Abstract
Recently, it was determined that representatives of the riboswitch candidates called ykkC and ykkC-III directly bind free guanidine. Guanidine-binding ykkC motif RNAs, now renamed guanidine-I riboswitches, were demonstrated to commonly regulate the expression of genes encoding guanidine carboxylases, as well as others encoding guanidine efflux proteins such as EmrE and SugE. Likewise, genes encoding similar efflux proteins are associated with ykkC-III motif RNAs, which have now been renamed guanidine-III riboswitches. Prior to the validation of guanidine as the ligand for these newly-established riboswitch classes, another RNA motif was discovered by comparative genomic analysis and termed mini-ykkC due to its small size and gene associations similar to the original ykkC motif. It was hypothesized that these distinct RNA structures might respond to the same ligand. However, the small size and repetitive nature of mini-ykkC RNAs suggested that it might respond to ligand via the action of a protein factor. Herein we demonstrate that, despite its extremely simple architecture, mini-ykkC motif RNAs constitute a distinct class of guanidine-sensing RNAs, called guanidine-II riboswitches. Surprisingly, each of the two stem loops that comprise the mini-ykkC motif appears to directly bind free guanidine in a cooperative manner. These findings reveal that bacteria make extensive use of diverse guanidine-responsive riboswitches to overcome the toxic effects of this compound.
Keywords: aptamer, guanidine-II riboswitch, in-line probing, noncoding RNA, mini-ykkC motif
INTRODUCTION
The ykkC motif is a conserved and highly-structured bacterial noncoding RNA that was first reported in 2004.1 For over a decade, the cognate ligand for the ykkC riboswitch remained unidentified, mostly due to the apparent lack of connection amongst genes regulated by this RNA. Despite the challenges associated with this orphan riboswitch candidate, the search for the ligand for the putative ykkC riboswitch class continued. This effort was driven by the fact that the unidentified ligand would have an important, but likely unknown or underappreciated, role in bacteria because this riboswitch candidate is so common and widespread.2
The importance of this mystery ligand was further implicated by the discovery of two additional RNA motifs with highly similar gene associations, termed mini-ykkC3 and ykkC-III.4 It was predicted that all three types of ykkC RNA motifs might respond to this same, yet-to-be-identified compound. Recently, representatives of the ykkC candidate riboswitch class were experimentally validated to sense and respond to free guanidine.5 Consequently, the subset of ykkC motif RNAs that have been demonstrated or are predicted to respond to this ligand have been renamed guanidine-I riboswitches. This work further revealed that free guanidine is naturally produced by bacteria and that multiple pathways exist to minimize its toxic effects. Similarly, in a companion paper6 to the current report, we provide evidence that members of the ykkC-III class also function as guanidine-sensing RNAs, named guanidine-III riboswitches.
Upon the experimental validation of guanidine-I riboswitches, we sought to assess the hypothesis that mini-ykkC motif RNAs might function as a distinct class of guanidine-sensing riboswitches. However, the mini-ykkC motif was initially considered a poor candidate for a riboswitch class due to its low structural complexity and its limited number of conserved nucleotides.3 Additionally, repetitive stem loops with identical sequences were likely to be more characteristic of binding sites for RNA-binding protein homo-complexes. Thus, it seemed more plausible that mini-ykkC RNAs would require a protein partner to perform their regulatory role.
The mini-ykkC motif commonly controls the expression of small multidrug resistance (SMR) family efflux pumps including those annotated as EmrE and SugE. In our previous work, we show that a representative of the SMR proteins typically controlled by guanidine-I riboswitches is able to bind guanidine selectively.5 Additionally, guanidine-I riboswitches and mini-ykkC RNAs are commonly located adjacent to genes originally annotated as urea carboxylase and associated proteins, and genes for allophanate hydrolase. Our previous work revealed that a representative urea carboxylase protein from O. sagaranensis prefers guanidine as a substrate, with a 40-fold higher catalytic efficiency for guanidine versus urea.5 Thus, mini-ykkC motif RNAs remained intriguing candidates as guanidine-responsive regulatory elements despite their simple architecture.
In this report, we present results confirming that mini-ykkC RNAs directly and selectively respond to guanidine. Also described are additional structural and biochemical attributes of mini-ykkC motif RNAs, which we rename as guanidine-II riboswitches. Most importantly, guanidine-II riboswitches appear to bind two guanidine molecules cooperatively, and therefore exhibit unusually complex characteristics for such a small and simple motif.
