Abstract
Realized kinship is a key statistic in analyses of genetic data involving relatedness of individuals or structure of populations. There are several estimators of kinship that make use of dense SNP genotypes. We introduce a class of estimators, of which some existing estimators are special cases. Within this class, we derive properties of the estimators and determine an optimal estimator. Additionally, we introduce an alternative marker weighting that takes allelic associations [linkage disequilibrium (LD)] into account, and apply this weighting to several estimators. In a simulation study, we show that improved estimators are obtained (1) by optimal weighting of markers, (2) by taking physical contiguity of genome into account, and (3) by weighting on the basis of LD.
Keywords: realized kinship, genomic relationship matrix (GRM), local identity by descent (IBD), linkage disequilibrium, locus weighting
GENES inherited from the same ancestral copy by related individuals are said to be identical by decent (IBD). At the locus level, a pair of individuals share 0–4 genes that are IBD. At the genome level, the kinship coefficient is often used to summarize the average amount of IBD sharing across all loci. The kinship coefficient is important in genetic data analyses that either use or adjust for pairwise relatedness. The pedigree kinship, the probability that genes segregating from each individual at a randomly chosen locus are IBD, is a deterministic function of the pedigree relationship. However, the realized kinship, the actual proportion of IBD genome between two individuals varies widely about its expectation as a consequence of Mendelian sampling (Hill and Weir 2011). Additionally, in samples ascertained on the basis of trait information or samples affected by artificial selection, there are biases in the levels of relatedness of individuals (Liu et al. 2003; Purcell et al. 2007). Thus, even when pedigree information is available, estimates of realized kinship are often preferred to pedigree kinship (Visscher et al. 2006; VanRaden 2007, 2008; Hayes et al. 2009).
The availability of dense SNP genotypes makes it possible to estimate realized kinship accurately without pedigree information. Such estimates are particularly useful in studies that involve population samples (Choi et al. 2009; Yang et al. 2010; Day-Williams et al. 2011). Improving the precision of realized-kinship estimators has value for human genetics applications involving gene mapping or genotypic disease risk prediction, and also for animal and plant breeding, where incremental improvements in predictive accuracy are of economic value in trait optimization.
The matrix of inferred pairwise realized kinship is a measure of genomic similarity and a kernel in the sense of Gianola and van Kaam (2008). It is therefore appropriate for phenotype prediction or whole genome prediction (de los Campos et al. 2010). The use of this matrix for phenotype or breeding-value prediction is the genomic best linear unbiased prediction methodology (for example, Bernardo 2008). In plant and animal breeding applications, pedigree information has been combined with inferred kinships (Crossa et al. 2010). In the mixed model context, the matrix is used for variance component estimation (Yang et al. 2010), or as a proxy for polygenic effects in the presence of major gene effects (Kang et al. 2010).
One group of existing estimators of realized kinship makes use only of the population allele frequencies at each SNP marker, and not of additional information such as the ordering of markers along the chromosome. Estimators in this group require only dense SNP genotypes and reliable sources of marker allele frequencies as input. The PLINK (Purcell et al. 2007) method-of-moments estimator estimates realized kinship from the k coefficients; the proportion of genome at which two noninbred individuals share 0, 1, or 2 IBD genes. Choi et al. (2009) adopted a maximum likelihood estimator (MLE) that estimates the k coefficients using an EM algorithm. The classic genomic relationship matrix (GRM) estimator estimates kinship through the empirical correlation of genotypes; see, for example Hayes et al. (2009). An alternative version of the GRM estimator is more robust to presence of rare alleles (VanRaden 2008). Day-Williams et al. (2011) proposed a method-of-moments estimator which estimates kinship by exploring the relationship between identity by state (IBS) and IBD.
Other estimators make use of various sources of additional information to improve accuracy of kinship estimation. A number of methods have been developed to estimate IBD sharing at the locus level; see for example Moltke et al. (2011) and methods cited in Brown et al. (2012). These location-specific IBD estimates in turn provide estimates of kinship; note that location-specific kinships are constrained to the values 0, 1/4, 1/2, and 1 (Day-Williams et al. 2011). Here, we consider estimators from two of such local IBD methods that have not been previously used to estimate genome-wide realized kinship. The local method of Day-Williams et al. (2011) with resulting kinship estimator (to be distinguished from requires information on the ordering of markers along the chromosomes. It predicts the amount of sharing at each marker by first estimating a neighborhood kinship for each marker and then applying a constrained smoothing algorithm on each chromosome. The hidden Markov model (HMM) proposed by Brown et al. (2012), with resulting kinship estimator estimates probabilities of IBD states between two or more individuals at each marker location. A genetic map is needed to compute transition probabilities along the Markov chain.
