Abstract
L1-seq is a high-throughput sequencing technique which is utilized to identify novel L1 insertions in genomic DNA samples of interest. Using special diagnostic nucleotides unique to the youngest and most active L1 sequence, we can amplify new somatic insertions. This technique has helped to establish the number of L1 insertions present in the general population as well as the variation among individuals with regard to their complement of active L1 elements. More recently, this technique has been employed to assess the level of retrotransposition occurring in various diseases such as cancer. These efforts try to establish a connection between the process of retrotransposition and disease development and/or progression.
Keywords: Non-LTR retrotransposon, Retroelement, LINE-1, L1, Next-Generation DNA sequencing
1 Introduction
Retrotransposons are nearly ubiquitous in eukaryotes from slime molds [1] to humans [2] and have contributed greatly to genome composition of these organisms. Retrotransposons make up 45 % of the human genome [2]. In particular, the LINE-1 (L1) element has contributed to approximately 17 % of the human genome and continues to add to it via a copy and paste mechanism with an RNA intermediate [2]. L1 is the only autonomous retrotransposon in the human genome because it encodes two proteins necessary for mobilization and reinsertion into the genome; however, these two proteins, once expressed can mobilize other types of retrotransposons as well as processed pseudogenes [3–5]. Each individual has a different complement of potentially active L1 elements, although the majority of the L1s in each individual’s genome are truncated and therefore inactive. L1-seq [6] was developed to help characterize L1 variation among individuals because L1s have contributed to a substantial fraction of the genome and are capable of inducing many types of mutations. L1-seq has since been used to evaluate several types of cancer to establish the level of retrotransposition occurring in colon cancer, lung cancer, breast cancer, and many other cancers [7, 8]. Additional sequencing techniques have confirmed the L1-seq data and demonstrated that L1 elements are active in many cancer types [8–13]. The results have demonstrated that L1s are active in a subset of patients with cancer; in addition, L1 elements are active in all epithelial cancers tested. The L1-seq technique consists of a DNA library prep as well as the validation of the predicted new insertions detected in the samples used. Although few of the insertions may be directly responsible for the development of the disease, it should be possible to utilize known insertions present in a cancer sample for monitoring the cancer’s progression to metastasis. To detect metastasis using a new L1 insertion, a PCR would be performed on serum DNA from a patient to determine whether or not the insertion was detectable in the blood and therefore potentially in a floating cancer cell. This technique is useful both for evaluating the overall complement of L1 elements in a genome as well as looking for new insertion events. L1-seq utilizes unique nucleotides, “ACA” 91–93 nucleotides from the 3′ end of the element, to selectively amplify the young and active subset of elements in the human genome [6]. Following the initial five cycles of the PCR, wherein the linear amplification of L1 elements occurs, degenerate primers are added to the mixture to exponentially amplify both polymorphic and potentially somatic insertions present in the genome.
2 Materials
Store all reagents as specified by manufacturers. Diligently follow all waste disposal regulations when disposing of waste materials. All primers need to be diluted to 100 µM upon receipt in diethylpurocarbonate (DEPC) water. Primers will be further diluted as specified later in the protocol.
2.1 DNA Isolation
DNeasy Blood and Tissue Kit (Qiagen).
500 mL of absolute ethanol (200 proof).
Qubit™ dsDNA BR Assay kit (Life Technologies).
2.2 Library Preparation
Promega GoTaq Flexi.
25 mM MgCl2.
10 mM dNTPs.
Diethylpurocarbonate (DEPC) water.
100 % DMSO.
Pfu polymerase.
1 µg of good quality DNA per sample at a concentration of 100 ng/µL (see Note 1).
LE agarose.
1× TAE; Prepare 50× solution by dissolving 242 g Tris base in 750 mL of deionized water. Carefully add 57.1 mL glacial acetic acid, and 100 mL of 0.5 M EDTA, pH 8.0 and adjust solution to final volume of 1 L. Dilute the 50× solution to 1× in deionized water.
Ethidium bromide (10 mg/mL).
QIAquick Gel Extraction Kit (Qiagen).
MinElute PCR Purification Kit (Qiagen).
Isopropanol (200 proof).
500 mL of Ethanol (200 proof).
Agilent DNA 1000 kit or high sensitivity DNA kit (choose as needed, see Note 1).
L1-seq primers (order HPLC grade for library preparation). See Table 1.