MATERIALS AND METHODS
Chemicals and Oligonucleotides
All chemicals and chemically synthesized oligonucleotides were purchased from Sigma-Aldrich with the exception of guanidinosuccinic acid (Alfa Chemistry). [γ-32P]-ATP was purchased from PerkinElmer. Enzymes were purchased from New England BioLabs, unless otherwise noted. A complete list of DNA and RNA oligonucleotides used in this study can be found in Table S1.
Bioinformatics Analyses
Additional examples of mini-ykkC motif RNAs were identified using Infernal 1.17 to search RefSeq version 63 plus additional environmental microbial databases as described previously.8 Iterative searches for new sequences were performed based on the previously published alignment of the mini-ykkC motif,3 revealing over 800 unique examples. The consensus sequence and secondary structure model was constructed using R2R software.9
RNA Oligonucleotide Preparation
Synthetic DNA oligonucleotides (Supplemental Table S1) containing a 5′-terminal T7 RNA polymerase (T7 RNAP) promoter followed by a template region coding for the aptamer of interest were combined in equimolar amount with their complementary strand to form a double-stranded DNA (dsDNA) template. The desired RNA constructs were prepared from these dsDNAs by in vitro transcription for four hours at 37°C in a 25 or 50 µL reaction volume containing laboratory-prepared T7 RNAP (2 U µL−1), 80 mM HEPES-KOH (pH 7.5 at 23°C), 24 mM MgCl2, 2 mM spermidine, 40 mM DTT, and 2 mM of each NTP.
The desired RNA products were purified via denaturing (8 M urea) 10% PAGE (National Diagnostics). For the truncated 30 emrE RNA construct, the synthetic RNA was chemically synthesized and purified via denaturing PAGE. The gel slice containing the appropriately-sized band was excised and the RNA extracted via the crush-soak method for 30 min at 23°C in 350 µL containing 200 mM NaCl, 10 mM Tris-HCl (pH 7.5 at 23°C) and 1 mM EDTA. RNA in the resulting supernatant was precipitated by the addition of 20 µL 3 M sodium acetate (pH 5.5 at 23°C) and 750 µL cold 100% ethanol, and incubated for up to 30 min at −20°C. The RNA was subsequently pelleted via centrifugation, the supernatant removed, and the remaining pellet was dried and resuspended in 20 µL of deionized water (dH2O). To generate 5′ 32P-labeled RNAs, the 5′-terminal phosphate was removed via rAPid alkaline phosphatase (Roche Life Sciences) according to manufacturer instructions. Subsequently, 20 pmol of dephosphorylated RNA was radiolabeled using T4 polynucleotide kinase in a 20 µL reaction containing 25 mM CHES (pH 9.0 at 23°C), 5 mM MgCl2, 3 mM DTT, and 20 µCi [γ-32P]-ATP and incubated for one hour. The resulting radiolabeled RNAs were purified before use by denaturing 10% PAGE as described above.
RNA In-line Probing Analyses
In-line probing assays were performed largely as previously described.10,11 Briefly, 5′-labeled RNAs were incubated at 23°C either with or without candidate ligands for 42 to 48 hours in 10 µL reactions containing 20 mM MgCl2, 100 mM KCl, 50 mM Tris-HCl (pH 8.3 at 23°C). The products of spontaneous cleavage were subsequently analyzed via denaturing 10% PAGE and visualized using a phosphorimager (GE Healthcare Life Sciences).
Fraction bound values were calculated by varying the ligand concentration in separate reactions and quantifying the changes in band intensity at nucleotides that exhibit structural modulation. Band intensities were corrected for loading differences based on the analysis of a band that does not alter its intensity upon ligand addition. The resulting band intensity values for specific regions were normalized to a fractional value between 0 and 1, the values for regions with band intensities that decrease with increasing ligand concentration were inverted, and all resulting values were plotted as ‘fraction bound’ using GraphPad Prism 7. The KD and Hill coefficients were determined by plotting the average of the fraction bound at the indicated sites of modulation as a function of the logarithm of ligand concentration and using a sigmoidal four parameter logistic fit.
RESULTS AND DISCUSSION
Additional mini-ykkC Motif RNA Representatives Form Highly-Conserved Symmetrical Loops
Initial bioinformatics revealed 208 representatives of the mini-ykkC motif RNA.3 An updated homology search was conducted using Infernal7 to further expand the number of representatives. Approximately 800 unique examples were found mainly in Proteobacteria, with additional examples in Actinobacteria, Chrysiogenetes, Cyanobacteria, Planctomycetes, and Verrucomicrobia.