Finally, we consider efficient estimation of realized kinship in the presence of linkage disequilibrium (LD). An increase in the density of SNP panels, by itself, will not improve precision without limit in the presence of LD, as additional SNPs are not independent sources of information. Speed et al. (2012) developed LDAK a weighted version of the GRM that takes LD into account. By analogy with methods introduced in the population structure context by Patterson et al. (2006) and Zou et al. (2010), LDAK equalizes contributions of linked SNPs by downweighting SNPs which make redundant contribution to the GRM as evidenced by off-diagonal terms in the (squared) SNP correlation matrix. We derive an analogous weighting by optimizing the weighted GRM estimator variance under tractable assumptions.
This article focuses on pedigree-free estimation of realized kinship in a homogenous population. In Methods, we first introduce the framework of a general class of GRM estimators, of which the classic GRM estimator the robust GRM estimator and the global Day-Williams estimator are all special cases. Under the assumptions of linkage equilibrium and absence of inbreeding, we propose a two-step GRM estimator that approximates the minimum variance estimator within this class. In the general case of LD, we derive the variance for a weighted GRM estimator, and construct an estimator with certain optimality properties given the LD structure of the population. We next describe the implementation of our simulation study which compares performance of the different kinship estimators detailed above. Additionally, the optimal weights derived for may also be applied to other estimators; specifically, we consider reweighted versions of and Results of the simulation study are presented in Results. We show that the proposed estimators are very competitive against existing estimators. We conclude with a Discussion.
Methods
A general class of GRM estimators
At any autosomal locus, there are 15 possible IBD states among the four genes of two individuals. These 15 IBD states fall into nine genotypically distinct classes (see, for example, Thompson 2000). When interest lies only in the amount of sharing between (as opposed to within) the individuals, the nine classes of states condense further into four groups of states characterized by local kinship, φ, which takes values in . Global realized kinship, measures the IBD proportion shared between the individuals across the genome, and so takes values in the range Since IBD results from the meiotic process, we measure this proportion in terms of genetic distance. However, since recombination is a coarse process relative to dense SNP markers, global kinship is well approximated by the average of local kinship over a large number of marker loci approximately evenly spaced in genetic distance throughout the genome.
We consider a general class of GRM estimators of the following form:
| (1) |
where L is the total number of marker loci, and are counts of reference alleles at each marker for the pair of individuals, are the population frequencies of the reference alleles, are multiplicative factors, and are nonnegative weights satisfying The vectors and are parameters that distinguish different GRM estimators, while and are the data random variables and is assumed known. Note that the denominator in (1) is as opposed to reflecting the difference between kinship coefficients and the relatedness coefficients of the numerator relationship matrix (Henderson 1976).
The classic GRM estimator is a special case of (1) with and for all l:
| (2) |
The robust GRM estimator is a special case with and for all l:
| (3) |
The global Day-Williams estimator can be reparametrized (see Appendix A) into the form of (1) with and for all l:
| (4) |
In the general form of (1), write
so that
Note that depends on the parameters but not on We make two basic assumptions throughout the article. First, we assume IBD genes have the same allelic types and non-IBD genes have independent allelic types. In addition, we assume exchangeability of parental lineage, so that either of the two genes from the first individual is equally likely to be IBD to either of the two genes of the second individual at the same locus. Under these assumptions, it can be shown that regardless the choice of (see Appendix B, General Case). Thus, for any and
Performance of unbiased estimators depend on their variances. For a GRM estimator in the general form of (1),
| (5) |
where . Computation of is intractable without simplifying assumptions. We next derive the and that minimize under different assumptions.
Linkage equilibrium
We first assume linkage equilibrium. Although in reality there is LD, empirical results show that the relative values of variances of estimators are well approximated by those derived under this assumption (see Discussion). When markers are in linkage equilibrium, the allelic types at different markers on the same haplotype are independent. Equation 5 then reduces to
| (6) |
To find the GRM estimator with minimal variance, Equation 6 suggests that one should first (for each l) choose that minimizes and then choose to minimize .
