Abstract
The immunological role of exosomes in allograft rejection remains unknown. We sought to determine whether exosomes are induced during lung allograft rejection and to define the antigenic compositions of human leukocyte antigen (HLA), lung associated self-antigens (SAgs), and microRNAs (miRNAs). Exosomes were isolated from sera and bronchoalveolar lavage fluid from 30 lung transplant recipients (LTxR) who were stable or had acute rejection (AR) or bronchiolitis obliterans syndrome (BOS). Exosomes were defined by flow cytometry for CD63 and Western blotting for Annexin V. SAgs, Collagen-V (Col-V), and Kα1 Tubulin were examined by electron microscopy; miRNAs were profiled by Affymetrix miRNA array. Donor HLA and SAgs were detected on exosomes from LTxRs with AR and BOS, but not from stable LTxRs. Exosomes expressing Col-V were isolated from LTxR sera 3 months before AR and 6 months before BOS diagnosis, suggesting that exosomes with SAgs may be a noninvasive rejection biomarker. Exosomes isolated from LTxRs with AR or BOS also contained immunoregulatory miRNAs. We conclude that exosomes expressing donor HLA, SAgs, and immunoregulatory miRNAs are present in the circulation and local site after human lung transplantation, playing an important role in the immune-pathogenesis of acute allograft rejection and BOS.
Introduction
Lung transplantation (LTx) is a treatment option for patients with end-stage lung disease such as idiopathic pulmonary fibrosis, chronic obstructive pulmonary disease, and cystic fibrosis (1-3). Although short-term survival has improved, acute rejection (AR) and bronchiolitis obliterans syndrome (BOS) remain major hurdles to long-term allograft function (4). The mechanisms that lead to AR and BOS i.e., immune responses to mismatched donor human leukocyte antigen (HLA) and non-HLA lung associated self-antigens (SAgs), Collagen-V (Col-V), and K alpha 1 Tubulin (Kα1T)) also contribute to lung allograft rejection. Previous studies have demonstrated that de novo development of antibodies (Abs) to donor-mismatched HLA (DSA) and SAgs is associated with increased risk of AR and BOS after LTx (5-8). We have also demonstrated that DSA-induced changes to microRNAs (miRNAs) may prompt development of BOS (9). Using an animal model of obliterative airway disease (OAD), we demonstrated that Abs administration to the SAg Col-V can induce immune responses not only to Col-V, but also to another SAg, Kα1T, and vice versa (10). Further, immune responses to SAgs can break self-tolerance, leading to airway inflammation and fibrosis in a syngeneic murine orthotopic LTx model (10).
Exosomes are membrane vesicles (40-100 nm in diameter) produced by endocytic pathways and secreted into body fluids (11) (12). They contain membrane, cytosolic proteins, mRNA and miRNA (13) (14). Major histocompatibility complex (MHC) class I and II have been detected on tumor cell exosomes and can induce anti-tumor immunity (15). Exosomes from epithelial cells can induce activation of macrophages during lung inflammation, suggesting that exosomes can trigger airway inflammation (16). The aim of this study was to determine whether exosomes are generated during lung allograft rejection, and to define the origin and antigenic makeup of these exosomes, including the presence of SAgs. Herein we demonstrate that exosomes containing SAgs are detectable both in the bronchoalveolar lavage (BAL) fluid and sera of lung transplant recipients (LTxRs) with AR and BOS, but not in stable patients. This indicates that the exosomes originate from the transplanted organ after immune injury. We also demonstrate that mismatched donor HLA and SAgs are expressed on exosome surfaces, and that exosomes contain miRNAs known to induce inflammation, endothelial activation, antibody-mediated chronic rejection, and Th17 differentiation.
METHODS
Patient population
In this case-control observational study, we identified 30 LTxRs who underwent bilateral LTx at Washington University/Barnes Hospital between 2012 and 2013. Ten of these patients developed BOS after LTx, 10 developed AR, and 10 remained stable. Upon approval from the Institutional Review Board at Washington University and informed consent from patients, sera and BAL fluid were collected at 1, 3, 6, and 12 months after LTx and when required for clinical diagnosis. LTxRs with no evidence of rejection or microbial infection were designated as stable; AR and BOS were defined by ISHLT criteria (17).
Histopathology and C4d evaluation of biopsy showed that, of the 10 patients with AR, 1 patient had A1 rejection, 8 had A2 rejection, and 1 had both A1 and A2B2 rejection. DSA was present in 3 patients (30%) in the BOS group, 4 patients (40%) in the AR group, and in no patients in the stable group. There were no significant demographic differences between the groups (Table 1). Immunotherapy protocol included cyclosporine, azathioprine, and prednisone. Sera and BAL fluid were stored at −80°C.
Table 1.
Patient Demographics (N=30)*
| Clinical features | BOS (n=10) |
AR (n=10) |
Stable (n=10) |
|---|---|---|---|
| Age, y, mean±SD | 54±12 | 61±7 | 57±10 |
| Sex, male | 6(60) | 8(80) | 7(70) |
| Ethnicity | |||
| African American | 0(0) | 0 | 1(10) |
| White | 10(100) | 10(100) | 9(90) |
| ESLD | |||
| IPF | 3(30) | 5(50) | 0 |
| CF | 4(40) | 1(10) | 1(10) |
| COPD | 2(20) | 3(30) | 6(60) |
| Other | 1(10) | 1(10) | 3(30) |
| HLA mismatch | |||
| A | 1.6±0.5 | 1.7±0.5 | 1.5±0.53 |
| B | 1.8±0.4 | 1.6±0.5 | 1.7±0.48 |
| DR | 1.6±0.5 | 1.7±0.5 | 1.6±0.70 |
| Bilateral LTx | 10(100) | 10(100) | 10(100) |
| Sampling from LTx, months | 16.8±6.3 | 1.44±0.7 | 22.4±9.0 |
| Time of BOS onset, months | 15.2±7.8 | N/A | N/A |
| Time of AR onset, months | N/A | 1.4±0.6 | N/A |
Values presented as n (%), unless otherwise noted.
Abbreviations: AR, acute rejection; BOS: bronchiolitis obliterans syndrome, CF, cystic fibrosis; COPD, chronic obstructive pulmonary disease; ESLD, end-stage lung disease; HLA, human leukocyte antigen; IPF, idiopathic pulmonary fibrosis; LTx, lung transplantation; N/A, not applicable; SD, standard deviation.
