Abstract
Background
MicroRNAs (miRNAs) were recently identified as modulators of immune responses after human lung transplantation (LTx). This study was undertaken to investigate the miRNAs’ contribution to the pathogenesis of primary graft dysfunction (PGD) after LTx.
Methods
Of the 39 recipients, 14 (35.9%) experienced grade 3 PGD (i.e., severe PGD) within the first 72 hours of LTx. The remaining 25 recipients (64.1%) had grade 2 or less PGD, served as control group. MiRNAs were isolated from the cells purified from bronchoalveolar lavage (BAL). Bioinformatic prediction and validation by Luciferase reporter assays were performed to identify targets regulated by miR-21. Transfection of human monocytic cell line (THP-1) was conducted to determine miR-21’s cellular function.
Results
Pilot miRNA profiling of donor BAL samples before implantation in PGD (n=6) revealed significant upregulation in 44 miRNAs and downregulation in 80 miRNAs compared with control (n=6). Validation using a separate cohort demonstrated significant under expression of miR-21 in patients with severe PGD. Furthermore, under expression of miR-21 levels was negatively correlated with clinical PGD grades (PGD2 vs PGD0: P=0.042; PGD3 vs PGD0: P=0.004). Molecular analysis demonstrated that miR-21 targeted key components in TLR signaling pathway, including TLR4, IRAK3, and CXCL10. Further, incubation of THP-1 cells with cell-free BAL from severe PGD resulted in transactivation of inflammatory cytokines IL1β and TNFα. In contrast, increased expression of miR-21 resulted in marked suppression of IL1β and TNFα production.
Conclusions
Under expression of miR-21 might lead to the development of severe PGD by activating key components of the TLR pathway.
Keywords: lung transplantation, microRNAs, primary graft dysfunction
Introduction
Primary graft dysfunction (PGD), which occurs in some patients after lung transplantation (LTx), is characterized by nonspecific diffuse alveolar damage, lung edema, and severe hypoxia (1, 2). The incidence of PGD after LTx ranges from 12% to 25% (1, 2), and it remains one of the leading causes for early death after LTx, accounting for roughly 30% of all mortality within 30 days of transplantation (1, 2). Although it is a known cause of early morbidity and mortality after transplant (3, 4), the mechanisms leading to its development remain unclear. The etiology of PGD is currently considered to be multifactorial, involving donor and recipient characteristics, organ preservation issues, and intraoperative factors (5). The risk for PGD is highest when the donor lungs have sustained a significant ischemia-reperfusion injury (IRI) (6). IRI has been shown to contribute to PGD by activating cellular pathways involved in immune activation, leading to a phenomenon similar to acute respiratory distress (7–9). Innate immune activation also plays a role in the propagation of IRI in transplanted solid organs (10–13). For instance, it has been reported that IRI in heart transplantation involves toll-like receptor (TLR) signaling pathways, complement protein activation, and natural killer cell infiltration (10, 11).
MicroRNAs (miRNAs) regulate genes by selectively silencing their target messenger RNAs by binding their 3′-untranslated region (3′UTR) (14). Several studies have demonstrated that miRNAs play an active role in regulating the development and function of immune cells, and aberrant expression of miRNAs has been reported in many immune system disorders (15, 16). MicroRNA-21 (miR-21) has been shown to protect against cardiac IRI via PTEN/Akt pathway (17) and attenuate renal IRI by targeting caspase signaling (18). In this manuscript, we determined the role of miRNAs in particular miR-21 for the development of PGD after human LTx.
Methods
Study population
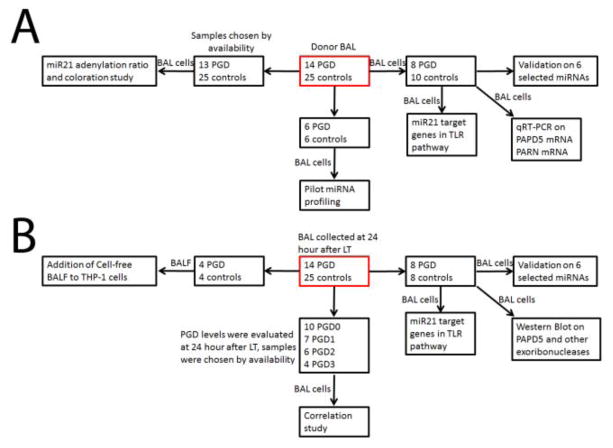
The Institutional Review Board at Washington University School of Medicine approved this study, and informed written consent was obtained from all participants. In total, 39 patients underwent LTx between July 2013 and August 2015. PGD grade was determined using the grading scale established by the International Society of Heart and Lung Transplantation (ISHLT) (19). Patients and samples were chosen according to the experimental design (Figure 1). All subjects and samples were collected by availability for this study.
Figure 1.
Flowchart of experimental design on how subjects and samples were chosen in this study.