We now make an additional assumption of no inbreeding. For general choices of it can be shown (see Appendix B, Linkage Equilibrium) that
| (7) |
where is the realized kinship and is the realized proportion of the genome that the pair of individuals share both genes IBD. Note that the value of is asymmetric about for general choices of and thus is sensitive to the choice of reference allele. However, when is any weighted average of and 1 [as in (8)], becomes invariant to the choice of reference allele. With such an attains its minimum at conditional on and (see Appendix B, Linkage Equilibrium).
It follows from (7) that the optimal multiplicative factors that minimize the unweighted single-marker variances have the form
| (8) |
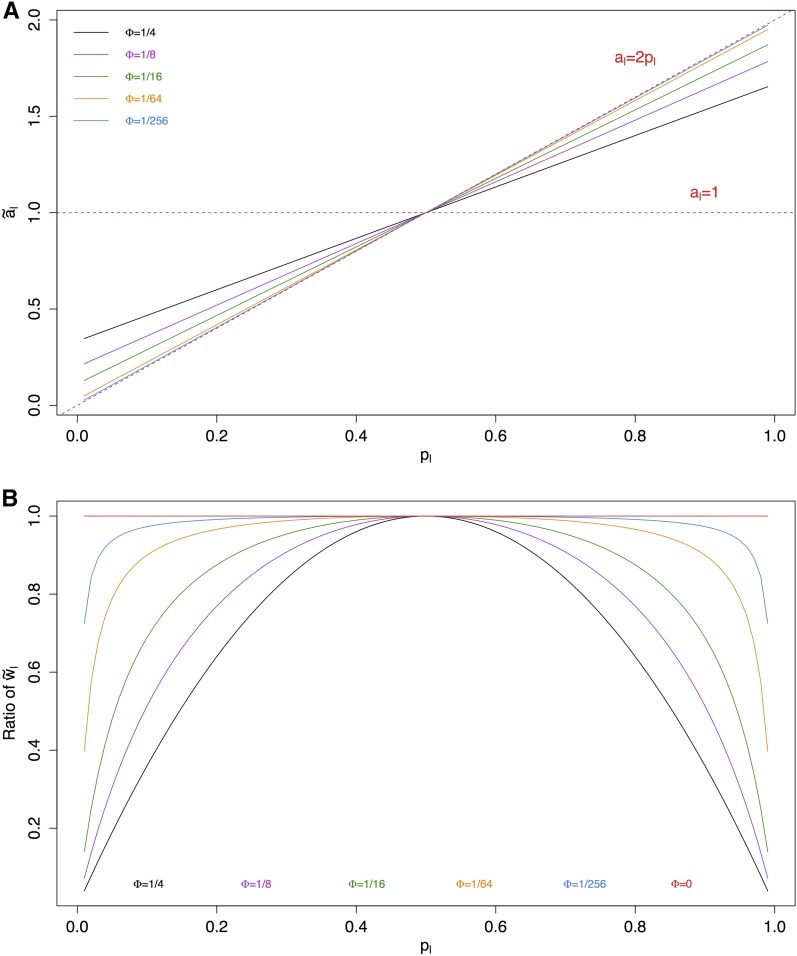
Interestingly, is a weighted average of and 1, which are the choices used by and respectively (compare Equations 2–4). Figure 1A shows how varies with for different Φ’s. We see that is optimal when whereas is far from optimal even when Since and use the same weights, is more efficient than for Φ < 1/2 (from Equation (7)) under the assumptions of this section.
Figure 1.
(A) as a function of for different with and as reference lines. (B) Ratio of weights between a marker with allele frequency and a reference marker with allele frequency 0.5, under the optimal weighting scheme for different combinations of In calculation of we used when and 0 otherwise. Note that these combinations of correspond to pedigree expectations of a pair of individuals that are full siblings, half siblings, first cousins, second cousins, or third cousins.
Conditional on the optimal weights that minimize have the form
| (9) |
The weights in (9) can be equivalently specified by the ratios
which are functions of allele frequencies at the corresponding pairs of markers, conditional on and the choice of For Figure 1B shows how a marker with frequency is weighted relative to a marker with frequency 0.5 under the optimal weighting scheme for different combinations of The optimal solution weights markers very differently, especially for large In this case the uniform weighting of is optimal when whereas the weighting of is optimal when and
The estimator would be an obvious choice if we knew and However, elements of are functions of the unknown and elements of are functions of the unknown and The closed form expressions given in Equations 7–9 motivate a two-step estimator, which approximates following these two steps:
Obtain initial estimates of and using existing methods.
