Abstract
Stabilization of mutant p53 (mutp53) in tumours greatly contributes to malignant progression. However, little is known about the underlying mechanisms and therapeutic approaches to destabilize mutp53. Here, through high-throughput screening we identify statins, cholesterol-lowering drugs, as degradation inducers for conformational or misfolded p53 mutants with minimal effects on wild-type p53 (wtp53) and DNA contact mutants. Statins preferentially suppress mutp53-expressing cancer cell growth. Specific reduction of mevalonate-5-phosphate by statins or mevalonate kinase knockdown induces CHIP ubiquitin ligase-mediated nuclear export, ubiquitylation, and degradation of mutp53 by impairing interaction of mutp53 with DNAJA1, a Hsp40 family member. Knockdown of DNAJA1 also induces CHIP-mediated mutp53 degradation, while its overexpression antagonizes statin-induced mutp53 degradation. Our study reveals that DNAJA1 controls the fate of misfolded mutp53, provides insights into potential strategies to deplete mutp53 through the mevalonate pathway–DNAJA1 axis, and highlights the significance of p53 status in impacting statins’ efficacy on cancer therapy.
The tumour suppressor p53 regulates transcription of numerous downstream target genes involved in cell cycle arrest, apoptosis and metabolism1,2. Loss of p53 activity by gene deletion or mutations in normal cells causes uncontrolled cell proliferation and death, leading to immortalization and ultimately cancer. Missense mutations in p53 frequently occur at hotspot amino acids in the DNA-binding domain, resulting in loss of function as a transcription factor3. These p53 mutants also show dominant-negative activities through hetero-oligomerization with wtp53. Additionally, mutp53 shows oncogenic gain-of-function (GOF) activities, such as enhanced tumour progression, metastatic potential and drug resistance4.
Increasing evidence suggests that mutp53 is stabilized in tumours but not in normal tissues5, and ubiquitin ligases other than MDM2 may play roles in mutp53 ubiquitylation6. Thus, mechanisms of mutp53 stabilization or degradation in tumours are not exactly the same as those for wtp53. Moreover, recent findings suggest that stabilization of mutp53 in tumours is crucial for its GOF activity, while its knockdown reduces oncogenic properties of mutp53-carrying cancers7,8, suggesting that malignant progression of cancer cells is dependent on mutp53 stabilization. Hence, it is crucial to determine the mechanisms specific to mutp53 stabilization/degradation and identify compounds that destabilize mutp53 without altering wtp53 levels. Given the high frequency of p53 mutations found in numerous human cancers9, compounds that specifically deplete mutp53 could be used for targeted cancer therapy with minimal side effects.
Statins induce mutp53 degradation by CHIP
To identify compounds that deplete mutp53, we used a Saos2 (p53null) cell line constitutively expressing a chimaeric fusion protein of p53R175H and luciferase (p53R175H-Luc) or luciferase alone (Luc). We screened chemical libraries comprising of ~9,000 compounds in p53R175H-Luc cells by performing luciferase assays and identified 44 compounds that decreased luciferase activity compared with dimethylsulfoxide (DMSO; first screening, Supplementary Fig. 1a). Next, both p53R175H-Luc and Luc cells were treated with these 44 compounds at 8 different concentrations to establish dose–response relationships and eliminate false positives including luciferase inhibitors and general transcription/translation inhibitors (second screening, Supplementary Fig. 1a). The top ten compounds showing dose-dependent reduction of luciferase activity preferentially in p53R175H-Luc cells were examined further for their effects on the levels of p53R175H and wtp53 by western blotting. Of these compounds, seven failed to deplete p53R175H or reduced wtp53 as well (Supplementary Fig. 1b). The remaining three compounds (lovastatin, atorvastatin and mevastatin) belong to a class of cholesterol-lowering drugs called statins that inhibit 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase10.
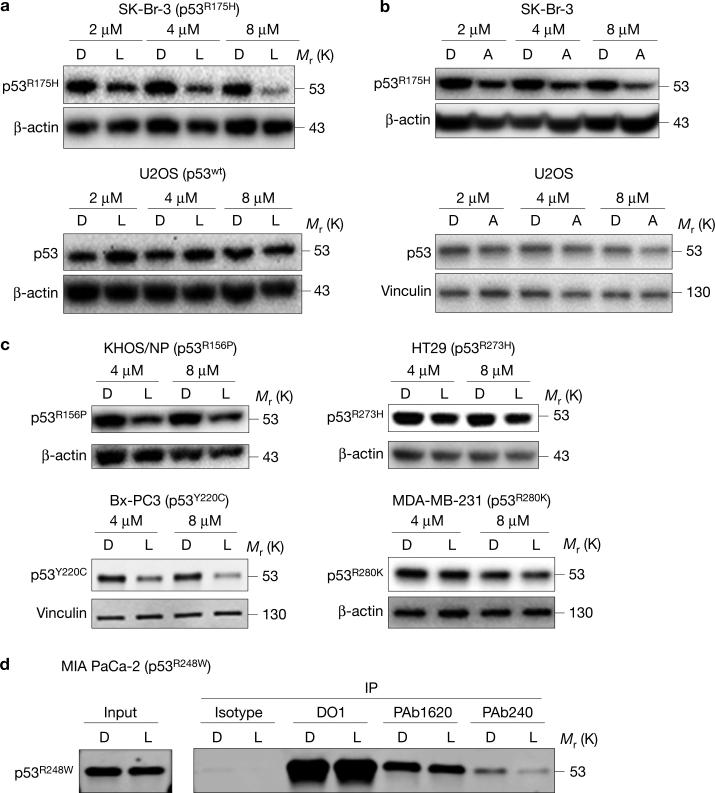
Next, we studied whether statins altered the levels of p53R175H and wtp53 using SK-Br-3 and U2OS cell lines, respectively. Lovastatin, a lipophilic statin, depleted p53R175H in a dose-dependent manner with no reduction in wtp53 (Fig. 1a). The efficient reduction of p53R175H was observed after 24 h of lovastatin treatment (Supplementary Fig. 1c). The specificity of mutp53 depletion by lovastatin was also demonstrated in SJSA-1 (p53wt) cells exogenously expressing p53R175H (Supplementary Fig. 1d). Similarly, another lipophilic statin, atorvastatin, depleted mutp53 in a dose-dependent manner without affecting wtp53 (Fig. 1b). Apart from p53R175H, lovastatin depleted other conformational p53 mutants including p53R156P (KHOS/NP), p53V157F (H-2087) and p53Y220C (BxPC-3), but showed minimal effects on DNA contact mutants, such as p53R273H (HT29) and p53R280K (MDA-MB-231) (Fig. 1c and Supplementary Fig. 1e). To confirm whether the effect of lovastatin was specific for conformational or misfolded mutp53, we performed immunoprecipitation studies, using MIA PaCa-2 (p53R248W) cells. The p53R248W mutant has both folded/native and misfolded/denatured conformations that can be detected with conformation-specific antibodies PAb1620 and PAb240, respectively. Results showed that lovastatin depleted predominantly the misfolded form of p53R248W (Fig. 1d). These results suggest that ‘statins’ deplete mainly the misfolded form of p53.
Figure 1.