Table 1.
Primers used for L1-seq
| Sequences 5′ to 3′ | |
|---|---|
| Primers with Illumina adapters: | |
| Adap 1 L 1HsG | CAAGCAGAAGACGGCATAOGAGCTCTTCCGATC TTGCACATGAOCCTAAAACTTAG |
| Adap2Seq1 | AATGATACGGCGACCACCGAGATCTACACTTTOCC TACACGACGACGCTCTTCCGATCT |
| L1 specific primers: | |
| L1HsSP1A2 | GGGAGATATACCTAATGCTAGATGACAC (specific for L 1Hs subset) |
| L1 “G” primer | TGCACATGTACOCTAAAACTTAG (specific for L 1Hs subset) |
| L1nt112out | GATGAACCCGTACCTCAGA |
| Degenerate primers: | |
| DEG Seq 1N5TCTGT | ACACTCTTTCCCTACACGACGACGCTCTTCCGA TCTNNNNNTCTGT |
| DEGSeq1N5CTTCT | ACACTCTTTCCCTACACGACGCTCTTCCGATCTNN NNNCTTCT |
| DEGSeq1N5TGCCT | ACACTCTTTCCCTACACGACGACGCTCTTCCGAT CTNNNNNTGCCT |
| DEGSeq1NTCTCA | ACACTCTTTCCCTACACGACGCTCTTCCGATCTN NNNNTCTCA |
| DEGSeq1N5CAGAG | ACACTCTTTCCCTACACGACGCTCTTCCGATCTNN NNNCAGAG |
| DEGSeq1N5TTGAA | ACACTCTTTCCCTACACGACGCTCTTCCGATCTNN NNNTTGAA |
| DEGSeq1N5CTTTG | ACACTCTTTCCCTACACGACGCTCTTCCGATCTNN NNNCTTTG |
2.3 Next-Generation Sequencing Data Analysis
Server with at least 4GB of RAM and L1-seq scripts properly formatted (https://github.com/adamewing/l1seq).
Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) and all relevant human genome files and indices as per Bowtie2 instructions.
2.4 Data Validation
GoTaq Green Master Mix (2×).
Diethylpurocarbonate (DEPC) water.
DNA at concentration of 12.5 ng/µL.
LE agarose.
1× TAE; Prepare 50× solution by dissolving 242 g Tris base in 750 mL of deionized water. Carefully add 57.1 mL glacial acetic acid, and 100 mL of 0.5 M EDTA (pH 8.0) and adjust solution to final volume of 1 L. Dilute the 50× solution to 1× in deionized water.
Ethidium bromide (10 mg/mL).
QIAquick Gel Extraction Kit (Qiagen).
Isopropanol (200 proof).
500 mL of Ethanol (200 proof).
L1SP1A2 primer, L1nt112out, L1 “G” primer (see Table 1).
3 Methods
3.1 Embedding and Cryosectioning Tissue
To begin, embed each piece of tissue to be assayed in OCT freezing medium. You can simply put a thin layer of the media onto a pre-chilled (≤20 °C) chuck.
Quickly placing the thawed tissue section onto the OCT.
Immediately cover the tissue in more OCT until it is barely visible through the medium. The OCT medium will change from clear to white, when the entire block of tissue/OCT is completely frozen, you can begin to cryosection the tissue for DNA extraction. It is best for the freezing to occur as rapidly as possible, to this end, a heat extractor can be used to enhance and shorten the freezing process and adherence to the chuck on which the tissue is being embedded in the OCT freezing medium.
Set the cryostat to slice sections of tissue between 10 and 30 µm.
During sectioning, carefully remove each roll of tissue and place 10–20 slices into a pre-chilled (≤20 °C) 1.5 mL microtube. See Note 2.
3.2 Extracting DNA from Sectioned Tissue
Remove microtubes with tissue slices from −80° freezer and place them on ice.
Add 360 µL of Buffer ATL (DNeasy Blood and Tissue kit). There is no need to further homogenize the tissue. Add 40 µL of proteinase K and mix thoroughly by vortexing (see Note 1).
Incubate at 55 °C overnight until the tissue is completely lysed. Vortex occasionally during incubation to help disperse samples.
Vortex for 15 s.
Add 400 µL of buffer AL (DNeasy Blood and Tissue Kit) to the sample and mix thoroughly by vortexing.