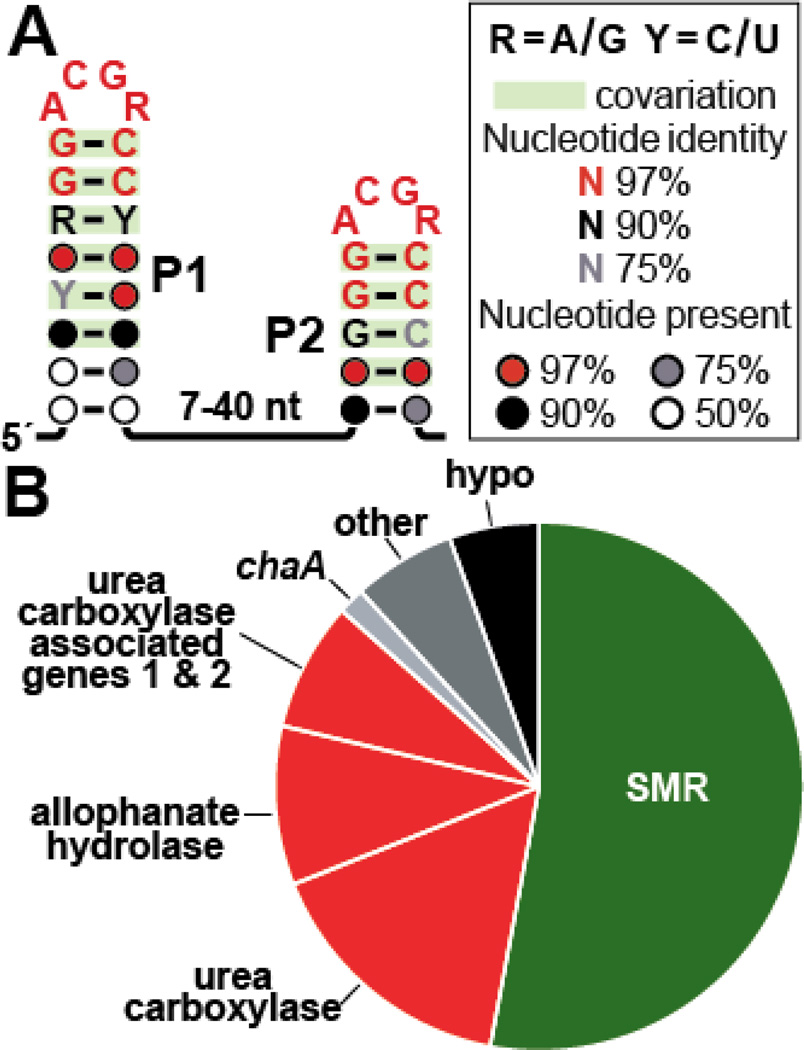
We used the updated homology search results to refine the consensus sequence and secondary structure model for this motif (Figure 1A). Overall, mini-ykkC motif RNAs are composed of two GC-rich stems, called pairing element 1 and 2 (P1 and P2), that are capped with ‘ACGR’ (R represents either G or A) tetraloops. These hairpins are connected by a non-conserved linker formed by a minimum of seven nucleotides. The presence of the repetitive loop sequences flanked by GC-rich stem sequences was suggestive of the presence of two binding sites for a possible riboswitch ligand or for a dimeric protein factor.
Figure 1.
Structure and function of guanidine-II riboswitches. (A) The consensus sequence and structural model for the ~850 unique examples of mini-ykkC motif (guanidine-II) RNAs. Covarying base-pairs (indicated by green shading) are found in the P1 and P2 stems. Each stem has an extremely highly conserved ‘ACGR’ loop. (B) Genes whose expression is controlled by guanidine-II riboswitches, including those encoding predicted small multidrug resistance (SMR) transporters (green), urea (guanidine) carboxylases and their associated proteins (red), and other (gray) or hypothetical (hypo, black) genes.
Examples of mini-ykkC motif RNAs frequently occur in close proximity (typically ~30 nucleotides) to the ribosome binding site (RBS) and start codon of each adjoining open reading frame (ORF). This arrangement is characteristic of regulatory RNA domains such as riboswitches. Moreover, if mini-ykkC motif RNAs enact their regulatory response in a manner consistent with guanidine-I riboswitches, then genes that alleviate guanidine toxicity should be expressed when guanidine concentrations are high. Accordingly, guanidine should interact with the ligand-binding aptamer domain in a manner that exposes the RBS and allows the translational machinery to initiate protein production.
