Abstract
Continuum-solvent models (CSMs) have successfully predicted many quantities, including the solvation-free energies (ΔG) of small molecules, but they have not consistently succeeded at reproducing experimental binding free energies (ΔΔG), especially for protein–protein complexes. Several CSMs break ΔG into the free energy (ΔGvdw) of inserting an uncharged molecule into solution and the free energy (ΔGel) gained from charging. Some further divide ΔGvdw into the free energy (ΔGrep) of inserting a nearly hard cavity into solution and the free energy (ΔGatt) gained from turning on dispersive interactions between the solute and solvent. We show that for 9 protein–protein complexes neither ΔGrep nor ΔGvdw was linear in the solvent-accessible area A, as assumed in many CSMs, and the corresponding components of ΔΔG were not linear in changes in A. We show that linear response theory (LRT) yielded good estimates of ΔGatt and ΔΔGatt, but estimates of ΔΔGatt obtained from either the initial or final configurations of the solvent were not consistent with those from LRT. The LRT estimates of ΔGel differed by more than 100 kcal/mol from the explicit solvent model’s (ESM’s) predictions, and its estimates of the corresponding component (ΔΔGel) of ΔΔG differed by more than 10 kcal/mol. Finally, the Poisson–Boltzmann equation produced estimates of ΔGel that were correlated with those from the ESM, but its estimates of ΔΔGel were much less so. These findings may help explain why many CSMs have not been consistently successful at predicting ΔΔG for many complexes, including protein–protein complexes.
Graphical Abstract
1. INTRODUCTION
Implicit solvent models, such as the Poisson–Boltzmann equation1 (PBE), generalized Born (GB) models,2 proximal distribution approaches,3–5 and integral equation methods,6,7 provide estimates of the solvation energies (ΔG) of biomolecules more quickly than explicit solvent models (ESMs). Implicit solvent models are faster than ESMs because they approximately integrate over the degrees of freedom of the aqueous solvent in the partition function. Once estimates of ΔG have been obtained, they can be used to obtain estimates of other free energies, such as binding (ΔΔG) and mutation free energies.8,9 Implicit solvent models may be divided into two classes, continuum solvent models (CSMs),1,2 such as the PBE and GB models that model water as a high-dielectric continuum, and those that approximate the distribution of the solvent,3–7 such as integral equations, density functional theories, and other structured approaches. CSMs have been fairly successful at predicting many quantities, such as ΔG, and the salt dependencies of various free energies,10–14 but they have not been uniformly successful at predicting some other quantities, such as ΔΔG.15–21
Typically, CSMs break ΔG into the free energy (ΔGvdw) required to insert an uncharged molecule into solution and the free energy (ΔGel) gained by turning on the partial atomic charges.22–24 Some researchers have then further divided ΔGvdw into the free energy (ΔGrep) required to insert a nearly hard cavity into solution and the free energy (ΔGatt) gained from turning on the dispersive interactions between the solute and solvent.25–33 The Methods contains formal definitions of these and related quantities.
In previous work, we demonstrated that the methods used to compute ΔGrep, ΔGatt, and ΔGvdw in some CSMs were unsatisfactory for alanine and glycine peptides.24,34 In this work, we extend this analysis to nine protein–protein complexes covering wide ranges of sizes, biological functions, and equilibrium binding constants. By doing so, we try to ensure that our results are generally applicable to protein–protein complexes and not artifacts of our choice of examples. The complexes with their PDB codes are bovine α-chymotrypsin with eglin c complex (1ACB),35 porcine pancreatic trypsin with soybean trypsin inhibitor complex (1AVX),36 bovine β-lactoglobulin, which is a homodimer (1BEB),37 barnase–barstar complex (1BRS),38 colicin E9 dnase domain with IM9 (1EMV),39 Pseudomonas aeruginosa exos toxin with human rac (1HE1),40 bovine β-trypsin with CMTI-I (1PPE),41 and uracil-dna glycosylase with uracil glycosylase inhibitor (1UDI).42 In the following sections, we present the theoretical framework followed by the computational methods. We then present our results and discuss their implications for CSMs.
2. THEORY
In principle, ΔGrep, ΔGatt, ΔGvdw, and ΔGel could be computed with thermodynamic integration by integrating over λ the derivatives (〈∂Urep(λ)/∂λ〉λ, 〈∂Uatt(λ)/∂λ 〉λ, 〈∂Uvdw(λ)/∂λ〉λ, and 〈∂Uel(λ)/∂λ〉λ) of λ-dependent potentials.43,44 Plots of 〈∂Uatt(λ)/∂λ〉λ and 〈∂Uel(λ)/∂λ〉λ for one of the complexes in the present study (1BRS) are shown in the Supporting Information. If these curves represented purely linear functions of λ, then ΔGatt and ΔGel could be computed from linear response theory (LRT)
| (1) |
| (2) |
where Uatt and Uel are attractive and electrostatic potential energies between the solute and solvent, respectively, as defined in the Methods, and 〈…〉0 and 〈…〉1 indicate that expectation values were computed in ensembles where λ = 0 and λ = 1. Some work has then gone farther by assuming that ΔGatt can be computed by averaging Uatt over an approximate solvent distribution.29,30,33 Natural choices for such a solvent distribution would be either the initial or final solvent configurations leading to single-step perturbation (SSP) estimates
| (3) |
| (4) |
of ΔGatt. Similarly, most CSMs assume that 〈Uel〉0 = 0,5,45 yielding a SSP approximation
| (5) |
In contrast, the LRT cannot be used to compute either ΔGrep or ΔGvdw because 〈∂Urep/∂λ〉λ and 〈∂Uvdw/∂λ〉λ typically contain large peaks or even poles.24,46 Instead, CSMs typically use formulas taken from macroscopic liquid theory, such as
| (6) |
| (7) |
where A is the solvent-accessible surface area of the molecule, and γvdw and γrep are positive constants analogous to the surface tension of macroscopic liquid interfaces.9,47–51
Alternatively, some studies have claimed that ΔGrep should increase linearly with the solvent-accessible volume (V) rather than A for sufficiently small cavities and that for larger cavities it should approach eq 7.52–55 In the present study, all of our proteins are large enough that this model would predict that ΔGrep should approximately obey eq 7. The interested reader is referred to ref 24 and its supporting information for the volume correlations.