Exosome isolation from BAL fluid by ultracentrifugation
BAL fluid was centrifuged at 3,000g for 30 minutes at 4ºC, then at 10,000g for 30 minutes at 4ºC. The supernatant was centrifuged at 100,000g for 70 minutes at 4ºC. Exosome-containing pellets were washed with 1X phosphate buffered saline (PBS) and centrifuged again at 100,000xg for 70 minutes at 4ºC. Exosome pellets were resuspended with 100μL of 1X PBS buffer.
Exosome isolation from sera by precipitation
Exosomes from 1 mL of sera were isolated using the Total Exosome Isolation Reagent kit (Catalog # 4478360, Invitrogen, ThermoFisher Scientific, Waltham, MA) (18). Sera were centrifuged at 2,000g for 20 minutes at room temperature, then at 10,000g for 20 minutes at room temperature. Sera were then diluted with 0.5 volume of 1X PBS and exosomes were precipitated with 0.2 volume of exosome precipitation reagent and centrifuged at 10,000g for 5 minutes at room temperature. Exosome pellets were solubilized with 1X PBS. Protein concentration of the exosomes was quantified using Protein Assay Kit (Catalog # 23225, Pierce, ThermoFisher Scientific, Waltham, MA). The mean concentration of exosomes isolated from LTxRs with BOS was 0.89 ng/μl (serum) and 0.33 ng/μl (BAL fluid). Mean concentration of exosomes isolated from LTxRs with AR was 0.34 ng/μl (serum) and 0.35 ng/μl (BAL fluid). Stable LTxRs also had exosomes with the CD63 marker, but neither sera nor BAL fluid contained the SAgs (serum 0.64 ng/μl, BAL fluid 0.45 ng/μl).
Exosome purification
Exosomes were purified with sucrose cushion (30%) ultracentrifugation to eliminate protein aggregates nonspecifically associated with exosomes (19, 20). Exosomes isolated by ultracentrifugation method were briefly resuspended in 25 mL of PBS. Four mL of sucrose cushions (Tris/sucrose/D2O solution) were added to the bottom of a SW 28 tube; diluted exosomes were loaded without disturbing the interface. The tubes were centrifuged at 100,000g for 75 minutes at 4°C. 3.5 mL of Tris/sucrose/D2O cushion were collected without disturbing the interface and the samples were made up to 60 mL with PBS. The exosome sample was centrifuged again for 70 minutes at 100,000g at 4°C, and the pelleted exosomes were resuspended with 100 μl of PBS for Western blotting.
Western blotting
Total exosome protein from BAL and sera isolated from AR, BOS and stable LTxR were analyzed for exosome specific marker Annexin-V, and lung associated SAg, Col-V, using immunoblot. Briefly, 10μg of total exosome protein were denatured in laemmli buffer (Bio-Rad Laboratories) and boiled at 95°C for 5 min. Exosome protein samples were resolved in 12% Bis-Tris polyacrylamide gel and transferred to polyvinylidene fluoride membrane. The membrane was probed with exosome-specific marker protein Annexin V, using anti-Annexin V Ab (ab14196 Abcam, Cambridge, United Kingdom), and Abs to SAg Col-V (ab7046, Abcam). Goat anti-rabbit horseradish peroxidase was used as a secondary Ab (sc-2004, Santa Cruz Biotechnology, Dallas, TX). The blots were developed and exposed to x-ray films. Col-V band intensity was quantified using ImageJ software and normalized with Annexin V.
Transmission electron microscopy
Exosomes were negatively stained after absorption onto formvar/carbon-coated copper grids for 2 minutes. Grids were washed twice for 1 minute each in dH2O and stained for 1 minute with 1% aqueous uranyl acetate (Ted Pella Inc., Redding, CA). Samples were viewed on a JEOL 1200EX transmission electron microscopy (EM) (JEOL USA, Peabody, MA) equipped with an AMT 8 megapixel digital camera (Advanced Microscopy Techniques, Woburn, MA). For immunogold labeling of exosomes, grids were incubated with mouse anti-CD63 Ab, anti-Col-V and Kα1T for 30 minutes, followed by secondary goat anti-mouse IgG Ab conjugated to colloidal gold (Jackson Immuno Research Laboratories, West Grove, PA) for 30 minutes. Grids were washed and stained with uranyl acetate and viewed by transmission EM as described above.
Flow cytometry
Serum exosomes were analyzed by human CD63 isolation/detection kit (Life Technologies, Carlsbad, CA) with the use of CD63-coated dynabeads (4.5μM) per manufacturer protocol. Exosomes bound to CD63 beads were briefly stained with PerCP/Cy5.5-conjugated, anti-HLA A2 (Catalog # 343315, Clone BB7.2, BioLegend, San Diego, CA) to track the donor- and allophycocyanin (APC)-conjugated anti-HLA A3 (Clone GAP.A3, eBioscience 17-5754) for recipient origin of the exosome. Baseline was established by staining the unbound CD63 dynabeads and was compared to serum exosomes isolated from 2 BOS LTxRs. Analysis of the presence of SAg (i.e., Col-V) on exosomes was evaluated by co-staining the exosomes bound to CD63 dynabeads with APC-conjugated anti-Col-V (Catalog # AC16-0069-03, Abcore, Ramona, CA) and Exo-FITC (System Biosciences, Palo Alto, CA). Exosome surface marker analysis was performed using BD FACSCANTO II system (BD Biosciences, San Jose, CA).
miRNA profiling
Exosomic miRNAs were isolated from pooled sera (1 sample from each patient) using a mirVana miRNA isolation kit (Applied Biosystems, Foster City, CA). Global miRNA profiling was performed using Affymetrix platform (GeneChip miRNA 3.0 array). Differentially expressed exosomal miRNAs were analyzed using Partek Genomics Suite 6.6 software (Partek, St. Louis, MO) and the heat map was created using R statistics software. miRNA target prediction and pathway analysis was performed using DIANA miRPath (http://microrna.gr/mirpath).
TaqMan miRNA real time polymerase chain reaction
TaqMan miRNA real time polymerase chain reaction (RT-PCR) was performed as described previously (9). Total RNA from exosomes isolated from sera and BAL fluid was briefly isolated using mirVana miRNA isolation kit (Applied Biosystems, Foster City, CA). Reverse transcription was performed using TaqMan miRNA Reverse Transcription Kit (Applied Biosystems). qRT-PCR was performed by TaqMan miRNA assays as previously described (9).