Genes Associated with mini-ykkC are Related to Guanidine Metabolism
The genes associated with mini-ykkC motif RNAs (Figure 1B) largely overlap with those controlled by guanidine-I riboswitches, although the SMR transporters are more highly represented. Comparative analysis suggests that these predicted SMR proteins fall into the category of SugE-like heterodimers (Randy Stockbridge, personal communication). The protein products from these genes are distinct from the well-characterized canonical EmrE protein from E. coli, which has broad specificity for small toxic compounds.12 Members of mini-ykkC are also associated with genes annotated as urea carboxylases, which are also commonly controlled by guanidine-I riboswitches. As noted above, we recently demonstrated that a representative of this enzyme class favors guanidine as a substrate over urea by 40 fold.5
Other gene associations, such as those encoding urea carboxylase associated proteins 1 and 2, and allophanate hydrolases, are also consistent with our hypothesis that mini-ykkC motif RNAs regulate gene expression in response to guanidine. An additional gene, chaA, whose expression is occasionally associated with mini-ykkC, encodes a predicted Ca2+/H+ antiporter. Perhaps this activity assists in maintaining the membrane potential of the cell as it ejects positively charged guanidinium from the cytoplasm. The remaining genes associated with mini-ykkC motif RNAs are either very rarely affiliated, or have unknown functions (Figure 1B).
Intriguingly, mini-ykkC motif RNAs are occasionally encoded within apparent operons for a putative guanidine degradation pathway. Such genomic arrangements typically consist of genes annotated as encoding nitrate/sulfate/bicarbonate transporters followed by urea carboxylase associated proteins, urea carboxylase and finally allophanate hydrolase. In many cases a guanidine-I riboswitch, which is a transcriptional ‘ON’ switch, appears in the 5′ UTR between the promoter and the first gene. Further into the operon, a mini-ykkC element is found in the intergenic region between the urea carboxylase associated genes and urea carboxylase. We predict that mini-ykkC is a translational ‘ON’ switch that regulates expression of the urea carboxylase gene immediately downstream by controlling access to the RBS and thereby translation initiation.
Initially, this seems like either an unnecessarily complex arrangement of two regulatory RNAs that respond to the same ligand, or hints at the existence of two distinct ligands for these two regulatory RNA structures. However, the RNA elements might demark the beginning of two different transcripts, or bacteria might be employing the mini-ykkC motif RNA as a secondary checkpoint before investing in the translation of the massive 125 kDa urea carboxylase protein long after the mRNA for the full operon has been transcribed.
Guanidine is Bound by mini-ykkC Motif RNAs with Affinity Similar to Guanidine-I Riboswitches
The experimental validation of guanidine as the ligand for the majority of ykkC motif RNAs5 strongly implicated this same ligand as the trigger for the predicted gene regulation mediated by mini-ykkC RNAs. The major question was whether members of this RNA class act independently to directly sense guanidine or whether they require an additional factor, protein or otherwise, to respond to guanidine.
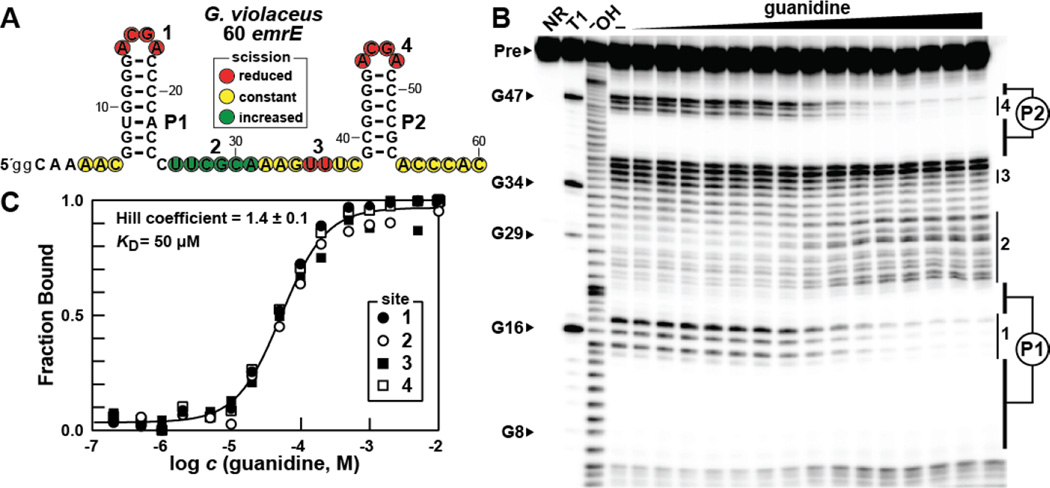
To assess ligand-binding potential, we prepared an RNA construct encompassing 60 nucleotides centered on the mini-ykkC motif of the emrE gene of Gloeobacter violaceus (Figure 2A). Note that, although this gene is annotated as emrE, it actually codes for a protein that is SugE-like, as do all genes regulated by guanidine riboswitches. This small RNA construct, called ‘G. violaceus 60 emrE’, was subjected to in-line probing assays10,11 to monitor changes in spontaneous RNA degradation in response to ligand binding. Surprisingly, guanidine directly causes robust modulation of spontaneous RNA cleavage throughout the RNA construct (Figure 2B), indicating that the RNA substantially alters its shape upon ligand binding.