For an equilibrium binding process between proteins A and B, given that estimates of ΔG of the components and the complex have been obtained, the desolvation energy (ΔΔGdesol = ΔGc − ΔGa − ΔGb, where ΔGa, ΔGb, and ΔGc are the ΔG’s of the first component of the complex, the second component, and the complex, respectively) can be defined. In turn, ΔΔGdesol can be broken into repulsive , attractive , total van der Waals cavity insertion , and electrostatic components, as described in the Methods.
Combining the definitions of and with eqs 6 and 7, we can write
| (8) |
| (9) |
where ΔA = Ac − Ab − Aa, where Ac, Aa, and Ab are the areas of the complex and its two components.
Given ΔΔGdesol, estimates of ΔΔG can be obtained by adding the binding free energy (ΔGvac) of the complex in vacuum. In turn, ΔΔG can be broken into repulsive (ΔΔGrep), attractive (ΔΔGatt), total van der Waals cavity insertion (ΔΔGvdw), and electrostatic (ΔΔGel) components, as described in the Methods.
Many of these assumptions underlying CSMs can be called into question for biomolecules with rough surfaces and charge distributions varying over lengths comparable to the size of a water molecule. We recently found that none of ΔGrep, ΔGatt, or ΔGvdw are simple functions of A for alkanes and short peptides, indicating that eqs 6 and 7 are not strictly valid, and that eqs 3 and 4 were not consistent with ESMs.24,34,46,56,57 Some other studies have examined the validity of eqs 2 and 5.5,45 Also, and are typically positive because they account for the loss of favorable solute–solvent interactions upon binding. In contrast, and are typically negative because and because binding partners usually have complementary charges at the binding interface. Therefore, ΔΔGatt and ΔΔGel can be much smaller than the ΔGatt and ΔGel of the complex and its components. Because of this cancellation of free energies, theories that generate estimates of ΔG that are correlated with experimental data are not guaranteed to produce similarly accurate estimates of ΔΔG.58
In the present study, we test the validity of eqs 1–9 for 9 protein–protein complexes35–42,59 and also whether the PBE1 produces estimates of ΔGel, , and ΔΔGel consistent with those obtained from ESMs for these complexes.
3. METHODS
All MD simulations were run with a modified version of NAMD 2.9.60 SHAKE was used to constrain the hydrogens. All simulations used the TIP3P water model61 modified for use with the CHARMM force field,62 a constant temperature of 300 K, a constant pressure of 1 atm, periodic boundary conditions, particle mesh Ewald for the electrostatics, and a 2 fs time step. In all simulations, all protein atoms were fixed. All A, V, and their derivatives with respect to the atomic coordinates were computed with the DAlphaBall program.63 The solvent-accessible surface was used64 with the van der Waals radii taken from the CHARMM36 force field.62,65,66 A probe radius of 1.7682 Å was used rather than the normal choice of 1.4 Å because it corresponds to the vdW radius of the oxygen atom in the water model, which for a chargeless protein is the only interaction with solvent via the Lennard-Jones force. This choice has been used in previous works.24,34
All PBE calculations were run with the Adaptive Poisson– Boltzmann Solver (APBS)67 with a temperature of 300 K, an interior dielectric constant of 1 (because the CHARMM36 force field is not polarizable), an exterior dielectric constant of 96.7 (to match the dielectric constant of TIP3P water68), the solvent-excluded surface defined with a probe radius of 1.4 Å, no salt, autofocusing with a fine grid 20 Å larger than the molecule in each dimension, a coarse grid 1.7 times the size of the molecule in each dimension, and a fine-grid spacing of either 0.5 or 0.55 Å (to check convergence with respect to grid spacing).
3.1. Structure Preparation
The coordinates of the 9 protein–protein complexes35–42,59 were taken from the RCSB protein databank69 with no minimization or equilibration of these initial structures. These atomic coordinates remained fixed through all of the remaining calculations. All crystal waters and nonprotein atoms were removed. The chains from these structure files used in each calculation are shown in Table 1.
Table 1.
Chains from the Structure Files Used in Each Calculationa
| molecule first chain second chain |
1ACB E I |
1AVX A B |
1BEB A B |
1BRS A D |
1EAW A B |
1EMV A B |
1HE1 A C |
1PPE E I |
1UDI E I |
|
|---|---|---|---|---|---|---|---|---|---|---|
| protein A | A | 48978 | 44830 | 35787 | 24965 | 48348 | 20160 | 29952 | 44429 | 50395 |
| γrep | 0.017 | 0.020 | 0.017 | 0.023 | 0.012 | 0.024 | 0.019 | 0.017 | 0.017 | |
| R2 | 0.51 | 0.55 | 0.46 | 0.61 | 0.40 | 0.64 | 0.53 | 0.49 | 0.47 | |
| γvdw | 0.040 | 0.042 | 0.034 | 0.051 | 0.027 | 0.048 | 0.040 | 0.041 | 0.039 | |
| R2 | 0.15 | 0.14 | 0.12 | 0.20 | 0.08 | 0.21 | 0.15 | 0.14 | 0.13 | |
| protein B | A | 15699 | 39342 | 35458 | 20705 | 14221 | 30953 | 39824 | 8048 | 20087 |
| γrep | 0.025 | 0.019 | 0.020 | 0.024 | 0.027 | 0.019 | 0.018 | 0.032 | 0.022 | |
| R2 | 0.64 | 0.54 | 0.54 | 0.64 | 0.66 | 0.55 | 0.50 | 0.73 | 0.55 | |
| γvdw | 0.051 | 0.041 | 0.043 | 0.051 | 0.051 | 0.038 | 0.035 | 0.062 | 0.042 | |
| R2 | 0.19 | 0.15 | 0.16 | 0.19 | 0.22 | 0.13 | 0.10 | 0.34 | 0.15 | |
| complex | A | 62232 | 81657 | 69623 | 43280 | 60246 | 48726 | 66480 | 49854 | 67477 |
| γrep | 0.016 | 0.012 | 0.015 | 0.013 | 0.012 | 0.016 | 0.012 | 0.018 | 0.013 | |
| R2 | 0.49 | 0.38 | 0.40 | 0.39 | 0.37 | 0.49 | 0.36 | 0.49 | 0.38 | |
| γvdw | 0.042 | 0.029 | 0.030 | 0.026 | 0.025 | 0.037 | 0.028 | 0.045 | 0.030 | |
| R2 | 0.15 | 0.08 | 0.09 | 0.07 | 0.06 | 0.13 | 0.09 | 0.16 | 0.08 | |
| binding | γrep | 0.017 | 0.014 | 0.017 | 0.017 | 0.014 | 0.019 | 0.015 | 0.019 | 0.016 |
| R2 | 0.50 | 0.40 | 0.47 | 0.47 | 0.40 | 0.52 | 0.42 | 0.50 | 0.44 | |
| γvdw | 0.041 | 0.031 | 0.034 | 0.037 | 0.028 | 0.039 | 0.033 | 0.044 | 0.036 | |
| R2 | 0.15 | 0.09 | 0.12 | 0.14 | 0.09 | 0.14 | 0.12 | 0.16 | 0.12 |
Horizontal sections labeled protein A, protein B, and complex give estimates of γrep and γvdw obtained by fitting ∂ΔGrep/∂xi and ∂ΔGvdw/∂xi versus ∂A/∂xi and the squares (R2) of the Pearson’s correlation coefficients of these plots. The horizontal section labeled binding contains estimates of γrep and γvdw obtained by fitting ∂ΔΔGrep/∂xi and ∂ΔΔGvdw/∂xi versus ∂ΔA/∂xi and the squares (R2) of the Pearson’s correlation coefficients of these plots. The plots are in the Supporting Information. All A are in units of Å2, and all γvdw and γrep are in units of kcal mol−1 Å−2.