Statins reduce levels of conformational or misfolded mutp53. (a) Western blotting (WB) for p53 and β-actin using SK-Br-3 (p53R175H) and U2OS (p53wt) cells treated with the indicated concentrations of DMSO (D) or lovastatin (L) for 24 h. (b) WB for p53 and β-actin or vinculin using SK-Br-3 and U2OS cells treated with the indicated concentrations of DMSO (D) or atorvastatin (A) for 24 h. (c) WB for the indicated proteins using KHOS/NP (p53R156P), BxPC-3 (p53Y220C), HT29 (p53R273H) and MDA-MB-231 (p53R280K) cells treated with D or L for 24 h. (d) Immunoprecipitation (IP) for total p53 (DO1), the folded/native form of p53R248W (PAb1620), and the misfolded/denatured form of p53R248W (PAb240), using MIA PaCa-2 cells treated with D or L, followed by WB for p53 (7F5). Additional results are shown in Supplementary Fig. 1. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
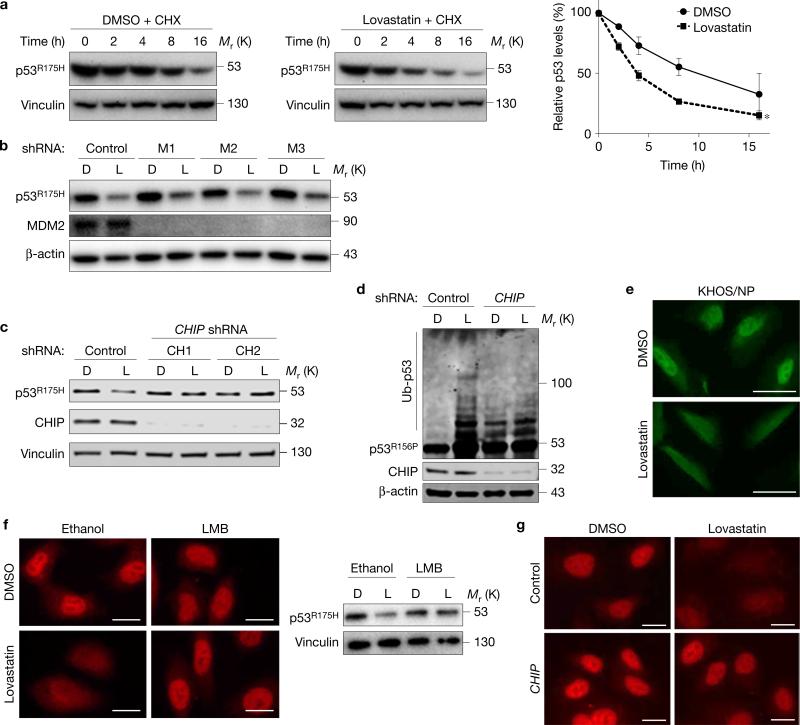
To understand mechanisms of statin-induced mutp53 depletion, we first examined whether statins altered messenger RNA expression of p53; no change was observed in both SK-Br-3 and U2OS cells (Supplementary Fig. 2a). We next tested whether statins decreased the half-life of mutp53 and found that lovastatin accelerated the degradation of p53R175H (Fig. 2a) and p53R156P (Supplementary Fig. 2b). This effect was due to enhanced ubiquitylation of mutp53 by lovastatin (Supplementary Fig. 2c). To identify ubiquitin ligases responsible for statin-induced mutp53 degradation, we first tested MDM2, a major ubiquitin ligase for p53, but knockdown of MDM2 did not nullify lovastatin-induced mutp53 degradation in SK-Br-3 and KHOS/NP cells (Fig. 2b and Supplementary Fig. 2d). However, statin-induced degradation and ubiquitylation of mutp53 were considerably nullified by downregulation of another ubiquitin ligase for p53, CHIP (C terminus of Hsc70-interacting protein, also known as STUB1, Fig. 1c,d and Supplementary Fig. 2e), which is known to ubiquitylate and induce degradation of misfolded proteins including mutp536.
Figure 2.
Statins induce mutp53 degradation by CHIP. (a) WB for p53 and vinculin using SK-Br-3 cells treated with cycloheximide (CHX, 50 nM) for the indicated time periods (h) following pre-incubation with DMSO or lovastatin (4 μM) for 12 h. Right: graph showing relative p53R175H levels compared with those without CHX. Error bars, means ± s.d. (n = 3 independent experiments), *P < 0.05; Student's t-test (two-tailed). (b) WB for the indicated proteins using SK-Br-3 cells infected with non-silencing control or MDM2 (M1, 2, 3) shRNA-encoding lentiviral vectors and treated with DMSO (D) or lovastatin (L) at 4 μM for 24 h. (c) WB for the indicated proteins using SK-Br-3 cells infected with lentiviral vectors encoding non-silencing control or CHIP (CH1, 2) shRNAs and treated with D or L (4 μM) for 24 h. (d) Ubiquitin assays for p53R156P and WB for the indicated proteins. KHOS/NP cells with or without CHIP knockdown were transfected with an ubiquitin-encoding plasmid, treated with D or L (4 μM) for 22 h, further incubated with 30 μM of MG132 for 6 h, and harvested with hot SDS lysis buffer. (e) Immunofluorescence for p53R175H in KHOS/NP cells treated with D or L for 18 h. Scale bars, 50 μm. (f) Immunofluorescence (left) and WB (right) for p53R175H using SK-Br-3 cells treated with D or L along with vehicle (ethanol) or leptomycin B (LMB, 50 nM) for 18 h. Scale bars, 50 μm. (g) Immunofluorescence for p53R175H using SK-Br-3 cells infected with control or CHIP shRNA-encoding lentiviral vectors and treated with DMSO or lovastatin. Scale bars, 50 μm. Statistics source data for a are provided in Supplementary Table 1. Additional results are shown in Supplementary Fig. 2. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
To examine the effect of statins on cellular localization of mutp53, we performed immunofluorescence studies for p53. Lovastatin treatment induced cytoplasmic localization and degradation of p53R156P in KHOS/NP cells (Fig. 2e). We next treated SK-Br-3 cells with lovastatin in the presence or absence of a nuclear export inhibitor leptomycin B (LMB). LMB treatment completely rescued statin-induced nuclear export and degradation of p53R175H (Fig. 2f), suggesting that nuclear export of mutp53 was required for statin-induced mutp53 degradation. Also, knockdown of CHIP almost completely abrogated the p53R175H nuclear export and degradation induced by lovastatin (Fig. 2g). Additionally, CHIP knockdown led to an overall increase in the half-life of p53R175H in SK-Br-3 cells, even in the presence of lovastatin, thus cancelling statin's effect on mutp53 degradation (Supplementary Fig. 2f). These results suggest that CHIP plays key roles in statin-induced molecular processes of nuclear export, ubiquitylation, and degradation of conformational/misfolded mutp53, reminiscent of the functional relationship between wtp53 and its ubiquitin ligase MDM211.
Reduced MVP mirrors statins’ effects on mutp53
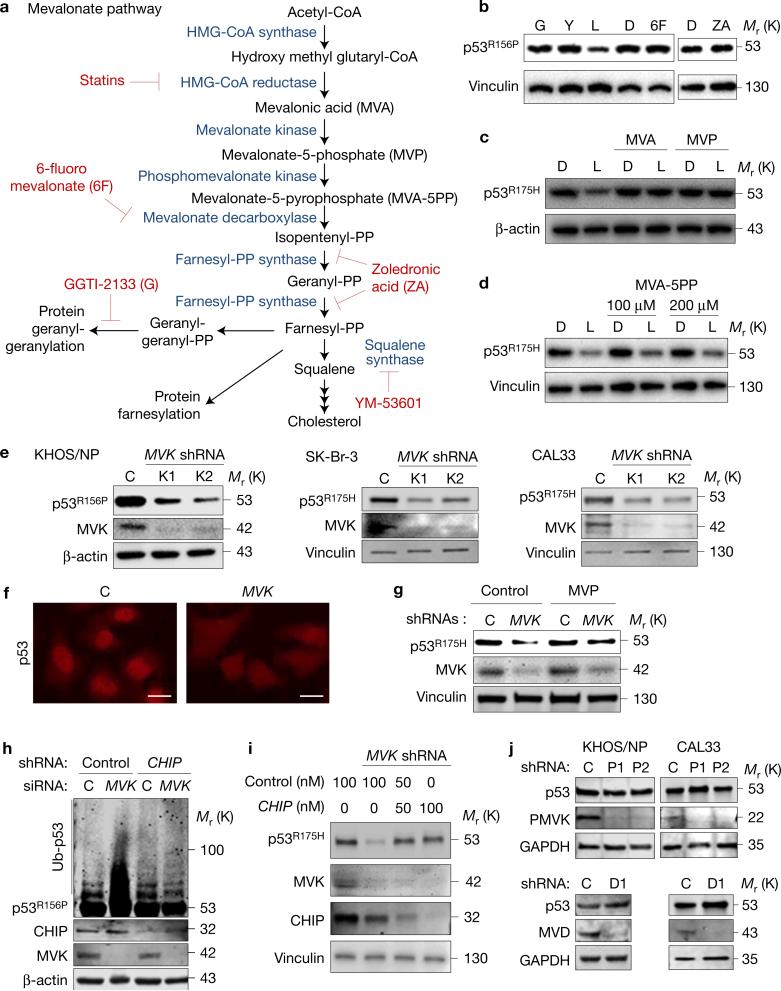
Statins inhibit HMG-CoA reductase activity that catalyses the first rate-limiting step in the mevalonate pathway (Fig. 3a), and hence are used to treat hypercholesterolaemia12. HMG-CoA reductase also regulates protein prenylation (farnesylation and geranyl-geranylation) that facilitates membrane attachment of target proteins involved in cell adhesion, migration and proliferation (for example, Rho, Rac, Ras)13. To examine whether mevalonate pathway inhibitors other than lovastatin also induced mutp53 degradation, we treated KHOS/NP cells with GGTI-2133, YM-53601, 6-fluoromevalonate and zoledronic acid (Fig. 3a). Surprisingly, none of these inhibitors depleted mutp53 (Fig. 3b).