Immediately add 400 µL of ethanol (200 proof) and mix again thoroughly by vortexing (notes).
Pipette 750 µL of the mixture (including any precipitate) into the DNeasy Mini spin column placed in a 2 mL collection tube (provided in the kit).
Centrifuge at ≥6000 g for 1 min. Discard flow through.
Repeat steps 7 and 8 until all of mixture has been run though the same column for each sample.
After the final spin with the aforementioned mixture, discard the collection tube and replace it with a new 2 mL collection tube.
Add 500 µL of Buffer AW1. (Ensure that ethanol has been added to Buffer AW1 before use.)
Centrifuge for 1 min at ≥6000 g.
Discard flow-through and collection tube and place the spin column in a new 2 mL collection tube.
Add 500 µL of Buffer AW2. (Ensure ethanol has been added to Buffer AW2 before use.)
Centrifuge for 3 min at 20,000 g to dry the DNeasy membrane and then discard flow-through and collection tube.
Place the DNeasy Mini spin column in a clean 1.5 mL micro-centrifuge tube and pipet 100 µL of pre-warmed (55 °C) Buffer AE onto the DNeasy membrane.
Incubate at room temperature for 10 min.
Centrifuge for 1 min at ≥6000 g to elute.
Pipette an additional 50 µL of pre-warmed AE buffer directly onto DNeasy membrane. (There is no need to replace the micro-centrifuge tube. The additional DNA elution can be collected into the same tube up to a volume of no more than 150 µL.)
Incubate for 10 min at room temperature.
Centrifuge for 1 min at 20,000 g.
This protocol is adapted from the Qiagen handbook for the DNeasy Blood and Tissue Kit.
3.3 Measuring DNA Concentration (Qubit™)
Use the Qubit™ fluorometer to measure DNA concentration because it is one of the most accurate methods. Follow manufacturer protocols exactly and see Note 1. (http://www.ebc.uu.se/digitalAssets/176/176882_3qubitquickrefcard.pdf).
3.4 L1-seq Library Preparation (see Note 3)
Before beginning the relevant library prep PCRs, it is necessary to determine which samples will be pooled together in the library preparation. As many as ten samples can be pooled together without using barcoding (notes). Equal amounts of each sample must be put into the DNA pool to be used in the library. A total of 24 µL of pooled DNA at a concentration of 100 ng/µL is needed. If there is not enough DNA available from one or more samples, a modified protocol can be used (notes).
-
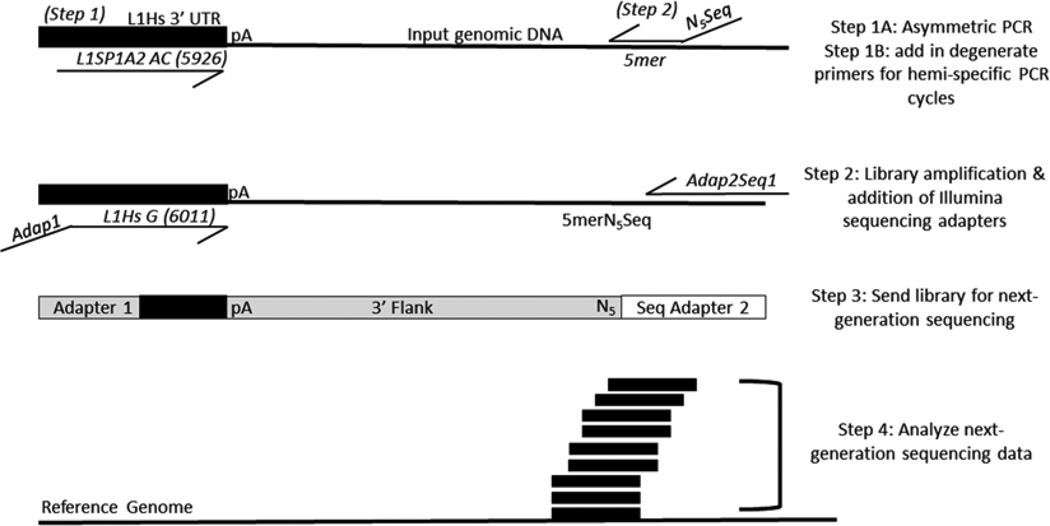
Round 1 PCR: Linear amplification of L1 flanks followed by a hemi-specific PCR incorporating the Illumina sequencing primer (Fig. 1). To prevent running out of master mix, make enough for nine reactions even though only eight reactions will be assembled. Master mix (per 1 reaction): 10 µL of Promega Go-Taq flexi buffer (5×), 6 µL of 25 mM MgCl2, 2 µL of 20 µM L1SP1A2 primer (the second primer for the reaction, the degenerate primers previously mentioned, will be added following the completion of five cycles of the PCR which consists of the linear amplification step), 0.5 µL of DMSO 100 %, 0.5 µL of FlexiTaq, 1 µL of 10 mM dNTPs, 2 µL of pooled DNA (at 100 ng/µL), 24 µL of DEPC water. The reaction should total 46 µL before the addition of 4 µL of the degenerate primers to be added after linear amplification is finished. When the linear amplification is finished, add 1 of each of the degenerate primers at 5 µM (e.g. DEGSeq1N5TCTGT) to each of the eight reactions. Use the following cycling program:
L1-Seq PCR 1- 95 °C for 2 min 30 s.