3.2. Potential and Free Energy Components
We defined ΔGvdw to be the free energy required to move from an ensemble where the solute and solvent did not interact to one where the interaction potential between an atom i in the solute and an atom j in the solvent was given by the Lennard-Jones potential
| (10) |
where εij is the well depth, and is the location of the minimum of . Our definitions of ΔGrep and ΔGatt followed the Weeks–Chandler–Andersen breakdown,25,26 where ΔGrep was the free energy required to move from an ensemble where the solute and solvent did not interact to one where the interaction potential between an atom i in the solute and an atom j in the solvent was given by
| (11) |
Next, we defined ΔGatt to be the free energy gained by moving from an ensemble where the solute–solvent potential was given by to one where it was given by . This process could also be described as turning on the attractive part of
| (12) |
Finally, we defined ΔGel to be the free energy gained by moving from an ensemble where the solute–solvent potential was given by to one where it was given by
| (13) |
where qi and qj are the charges on atoms i and j, respectively, and ε0 is the permittivity of free space.
We then defined , and , where the summations were taken over all solute–solvent atom pairs.
Similarly to the definition of ΔΔGdesol in the Introduction, , and can be defined by
| (14) |
| (15) |
| (16) |
| (17) |
where , and are the ΔGrep of the complex and its first and second components, , and are the ΔGatt of the complex and its first and second components, , and are the ΔGvdw of the complex and its first and second components, and , and are the ΔGel of the complex and its first and second components, respectively.
ΔΔG and its components were defined as follows
| (18) |
| (19) |
| (20) |
| (21) |
| (22) |
where and , where these summations were taken over atom pairs with one atom in each component of the complex.
3.3. Free Energy Calculations
For the 1BRS complex, we computed ΔGatt and ΔGel by backward and forward free energy perturbation (FEP) and thermodynamic integration (TI).43,44 To do so, we defined the λ-dependent potentials
| (23) |
| (24) |
To compute ΔGatt for each component and the complex, 11 1 ns simulations were run at λ values ranging from 0 to 1 in intervals of 0.1. To compute ΔGel for the components of the complex, 21 1 ns simulations were run at λ values ranging from 0 to 1 in intervals of 0.05. To compute ΔGel for the complex 1 ns simulations were run at the following λ values: 0, 0.025, 0.05, 0.075, 0.1, 0.125, 0.15, 0.175, 0.2, 0.225, 0.25, 0.275, 0.3, 0.325, 0.35, 0.375, 0.4, 0.425, 0.45, 0.475, 0.5, 0.525, 0.55, 0.575, 0.6, 0.625, 0.65, 0.675, 0.7, 0.725, 0.75, 0.775, 0.8, 0.825, 0.85, 0.8625, 0.875, 0.8875, 0.9, 0.9125, 0.925, 0.9375, 0.95, 0.9625, 0.975, 0.9875, and 1.
For the other 8 complexes, and were computed using eqs 1 and 2 for both the components and the complexes by running 1 ns simulations where the interaction potential between the solute and solvent was , and Uij. Estimates of , and were extracted from these same simulations.
Initial structures for all of these simulations were obtained by immersing the structure in a water box that was 20 Å longer in each dimension than the molecule and adding either Na+ or Cl− ions to neutralize the system. The structures were then minimized for 500 steps with the solute–solvent potential set as Uij. Copies of these minimized structure then underwent equilibration at each value of λ. The temperature of each of these systems was increased from 0 to 300 K in units of 25 K with 1000 steps of simulation time at each temperature.
3.4. Free Energy Derivatives
Computing ΔGrep and ΔGvdw for systems as large as these protein–protein complexes would require a great deal of computational time due to the drying of the molecular interior, so we instead adopted a procedure used in our previous publications,24,34 computing the derivatives of ΔGrep and ΔGvdw with respect to the coordinates (xi) of the centers of the fixed protein atoms
| (25) |
where this average was taken in the ensemble defined by Urep, and
| (26) |
where these derivatives were computed from the same simulations used to compute the LRT and SSP estimates of ΔGatt. These derivatives were computed for each Cartesian coordinate of each protein atom with the rest of the coordinates held fixed.
If ΔGrep and ΔGvdw were linear functions of either A or V, as expected from eqs 6 and 7, then the slopes of plots of these quantities versus A would be γrep and γvdw, respectively. Similarly, if and were linear in ΔA, then plots of
| (27) |
| (28) |
versus ΔA would have slopes of γrep and γvdw, respectively. Estimates of γrep and γvdw obtained from such plots are shown in Table 1.