Statistical analysis
The 10 patients in each group were selected based on sample availability and statistical test power. The efficiency of sample pooling method was tested with efficient statistical test power (21, 22). Exosome levels were compared between cohorts and correlated with clinical outcomes (Mann-Whitney two-tailed Student t-test). Statistical analysis was performed with GraphPad software (GraphPad Prism, La Jolla, CA). Statistical data were expressed as mean±standard deviation. P-values less than 0.05 were considered statistically significant.
Results
Exosomes containing SAg Col-V are induced by alloimmune responses after human LTx
To determine whether exosomes generated from the transplanted organ are induced by alloimmune responses after LTx, and to determine their presence both locally and in the circulation, exosomes were isolated from BAL fluid and sera of LTxRs who were stable (n=10, no detectable DSA, rejection-free), LTxRs with AR (n=10, 4 with DSA and 6 without), and LTxRs with BOS (n=10, 3 with DSA and 7 without). The isolated exosomes were pooled and Western blotting was performed using Abs specific to Col-V. Exosomes derived from BOS patients sera contained the SAg Col-V, but serum supernatant did not (Figure S1A). Further, Western blotting demonstrated that the purified exosomes from the sucrose cushion method contained exosome marker protein Annexin V and SAg Col-V (Figure S1B). Density metric analyses of Col-V band intensity were performed and normalized with Annexin V intensity (Figure S1C). This clearly indicates that exosomes derived from LTxRs with BOS contained SAg in exosome fraction purified by sucrose cushion method, and presence of Col-V did not result from protein aggregates.
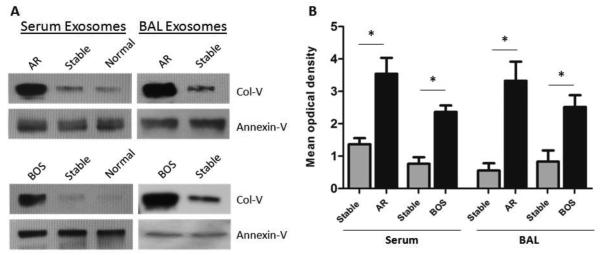
Western blotting demonstrated SAg Col-V in exosomes isolated from AR and BOS LTxRs, significantly less in the stable group or in healthy subjects (Figure 1A). Figure 1B shows a relative densitometric reading that shows that exosomes containing Col-V are detectable in AR and BOS LTxRs, significantly less in stable LTxRs. Exosomes isolated from sera of BOS LTxRs contained significantly increased levels of SAg Col-V (p=0.003), compared to stable LTxRs. Similarly, increased Col-V (p=0.001) was also seen in AR LTxRs compared to stable LTxRs. Western blotting of exosomes derived from BAL fluid showed a significant increase in SAg, Col-V in BOS and AR LTxRs compared to the stable group (Figure 1A). Densitometry of exosome containing SAg of individual BOS patients showed significant increase in Col-V (p=0.014) compared to stable patients. Similarly, a significant increase in exosomes containing Col-V was also noted in AR LTxRs compared with the patients in the stable group (p=0.001; Figure 1B). These results clearly demonstrated that exosome containing SAg Col-V could be a noninvasive biomarker of impending rejection.
Figure 1. Lung associated SAgs demonstrable in sera and BAL fluid exosomes of BOS, AR, and stable LTx recipients.
(A) Representative Western blotting of pooled exosomes derived from serum and BAL fluid show significant increase of Col-V protein level in BOS (n=10) and AR (n=10) LTxRs compared with stable (n=10) LTxRs and normal subjects (n=7). (B). Densitometry analysis of Col-V optical density in individual BOS (n=10) and AR (n=10) LTxR cohorts compared with stable LTxRs (n=10) (*, p<0.05). Exosome marker Annexin-V was used as loading control. AR, acute rejection; BAL, bronchoalveolar lavage; BOS, bronchiolitis obliterans syndrome; Col-V, collagen V; LTxR, lung transplant recipient; SAg, self-antigen.
SAgs Col-V and Kα1T were present in surface of exosomes isolated from BOS and AR LTxRs
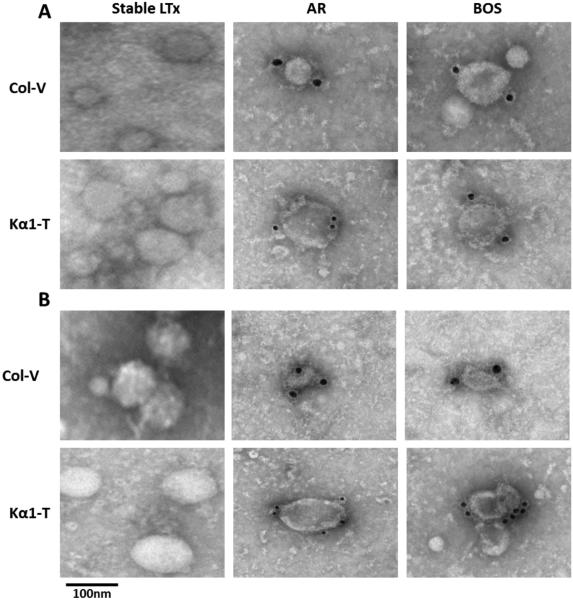
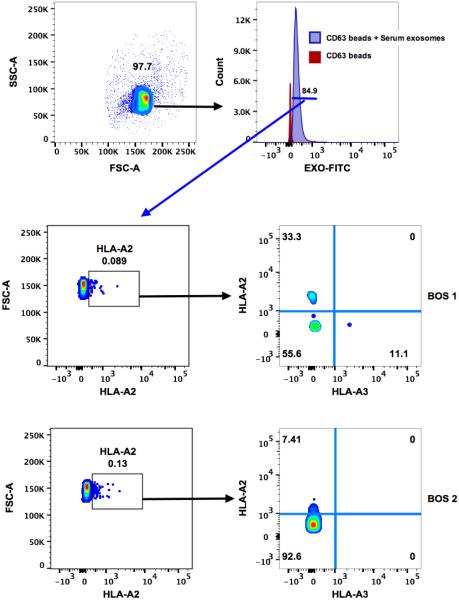
Exosomes derived from sera of BOS and AR LTxRs expressed SAgs, which were undetectable on the exosomes isolated from stable LTxRs (Figure 2A). Similarly, exosomes isolated from BAL fluid from both AR and BOS LTxRs demonstrated expression of Col-V and Kα1T (Figure 2B), whereas exosomes isolated from stable LTxRs did not. Further, to determine that serum derived exosomes are of donor origin, we targeted mismatched donor HLA (HLA-A2) and recipient HLA (HLA-A3) from the exosomes of 2 patients with BOS by flow cytometry. The donor HLA-A2 was expressed on the exosomes, but there was no expression on recipient HLA-A3. This confirms that the exosomes were released from the transplanted lung after allo-immune responses (Figure 3). The expression of SAgs and donor HLA on the surface of the exosomes isolated from BOS and AR LTxRs suggests that exosome mediated immune responses may contribute to AR and BOS pathogenesis.