- 95 °C for 30 s.
- 58 °C for 1 min.
- 72 °C for 2 min.
- Go to step b (5×).
- 60 °C (pause and add 4 µL of degenerate primer into each of the eight reactions, one primer per reaction for each of the eight different degenerate primers).
- 95 °C for 30 s.
- 55 °C for 30 s.
- 72 °C for 1 min and 30 s.
- Go to step g (14×).
- 72 °C for 10 min.
- 4 °C hold.
Purify all eight reactions on eight separate Qiagen PCR Clean-up columns following the Qiagen protocol, eluting in 50 µL of pre-warmed (55 °C) EB (do a 10 min final incubation before elution to optimize DNA eluted from column).
-
L1-Seq PCR 2: Amplification of library and addition of the Illumina sequencing adapters (Fig. 1). Master mix (for 1 reaction): 12.5 µL of master mix (Promega GoTaq Green 2×), 1.5 µL of 20 mM primer Adap1L1HsG, 1.5 µL of 20 µM Adap2Seq1, 2.5 µL of purified round 1 product (1 degenerate primer per reaction), 7 µL of DEPC ddH2O. Again, make enough for nine reactions to prevent running out of master mix for the samples to be amplified. The reactions will each have a total of 25 µL. Use the following cycling program:
L1-Seq PCR 2- 95 °C for 2 min.
- 95 °C for 30 s.
- 62 °C for 30 s.
- 72 °C for 1 min.
- Go to step b (19×).
- 72 °C for 5 min.
- 4 °C Hold.
Resolve products on a 1 % TAE gel.
Excise the constellation of bands between 200 and 500 nucleotides with a sterile scalpel (using a different scalpel for each reaction) and purify the DNA using the Qiagen Gel Purification protocol.
Elute the library in 50 µL of pre-warmed EB buffer (55 °C with a 10 min incubation before elution).
Run each DNA sample on the Agilent Bioanalyzer with the DNA 1000 kit to get an accurate measure of concentration and the average size of the DNA amplified. Using the concentration and average size of the molecules, calculate how to add the DNA from all eight reactions in equimolar ratios to one tube. See Note 4.
After mixing the DNAs together, purify the entire mixture with the Qiagen MinElute PCR purification kit eluting in 50 µL of pre-warmed EB (55 °C and 10 min incubation at room temperature before elution).
End-polishing must be performed on the library because Taq leaves adenine overhangs which could cause problems when the library is annealed to the Illumina flow cell. To accomplish the end-polishing: mix 6 µL of 10× Pfu buffer, 2.5 µL of Pfu polymerase, 2.5 µL of 10 mM dNTPs, and 49 µL of library. Incubate for 1 h at 72 °C.
Purify reaction on a Qiagen MinElute column and elute in 10 µL of pre-warmed (55 °C) EB following a 10 min room-temperature incubation before elution.
Measure final DNA concentration with Qubit™ fluorometer to get an accurate concentration for sending samples for next-generation sequencing on the Illumina HiSeq 2500. Opt for single end sequencing and at least 100 bp reads. See Notes 3 and 5.
Fig. 1.