Figure 2.
Guanidine is bound by a guanidine-II riboswitch aptamer in vitro. (A) The sequence and observed secondary structure of the 60 emrE RNA from G. violaceus. Lowercase nucleotides on the 5′ terminus designate G residues added to enable transcription by T7 RNA polymerase. Red, yellow, and green circles indicate nucleotides that undergo less, constant, or more spontaneous cleavage, respectively, upon addition of the ligand as determined by their relative band intensities in B. Four regions of guanidine-mediated structural modulation as identified in B are numbered accordingly. (B) Polyacrylamide gel electrophoresis (PAGE) analysis of in-line probing reactions of 60 emrE G. violaceus RNA in the absence (−) and presence of guanidine ranging from 200 nM to 10 mM. NR, T1, and −OH indicate no reaction, RNA partially digested with T1 ribonuclease (cleaves after each G), and RNA partially digested under alkaline conditions (cleaves after every nucleotide). Precursor RNA (Pre) as well as certain G residues within the RNA sequence are annotated, and regions of modulation (vertical bars) are numbered as designated in A. Regions that are devoid of spontaneous cleavage and that correspond to base-paired substructures P1 and P2 stems are identified. (C) Plot of the fraction of RNA bound to ligand versus the logarithm of the guanidine concentration as determined by quantification of the PAGE data in B (see Materials and Methods for details). The KD and Hill coefficient values were determined by the sigmoidal curve fit, and the reported error represents the standard deviation reported in the fitting parameters.
Most notably, both conserved ACGR loops of this RNA construct undergo dramatic reductions in spontaneous cleavage, which is consistent with guanidine-induced structure formation. Given the near identical sequence and structural features of P1 and P2, we considered the possibility that these repeated substructures might collaborate to form two binding pockets for guanidine. Some structural modulation is also observed in the non-conserved linker between the P1 and P2 stems, which might be required to allow crosstalk between the two, nearly-identical stem-loop substructures.
By plotting the relative increase or decrease in RNA scission at positions in four distinct regions of the construct versus the concentration of guanidine, we observe a guanidine-dependent dose-response curve with a Hill coefficient of ~1.4 (Figure 2C). A value greater than 1 suggests that multiple processes are being coupled, which is likely to be two or more ligand binding events. We anticipate the existence of two binding sites for guanidine that function cooperatively such that binding at the first site improves binding affinity at the second site, and vice versa. It seems likely that an individual guanidine binding pocket will be formed in large part by a single highly-conserved ACGR loop sequence, presumably with some contact with the neighboring stem-loop structure. The apparent overall dissociation constant (KD) of 50 µM also compares favorably to those exhibited by guanidine-I5 and guanidine-III riboswitches.6 These findings are consistent with riboswitch function for mini-ykkC motif RNAs, and hereafter we call members of this class ‘guanidine-II’ riboswitches.
Two additional representative guanidine-II riboswitch aptamer sequences were selected to confirm that mini-ykkC sequences from a distantly related organism, or with different gene associations, also respond cooperatively to guanidine. Both the 60-nucleotide aptamer associated with the sugE gene from Escherichia coli (E. coli 60 sugE, Figure S1) and the 60-nucleotide aptamer associated with the urea carboxylase gene from Klebsiella pneumoniae (60 uca, Figure S2) exhibit similar guanidine-dependent patterns of structure modulation. Specifically, each construct undergoes dramatic stabilization of both conserved ACGR loops with Hill coefficients greater than 1. Moreover, the E. coli 60 sugE and the 60 uca constructs exhibit KD values for guanidine of 300 µM and 200 µM, respectively. Again, the binding affinities for these diverse guanidine-II riboswitch aptamers are similar those of guanidine-I5 and guanidine-III6 riboswitches. These results demonstrate that guanidine-II riboswitches are present in many bacterial species where they sense and respond cooperatively to guanidine.