Bronchoalveolar lavage
Bronchoalveolar lavage (BAL) was performed in the right middle lobe or lingula before donor organ retrieval. The BAL consisted of 2 aliquots of 50 mL of normal saline instilled and aspirated manually using a 60 mL syringe. Return fluid from the first aliquot was discarded, and only the return fluid from the second aliquot was used for these analyses. The BAL fluid was centrifuged at 1600 rpm for 10 minutes at 4°C. The supernatant was aliquoted and the cell pellet was stored at −80°C for analysis. Total RNA was extracted using the mirVana miRNA isolation kit (Ambion, Austin, TX).
miRNA arrays
Affymetrix GeneChip miRNA 4.0 Arrays (Affymetrix, Santa Clara, CA) were used. The microRNA analysis was performed at the Genome Technology Access Center at Washington University School of Medicine. Selected individual miRNAs were further quantitated by TaqMan miRNA assays (Applied Biosystems).
Quantification of messenger RNAs
cDNA was synthesized using Super-Script II reverse transcriptase with random hexamers (Life Technologies, Carlsbad, CA). Real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA).
Luciferase assay
Luciferase assay with TLR4, CXCL10, and IRAK3 3′UTRs was performed in MRC-5 cells as previously described (20). MRC5 was chosen for luciferase report assay because of its high transfection efficiency.
Western blotting
Lysates from human monocytic cell line THP-1 and lung transplant BAL cells were subjected to western blot based on a previously published protocol (21).
Validation of miR-21 isoforms by qRT-PCR
miR-ID@ miRNA quantification assays was used to detect the relative expression level of the miR-21+CA and miR-21+C isomiRs. In brief, 100 ng total RNA was used as input to a T4 RNA ligase circularization reaction. Two microliters of each reaction were used for miR-specific RT. The specific RT primers used were TGATAAGCTATGT for miR-21+CA and CTGATAAGCTAGT for miR-21C. PCR primers were CTTATCAGACTGATGTTGACAT (forward) and CATCAGTCTGATAAGCTATGTC (reverse) for miR-21+CA and AGCTTATCAGACTGATGTTGACT (forward) and AACATCAGTCTGATAAGCTAGTC (reverse) for miR-21+C. A single circulation/RT reaction was performed and two PCR reactions were performed on each RT. The specificity of the detection by this method was confirmed using synthetic miR-21+CA and synthetic miR-21+C. Statistical analysis of miR-21 adenylation/degradation ratios was performed as previously described (22). Adenylation ratio = (count of miR-21+CA)/(count of miR-21+C).
Statistical analysis
Results are expressed as mean ± SD and were compared by Mann-Whitney U test. Differences were considered significant at P-value less than 0.05.
Results
Patient demographics
In total, 39 donors and 39 recipients were included in this study (Table 1). All the donors were brain dead donors. Fourteen patients (35.9%) had severe PGD within 72 hours after LTx (Table S1). The analysis of standard donor-related risk factors with respect to clinical outcomes revealed no significant differences between the patients who developed severe PGD and those who did not. There were no significant differences among the donor-related risk factors, including donor age, smoking history, ABO status, traumatic injury, last PaO2/FiO2 ratio, duration of mechanical ventilation and the ischemic time. All non-PGD recipients were white, the underlying diseases of them are 9 COPD, 8 IPF, 6 ILD and 2 Sarcoidosis. The gender, age, race and underlying disease were matched between PGD and non-PGD groups. No patients received extracorporeal membrane oxygenation support after LTx. All of the donor samples were negative for BAL cultures for Pseudomonas aeruginosa and Aspergillus. None of the patients had episodes of acute rejection during the early stage after LTx. No significant difference of peri-operative transfusions between PGD and control group.
Table 1.
Clinical donor characteristics and operative factors.a
| Characteristic | PGD (n=14) | Control (n=25) |
|---|---|---|
| Age, y | 50±11 | 59±8 |
| Sex, male, n(%) | 11(78.6) | 15(60) |
| Cause of death | 64% trauma, 36% non-trauma | 72% trauma, 28% non-trauma |
| Smoking history, n(%) | 9(64.2) | 17(68) |
| >20 pack-years, n(%) | 5(35.7) | 8(32) |
| Last PaO2/FiO2 ratio <300 | 438±55 | 477±83 |
| Time on ventilator >72 h | 5(25.7) | 8(32) |
| Organ ischemic time | ||
| 1st lung ischemic period, min | 222±38 | 198±60 |
| 2nd lung ischemic period, min | 295±54 | 277±57 |
| Cardiopulmonary bypass | 4(28.4) | 7(28) |
| ABO-identical compatibility, n(%) | 14(100) | 25(100) |
| CMV mismatch (Donor (+) recipient (−)) | 5(35.7) | 10 (40) |
| Extracorporeal membrane oxygenation | 0/14 | 0/25 |
Values expressed as mean ± standard deviation, unless indicated otherwise.
Abbreviation: CMV, cytomegalovirus; PGD, primary graft dysfunction.