Scheme of L1-seq. The first PCR consists of five cycles which enrich for sequences containing human-specific L1 sequences via primer extension with the L1Sp1A2 primer using diagnostic nucleotides for the human specific subfamily of L1. After enrichment for human-specific L1 flanks in the first five cycles, each reaction has one degenerate primer added. There are eight degenerate primers, each with a specified 5-mer at the 3′ end preceded by five degenerate bases (NNNNN) and a sequencing primer used for the Illumina platform. Eight different reactions are performed, each with a unique 5mer. The second PCR enriches for human-specific L1 3′ flanks by utilizing another diagnostic nucleotide in the 3′ UTR of L1 and adds the necessary adapter sequences via primer overhangs. The products of this reaction are mixed in equimolar ratios before sequencing on the Illumina 2500 platform. Following sequencing and initial processing, the reads are aligned to the human genome (hg19) and L1 insertions are identified for validation
3.5 Analyzing the Next-Generation Sequencing Data (see Note 6)
Obtain the sequencing reads from the core or center where the samples were sequenced and transfer them into the L1-seq directory on the server in which all the correctly formatted L1-seq scripts reside. These scripts can be acquired from https://github.com/adamewing/l1seq.
Obtain the contents of a database with all reference L1 insertions and polymorphic L1 insertions which have been previously published to use for filtering sequencing data. A database of L1 insertions may be obtained at: http://nar.oxfordjournals.org/content/43/D1/D43 [14].
Once the documents are downloaded through the terminal, they must be unzipped. To unzip the fastq.gz files type “gunzip –d FILE_NAME.fastq.gz &” (the & symbol allows the unzipping to run in the background so that you can set all the files and can unzip simultaneously by typing this command for each file in turn.)
After the files are unzipped, use the script to run bowtie and create indices for your data. You can execute this process with the command “./run_bowtie.py/whatever_fastq/directions/to/bowtie directions/to/hg19.fa &”. For any process which takes more than 10 min, it is helpful to use the screen function by typing “screen –rAad” which will allow for the monitoring of all the processes simultaneously running. It also enables the user to monitor the total memory being used for analysis and the length of time each process has been running.
Once the run_bowtie script finishes, run the l1seq.py script as follows: “./l1seq.py –bam whatever.ba, > whatever.l1seq.txt &”.
Once all of the L1-seq.txt files are made, all the files need to be compressed for sorting. To compress the files, type “bgzip whatever_l1seq.txt &”.
To sort the files, type “tabix –s 1 –b 3 –e 4 watever.l1seq.txt.gz &”.
All the files must be compared to one another (e.g. normal compared with tumor, etc.) to do this analysis, type “./compare. py group1_L1seq.txt.gz group2_l1seq.txt.gz group3_l1seq.txt.gz > filename_for_comparisons.tsv &”.
Finally, after the comparison file has been made, primers must be designed for validation of the data. It is best to run the makeprimers.pl script on the entire comparison file before looking at the data because the script does not take long to run and having the primer sequences ready to order is very useful. To run this final script, type “./makeprimers.pl filename_for_comparisons.tsv > filename_for_comparisons_with_primers.tsv &”. Use sftp to transfer the files back to your local computer if desired.
3.6 Validating the Predicted Insertions from L1-Seq with Site-Specific PCR
The presence of nonreference insertions is validated with site-specific PCR (Fig. 2). If the samples are not barcoded (see Note 6), all samples in a pool must be evaluated for the presence or absence of a predicted insertion. The DNA from each input sample from a pool needs to be at 12.5 ng/µL and 2 µL of DNA used per reaction. The primers will be named by the makeprimers.pl script as “filled site” or FS and “empty site” ES refer to Fig. 2 for orientation of primers with regard to the potential insertion. For each validation to be complete, the FS and L1SP1A2 primer reaction needs to be performed on all samples in the pool from which the prediction came. If comparing two states of the same tissue such as tumor and normal and the insertion is predicted only in one, the reaction must also be performed on both DNA samples. A control reaction can also be performed with the “empty site” or ES primer and the FS primer. Both the FS and ES primers are genomic and will produce a product of predetermined size in any DNA sample regardless of presence or absence of an insertion.
-
For the FS/L1SP1A2 (filled site PCR) use the following master mix (1×): 12.5 µL of Promega GoTaq green (2×) master mix, 0.8 µL of 20 µM FS primer, 1.6 µL of 20 µM L1SP1A2, 2 µL of genomic DNA (12.5 ng/µL), 8.2 µL of DNA-free H2O. For the FS/ES primers (empty site PCR) use the following master mix (1×): 12.5 µL of Promega GoTaq green (2×) master mix, 1 µL of 20 µM FS primer, 1 µL of 20 µM ES primer, 2 µL of genomic DNA (12.5 ng/µL), 9.5 µL of DNA-free H2O. Use the following parameters for the PCR:
3′ L1 Validation PCR- 95 °C for 2 min.
