is reflected in an ∼4- to 5-fold enhancement in kinase activity. Such a two-pronged parallel regulation operates to integrate the uncoupled lipid-regulated phosphotransferase activity with stimulus-sensing functions. This enhances the sensitivity of the stimulus response in membrane-associated enzymes. Therefore, this study has broader implications for all membrane-embedded protein receptor kinases.
Materials and Methods
Materials
All chemicals and reagents were obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. E. coli BL21 (DE3) strain was purchased from Invitrogen (Carlsbad, CA). Lysogeny broth media for bacterial cell cultures were prepared from tryptone and yeast extract obtained from BD Biosciences (Franklin Lakes, NJ), and isopropyl β-D-1-thiogalactopyranoside (IPTG) was purchased from Bio Basic (Ontario, Canada). N,N,N′,N′-tetramethylethane-1,2-diamine (electrophoresis grade) was obtained from MP Biomedicals (Solon, OH), ammonium persulfate and 30% acrylamide/bis-acrylamide were purchased from Bio-Rad Laboratories (Hercules, CA), and Coomassie Blue G 250 was obtained from US Biological (Swampscott, MA). Ethylenediaminetetraacetic acid (EDTA)-free complete protease inhibitor cocktail tablets were purchased from Roche Diagnostics (Mannheim, Germany). TALON cobalt affinity resin was obtained from Clontech Laboratories (Mountain View, CA). E. coli total lipid extract, 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), and n-dodecyl-β-D-maltoside (DDM) were purchased from Avanti Polar Lipids (Alabaster, AL). Recombinant membrane scaffold protein (MSP1D1) was purchased from Sigma-Aldrich.
Expression and purification of EnvZ and EnvZc
A gene encoding full-length E. coli EnvZ (residues 1–450, Uniprot: P0AEJ4) was obtained from DNA 2.0 (Newark, CA) with an N-terminal 6xHis-tag and inserted into the ampicillin-resistant vector pJexpress414. The plasmid was transformed in E. coli BL21(DE3)-competent cells for protein overexpression, and the cells were grown in lysogeny broth medium (10 g tryptone, 5 g yeast extract, and 10 g NaCl per liter) containing a final concentration of 100 μg/mL ampicillin at 37°C until an OD600 of ∼0.6–0.8 was reached. Expression was induced by the addition of IPTG to a final concentration of 250 μM with overnight shaking at 18°C. The cells were harvested at 7500 × g for 15 min and stored at −30°C. The frozen pellet was resuspended in ice-cold lysis buffer (20 mM Tris-HCl, 200 mM NaCl, 10 μM EDTA, 5% (v/v glycerol, pH 7.6) supplemented with EDTA-free protease inhibitor cocktail tablets. The cells were lysed by sonication for 15 min (1 s pulse every 1 s) followed by centrifugation of the lysate at 18,000 × g for 1 h at 4°C to remove cell debris and inclusion bodies. To isolate the membrane fraction, the supernatant was subjected to ultracentrifugation at 80,000 × g for 90 min at 4°C in a Beckman Coulter XL-90 preparative ultracentrifuge. The pellet containing the membrane fraction was solubilized in equilibration buffer (20 mM Tris-HCl, 0.1% DDM, pH 7.6) and added to TALON cobalt affinity resin equilibrated with two bed volumes of equilibration buffer and incubated overnight at 4°C with mild shaking. After removal of unbound proteins in the flow-through, the resin was washed with two bed volumes of wash buffer (equilibration buffer containing 5 mM imidazole) for removal of nonspecifically bound proteins. His-tagged EnvZ was eluted with two bed volumes of elution buffer (equilibration buffer containing 250 mM imidazole). The eluate was further subjected to size-exclusion chromatography in a Superdex S200 10/300 GL column preequilibrated with two column volumes of equilibration buffer in an AKTA FPLC system (General Electric Healthcare, Chicago, IL) to obtain DDM-solubilized EnvZ. Protein purity was ascertained at every purification step on a 12% SDS-PAGE gel.
Reconstitution of EnvZ in phospholipid bilayer nanodiscs
DDM-solubilized EnvZ was reconstituted into phospholipid bilayer nanodiscs following published protocol (31). EnvZ was embedded in nanodiscs prepared from either E. coli total lipid extract or synthetic DOPC and MSP1D1. Lipid stock solutions were prepared by dissolving lyophilized lipids in chloroform to a final concentration of 250 mM and stored at −30°C for long-term storage. Before use for nanodisc self-assembly, the chloroform was removed by evaporation and the lipids were solubilized in sodium deoxycholate to a final concentration of 100 mM lipids and 200 mM sodium deoxycholate. DDM-solubilized EnvZ, MSP1D1, and lipids were mixed in ratios of 1:5:200 for E. coli total lipids and 1:5:350 for DOPC, and the mixture was incubated at 4°C with mild shaking for 30 min. This was followed by dialysis of the mixture in a buffer containing 20 mM Tris-HCl, pH 7.6, for 36 h to remove the detergents and promote self-assembly of the nanodiscs. The dialyzed mixture was then subjected to size-exclusion chromatography in a HiLoad 16/60 Superdex S200 prep grade column in 20 mM Tris-HCl, pH 7.6, to resolve EnvZ-embedded nanodiscs from empty ones. Nonembedded nanodiscs were separately assembled using the same MSP1D1/lipid ratios, and the same protocol was followed for both E. coli lipids and DOPC. Purification of EnvZc was performed as previously described (22).