MicroRNA-21 expression was significantly decreased in lung transplant recipients with severe PGD and inversely correlated with PGD grades
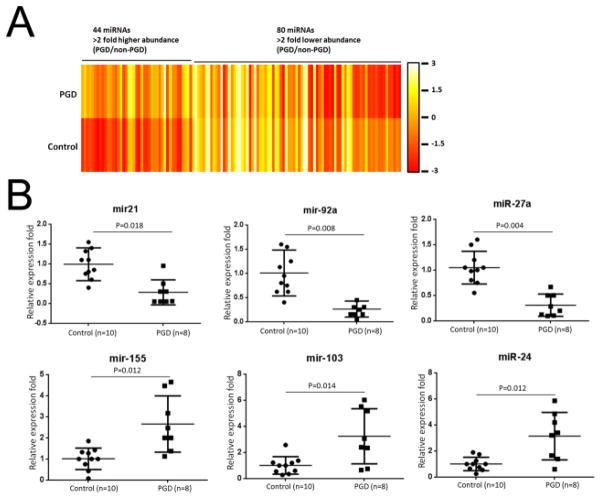
To identify differentially expressed miRNA in lungs with PGD, we profiled the expression of all human matured miRNAs in BAL cells collected using the Affymetrix miRNA 4.0 arrays. We analyzed 2 pooled RNA samples from the BAL cells of 6 severe PGD donors and 6 controls and found differential expression of 44 upregulated miRNAs and 80 downregulated miRNAs (relative fold > 2, Figure 2A, Table S2) in severe PGD donors compared with control donors. We used TaqMan RT-PCR to validate the expression level of six differentially expressed miRNAs (miR-21, miR-92a, miR-27a, miR-155, miR-103, and miR-24) using an independent cohort of 8 severe PGD donors and 10 control donors. These six miRNAs were selected for validation based on previous reports of association between ischemic and inflammatory injury (17, 18, 23–28). The TaqMan qRT-PCR assay showed under expression of miR-21, -92a, -27a and over expression of miR-155, -103, -24 in donor BALs (Figure 2B) and BALs collected 24 hours after transplantation (Figure S1) from 8 patients with severe PGD compared to 8 control patients. Among these miRNAs, miR-21 in particular has been reported to negatively regulate the innate immune response by targeting interleukin-1 receptor-associated kinase 1 (IRAK1) (29), myeloid differentiation primary response gene 88 (MYD88) (29), programmed cell death 4 (PDCD4) (30), and peroxisome proliferator-activated receptor alpha (PPARA) (31). We reasoned that defects in the negative regulation of miR-21 could result in uncontrolled innate immune activation and therefore that miR-21 may play an important role in the immuno-pathogenesis of PGD.
Figure 2.
MicroRNAs (miRNAs) were differentially expressed in the donor bronchoalveolar lavage (BAL) fluid of patients with severe primary graft dysfunction (PGD) and compared with the miRNAs found in the BAL fluid in the control group. (A) Heatmap representation of the mean fold change in PGD donor-related differential miRNA signature. Two-dimensional grid matrix displaying 124 differential miRNAs (44 upregulated and 80 downregulated miRNAs) in PGD and controls was obtained by the functional heatmap in R. Each entry of the grid refers to relative fold (log2) between the expression level of a given miRNA in a pool of 6 lungs with severe PGD lungs relative to those of 6 lungs in the control group. The color of each entry is determined by the value of that fold difference, ranging from red (negative values) to yellow (positive values). (B) Quantitative expression of miR-21, miR-92a, miR-27a, miR-155, miR-103, and miR-24 was assessed by TaqMan real-time RT-PCR on RNA samples of BAL cells from an independent cohort of 10 control and 8 PGD donor lungs. Relative expression of these miRNAs was calculated and observed statistically differentially expressed compared to controls (p<0.05, Mann Whitney U-test).
Validation experiments were carried out to determine any correlation between miR-21 levels and the clinical features of PGD severity. As shown in Figure 3, under expression of miR-21 levels negatively correlated with clinical PGD grades (PGD2 vs PGD0: P=0.042; PGD3 vs PGD0: P=0.004). Based on these results, we proposed that downregulation of miR-21 could be significantly associated with development of PGD after human LTx.
Figure 3.
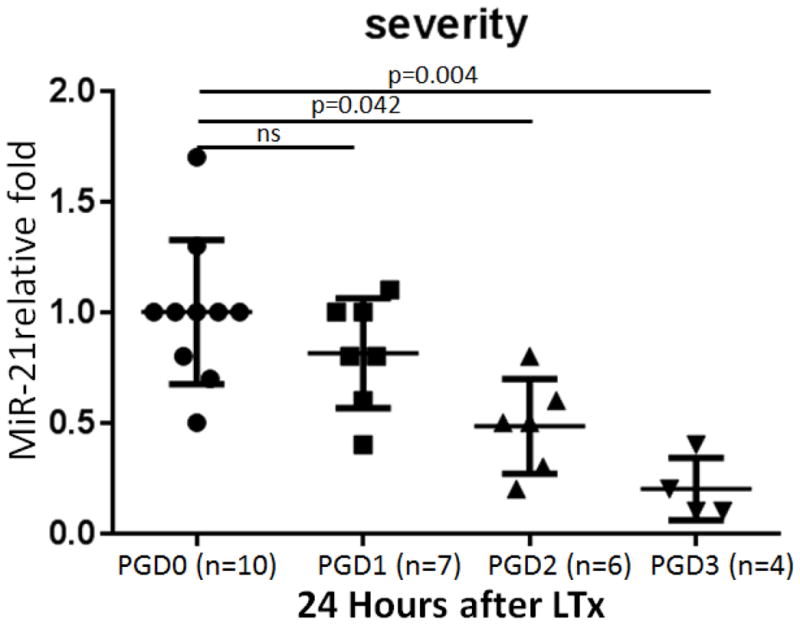
Under expression of miR-21 in patients with severe primary graft dysfunction (PGD) was negatively associated with PGD severity. Quantitative expression of miR-21 was assessed by TaqMan real-time RT-PCR on RNA samples of bronchoalveolar lavage cells collected from 10 PGD0, 7 PGD1, 6 PGD2, and 4 PGD3 samples 24 hours after lung transplantation (LTx). Comparisons were made using the Mann Whitney U-test.