Figure 3.
Reduced MVP mirrors statins’ effects on mutp53. (a) The mevalonate pathway and its inhibitors. (b) WB for p53R156P and vinculin using KHOS/NP cells treated with DMSO (D) or different mevalonate pathway inhibitors including GGTI-2133 (G, 4 μM), YM-53601 (Y, 4 μM), lovastatin (L, 4 μM), 6-fluoromevalonate (6F, 100 μM) and zoledronic acid (ZA, 25 μM) for 24 h. (c,d) WB for p53 and loading controls (β-actin or vinculin) using SK-Br-3 cells treated with D or L (4 μM) in the presence or absence of mevalonic acid (MVA, 200 μM; c), mevalonate-5-phosphate (MVP, 200 μM, c), or MVA-5PP (100 and 200 μM; d) for 24 h. (e) WB for p53, MVK and loading controls using KHOS/NP, SK-Br-3 and CAL33 (p53R175H) cells infected with lentiviral vectors encoding non-silencing control (C) or MVK shRNAs (K1, K2). (f) Immunofluorescence for p53 using SK-Br-3 infected with lentiviral vectors encoding control (C) or MVK shRNAs. Scale bars, 50 μm. (g) WB for the indicated proteins using SK-Br-3 cells with or without MVK knockdown supplemented with vehicle (water, control) or MVP (200 μM) for 24 h. (h) Ubiquitin assays for p53R156P and WB for the indicated proteins using KHOS/NP cells with or without knockdown of CHIP and/or MVK. (i) WB for the indicated proteins using SK-Br-3 cells with or without knockdown of MVK, along with transfection of control or CHIP siRNAs (50 or 100 nM). (j) WB for the indicated proteins using KHOS/NP and CAL33 cells infected with lentiviral vectors encoding control (C), PMVK (P1, P2) or MVD (D1) shRNAs. Additional results are shown in Supplementary Fig. 3. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
To determine the specific effectors in the mevalonate pathway critical for statin-induced mutp53 degradation and to mitigate the possibility of statins’ off-target effects, we treated SK-Br-3 cells with statins along with or without supplementation of mevalonic acid (MVA) or mevalonate-5-phosphate (MVP). Supplementation of either MVA or MVP completely rescued lovastatin-induced p53R175H degradation (Fig. 3c), suggesting that statin-induced mutp53 degradation was specifically caused by inhibition of the mevalonate pathway. However, supplementation of mevalonate-5-pyrophosphate (MVA-5PP) failed to rescue statin-induced mutp53 degradation in both SK-Br-3 and KHOS/NP cells (Fig. 3d and Supplementary Fig. 3a). Moreover, supplementation with MVP, but not MVA-5PP, abrogated the accelerated degradation of p53R175H by statins (Supplementary Fig. 3b). We confirmed that both MVP and MVA-5PP were successfully delivered to the cells, as they rescued cholesterol production in these cells treated with statins, thus restoring the mevalonate pathway (Supplementary Fig. 3c). These observations suggest that specific reduction of cellular MVP by statins triggers mutp53 degradation.
An alternative way to reduce cellular MVP levels is to downregulate mevalonate kinase (MVK, Fig. 2a). Knockdown of MVK by its specific short hairpin RNAs (shRNAs; K1, K2) also led to significant reduction in mutp53 levels in KHOS/NP, SK-Br-3 and CAL33 (p53R175H) cells (Fig. 3e). Similar to statin treatment, MVK knockdown induced nuclear export and degradation of p53R175H in SK-Br-3 cells (Fig. 3f), and supplementation with MVP rescued MVK knockdown-induced mutp53 degradation (Fig. 3g). Moreover, ubiquitylation and degradation of mutp53 induced by MVK knockdown were nullified by simultaneous knockdown of CHIP (Fig. 3h,i), showing that MVK knockdown phenocopied statins’ effects on mutp53. We also showed that supplementation with MVP, but not MVA, substantially rescued the inhibited cholesterol production induced by MVK knockdown in CAL33 cells (Supplementary Fig. 3d), confirming the expected reduction in MVP levels following MVK knockdown. Moreover, downregulation of phosphomevalonate kinase (PMVK) or mevalonate decarboxylase (MVD), immediate downstream enzymes of MVK in the mevalonate pathway, did not reduce levels of p53R156P and p53R175H (Fig. 3j). These results indicate an unappreciated role of MVP in regulating mutp53 stability, where reduced MVP by statins or MVK knockdown induces CHIP-mediated nuclear export, ubiquitylation, and degradation of mutp53 in a manner independent of protein prenylation.
Statins favourably impede mutp53–tumour growth
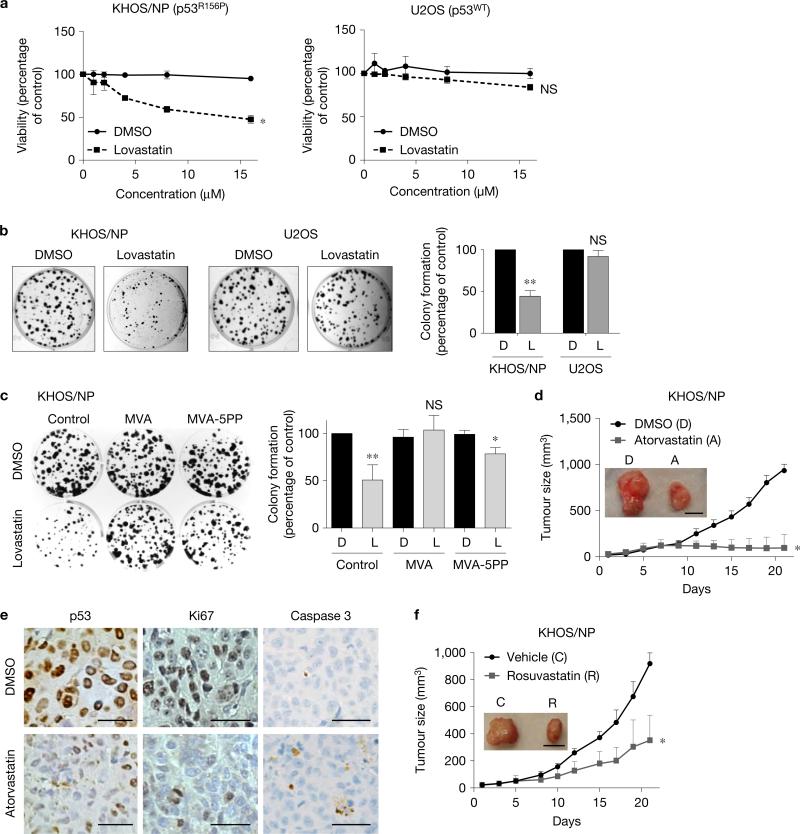
Statins suppress malignant properties of cancer cells via inhibition of prenylation of proteins involved in tumour progression13. Intriguingly, mutp53 upregulates enzymes involved in the mevalonate pathway, while inhibition of protein prenylation by statins impairs growth of mutp53-expressing breast cancer cells in three-dimensional culture14. We also found that statins preferentially suppressed viable cell proliferation and colony formation of KHOS/NP (p53R156P) cells with minimal effects on U2OS (p53wt) cells (Fig. 4a,b). Additionally, we used SK-Br-3 (p53R175H), HT29 (p53R273H) and HCT116 subcell lines with different p53 status (p53wt, p53null, p53null+R175H, p53null+R273H), and found that statins showed robust effects on mutp53 levels, cell viability and colony formation in cells expressing conformational p53R175H, and minimal effects on those expressing wtp53, DNA contact p53R273H, or null for p53 (Supplementary Fig. 4a, b). Reduced colony formation of KHOS/NP cells by statins was rescued completely by supplementation with MVA, but only partially by MVA-5PP (Fig. 4c), suggesting that reduced colony formation by statins was due to combinatory effects of inhibition of protein prenylation and mutp53 degradation.
