SUMMARY
Completion of a plant’s life cycle depends on successful prioritization of signaling favoring either growth or defense. Although hormones are pivotal regulators of growth–defense tradeoffs, the underlying signaling mechanisms remain obscure. The unfolded protein response (UPR) is essential for physiological growth as well as management of endoplasmic reticulum (ER) stress in unfavorable growth conditions. The plant UPR transducers are the kinase and ribonuclease IRE1 and the transcription factors bZIP28 and bZIP60. We analyzed management of the tradeoff between growth and ER stress defense by the stress response hormone salicylic acid (SA) and the UPR, which is modulated by SA via unknown mechanisms. We show that the plant growth and stress regulator CPR5, which represses accumulation of SA, favors growth in physiological conditions through inhibition of the SA-dependent IRE1–bZIP60 arm that antagonizes organ growth; CPR5 also favors growth in stress conditions through repression of ER stress-induced bZIP28/IRE1–bZIP60 arms. By demonstrating a physical interaction of CPR5 with bZIP60 and bZIP28, we provide mechanistic insights into CPR5-mediated modulation of UPR signaling. These findings define a critical surveillance strategy for plant growth–ER stress defense tradeoffs based on CPR5 and SA-modulated UPR signaling, whereby CPR5 acts as a positive modulator of growth in physiological conditions and in stress by antagonizing SA-dependent growth inhibition through UPR modulation.
Keywords: CPR5, ER stress response, plant defense tradeoffs, salicylic acid, unfolded protein response, Arabidopsis thaliana
INTRODUCTION
The ability of plants to control the balance between growth and defense is vital for survival and adaptation to the environment (Huot et al., 2014; Smakowska et al., 2016). As defense from biotic and abiotic stresses causes energy to be diverted from growth, plants have evolved inducible strategies such as hormone production to direct metabolic expenditure toward either growth or defense (Clarke et al., 2000). Salicylic acid (SA) is a prime example of an inducible response in conditions of biotic and abiotic stresses (Nguyen et al., 2016; Verma et al., 2016). The synthesis of SA in response to stress leads to the utilization of defense pathways and inhibition of growth (Rivas-San and Plasencia, 2011). Therefore, a monitoring system must be in place to control the induction and amplitude of the SA-induced stress responses and maintain growth. CPR5 (constitutive expresser of pathogenesis-related genes-5), a plant-specific master regulator of growth and defense, is known as a negative modulator of SA, acting just downstream of pathogen recognition and upstream of SA in a resistance pathway dependent on NPR1 (non-expressor of pathogenesis-related genes 1) (Bowling et al., 1997). cpr5 was indeed originally isolated as a pathogen-resistant mutant with constitutive expression of pathogenesis-related gene (PR-1) and a high SA content (Bowling et al., 1997). Importantly however, additional functions in plant defense and growth have been attributed to CPR5, including cell cycle-related effector triggered immunity (ETI)-induced programmed cell death (PCD) (Wang et al., 2014), cell proliferation and expansion (Kirik et al., 2001), cell wall biogenesis (Kirik et al., 2001; Brininstool et al., 2008;) and redox balance (Jing et al., 2008), indicating that CPR5 is a master regulator of a number of processes that can impose stress on the plant independently of pathogen attack. Although genetic analyses support the suggestion that CPR5 controls several of these processes independently (Clarke et al., 2000; Jing et al., 2008; Perazza et al., 2011; Bao and Hua, 2014), the underlying regulatory mechanisms exerted by CPR5 via SA in growth and defense are still largely unknown.
Growth and stress adaptation rely on the biosynthetic capacity of the endoplasmic reticulum (ER) for the production of approximately one-third of the cellular proteome (Ghaemmaghami et al., 2003). In specific stress and development situations requiring enhanced secretory protein synthesis and folding (e.g. pathogen attack, but also heat stress, and cell growth; Howell, 2013), the biosynthetic capacity of the ER can be overwhelmed leading to a potentially lethal condition known as ER stress, which results from the accumulation of misfolded and unfolded proteins in the ER (Chen and Brandizzi, 2013; Liu and Howell, 2016). During ER stress, a sophisticated signaling pathway, known as the unfolded protein response (UPR), is actuated to modulate gene expression and restore ER homeostasis through chaperone-assisted protein folding, translational attenuation and ER-associated protein degradation (ERAD) (Wan and Jian 2016). In plants, the UPR alters the cell’s transcriptional programs mainly through the action of two ER-associated sensors: the membrane-tethered transcription factor (TF) bZIP28 and the protein kinase and ribonuclease IRE1, which are functionally highly conserved in eukaryotes and operate to modulate UPR gene expression in the nucleus (Ruberti and Brandizzi, 2014). In ER stress-inducing conditions, bZIP28 is activated by intramembrane proteolytic cleavage mediated by the canonical site 1 and site 2 proteases (S1P/S2P) in the Golgi, an event that leads to release of the functional basic leucine zipper (bZIP) domain and translocation to the nucleus for modulation of UPR gene expression (Liu et al., 2007). The protein kinase and ribonuclease IRE1 cleaves the mRNA of bZIP60 leading to a frameshift translation of a potent TF that modulates UPR gene expression in the nucleus (Nagashima et al., 2011). As an additional UPR gene modulation mechanism, IRE1 can cleave mRNAs through a process known as regulated IRE1-dependent decay (RIDD) (Mishiba et al., 2013). There are two homologs of IRE1 in Arabidopsis, IRE1A and IRE1B, which are believed to share a largely overlapping function in the UPR (Chen and Brandizzi, 2012; Deng et al., 2013). Similarly, bZIP28 and bZIP60 appear to have partially redundant functions in ER stress and in the modulation of the transcription of downstream UPR target genes such ER-resident molecular chaperones (Ruberti et al., 2015).
The UPR transducers play positive roles in the UPR signaling pathways by protecting cells from ER stress (Chen and Brandizzi, 2012; Deng et al., 2013). However, IRE1 is necessary not only for response to proteotoxic stress in the ER but also for growth in the absence of induced ER stress. In mammalian and plant cells, IRE1 has critical, possibly essential, roles, as demonstrated by the lethality of knockout mutations of a mammalian IRE1α in mouse (Iwawaki et al., 2009) and loss of gametophytic transmission of a knockout allele of IRE1B in Arabidopsis (Lu and Christopher, 2008; Chen and Brandizzi, 2012; Deng et al., 2013). Furthermore, functional studies in Arabidopsis with a double IRE1 mutant bearing an IRE1A knockout allele and an IRE1B partial loss-of-function allele (ire1a ire1b) demonstrated the occurrence of a short root phenotype in normal conditions of growth (Chen and Brandizzi, 2012; Deng et al., 2013). This phenotype is independent of the function of IRE1–bZIP60 and bZIP28 in ER stress responses, as single or double loss-of-function mutants of bZIP28 and bZIP60 do not affect growth in physiological growth conditions (Chen and Brandizzi, 2012; Sun et al., 2013). These results highlight that IRE1 controls a signaling pathway that has a critical role in growth in physiological conditions but is independent of the IRE1–bZIP60 signaling mechanisms that operate in ER stress-activated UPR.
Because of its critical biological roles in stress defense and growth, the UPR must be tightly regulated. For example, a misregulated UPR is potentially lethal, as demonstrated by the evidence that loss of function of IRE1 in plants accelerates progression to death in conditions of unresolved ER stress (Chen and Brandizzi, 2012; Deng et al., 2013). Furthermore, upon resolution of ER stress, the stress-induced UPR signaling must be attenuated, and in conditions of unresolvable (chronic) ER stress the protective UPR needs to be terminated for PCD to take place (Hetz, 2012; Chen and Brandizzi, 2013). Negative modulators of the UPR have been identified in mammalian cells. For example, Bax inhibitor-1 (BI-1) can physically interact with IRE1α and decrease IRE1 endoribonuclease activity (Hetz, 2012; Chen and Brandizzi, 2013). When IRE1 activity is attenuated, the abundance of the mature form of its downstream ribonuclease TF target, XBP1, is decreased through formation of a stable complex between spliced and unspliced XBP1 that is degraded by the proteasome (Yoshida et al., 2006). Additionally, the ERAD factor Derlin1 can recognize unspliced XBP1 and initiate degradation together with the E3 ubiquitin ligase TRC8 and signal peptidase (SPP) (Chen et al., 2014a). Similar to XBP1, ATF6, which is the metazoan counterpart of bZIP28, can also be reduced by WFS1, which stabilizes the E3 ubiquitin ligase HRD1 and recruits ATF6 to the ubiquitin-proteasome for degradation (Fonseca et al., 2010). Other negative regulators such as GADD34 and P58IPK have been identified for the PERK–eIF2α–ATF4 signaling branch, which is a UPR signaling arm in metazoan cells that has not been identified in plants (Novoa et al., 2001; Van Huizen et al., 2003). Intriguingly, even if few determinants for plant survival under ER stress have been determined (Watanabe and Lam, 2008; Yang et al., 2014), modulators of the plant UPR transducers analogous to the metazoan regulators of the UPR transducers have yet to be discovered.
Because of its relevance to growth and the stress response, the UPR is intertwined with hormonal pathways that control growth and stress responses. For example, it has been demonstrated that ER stress leads to hyper-accumulation of the phytohormone auxin and that UPR activation is reduced in mutants of ER-localized auxin transporters (Chen et al., 2014a,b). Furthermore, the stress hormone SA has been shown to induce the UPR through IRE1–bZIP60 and bZIP28 when exogenously added to the growth medium (Nagashima et al., 2014). While these results imply that plants hormones participate in defense to ER stress, the underlying mechanisms connecting hormone pathways and the UPR transducers are yet unknown at a mechanistic level.