- 95 °C for 30 s.
- 57 °C for 30 s.
- 72 °C for 1 min and 30 s.
- Go to step b (29×).
- 72 °C for 5 min.
- 4 °C Hold.
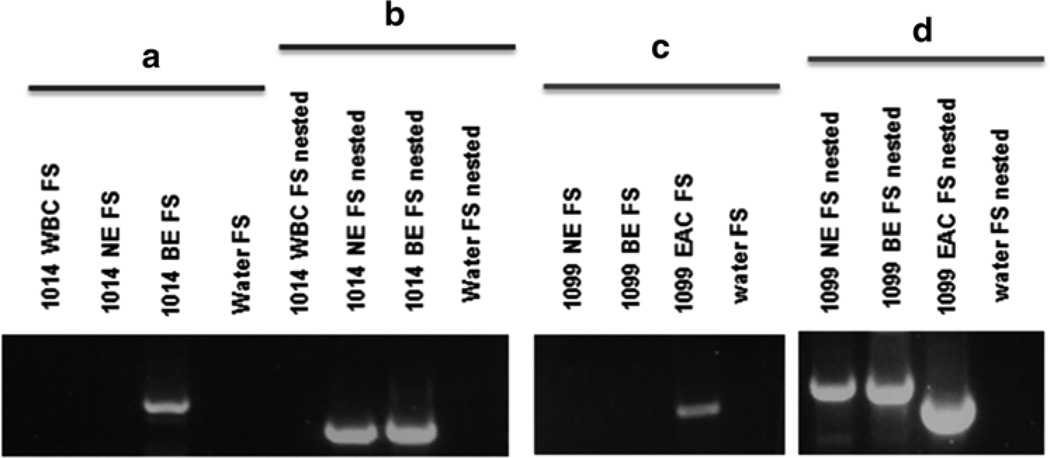
Run the PCR products on a 1.5 % TAE gel to resolve the products (see Note 7). Take images of the gel while it is exposed to UV to visualize the products. Excise fragments which are unique to only one of the samples upon which the PCR was run (Fig. 2b). Isolate the DNA from the band and send for sequencing. If no clear filled site band is uniquely present in one of the samples tested, a nested PCR may be necessary (Fig. 2a). Alternatively, the PCR conditions can be further optimized to attempt to amplify the insertion (see Notes 7 and 8).
Finally, send the DNA for Sanger sequencing to ensure it is the correct product. Sequence the product with both the FS primer as well as the L1SP1A2 primer. When the sequence from the FS is aligned to the genome with BLAT or another alignment algorithm, part of the sequence should align to the genome and a poly T tract should also be visible adjacent to the aligning sequence. For the sequence from the reaction performed with the L1-specific primer, the 3′ end of the L1 should be visible in addition to the poly-A tail (Fig. 2c) (see Notes 7 and 8).
-
To find the 5′ end of the insertion, several different methods can be utilized. Because many new L1 insertions are truncated on the 5′ end of the element, it is frequently possible to detect the 5′ end of the element by using the reverse complement of the L1SP1A2 primer (L1 GTG primer) with the ES primer. To do this, make the master mix as follows: mix (1×): 12.5 µL of Promega GoTaq green (2×) master mix, 0.8 µL of 20 µM ES primer, 1.6 µL of 20 µM L1 GTG primer, 2 µL of genomic DNA (12.5 ng/µL), 8.2 µL of DNA-free H2O. For insertions with a longer 5′ end present, this PCR will likely fail; however, it is possible to tile across the L1 element with various primers at different locations (e.g. L1nt112out) in the element accompanied by the empty site primer to find the 5′ end. For this PCR, use the following master mix (1×): 12.5 µL of Promega GoTaq green (2×) master mix, 0.8 µL of 20 µM ES primer, 1.6 µL of 20 µM L1 internal primer, 2 µL of genomic DNA (12.5 ng/µL), 8.2 µL of DNA-free H2O.
5′ L1 GTG PCR Parameters- 95 °C for 2 min.