Figure 4.
Statins favourably impede mutp53-tumour growth. (a) MTT assays using KHOS/NP and U2OS cells treated with various concentrations of DMSO or lovastatin for 48 h. Error bars, means ± s.d. (n = 3 independent experiments), *P < 0.05; NS, not significant; Student's t-test (two-tailed). (b) Colony formation assays using KHOS/NP and U2OS cells (500) seeded onto 6-well plates and treated with DMSO or lovastatin (4 μM) every other day for 10 days. Representative images (left) and a summarized graph (right). Error bars, means ± s.d. (n = 3 independent experiments), **P <0.01; Student's t-test (two-tailed). (c) Colony formation assays with or without supplementation of vehicle control, mevalonic acid (MVA) or mevalonate-5-pyrophosphate (MVA-5PP). Representative images (left) and summarized graph (right). Error bars, means ± s.d. (n = 3 independent experiments), *P < 0.05, **P < 0.01; Student's t-test (two-tailed). (d) Tumour formation assays in mice subcutaneously injected with KHOS/NP cells (1×106). When tumours reached 3 mm in diameter, mice were intraperitoneally injected with DMSO (D) or atorvastatin (A, 30 mg kg−1) daily. Tumour sizes were measured every 2–3 days. The graph shows tumour growth with representative images of tumours (inset). Error bars, means ± s.d. (n = 5 animals for each group), *P <0.05; Student's t-test (two-tailed). Scale bar, 1 cm. (e) Representative images of IHC for p53, Ki67 and cleaved caspase 3 using KHOS/NP tumours treated with D or A (magnification, ×40). Scale bars, 50 μm. (f) Tumour formation assays in KHOS/NP-injected mice treated with vehicle control (C) or 10 mg kg−1 of rosuvastatin (R) daily. Scale bar, 1 cm. Error bars, means ± s.d. (n = 5 animals for each group), *P < 0.05; Student's t-test (two-tailed). Statistics source data for a–c are provided in Supplementary Table 1. Additional results are shown in Supplementary Fig. 4.
To better understand the mechanisms behind statin-induced growth suppression of mutp53-expressing cancer cells, we performed propidium iodide staining and flow cytometry. Lovastatin treatment resulted in increased sub-G0/G1 fraction and G1/S ratio in KHOS/NP cells, but minimal changes were observed in U2OS cells (Supplementary Fig. 4c).
We also examined the effects of statins on in vivo tumour growth. For these experiments, we used atorvastatin with a longer half-life (~14 h) than lovastatin (~4.5 h). Administration of atorvastatin (30 mg kg−1, once daily, intraperitoneally) significantly reduced subcutaneous tumour growth of both KHOS/NP and CAL33 cells (Fig. 4d and Supplementary Fig. 4d). No adverse effect was observed in mice treated with atorvastatin including changes in body weight during the course of the experiments (Supplementary Fig. 4e). Immunohistochemistry (IHC) analyses of formed tumours revealed that statin treatment reduced mutp53 levels in KHOS/NP and CAL33 tumours with significant decrease in Ki67 staining and slight increase in cleaved caspase 3 staining (Fig. 4e and Supplementary Fig. 4f). We also found that rosuvastatin, a hydrophilic statin with a relatively long half-life (~19 h), inhibited KHOS/NP tumour growth at a dose as low as 10 mg kg−1 (Fig. 4f).
Statins reduce growth of p53R172H MEFs
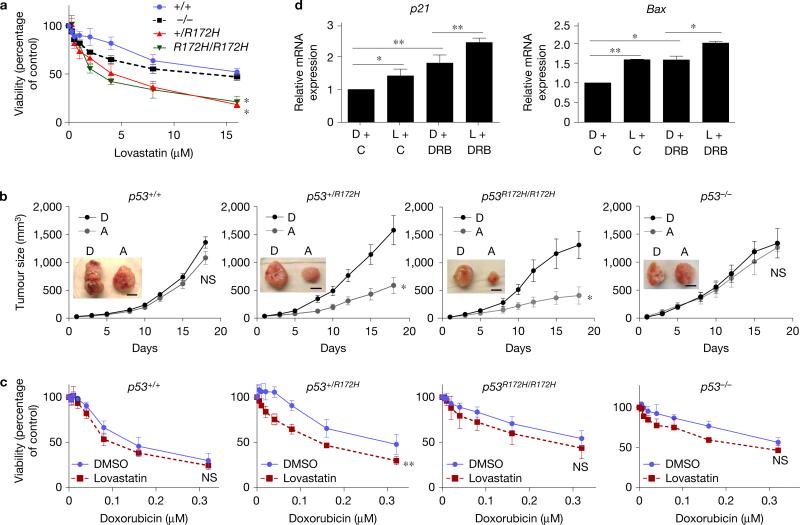
During early stages of tumour development, tumours frequently carry both the wtp53 and mutp53 alleles15,16. We hence wanted to examine the effects of the remaining wtp53 allele on tumour growth suppression by statins using cancer cells heterozygous for wtp53 and mutp53. Since these cell lines were not readily available, we generated mouse embryonic fibroblasts (MEFs) from C57BL/6 mice with different p53 genotypes—p53+/+, p53−/−, p53+/R172H and p53R172H/R172H (p53R172H is equivalent to human p53R175H), transformed them with adenovirus E1A and H-RasG12V, and allowed them to form foci that were then isolated and cultured for further experiments. We first confirmed that both lovastatin and atorvastatin reduced p53R172H levels, but not wtp53, in these transformed MEFs (Supplementary Fig. 5a). We next examined mRNA expression of p53 target genes, p21 and PUMA, following treatment of MEFs with doxorubicin. Expression of both genes was upregulated depending on the wtp53 gene dosage (Supplementary Fig. 5b), suggesting maintenance of the wtp53 allele(s) in the transformed p53+/+ and p53+/R172H MEFs.
We then examined viability and proliferation of these MEFs following treatment with various concentrations of lovastatin. As expected, lovastatin preferentially reduced viability (Fig. 5a) and colony formation (Supplementary Fig. 5c) of MEFs carrying p53R172H. Tumour formation assays using transformed MEFs demonstrated that administration of atorvastatin (30 mg kg−1, daily, intraperitoneally) significantly inhibited tumour growth of p53R172H-expressing MEFs, but not p53+/+ or p53−/− MEFs (Fig. 5b). IHC analyses confirmed significant reduction in the levels of p53 and Ki67 with slight increase in cleaved caspase 3 staining in statin-treated p53+/R172H and p53R172H/R172H tumours (Supplementary Fig. 5d). Intriguingly, in all of the experiments performed above, no obvious difference was observed between p53+/R172H and p53R172H/R172H MEFs, suggesting that depletion of mutp53 by statins might not be sufficient to restore wtp53 activity in p53+/R172H MEFs.
Figure 5.
Statins reduce growth of p53R172H MEFs. (a) MTT assays using E1A/H-RasG12V-transformed p53+/+, p53−/−, p53+/R172H, and p53R172H/R172H MEFs treated with various concentrations of lovastatin for 48 h. Error bars, means ± s.d. (n = 3 independent experiments), *P < 0.05; Student's t-test (two-tailed). (b) Tumour formation assays in mice subcutaneously injected with transformed MEFs (1×106) and treated with DMSO (D) or atorvastatin (A) (30 mg kg−1). Scale bars, 1 cm. Error bars, means ± s.d. (n = 5 animals for each group), *P <0.05; NS, not significant; Student's t-test (two-tailed). (c) MTT assays using transformed MEFs treated with various concentrations of doxorubicin together with DMSO or lovastatin (4 μM) for 48 h. Error bars, means ± s.d. (n = 5 independent experiments), **P <0.01; Student's t-test (two-tailed). (d) Quantitative RT–PCR for p21 and BAX using transformed p53+/R172H MEFs pre-treated with DMSO (D) or lovastatin (L) (4 μM) for 24 h and further exposed to vehicle control (C) or doxorubicin (DRB, 0.1 μM near IC50 in p53+/+ MEFs) for 24 h. Error bars, means ± s.d. (n = 3 independent experiments), *P <0.05, **P <0.01; Student's t-test (two-tailed). Statistics source data for a and d are provided in Supplementary Table 1. Additional results are shown in Supplementary Fig. 5.