Here we investigated the mechanisms underlying plant growth–ER stress defense tradeoffs by dissecting the functional connection of CPR5, SA and the UPR in the control of growth and defense. We demonstrate that CPR5 exerts a critical role in growth–defense tradeoffs by favoring growth through suppression of SA-mediated growth inhibition operated through IRE1–bZIP60 signaling. We also demonstrate that CPR5 functions to suppress ER stress responses to favor growth by antagonizing SA and the downstream UPR signaling function of bZIP28 and bZIP60. By demonstrating an interaction of CPR5 with bZIP28 and bZIP60 at the protein level, we propose that in addition to modulation of SA levels, CPR5 monitors UPR signaling directly through physical interaction with the UPR bZIP TFs.
RESULTS
The IRE1–bZIP60 arm is required for growth inhibition in conditions of elevated levels of SA
In this study, we adopted a complete loss-of-function mutant of CPR5 (Boch et al., 1998; Borghi et al., 2011). Compared with the wild type (Wt), this mutant has elevated levels of free and conjugated SA (Bowling et al., 1997; Boch et al., 1998) (Figure 1a), a significant reduction in the length of the primary root (LOPR; Figure 1b,d) and the number of lateral roots (NOLR; Figure 1d), as well as differences in appearance of the shoot, as shown by reduced fresh weight (SFW; Figure 1c,d), and accelerated senescence of the cotyledons (Figure 1c; Bowling et al., 1997; Boch et al., 1998). To test whether these phenotypes are due to a high SA content in cpr5, we generated a cpr5 sid2 double mutant that lacks SA synthase SA-induction deficient 2 (SID2) (Bowling et al., 1997; Boch et al., 1998). We established that the cpr5 sid2 double mutant lost the elevated SA levels (Figure 1a) but restored LOPR (Figure 1c,d) and NOLR values (Figure 1d) to Wt levels; however, the cotyledon phenotype (Figure 1c) and SFW defects (Figure 1d) were not significantly different from cpr5. We deduce that the elevated levels of SA in cpr5 cause defects in growth at a tissue-specific level (i.e. LOPR and NOLR).
Figure 1.
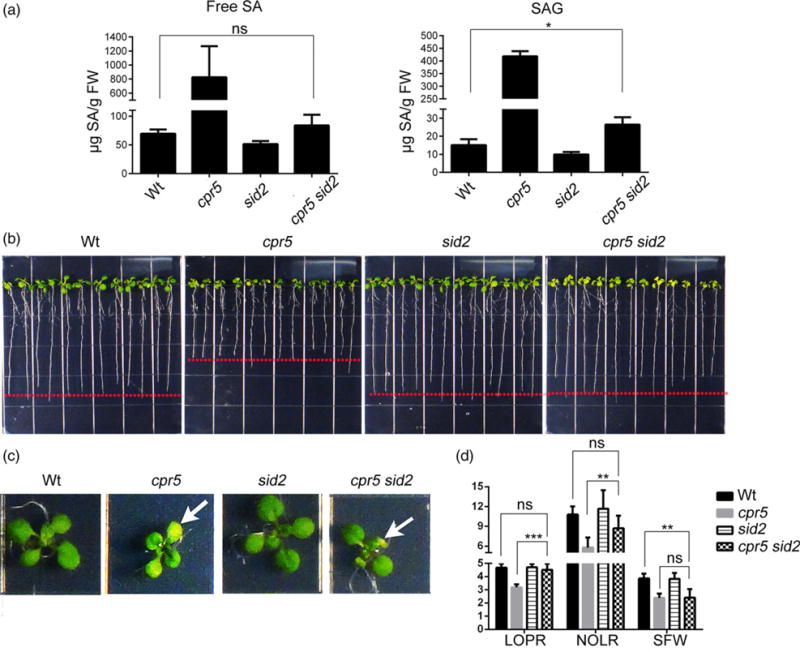
Enhanced levels of endogenous SA cause the growth defects of cpr5.
(a) Measurement of SA in wild type (Wt), cpr5, sid2 and cpr5 sid2. The levels of free SA and SA glucosides (SAG) in 12-day-old seedlings were analyzed using HPLC. Values represent mean ± SD from three biological replicates.
(b), (c) Seedlings were germinated on 1/2 Linsmaier and Skoog medium for 12 days vertically (b) and horizontally (c). The arrow in (c) indicates the occurrence of necrosis in the cotyledons that is typical of cpr5.
(d) Absolute growth values of seedlings grown as indicated in (b). LOPR, length of primary root; NOLR, number of lateral roots; SFW, shoot fresh weight. Error bars represent SD; n = 24 for each genotype. The P-values were obtained using Student’s t-test for each comparison. **P < 0.001; ***P < 0.0001; ns, not significant.
Because of the SA-dependent root phenotype of cpr5 and the reported short root phenotype of IRE1 loss-of-function mutants (Gao et al., 2011; Chen and Brandizzi, 2012), we tested whether a genetic relationship could exist between CPR5 and IRE1 in the control of root growth. Therefore, we compared the plant phenotype of Wt, CPR5 and IRE1 single and higher-order mutants in physiological conditions of growth. Compared with the Wt, single gene mutations in IRE1A or IRE1B did not result in a significant reduction in LOPR (Figure 2a,c,d,f) and NOLR (Figure 2c,f) or overall SFW (Figure 2b,c,e,f). These results support earlier findings that, in physiological conditions of growth, the IRE1 isoforms have overlapping roles in root length growth (Chen and Brandizzi, 2012; Deng et al., 2013). Notably, when we compared the root of cpr5 with either cpr5 ire1a or cpr5 ire1b double mutant, we found that the reduced LOPR and NOLR phenotype of cpr5 was restored by mutation of either IRE1A or IRE1B (Figure 2a,c,d,f). Furthermore, the SFW of cpr5 ire1a and cpr5 ire1b mutants was significantly greater than that of cpr5 yet lower than Wt (Figure 2c,f). However, compared with cpr5 and ire1a ire1b, the cpr5 ire1a ire1b triple mutant showed similar LOPR and NOLR values to ire1a ire1b (Figure 2g,i). Also, the SFW of cpr5 ire1a ire1b was partially recovered compared with cpr5 (Figure 2i). These results indicate that in conditions of elevated SA IRE1 and CPR5 have an epistatic interaction for the control of plant growth and that the SA-induced inhibition of growth requires IRE1 signaling to take place. Such a relationship is predominant in the primary and lateral root and to a minor extent in the aerial tissues. These results also indicate that although the IRE1 isoforms have overlapping roles in root growth in physiological conditions of growth, in situations of elevated SA they assume roles that are interdependent.
Figure 2.
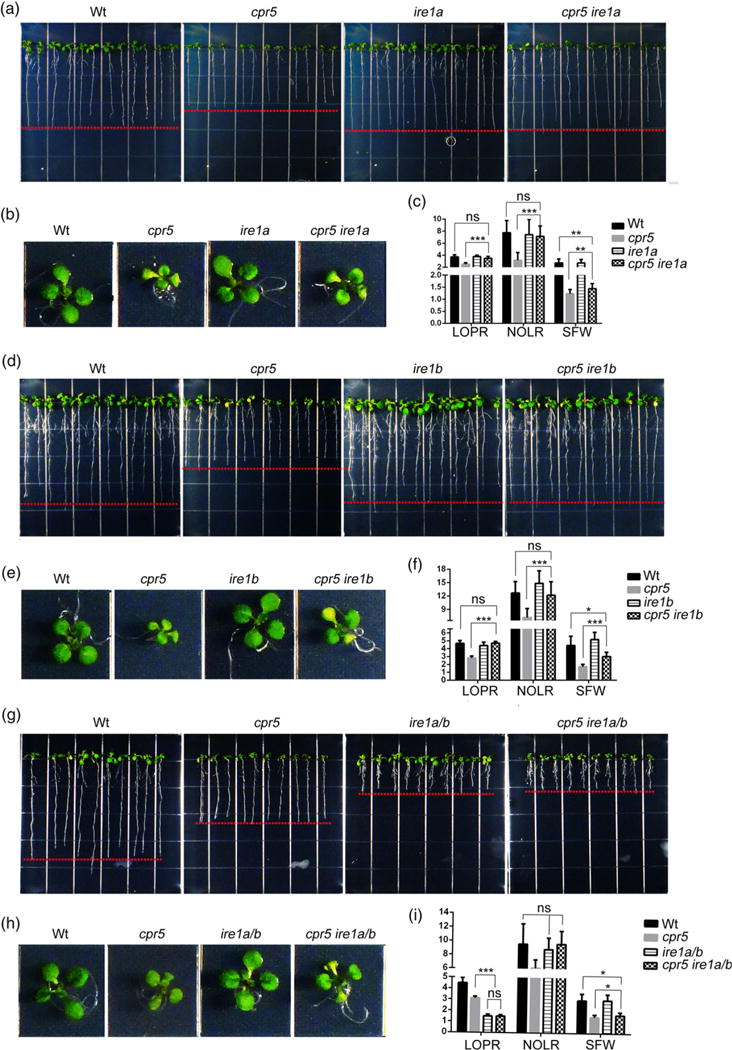
CPR5 acts upstream of IRE1 in the control of primary root growth.