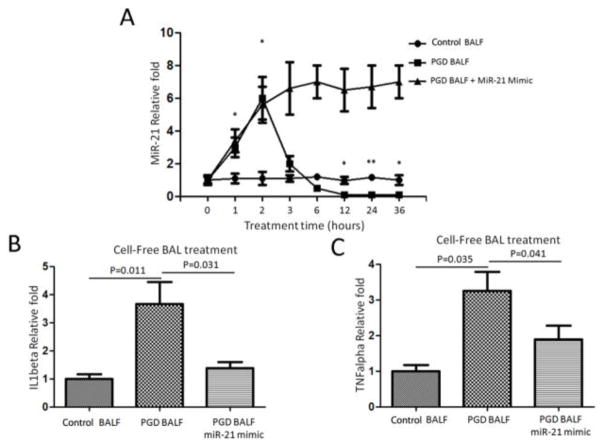
Addition of cell-free BAL to human monocyte cells induced transient miR-21 expression followed by rapid decrease, leading to increases in IL1β and TNFα
To determine the effect of BAL fluid on miR-21 expression levels, we analyzed the miR-21 expression in THP-1 after cells were incubated for 1, 2, 3, 6, 12, 24, and 36 hours with cell-free BAL fluid obtained from severe PGD or control LTx recipients. Time-course kinetics of stimulation demonstrated that PGD-derived, cell-free BAL induced miR-21 expression at 1 hour. miR-21 expression went on to peak at 2 hours, rapidly decreased at 3 hours, and was nearly undetectable at 12 hours. In contrast, there was no induction of miR-21 expression with incubation with cell-free BAL fluid obtained from the control group (Figure 4A). Further, we found that forced expression with miR-21 mimic could reverse the downregulation of miR-21 expression (Figure 4A). IL1β and TNFα mRNAs were significantly upregulated only in cell-free BAL derived from severe PGD patients; there was no upregulation in that incubated from the control group (Figures 4B and 4C). Forced expression of miR-21 could downregulate the expression levels of IL1β and TNFα mRNA. These results strongly support that miR-21 was significantly downregulated in severe PGD and remains at undetectable levels after incubation with PGD-derived BAL fluid. This also results in induction of proinflammatory cytokines IL1β and TNFα, which can contribute to the immuno-pathogenesis of PGD.
Figure 4.
Cell-free bronchoalveolar lavage (BAL) treatment induced miR21 expression followed by a rapid decrease, and promoted expression of IL1β and TNFα. (A) qRT-PCR analysis of the expression of miR-21 in THP-1 cells after a time-course stimulation treatment of cell-free BAL from primary graft dysfunction (PGD) and control LTx recipients. (B and C) Quantitative expression of IL1β and TNFα mRNAs after 24-hour treatment of cell-free BAL fluid from severe PGD and control groups.