To further address this, we treated the transformed p53+/R172H MEFs with lovastatin along with doxorubicin to activate wtp53. When cells were exposed to various concentrations of doxorubicin together with 4 μM of lovastatin, lovastatin sensitized p53+/R172H MEFs to doxorubicin. However, there was no significant effect on the viability of p53+/+, p53R172H/R172H and p53−/− MEFs (Fig. 5c). We also confirmed additive effects of lovastatin and doxorubicin on the mRNA expression of p53 target genes, p21 and BAX, in the transformed p53+/R172H MEFs (Fig. 5d). These results suggest that mutp53 depletion by statins, in combination with doxorubicin, restores wtp53 activity and enhances chemosensitivity in cells expressing both wtp53 and mutp53.
DNAJA1 knockdown induces mutp53 degradation
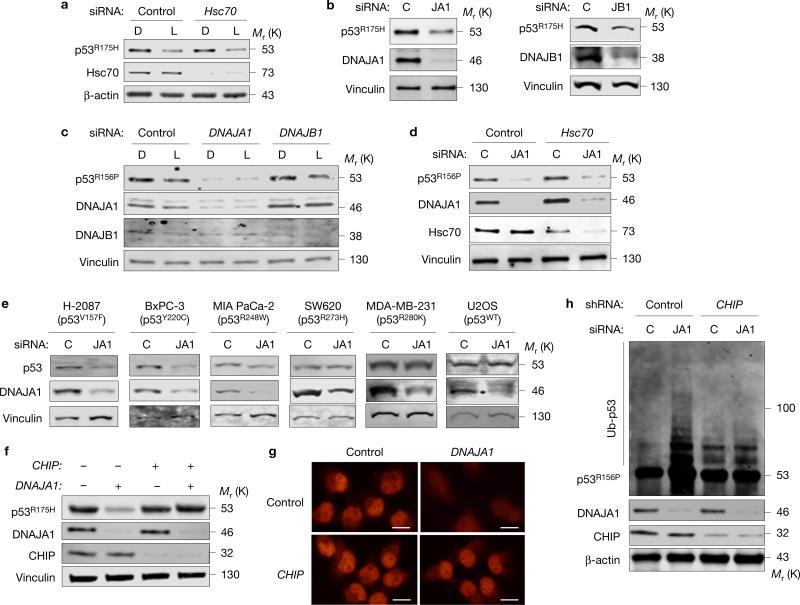
A crucial question to address further is how reduced MVP triggers CHIP-mediated degradation of mutp53. Since CHIP is a Hsc70-binding protein and is involved in degradation of misfolded proteins17,18, we hypothesized that Hsc70 is involved in statin-induced p53R175H degradation. Surprisingly, Hsc70 knockdown in CAL33 cells neither altered p53R175H levels nor affected lovastatin-induced p53R175H degradation (Fig. 6a).
Figure 6.
DNAJA1 knockdown induces mutp53 degradation. (a) WB for p53, Hsc70 and β-actin using CAL33 cells transfected with control or Hsc70 siRNAs and treated with DMSO (D) or lovastatin (L, 4 μM). (b) WB for the indicated proteins using SK-Br-3 cells transfected with siRNAs specific for DNAJA1 (JA1) or DNAJB1 (JB1). (c) WB for the indicated proteins using KHOS/NP cells downregulated for DNAJA1 or DNAJB1 by siRNAs and treated with D or L (4 μM) for 24 h. (d) WB for the indicated proteins using CAL33 cells downregulated for DNAJA1 (JA1) and/or Hsc70. (e) WB for p53, DNAJA1 and vinculin using cell lines with different p53 status transfected with control (C) or DNAJA1 (JA1) siRNAs. (f) WB for the indicted proteins using SK-Br-3 cells downregulated for CHIP and/or DNAJA1. (g) Immunofluorescence for p53R156P using KHOS/NP cells downregulated for DNAJA1 and/or CHIP. Scale bars, 50 μm. (h) Ubiquitin assays for p53R156P and WB for the indicated proteins using KHOS/NP cells downregulated for DNAJA1 and/or CHIP. Additional results are shown in Supplementary Fig. 6. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
The molecular chaperone Hsp90 assists folding of p53 mutants and inhibits activities of MDM2 and CHIP19,20. Inhibition of Hsp90 causes degradation of both conformational and DNA contact p53 mutants via MDM2 and CHIP21. However, we found that neither the expression level of Hsp90 itself nor its activity (indicated by expression of Hsp90 client proteins, such as EGFR, Raf-1 and ErbB2) was reduced by statin treatment (Supplementary Fig. 6a). Since ErbB2 is ubiquitylated and degraded by CHIP22, these results suggest that statin-induced mutp53 degradation is not simply due to inhibition of Hsp90 or nonspecific activation of CHIP.
Another molecular chaperone known to bind to mutp53 and implicated in misfolded protein degradation by CHIP is Hsp40, also known as DNAJ23–26. We therefore examined the involvement of Hsp40 in statin-induced mutp53 degradation. Since Hsp40 has 41 isoforms in humans, we chose relatively well-characterized Hsp40, DNAJA1 (HDJ-2, type I Hsp40) and DNAJB1 (HDJ-1, type II Hsp40). Knockdown of DNAJA1, as well as DNAJB1, resulted in depletion of p53R175H (Fig. 6b). However, depletion of p53R156P was observed only when DNAJA1 was downregulated, but not DNAJB1 (Fig. 6c). Lovastatin could not further reduce p53R156P levels when DNAJA1 was downregulated, whereas it depleted p53R156P independent of DNAJB1 knockdown (Fig. 6c), suggesting that DNAJA1 knockdown caused similar effects to statins on mutp53. Although Hsp40 often functions as a co-chaperone of Hsc70/Hsp70 (ref. 27), our results in Fig. 6a suggest that statins’ effects on mutp53 are independent of Hsc70. Similarly, p53R156P depletion by DNAJA1 knockdown was not affected by Hsc70 knockdown (Fig. 6d).
Next, we examined the effects of DNAJA1 knockdown on other p53 mutants. Similar to statins, DNAJA1 knockdown reduced protein levels of conformational mutants (p53V157F, p53Y220C), but did not alter the levels of DNA contact mutants (p53R273H, p53R280K) or wtp53; p53R248W was modestly affected (Fig. 6e). We also found that mutp53 depletion by DNAJA1 knockdown was substantially nullified by CHIP knockdown (Fig. 6f). Immunofluorescence analyses revealed that DNAJA1 knockdown-mediated nuclear export and depletion of mutp53 were rescued by CHIP knockdown and LMB treatment (Fig. 6g and Supplementary Fig. 6b). Moreover, increased ubiquitylation of p53R156P by DNAJA1 knockdown was attenuated by simultaneous knockdown of CHIP (Fig. 6h). Thus, DNAJA1 knockdown mirrored statins’ effects on mutp53, suggesting that DNAJA1 could be a crucial downstream effector of statin-induced mutp53 degradation via CHIP.
DNAJA1 nullifies statins’ effects on mutp53
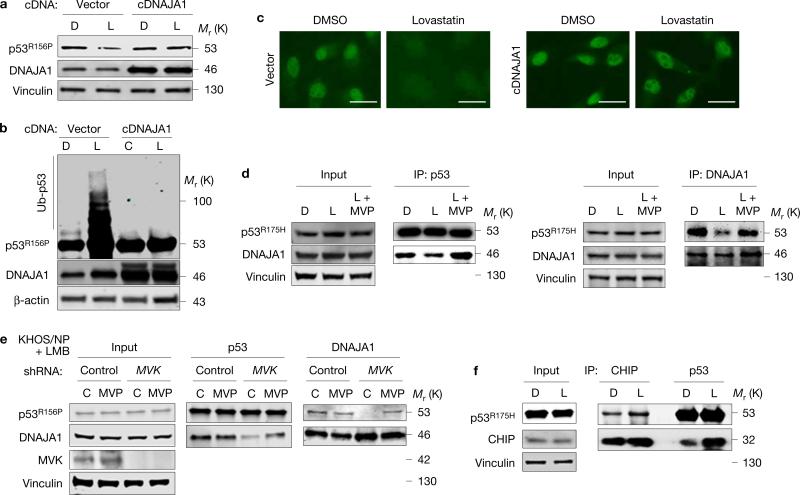
To determine whether DNAJA1 played a key role in CHIP-mediated mutp53 degradation induced by statins or reduced MVP, we examined the effects of DNAJA1 overexpression on statin-induced mutp53 degradation. Overexpression of DNAJA1 almost completely nullified statin-induced mutp53 degradation, ubiquitylation and nuclear export (Fig. 7a–c).