(a), (b), (d), (e), (g), (h) Seedlings were germinated on 1/2 Linsmaier and Skoog medium for 12 days vertically (a, d, g) and horizontally (b, e, h). (c), (f), (i) Absolute growth values of single and high-order mutants of cpr5 and ire1 as indicated in (a, d, g). LOPR, length of primary root; NOLR, number of lateral roots; SFW, shoot fresh weight; Wt, wild type. Error bars represent SD; n = 24 for each genotype. The P-values were obtained using Student’s t-test for each comparison.
*P < 0.05; **P < 0.001; ***P < 0.0001; ns, not significant.
We next aimed to test the involvement of bZIP28 and bZIP60 in CPR5-controlled processes for organ growth by comparing growth parameters (i.e. LOPR, NOLR and SFW) in Wt, single and higher-order mutants of CPR5, bZIP28 and bZIP60 in physiological growth conditions. Compared with cpr5, the bzip28 and bzip60 single mutants and the bzip28 bzip60 double mutant had significantly increased LOPR, NOLR and SFW values but no differences from Wt (Figure 3). When we compared double and triple mutations of CPR5 with the bZIP-transcription factors, we found that although the necrotic lesions of the cpr5 cotyledons were maintained (Figure 3h), the cpr5 bzip60 double mutant and the cpr5 bzip28 bzip60 triple mutant were otherwise indistinguishable from bzip60 and Wt (Figure 3a–c,g–i). In complete contrast, the cpr5 bzip28 double mutant was indistinguishable from cpr5 and had significantly lower LOPR, NOLR and SFW values compared with Wt (Figure 3d–f). These results indicate that for SA-induced inhibition of growth to occur bZIP60 must be present. Therefore, in situations of elevated SA, in contrast to bZIP28, bZIP60 is necessary for root growth, which is consistent with a functional connection of bZIP60 and IRE1 (Deng et al., 2013) and the verified genetic relationship of IRE1 with CPR5 in growth (Figure 2).
Figure 3.
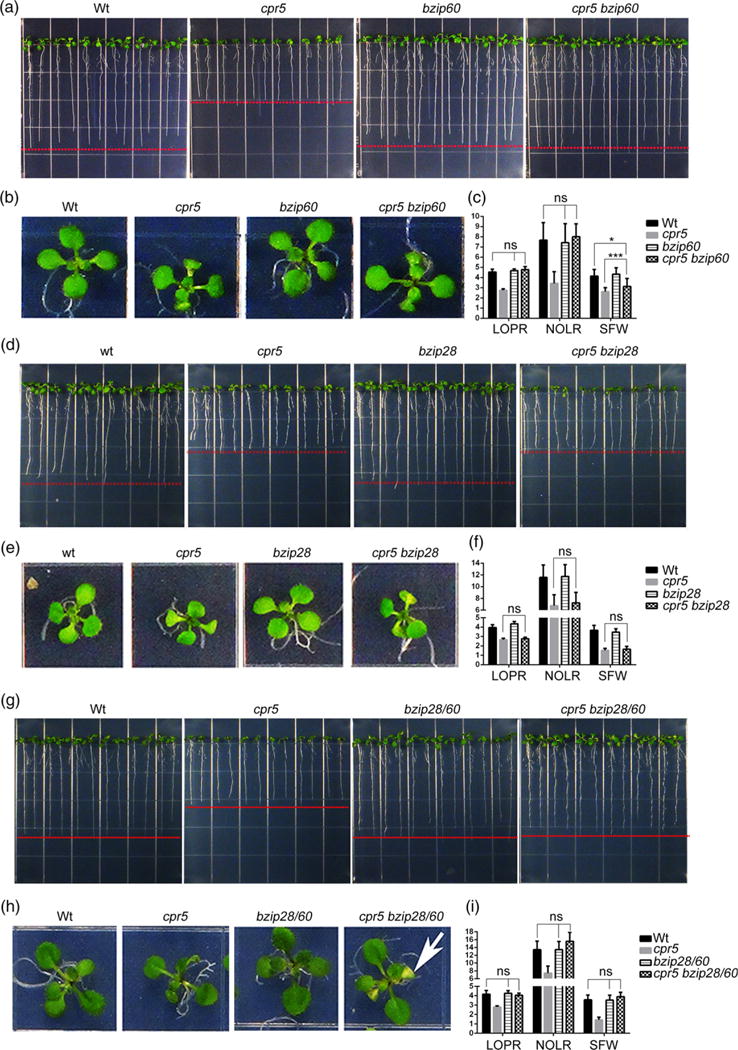
The role of CPR5 in primary root growth is affected by bZIP60, while it is independent from bZIP28.
(a), (b), (d), (e), (g), (h) Seedlings were germinated on 1/2 Linsmaier and Skoog medium for 12 days vertically (a, d, g) and horizontally (b, e, h). (c), (f), (i) Absolute growth values of single and high-order mutants of cpr5, bzip28 and bzip60 as indicated in (a, d, g). LOPR, length of primary root; NOLR, number of lateral roots; SFW, shoot fresh weight; Wt, wild type. Error bars represent SD; n = 24 for each genotype. The P-values were obtained using Student’s t-test for each comparison. *P < 0.05; ***P < 0.0001; ns, not significant. The arrow in (h) indicates the occurrence of necrosis in the cotyledons that is typical of cpr5.
The loss of CPR5 alters the basal levels of the UPR independently of SA
Because exogenous SA has been reported to induce UPR by activating both IRE1–bZIP60 and bZIP28 arms (Nagashima et al., 2014) and cpr5 has elevated endogenous levels of SA (Figure 1a), we tested whether CPR5 could be involved in the management of UPR by assaying the expression of well-established UPR biomarker genes in normal conditions of growth using quantitative real-time RT-PCR (qRT-PCR) in Wt and cpr5. We found that compared with the Wt the basal levels of BiP3 were significantly lower in the UPR mutants bzip28, bzip60, bzip28 bzip60 (Figure 4a). Similarly the basal levels of the splicing of bZIP60 (sbZIP60) were significantly lower in bzip28, while, as expected, sbZIP60 was undetectable in bzip60 single and higher-order mutant backgrounds (Figure 4b). Interestingly, adding the cpr5 mutation onto the bzip28 or bzip60 mutation caused a massive and statistically significant reduction of the BiP3 (in both backgrounds) and sbZIP60 transcript levels (in bzip28) (Figure 4a,b). These results, together with the evidence that BiP3 levels were significantly reduced in a cpr5 bzip28 bzip60 triple mutant compared with Wt, strongly indicate that the loss of CPR5 evokes UPR gene induction through both the IRE1–bZIP60 arm and the bZIP28 arm (Figure 4a,b).
Figure 4.
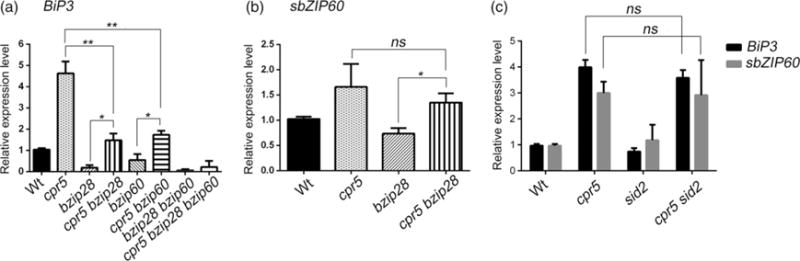
Loss of CPR5 affects the basal levels of the unfolded protein response (UPR).
(a) Relative expression of the UPR marker gene BiP3 in the wild type (Wt), single and high-order mutants of cpr5 and the UPR sensors estimated by quantitative real-time RT-PCR (qRT-PCR) analyses in 12-day-old seedlings under normal growth conditions. The relative gene expression represents the expression level of the gene in the mutants divided by that in Wt, both of which were normalized to the expression of UBQ10. The values shown were relative to Wt which was set to 1. Note that the induction of BiP3 is significantly lower in bzip28, bip60 and bzip28 bzip60 mutant backgrounds than in Wt. Error bars represent SD from three biological replicates. *P < 0.05; **P < 0.001; ***P < 0.0001; ns, not significant.
(b) Relative expression of the spliced bZIP60 (sbZIP60) in Wt, cpr5, bzip28 and cpr5 bzip28 estimated by qRT-PCR analyses in 12-day-old seedlings under normal growth conditions. The relative gene expression represents the expression level of the gene in the mutants divided by that in Wt, both of which were normalized to the expression of UBQ10. The values shown are relative to Wt, which was set to 1. Error bars represent SD from three biological replicates. *P < 0.05; **P < 0.001; ***P < 0.0001; ns, not significant.
(c) The qRT-PCR analyses of BiP3 and sbZIP60 transcripts in the indicated genetic backgrounds in 12-day-old seedlings. Error bars represent SD from three biological replicates. ns, not significant.
We next wanted to test whether the induced UPR levels in cpr5 were linked to a high content of SA. Therefore, we assayed the transcription levels of the UPR marker genes BiP3 and sbZIP60 in normal conditions of growth in the SA-defective mutant backgrounds cpr5 sid2 (Figure 1a). Interestingly, we found that in the cpr5 sid2 mutant the transcripts levels of the UPR indicators BiP3 and sbZIP60 were similar to cpr5 (Figure 4c). We deduce that the observed UPR gene induction in cpr5 is independent of the elevated levels of endogenous SA in this mutant. We hypothesize therefore that CPR5 may harness unique strategies to modulate the UPR. This hypothesis is further supported by the evidence that other known mutants with a high SA content, such as bon1-1 (Yang and Hua, 2004), snc1-1 (Yang and Hua, 2004) and siz1-2 (Castro et al., 2015), did not show enhanced levels of the basal UPR unlike cpr5 (Figure S1e in the Supporting Information).