Figure 2. Transmission Electron Microscopy (TEM) for surface expression of SAgs, Col-V and Kα1T.
(A). TEM images of exosomes derived from sera of AR and BOS LTxRs show surface expression of SAgs Col-V and Kα1T, but not in stable LTxRs. (B). Detection of SAgs on the surface of exosomes from BAL fluid from AR and BOS LTxRs, but not in stable LTxRs. Exosomes were stained with Col-V and Kα1T Abs and detected using gold conjugated Abs and analyzed by EM. Scale bar=100 nm. Results shown are from one experiment (2 were performed). AR, acute rejection; BAL, bronchoalveolar lavage; BOS, bronchiolitis obliterans syndrome; Col-V, collagen V; Kα1T, K-alpha 1 tubulin; LTxR, lung transplant recipient; SAg, self-antigen.
Figure 3. Flow cytometric analysis on donor derived exosomes.
Serum derived exosomes bound to CD63-coated dynabeads were stained with anti-HLA-A2-PerCP/Cy5.5 and anti-HLA-A3-APC. Positive staining for HLA-A2 (LTx donor HLA) and negative staining for HLA-A3 (LTx recipient HLA) identifies donor-derived exosomes. LTx, lung transplantation.
Exosomes containing the SAg Col-V were demonstrable in the sera of LTxRs with AR and BOS before clinical diagnosis
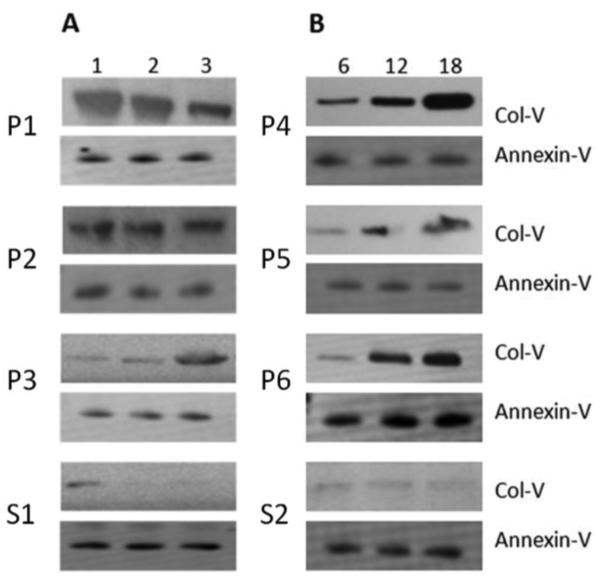
To determine whether exosomes are induced ahead of clinical diagnosis of acute and chronic rejection and are present in the circulation, we isolated exosomes from the sera collected serially from 3 patients at 1, 3, and 6 months after LTx. AR was clinically diagnosed in these patients at 1.9 months, 2.8 months, and 1.5 months post-LTx. Exosomes isolated from stable LTxRs at similar time points were used as control. As shown in Figure 4A, exosomes containing Col-V were detected in the exosomes isolated from the sera just 1 month after LTx in all patients with AR, much earlier than clinically diagnosed rejection (at 3 months). Similarly, exosomes isolated from sera at 6, 12, and 18 months from 3 LTxRs with BOS also contained Col-V, detectable at 6 months post-LTx. BOS was clinically diagnosed in these patients at 19.8 months, 24.4 months, and 13.6 months post-LTx (Figure 4B). These results demonstrated that exosomes containing Col-V present in the sera of LTxRs with AR and BOS are detectable before clinical diagnosis of acute and chronic rejection. In contrast, Col-V signal noted in stable LTx at 1 month went away to undetectable levels in 3 and 6 months.
Figure 4. Exosomes containing lung associated SAg, Col-V are demonstrable in the serial sera from LTxRs with AR and BOS.
(A). Western blotting shows early detection of lung associated SAg Col-V in exosomes isolated from serial sera (1, 3, and 6 months) from AR LTxRs compared with stable LTxRs. (B). Exosomes derived from serial sera of LTxRs diagnosed with BOS (6, 12, and 18 months) show Col-V expression as early as 6 months, compared with stable LTx. Annexin-V was used as loading control. P= AR/BOS patients, S=Stable LTx. AR, acute rejection; BOS, bronchiolitis obliterans syndrome; Col-V, collagen V; LTx, lung transplantation; LTxR, lung transplant recipient; SAg, self-antigen.
miRNAs were differentially expressed in exosomes from BOS and AR LTxRs compared to stable LTxRs
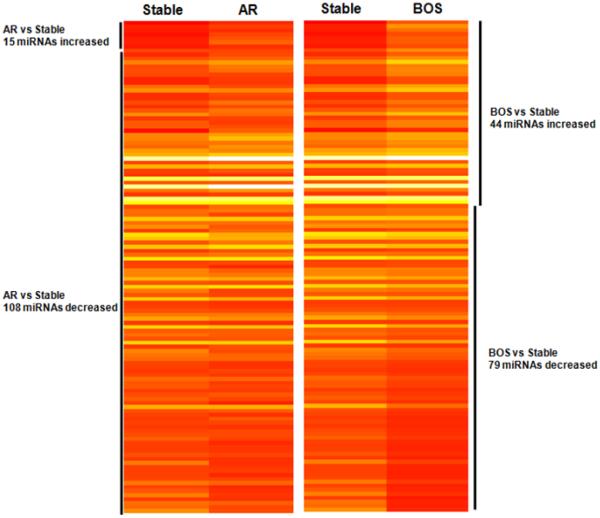
To investigate the contribution of serum derived exosomic miRNA in the immunopathogenesis of AR and BOS, we determined the miRNA of the exosomes isolated from pooled sera collected from each of the 30 patients. The graphical representation of heat maps indicates differentially expressed miRNAs, which includes lower and higher miRNA signals identified by gene chip miRNA analysis. The heat map represents relative fold change of differentially expressed miRNA compared between AR, BOS, and stable LTxRs (Figure 5). The global miRNA analysis demonstrated 123 detectable miRNAs (i.e., at least 1 pool with positive detection signal) in sera exosomes. Of these, 15 miRNAs were higher and 108 miRNAs were lower in AR LTxRs compared to stable LTxRs (Table S1). In contrast, 44 miRNAs were higher and 79 miRNAs were lower in BOS LTxRs compared to stable LTxRs (Table S1).