Figure 7.
DNAJA1 nullifies statins’ effects on mutp53. (a) WB for the indicated proteins using KHOS/NP cells infected with empty (vector) or DNAJA1 cDNA (cDNAJA1)-encoding lentiviral vectors and treated with DMSO (D) or lovastatin (L, 4 μM) for 24 h. (b) Ubiquitin assays for p53R156P and WB for the indicated proteins using KHOS/NP cells with or without overexpression of DNAJA1 (cDNAJA1) and treated with D or L for 24 h in the presence of MG132 for 6 h. (c) Immunofluorescence for p53R156P using KHOS/NP cells with or without overexpression of DNAJA1 and treated with DMSO or lovastatin. Scale bars, 50 μm. (d) Co-immunoprecipitation and WB for DNAJA1 and p53R175H using CAL33 cells treated with D, L or L + mevalonate-5-phosphate (MVP) for 11 h before mutp53 degradation. (e) Co-immunoprecipitation and WB for p53R156P and DNAJA1 using KHOS/NP cells infected with lentiviral vectors encoding control or MVK shRNAs and treated for 18 h with control (water) or MVP (200 μM) along with leptomycin B (LMB, 50 nM). (f) Co-immunoprecipitation and WB for p53R175H and CHIP using CAL33 cells treated with D or L for 11 h. Additional results are shown in Supplementary Fig. 7. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
We furthermore addressed the potential mechanisms of how DNAJA1 inhibited statin-induced mutp53 degradation. We hypothesized that statins inhibit the binding of DNAJA1 with mutp53, allowing CHIP to bind with and ubiquitylate mutp53. Indeed, co-immunoprecipitation studies revealed that treatment of CAL33 cells with lovastatin for 11 h, before degradation of mutp53, inhibited the DNAJA1-p53R175H interaction (Supplementary Fig. 7a). The inhibited DNAJA1-mutp53 interaction by statins was rescued by supplementation with MVP in both CAL33 and BxPC-3 cells (Fig. 7d and Supplementary Fig. 7b). Additionally, MVK knockdown also reduced interaction between DNAJA1 and p53R156P, even when the nuclear export of mutp53 was blocked by LMB, which was rescued by MVP supplementation as expected (Fig. 7e). These results indicate that the inhibited DNAJA1–mutp53 interaction following reduction in MVP levels was not due to altered subcellular localization of mutp53. Furthermore, lovastatin treatment increased the interaction of mutp53 (p53R175H and p53Y220C) with CHIP (Fig. 7f and Supplementary Fig. 7c). Taken together, our results strongly suggest that reduction of cellular MVP causes inhibition of DNAJA1–mutp53 interaction, allowing CHIP to mediate nuclear export, ubiquitylation, and hence degradation of conformational/misfolded mutp53 (Supplementary Fig. 7d).
DISCUSSION
Our results indicate that specific reduction of MVP induces degradation of conformational or misfolded mutp53 independent of protein prenylation. Previously, mutp53 was shown to bind with and activate SREPB transcription factors and hence upregulate multiple enzymes involved in the mevalonate pathway as its GOF activity14. In line with our study, increased MVP production in cancer cells expressing a GOF mutp53 could in turn stabilize mutp53 and exacerbate the GOF activity, thus forming a positive feedback loop. As a result, cancer cells carrying conformational mutp53 may be more susceptible to statin treatment, since statins inhibit two cellular activities crucial for cancer progression: protein prenylation and mutp53 stabilization. Also, statins sensitize p53+/R172H MEFs to doxorubicin. These observations propose the potential use of statins as a standalone or combinatorial treatment with other chemotherapeutics. However, daily doses of atorvastatin (30 mg kg−1) and rosuvastatin (10 mg kg−1) used in our mouse studies are equivalent to human doses of ~140 mg and ~50 mg, respectively, based on body surface area using allometric scaling, which are slightly higher than the prescribed clinical dosage in adults, ranging from 10 mg to 80 mg for atorvastatin and from 5 mg to 40 mg for rosuvastatin. Although this dosage regimen is within the range of doses demonstrating antilipidemic effects in mouse models28,29, it would be important to discover methods to lower the dose of statins, while still retaining its potential to induce mutp53 degradation. Nonetheless, our study could provide a reason why outcomes of clinical trials testing the efficacy of statins on cancer therapy are controversial30–32, because no correlative study to date is conducted based on p53 status in tumours and controlled doses of statins used in patients.
We also demonstrate that knockdown of MVK, but not PMVK or MVD, induces mutp53 degradation, suggesting involvement of MVP in the mutp53 stability. Meanwhile, we show that reduced MVP inhibits the interaction between DNAJA1 and mutp53. Yet, it is unclear how MVP, a specific metabolite in the mevalonate pathway, affects this specific interaction as its non-canonical function. This might be caused by changes in the protein folding machinery or post-translational modifications of DNAJA1 and/or mutp53, following reduction in cellular MVP. It is also possible that MVP alters levels or activities of yet unidentified proteins that could regulate DNAJA1–mutp53 binding. Additionally, the role of other Hsp40/DNAJ members in mutp53 degradation is unexplored. Further studies are required to clarify these remaining issues.
The Hsp40/DNAJ family is involved in diverse cellular activities including protein translation, folding/unfolding/refolding, translocation and degradation33. Our data demonstrate that knockdown of DNAJA1 induces CHIP-mediated nuclear export, ubiquitylation, and degradation of conformational/misfolded mutp53, while its overexpression prevents statin-induced mutp53 degradation. Given that Hsp40 binds to misfolded proteins for refolding34, misfolded mutp53 that fails to bind to DNAJA1 may be designated to be degraded by CHIP instead of being refolded. Thus, our results may suggest that DNAJA1 inhibits the activities of CHIP on mutp53 by competitively binding with mutp53 and determines the fate of misfolded mutp53 (Supplementary Fig. 7d). Induction of mutp53 degradation through inhibition of the mevalonate pathway–DNAJA1 axis may represent a promising therapeutic strategy for cancers expressing mutp53.
METHODS
Cell lines
All of the following human cell lines (with different p53 status) were maintained in Dulbecco's modified Eagle's medium (DMEM) or Roswell Park Memorial Institute (RPMI) 1640 with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin: human osteosarcoma KHOS/NP (p53R156P), U2OS (p53 wild-type: p53wt), SJSA-1 (p53wt), Saos2 (p53null), colorectal carcinoma HCT116 (p53wt and p53null, provided by B. Vogelstein at Johns Hopkins Medicine, USA), tongue squamous cell carcinoma CAL33 (p53R175H, provided by S. Thomas at University of Kansas Medical Center, USA), pancreatic carcinoma MIA PaCa-2 (p53R248W, provided by D. R. Welch at University of Kansas Medical Center, USA), pancreatic adenocarcinoma BxPC-3 (p53Y220C, provided by D. R. Welch at University of Kansas Medical Center, USA), colorectal adenocarcinoma HT29 and SW620 (p53R273H, provided by D. A. Dixon and S. Anant, respectively, at University of Kansas Medical Center, USA), breast adenocarcinoma SK-Br-3 (p53R175H, provided by D. R. Welch at University of Kansas Medical Center, USA), MDA-MB-231 (p53R280K, provided by J. Lewis-Wambi at University of Kansas Medical Center, USA), and non-small cell lung cancer adenocarcinoma H-2087 (p53V157F, provided by T. Komiya at University of Kansas Medical Center, USA). All cell lines except for HCT116 and CAL33 were from ATCC. Saos2 p53R175H-Luc and Saos2 Luc cell lines were generated by infecting Saos2 cells with lentiviral vectors encoding a p53R175H-luciferase chimaeric fusion protein and a luciferase alone, respectively. HCT116 subcell lines, p53null+R175H and p53null+R273H, were generated by infecting HCT116 p53null cells with retroviral vectors encoding p53R175H and p53R273H cDNAs, respectively. Mouse embryonic fibroblasts (MEFs) with different genotypes (p53+/+, p53+/R172H, and p53R172H/R172H) in the C57BL/6 background were generated from day 13.5 mouse embryos. p53−/− MEFs in the C57BL/6 background were provided by G. Lozano at MD Anderson Cancer Center, USA. All cell lines were authenticated by autosomal STR profiles provided by the University of Arizona Genetics Core. All cell lines were tested negative for mycoplasma. None of the cell lines used was found in the database of commonly misidentified cell lines that are maintained by ICLAC and NCBI Biosample.