Kinase autophosphorylation assay
Autophosphorylation by EnvZ was measured using the in vitro ADP-Glo kinase assay (Promega, Madison, WI), which quantifies the amount of ADP formed as a product in a kinase reaction using a luminescence-based detection method (32). All reactions were carried out according to the manufacturer’s recommendations and were performed at least in duplicates. Kinase reactions were performed for the following samples: 1) full-length EnvZ reconstituted in E. coli total lipid extract nanodiscs, 2) full-length EnvZ reconstituted in DOPC nanodiscs, 3) full-length EnvZ solubilized in DDM micelles, 4) EnvZc in the presence of nonembedded E. coli total lipid extract nanodiscs, and 5) EnvZc under both low- and high-osmolality conditions. The protein samples were diluted to a final concentration of 5 μM EnvZ in a low-osmolality buffer containing 20 mM Tris-HCl, 2 mM MgCl2, 100 μM ATP, pH 7.6, to a final volume of 10 μL. The high-osmolality buffer additionally contained a final concentration of 20% sucrose. Standard curves were plotted with the same buffer containing 5 μM of nonembedded nanodiscs for the nanodisc samples. The buffers for full-length EnvZ solubilized in DDM micelles contained 0.1% DDM along with the other components. Luminescence recordings were obtained with a Glomax Discover Multimode Detection System (Promega). The amount of ADP produced was calculated based on standard curves constructed according to the manufacturer’s recommendations. Enzyme activity is represented as moles of ADP produced per mole of enzyme per minute (Table 1).
Table 1.
Embedded and Peripheral Lipid Interactions Enhance EnvZ His243 Autophosphorylation
| ADP Produced (Mole/Min per Mole of Enzyme) |
||
|---|---|---|
| Low Osmolality | High Osmolality | |
| EnvZc | 0.09 ± 0.04 | 0.32 ± 0.08 |
| EnvZ-E. coli nanodiscs | 1.23 ± 0.17 | 4.9 ± 0.01 |
| EnvZ-DOPC nanodiscs | 0.93 ± 0.06 | 1.36 ± 0.05 |
| EnvZc + nonembedded E. coli lipid nanodiscs | 2.2 ± 0.33 | 4.4 ± 0.4 |
| EnvZ-DDM | 2.5 ± 0.1 | 7.3 ± 0.27 |
EnvZ His243 autophosphorylation in different membrane environments was measured using an in vitro ADP Glo kinase assay, which quantitates the amount of ADP produced in a kinase reaction via a luminescence-based detection method. These measurements were performed under conditions of both low (100 mOsm/kg) and high (20% sucrose/780 mOsm/kg) osmolality. Full-length EnvZ exhibited higher basal activity relative to EnvZc in E. coli lipid and DOPC nanodiscs, as well as DDM micelles. Increased autophosphorylation was also observed in EnvZc in the presence of nonembedded E. coli lipid nanodiscs. A ∼4-fold increase in kinase autophosphorylation was observed with increased osmolality in all environments. Activity is expressed in mol ADP/mol EnvZ per min.
HDXMS
HDXMS was performed in accordance with an established protocol (22) on the following samples: 1) full-length EnvZ reconstituted in E. coli total lipid extract nanodiscs, 2) full-length EnvZ reconstituted in DOPC nanodiscs, 3) full-length EnvZ solubilized in DDM micelles, and 4) EnvZc in the presence of empty E. coli total lipid extract nanodiscs. Deuterium exchange was initiated by diluting ∼100 pmol of protein sample in buffer reconstituted in 99.9% D2O to a final concentration of 90% D2O and incubated at 25°C for different time points. Deuterium exchange was performed for all full-length EnvZ samples for 1, 2, 5, 10, and 30 min, whereas the EnvZc samples were subjected to deuterium exchange for 1 and 5 min only. The low-osmolality buffer consisted of 20 mM Tris-HCl, pH 7.6, and the high-osmolality buffer contained an additional 20% sucrose. All deuterium-exchange experiments were performed at least in duplicates. Undeuterated samples were processed in an identical manner and served as the negative control for mass spectral analysis.
After deuteration, the exchange reaction was quenched with prechilled 0.1% TFA to bring the final pHread to ∼2.5. All steps after the exchange reaction quench step were performed on ice or at 4°C to minimize back-exchange of exchanged amide deuterons. An additional 0.5% DDM was added to the quench solution for all nanodisc samples to allow disassembly of the nanodiscs, thereby releasing the embedded protein and facilitating subsequent pepsin digestion. The ionic nature of phospholipids and their high concentration in nanodisc samples tend to suppress peptide signal intensity in a mass spectrometer. To circumvent this problem, it is essential to remove the phospholipids from a quenched sample before injecting it. The quenched nanodisc samples containing DDM were therefore incubated on ice for 4 min to facilitate nanodisc disassembly before 5 mg of powdered TiO2 was added to adsorb the phospholipids. After 1 min of incubation with TiO2, the sample was filtered by centrifugation for 1 min at 13,000 × g and then injected into a chilled nanoACQUITY UltraPerformance LC (UPLC) system (Waters, Milford, MA). The sample was first subjected to proteolysis by a 2.1 × 30 mm2 Poroszyme immobilized pepsin column with 0.1% (v/v) formic acid in water at pH ∼2.5 at a flow rate of 100 μL/min, serving as the digestion buffer. Peptides were initially trapped in an ACQUITY UPLC BEH C18 VanGuard precolumn (130 Å, 1.7 μm, 2.1 × 5 mm2) for 3 min, followed by elution with a water-acetonitrile gradient of 8–40% (v/v) acetonitrile in a 1.0 × 100 mm2 ACQUITY UPLC BEH C18 reversed-phase column (Waters), with both solvents containing 0.1% (v/v) formic acid and flowing at 40 μL/min. The eluted peptides were detected by a SYNAPT G2-Si high-definition mass spectrometer (Waters, Manchester, UK), and data were acquired in MSE data acquisition mode. Continuous calibration of the mass spectrometer was performed with 200 fmol/μL of [Glu1]-fibrinogen peptide B as a standard, at a flow rate of 1 μL/min.