Figure 5. Heat map representation of differentially expressed miRNA in AR, BOS and stable LTxRs.
Global miRNA analysis with differential miRNA expression in AR, BOS and stable LTxRs is represented in the heat map (generated using R statistics software). Each entry of the grid refers to relative fold (log2) between the expression level of a given exosomic miRNA on AR or BOS pools relative to stable pools. The color of each entry is determined by the value of that fold difference, ranging from red (negative values) to yellow (positive values). AR, acute rejection; BOS, bronchiolitis obliterans syndrome; Col-V, collagen V; LTxR, lung transplant recipient.
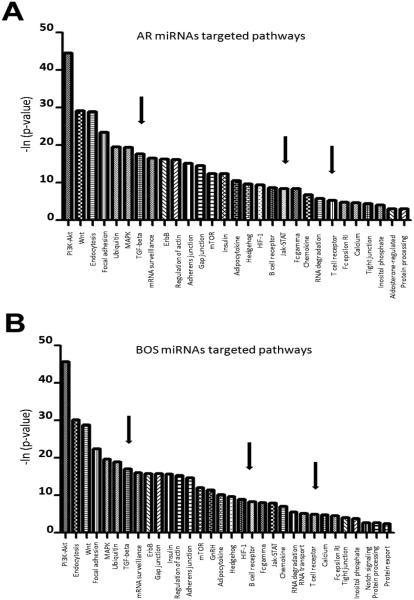
We also performed comparative pathway analysis using DIANA-mirPath on differentially expressed miRNAs from AR and BOS exosomes. The results demonstrated that PI3K-Akt, Wnt, endocytosis, focal adhesion, ubiquitin-mediated proteolysis, MAPK, TGF-β, and mRNA surveillance pathways were significantly enriched in exosomic miRNAs from AR and BOS LTxRs (p<0.0001; Figure 6). The differentially expressed miRNAs associated with various molecular pathways are summarized in Table S2.
Figure 6. miRNA-mediated pathway analysis for differentially expressed miRNAs in AR and BOS LTxRs compared with stable LTxR.
(A) Graphical representation of AR-mediated targeted pathways and (B) BOS-mediated targeted pathways generated using DIANA miRPATH (y axis: p-values; x axis: miRNA-associated pathways). AR, acute rejection; BOS, bronchiolitis obliterans syndrome; LTxR, lung transplant recipient.
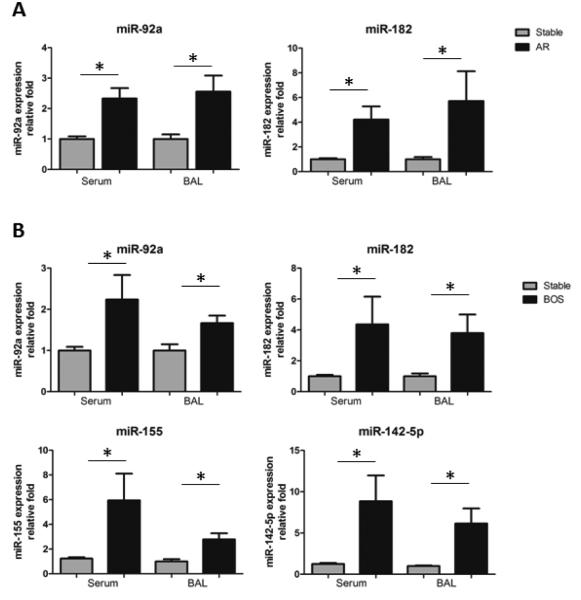
Global miRNA profiling showed high expression of miR-92a (involved in endothelial activation) and miR-182 (involved in inflammation) in AR LTxRs. Similarly, differentially expressed miRNA in BOS LTxRs demonstrated miR-92a, miR-182, and miRNA involved in Th-17 activation (miR-155), and antibody-mediated rejection (AMR) (miR-142-5p). We carried out qRT-PCR with independent cohorts (Table S3) for miR-92a, -182, -142-5p, and -155. Clinical parameters for these independent cohorts were similar (e.g., age, gender, disease, and transplantation type). qRT-PCR demonstrated significant upregulation of miR-92a (p<0.013) and miR-182 (p<0.020) in AR LTxRs compared with stable LTxRs, and significant upregulation of miR-92a (p<0.041), miR-182 (p<0.012), miR-142-5p (p<0.015), and miR-155 (p<0.002) in BOS LTxRs compared with stable LTxRs. Similarly, exosomes derived from BAL fluid showed significant upregulation of miR-92a (p<0.014) and miR-182 (p<0.026) in AR LTxRs compared with stable LTxRs, and significant upregulation of miR-92a (p<0.024), miR-182 (p<0.026), miR-142-5p (p<0.026), and miR-155 (p<0.008) in BOS LTxRs compared with stable LTxRs (Figure 7). This shows that miRNAs involved in inflammation and endothelial activation were significantly increased in both AR and BOS LTxRs, and that the IL17 pathway was activated in BOS LTxRs.
Figure 7. qRT-PCR for higher expressed miRNA in independent AR (n=10), BOS (n=10), and stable (n=10) LTxRs.
(A). MiRNAs involved in endothelial activation miR-92a and inflammation miR-182 in exosomes derived from sera and BAL fluid from AR LTxRs compared with stable LTxRs. (B). MiRNAs involved in endothelial activation miR-92a and inflammation miR-182, AMR miR-142-5p, and Th 17 activation miR-155 in exosomes derived from sera and BAL fluid from BOS LTxRs compared with stable LTxRs. (y axis: relative fold change; x axis: sample source). Differentially regulated miRNA was compared between each cohort by Mann-Whitney two-tailed Student t test (GraphPad software). P-value less than 0.05 was considered statistically significant. AMR, antibody-mediated rejection; AR, acute rejection; BAL, bronchoalveolar lavage; BOS, bronchiolitis obliterans syndrome; Col-V, collagen V; LTxR, lung transplant recipient.