Chemicals and compounds
Lovastatin, atorvastatin, rosuvastatin calcium salt and doxorubicin were purchased from Cayman Chemical. Mevalonic acid 5-phosphate trilithium salt hydrate (MVP), mevalonolactone (MVA), mevalonic acid 5-pyrophosphate tetralithium salt (MVA-5PP), cycloheximide, MG132, leptomycin B, adenosylcobalamine, emetine dihydrochloride hydrate, 1-hydroxypyridine-2-thione zinc salt and plicamycin were obtained from Sigma-Aldrich. Celastrol, pristimerin and mycophenolate mofetil were purchased from Tocris Bioscience.
siRNA transfection
Transfection of siRNAs (25–80 nM) was performed with INTERFERin according to the manufacturer's protocol (Polyplus-transfection). Double-strand siRNAs for CHIP (SR306955), DNAJA1 (SR302250), Hsc70 (SR302258) and MVK (SR303014) were purchased from Origene Technologies. In all experiments, non-target no. 1 siRNA (GE Healthcare Life Sciences) was used as a negative control.
Western blotting (WB)
Cells were lysed with radioimmunoprecipitation assay (RIPA) buffer containing phosphatase and protease inhibitors (EMD Chemicals). Cell lysate containing 20–100 μg protein was loaded onto 4–12% tris-glycine gel (Bio-Rad Laboratories), separated by electrophoresis, transferred to polyvinylidene fluoride (PVDF) membrane (GE Healthcare Life Sciences), blotted with primary antibodies against specific proteins, and appropriate secondary antibodies conjugated with fluorescence. All blots were analysed with the Li-Cor Odyssey infrared imaging systems. The following antibodies were used: p53 (sc-126, clone no. DO1, 1:3,000 dilution; sc-1313, R19, 1:2,000 dilution), DNAJA1 (sc-59554, clone no. KA2A5.6, 1:2,000 dilution; sc-135152, H-53, 1:500 dilution; sc-47051, C-14, 1:1,000 dilution), CHIP (sc-66830, H-231, 1:1,000 dilution; sc-133083, clone no. C-10, 1:500 dilution), MVK (sc-27587,N-20, 1:200 dilution), MDM2 (sc-813, N-20, 1:1,000 dilution), DNAJB1 (sc-1800, C-20, 1:500 dilution), EGFR (sc-377229, clone no. C-2, 1:2,000 dilution), RAF-1 (sc-133, C-12, 1:1,000 dilution), Hsp90 (sc-69703, clone no. 4F10, 1:1,000 dilution), Hsc70 (sc-7298, clone no. B-6, monoclonal, 1:5,000 dilution), MVD (sc-160550, E-20, 1:500 dilution), GAPDH (sc-166574, clone no. H-12, 1:3,000 dilution, Santa Cruz Biotechnology), vinculin (10R-C105a, clone no. V284, 1:1,000 dilution, Fitzgerald), β-actin (A2228, clone no. AC-74, 1:20,000 dilution, Sigma-Aldrich), p53 (mAb no. 2527, clone no. 7F5, 1:1,000 dilution), ERBB2 (2242, 1:2,000 dilution, Cell Signaling Technology), PMVK (EPR15029, clone no. EPR15029, 1:200 dilution, Abcam), IRDye 680RD goat anti-rabbit IgG (926-68073, 1:3,000 dilution), and IRDye 800CW goat anti-mouse IgG (926-32212, 1:5,000 dilution, LI-COR).
Co-immunoprecipitation
Cells were lysed with IP lysis buffer (Pierce) in the presence of protease inhibitor cocktail (Roche). Approximately 500 μg of whole-cell lysates were incubated with antibodies against p53 (OP33, clone no. PAb1620, 2 μg per sample, EMD Millipore; sc-99, clone no. PAb240, 2 μg per sample; clone no. DO1, 2 μg per sample, Santa Cruz Biotechnology), DNAJA1 (clone no. KA2A5.6, 2 μg per sample; clone no. SPM251, 2 μg per sample) or CHIP (H-231, 3 μg per sample) overnight at 4 °C and then precipitated with the antibody–protein complex using protein A/G plus-agarose beads (Santa Cruz Biotechnology). Matched isotype antibodies were used as negative controls (mouse IgG, sc-2025, 2 μg per sample; rabbit IgG, sc-2027, 3 μg per sample, Santa Cruz Biotechnology). The precipitants were resolved on SDS–PAGE for western blotting (WB) with antibodies against p53 (7F5), CHIP (C-10) or DNAJA1 (C-14).
Ubiquitin assay
Cells transfected with a ubiquitin-encoding plasmid were incubated with DMSO or MG132 (30 μM) for 6 h, followed by harvesting cells with hot SDS lysis buffer and western blotting for p53 (DO1).
MTT assay
Cells (10,000 cells) were seeded onto a 96-well plate. Twenty-four hours later, cells were treated with varying concentrations of lovastatin (0–16 μM), with or without doxorubicin (0–0.32 μM) for 48 h, followed by standard MTT assays. Briefly, after cells were incubated with 5 mg ml−1 of MTT for 3 h, the medium was replaced with DMSO to dissolve blue formazan crystals, and plates were shaken for 15 min in the dark. Results were obtained by reading the plate at 570 nm.
Colony formation assay
One day after seeding cells (500) on a 6-well plate, cells were treated with DMSO or lovastatin every other day for 10 days. When indicated, cells were supplemented with MVA (200 μM) or MVA-5PP (200 μM). Colonies were fixed with methanol and stained with 0.1% of crystal violet and counted.
Quantitative PCR with reverse transcription (qRT–PCR)
RNA was isolated using the RNA-Quick MiniPrep (Zymo Research). Total RNA (1 μg) was reversed transcribed to cDNA using M-MLV reverse transcriptase (Amresco), according to the manufacturer's instructions, and TaqMan assays were performed with ViiA7 (Life Technologies). TaqMan assay primers and probes were purchased from Life Technologies using the following assay numbers: human p53, Hs00153349_m1; mouse p21 Mm00432448_m1; Bax Mm00432050_m1, Puma Mm00519268_m1; Gapdh, Mm99999915_g1. Taqman assay for human GAPDH was purchased from Integrated DNA Technologies (Hs.PT.58.40035104). The mRNA levels were normalized to those of GAPDH.
Immunofluorescence
Cells grown onto poly-D-lysine/laminin-coated glass cover-slips (BD Biosciences) were fixed with 4% paraformaldehyde for 20 min and then permeabilized with 0.3% Triton X-100 for 5 min, followed by blocking in 1% BSA in PBS-T for 1 h and incubation with PAb240 p53 antibody (1:2,000 dilution) overnight at 4 °C. Goat anti-mouse IgG was used as a secondary antibody. Samples were mounted in the ProLong Gold Antifade Reagent with DAPI (Invitrogen), followed by analysis with a Nikon epifluorescence microscope.
Mice and tumour growth assay
All mice were maintained under specific-pathogen-free conditions, and all experimental procedures were conducted in accordance with the institutional animal welfare guidelines of the University of Kansas Medical Center. Cells (1×106) were subcutaneously injected into 6-week-old female NIH-III nude mice (Charles River). When tumours became 3 mm in diameter, atorvastatin (30 mg kg−1) or rosuvastatin (10 mg kg−1) was intraperitoneally injected daily. Tumour sizes were measured every 2–3 days for about 3 weeks.
Immunohistochemistry (IHC)
The tumour tissues were sectioned at 4 μm and subjected to IHC by standard procedures using the following antibodies: p53 (clone no. DO1, 1:2,000), p53 (VP-P956, CM5, 1-2500, Vector Laboratories), Ki67 (ab15580, 1:2,500, Abcam), and cleaved caspase 3 (9579, clone no. D3E9, 1:500, Cell Signaling Technologies).