Mutation of Conserved Nucleotides Confirms Multiple Guanidine Binding Sites
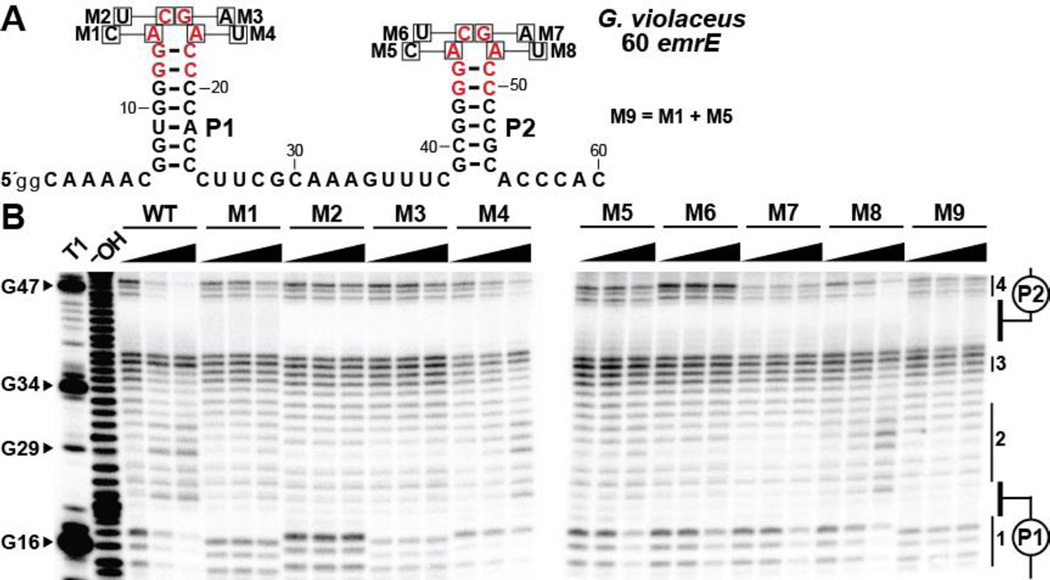
To assess the importance of highly conserved nucleotides, and to further evaluate the multiple binding site hypothesis, we constructed a series of mutant G. violaceus 60 emrE RNA constructs (Figure 3A) and assessed the effects of these mutations on guanidine binding. Specifically, eight mutant constructs (M1 through M8) were prepared that each carry a single nucleotide change in one of the two ACGR loops. A ninth construct (M9) was prepared that carries a single mutation in both of the ACGR loops.
Figure 3.
Mutations of conserved loop nucleotides affect structural modulation of both loops. (A) Sequence and secondary structure of the 60 emrE RNA from G. violaceus. Red letters denote highly conserved (>97%) nucleotide positions of the consensus sequence. Single nucleotide positions altered to create each RNA are identified with boxes, annotated with the mutant nucleotide identity, and labeled M1 through M8. M9 contains mutations at both positions specified by M1 and M5. (B) PAGE analysis of in-line probing assays with WT and mutated sequences of the 60 emrE RNA in the presence of 0, 0.1, and 1 mM guanidine. The cropped gel image depicts the regions spanning from loop 1 (nucleotides 14–17) to loop 2 (nucleotides 45–48). Additional annotations are as described in the legend to Figure 2B.
Mutations M1 through M3, which are located in the loop of P1, completely eliminate guanidine-dependent structural modulation in this loop (Figure 3B). Moreover, structural modulation is greatly diminished in the unaltered loop of P2, even at a guanidine concentration as high as 1 mM, which would be expected if two cooperative ligand binding sites are present. Reduced structure modulation is detected at 1 mM guanidine for M4 versus WT, although it is worth noting that this mutation also weakens the apparent affinity for guanidine at the second putative binding site. Additionally, this fourth position of the tetraloop is the least conserved, in that either purine nucleotide is acceptable.
Accordingly, near symmetrical results are observed for mutations M5 through M7, which exhibit little or no modulation in the loop of P2 upon guanidine addition. However, these mutations more modestly weaken ligand binding at the opposing site than do direct mutations in the loop of P1. Further analysis of M1 and M5 revealed that a mutation in one binding site decreases the affinity at that site by ~100 fold, but the other site only experiences a ~10-fold decrease in affinity for guanidine binding (Figure S3). Additionally, the Hill slope for these interactions is approximately 1, indicating that a single binding event at each site is being observed. When a mutation is simultaneously introduced into both loops (M9), binding is not observed at either site (Figure 3B).