4. RESULTS
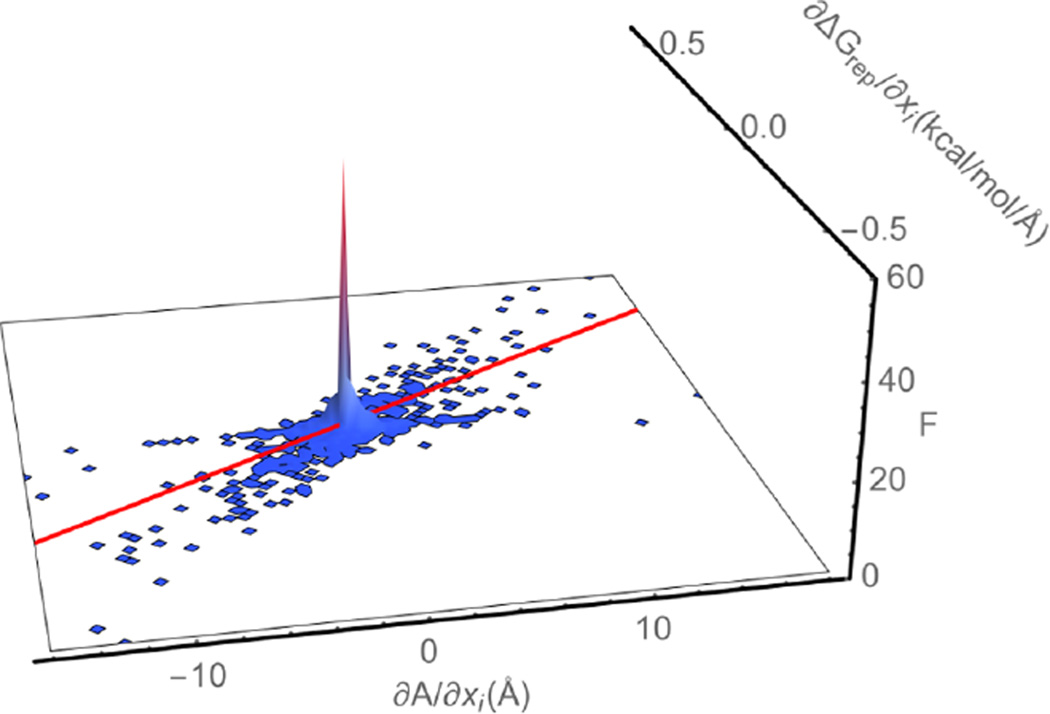
Table 1 gives estimates of γrep and γvdw obtained by fitting least-squares lines to plots of ∂ΔGrep/∂xi and ∂ΔGvdw/∂xi versus ∂A/∂xi and and versus ∂ΔA/∂xi. The Supporting Information contains these plots and analyses of the precision of the underlying quantities. If eqs 6–9 were valid, then the squares of the Pearson’s correlation coefficients (R2) would be close to 1, and the estimates of γrep and γvdw would be consistent from molecule to molecule as well as between estimates obtained from solvation free energies and those obtained from binding free energies. Instead, plots of ∂ΔGrep/∂xi and ∂ΔΔGrep/∂xi versus ∂A/∂xi and ∂ΔA/∂xi revealed very weak correlations between these quantities. Figure 1 shows a typical example of a plot of ∂ΔGrep/∂xi versus ∂A/∂xi for one protein molecule. These findings are in agreement with our previous findings that the geometry and chemical environment of an atom’s surroundings affects how changes in that atom’s position change ΔGrep.24,34
Figure 1.
A plot of the probability density (F) of a given combination of the derivative (∂ΔGrep/∂xi) of the repulsive component (ΔGrep) of the free energy required to insert an uncharged molecule into solution with respect to the atomic coordinates (xi) and the derivative (∂A/∂xi) of the solvent-accessible surface area (A) with respect to xi for the second chain of the 1UDI complex. The red line is a least-squares line drawn through all of the points. Its slope provides an estimate of γrep for this protein, and it is equal to 0.022 kcal mol−1 Å−2. Those atoms for which both ∂ΔGrep/∂xi and ∂A/∂xi were equal to 0 were not included when computing F. If they had been included, the peak in F would have been significantly larger.
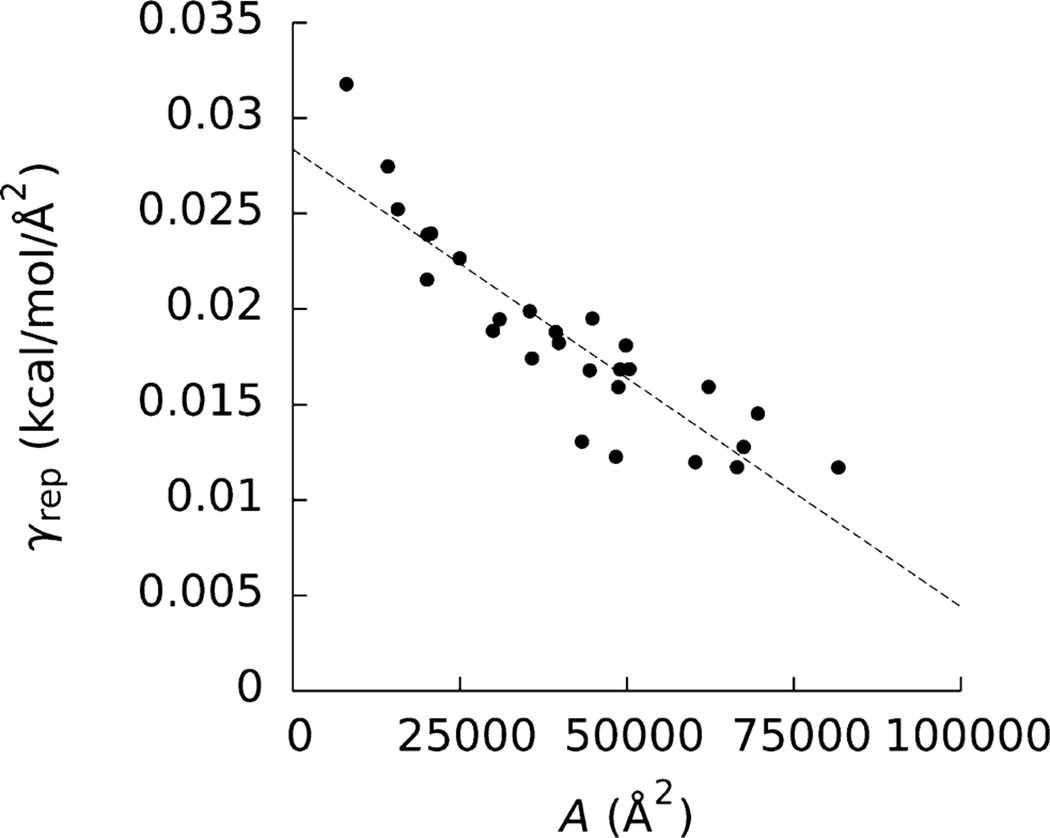
The estimates of γrep obtained from plots of ∂ΔGrep/∂xi versus ∂A/∂xi not only differed between molecules but decreased with increasing A (Table 1 and Figure 2). These estimates of γrep ranged from 0.012 to 0.032 kcal mol−1 Å−2, and these values were generally smaller than those we found previously for decaalanine, decaglycine, and alkanes, which generally had smaller A than the proteins examined here.24,34 This observation appears to contradict the notion that a critical A exists for which molecules with larger A have ΔGrep that are linear in A and below which ΔGrep is linear in V. This finding may indicate that the vicinity of the biomolecule becomes progressively “dryer” as the size of the cavity increases. Eq 7 is not consistent with these findings.