Enhanced levels of endogenous SA render cpr5 growth insensitive to unresolved ER stress
Having established that CPR5 modulates the basal levels of the UPR in normal conditions of growth, we next aimed to investigate its role in conditions of ER stress. To do so, Wt and cpr5 seedlings were grown on medium containing tunicamycin (Tm), an inducer of ER stress (Chen and Brandizzi, 2012), or DMSO (solvent control). While the growth of Wt seedlings was visibly compromised by Tm (Figure 5a; Chen and Brandizzi, 2012), the growth of cpr5 was largely unaffected (Figure 5a). To provide quantification of the Tm sensitivity for each genetic background, we calculated the relative growth values (i.e. we estimated the LOPR, NOLR and SFW values on seedlings grown on Tm divided by measurements on seedlings grown on control plates). Compared with Wt, cpr5 had no significant reduction of LOPR and NOLR and only a modest decrease of SFW (Figure 5b). These analyses of the relative growth values show a sensitivity of the Wt but resistance of cpr5 to Tm (Figure 5b). These results indicate that sensitivity of the Wt to unresolved ER stress is linked to CPR5, because the loss of CPR5 confers resistance to Tm. Therefore, to gain more insight into how cpr5 could resist prolonged ER stress, we aimed to test whether the Tm resistance of cpr5 could depend on SA. To do this, we analyzed the Tm sensitivity of a cpr5 sid2 mutant and found that the LOPR and NOLR values of cpr5 sid2 were similar to those of the Wt, but that the SFW values were much like those of cpr5 (Figure 5a,b). Therefore, the root of cpr5 sid2 was as sensitive to chronic ER stress as the Wt; however, the shoot was as insensitive to chronic ER stress as cpr5 (Figure 5b). These data support that enhanced SA levels in cpr5 render root growth in this mutant largely insensitive to chronic ER stress. This seems to be a phenotype uniquely linked to cpr5, as other known high-SA-content mutants showed similar Tm sensitivity to the Wt (Figure S1a–d), suggesting that, among high-SA-content mutants, cpr5 harnesses unique strategies to survive chronic ER stress.
Figure 5.
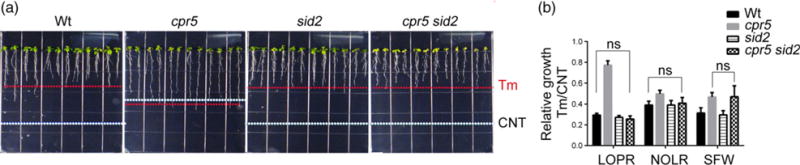
Enhanced levels of endogenous SA render cpr5 insensitive to chronic endoplasmic reticulum stress. (a) Seedlings were germinated on 1/2 Linsmaier and Skoog medium containing 50 ng ml−1 tunicamycin (Tm) for 12 days vertically. White dashed line, length of the primary roots grown in normal growth conditions as shown in Figure 1(b). Red dashed line, length of the primary roots grown with Tm treatment.
(b) Relative growth values of seedlings as indicated in (a). LOPR, length of primary root; NOLR, number of lateral roots; SFW, shoot fresh weight. Error bars represent SD; n = 24 for each genotype. ns, not significant.
ER stress resistance promoted by high levels of endogenous SA relies on canonical UPR signaling pathways
Having established that the high levels of endogenous SA render the growth of cpr5 insensitive to unresolved ER stress (Figure 5), we next aimed to test which UPR arms were harnessed by endogenous SA for resistance to chronic ER stress in cpr5. To do so, we analyzed the root phenotype of Wt, single and higher-order mutants of CPR5 and the UPR sensors. We first analyzed IRE1 mutants. Consistent with previous reports (Chen and Brandizzi, 2012), the loss of either IRE1 isoform did not affect the sensitivity of plants to Tm compared with the Wt (Figure 6a–d), supporting largely overlapping roles of IRE1A and IRE1B in sustaining prolonged ER stress. However, when we analyzed the relative growth values of cpr5 ire1a and cpr5 ire1b grown on Tm-containing plates compared with control plates, we found that the Tm insensitivity of cpr5 ire1a and cpr5 ire1b was significantly reduced compared with cpr5 (Figure 6b,d). Mutation of both IRE1 genes in cpr5 led to a reduced Tm sensitivity compared with the ire1a ire1b mutant (Figure 6e,f). Together these results support that although IRE1 isoforms have largely overlapping roles in root growth in physiological conditions of growth, in conditions of prolonged ER stress and high levels of endogenous SA linked to the lack of CPR5, each IRE1 isoform is required for survival of ER stress. These results also indicate that endogenous SA relies on the IRE1 arm for supporting survival to chronic ER stress.
Figure 6.
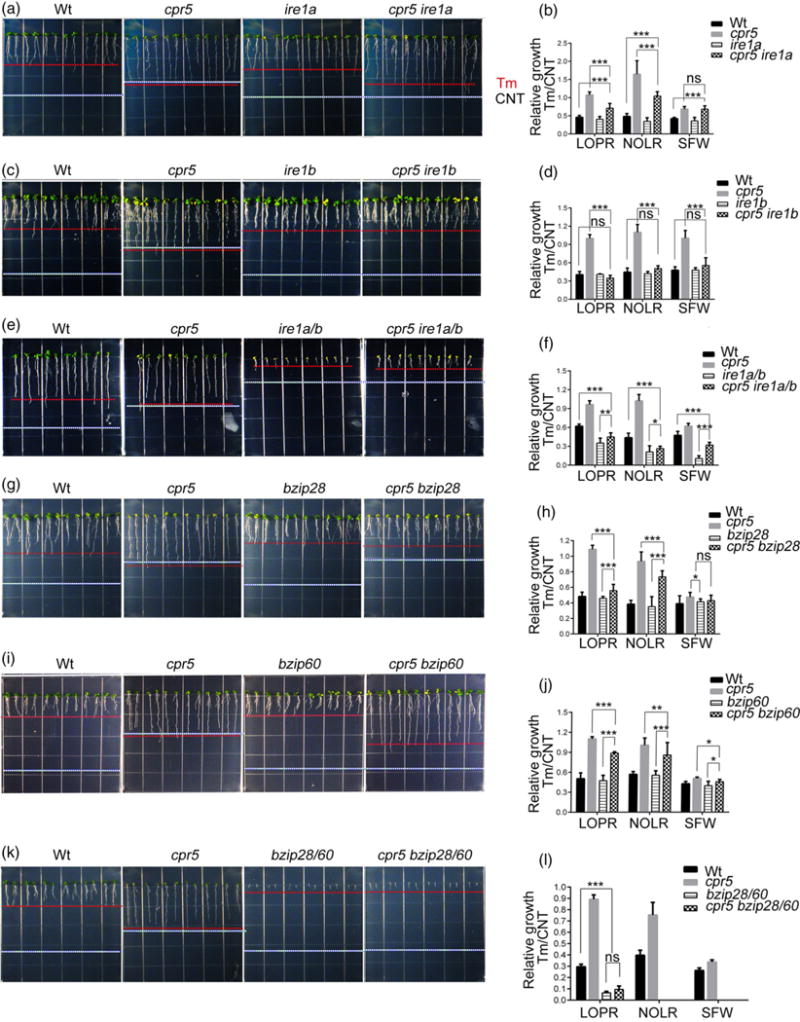
CPR5 acts upstream of IRE1, bZIP60 and bZIP28 in the control of plant growth under the endoplasmic reticulum stress response.
(a), (c), (e), (g), (i), (k) Seedlings were germinated on 1/2 Linsmaier and Skoog medium containing 50 ng ml−1 tunicamycin (Tm) for 12 days vertically. White dashed line, length of the primary roots grown under normal growth conditions as shown in Figure 2(a, d, g) and Figure 3(a, d, h). Red dashed line, length of the primary roots grown with Tm treatment.
(b), (d), (f), (h), (j), (l)) Relative growth values of seedlings as indicated in (a, c, e, g, i, k). LOPR, length of primary root; NOLR, number of lateral roots; SFW, shoot fresh weight; Wt, wild type. Error bars represent SD; n = 24 for each genotype. *P < 0.05; **P < 0.001; ***P < 0.0001; ns, not significant.
We next tested the genetic interaction of CPR5 with bZIP28 and bZIP60 in response to chronic ER stress. As reported earlier, compared with the Wt, single bzip60 or bzip28 mutants exhibited similar sensitivity to chronic ER stress conditions; however, the loss of both transcription factors accelerated pro-death responses (Figure 6g–l; Chen and Brandizzi, 2012). Analyses of cpr5 bzip28, cpr5 bzip60 and cpr5 bzip28 bzip60 showed an enhanced Tm sensitivity of the high-order mutants compared with cpr5 (Figure 6g–l), indicating the occurrence of a genetic interaction of CPR5 with bZIP60 and bZIP28 whereby these transcription factors function downstream of CPR5 in management of the UPR in chronic ER stress.