Compute (Equation 8) and then (Equations 7 and 9) using these estimates of and
Then .
In practice, existing kinship estimators such as and may produce negative estimates of in Step 1. Our implementation uses to obtain an initial estimate of When this initial estimate is negative, we simply retain it as the estimate. For simplicity, we set when computing for all l. In principle, this affects the calculation of for bilateral relatives, but it makes no practical difference (see Supplemental Material, File S1, section A).
Linkage disequilibrium
We now drop the assumptions of linkage equilibrium and absence of inbreeding. To calculate variances, we follow the approach of Sverdlov (2014), and consider the case where the pair of individuals are unrelated. Under these assumptions, it can be shown (see Appendix B, Linkage Disequilibrium) that
However, there generally does not exist an that jointly minimizes each of the unweighted covariance terms in Equation 5. Thus, we conveniently set in the remainder of this section.
Let and denote the inbreeding coefficients of the two individuals respectively. We have
| (10) |
and
| (11) |
where is the genotype dosage correlation between locus l and locus m. Conveniently, only enters the expression in a squared form, so the covariance is invariant to the choice of reference allele. Combining (10) and (11), Equation 5 becomes
| (12) |
where when We assume that the matrix of squared LD correlations, is known. The goal is to find the that minimizes . Note that the inbreeding coefficients, and are part of a fixed scaling factor.
The optimization problem reduces to
| (13) |
Presence of the second term in the objective function forces a solution that satisfies for some which can be rescaled by to obtain the final solution The LD weighted GRM estimator is then
The matrix is positive semidefinite (see Appendix B, Linkage Disequilibrium). The above minimization problem can be solved for general using standard quadratic programming procedures, and closed-form solutions exist for special cases of (see File S1, section B). However, in practice, it will be necessary to divide the large set of genome-wide SNPs into blocks. In the case where has a block-diagonal structure, the optimization problem can be solved for each block. The final solutions will be a concatenation of the rescaled block solutions. Additional details are given in Appendix B, Linkage Disequilibrium.
Methods of analysis
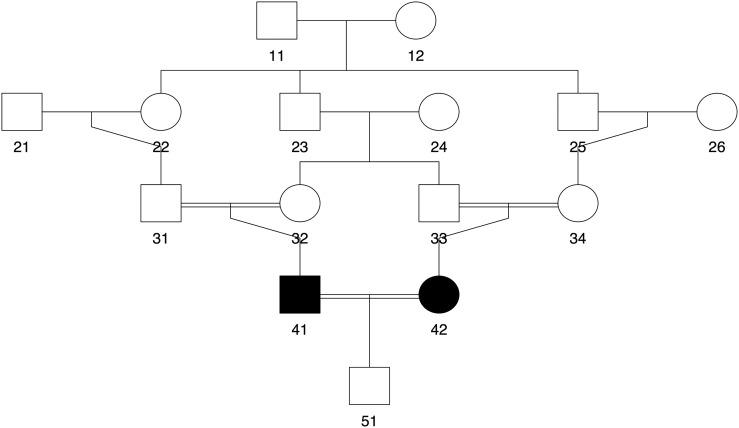
In the simulation study, we considered six relationship types: full siblings (FS), half siblings (HS), first cousins (C1), second cousins (C2), third cousins (C3), and inbred cousins (IN) from the complex JV pedigree (Goddard et al. 1996) shown in Figure 2. Dense SNP genotypes were generated for 1000 independent pairs of each relationship type as follows:
Figure 2.
The JV pedigree. Individual 41 and 42 are the inbred quadruple cousins (IN) considered in the simulation study. Double lines indicate consanguineous marriages.
Simulate recombination breakpoints and Mendelian sampling for all meioses in the smallest complete pedigree that contains the pair of relatives.
Assign founder genome labels (FGLs) (Sobel and Lange 1996) to founder haplotypes, and determine the inherited FGLs at all marker positions for all nonfounders with respect to the inheritance pattern simulated in step 1.
Sample founder haplotypes from a reference pool, and assign alleles to nonfounders with respect to both the sampled founder haplotypes and the inherited FGLs determined in step 2.