The raw spectra were analyzed in the same manner as described previously (22). The MSE data for undeuterated samples were searched against a database containing the primary sequence of the protein and specific experimental conditions, using ProteinLynx Global SERVER software (PLGS v3.0; Waters). Peptides identified by PLGS were further filtered and analyzed in the semiautomated analysis software DynamX (v3.0; Waters) to map the deuteration profiles of experimental samples.
Results
Nanodisc reconstitution enhanced EnvZ autophosphorylation
Full-length EnvZ was expressed and purified as described in Materials and Methods and solubilized in DDM, a nonionic detergent, and also reconstituted into phospholipid bilayer nanodiscs. DDM was specifically chosen because it is a nonionic detergent that is commonly used for membrane protein analysis and is amenable to mass spectrometry (23). Two compositions of lipids were selected to prepare EnvZ-incorporated nanodiscs. The condition that most closely mimicked native EnvZ was achieved by preparing nanodiscs with E. coli total lipid extracts. To test the effect of lipid composition on EnvZ function, we further prepared an alternative nanodisc with DOPC, a phospholipid that is not native to E. coli. Both detergent and nanodisc preparations of EnvZ were dimeric, as monitored by size-exclusion chromatography (Figs. S1 in the Supporting Material). EnvZ in DDM, as well as in both nanodisc preparations, was active in in vitro kinase assays (described in Materials and Methods), as evidenced by an osmolality-dependent increase in autophosphorylation (Table 1). The intrinsic autophosphorylation activity of full-length EnvZ at low osmolality was significantly greater (by ∼15-fold) in nanodisc samples prepared with E. coli total lipid extracts and DOPC in comparison with EnvZc. Detergent-solubilized and nanodisc-embedded EnvZ samples showed equivalent increases in autophosphorylation. All EnvZ samples displayed a ∼4-fold enhancement in autophosphorylation at high osmolality compared with low osmolality (Table 1), as described previously (22).
Membrane anchoring increased deuterium exchange across the protein
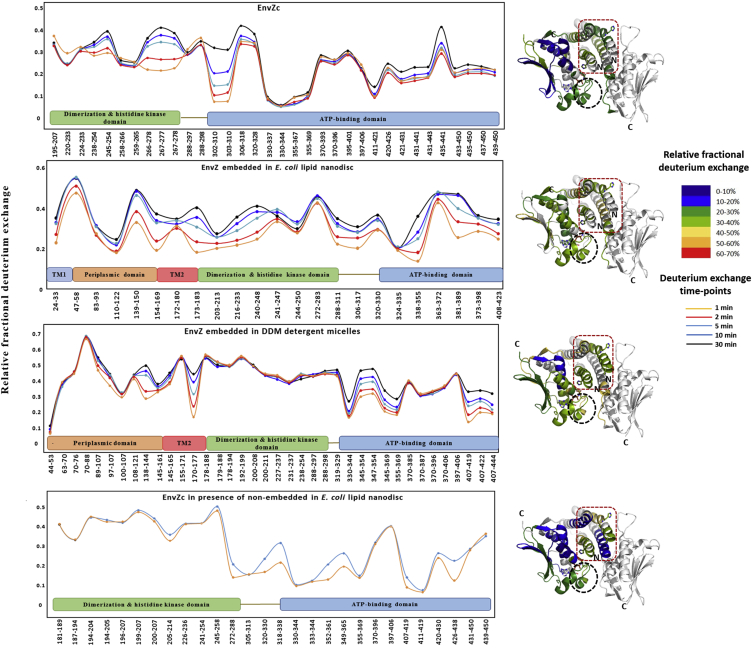
HDXMS was used to probe the dynamics of EnvZ and EnvZc in the presence of lipid. HDXMS of soluble EnvZc in the absence of lipid was used as a reference to map the effects of 1) N-terminal residues, 2) membrane anchoring, and 3) the lipid-protein interface on the dynamics of EnvZ. Consequently, HDXMS was performed on separate samples of EnvZ and EnvZc in different environments. HDXMS of full-length EnvZ embedded in nanodiscs prepared with E. coli lipids showed higher relative deuterium exchange across the entire protein compared with EnvZc at low osmolality (Fig. 4, A and B). Movies S1 and S2 show the deuterium exchange at every time point and in every state of E. coli lipid-embedded EnvZ and all other conditions. Overall, full-length EnvZ embedded in nanodiscs showed higher exchange in both the kinase and four-helix bundle peptides relative to EnvZc. Thus, the N-terminal 179 residues encompassing the periplasmic region, TM helices, the HAMP domain, and the lipid-protein interface in the nanodisc contributed to increasing deuterium exchange throughout the protein (Fig. 4 B). HDXMS of full-length EnvZ solubilized in DDM also showed increased overall exchange in both the His243 osmosensing core and kinase subdomains (Fig. 4 C).