CPR5 physically interacts with bZIP60 and bZIP28
The evidence that CPR5 harnesses unique strategies to modulate the UPR in normal growth conditions (Figures 4 and S1) and in chronic ER stress (Figures 5 and 6) supported the possibility that CPR5 could rely on unique mechanisms to manage the UPR. Based on these considerations and the genetic relationship between CPR5 and the bZIP TFs established above, we hypothesized that CPR5 could act simultaneously on bZIP60 and bZIP28, possibly at the protein level. To test this hypothesis, we aimed to assay whether CPR5 could have a similar subcellular distribution to the bZIP TFs. To do this we generated a translational fusion of the yellow fluorescent protein to CPR5 (YFP–CPR5), which complements the growth phenotype of cpr5 and is therefore functional (Figure S2c). Based on the presence of membrane domains and a bipartite nuclear localization signal (Figure S2a), we expected localization of CPR5 to multiple compartments in live cells, as also reported earlier (Kirik et al., 2001; Yoshida et al., 2002; Wang et al., 2014). However, we found a weak fluorescence signal barely above the auto-fluorescence levels in the complemented lines despite the high levels of expression of the transgene (Figure S2d), which suggests translational modulation of CPR5 in controlling protein abundance in stable transformants. As an alternative approach, we used Agrobacterium-mediated transient expression analyses, which is extensively used for subcellular protein localization analyses (Denecke et al., 2012). We found that CPR5 was localized to the ER, nuclear envelope, Golgi stacks and nucleoplasm (Figure S2b). The results are consistent with the microsomal and nuclear localization of CPR5 established in fractionation studies with CPR5 antibodies (Wang et al., 2014). Intriguingly, the subcellular localization of CPR5 is similar to the subcellular localization of bZIP28 (Gao et al., 2008), suggesting a possible mechanism of action of CPR5 that relies on proteolytic cleavage of CPR5 and membrane release of the protein region containing the NLS for traffic to the nucleus.
Given the nuclear distribution of the protein, we hypothesized that CPR5 could interact directly with bZIP60 and bZIP28 in the nucleus where these proteins exert their transcriptional activity. To test the interaction, we used the yeast two-hybrid (Y2H) approach. The N-terminus of the CPR5 coding sequence devoid of the transmembrane domains (CPR5DTMD) was fused to LexA-binding domain (LexA-BD), and the LexA-activation domain (LexA-AD) was fused to the truncated form of spliced bZIP60 that was missing the activation domain (sbZIP60 ΔAD). The yeast co-transformed with the AD/BD constructs detailed above was grown on selective medium containing X-Gal. In the event of an interaction, we expected that the galactosidase reporter gene would be expressed and the yeast cells would turn blue. Indeed, we found interaction of CPR5 with bZIP60 but not with the negative controls (Figure 7a). To further verify this interaction, acceptor photobleaching fluorescence resonance energy transfer (APB-FRET) assays were performed in vivo. Cyan fluorescent protein (CFP)– sbZIP60 and YFP–CPR5 were co-expressed in tobacco epidermal cells. The nuclei were selected as the region of interest (ROI) to monitor the FRET. Multichannel images were collected before and after photo-bleaching of YFP (acceptor), and the relative increase in CFP (donor) fluorescence was quantified to determine the FRET efficiency (Figure 7b,c). The mean FRET efficiency between YFP– CPR5 and CFP–sbZIP60 was around 10%, which is statistically significant compared with the negative control in the nucleoplasm (i.e. untargeted/cytosolic YFP that diffuses to the nucleoplasm) (Figure 7b,c). Therefore, we confirmed that CPR5 and bZIP60 can physically associate with each other in plant cells.
Figure 7.
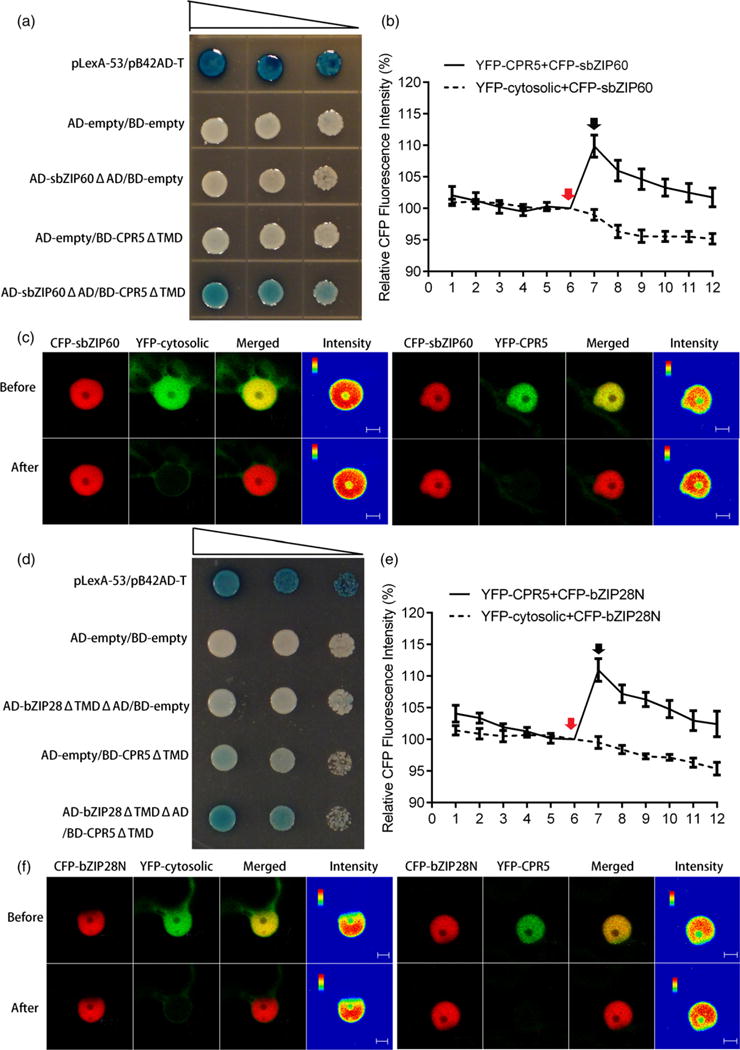
CPR5 interacts with bZIP60 and bZIP28.
(a), (d) Interaction of CPR5 with bZIP60 (a) and bZIP28 (d) in a yeast two-hybrid system. The bait is the N-terminal segment of CPR5 without transmembrane domains (TMD). The N-terminal segments of bZIP transcription factors (TFs) without both transcription activation (AD) and transmembrane domains (TMD) are tested as prey. Yeast co-transformed with the test constructs was grown on SD-Ura-His-Trp plates containing X-Gal. Expression of the lacZ gene (shown by the blue color) was used to indicate of interaction. pLexA-53/pB42AD-T is the positive control. Serial dilutions of transformed cells are shown by narrowing triangles. (b), (e) Quantification of fluorescence resonance energy transfer (FRET) efficiency between YFP–CPR5 and CFP–sbZIP60 (b) or CFP–bZIP28N (e). The percentage of increase in CFP fluorescence after photobleaching YFP (FRET efficiency) was determined. Red arrows indicate pre-bleaching points, and black arrows indicate post-bleaching points. The x-axis indicates the series of images scanned every 10 sec before and after photobleaching. Co-expression of cytosolic YFP and CFP–bZIP was used as negative control. Error bars represent SEM (n = 15). (c), (f) Single-scan confocal images of a representative nucleus taken before and after YFP photobleaching marked by arrows in (b) and (e). For the qualitative detection of FRET, CFP fluorescence images are provided as pseudo-colored intensity maps. Bars = 5 μm.
We next tested whether CPR5 could also interact with bZIP28, which can form heterodimers with bZIP60 (Liu and Howell, 2010). Therefore, we used the Y2H system to examine whether CPR5DTMD could interact with a form of bZIP28 devoid of the activation domain and transmembrane anchor (bZIP28ΔTMDΔAD). The yeast colonies transformed with BD-CPR5ΔTMD and AD-bZIP28ΔTMDΔAD turned blue in the selective plates but not in the controls (Figure 7d), indicating interaction of CPR5 and bZIP28. In FRET analyses, the bleaching of YFP-CPR5 resulted in an increase of about 12% in the emission from CFP-bZIP28ΔTMD, but no increase in the negative control (i.e. CFP-bZIP28ΔTMD and cytosolic YFP), confirming the Y2H results that an interaction can occur between CPR5 and bZIP28 (Figure 7e,f). These results support that CPR5 interacts with bZIP28 and bZIP60, and provide insights into understanding at a mechanistic level how CPR5 may exert a modulatory role in the management of UPR signaling.
DISCUSSION
To thrive in their environment, plants have evolved efficient strategies to prioritize signaling for growth or defense and maximize resource allocations where needed. Although a role for hormones is emerging, how the growth–defense tradeoffs are intertwined with regulatory signaling pathways to manage the balance between growth and stress responses is largely unknown. Here we have investigated the role of CPR5, a critical regulator of SA-mediated growth and defense, in conjunction with the UPR, an essential signaling pathway for growth and stress responses. We have demonstrated that in physiological conditions CPR5 acts as a positive modulator of growth by suppressing the levels of endogenous SA, which in turn harnesses the IRE1–bZIP60 signaling pathway to repress growth. In addition, CPR5 negatively modulates the basal levels of UPR gene induction independently from SA. We have also shown that CPR5 monitors the UPR under induced ER stress, CPR5 being a negative modulator of the UPR by modulating the bZIP60/bZIP28 arms dependently from SA. Together these findings support that CPR5 acts as a modulator of growth through: (i) the monitoring of SA and the downstream UPR arms and (ii) suppressing inhibition of growth by SA in physiological conditions and in situations causing ER stress. Moreover, based on enhanced UPR gene induction dependent on IRE1–bZIP60 and bZIP28 in the cpr5 mutant and on the established interaction of CPR5 with the bZIP60 and bZIP28 at the protein level, we suggest that the negative role of CPR5 on UPR signaling is probably executed by suppression of the transcriptional function of bZIP28 and bZIP60 at the protein level.