At each locus, combine the two alleles inherited by the related pair of individuals to create genotype data.
Data generation was implemented using the ibd_create program of MORGAN version 3.3.1 (Thompson and Lewis 2016). Marker and haplotype information used in data generation were extracted from the 1000 Genomes Project Phase 3 data (1000 Genomes Project Consortium 2015). All 5008 phased haplotypes from the combined population were made available for sampling of founder haplotypes. This use of real haplotypes preserves the natural patterns of LD in the combined population. The locations of markers in Haldane cM were obtained from the Rutgers Map version 3a (Matise et al. 2007). A total of 169,751 markers were selected from the 22 autosomes based on spacing (∼50 markers per cM), minor allele frequency (≥0.05), and complete genotype information (no missing genotypes). The distribution of allele frequencies in the marker panel thus reflects real studies using common SNPs. The marker density was chosen to be dense enough to show patterns of LD, yet sparse enough to be attainable by older SNP arrays in human genetics and by modern SNP arrays in animal genetics.
All the kinship estimators were evaluated on the simulated data. We used PLINK version 1.07 to implement the PLINK estimator, ibd_haplo program (Brown et al. 2012) of MORGAN version 3.3.1 to implement the HMM estimator, and our own code to implement all other estimators. Marker allele frequencies used in analysis were estimated by PLINK version 1.07 for each relationship type separately. For the MLE estimator, we adopted a very stringent convergence criterion (order of 10−8 of the final log-likelihood) and used pedigree k coefficients as the starting configurations of the EM algorithm. The GRM estimators were computed both unconstrained to the range (consistent with the theory in Methods), and also constrained to this range.
The local Day-Williams method imputes local kinship directly, whereas the HMM method estimates probabilities of local IBD states. For ease of comparison, we used the most probable state from the HMM output to impute local kinship. For these two local methods, estimates of realized (global) kinships were calculated as the average of imputed local kinship across all marker loci. We simulated genotype data on an additional 300 independent pairs of cousins (100 first cousins, 100 second cousins, and 100 third cousins) for tuning purposes. For each of the two local IBD methods, a sparse grid search was implemented to find the set of tuning parameters that maximizes local kinship imputation rate over the tuning data set. The selected sets of tuning parameters were subsequently used in the actual analysis.
All LD-adjusted methods used the reference genotypes of all 2504 individuals from the 1000 Genomes Project Phase 3 data to obtain LD information. A naive LD-pruned GRM estimator is included as a baseline for comparison. For this estimator, LD pruning was done using PLINK version 1.07, where we sequentially threw out one of the pairs of markers that had a genotypic dosage correlation (option–r2 in PLINK). This pruning step reduced the marker set to about half of the original size, and the classic GRM was applied to the reduced marker set to obtain For the LD-weighted GRM estimator weights were computed in blocks of 2000 SNPs using the optimization package JuMP version 0.14.1 (Lubin and Dunning 2015) in the Julia language. Weights of the LDAK estimator were computed using LDAK version 4.9 with the default options.
Using a single processor on a standard desktop, the PLINK estimates or any of the GRM estimates can be computed for 1000 pairs of individuals with the selected genome-wide marker panel in a couple of minutes. The MLE estimates can take many hours to compute depending on run conditions (see Results). Either of the two local IBD estimates takes several hours to compute, but this is still computationally feasible. With reference genotypes from 2504 individuals as the basis for LD adjustment, LD pruning takes several minutes to implement. Weights of the LD-weighted GRM take <15 min to compute, whereas weights of LDAK take >15 hr to compute.
The performances of estimators were compared on the simulated data. To compare performance both across estimators and across relationship types, we summarize estimation accuracy by the ratio of the square root of mean squared error (MSE) and the average realized kinship,
| (14) |
where are realized kinship and are their estimates.
Data availability
The 1000 Genomes Project Phase 3 data are available at http://www.1000genomes.org/data. The Rutgers Map version 3a is available at http://compgen.rutgers.edu/download_maps.shtml. Information on the set of 169,751 SNP markers used in the simulation study is provided as File S2.