Figure 2.
Slopes (γrep) of the least-squares fit of the derivatives (∂Grep/∂xi) of the repulsive (ΔGrep) component of the cavity-insertion energy (ΔGvdw) with respect to the coordinates (xi) of the atomic centers versus derivatives (∂A/∂xi) of the solvent-accessible surface areas (A) with respect to xi versus A for all complexes and components in this study.
Table 2 shows estimates of ΔGatt and ΔGel for one of the complexes (1BRS) and its components and , and ΔΔGel for this complex given by FEP, TI, LRT, and SSP. These data show that the LRT gives good estimates of , and ΔΔGatt for this complex, confirming the validity of eq 1 for this complex. However, SSP gave estimates of , and ΔΔGatt that differed significantly from those given by FEP, indicating that eqs 3 and 4 were not valid for this system.
Table 2.
Attractive (ΔGatt) and Electrostatic Components (ΔGel) of the Solvation Energy of the 1BRS Complex and Its Two Components and the Attractive ( and ΔΔGatt) and Electrostatic ( and ΔΔGel) Components of the Desolvation and Binding Free Energiesa
|
|
ΔΔGatt | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| forward FEP | −315 | −259 | −458 | 116 | 2 | ||||
| backward FEP | −312 | −259 | −458 | 113 | −1 | ||||
| TI | −311 | −260 | −457 | 114 | −1 | ||||
| LRT | −325 | −267 | −480 | 112 | −2 | ||||
| SSP λ = 0 | −165 | −143 | −218 | 89 | −25 | ||||
| SSP λ = 1 | −485 | −392 | −742 | 135 | 21 | ||||
| ΔΔGel | |||||||||
| forward FEP | −1385 | −1556 | −2449 | 492 | −17 | ||||
| backward FEP | −1387 | −1558 | −2443 | 501 | −9 | ||||
| TI | −1388 | −1562 | −2448 | 501 | −9 | ||||
| LRT | −1517 | −1694 | −2691 | 520 | 10 | ||||
| SSP λ = 1 | −1528 | −1670 | −2678 | 520 | 10 | ||||
These energies were obtained by forward and backward free energy perturbation (FEP), thermodynamic integration (TI), linear response theory (LRT), and single-step perturbation (SSP) from either the beginning (λ = 0) or ends (λ = 1) of the integration. All energies are in units of kcal/mol.
The data in Table 2 show that the LRT yielded estimates of ΔGel that were more than 100 kcal/mol different from those obtained with FEP, indicating that eq 2 is poor for this system. FEP estimates of and ΔΔGel also differed significantly from those given by LRT.
Table 3 shows the R2, slopes, and y-intercepts of least-squares lines comparing various computed quantities for the 9 protein–protein complexes. All of these plots and analyses of the precisions of the resulting quantities are in the Supporting Information. These data show that the estimates of ΔGatt obtained from the LRT were highly correlated with A, but its estimates of and ΔΔGatt were not strongly correlated with ΔA, demonstrating that two theories can yield similar results for ΔG and very different results for ΔΔG.
Table 3.
Slopes (m), y-Intercepts (b), and the Squares of Pearson’s Correlation Factors (R2) of Least-Squares Lines of the Quantities Listed in the y-Column versus Those in the x-Columna
| x | y | R2 | m | b | ||
|---|---|---|---|---|---|---|
| A | 0.992 | −0.050 | −19 | |||
| ΔA | 0.64 | −0.077 | −33 | |||
| ΔA | 0.25 | −0.040 | −77 | |||
| 0.97 | 3.4 | 203 | ||||
| 0.64 | 1.60 | −37 | ||||
| 0.38 | 0.89 | 19 | ||||
| 0.9991 | 0.58 | −38 | ||||
| 0.97 | 0.60 | 35 | ||||
| 0.89 | 0.57 | −10.5 | ||||
| 0.9997 | 1.01 | 16 | ||||
| 0.99997 | 1.0007 | 2.0 | ||||
| 0.9999 | 1 | 1.6 | ||||
| 0.96 | 1.03 | −138 | ||||
| 0.92 | 0.40 | 125 | ||||
| 0.42 | −7.57 | 510 |
The corresponding plots are in Supporting Information. All energies are in kcal/mol. All areas and changes in area are in units of Å2. All m and b are in the corresponding units.
Additionally, the data in Table 3 show that both eqs 3 and 4 produced estimates of ΔGatt that were highly correlated with those obtained from the LRT (R2 > 0.97), but eq 3 systematically underestimated ΔGatt whereas eq 4 overestimated it. Table 3 also shows that the estimates of obtained from eq 3 were not highly correlated with those obtained from LRT. Conversely, the predictions of and ΔΔGatt obtained from eq 4 were reasonably well-correlated with those obtained from LRT, but these estimates were significantly larger than those obtained from LRT, indicating that combining these energy terms with the other components of ΔΔG could lead to unexpected results.