Both Stem Loops are Necessary for Guanidine Binding
The findings described above are consistent with the presence of two distinct guanidine binding pockets, and indicate that these two sites are intimately linked to cooperatively affect ligand binding. We considered the possibility that a single stem loop might be able to bind guanidine when tested in the context of a truncated RNA that lacks the second site and any possible negative-acting structures that compete with the ligand-bound architecture of the riboswitch aptamer.
To examine this possibility, we created a truncated 30 emrE RNA from G. violaceus and conducted in-line probing with guanidine. Ligand binding is evident only at relatively high guanidine concentrations when the RNA concentration in the in-line probing reaction is ~100 nM (Figure S4). However, increasing the RNA concentration in the in-line probing reaction to 2 µM improves the affinity for guanidine by ~5 fold, which is consistent with a model wherein two hairpin substructures need to assemble to achieve high-affinity ligand binding. The natural tandem arrangement of the stem loops in the guanidine-II RNA greatly increases the effective local concentration of the two hairpin substructures, and thereby allows the riboswitch to achieve the desired affinity and cooperativity characteristics for guanidine binding.
Guanidine-II Binds its Ligand Selectively Albeit with Less Stringency than Guanidine-I
A remarkable feature of the guanidine-I riboswitch class is its ability to strongly discriminate against a diversity of close analogs of guanidine, and generate differences in affinity that are typically greater than 100 fold.5 Based on the atomic-resolution x-ray structural model for a representative guanidine-I RNA,13 this high level of selectivity is accomplished by utilizing complex tertiary structures, cation-π interactions, and a putative Mg2+ ion binding site to prevent recognition of any compound with modifications to the guanidine molecule. The guanidine-II riboswitch aptamers described in this report employ less than half the number of conserved nucleotides compared to guanidine-I riboswitches, yet likely form double the number of binding pockets. Therefore, we anticipated that the binding pockets of guanidine-II riboswitches would not be as proficient at discriminating against certain guanidine analogs.
Ligand binding specificity of the G. violaceus 60 emrE RNA was investigated by conducting in-line probing assays using a variety of guanidine analogs (Figure S5). Guanidine analogs that carry small additions to the guanidine moiety core, such as the addition of a single methyl or amino group, induce structural modulation in a manner that is similar that observed for guanidine. However, the apparent affinity for these analogs is modestly poorer than for the parent compound. Specifically, methyl- and aminoguanidine bind the RNA nearly as well as guanidine, and exhibit dissociation constants of 80 µM and 200 µM, respectively (Figure S6). However, analogs with larger substituents exhibit far poorer binding affinity, suggesting that guanidine-II riboswitches might use steric effects to block larger natural compounds that carry a guanidyl moiety.
It is unlikely that these guanidine-dependent structural changes are not merely due to a general RNA-cation interaction, since compounds such as spermidine have no significant effect. Furthermore, it was previously shown that guanidine does not induce structural modulation in RNAs from other riboswitch classes.5 Therefore it is unlikely that guanidine is non-specifically altering the structure of guanidine-II aptamers.
Perhaps the most important compound for a guanidine riboswitch to discriminate against is urea, which has numerous roles in metabolism. Urea differs from guanidine in several ways that should make it possible for an RNA to strongly favor guanidine binding and exclude urea from the binding pocket. Importantly, urea carries a keto oxygen atom in place of an imine nitrogen atom. This difference converts a hydrogen bond donor location to a hydrogen bond acceptor. In addition, urea is neutral, whereas the imine nitrogen of the protonated form of guanidine (the predominant form under biological pH conditions) is positively charged. This makes possible several mechanisms by which a riboswitch can selectively bind guanidine even in the presence of high concentrations of urea. We observe no evidence of RNA structure modulation even when 10 mM urea is present, indicating that this highly similar molecule is actively discriminated against by the binding pocket.
Concluding Remarks
The individual aptamer domains of guanidine-II riboswitches are the simplest of all known riboswitch aptamers both in terms of structural complexity and number of conserved nucleotides. The minimal aptamer length of a functional guanidine-II aptamer substructure is no longer than the 30 emrE construct tested in this study (Figure S4), and can probably be less than 20 nucleotides. For comparison, the next-simplest natural riboswitch aptamer is ~34 nucleotides and belongs to the preQ1-I class.14 However, preQ1-I aptamers exploit a pseudoknot architecture that is more complex that a simple hairpin,15,16 and can function without another aptamer as a partner.