Figure 4.
Membrane interactions alter deuterium exchange across the cytoplasmic domain. (A–D) The relative fractional uptake in deuterium exchange for each peptic digest fragment is displayed in a modified mirror plot (left panels) for (B) EnvZ embedded in E. coli lipid nanodiscs, (A) EnvZc, (C) EnvZ solubilized in DDM micelles, and (D) EnvZc in the presence of nonembedded E. coli lipid nanodiscs. Deuterium exchange was performed for different time points: 1 min (yellow), 2 min (red), 5 min (light blue), 10 min (dark blue), and 30 min (black). The relative fractional uptake of different regions for a 5 min exchange is mapped onto the x-ray crystal structure of a chimeric construct of the cytoplasmic domain of EnvZ (PDB: 4KP4) in accordance with the key to the right. EnvZ embedded in E. coli lipid nanodiscs exhibits higher deuterium exchange in comparison with EnvZc across the cytoplasmic domain and particularly in the osmosensing core, as highlighted by the red dashed box. Further, the His243-containing region exhibits increased deuterium exchange in EnvZ solubilized in DDM micelles and in EnvZc in the presence of E. coli lipid nanodiscs. This demonstrates a membrane-mediated modulation of the dynamics of the cytoplasmic domain, particularly at the osmosensing core. The ATP-binding pocket shows increased deuterium exchange in all samples (highlighted by the black dashed circle) when compared with EnvZc.
The presence of nonembedded nanodiscs increased EnvZc autophosphorylation
To probe the role of peripheral, nonembedded lipid-protein interactions in function and dynamics, we carried out a control experiment, measuring autophosphorylation of EnvZc in the presence of equimolar concentrations of nonembedded nanodiscs in solution. Surprisingly, the basal autophosphorylation activity was ∼25-fold greater in the presence of nanodiscs than with EnvZc alone (Table 1). The HDXMS results showed highly similar patterns in EnvZc in the absence and presence of nanodiscs (Fig. 4 D). Interestingly, one locus that showed increased exchange in EnvZc in the presence of nonembedded nanodiscs was the glycine-rich loop (Fig. 4 D), a critical motif that is associated with ATP binding and phosphotransfer in all kinases (33). Importantly, the glycine-rich motif also exhibited increased intrinsic exchange in both nanodisc and detergent-solubilized EnvZ. This glycine-rich motif is discussed in greater detail in the Discussion section (Fig. 6).
Figure 6.
Peripheral membrane interactions are essential for lipid allostery in EnvZ. (A) The lipid membrane results in a significant increase (∼15-fold) in the in vitro kinase activity of EnvZ. We propose that the increased basal kinase activity of EnvZ is due to lipid allosteric effects mediated by local conformational changes within the kinase subdomain, particularly the ATP-binding pocket. The increase in His243 autophosphorylation due to increased environmental osmolality (denoted by the purple arrow) is independent of membrane effects and is achieved entirely through stabilization of the locally disordered osmosensing core (purple dashed circle) within the four-helix bundle. (B) The ATP-binding pocket (highlighted in red) and the glycine-rich loop (yellow) integrate lipid allosteric effects to enhance catalysis of phosphate group transfer from the kinase subdomain to the His243 residue. The yellow arrow denotes phosphotransfer from the ATP-binding pocket to His243. (C) Lipid interactions enhance the intrinsic dynamics of the glycine-rich loop, as revealed by increased deuterium exchange. We postulate that this enhancement in intrinsic dynamics forms the basis for a faster phosphotransfer rate.
The greatly enhanced intrinsic activity of EnvZ in either nanodisc or detergent micelles, as well as EnvZc in the presence of nonembedded nanodiscs, correlates with the increased exchange at the glycine-rich motif and indicates that lipid or micelle interactions converge at the glycine-rich motif to greatly enhance the basal phosphotransferase activity of the kinase subdomain. Together, these results further confirm the importance of nonembedded lipid interactions, which impacted the dynamics in the ATP-binding pocket, leading to a large enhancement in basal autophosphorylation activity.
Osmolality-dependent enhancement of autophosphorylation activity in full-length EnvZ
In the presence of osmolytes, EnvZ in all of the different conditions showed a ∼4-fold increase in autophosphorylation activity comparable to that observed with EnvZc alone (Table 1). A comparison of deuterium exchange in full-length EnvZ at low and high osmolality showed osmolality-dependent decreases in exchange across the protein, i.e., within the four-helix bundle and kinase subdomains, as well as in the periplasmic and TM regions in the N-terminal region (Fig. 5). Importantly, the four-helix bundle subdomain peptides showed a similar magnitude of decreased exchange with osmolality in EnvZ in all conditions. These results confirmed earlier reports that the osmosensing core is localized to the four-helix subdomain and that membrane association is uncoupled from osmosensing.
Figure 5.