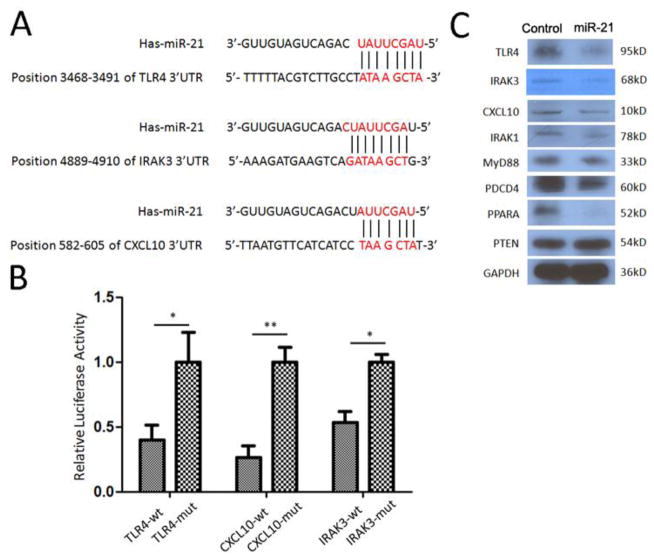
MiR-21 targets TLR signaling pathways
To determine the functional consequences of miR-21 downregulation in PGD development, we analyzed the predicted mRNA target sites. Using reporter assays, it has been shown that 3′UTRs of mRNAs encoding IRAK1, MyD88, PDCD4, PPARA, and PTEN proteins all have miR-21 target sequences (29–31). Computational algorithms (TargetScan and miRanda) were performed and demonstrated the potential binding sites of miR-21 to the 3′UTR of TLR4, IRAK3, and CXCL10 (Figure 5A). To determine whether TLR4, IRAK3, and CXCL10 were direct targets of miR-21, fragments of the 3′UTRs of TLR4, IRAK3, and CXCL10 containing wild-type or mutated miR-21 complementary sites were cloned into phRL-TK renilla luciferase reporter plasmid. Luciferase reporters were co-transfected with miRNA mimic of miR-21 into MRC-5 lung fibroblast cells. We then examined binding of human miR-21 to the 3′UTRs of TLR4, IRAK3, and CXCL10 mRNA using a luciferase assay. As the 3′UTRs of TLR4, IRAK3, and CXCL10 are inserted downstream of the luciferase open reading frame, specific binding of miR-21 prevented luciferase reporter gene expression (Figure 5B). Mutations of the TLR4, IRAK3, and CXCL10 binding sites also decreased specific binding of miR-21 and restored luciferase activity (Figure 5B). This indicates that TLR4, IRAK3, and CXCL10 were indeed targets of miR-21. Western blot data presented in Figure 5C clearly demonstrates that TLR4, IRAK3, CXCL10, IRAK1, MyD88, PDCD4, and PPARA protein levels were downregulated after over expression of miR-21. These results strongly support the concept that miR-21 can indeed regulate lung IRI via TLR signaling.
Figure 5.
MiR-21 targets the key factors in toll-like receptor pathways. (A) TLR4, IRAK3, and CXCL10 as candidate targets of miR-21 posttranscriptional repression. Shown is a sequence alignment of miR-21 and its target sites in 3′ UTRs of TLR4, IRAK3, and CXCL10. (B) The panels show the analysis of expression of TLR4-3′UTR, IRAK3-3′UTR, and CXCL10-3′UTR luciferase reporters in the presence of miR-21. (C) Western blot assay to examine the protein levels of TLR4, IRAK3, CXCL10, IRAK1, MyD88, PDCD4, PPARA, and PTEN when over expression of miR-21 in THP-1 cells. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as loading control.
MiR-21 target genes for TLR signaling were overexpressed in BAL cells obtained from lungs with severe PGD
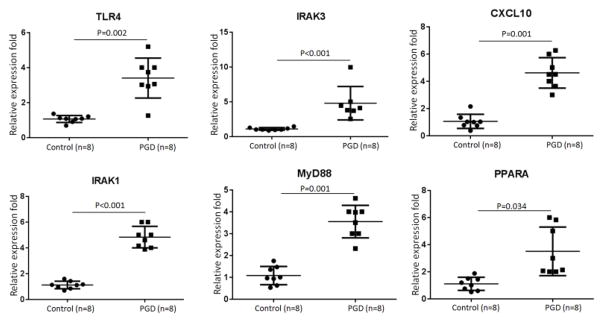
We examined the expression levels of TLR4, IRAK3, CXCL10, IRAK1, MyD88, and PPARA genes in the BAL cells of donor lungs of 8 severe PGD LTx recipients and 8 control LTx recipients. Compared to those of the controls, the BAL cells from the 8 patients with severe PGD showed significantly higher expression levels of TLR4 (P=0.002), IRAK3 (P<0.001), CXCL10 (P=0.001), IRAK1 (P<0.001), MyD88 (P=0.001), and PPARA mRNA (P=0.034) (Figure 6). There was also significant over expression of TLR4, IRAK3, CXCL10, IRAK1, MyD88, and PPARA mRNA levels in the BAL cells from patients with severe PGD collected 24 hours postoperatively compared to the BAL cells collected from the control recipients at the same time point (Figure S2). Due to limited BAL samples, we didn’t measure the protein levels for these target genes. These results conclusively demonstrate that TLR signaling cascades were activated in patients with severe PGD after transplantation.
Figure 6.
MiR-21 target genes were over expressed in donor lungs with primary graft dysfunction (PGD). Quantitative expression of TLR4, IRAK3, CXCL10, IRAK1, MyD88, and PPARA was assessed by qRT-PCR on RNA samples of bronchoalveolar lavage cells from 8 control lungs and 8 lungs with PGD. Comparisons were performed using the Mann Whitney U-test.