Discussion
Exosomes are nano-vesicles of endocytic origin, roughly 40-100nm in diameter. Previous studies have demonstrated their role in intercellular (23), cell-to-cell communication (24) and pathogenesis of cancer and various kidney diseases (14, 25). Exosomes can be released from different types of cells, including epithelial cells (26), B cells (27, 28), T cells (29), and macrophages (30); all of which can be isolated from body fluids such as urine (31), serum (32), and BAL fluid (33).
The surface expression of tumor-specific protein and human epidermal growth factor receptor 2 derived from a human breast cancer cell line strongly support the notion that tumor-derived exosomes can increase tumor cell proliferation (34), but the role of exosomes in acute or chronic rejection after organ transplantation remains unknown. We attempted to define the role of exosomes as potential biomarkers in rejection after human LTx. LTx has the highest incident of acute and chronic rejection of any organ, and LTxRs can be tested for presence of exosomes not only in the circulation, but also at the local site. In this study, exosomes isolated from BAL fluid and sera showed nano-vesicular structure by EM, and expression of exosome-specific surface marker CD63 in exosomes derived from AR and BOS LTxRs (Figure S2). The mean size of the vesicles isolated from the sera was 72 nm in diameter, and 75 nm from BAL fluid. SAg Col-V and exosome marker protein Annexin V in sucrose cushion demonstrated that Col-V is derived from exosomes. Further, the presence of Col-V in exosome pellets, but not in the supernatant, confirms that exosomes express Col-V.
Exosomes isolated from AR and BOS LTxRs contained SAgs (Figure 2) and donor HLA on their surface (Figure 3), suggesting that the exosomes in rejection originate from the transplanted organ and express the SAgs Col-V and Kα1T. This data supports the notion that both cellular and humoral rejection can induce formation of exosomes that are released into the circulation. We theorize that these exosomes may contribute to augmentation of immune responses against donor lungs, because of the presence of both alloantigens and SAgs. Exosomes have also been reported to transfer functional MHC complexes to dendritic cells, which can express internalized protein and initiate antigen presentation (35, 36). Preliminary results using flow cytometry demonstrated that exosomes from the sera of BOS LTxRs showed donor HLA-A2 antigen, but not recipient HLA-A3, indicating the donor origin of the exosomes following alloimmune responses (Figure 3). The number of exosomes in the circulation that expressed donor HLA-A2 were low, which may be due to the fact that isolated exosomes are the sum total of the exosomes released from different organs. Serial analysis of exosomes in the circulation demonstrated that exosomes containing Col-V were detectable 1 month post-LTx, 2 months before clinical diagnosis of AR and 6 months before clinical diagnosis of BOS.
The mechanism of exosome contribution to acute and chronic rejection post-LTx is yet to be determined. However, based on our previous findings that Ab-directed therapy and DSA clearance offers freedom from BOS (37), as well as clearance of both DSA and SAg Abs offers greater freedom from BOS development (38), it is possible that exosomes released from the donor lungs during rejection may support the immune responses that ultimately lead to BOS.
Our group has documented that development of DSA in LTxRs with BOS is associated with significant increases in miRNAs, which in turn induces TGF-β and B cell receptor signal pathways, ultimately leading to the BOS pathogenesis (9). We therefore hypothesize that the miRNA in exosomes might be involved in regulation of the TGF-β signaling pathway, which induces airway inflammation and fibrosis in transplanted lungs. Differential expression of miRNA observed in a global miRNA profile suggested that exosomes contain miRNA, which is involved in the B cell, T cell and TGF-β signaling pathways that regulate significant cellular processes and could contribute to immune mediated development of AR and BOS (Figure 6 and Table S2). miRNA qRT-PCR analysis of independent cohorts of exosomes isolated from sera and BAL fluid demonstrated similar findings (i.e., upregulation of miR-92a [endothelial activation], miR-182 [inflammation], miR-142-5p [antibody-mediated chronic rejection], and miR-155 [T cell activation]) (39-42), suggesting that these miRNAs may regulate a substantial proportion of the genes involved in auto- and alloimmune responses that resulted in the pathogenesis of AR and BOS.
One of the limitations of this study is relatively small samples size analyzed for AR, BOS, and stable LTxRs. However, consistent results across serial studies suggest that exosomes may contribute to lung allograft rejection. Another potential shortcoming is that we have not yet defined the mechanism by which exosomes may participate in augmenting immune responses leading to rejection. However, demonstration of SAgs on the surface of exosomes derived from BAL fluid and sera, coupled with donor HLA and miRNA involved in immune regulation, strongly suggest the role of exosomes in the immunopathogenesis of AR and BOS after LTx. A recent report by Gregson et al. described differentially expressed exosomal shuttle RNA in BAL fluid and suggested that BAL-exosomic RNA can be a biomarker of AR (43). We also demonstrated that exosome-containing SAgs are present in the exosomes derived from BAL fluid and sera, suggesting that exosome containing SAgs could be a noninvasive biomarker of impending rejection.
In conclusion, these results demonstrate that exosomes are released into the circulation during AR and BOS, expressing not only the alloantigens of the donor, but also SAgs. We also show that Col-V expressing exosomes can be detected before AR or BOS can be clinically diagnosed, and that these exosomes originated from the transplanted lung. Exosomes isolated from LTxRs with AR and BOS also contained miRNAs known to be involved in immune regulation. Therefore, exosomes seem to play a significant role in augmenting immune responses to donor alloantigens and SAgs, ultimately leading to allograft rejection.
Supplementary Material
Acknowledgments
This work was supported by NIH R21AI123034, HL056643 (TM). The authors would like to acknowledge Clare Prendergast and Billie Glasscock for assistance with preparation and submission of the manuscript.