The data in Table 3 also show that the computations of , and ΔΔGel given by SSP are very highly correlated (R2 > 0.999) with those obtained from LRT. Therefore, if the LRT is sufficient to predict these quantities, then the SSP would be sufficient as well. Unfortunately, the results in Table 2 indicate that the LRT may not be sufficient for computing these quantities.
Table 3 also shows that the PBE yielded predictions ( and ) of ΔGel and that were highly correlated with those obtained from the LRT, but its estimates of ΔΔGel were not correlated with those obtained from LRT (R2 = 0.43). Once again, this finding shows that strong correlations between the predictions of ΔG given by two theories do not guarantee that the theories will produce highly correlated estimates of ΔΔG.
5. CONCLUSIONS
The calculations on protein–protein complexes presented in this paper allow us to draw some conclusions about the assumptions underlying CSMs. As in our previous work, eqs 6 and 7 were not consistent with the results. Neither ΔGrep nor ΔGvdw was proportional to A. Eqs 8 and 9 were inconsistent with our calculations. Neither nor was proportional to ΔA. Apparently, the idea that either ΔGvdw or ΔGrep is linear in A and that the resulting ΔGvdw can be combined with the ΔGel obtained from CSMs to obtain good estimates of ΔG is not valid for protein–protein complexes.
Additionally, although these results show that none of ΔGrep, ΔGvdw, are truly linear in A or ΔA, if we extract estimates of apparent γrep from the derivative plots described above, we find our estimates of γrep decrease with A. This finding contradicts the hypothesis that there is a well-defined A above which ΔGrep is linear in A and below which ΔGrep is linear in V for realistic biomolecular solutes.
We found that LRT was sufficient for estimating , and ΔΔGatt. However, although the SSP using either eq 3 or 4 yielded estimates of ΔGatt that were correlated with those given by the LRT, and the predictions of and ΔΔGatt given by eq 4 were also correlated to those given by LRT, the magnitudes of these quantities were significantly different from those given by LRT. Whether such estimates would be sufficient for in turn estimating ΔΔG is therefore much less clear. Additionally, whereas ΔGatt was correlated with A, neither nor ΔΔGatt was correlated with ΔA, highlighting the observation that two theories could produce correlated estimates of solvation free energies while producing uncorrelated estimates of binding free energies.
For the one protein in the data set where ΔGel was obtained with FEP, this estimate differed by more than 100 kcal/mol from those given by SSP and LRT, and the corresponding estimate of ΔΔGel obtained from FEP differed by more than 10 kcal/mol from those obtained from the LRT and SSP, implying that the LRT and SSP approximations may be problematic in some situations. However, for all of the complexes in the data set, the SSP gave estimates of , and ΔΔGel that were highly correlated with those given by the LRT. Therefore, if the LRT is a reasonable approximation for a system, the SSP is probably reasonable as well. These calculations will have to be repeated for other systems to determine how general are these conclusions.
In summary, many of the assumptions underlying common CSMs are brought into question by this work. The predictions yielded by often-used hydrophobic models disagreed with the results from FEP. LRT did not yield estimates of electrostatic energies that were consistent with those obtained with FEP. Furthermore, although the PBE yielded estimates of ΔGel that were highly correlated with those obtained from FEP, its estimates of ΔΔGel were not in good agreement with those obtained by FEP. In combination with our observation that ΔGatt was correlated with A but that ΔΔGatt was not, we can see that we cannot conclude that a theory will give good estimates of ΔΔG simply because its estimates of ΔG have some agreement with experimental data. Attempting to improve the estimates of ΔΔG given by CSMs by comparing their predictions of ΔG to experimental measurements may therefore not be productive.
Supplementary Material
Acknowledgments
The Robert A. Welch Foundation (H-0037), the National Science Foundation (CHE-1152876), and the National Institutes of Health (GM-037657) are thanked for partial support of this work. This research was performed in part using Xsede resources provided by the National Science Foundation.
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jctc.5b00684.
Scatter plots and data analyses (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Grochowski P, Trylska J. Continuum molecular electrostatics, salt effects, and counterion binding-A review of the Poisson-Boltzmann theory and its modifications. Biopolymers. 2008;89:93–113. doi: 10.1002/bip.20877. [DOI] [PubMed] [Google Scholar]
- 2.Bashford D, Case DA. Generalized Born models of macromolecular solvation effects. Annu. Rev. Phys. Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 3.Lounnas V, Pettitt BM, Phillips GN., Jr A global model of the protein-solvent interface. Biophys. J. 1994;66:601–614. doi: 10.1016/s0006-3495(94)80835-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarov V, Pettitt BM, Feig M. Solvation and hydration of proteins and nucleic acids: a theoretical view of simulation and experiment. Acc. Chem. Res. 2002;35:376–384. doi: 10.1021/ar0100273. [DOI] [PubMed] [Google Scholar]
- 5.Lin B, Pettitt BM. Electrostatic solvation free energy of amino acid side chain analogs: Implications for the validity of electrostatic linear response in water. J. Comput. Chem. 2011;32:878–85. doi: 10.1002/jcc.21668. [DOI] [PubMed] [Google Scholar]
- 6.Hirata F, editor. Understanding chemical reactivity: Molecular theory of solvation. Norwell, MA, USA: Kluwer Academic Publishers; 2003. [Google Scholar]
- 7.Truchon J-F, Pettitt BM, Labute P. A cavity corrected 3D-RISM functional for accurate solvation free energies. J. Chem. Theory Comput. 2014;10:934–941. doi: 10.1021/ct4009359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babu CS, Tembe BL. The role of solvent models in stabilizing nonclassical ions. Proc. - Indian Acad. Sci., Chem. Sci. 1987;98:235–240. [Google Scholar]