MiR-21 degradation pathway was significantly increased in lungs with severe PGD
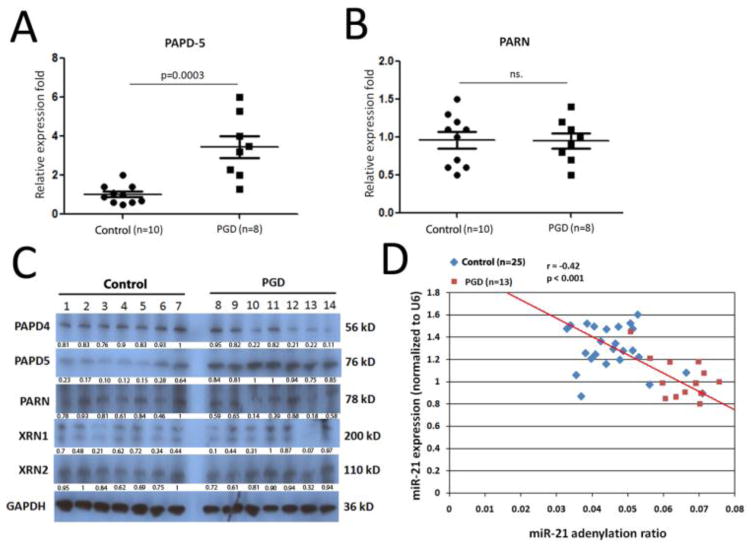
Almost half of the overall miR-21 expression was due to a 23-nt isomiR, commonly referred to as miR-21+C (32). PAPD5 added adenosines to miR-21+C, stimulating degradation of miR-21 by nuclease PARN (22). Therefore, we analyzed for dysregulation of PAPD5 and PARN in the BAL cells of patients with severe PGD. These patients (n=8) demonstrated significant elevation of PAPD5 mRNA level in donor BAL cells compared to the levels from the control patients (n=10) (P=0.0003, Figure 7A). No difference was observed in expression of PARN mRNA between severe PGD and control patients (Figure 7B). Further western blot by 24 hours BAL cell lysates from severe PGD and control patients (Figure 7C) demonstrated that PAPD-5 protein levels were significantly over expressed in patients with severe PGD, whereas there was no differences in the expression of other exoribonucleases, including XRN-1 and XRN-2. A significant negative correlation also existed between the miR-21 adenylation ratio and miR-21 expression levels (P<0.001) (Figure 7D).
Figure 7.
Enhancement of the miR-21 degradation pathway activation in lungs with primary graft dysfunction (PGD). (A and B) Quantitative expression of PAPD-5 and PARN mRNAs was assessed by qRT-PCR on RNA samples of bronchoalveolar lavage (BAL) cells from 10 control lungs and 8 lungs with severe PGD. Comparisons were performed with the Mann Whitney U-test. (C). Western blot assay on PAPD-4, PAPD-5, PARN, XRN-1, and XRN-2 protein levels in BAL cell lysates from 7 control and 7 donor lungs with severe PGD. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as loading control. Protein quantification was performed by bandscan and densitometry analysis with optical density for PAPD-4, PAPD-5, PARN, XRN-1, and XRN-2. (D). miR-ID® miRNA quantification assays was used to specifically detect the relative expression level of the miR-21+CA and miR-21+C isomiRs. Donor BAL samples from 25 control and 13 PGD were included here. The P-values were calculated using Pearson’s correlation tests.
Discussion
Lungs are often exposed to environmental antigens that activate an innate immune response, which modulates future alloimmune responses. After transplantation, therefore, the lungs can induce an innate immune activation that may cause post-transplant complications. Previous studies have reported the effects of alterations in human donor lung gene expression early after LTx (33, 34), but the role of miRNAs in this process remains elusive. Therefore, in this study, we identified which miRNAs were dysregulated in patients who developed severe PGD after LTx to more clearly define the role of miRNA in the development of PGD. Especially, miR-21 was significantly negatively correlated with PGD grades. We further demonstrated that miR-21 targets several genes known to control early inflammation, including TLR4, IRAK3, and CXCL10. Finally, in vitro experiments have demonstrated that forced expression of miR-21 in human monocyte cells repressed downstream transactivation of IL1β and TNFα following incubation with cell-free BAL fluid from patients with severe PGD. These results support the conclusion that loss of miR-21 can result in abnormal activation of the TLR pathway, ultimately leading to PGD by targeting its key signaling proteins.
The fine-tuning of the innate immune response by miRNAs has been documented by several recent reports (16, 35, 36). Functionally, miRNAs may modulate epithelial immune responses at many different phases of the innate immune network, including the production and release of cytokines and chemokines, release of exosomes, and feedback regulation of immune homeostasis (36). In this study, we demonstrated that miR-21 targets TLR4, IRAK1, IRAK3, CXCL10, MyD88, PPARA, and PDCD4 in patients with severe PGD. Given the essential characteristics of these proteins, the negative regulation by miR-21 on the activation of the TLR pathway could be effective and may affect multiple targets. This is similar to the reported role of miR-181a in T cells, which can result in modest reductions of multiple phosphatases affecting the excitation threshold of T cells (37). Deficiency of miR-21, therefore, is deleterious and can lead to accumulation of its target proteins, albeit with varied concentrations. This, in turn, can result in sustained overproduction of TLR components and downstream activation. Therefore, we propose that miR-21 deficiency may be a critical causal factor in the abnormal activation of the TLR pathway that results in PGD after LTx. The strong correlation we identified between miR-21 levels and activation of the TLR pathways in patients with severe PGD supports this idea.