Although the single aptamer construct present in the 30 emrE RNA does respond to guanidine, likely when forming a homodimer, this bimolecular complex must have an affinity that is far poorer than the natural tandem aptamers. Previously,3 we proposed that single hairpins similar to those in mini-ykkC motif RNAs might exist, but that it would be challenging to sort false positives from functional examples given that the motif is so simple. Now that it is apparent that the tandem hairpin arrangement of mini-ykkC motif RNAs cooperatively binds multiple guanidine molecules, it seems possible that sequence or structural adaptations have occurred to permit variants of this riboswitch class to sense a single guanidine molecule.
A simple adaptation might be for a hairpin closed by an ACGR loop to reside near another hairpin that lacks this conserved loop sequence. This arrangement might provide the accessory contacts that permit tight binding of guanidine by the conserved hairpin, in a manner analogous to the singlet versions of glycine riboswitches.17 Intriguingly, the ykkC-III motif that forms the aptamer of guanidine-III riboswitches might represent a distinctly-structured version of this proposed arrangement because it also contains an ‘ACGR’ loop, and appears to form only a single guanidine binding site.6 The proposed pseudoknot structure of guanidine-III riboswitches might use this more complex structure to permit the ACGR sequence to contribute to guanidine binding without forming a tandem arrangement.
The validation of mini-ykkC motif RNAs as members of a novel guanidine-binding riboswitch class called guanidine-II further highlights the importance of guanidine metabolism in many bacterial species. Also, this small yet surprisingly sophisticated riboswitch class provides evidence that riboswitches and other noncoding RNAs can be formed from unusually short and simple structures formed by very few conserved nucleotides. Thus, some small functional RNA domains might easily escape detection by existing bioinformatics approaches that rely on the conservation of extensive sequence and structural domains.
Acknowledgments
We thank Adam Roth, Shira Stav, Zasha Weinberg, and other members of the Breaker laboratory as well as Caroline Reiss of Yale University and Randy Stockbridge of the University of Michigan for helpful discussions. M.E.S. was supported by an NIH Cellular and Molecular Biology Training Grant (T32GM007223). S.N.M. was supported by the NSF Graduate Research Fellowship Program. This work was also supported by NIH grants (GM022778, DE022340) as well as funds from Howard Hughes Medical Institute to R.R.B.
Footnotes
ASSOCIATED CONTENT
Supporting Information (.doc)
REFERENCES
- 1.Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer MM, Hammond MC, Salinas Y, Roth A, Sudarsan N, Breaker RR. Challenges of ligand identification for riboswitch candidates. RNA Biol. 2011;8:5–10. doi: 10.4161/rna.8.1.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010;11:31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson JW, Atilho RM, Sherlock ME, Stockbridge RB, Breaker RR. Metabolism of free guanidine in bacteria is regulated by a widespread riboswitch class. Mol Cell. 2017;65:1–11. doi: 10.1016/j.molcel.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherlock ME, Breaker RR. Biochemical validation of a third guanidine riboswitch class in bacteria. Biochemistry. 2017 doi: 10.1021/acs.biochem.6b01271. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinberg Z, Kim PB, Chen TH, Li S, Harris KA, Lünse CE, Breaker RR. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol. 2015;11:606–610. doi: 10.1038/nchembio.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinberg Z, Breaker RR. R2R-software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics. 2011;12:1. doi: 10.1186/1471-2105-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 11.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuldiner SS. EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim Biophys Acta. 2009;1794:748–762. doi: 10.1016/j.bbapap.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 13.Reiss CW, Xiong Y, Strobel SA. Structural basis for ligand binding to the ykkC guanidine riboswitch. Structure. 2017 doi: 10.1016/j.str.2016.11.020. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roth A, Winkler WC, Regulski EE, Lee BW, Lim J, Jona I, Barrick JE, Ritwik A, Kim JN, Welz R, et al. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat Struct Mol Biol. 2007;14:308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- 15.Klein DJ, Edwards TE, Ferre-D’Amare AR. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spitale RC, Torelli AT, Krucinska J, Bandarian V, Wedekind JE. The structural basis for recognition of the PreQ0 metabolite by an unusually small riboswitch aptamer domain. J Biol Chem. 2009;284:11012–11016. doi: 10.1074/jbc.C900024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruff KM, Muhammad A, McCown PJ, Breaker RR, Strobel SA. Singlet glycine riboswitches bind ligand as well as tandem riboswitches. RNA. 2016;22:1728–1738. doi: 10.1261/rna.057935.116. [DOI] [PMC free article] [PubMed] [Google Scholar]