Statistics and reproducibility
The differences in all assays including cell proliferation, migration, survival, gene expression, protein stability, flow cytometry, and tumour growth between different samples and/or treatments were analysed by two-tailed Student's t-tests using GraphPad Prism 5 (GraphPad Software). Statistical significance was set at P < 0.05, unless otherwise stated in the text. No statistical methods were used to predetermine sample size. All experiments were carried out with at least three biological replicates. The numbers of animals used are described in the corresponding figure legends. We chose the appropriate tests according to the data distributions. The experiments were not randomized and we did not exclude any samples. The investigators were not blinded to allocation during experiments and outcome assessment. Panels in Figs 1a–d, 2a–g, 3b–j, 4b–f, 5b, 6a–h and 7a–f and Supplementary Figs 1b–e, 2b–f, 3a,b, 4a–d,f, 5a,c,d, 6a,b and 7a–c show a representative image of at least three independent experiments.
Data availability
Statistics source data for Figs 2a, 4a–c and 5a,d and Supplementary Figs 2a,f, 3b–d, 4a–c and 5b,c have been provided in Supplementary Table 1. All the data supporting the findings of this study are available from the corresponding author on request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank B. Vogelstein (Johns Hopkins Medicine, USA), D. R. Welch (University of Kansas Medical Center, USA), S. Anant (University of Kansas Medical Center, USA), D. A. Dixon (University of Kansas Medical Center, USA), J. Lewis-Wambi (University of Kansas Medical Center, USA), S. Thomas (University of Kansas Medical Center, USA), T. Komiya (University of Kansas Medical Center, USA) and G. Lozano (MD Anderson Cancer Center, USA) for providing cell lines. We also thank A. K. Godwin, S. Hyter, R. Perez, N. K. Sharma, R. Pradhan, M. Danley, T. Izumi and R. Stein for technical assistance and helpful discussion. Research reported in this publication was supported by NIH R01-CA174735-01A1 (T.I.), P30-GM103495 (T.I.) and P30-CA168524-02 (T.I.) grants, and utilized the Lead Development and Optimization Shared Resource.
Footnotes
AUTHOR CONTRIBUTIONS
T.I. supervised the project. A.P., A.Ranjan, S.V.I. and A.Roy performed the experiments. A.Roy performed high-throughput analyses. T.I. and A.P. wrote the manuscript. A.Ranjan, S.V.I., S.P., S.J.W. and A.Roy commented on experiments and edited the manuscript.
Supplementary Information is available in the online version of the paper
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Lane D, Levine A. p53 research: the past thirty years and the next thirty years. Cold Spring Harb. Perspect. Biol. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levav-Cohen Y, et al. The p53-Mdm2 loop: a critical juncture of stress response. Subcell. Biochem. 2014;85:161–186. doi: 10.1007/978-94-017-9211-0_9. [DOI] [PubMed] [Google Scholar]
- 3.Rivlin N, Koifman G, Rotter V. p53 orchestrates between normal differentiation and cancer. Semin. Cancer Biol. 2015;32:10–17. doi: 10.1016/j.semcancer.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Boeckler FM, et al. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc. Natl Acad. Sci. USA. 2008;105:10360–10365. doi: 10.1073/pnas.0805326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terzian T, et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22:1337–1344. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lukashchuk N, Vousden KH. Ubiquitination and degradation of mutant p53. Mol. Cell. Biol. 2007;27:8284–8295. doi: 10.1128/MCB.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexandrova EM, et al. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature. 2015;523:352–356. doi: 10.1038/nature14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masciarelli S, et al. Gain-of-function mutant p53 downregulates miR-223 contributing to chemoresistance of cultured tumor cells. Oncogene. 2014;33:1601–1608. doi: 10.1038/onc.2013.106. [DOI] [PubMed] [Google Scholar]
- 9.Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2:466–474. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong S, et al. Statin use and mortality in cancer patients: Systematic review and meta-analysis of observational studies. Cancer Treat. Rev. 2015;41:554–567. doi: 10.1016/j.ctrv.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Li M, et al. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 12.Altwairgi AK. Statins are potential anticancerous agents (review). Oncol. Rep. 2015;33:1019–1039. doi: 10.3892/or.2015.3741. [DOI] [PubMed] [Google Scholar]
- 13.Shimoyama S. Statins are logical candidates for overcoming limitations of targeting therapies on malignancy: their potential application to gastrointestinal cancers. Cancer Chemother. Pharmacol. 2011;67:729–739. doi: 10.1007/s00280-011-1583-2. [DOI] [PubMed] [Google Scholar]
- 14.Freed-Pastor WA, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varley JM, et al. A detailed study of loss of heterozygosity on chromosome 17 in tumours from Li-Fraumeni patients carrying a mutation to the TP53 gene. Oncogene. 1997;14:865–871. doi: 10.1038/sj.onc.1201041. [DOI] [PubMed] [Google Scholar]
- 16.Venkatachalam S, et al. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO J. 1998;17:4657–4667. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8:303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edkins AL. CHIP: a co-chaperone for degradation by the proteasome. Subcell. Biochem. 2015;78:219–242. doi: 10.1007/978-3-319-11731-7_11. [DOI] [PubMed] [Google Scholar]
- 19.Peng Y, Chen L, Li C, Lu W, Chen J. Inhibition of MDM2 by hsp90 contributes to mutant p53 stabilization. J. Biol. Chem. 2001;276:40583–40590. doi: 10.1074/jbc.M102817200. [DOI] [PubMed] [Google Scholar]
- 20.Li D, et al. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol. Cancer Res. 2011;9:577–588. doi: 10.1158/1541-7786.MCR-10-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller P, Hrstka R, Coomber D, Lane DP, Vojtesek B. Chaperone-dependent stabilization and degradation of p53 mutants. Oncogene. 2008;27:3371–3383. doi: 10.1038/sj.onc.1211010. [DOI] [PubMed] [Google Scholar]
- 22.Zhou P, et al. ErbB2 degradation mediated by the co-chaperone protein CHIP. J. Biol. Chem. 2003;278:13829–13837. doi: 10.1074/jbc.M209640200. [DOI] [PubMed] [Google Scholar]
- 23.Murata S, Minami Y, Minami M, Chiba T, Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2001;2:1133–1138. doi: 10.1093/embo-reports/kve246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiraki M, et al. Small-molecule reactivation of mutant p53 to wild-type-like p53 through the p53-Hsp40 regulatory axis. Chem. Biol. 2015;22:1206–1216. doi: 10.1016/j.chembiol.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King FW, Wawrzynow A, Hohfeld J, Zylicz M. Co-chaperones Bag-1, Hop and Hsp40 regulate Hsc70 and Hsp90 interactions with wild-type or mutant p53. EMBO J. 2001;20:6297–6305. doi: 10.1093/emboj/20.22.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yue X, et al. BAG2 promotes tumorigenesis through enhancing mutant p53 protein levels and function. Elife. 2015;4:e08401. doi: 10.7554/eLife.08401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan CY, Lee S, Cyr DM. Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones. 2003;8:309–316. doi: 10.1379/1466-1268(2003)008<0309:mfrohf>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gandelman K, Malhotra B, LaBadie BB, Crownover P, Bergstrom T. Analytes of interest and choice of dose: two important considerations in the design of bioequivalence studies with atorvastatin. Bioequiv. Bioavailab. 2011;3:62–68. [Google Scholar]
- 29.Bisgaier CL, et al. Attenuation of plasma low density lipoprotein cholesterol by select 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitors in mice devoid of low density lipoprotein receptors. J. Lipid Res. 1997;38:2502–2515. [PubMed] [Google Scholar]
- 30.Moon H, Hill MM, Roberts MJ, Gardiner RA, Brown AJ. Statins: protectors or pretenders in prostate cancer? Trends Endocrinol. Metab. 2014;25:188–196. doi: 10.1016/j.tem.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Baandrup L, Dehlendorff C, Friis S, Olsen JH, Kjaer SK. Statin use and risk for ovarian cancer: a Danish nationwide case-control study. Br. J. Cancer. 2015;112:157–161. doi: 10.1038/bjc.2014.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang XL, et al. Statin use and risk of kidney cancer: a meta-analysis of observational studies and randomized trials. B. J. Clin. Pharmacol. 2014;77:458–465. doi: 10.1111/bcp.12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol. Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kota P, Summers DW, Ren HY, Cyr DM, Dokholyan NV. Identification of a consensus motif in substrates bound by a Type I Hsp40. Proc. Natl Acad. Sci. USA. 2009;106:11073–11078. doi: 10.1073/pnas.0900746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Statistics source data for Figs 2a, 4a–c and 5a,d and Supplementary Figs 2a,f, 3b–d, 4a–c and 5b,c have been provided in Supplementary Table 1. All the data supporting the findings of this study are available from the corresponding author on request.