Endogenous SA controls primary root growth through IRE1–bZIP60-mediated signaling
Our current understanding of the plant UPR is mainly based on the study of the induction of ER stress in adverse growth conditions, such as salt and heat stress (Deng et al., 2011; Henriquez-Valencia et al., 2015), or in the presence of chemicals, such as Tm and dithiothreitol that induce the UPR by altering the homeostasis of the ER proteome (Chen and Brandizzi, 2012; Deng et al., 2013). There is limited information about the physiological roles of the UPR transducers under normal conditions of growth. Yet it is well established that IRE1 has a critical role in plant growth because ire1b knockout is likely lethal and the ire1a ire1b mutant used in this work and others has a short root phenotype (Chen and Brandizzi, 2012; Deng et al., 2013). It has been established that the root growth defects of an ire1a ire1b mutant are linked to misregulation of cell length (Chen and Brandizzi, 2012), but how IRE1 controls growth of the primary root at a mechanistic level is still unknown. Although activation of bZIP60 depends on IRE1, the evidence that bzip28 and bzip60 single mutants and a bzip28 bzip60 double mutant do not have a short root phenotype argues that IRE1-controlled growth in physiological conditions is independent of these bZIP TFs. As a bzip28 bzip60 mutant is lethal in conditions of chronic ER stress, it has been concluded that these transcription factors are essential to survive chronic ER stress rather than for controlling growth (Deng et al., 2013; Sun et al., 2013). Although these findings provide some insights into the pathways controlled by IRE1 in growth, not much is known about the upstream regulatory mechanisms in physiological conditions of growth. In this work we have shown that the cpr5 ire1a or the cpr5 ire1b mutations restore the root growth defects of cpr5 to Wt levels. These results support an epistatic interaction between CPR5 and IRE1, as the root growth defects due to the elevated levels of endogenous SA caused by the cpr5 mutation depend on the IRE1 isoform. The evidence that deletion of either IRE1 isoform is sufficient to restore the cpr5 root phenotype argues that in conditions of elevated endogenous SA levels IRE1A and IRE1B have interdependent roles in root growth. These results suggest that the functional requirements of IRE1 in growth are different between unchallenged plants and plants challenged by conditions requiring SA defense. In line with this argument, our results have also provide significant insights into the signaling underlying primary root growth controlled by IRE1 and attribute a function of bZIP60 but not bZIP28 in such a process. Indeed, the evidence that a cpr5 bzip28 mutation does not affect the cpr5 phenotype in normal conditions of growth supports the notion that bZIP28 is not involved in root growth (Chen and Brandizzi, 2012; Deng et al., 2013; Sun et al., 2013). Nonetheless, we also showed that the simultaneous loss of bZIP60 and CPR5 in cpr5 bzip60 and cpr5 bzip28 bzip60 causes suppression of the cpr5 primary root phenotype to bzip60 and Wt levels of growth. These results can be explained by a role for bZIP60 in primary root growth, whereby the signaling mediated by IRE1–bZIP60 exerts a negative role on growth in situations causing an elevated SA content (Figure S3). The evidence that the cpr5 ire1a ire1b root phenocopies the ire1a ire1b root suggests that IRE1 also assumes roles in growth that are independent of SA. Therefore, although we cannot exclude that other IRE1-dependent factors may control primary root growth, our results that a cpr5 and cpr5 bzip60 or cpr5 bzip28 bzip60 have opposite primary root growth phenotypes support the suggestion that such factors are unlikely to be dependent on the RIDD function of IRE1. We speculate that in stress situations causing high endogenous SA levels bZIP60 may interact with SA-responsive factors that function with bZIP60 as a negative growth modulators. Although this hypothesis is yet to be experimentally tested, our results that CPR5 exerts a homeostatic control of growth through SA and IRE1–bZIP60 provide critical insights into the modulation of the SA pathway in organ growth. The evidence in this work that other mutants with a high SA content do not show growth phenotypes similar to cpr5 argues that the SA-dependent mechanisms of growth modulation mediated by SA are specific to CPR5. We hypothesize the scenario whereby downstream effectors of CPR5 are activated or inhibited by SA. These could be the factors necessary for a CPR5-specific response to SA among the various mutants with a high SA content.
Endogenous SA is a positive modulator of the UPR bZIP TFs in chronic ER stress
In metazoans, several factors are in place to monitor the amplitude and duration of the UPR induced by proteotoxic stress in the ER (Yoshida et al., 2006; Lisbona et al., 2009; Fonseca et al., 2010). The identity of such factors in plants is largely unknown. In this work we have shown that that the loss of CPR5 confers resistance to chronic ER stress. We demonstrated that in cpr5 the high SA content is necessary to support resistance to chronic ER stress, given that a cpr5 sid2 mutant, which restores SA to Wt levels, has root growth values similar to the Wt in conditions of prolonged ER stress. These results provide support for a negative role of CPR5 with SA to favor growth in conditions of ER stress. Furthermore, the evidence that the loss of the UPR arms controlled by bZIP28 and/or IRE1–bZIP60 reduces the ER stress-resistant growth phenotype of a cpr5 mutant to prolonged ER stress conditions argues for a negative modulatory role of CPR5 on the function of these UPR transducers in ER stress (Figure S3). Together, these results indicate that CPR5 functions to suppress SA-dependent defense to ER stress mediated by bZIP28 and bZIP60 and favor growth. Hence, in growth–defense tradeoffs, CPR5 emerges as a critical upstream UPR modulator to reduce the allocation of energy resources to defense in support of growth.
We have also shown that the loss of CPR5 is associated with elevated basal levels of the UPR gene, which is attenuated by high-order mutations of cpr5 with the UPR transducers and that is independent of SA. Based on the demonstrated protein–protein interaction of bZIP28 and bZIP60 with CPR5 we propose that the genetic interaction of bZIP TFs and CPR5 in management of UPR signaling is functionally related at a protein level. CPR5 is predicted to contain a cytosol-exposed protein domain with a putative nuclear localization sequence followed by five transmembrane domains. Localization analyses of a YFP–CPR5 fusion protein driven by a 35S promoter in a cpr5 mutant have indicated a nuclear and cytosolic distribution of CPR5 in roots (Gao et al., 2011). However, a CPR5 devoid of the TMD regions was unable to complement the cpr5 phenotype, suggesting that a membrane association is required for the function of CPR5 (Gao et al., 2011), which may serve for post-translational modification of this protein (e.g. phosphorylation). We have not been able to define the distribution of a 35S::YFP–CPR5 fusion in our transgenic lines, despite the demonstrated ability of this protein chimera to complement a cpr5 mutant. These results can be explained on the basis of post-translational suppression of CPR5 to levels that prevent clear detection of fluorescence signal over the background. Imaging of the roots of our cpr5/35S::YFP-CPR5 lines led to detection of high levels of auto-fluorescence of the root tissue, which is similar to the proposed distribution of GFP–CPR5 (Gao et al., 2011). However, in transient expression we verified a ER–Golgi and nuclear localization of CPR5, which is consistent with that of several other transcriptional modulators such as ATF6 and bZIP28, as well as the sterol regulatory element-binding protein (SREBP), which are normally resident in the ER but are translocated to the Golgi where the portion of the protein destined for the nucleus is cleaved by proteases (Bartz et al., 2008; Gao et al., 2011; Sato et al., 2011). Our results are also consistent with subcellular fractionation results using CPR5 antibodies of a localization of endogenous CPR5 with microsomes and the nuclear fraction (Wang et al., 2014). Based on these results, and the proven requirement of CPR5 to associate with the membrane for its function (Gao et al., 2011; Perazza et al., 2011) and our microscopy results and earlier fractionation results (Wang et al., 2014), we propose that CPR5 follows the path of other membrane-associated transcriptional regulators that are proteolytically activated to control gene expression in the nucleus. Based on an antagonistic role of CPR5 on bZIP TFs in response to ER stress, we speculate that the interaction of CPR5 with bZIP TFs suppresses their transcriptional activity. CPR5 may suppress the transcriptional activation of target genes of the bZIP TFs by facilitating selective degradation of bZIP28 and bZIP60 analogously to the role of WFS1-mediated degradation of ATF6 in mammals (Fonseca et al., 2010; Chen et al., 2014a,b). Of note, using the ELM tool designed to recognize linear motifs in eukaryotic proteins (http://elm.eu.org/; Dinkel et al., 2016), we identified a conserved ‘WDR5-binding motif’ (amino acids 318–321) in CPR5, which is also present in Ash2 and in Rbl, two core proteins of the COMPASS-like complex along with WDR5A (Jiang et al., 2011). Intriguingly, bZIP28 and bZIP60 physically interact with the COMPASS-like complex components Ash2 and WDR5a to promote the formation of the pre-initiation complex and generate the deposition of the histone trimethyl H3K4 at specific promoters of the UPR genes, thereby regulating stress-responsive gene expression (Song et al., 2015). We therefore speculate that CPR5 may control the transcriptional activity of bZIP28 and bZIP60 by modulating their binding affinity to the COMPASS-like complex.
In conclusion, CPR5 functions as a master regular of seemingly unrelated processes in growth and stress defense. Our results that misregulation of UPR signaling affects growth of cpr5 root, and to a limited extent the shoot, which in turn can be restored by mutations of regulators of effector-triggered PCD and immunity responses (Wang et al., 2014), support this notion. In this work, we have demonstrated a role for CPR5 in the UPR, whereby CPR5 suppresses the functions of the IRE1 arm in the homeostatic control of growth mediated by SA and serves as a negative modulator of the UPR bZIP TFs, probably through protein–protein interactions. Together our results define unpredicted but critical mechanisms for the control of resource allocation in plant growth and defense responses that are executed in a SA-dependent stress defense pathway controlled by CPR5 via critical UPR signaling components.