- 9.Baldwin R. Energetics of Protein Folding. J. Mol. Biol. 2007;371:283–301. doi: 10.1016/j.jmb.2007.05.078. [DOI] [PubMed] [Google Scholar]
- 10.Mohan V, Davis ME, McCammon JA, Pettitt BM. Continuum model calculations of solvation free energies: Accurate evaluation of electrostatic contributions. J. Phys. Chem. 1992;96:6428–6431. [Google Scholar]
- 11.Simonson T, Brünger AT. Solvation free energies estimated from macroscopic continuum theory: An accuracy assessment. J. Phys. Chem. 1994;98:4683–4694. [Google Scholar]
- 12.Nicholls A, Mobley DL, Guthrie JP, Chodera JD, Bayly CI, Cooper MD, Pande VS. Predicting small-molecule solvation free energies: An informal blind test for computational chemistry. J. Med. Chem. 2008;51:769–779. doi: 10.1021/jm070549+. [DOI] [PubMed] [Google Scholar]
- 13.Guthrie JP. A blind challenge for computational solvation free energies: Introduction and overview. J. Phys. Chem. B. 2009;113:4501–4507. doi: 10.1021/jp806724u. [DOI] [PubMed] [Google Scholar]
- 14.Mobley DL, Wymer KL, Lim NM, Guthrie JP. Blind prediction of solvation free energies from the SAMPL4 challenge. J. Comput.-Aided Mol. Des. 2014;28:135–150. doi: 10.1007/s10822-014-9718-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Morin P, Wang W, Kollman PA. Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J. Am. Chem. Soc. 2001;123:5221–5230. doi: 10.1021/ja003834q. [DOI] [PubMed] [Google Scholar]
- 16.Hou T, Wang J, Li Y, Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adler M, Beroza P. Improved ligand binding energies derived from molecular dynamics: Replicate sampling enhances the search of conformational space. J. Chem. Inf. Model. 2013;53:2065–2072. doi: 10.1021/ci400285z. [DOI] [PubMed] [Google Scholar]
- 18.Harris RC, Mackoy T, Fenley MO. A stochastic solver of the generalized Born model. Mol. Based Math. Biol. 2013;1:63–74. [Google Scholar]
- 19.Li M, Petukh M, Alexov E, Panchenko AR. Predicting the impact of missense mutations on protein-protein binding affinity. J. Chem. Theory Comput. 2014;10:1770–1780. doi: 10.1021/ct401022c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muddana HS, Fenley AT, Mobley DL, Gilson MK. The SAMPL4 host-guest blind prediction challenge: An overview. J. Comput.-Aided Mol. Des. 2014;28:305–317. doi: 10.1007/s10822-014-9735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y-L, Beroza P, Artis DR. Including explicit water molecules as part of the protein structure in MM/PBSA calculations. J. Chem. Inf. Model. 2014;54:462–469. doi: 10.1021/ci4001794. [DOI] [PubMed] [Google Scholar]
- 22.Sharp KA, Honig B. Electrostatic interactions in macromolecules: Theory and applications. Annu. Rev. Biophys. Biophys. Chem. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- 23.Cramer CJ, Truhlar DG. Implicit Solvation Models: Equilibria, Structure, Spectra, and Dynamics. Chem. Rev. 1999;99:2161–2200. doi: 10.1021/cr960149m. [DOI] [PubMed] [Google Scholar]
- 24.Harris RC, Pettitt BM. Effects of geometry and chemistry on hydrophobic solvation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14681–14686. doi: 10.1073/pnas.1406080111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weeks JD, Chandler D, Andersen HC. Role of repulsive forces in determining the equilibrium structure of simple liquids. J. Chem. Phys. 1971;54:5237–5247. [Google Scholar]
- 26.Chandler D, Weeks JD, Andersen HC. Van der Waals picture of liquids, solids, and phase transformations. Science. 1983;220:787–794. doi: 10.1126/science.220.4599.787. [DOI] [PubMed] [Google Scholar]
- 27.Ashbaugh HS, Kaler EW, Paulaitis ME. A “universal” surface area correlation for molecular hydrophobic phenomena. J. Am. Chem. Soc. 1999;121:9243–9244. [Google Scholar]
- 28.Gallicchio E, Kubo MM, Levy RM. Enthalpy-entropy and cavity decomposition of alkane hydration free energies: Numerical results and implications for theories of hydrophobic solvation. J. Phys. Chem. B. 2000;104:6271–6285. [Google Scholar]
- 29.Gallicchio E, Zhang LY, Levy RM. The SGB/NP hydration free energy model based on the surface generalized Born solvent reaction field and novel nonpolar hydration free energy estimators. J. Comput. Chem. 2002;23:517–529. doi: 10.1002/jcc.10045. [DOI] [PubMed] [Google Scholar]
- 30.Zacharias M. Continuum solvent modeling of nonpolar solvation: Improvement by separating surface area dependent cavity and dispersion contributions. J. Phys. Chem. A. 2003;107:3000–3004. [Google Scholar]
- 31.Choudhury N, Pettitt BM. On the mechanism of hydrophobic association of nanoscopic solutes. J. Am. Chem. Soc. 2005;127:3556–3567. doi: 10.1021/ja0441817. [DOI] [PubMed] [Google Scholar]
- 32.Choudhury N, Montgomery Pettitt B. Local density profiles are coupled to solute size and attractive potential for nanoscopic hydrophobic solutes. Mol. Simul. 2005;31:457–463. [Google Scholar]
- 33.Wagoner JA, Baker NA. Assessing implicit models for nonpolar mean solvation forces: The importance of dispersion and volume terms. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8331–8336. doi: 10.1073/pnas.0600118103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris RC, Drake JA, Pettitt BM. Multibody correlations in the hydrophobic solvation of glycine peptides. J. Chem. Phys. 2014;141:22D525. doi: 10.1063/1.4901886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frigerio F, Coda A, Pugliese L, Lionetti C, Menegatti E, Amiconi G, Schnebli HP, Ascenzi P, Bolognesi M. Crystal and molecular structure of the bovine α-chymotrypsin-eglin c complex at 2.0 Å resolution. J. Mol. Biol. 1992;225:107–123. doi: 10.1016/0022-2836(92)91029-o. [DOI] [PubMed] [Google Scholar]
- 36.Song HK, Suh SW. Kunitz-type soybean trypsin inhibitor revisited: Refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator. J. Mol. Biol. 1998;275:347–363. doi: 10.1006/jmbi.1997.1469. [DOI] [PubMed] [Google Scholar]