Individual miRNAs may display rapid decay dynamics in some situations (38–41). The mechanism by which specific miRNAs are marked for decay as well as ribonucleases that can catalyze the degradation reaction has been recently identified (42). PAP associated domain containing 4 (PAPD4, also known as GLD2) catalyzes polyadenylation of some noncoding RNAs and mRNAs. Mature miR-21 was produced in two prevalent isomiR forms that differ by 1 nt at their 3′ end, and the 3′ end of miR-21 was post-transcriptionally adenylated by the non-canonical poly(A) polymerase PAPD5 (22). Exoribonuclease knockdown experiments followed by small-RNA sequencing have also suggested that PARN degrades miR-21 in the 3′-to-5′ direction (22). Our results in this study for the first time demonstrated significant elevation of PAPD5 mRNA and protein levels in lungs with severe PGD after transplantation (Figure 7A and 7C). We have also demonstrated a significant negative correlation between the adenylation ratio of miR-21 and its expression level in PGD lungs (Figure 7D).
This study is limited by a relatively small sample size. In addition, in vivo experiments involving overexpression of miR-21 in animal models of IRI will be needed to prove its inhibitory role on lung injury after LTx. Future study is also required for miR-21’s effect on neutrophils during PGD development.
In conclusion, our data provide further evidence that dysregulated miRNAs can be a novel regulator of TLR signaling in the development of lung injury early after human LTx. We demonstrated that under expression of miR-21 might lead to the development of severe PGD by activating key components of the TLR pathway.
Supplementary Material
Footnotes
Disclosure Statement
This research was supported by the National Institutes of Health grant RO1 HL056643-18 (TM) and the BJC Foundation (TM). We thank Ms. B. Glasscock and C. Prendergast for their assistance in preparing this manuscript. None of the authors has a financial relationship with a commercial entity that has an interest in the subject of the presented manuscript or other conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Christie JD, Bavaria JE, Palevsky HI, et al. Primary graft failure following lung transplantation. Chest. 1998;114:51–60. doi: 10.1378/chest.114.1.51. [DOI] [PubMed] [Google Scholar]
- 2.Christie JD, Carby M, Bag R, Corris P, Hertz M, Weill D. Report of the ISHLT Working Group on Primary Lung Graft Dysfunction part II: definition. A consensus statement of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant. 2005;24:1454–9. doi: 10.1016/j.healun.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 3.Christie JD, Kotloff RM, Ahya VN, et al. The effect of primary graft dysfunction on survival after lung transplantation. Am J Respir Crit Care Med. 2005;171:1312–6. doi: 10.1164/rccm.200409-1243OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christie JD, Kotloff RM, Pochettino A, et al. Clinical risk factors for primary graft failure following lung transplantation. Chest. 2003;124:1232–41. doi: 10.1378/chest.124.4.1232. [DOI] [PubMed] [Google Scholar]
- 5.Diamond JM, Lee JC, Kawut SM, et al. Clinical risk factors for primary graft dysfunction after lung transplantation. Am J Respir Crit Care Med. 187:527–34. doi: 10.1164/rccm.201210-1865OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JC, Christie JD. Primary graft dysfunction. Proc Am Thorac Soc. 2009;6:39–46. doi: 10.1513/pats.200808-082GO. [DOI] [PubMed] [Google Scholar]
- 7.Iwata T, Philipovskiy A, Fisher AJ, et al. Anti-type V collagen humoral immunity in lung transplant primary graft dysfunction. J Immunol. 2008;181:5738–47. doi: 10.4049/jimmunol.181.8.5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno I, Vicente R, Ramos F, Vicente JL, Barbera M. Determination of interleukin-6 in lung transplantation: association with primary graft dysfunction. Transplant Proc. 2007;39:2425–6. doi: 10.1016/j.transproceed.2007.07.056. [DOI] [PubMed] [Google Scholar]
- 9.Diamond JM, Christie JD. The contribution of airway and lung tissue ischemia to primary graft dysfunction. Curr Opin Organ Transplant. 15:552–7. doi: 10.1097/MOT.0b013e32833e1415. [DOI] [PubMed] [Google Scholar]
- 10.Millington TM, Madsen JC. Innate immunity and cardiac allograft rejection. Kidney Int Suppl. :S18–21. doi: 10.1038/ki.2010.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Millington TM, Madsen JC. Innate immunity in heart transplantation. Curr Opin Organ Transplant. 2009;14:571–6. doi: 10.1097/MOT.0b013e32832e7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosieradzki M, Rowinski W. Ischemia/reperfusion injury in kidney transplantation: mechanisms and prevention. Transplant Proc. 2008;40:3279–88. doi: 10.1016/j.transproceed.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Uchida Y, Ke B, Freitas MC, et al. T-cell immunoglobulin mucin-3 determines severity of liver ischemia/reperfusion injury in mice in a TLR4-dependent manner. Gastroenterology. 139:2195–206. doi: 10.1053/j.gastro.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Sarma NJ, Tiriveedhi V, Ramachandran S, Crippin J, Chapman W, Mohanakumar T. Modulation of immune responses following solid organ transplantation by microRNA. Exp Mol Pathol. 2012;93:378–85. doi: 10.1016/j.yexmp.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun. 2009;32:189–94. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu Y, Wan L, Fan Y, et al. Ischemic postconditioning-mediated miRNA-21 protects against cardiac ischemia/reperfusion injury via PTEN/Akt pathway. PloS one. 2013;8:e75872. doi: 10.1371/journal.pone.0075872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu H, Jiang W, Xi X, Zou C, Ye Z. MicroRNA-21 attenuates renal ischemia reperfusion injury via targeting caspase signaling in mice. American journal of nephrology. 2014;40:215–23. doi: 10.1159/000368202. [DOI] [PubMed] [Google Scholar]