EXPERIMENTAL PROCEDURES
Plant materials
The Arabidopsis T-DNA mutants bzip60 (SALK_050203) (Chen and Brandizzi, 2012), bzip28 (SALK_132285) (Chen and Brandizzi, 2012), ire1a (WISCDSLOX420D09) (Chen and Brandizzi, 2012), ire1b (SAIL_238_F07) (Chen and Brandizzi, 2012), siz1 (SALK_065397) (Castro et al., 2015) and the cpr5 loss-of-function allele based on a single base substitution cpr5 (CS3770) were acquired from the Arabidopsis Biological Resource Center. The sid2-2 and snc1-1 and bon1-1 mutants have been previously characterized (Wildermuth et al., 2001; Yang and Hua, 2004). The Col-0 ecotype was used as the Wt control. The high-order mutants were generated by crossing the mutants above. Genotyping the homozygous T-DNA insertion mutants was performed as described earlier (Slabaugh et al., 2011). Genotyping the homozygous mutants with a point mutation (cpr5) was accomplished using the allele-specific PCR method described earlier (Hayashi et al., 2004). The mutant sid2-2 with a small deletion was genotyped using the primers which were designed based on the missing sequence. The primers for genotyping are listed in Table S1.
Plant growth conditions and Tm treatment
Surface-sterilized seeds were directly plated on half-strength Linsmaier and Skoog (LS) medium containing 1.5% w/v sucrose and 1.2% Agar (Acumedia, http://foodsafety.neogen.com/pdf/Acumedia_PI/7558_PI.pdf) at 4°C for 2 days in the dark and then grown at 21°C under a 16-h light/8-h dark cycle for 12 days before photographed. For chronic ER stress assays, 50 ng ml−1 Tm (Sigma-Aldrich T7765, http://www.sigmaaldrich.com/) or an equivalent volume of DMSO (Tm solvent) was added to the medium above.
Phenotypical analysis
The parameters of LOPR, NOLR and SFW were averaged from 24 plants for each genotype. The relative growth rate was calculated using the growth value in the presence of Tm divided by the growth value in the absence of Tm to estimate the sensitivity to chronic ER stress. Statistical significance was estimated by Student’s two-tailed t-test; data with a P-value <0.05 were considered significant. At least three independent experiments were performed.
Gene expression analysis
Total RNA was extracted according to the manufacturer’s instructions (Macherey-Nagel, http://www.mn-net.com/). The qRT-PCR analyses were conducted as described earlier (Chen and Brandizzi, 2012). Data were analyzed using the ΔΔCt method. Relative transcript levels of each gene were normalized to that of UBQ10 and the values shown were relative to Wt, which was set to 1. Values of the fold change were averages from three biological replicates (mean±SD). P-values were calculated with the Student’s two-tailed t-test to establish statistical significance. Data with a P-value <0.05 were considered significant.
Yeast two-hybrid assay
The Y2H experiments were performed using a matchmaker LexA system (Clontech, http://www.clontech.com/) with yeast strain EGY48. We cloned the CPR5 coding sequence without a transmembrane domain sequence into the yeast expression vector pGlida (bait) and fused the truncated forms of sbZIP60 or bZIP28 without activation and transmembrane domains into vector pB42AD (prey) (see Table S1 for primers). The bait and prey proteins were co-transformed to the yeast cells which were later plated on the SD/GAL/RAF medium (Clontech) lacking Ura, Trp and His supplemented with 20 μg ml−1 X-Gal. The positive colonies in which the proteins interacted would turn into blue because of activation of LacZ reporter gene.
Fluorescence resonance energy transfer-acceptor photo-bleaching analysis
The coding sequence of CPR5 was cloned into the pEarleygate104 vector to construct 35S::YFP–CPR5. The coding regions of spliced full-length bZIP60 and the bZIP28 without a transmembrane domain were, respectively, subcloned into the pVKH18En6-gw vector to construct 35S::CFP-sbZIP60 and 35S::CFP-bZIP28ΔTMD. The cytosolic YFP was constructed previously (Chen and Brandizzi, 2012). The CFP-tagged donor and the YFP-tagged acceptor constructs were introduced into tobacco epidermal cells by Agrobacterium- mediated infiltration and incubated for 2 days. FRET-APB was analyzed using an inverted Zeiss LSM510 META laser scanning confocal microscope (Carl Zeiss, http://www.zeiss.com/). CFP was excited at 405 nm, and emission was detected between 460 and 480 nm. YFP was excited at 514 nm, and emission was detected between 530 and 570 nm. Time series of CFP and YFP fluorescence were collected simultaneously every 10 s for a single scan at the low laser intensities before and after photobleaching. The selected region of interest was limited to nuclei, which were irradiated by the 514-nm laser (100% intensity) to photobleach the acceptor YFP. Increased emission from the donor CFP indicated that FRET had occurred between the two proteins prior to the bleaching. The relative CFP fluorescence intensity (%) was quantified using the equation (CFP post-bleach/CFP pre-bleach) × 100. At least 15 different nuclei were analyzed and average intensities before and after bleaching were plotted on the graph (mean ± SEM) from one representative data set are presented. Three independent experiments were performed.
SA measurements
The measurements of free SA and SA glucoside were performed on 10-day-old seedlings, as described previously (Zeng et al., 2011).
Supplementary Material
Figure S1. Mutants with a high SA content do not show the endoplasmic reticulum stress tolerance phenotype of cpr5.
Figure S2. The subcellular localization of CPR5 in the endoplasmic reticulum, Golgi and nucleoplasm is suggestive of activation of CPR5 by proteolysis.
Figure S3. Proposed working model of roles of CPR5 in growth–endoplasmic reticulum stress defense tradeoff through SA and UPR.
Table S1. List of primers.
Acknowledgments
We thank Drs Yuti Chen and ShengYang He (Michigan State University) for useful suggestions on this work and the gift of sid2-2 seeds. We thank Dr Shuhua Yang (China Agriculture University) for the gifts of snc1-1 and bon1-1 seeds. We thank Dr Michael F. Thomashow (Michigan State University) for helpful comments on the manuscript and for the gift of siz1 seeds. The authors declare that there are no conflicts of interest. This work was primarily supported by the National Institutes of Health (R01 GM101038) with contributing support from the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, US Department of Energy (award no. DE-FG02-91ER20021) and AgBioResearch.
Footnotes
Accession numbers
CPR5: At5g64930; UBQ10: At4g05320; BiP3: At1g09080; ERdj3A: At3g08970; ERdj3B: At3g62600; PDI6: At1g77510; PDI1-3: At3g54960; bZIP60: At1g42990; IRE1A: At2g17520; IRE1B: At5g24360; bZIP28: At3g10800; SID2: At1g74710; BON1: At5g61900; SNC1: At4g16890; SIZ1: At5g60410.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
References
- Bao Z, Hua J. Interaction of CPR5 with cell cycle regulators UVI4 and OSD1 in Arabidopsis. PLoS ONE. 2014;9:e100347. doi: 10.1371/journal.pone.0100347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz R, Sun LP, Bisel B, Wei JH, Seemann J. Spatial separation of Golgi and ER during mitosis protects SREBP from unregulated activation. EMBO J. 2008;27:948–955. doi: 10.1038/emboj.2008.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Verbsky ML, Robertson TL, Larkin JC, Kunkel BN. Analysis of resistance gene-mediated defense responses in Arabidopsis thaliana plants carrying a mutation in CPR5. Mol Plant Microbe Interact. 1998;11:1196–1206. [Google Scholar]
- Borghi M, Rus A, Salt DE. Loss-of-function of constitutive expresser of pathogenesis related genes5 affects potassium homeostasis in Arabidopsis thaliana. PLoS ONE. 2011;6:e26360. doi: 10.1371/journal.pone.0026360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brininstool G, Kasili R, Simmons LA, Kirik V, Hulskamp M, Larkin JC. Constitutive expressor of pathogenesis-related genes5 affects cell wall biogenesis and trichome development. BMC Plant Biol. 2008;8:58. doi: 10.1186/1471-2229-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro PH, Verde N, Lourenco T, Magalhaes AP, Tavares RM, Bejarano ER, Azevedo H. SIZ1-dependent post-translational modification by SUMO modulates sugar signaling and metabolism in Arabidopsis thaliana. Plant Cell Physiol. 2015;56:2297–2311. doi: 10.1093/pcp/pcv149. [DOI] [PubMed] [Google Scholar]