- 95 °C for 30 s.
- 57.5 °C for 1 min and 30 s.
- 72 °C for 3 min.
- Go to step b (29×).
- 72 °C for 5 min.
- 4 °C Hold.
5′ L1 Internal Primer (e.g. L1nt112out) PCR Parameters- 95 °C for 2 min.
- 95 °C for 30 s.
- 57 °C for 30 s.
- 72 °C for 45 s.
- Go to step b (29×).
- 72 °C for 5 min.
- 4 °C Hold.
Fig. 2.
Validation of results. (a) Diagram of the PCR validation scheme for putative insertions, the 3′ end of the LINE-1 insertion is pictured adjacent to a poly A tail. The nested empty site and filled site primers are flanking the empty and filled site primers. In a nested PCR, the nested primers are used in the first of two reactions. One and a half µL of product from the first reaction (with ES and FS primers) are used as template in a second PCR with the nested primers to amplify difficult or rare products. (b) Two examples of validations for insertions present in only tumor and absent from normal DNA. On the left, a PCR result depicting both the empty site (ES) and filled site (FS) products for both the normal and tumor DNA samples from patients. Only in the tumor of patient 11 is a filled site band present confirming the insertion is present. In the image on the right side on b, a PCR depicting another validation of a somatic insertion present in BE and absent from normal esophageal and white blood cell DNA. There is only a band present in the BE sample for the FS PCR; however, the ES PCR has bands for all three DNA samples as a positive control. (c) An insertion sequence with the unique genomic DNA (blue), target site duplications (purple), LINE-1 sequence (red), and the poly-A tail sequence (orange) [15]
Acknowledgments
This work was funded by a P-50 grant awarded to H.H.K. Jr.
Footnotes
If very little DNA is available for both library prep and validation PCRs, L1-seq can still be successfully performed. L1-seq has successfully been executed with as little as 25 ng of input per sample for the library prep. For the steps following next-generation sequencing, whole genome amplification can be used (e.g. the Qiagen Repli-G kit) to provide more DNA to use for the validation PCRs. If adjusting the amount of DNA used, be sure to account for volume changes and the concentrations of the other reagents to ensure all final concentrations are the same as described in the original technique.
If the tissue sample is large enough, more than one tube of tissue slices can be made. Following the sectioning, tissue slices should be stored at −80 °C until it is time to isolate DNA. Embedding tissue in OCT freezing medium is only one way of extracting DNA from frozen tissue.
When first performing L1-seq it is prudent to execute a TA cloning step after completing the libraries and mixing them in equimolar ratios, but before completing the end-polishing step. To do this, simply take 1 µL from the mixed libraries and use it in a Topo TA cloning reaction. Follow kit instructions and after growing colonies overnight, select 12 or more from each plate for colony PCR. Following colony PCR, run the product on a gel to be sure the cloning worked effectively, select some or all of the successful clones for Sanger sequencing. When analyzing the Sanger sequencing, look for different L1Ta elements from many different areas of the genome. Essentially, this is a step to check that the library does not consist of amplicons of only a handful of LINE-1 elements in the genome and that elements in the genome are equally represented in the library. This step does not need to be performed for every library prep; however, if a problem occurs with next-generation sequencing, this step could consequently be taken to determine whether or not overrepresentation of a few elements precluded successful sequencing.
Occasionally, one of the degenerate primer reactions will not be as robust as the other reactions and when the libraries are run on a gel, the amount of DNA present is variable between reactions. This may not be an issue if there is enough DNA present after the gel purification for the samples to easily be mixed in equal amounts. However, if the concentration of the DNA isolated from the gel purification step is too little to continue without grossly diminishing the amount of total DNA in the combined library, simply repeat the second reaction of L1-seq and combine the isolated DNA from both gel purifications and concentrate the DNA. If the DNAs from the respective degenerate primer reactions are run on the Bioanalyzer and produce very different size distributions of products, it may be necessary to repeat the second L1 PCR again on that DNA sample as well. Ideally, the average product size for each degenerate primer reaction should be within one standard deviation of 350 nucleotides. If the size varies more than one standard deviation from 350, the reaction should be repeated and rerun on a gel. If the size is wrong, it is likely that the excision was initially imprecise.
If the DNA being measured at any point in the library prep is at a low concentration and undetectable with the standard Qubit™ broad range kit or the Agilent 1000 DNA chip, there are low concentration versions of these reagents available.