Abbreviations
- AR
acute rejection
- APC
allophycocyanin
- Ab
antibody
- AMR
antibody-mediated rejection
- BOS
bronchiolitis obliterans syndrome
- BAL
bronchoalveolar lavage
- Col-V
collagen V
- DSA
donor specific antibodies
- EM
electron microscopy
- HLA
human leukocyte antigen
- Kα1T
K-alpha 1 tubulin
- LTxR
LTx recipient
- LTx
Lung transplantation
- MHC
major histocompatibility complex
- miRNA
microRNA
- OAD
obliterative airway disease
- PBS
phosphate buffered saline
- qRT-PCR
quantitative real time polymerase chain reaction
- SAg
self-antigen
Footnotes
Author Contributions
MG, TM: Participated in Research Design
MG, ZX, DN, MN, RW, RMB, MAS, TM: Participated in the writing of the paper
MG, DN, TM: Participated in the performance of the research
MG, ZX, TM: Participated in data analysis
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Table S1. Differentially regulated miRNA comparison in BOS, AR, and stable LTxR cohorts.
Table S2. Comparative molecular pathway analysis using DIANA-mirPath on differentially expressed miRNAs from AR or BOS exosomes.
Table S3. LTxRs demographics of replicative independent cohort for miRNA validation.
Figure S1. Exosomes isolated from the sera from BOS LTxRs contained lung-associated self-antigen Col-V. Supernatant of serum samples after exosome isolation did not contain Col-V. Since analyses were done using both exosomes and supernatants, for this experiment we employed B-actin as loading control (A). Results of western blot (B) and densitometric analysis of Col-V band intensity (C) showed lung-associated self-antigen Col-V in exosome fraction purified by the sucrose cushion method from 3 different LTxRs diagnosed with BOS. Col-V band intensity was normalized with loading control Annexin-V.
Figure S2. Exosome Validation. Exosomes isolated from serum samples were stained with 1% uranyl acetate. Vesicular structures, ranging from 40nm-100 nm in diameter, were detected using TEM (A). Immunogold EM demonstrates exosome-specific surface marker CD-63 in exosomes isolated from serum (B) and BAL (C). EM images are from one of the 2 experiments performed for serum and BAL fluid from LTxRs classified as stable or diagnosed with AR or BOS.
References
- 1.Verleden GM. Chronic allograft rejection (obliterative bronchiolitis) Semin Respir Crit Care Med. 2001;22(5):551–558. doi: 10.1055/s-2001-18427. [DOI] [PubMed] [Google Scholar]
- 2.Trulock EP, Edwards LB, Taylor DO, Boucek MM, Keck BM, Hertz MI, et al. Registry of the International Society for Heart and Lung Transplantation: twenty-third official adult lung and heart-lung transplantation report--2006. J Heart Lung Transplant. 2006;25(8):880–892. doi: 10.1016/j.healun.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Arcasoy SM, Kotloff RM. Lung transplantation. N Engl J Med. 1999;340(14):1081–1091. doi: 10.1056/NEJM199904083401406. [DOI] [PubMed] [Google Scholar]
- 4.Husain AN, Siddiqui MT, Holmes EW, Chandrasekhar AJ, McCabe M, Radvany R, et al. Analysis of risk factors for the development of bronchiolitis obliterans syndrome. Am J Respir Crit Care Med. 1999;159(3):829–833. doi: 10.1164/ajrccm.159.3.9607099. [DOI] [PubMed] [Google Scholar]
- 5.Goers TA, Ramachandran S, Aloush A, Trulock E, Patterson GA, Mohanakumar T. De novo production of K-alpha1 tubulin-specific antibodies: role in chronic lung allograft rejection. Journal of immunology. 2008;180(7):4487–4494. doi: 10.4049/jimmunol.180.7.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saini D, Weber J, Ramachandran S, Phelan D, Tiriveedhi V, Liu M, et al. Alloimmunity-induced autoimmunity as a potential mechanism in the pathogenesis of chronic rejection of human lung allografts. J Heart Lung Transplant. 2011;30(6):624–631. doi: 10.1016/j.healun.2011.01.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwata T, Philipovskiy A, Fisher AJ, Presson RG, Jr., Chiyo M, Lee J, et al. Anti-type V collagen humoral immunity in lung transplant primary graft dysfunction. Journal of immunology. 2008;181(8):5738–5747. doi: 10.4049/jimmunol.181.8.5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burlingham WJ, Love RB, Jankowska-Gan E, Haynes LD, Xu Q, Bobadilla JL, et al. IL-17-dependent cellular immunity to collagen type V predisposes to obliterative bronchiolitis in human lung transplants. J Clin Invest. 2007;117(11):3498–3506. doi: 10.1172/JCI28031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Z, Nayak D, Yang W, Baskaran G, Ramachandran S, Sarma N, et al. Dysregulated MicroRNA Expression and Chronic Lung Allograft Rejection in Recipients With Antibodies to Donor HLA. Am J Transplant. 2015;15(7):1933–1947. doi: 10.1111/ajt.13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subramanian V, Ramachandran S, Banan B, Bharat A, Wang X, Benshoff N, et al. Immune response to tissue-restricted self-antigens induces airway inflammation and fibrosis following murine lung transplantation. Am J Transplant. 2014;14(10):2359–2366. doi: 10.1111/ajt.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2014;86(2):433–444. doi: 10.1038/ki.2013.502. [DOI] [PubMed] [Google Scholar]
- 12.Manterola L, Guruceaga E, Gallego Perez-Larraya J, Gonzalez-Huarriz M, Jauregui P, Tejada S, et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol. 2014;16(4):520–527. doi: 10.1093/neuonc/not218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melo SA, Sugimoto H, O'Connell JT, Kato N, Villanueva A, Vidal A, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26(5):707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8(19):4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 15.Wolfers J, Lozier A, Raposo G, Regnault A, Thery C, Masurier C, et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7(3):297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- 16.Kulshreshtha A, Ahmad T, Agrawal A, Ghosh B. Proinflammatory role of epithelial cell-derived exosomes in allergic airway inflammation. J Allergy Clin Immunol. 2013;131(4):1194–1203. doi: 10.1016/j.jaci.2012.12.1565. 1203 e1191-1114. [DOI] [PubMed] [Google Scholar]