- Chen Y, Brandizzi F. AtIRE1A/AtIRE1B and AGB1 independently control two essential unfolded protein response pathways in Arabidopsis. Plant J. 2012;69:266–277. doi: 10.1111/j.1365-313X.2011.04788.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Brandizzi F. IRE1: ER stress sensor and cell fate executor. Trends Cell Biol. 2013;23:547–555. doi: 10.1016/j.tcb.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Malchus NS, Hehn B, Stelzer W, Avci D, Langosch D, Lemberg MK. Signal peptide peptidase functions in ERAD to cleave the unfolded protein response regulator XBP1u. EMBO J. 2014a;33:2492–2506. doi: 10.15252/embj.201488208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Aung K, Rolčík J, Walicki K, Friml J, Brandizzi F. Inter-regulation of the unfolded protein response and auxin signaling. Plant J. 2014b;77:97–107. doi: 10.1111/tpj.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X. Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell. 2000;12:2175–2190. doi: 10.1105/tpc.12.11.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke J, Aniento F, Frigerio L, Hawes C, Hwang I, Mathur J, Neuhaus JM, Robinson DG. Secretory pathway research: the more experimental systems the better. Plant Cell. 2012;24:1316–1326. doi: 10.1105/tpc.112.096362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Humbert S, Liu JX, Srivastava R, Rothstein SJ, Howell SH. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:7247–7252. doi: 10.1073/pnas.1102117108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Howell SH. Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc Natl Acad Sci USA. 2013;110:19633–19638. doi: 10.1073/pnas.1314749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkel H, Van Roey K, Michael S, et al. ELM 2016–data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res. 2016;44:D294–D300. doi: 10.1093/nar/gkv1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca SG, Ishigaki S, Oslowski CM, et al. Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J Clin Invest. 2010;120:744–755. doi: 10.1172/JCI39678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Brandizzi F, Benning C, Larkin RM. A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:16398–16403. doi: 10.1073/pnas.0808463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Zhang S, Wang C, Yang X, Wang Y, Su X, Du J, Yang C. Arabidopsis CPR5 independently regulates seed germination and postgermination arrest of development through LOX pathway and ABA signaling. PLoS ONE. 2011;6:e19406. doi: 10.1371/journal.pone.0019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Hashimoto N, Daigen M, Ashikawa I. Development of PCR-based SNP markers for rice blast resistance genes at the Piz locus. Theor Appl Genet. 2004;108:1212–1220. doi: 10.1007/s00122-003-1553-0. [DOI] [PubMed] [Google Scholar]
- Henriquez-Valencia C, Moreno AA, Sandoval-Ibanez O, Mitina I, Blanco-Herrera F, Cifuentes-Esquivel N, Orellana A. bZIP17 and bZIP60 regulate the expression of BiP3 and other salt stress responsive genes in an UPR-independent manner in Arabidopsis thaliana. J Cell Biochem. 2015;116:1638–1645. doi: 10.1002/jcb.25121. [DOI] [PubMed] [Google Scholar]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- Howell SH. Endoplasmic reticulum stress responses in plants. Annu Rev Plant Biol. 2013;64:477–499. doi: 10.1146/annurev-arplant-050312-120053. [DOI] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY. Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant. 2014;7:1267–1287. doi: 10.1093/mp/ssu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Yamanaka S, Kohno K. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc Natl Acad Sci USA. 2009;106:16657–16662. doi: 10.1073/pnas.0903775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Kong NC, Gu X, Li Z, He Y. Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet. 2011;7:e1001330. doi: 10.1371/journal.pgen.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing HC, Hebeler R, Oeljeklaus S, Sitek B, Stuhler K, Meyer HE, Sturre MJ, Hille J, Warscheid B, Dijkwel PP. Early leaf senescence is associated with an altered cellular redox balance in Arabidopsis cpr5/old1 mutants. Plant Biol. 2008;10(Suppl 1):85–98. doi: 10.1111/j.1438-8677.2008.00087.x. [DOI] [PubMed] [Google Scholar]
- Kirik V, Bouyer D, Schobinger U, Bechtold N, Herzog M, Bonneville JM, Hulskamp M. CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein. Curr Biol. 2001;11:1891–1895. doi: 10.1016/s0960-9822(01)00590-5. [DOI] [PubMed] [Google Scholar]
- Lisbona F, Rojas-Rivera D, Thielen P, et al. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol Cell. 2009;33:679–691. doi: 10.1016/j.molcel.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22:782–796. doi: 10.1105/tpc.109.072173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH. Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 2016;211:418–428. doi: 10.1111/nph.13915. [DOI] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH. An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell. 2007;19:4111–4119. doi: 10.1105/tpc.106.050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DP, Christopher DA. Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Mol Genet Genomics. 2008;280:199–210. doi: 10.1007/s00438-008-0356-z. [DOI] [PubMed] [Google Scholar]
- Mishiba K, Nagashima Y, Suzuki E, Hayashi N, Ogata Y, Shimada Y, Koizumi N. Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc Natl Acad Sci USA. 2013;110:5713–5718. doi: 10.1073/pnas.1219047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima Y, Mishiba K, Suzuki E, Shimada Y, Iwata Y, Koizumi N. Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci Rep. 2011;1:29. doi: 10.1038/srep00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima Y, Iwata Y, Ashida M, Mishiba K, Koizumi N. Exogenous salicylic acid activates two signaling arms of the unfolded protein response in Arabidopsis. Plant Cell Physiol. 2014;55:1772–1778. doi: 10.1093/pcp/pcu108. [DOI] [PubMed] [Google Scholar]
- Nguyen D, Rieu I, Mariani C, van Dam NM. How plants handle multiple stresses: hormonal interactions underlying responses to abiotic stress and insect herbivory. Plant Mol Biol. 2016;91:727–740. doi: 10.1007/s11103-016-0481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D, Laporte F, Balague C, Chevalier F, Remo S, Bourge M, Larkin J, Herzog M, Vachon G. GeBP/GPL transcription factors regulate a subset of CPR5-dependent processes. Plant Physiol. 2011;157:1232–1242. doi: 10.1104/pp.111.179804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-San VM, Plasencia J. Salicylic acid beyond defence: its role in plant growth and development. J Exp Bot. 2011;62:3321–3338. doi: 10.1093/jxb/err031. [DOI] [PubMed] [Google Scholar]
- Ruberti C, Brandizzi F. Conserved and plant-unique strategies for overcoming endoplasmic reticulum stress. Front Plant Sci. 2014;5:69. doi: 10.3389/fpls.2014.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberti C, Kim SJ, Stefano G, Brandizzi F. Unfolded protein response in plants: one master, many questions. Curr Opin Plant Biol. 2015;27:59–66. doi: 10.1016/j.pbi.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Nadanaka S, Okada T, Okawa K, Mori K. Luminal domain of ATF6 alone is sufficient for sensing endoplasmic reticulum stress and subsequent transport to the Golgi apparatus. Cell Struct Funct. 2011;36:35–47. doi: 10.1247/csf.10010. [DOI] [PubMed] [Google Scholar]
- Slabaugh E, Held M, Brandizzi F. Control of root hair development in Arabidopsis thaliana by an endoplasmic reticulum anchored member of the R2R3-MYB transcription factor family. Plant J. 2011;67:395–405. doi: 10.1111/j.1365-313X.2011.04602.x. [DOI] [PubMed] [Google Scholar]
- Smakowska E, Kong J, Busch W, Belkhadir Y. Organ-specific regulation of growth-defense tradeoffs by plants. Curr Opin Plant Biol. 2016;29:129–137. doi: 10.1016/j.pbi.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Song ZT, Sun L, Lu SJ, Tian Y, Ding Y, Liu JX. Transcription factor interaction with COMPASS-like complex regulates histone H3K4 trimethylation for specific gene expression in plants. Proc Natl Acad Sci USA. 2015;112:2900–2905. doi: 10.1073/pnas.1419703112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Lu SJ, Zhang SS, Zhou SF, Sun L, Liu JX. The lumen-facing domain is important for the biological function and organelle-to-organelle movement of bZIP28 during ER stress in Arabidopsis. Mol Plant. 2013;6:1605–1615. doi: 10.1093/mp/sst059. [DOI] [PubMed] [Google Scholar]
- Van Huizen R, Martindale JL, Gorospe M, Holbrook NJ. P58IPK, a novel endoplasmic reticulum stress-inducible protein and potential negative regulator of eIF2alpha signaling. J Biol Chem. 2003;278:15558–15564. doi: 10.1074/jbc.M212074200. [DOI] [PubMed] [Google Scholar]
- Verma V, Ravindran P, Kumar PP. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016;16:86. doi: 10.1186/s12870-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan S, Jiang L. Endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) in plants. Protoplasma. 2016;253:753–764. doi: 10.1007/s00709-015-0842-1. [DOI] [PubMed] [Google Scholar]
- Wang S, Gu Y, Zebell SG, Anderson LK, Wang W, Mohan R, Dong X. A noncanonical role for the CKI-RB-E2F cell-cycle signaling pathway in plant effector-triggered immunity. Cell Host Microbe. 2014;16:787–794. doi: 10.1016/j.chom.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Lam E. BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J Biol Chem. 2008;283:3200–3210. doi: 10.1074/jbc.M706659200. [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- Yang S, Hua J. A haplotype-specific resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell. 2004;16:1060–1071. doi: 10.1105/tpc.020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZT, Wang MJ, Sun L, Lu SJ, Bi DL, Sun L, Song ZT, Zhang SS, Zhou SF, Liu JX. The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet. 2014;10:e1004243. doi: 10.1371/journal.pgen.1004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Ito M, Nishida I, Watanabe A. Identification of a novel gene HYS1/CPR5 that has a repressive role in the induction of leaf senescence and pathogen-defence responses in Arabidopsis thaliana. Plant J. 2002;29:427–437. doi: 10.1046/j.0960-7412.2001.01228.x. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Oku M, Suzuki M, Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol. 2006;172:565–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Brutus A, Kremer JM, Withers JC, Gao X, Jones AD, He SY. A genetic screen reveals Arabidopsis stomatal and/or apoplastic defenses against Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 2011;7:e1002291. doi: 10.1371/journal.ppat.1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Mutants with a high SA content do not show the endoplasmic reticulum stress tolerance phenotype of cpr5.
Figure S2. The subcellular localization of CPR5 in the endoplasmic reticulum, Golgi and nucleoplasm is suggestive of activation of CPR5 by proteolysis.
Figure S3. Proposed working model of roles of CPR5 in growth–endoplasmic reticulum stress defense tradeoff through SA and UPR.
Table S1. List of primers.