Barcoding may also be utilized with this technique; however, results may vary. In 2012, Evrony et al. performed L1-seq using barcoding and were able to validate some new LINE-1 insertions following sequencing analysis. However, other groups have had more difficulty getting the technique to work well and seem to have more success with pooling samples without barcodes. Pooling samples without barcodes does create more work for the validation steps of the technique; however, it seems to have more reproducible results.
If validation PCRs are unsuccessful after many attempts, be sure to check the specificity of the primers being used in the amplification. Oftentimes, it is helpful to perform a nested PCR following the first conventional PCR to amplify difficult or low-copy insertions which may have been easily detectable with next-generation sequencing and not with Sanger sequencing. You can nest both the filled site primers as well as the L1Ta-specific primers to increase the specificity of the reaction greatly. Nested PCR along with an increase in cycle numbers and/or altering the melting temperature of the PCR often alleviates validation PCR issues.
With regard to choosing predicted insertions for validation, one of two main methods may be employed. A random number generator can be used to select putative somatic insertions for validation which will potentially give a good estimate of the number of true somatic insertions in the data set. Alternatively, putative somatic insertions with unique read counts above 5, map scores of 1, and alignment windows of at least 100 base pairs can be selected for validation. Depending on the validation rate with the primary insertions selected, the level of stringency can be altered until the ideal validation rate is achieved. A validation rate above 60 % is generally acceptable for this technique; however, PCR optimization, good primer design, and good DNA are key to successful validations.
References
- 1.Voytas DF, Cummings MP, Konieczny A, Ausubel FM, Rodermel SR. copia-like retrotransposons are ubiquitous among plants. Proc Natl Acad Sci. 1992;89(15):7124–7128. doi: 10.1073/pnas.89.15.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22(3):191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35(1):41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 4.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet. 2003;73(6):1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24(4):363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 6.Ewing AD, Kazazian HH., Jr High-throughput sequencing reveals extensive variation in human specifi c L1 content in individual human genomes. Genome Res. 2010;20(9):1262–1270. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solyom S, Ewing AD, Rahrmann EP, Doucet T, Nelson HH, Burns MB, Harris RS, Sigmon DF, Casella A, Erlanger B, Wheelan S, Upton KR, Shukla R, Faulkner GJ, Largaespada DA, Kazazian HH., Jr Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012;22(12):2328–2338. doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, Lohr JG, Harris CC, Ding L, Wilson RK, Wheeler DA, Gibbs RA, Kucherlapati R, Lee C, Kharchenko PV, Park PJ. Landscape of somatic retrotransposition in human cancers. Science. 2012;337(6097):967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shukla R, Upton KR, Muñoz-Lopez M, Gerhardt DJ, Fisher ME, Nguyen T, Brennan PM, Baillie JK, Collino A, Ghisletti S, Sinha S, Iannelli F, Radaelli E, Dos Santos A, Rapoud D, Guettier C, Samuel D, Natoli G, Carninci P, Ciccarelli FD, Garcia-Perez JL, Faivre J, Faulkner GJ. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153(1):101–111. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helman E, Lawrence MS, Stewart C, Sougnez C, Getz G, Meyerson M. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014;24(7):1053–1063. doi: 10.1101/gr.163659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, Talbot RT, Gustincich S, Freeman TC, Mattick JS, Hume DA, Heutink P, Carninci P, Jeddeloh JA, Faulkner GJ. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479(7374):534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, Park PJ, Walsh CA. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151(3):483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tubio JM, Li Y, Ju YS, Martincorena I, et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014;345(6196):1251343. doi: 10.1126/science.1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mir AA, Philippe C, Cristofari G. euL1db: the European database of L1HS retrotransposon insertions in humans. Nucleic Acids Res. 2014;43(Database issue):D43–D47. doi: 10.1093/nar/gku1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doucet-O’Hare T, Rodic N, Sharma R, Darbari I, Abril G, Choi JA, Young Ahn J, Cheng Y, Anders RA, Burns KH, Meltzer SJ, Kazazian HH., Jr LINE-1 expression and retrotransposition in Barrett’s esophagus and esophageal carcinoma. Proc. Natl. Acad Sci. 2015;112(35):4894–4900. doi: 10.1073/pnas.1502474112. [DOI] [PMC free article] [PubMed] [Google Scholar]