Osmolyte-induced stabilization of EnvZ embedded in E. coli lipid nanodiscs. (A) A comparison of deuterium exchange between conditions of low and high osmolality in EnvZ embedded in E. coli lipid nanodiscs is demonstrated by a difference plot. A threshold difference of 0.5 Da is considered to be a significant difference and is indicated by the red dashed line on the difference plot. Positive changes denote decreased deuterium exchange, and negative changes denote increased exchange in the presence of high osmolality. EnvZ embedded in E. coli lipid nanodiscs responds to osmolality through stabilization of multiple regions (boxed in red) of the protein, including the TM and periplasmic and kinase domains, besides the dimerization and autophosphorylation domains. (B) The relative fractional deuterium uptake is mapped onto the crystal structure of EnvZc (PDB: 4KP4) under conditions of both low and high osmolality. EnvZc responds to increased osmolality though stabilization of the His243-containing four-helix bundle, but no significant stabilization of the other regions of the protein is observed. In EnvZ embedded in E. coli lipid nanodiscs, osmolyte-induced stabilization is observed in the four-helix bundle along with stabilization of regions within the kinase domain. (C) Electrospray ionization quadrupole time-of-flight mass spectra for different peptides (green dashed boxes in the difference plot) of EnvZ embedded in E. coli lipid nanodiscs are compared under conditions of low and high osmolality.
Significantly, the glycine-rich motif within the ATP-binding pocket showed no differences in deuterium exchange at high osmolality. This indicated that osmolality did not impact lipid allosteric control of phosphotransferase activity. The four-helix bundle subdomain remains the essential osmosensing core in full-length EnvZ, wherein osmolality-dependent stabilization of local unfolding in the His243 and flanking peptides resulted in enhanced autophosphorylation. Lipid-mediated allostery, on the other hand, altered autophosphorylation catalysis mediated by the kinase subdomain.
HDXMS of TM regions indicates conformational fluidity
One of the important advantages of HDXMS is that it offers insights into the conformational dynamics of TM segments in membrane proteins. Of the two predicted TM helical segments, deuterium exchange across residues 24-33 (TM1) and residues 154-169, 172-180 and 173-183 (TM2), respectively were monitored. All of these peptides showed high deuterium exchange, with a relative fractional deuterium uptake across these peptides of ∼40% at low osmolality (Fig. 4, B and C). Factoring in deuterium back-exchange and a maximum theoretical deuterium exchange of 90% in our experimental conditions, this indicated a high exchange in the TMs (∼60%). This surprising result shows that the TMs are highly solvent accessible and display a high relative exchange, indicating a fluid conformational ensemble character of the TMs. Both TMs exhibited osmolality-dependent decreases in deuterium exchange, suggesting osmolality-dependent stabilization of EnvZ TMs.
The scaffold protein MSP1D1, which is essential for the integrity of nanodiscs, served as a suitable control to determine the effects of lipid composition on scaffold protein composition (34). We observed only minor effects of lipid composition on exchange across MSP1D1 peptides (Table S1).
Discussion
Lipids as allosteric modulators of membrane protein function
Lipids are important matrices in cellular membranes that contribute various physical and chemical properties to associated membrane proteins, including fluidity, curvature, surface charge, and hydrophobicity (35, 36). Lipids are thus increasingly being recognized for mediating divergent regulatory roles in controlling associated embedded membrane protein function (37, 38, 39, 40). In this study, we demonstrate how lipid-mediated allostery has a profound impact on receptor function, which has direct implications for understanding TM receptor signaling mechanisms.
The most significant result from this study is the allosteric effect of the lipid environment on EnvZ’s conformational dynamics and autophosphorylation function. Similar allosteric modulation of receptor function has been demonstrated in diverse membrane protein families, including G-protein-coupled receptors, receptor tyrosine kinases, and proton channels (37, 39, 41, 42, 43, 44), as well as soluble cytoplasmic proteins containing lipid-targeting domains (45, 46). Lipids impact the structure and function of membrane-embedded or membrane-associated proteins either through bulk effects mediated by the phospholipid bilayer or through binding of lipid molecules at specific sites (40, 47, 48, 49, 50). Interestingly, from our study, we conclude that lipid effects in EnvZ operate predominantly through peripheral interactions between nonembedded cytoplasmic domains and the lipid membrane. Peripheral interactions of soluble cytoplasmic domains with the lipid environment and the resulting modulation in kinase activity have been observed in other lipid-dependent kinases as well (45, 51, 52).
The lipid composition of the membrane is a key determinant of the allosteric modulation of receptor function for some receptors (37, 39, 53). However, in the case of EnvZ, we observed no significant differences in enzymatic activity between the physiological E. coli lipids and the nonbacterial phospholipid DOPC. In fact, the receptor activity was retained even in detergent micelles, signifying that the impact of the hydrophobic membrane on EnvZ function is independent of its chemical composition. Although basal autophosphorylation and osmolyte-induced enhancement of activity were comparable in all membrane-mimetic environments, some differences in conformational dynamics were observed between the nanodisc and micellar samples. These differences in the HDXMS results may be a consequence of clustering and lateral diffusion of multiple EnvZ dimers within a detergent micelle. The MSP scaffold protein restricts incorporation of EnvZ to only one functional dimer in a nanodisc, thereby eliminating the possibility of clustering and lateral diffusion.
Dynamics of TM segments and implications for TM signaling
Structural insights into TMs in membrane proteins have been gleaned from x-ray crystallographic studies of membrane proteins, where they were predominantly observed to adopt α-helical conformations (54). However, these conformations are stabilized endpoint states generated under crystallization conditions in the presence of high concentrations of salts and precipitants, and thus are likely to promote and stabilize ordered structures. Our studies demonstrate the ability of HDXMS to monitor conformations of TM segments in solution and in different membrane-mimicking environments.