- 17.Cooper JD, Billingham M, Egan T, Hertz MI, Higenbottam T, Lynch J, et al. A working formulation for the standardization of nomenclature and for clinical staging of chronic dysfunction in lung allografts. International Society for Heart and Lung Transplantation. J Heart Lung Transplant. 1993;12(5):713–716. [PubMed] [Google Scholar]
- 18.Li M, Zeringer E, Barta T, Schageman J, Cheng A, Vlassov AV. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos Trans R Soc Lond B Biol Sci. 2014;369(1652) doi: 10.1098/rstb.2013.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current protocols in cell biology / editorial board, Juan S Bonifacino [et al] 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3:Unit 3 22. [DOI] [PubMed] [Google Scholar]
- 20.Kadiu I, Narayanasamy P, Dash PK, Zhang W, Gendelman HE. Biochemical and biologic characterization of exosomes and microvesicles as facilitators of HIV-1 infection in macrophages. Journal of immunology. 2012;189(2):744–754. doi: 10.4049/jimmunol.1102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kendziorski CM, Zhang Y, Lan H, Attie AD. The efficiency of pooling mRNA in microarray experiments. Biostatistics. 2003;4(3):465–477. doi: 10.1093/biostatistics/4.3.465. [DOI] [PubMed] [Google Scholar]
- 22.Kainkaryam RM, Bruex A, Woolf PJ, Schiefelbein J. Smart pooling of mRNA samples for efficient transcript profiling. Methods Mol Biol. 2012;876:189–194. doi: 10.1007/978-1-61779-809-2_15. [DOI] [PubMed] [Google Scholar]
- 23.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baixauli F, Lopez-Otin C, Mittelbrunn M. Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front Immunol. 2014;5:403. doi: 10.3389/fimmu.2014.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mishra J, Dent C, Tarabishi R, Mitsnefes MM, Ma Q, Kelly C, et al. Neutrophil gelatinase-associated lipocalin (NGAL) as a biomarker for acute renal injury after cardiac surgery. Lancet. 2005;365(9466):1231–1238. doi: 10.1016/S0140-6736(05)74811-X. [DOI] [PubMed] [Google Scholar]
- 26.Park JA, Sharif AS, Tschumperlin DJ, Lau L, Limbrey R, Howarth P, et al. Tissue factor-bearing exosome secretion from human mechanically stimulated bronchial epithelial cells in vitro and in vivo. J Allergy Clin Immunol. 2012;130(6):1375–1383. doi: 10.1016/j.jaci.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183(3):1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. 1998;273(32):20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 29.Blanchard N, Lankar D, Faure F, Regnault A, Dumont C, Raposo G, et al. TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. Journal of immunology. 2002;168(7):3235–3241. doi: 10.4049/jimmunol.168.7.3235. [DOI] [PubMed] [Google Scholar]
- 30.Mitsuhashi M, Taub DD, Kapogiannis D, Eitan E, Zukley L, Mattson MP, et al. Aging enhances release of exosomal cytokine mRNAs by Abeta1-42-stimulated macrophages. FASEB J. 2013;27(12):5141–5150. doi: 10.1096/fj.13-238980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101(36):13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47:e184. doi: 10.1038/emm.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Admyre C, Grunewald J, Thyberg J, Gripenback S, Tornling G, Eklund A, et al. Exosomes with major histocompatibility complex class II and co-stimulatory molecules are present in human BAL fluid. Eur Respir J. 2003;22(4):578–583. doi: 10.1183/09031936.03.00041703. [DOI] [PubMed] [Google Scholar]
- 34.Koga K, Matsumoto K, Akiyoshi T, Kubo M, Yamanaka N, Tasaki A, et al. Purification, characterization and biological significance of tumor-derived exosomes. Anticancer Res. 2005;25(6A):3703–3707. [PubMed] [Google Scholar]
- 35.Thery C, Duban L, Segura E, Veron P, Lantz O, Amigorena S. Indirect activation of naive CD4+ T cells by dendritic cell-derived exosomes. Nat Immunol. 2002;3(12):1156–1162. doi: 10.1038/ni854. [DOI] [PubMed] [Google Scholar]
- 36.Denzer K, van Eijk M, Kleijmeer MJ, Jakobson E, de Groot C, Geuze HJ. Follicular dendritic cells carry MHC class II-expressing microvesicles at their surface. Journal of immunology. 2000;165(3):1259–1265. doi: 10.4049/jimmunol.165.3.1259. [DOI] [PubMed] [Google Scholar]
- 37.Hachem RR, Yusen RD, Meyers BF, Aloush AA, Mohanakumar T, Patterson GA, et al. Anti-human leukocyte antigen antibodies and preemptive antibody-directed therapy after lung transplantation. J Heart Lung Transplant. 2010;29(9):973–980. doi: 10.1016/j.healun.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hachem RR, Tiriveedhi V, Patterson GA, Aloush A, Trulock EP, Mohanakumar T. Antibodies to K-alpha 1 tubulin and collagen V are associated with chronic rejection after lung transplantation. Am J Transplant. 2012;12(8):2164–2171. doi: 10.1111/j.1600-6143.2012.04079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loyer X, Potteaux S, Vion AC, Guerin CL, Boulkroun S, Rautou PE, et al. Inhibition of microRNA-92a prevents endothelial dysfunction and atherosclerosis in mice. Circ Res. 2014;114(3):434–443. doi: 10.1161/CIRCRESAHA.114.302213. [DOI] [PubMed] [Google Scholar]
- 40.Kelada S, Sethupathy P, Okoye IS, Kistasis E, Czieso S, White SD, et al. miR-182 and miR-10a are key regulators of Treg specialisation and stability during Schistosome and Leishmania-associated inflammation. PLoS Pathog. 2013;9(6):e1003451. doi: 10.1371/journal.ppat.1003451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danger R, Paul C, Giral M, Lavault A, Foucher Y, Degauque N, et al. Expression of miR-142-5p in peripheral blood mononuclear cells from renal transplant patients with chronic antibody-mediated rejection. PLoS One. 2013;8(4):e60702. doi: 10.1371/journal.pone.0060702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng Z, Xia Y, Zhang M, Zheng J. MicroRNA-155 regulates T cell proliferation through targeting GSK3beta in cardiac allograft rejection in a murine transplantation model. Cell Immunol. 2013;281(2):141–149. doi: 10.1016/j.cellimm.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 43.Gregson AL, Hoji A, Injean P, Poynter ST, Briones C, Palchevskiy V, et al. Altered Exosomal RNA Profiles in Bronchoalveolar Lavage from Lung Transplants with Acute Rejection. Am J Respir Crit Care Med. 2015 doi: 10.1164/rccm.201503-0558OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.