- 37.Brownlow S, Cabral JHM, Cooper R, Flower DR, Yewdall SJ, Polikarpov I, North ACT, Sawyer L. Bovine β-lactoglobulin at 1.8 Å resolution-Still an enigmatic lipocalin. Structure. 1997;5:481–495. doi: 10.1016/s0969-2126(97)00205-0. [DOI] [PubMed] [Google Scholar]
- 38.Buckle AM, Schreiber G, Fersht AR. Protein-protein recognition: Crystal structural analysis of a barnase-barstar complex at 2.0-Å resolution. Biochemistry. 1994;33:8878–8889. doi: 10.1021/bi00196a004. [DOI] [PubMed] [Google Scholar]
- 39.Kühlmann UC, Pommer AJ, Moore GR, James R, Kleanthous C. Specificity in protein-protein interactions: The structural basis for dual recognition in endonuclease colicin-immunity protein complexes. J. Mol. Biol. 2000;301:1163–1178. doi: 10.1006/jmbi.2000.3945. [DOI] [PubMed] [Google Scholar]
- 40.Würtele M, Wolf E, Pederson KJ, Buchwald G, Ahmadian MR, Barbieri JT, Wittinghofer A. How the Pseudomonas aeruginosa ExoS toxin downregulates Rac. Nat. Struct. Biol. 2001;8:23–26. doi: 10.1038/83007. [DOI] [PubMed] [Google Scholar]
- 41.Bode W, Greyling HJ, Huber R, Otlewski J, Wilusz T. The refined 2.0 Å X-ray crystal structure of the complex formed between bovine β-trypsin and CMTI-I, a trypsin inhibitor from squash seeds (Cucurbita maxima) Topological similarity of the squash seed inhibitors with the carboxypeptidase A inhibitor from potatoes. FEBS Lett. 1989;242:285–292. doi: 10.1016/0014-5793(89)80486-7. [DOI] [PubMed] [Google Scholar]
- 42.Savva R, Pearl LH. Nucleotide mimicry in the crystal structure of the uracil-DNA glycosylase-uracil glycosylase inhibitor protein complex. Nat. Struct. Biol. 1995;2:752–757. doi: 10.1038/nsb0995-752. [DOI] [PubMed] [Google Scholar]
- 43.Beveridge DL, DiCapua FM. Free energy via molecular simulation: Applications to chemical and biomolecular systems. Annu. Rev. Biophys. Biophys. Chem. 1989;18:431–492. doi: 10.1146/annurev.bb.18.060189.002243. [DOI] [PubMed] [Google Scholar]
- 44.Straatsma TP, McCammon JA. Computational alchemy. Annu. Rev. Phys. Chem. 1992;43:407–435. [Google Scholar]
- 45.Åqvist J, Hansson A. On the validity of electrostatic linear response in polar solvents. J. Phys. Chem. 1996;100:9512–9521. [Google Scholar]
- 46.Kokubo H, Harris RC, Asthagiri D, Pettitt BM. Solvation free energies of alanine peptides: The effect of flexibility. J. Phys. Chem. B. 2013;117:16428–16435. doi: 10.1021/jp409693p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young T. An essay on the cohesion of fluids. Philos. Trans. R. Soc. 1805;95:65–87. [Google Scholar]
- 48.Stillinger FH. Structure in aqueous solutions of nonpolar solutes from the standpoint of scaled-particle theory. J. Solution Chem. 1973;2:141–158. [Google Scholar]
- 49.Pierotti RA. A scaled particle theory of aqueous and nonaqueous solutions. Chem. Rev. 1976;76:717–726. [Google Scholar]
- 50.Sharp KA, Nicholls A, Fine RF, Honig B. Reconciling the magnitude of the microscopic and macroscopic hydrophobic effects. Science. 1991;252:106–109. doi: 10.1126/science.2011744. [DOI] [PubMed] [Google Scholar]
- 51.Sitkoff D, Sharp KA, Honig B. Accurate calculation of hydration free energies using macroscopic solvent models. J. Phys. Chem. 1994;98:1978–1988. [Google Scholar]
- 52.Lum K, Chandler D, Weeks JD. Hydrophobicity at small and large length scales. J. Phys. Chem. B. 1999;103:4570–4577. [Google Scholar]
- 53.Huang DM, Chandler D. Temperature and length scale dependence of hydrophobic effects and their possible implications for protein folding. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8324–8327. doi: 10.1073/pnas.120176397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hummer G, Garde S, García AE, Pratt LR. New perspectives on hydrophobic effects. Chem. Phys. 2000;258:349–370. [Google Scholar]
- 55.Rajamani S, Truskett TM, Garde S. Hydrophobic hydration from small to large lengthscales: Understanding and manipulating the crossover. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9475–9480. doi: 10.1073/pnas.0504089102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu CY, Kokubo H, Lynch GC, Bolen DW, Pettitt BM. Backbone additivity in the transfer model of protein solvation. Protein Sci. 2010;19:1011–1022. doi: 10.1002/pro.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kokubo H, Hu CY, Pettitt BM. Peptide conformational preferences in osmolyte solutions: Transfer free energies of decaalanine. J. Am. Chem. Soc. 2011;133:1849–1858. doi: 10.1021/ja1078128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harris RC, Mackoy T, Fenley MO. Problems of robustness in Poisson-Boltzmann binding energies. J. Chem. Theory Comput. 2015;11:705–712. doi: 10.1021/ct5005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Friedrich R, Fuentes-Prior P, Ong E, Coombs G, Hunter M, Oehler R, Pierson D, Gonzalez R, Huber R, Bode W, Madison EL. Catalytic domain structures of MT-SP1/matriptase, a matrix-degrading transmembrane serine proteinase. J. Biol. Chem. 2002;277:2160–2168. doi: 10.1074/jbc.M109830200. [DOI] [PubMed] [Google Scholar]
- 60.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 62.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiókiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 63.Edelsbrunner H, Koehl P. The weighted-volume derivative of a space-filling diagram. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2203–2208. doi: 10.1073/pnas.0537830100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee B, Richards FM. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 65.MacKerell AD, Jr, Feig M, Brooks CL., III Improved treatment of the protein backbone in empirical force fields. J. Am. Chem. Soc. 2004;126:698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 66.Best RB, Zhu X, Shim J, Lopes PEM, Mittal J, Feig M, MacKerell AD., Jr Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Höchtl P, Boresch S, Bitomsky W, Steinhauser O. Rationalization of the dielectric properties of common three-site water models in terms of their force field parameters. J. Chem. Phys. 1998;109:4927–4937. [Google Scholar]
- 69.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.