HDXMS revealed high relative exchange across both putative TMs. This observation indicates that although these regions have a propensity to adopt helical conformations, they are not stable helices. It is also likely that the stability of helical TMs is dependent upon factors such as the primary sequence, oligomeric state, and local lipid composition (55, 56). This work highlights that the TMs in membrane proteins are nonuniform, and emphasizes the importance of mapping the conformational dynamics of TMs to describe TM signal transduction and propagation. It is significant that osmolality-dependent changes in deuterium exchange were also observed across both TMs, identifying TMs as distinct loci for allosteric coupling of lipids to EnvZ function.
Multiple allosteric relays couple lipids to EnvZ autophosphorylation
EnvZc retains full functionality and osmosensing (22) in vitro and in vivo. The results from EnvZc highlighted the modularity built into a membrane-associated signaling sensor kinase. Three broad functional modules in EnvZ can be identified: 1) the membrane-spanning segments, 2) the kinase subdomain necessary for phosphotransfer and ATP binding, and 3) the autophosphorylation site His243 within the four-helix subdomain. Our results in this study reaffirm previous reports (22) that the osmosensing core is localized entirely within the four-helix subdomain, and its functionality is uncoupled from the rest of the protein. However, here we report a significant increase in basal autophosphorylation in the presence of both lipids and membrane anchoring, which is indicative of cross talk between these independently functioning modules.
To probe this cross talk and correlate changes in function in the presence of lipids with lipid-mediated changes in dynamics, we compared HDXMS results for EnvZc with EnvZc in the presence of non-embedded nanodiscs as well as EnvZ embedded in nanodiscs. Our results show that EnvZ in detergent or lipids showed an extensive increase in deuterium exchange all across the kinase subdomain (Fig. 4). Interestingly, EnvZc also exhibited smaller-magnitude changes in deuterium exchange in the presence of nonembedded nanodiscs across a few peptides in the kinase subdomain. Importantly, the one hotspot that exhibited increased exchange in all samples of EnvZ/EnvZc in the presence of lipids was the glycine-rich motif (57) that is critical for mediating phosphotransfer of the γ-phosphate to the acceptor histidine (His243). Our results reveal that the allosteric effects of lipids can be propagated through peripheral nonembedded interactions in the absence of anchoring and facilitate allosteric modulation of associated kinases. This highlights the importance of parallel and multistate allosteric relays coupling lipids to downstream signaling modules in membrane proteins (58). We postulate that a combination of anchoring and peripheral lipid interactions is allosterically coupled to the glycine-rich loop, which serves to integrate both membrane anchoring of N-terminal segments of EnvZ and peripheral interactions to facilitate autophosphorylation of His243 (Fig. 6).
Lipid allostery operates to alter the ATP-binding pocket and kinase activity, while stimulus alters substrate activity
In our previous study (22), we discovered that the four-helix bundle subdomain is sensitive to osmolality and insensitive to nucleotide binding, and the ATP-binding domain is sensitive to nucleotide binding but insensitive to osmolality. Thus, the two subdomains are uncoupled from one another. Our results reported herein allow us to propose a model of EnvZ receptor function in which the kinase and osmosensing functions of EnvZ receptors are uncoupled and regulated by independent mechanisms. Phosphorylation at His243 is controlled primarily by the kinase subdomain, and the dynamics of the ATP-binding pocket governs the rate of phosphotransfer. In the absence of the membrane, as in EnvZc, the dynamics of the kinase subdomain is altered and phosphorylation is reduced. In the full-length receptor, or in the presence of lipids, the kinase activity is higher due to lipid-mediated allosteric effects propagated through alternate routes. Lipid allosteric effects are also propagated through peripheral interactions between the cytoplasmic domain and the membrane, and through conformational changes within the TM and periplasmic segments. The glycine-rich loop within the ATP-binding pocket acts as a critical node in integrating parallel allosteric relays, resulting in increased phosphorylation in the full-length receptor (Fig. 6). At high osmolality, enhanced phosphorylation is achieved through stabilization of the His243-containing osmosensing locus in the four-helix bundle. This stabilization is independent of the membrane or the N-terminal TM and periplasmic segments, and is likely to be the key event in stimulus-dependent activation of downstream signaling processes such as OmpR recruitment and phosphorylation.
Author Contributions
M.G., L.C.W., and G.S.A. designed the experiments. M.G., L.C.W., and L.K.M. performed experiments. M.G. and G.S.A. interpreted the results. M.G., G.S.A., and L.J.K. wrote the manuscript.
Acknowledgments
This work was supported by grants from the Singapore Ministry of Education Academic Research Fund Tier 3 to G.S.A. (MOE2012-T3-1-008), the Research Centre of Excellence in Mechanobiology at the National University of Singapore (funded by a grant from the Ministry of Education) to G.S.A., and the U.S. Department of Veterans Affairs (5IOBX000372) and National Institutes of Health (NIH R21-123640) to L.J.K.
Editor: Kalina Hristova.
Footnotes
Two figures, one table, and two movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)34332-6.
Contributor Information
Linda J. Kenney, Email: kenneyl@uic.edu.
Ganesh S. Anand, Email: dbsgsa@nus.edu.sg.