- 19.Christie JD, Bellamy S, Ware LB, et al. Construct validity of the definition of primary graft dysfunction after lung transplantation. J Heart Lung Transplant. 29:1231–9. doi: 10.1016/j.healun.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Z, Ramachandran S, Gunasekaran M, et al. MicroRNA-144 dysregulates the transforming growth factor-beta signaling cascade and contributes to the development of bronchiolitis obliterans syndrome after human lung transplantation. The Journal of heart and lung transplantation the official publication of the International Society for Heart Transplantation. 2015;34:1154–62. doi: 10.1016/j.healun.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goers TA, Ramachandran S, Aloush A, Trulock E, Patterson GA, Mohanakumar T. De novo production of K-alpha1 tubulin-specific antibodies: role in chronic lung allograft rejection. J Immunol. 2008;180:4487–94. doi: 10.4049/jimmunol.180.7.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boele J, Persson H, Shin JW, et al. PAPD5-mediated 3′ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc Natl Acad Sci U S A. 2014;111:11467–72. doi: 10.1073/pnas.1317751111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinkel R, Penzkofer D, Zuhlke S, et al. Inhibition of microRNA-92a protects against ischemia/reperfusion injury in a large-animal model. Circulation. 2013;128:1066–75. doi: 10.1161/CIRCULATIONAHA.113.001904. [DOI] [PubMed] [Google Scholar]
- 24.Liang S, Wang W, Gou X. MicroRNA 26a modulates regulatory T cells expansion and attenuates renal ischemia-reperfusion injury. Mol Immunol. 2015;65:321–7. doi: 10.1016/j.molimm.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Lorenzen JM, Kaucsar T, Schauerte C, et al. MicroRNA-24 antagonism prevents renal ischemia reperfusion injury. J Am Soc Nephrol. 2014;25:2717–29. doi: 10.1681/ASN.2013121329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang JX, Zhang XJ, Li Q, et al. MicroRNA-103/107 Regulate Programmed Necrosis and Myocardial Ischemia/Reperfusion Injury Through Targeting FADD. Circ Res. 2015;117:352–63. doi: 10.1161/CIRCRESAHA.117.305781. [DOI] [PubMed] [Google Scholar]
- 27.Liu P, Zhao H, Wang R, et al. MicroRNA-424 protects against focal cerebral ischemia and reperfusion injury in mice by suppressing oxidative stress. Stroke. 2015;46:513–9. doi: 10.1161/STROKEAHA.114.007482. [DOI] [PubMed] [Google Scholar]
- 28.Li XQ, Lv HW, Wang ZL, Tan WF, Fang B, Ma H. MiR-27a ameliorates inflammatory damage to the blood-spinal cord barrier after spinal cord ischemia: reperfusion injury in rats by downregulating TICAM-2 of the TLR4 signaling pathway. J Neuroinflammation. 2015;12:25. doi: 10.1186/s12974-015-0246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Chen J, Wang H, et al. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog. 2013;9:e1003248. doi: 10.1371/journal.ppat.1003248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–7. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 31.Zhou J, Wang KC, Wu W, et al. MicroRNA-21 targets peroxisome proliferators-activated receptor-alpha in an autoregulatory loop to modulate flow-induced endothelial inflammation. Proc Natl Acad Sci U S A. 2011;108:10355–60. doi: 10.1073/pnas.1107052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anraku M, Cameron MJ, Waddell TK, et al. Impact of human donor lung gene expression profiles on survival after lung transplantation: a case-control study. Am J Transplant. 2008;8:2140–8. doi: 10.1111/j.1600-6143.2008.02354.x. [DOI] [PubMed] [Google Scholar]
- 34.Ray M, Dharmarajan S, Freudenberg J, Zhang W, Patterson GA. Expression profiling of human donor lungs to understand primary graft dysfunction after lung transplantation. Am J Transplant. 2007;7:2396–405. doi: 10.1111/j.1600-6143.2007.01918.x. [DOI] [PubMed] [Google Scholar]
- 35.He X, Jing Z, Cheng G. MicroRNAs: new regulators of Toll-like receptor signalling pathways. Biomed Res Int. 2014;2014:945169. doi: 10.1155/2014/945169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou R, O’Hara SP, Chen XM. MicroRNA regulation of innate immune responses in epithelial cells. Cell Mol Immunol. 2011;8:371–9. doi: 10.1038/cmi.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li QJ, Chau J, Ebert PJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–61. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 39.Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neuroscience letters. 2009;459:100–4. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 40.Rajasethupathy P, Fiumara F, Sheridan R, et al. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63:803–17. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krol J, Busskamp V, Markiewicz I, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–31. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 42.Katoh T, Sakaguchi Y, Miyauchi K, et al. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23:433–8. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.