Supporting Material
References
- 1.Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu. Rev. Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 2.Lee A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Phillips R., Ursell T., Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stock A.M., Robinson V.L., Goudreau P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 5.Laub M.T., Goulian M. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 6.Alphen W.V., Lugtenberg B. Influence of osmolarity of the growth medium on the outer membrane protein pattern of Escherichia coli. J. Bacteriol. 1977;131:623–630. doi: 10.1128/jb.131.2.623-630.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forst S., Delgado J., Inouye M. Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1989;86:6052–6056. doi: 10.1073/pnas.86.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forst S.A., Roberts D.L. Signal transduction by the EnvZ-OmpR phosphotransfer system in bacteria. Res. Microbiol. 1994;145:363–373. doi: 10.1016/0923-2508(94)90083-3. [DOI] [PubMed] [Google Scholar]
- 9.Aiba H., Nakasai F., Mizuno T. Evidence for the physiological importance of the phosphotransfer between the two regulatory components, EnvZ and OmpR, in osmoregulation in Escherichia coli. J. Biol. Chem. 1989;264:14090–14094. [PubMed] [Google Scholar]
- 10.Brissette R.E., Tsung K.L., Inouye M. Suppression of a mutation in OmpR at the putative phosphorylation center by a mutant EnvZ protein in Escherichia coli. J. Bacteriol. 1991;173:601–608. doi: 10.1128/jb.173.2.601-608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harlocker S.L., Rampersaud A., Inouye M. Phenotypic revertant mutations of a new OmpR2 mutant (V203Q) of Escherichia coli lie in the envZ gene, which encodes the OmpR kinase. J. Bacteriol. 1993;175:1956–1960. doi: 10.1128/jb.175.7.1956-1960.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russo F.D., Silhavy T.J. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J. Mol. Biol. 1991;222:567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- 13.Tokishita S., Kojima A., Mizuno T. Transmembrane signal transduction and osmoregulation in Escherichia coli: functional importance of the transmembrane regions of membrane-located protein kinase, EnvZ. J. Biochem. 1992;111:707–713. doi: 10.1093/oxfordjournals.jbchem.a123823. [DOI] [PubMed] [Google Scholar]
- 14.Waukau J., Forst S. Molecular analysis of the signaling pathway between EnvZ and OmpR in Escherichia coli. J. Bacteriol. 1992;174:1522–1527. doi: 10.1128/jb.174.5.1522-1527.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y., Inouye M. Intermolecular complementation between two defective mutant signal-transducing receptors of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1991;88:11057–11061. doi: 10.1073/pnas.88.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Head C.G., Tardy A., Kenney L.J. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J. Mol. Biol. 1998;281:857–870. doi: 10.1006/jmbi.1998.1985. [DOI] [PubMed] [Google Scholar]
- 17.Forst S., Comeau D., Inouye M. Localization and membrane topology of EnvZ, a protein involved in osmoregulation of OmpF and OmpC in Escherichia coli. J. Biol. Chem. 1987;262:16433–16438. [PubMed] [Google Scholar]
- 18.Aravind L., Ponting C.P. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 19.Tomomori C., Tanaka T., Ikura M. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat. Struct. Biol. 1999;6:729–734. doi: 10.1038/11495. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka T., Saha S.K., Ikura M. NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ. Nature. 1998;396:88–92. doi: 10.1038/23968. [DOI] [PubMed] [Google Scholar]
- 21.Casino P., Miguel-Romero L., Marina A. Visualizing autophosphorylation in histidine kinases. Nat. Commun. 2014;5:3258. doi: 10.1038/ncomms4258. [DOI] [PubMed] [Google Scholar]
- 22.Wang L.C., Morgan L.K., Anand G.S. The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J. 2012;31:2648–2659. doi: 10.1038/emboj.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seddon A.M., Curnow P., Booth P.J. Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Nath A., Atkins W.M., Sligar S.G. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 25.Hebling C.M., Morgan C.R., Engen J.R. Conformational analysis of membrane proteins in phospholipid bilayer nanodiscs by hydrogen exchange mass spectrometry. Anal. Chem. 2010;82:5415–5419. doi: 10.1021/ac100962c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker C.H., Morgan C.R., Stafford D.W. A conformational investigation of propeptide binding to the integral membrane protein γ-glutamyl carboxylase using nanodisc hydrogen exchange mass spectrometry. Biochemistry. 2014;53:1511–1520. doi: 10.1021/bi401536m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treuheit N.A., Redhair M., Atkins W.M. Membrane interactions, ligand-dependent dynamics, and stability of cytochrome P4503A4 in lipid nanodiscs. Biochemistry. 2016;55:1058–1069. doi: 10.1021/acs.biochem.5b01313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woodward C.K. Dynamic solvent accessibility in the soybean trypsin inhibitor--trypsin complex. J. Mol. Biol. 1977;111:509–515. doi: 10.1016/s0022-2836(77)80066-1. [DOI] [PubMed] [Google Scholar]
- 29.Englander S.W., Kallenbach N.R. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 30.Konermann L., Pan J., Liu Y.H. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 31.Bayburt T.H., Sligar S.G. Membrane protein assembly into nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zegzouti H., Zdanovskaia M., Goueli S.A. ADP-Glo: a bioluminescent and homogeneous ADP monitoring assay for kinases. Assay Drug Dev. Technol. 2009;7:560–572. doi: 10.1089/adt.2009.0222. [DOI] [PubMed] [Google Scholar]
- 33.Hemmer W., McGlone M., Taylor S.S. Role of the glycine triad in the ATP-binding site of cAMP-dependent protein kinase. J. Biol. Chem. 1997;272:16946–16954. doi: 10.1074/jbc.272.27.16946. [DOI] [PubMed] [Google Scholar]
- 34.Morgan C.R., Hebling C.M., Engen J.R. Conformational transitions in the membrane scaffold protein of phospholipid bilayer nanodiscs. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M111.010876. M111.010876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouritsen O.G., Bloom M. Models of lipid-protein interactions in membranes. Annu. Rev. Biophys. Biomol. Struct. 1993;22:145–171. doi: 10.1146/annurev.bb.22.060193.001045. [DOI] [PubMed] [Google Scholar]
- 36.Nicolson G.L. The fluid-mosaic model of membrane structure: still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim. Biophys. Acta. 2014;1838:1451–1466. doi: 10.1016/j.bbamem.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 37.Amin D.N., Hazelbauer G.L. Influence of membrane lipid composition on a transmembrane bacterial chemoreceptor. J. Biol. Chem. 2012;287:41697–41705. doi: 10.1074/jbc.M112.415588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bechara C., Robinson C.V. Different modes of lipid binding to membrane proteins probed by mass spectrometry. J. Am. Chem. Soc. 2015;137:5240–5247. doi: 10.1021/jacs.5b00420. [DOI] [PubMed] [Google Scholar]
- 39.Dawaliby R., Trubbia C., Govaerts C. Allosteric regulation of G protein-coupled receptor activity by phospholipids. Nat. Chem. Biol. 2016;12:35–39. doi: 10.1038/nchembio.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee A.G. Lipid-protein interactions in biological membranes: a structural perspective. Biochim. Biophys. Acta. 2003;1612:1–40. doi: 10.1016/s0005-2736(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 41.Coskun Ü., Grzybek M., Drechsel D., Simons K. Regulation of human EGF receptor by lipids. Proc. Natl. Acad. Sci. USA. 2011;108:9044–9048. doi: 10.1073/pnas.1105666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ekanayake E.V., Fu R., Cross T.A. Structural influences: cholesterol, drug, and proton binding to full-length influenza a M2 protein. Biophys. J. 2016;110:1391–1399. doi: 10.1016/j.bpj.2015.11.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu S., Jang H., Zhang J. Ras conformational ensembles, allostery, and signaling. Chem. Rev. 2016;116:6607–6665. doi: 10.1021/acs.chemrev.5b00542. [DOI] [PubMed] [Google Scholar]
- 44.Nussinov R., Jang H., Tsai C.J. Oligomerization and nanocluster organization render specificity. Biol. Rev. Camb. Philos. Soc. 2015;90:587–598. doi: 10.1111/brv.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newton A.C. Lipid activation of protein kinases. J. Lipid Res. 2009;50(Suppl.):S266–S271. doi: 10.1194/jlr.R800064-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leonard T.A., Hurley J.H. Regulation of protein kinases by lipids. Curr. Opin. Struct. Biol. 2011;21:785–791. doi: 10.1016/j.sbi.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu W., Chun E., Stevens R.C. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oates J., Faust B., Watts A. The role of cholesterol on the activity and stability of neurotensin receptor 1. Biochim. Biophys. Acta. 2012;1818:2228–2233. doi: 10.1016/j.bbamem.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 49.Brannigan G., Hénin J., Klein M.L. Embedded cholesterol in the nicotinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA. 2008;105:14418–14423. doi: 10.1073/pnas.0803029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Contreras F.X., Ernst A.M., Wieland F., Brügger B. Specificity of intramembrane protein-lipid interactions. Cold Spring Harb. Perspect. Biol. 2011;3:a004705. doi: 10.1101/cshperspect.a004705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goñi F.M. Non-permanent proteins in membranes: when proteins come as visitors (review) Mol. Membr. Biol. 2002;19:237–245. doi: 10.1080/0968768021000035078. [DOI] [PubMed] [Google Scholar]
- 52.Tsaloglou M.N., Attard G.S., Dymond M.K. The effect of lipids on the enzymatic activity of 6-phosphofructo-1-kinase from B. stearothermophilus. Chem. Phys. Lipids. 2011;164:713–721. doi: 10.1016/j.chemphyslip.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 53.daCosta C.J., Medaglia S.A., Baenziger J.E. Anionic lipids allosterically modulate multiple nicotinic acetylcholine receptor conformational equilibria. J. Biol. Chem. 2009;284:33841–33849. doi: 10.1074/jbc.M109.048280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim S., Cross T.A. Uniformity, ideality, and hydrogen bonds in transmembrane alpha-helices. Biophys. J. 2002;83:2084–2095. doi: 10.1016/S0006-3495(02)73969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bondar A.N., White S.H. Hydrogen bond dynamics in membrane protein function. Biochim. Biophys. Acta. 2012;1818:942–950. doi: 10.1016/j.bbamem.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scholtz J.M., Baldwin R.L. The mechanism of alpha-helix formation by peptides. Annu. Rev. Biophys. Biomol. Struct. 1992;21:95–118. doi: 10.1146/annurev.bb.21.060192.000523. [DOI] [PubMed] [Google Scholar]
- 57.Saraste M., Sibbald P.R., Wittinghofer A. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 58.Moorthy B.S., Anand G.S. Multistate allostery in response regulators: phosphorylation and mutagenesis activate RegA via alternate modes. J. Mol. Biol. 2012;417:468–487. doi: 10.1016/j.jmb.2012.01.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.