Results
Table 1 summarizes the performance of different kinship estimators. The estimators are divided into two groups: Group-1 uses only marker-allele frequencies, whereas those in Group-2 use additional information. First, every estimator works better on closer relatives. This is expected given we are measuring estimation accuracies relative to the average amount of sharing (Equation 14), and the coefficients of variation are higher for remote relatives. For each estimator, the raw MSEs are in fact smaller on remote relatives (Table S4). Second, estimators that make use of additional sources of information (Group-2) generally do better than estimators that do not (Group-1). This is also expected as chromosomes are inherited as segments. Information such as marker order, genetic positions, and LD pattern are informative of the joint inheritance across markers.
Table 1. Estimation accuracies from simulation study as measured by the ratio of the square root of MSE and the average realized kinship (Equation 14).
| Additional information | Estimator | Relationship | ||||||
|---|---|---|---|---|---|---|---|---|
| FS | HS | C1 | C2 | C3 | IN | |||
| None | PLINK | 1.64 | 4.66 | 10.81 | 50.76 | 178.78 | 30.30 | |
| MLE | 1.51 | 3.55 | 7.26 | 27.00 | 78.66 | 12.79 | ||
| Global Day-Williams | 3.12 | 6.38 | 12.34 | 49.55 | 187.68 | 7.33 | ||
| Classic GRM | 3.00 | 4.85 | 8.32 | 30.83 | 116.22 | 5.92 | ||
| Robust GRM | 2.42 | 4.33 | 7.99 | 31.64 | 120.56 | 5.56 | ||
| Two-step GRM | 2.19 | 4.26 | 7.96 | 30.84 | 116.24 | 5.50 | ||
| LD pattern | Naive LD-pruned GRM | 3.01 | 4.70 | 7.87 | 28.20 | 105.46 | 5.82 | |
| LD pattern | LDAK | 2.64 | 4.10 | 7.21 | 23.74 | 85.57 | 6.31 | |
| LD pattern | LD-weighted GRM | 1.59 | 2.60 | 4.44 | 17.33 | 65.60 | 4.82 | |
| Marker order | Local Day-Williams | 2.04 | 2.76 | 6.52 | 10.79 | 24.26 | 8.31 | |
| LD pattern + marker order | LD-weighted local Day-Williams | 2.10 | 2.85 | 6.70 | 11.27 | 23.56 | 8.39 | |
| Genetic position | HMM | 1.77 | 3.36 | 6.04 | 17.90 | 64.97 | 4.84 | |
| LD pattern + genetic position | LD-weighted HMM | 1.57 | 2.83 | 5.16 | 14.55 | 52.19 | 4.52 | |
| Pedigree | Pedigree kinship | 7.80 | 10.49 | 17.63 | 37.30 | 70.16 | 14.73 | |
All estimators compared require dense SNP genotypes and reliable sources of marker allele frequencies as input. Pedigree kinship for the six relationship types are 0.25 (FS), 0.125 (HS), 0.0625 (C1), 0.0156 (C2), 0.0039 (C3), and 0.1094 (IN), respectively.
Relative performances of the GRM estimators in Group-1 match our expectations under the assumption of linkage equilibrium (Figure 1, A and B). The two-step GRM estimator compares favorably to others on all relationship types. The classic GRM estimator works well when (e.g., third cousins). The robust GRM estimator is preferred to on close relatives, and it dominates the global Day-Williams estimator on all relationship types.
The MLE estimator stands out among estimators in Group-1. Likelihood estimators are often known to be more accurate than method-of-moment estimators (Milligan 2003; Anderson and Weir 2007). However, accuracy and computational efficiency of is extremely sensitive to the starting configurations and the convergence criterion of the EM algorithm. Our implementation of adopted very favorable conditions (see Methods of analysis). Otherwise, the results were much less accurate and computation time much longer (results not shown). For full siblings, the PLINK estimator is more accurate than any Group-1 GRM estimator. Since estimates the k coefficients directly, this provides higher resolution on full siblings who share two IBD genes over, on average, 25% of the genome. When inbreeding is present, and perform poorly relative to the other Group-1 estimators. These two estimators estimate the zero-inbreeding k coefficients directly and are thus more sensitive to violation of the no-inbreeding assumption.
Among the estimators of Group-2, the local Day-Williams estimator performs better than the others on remote relatives. However, the smoothing algorithm of tends to produce downward bias (see File S1, section C, and Table S5) so that must perform well on remote relatives where there is not much room for underestimation. In contrast, the HMM estimator generally overestimates IBD in the presence of LD (Brown et al. 2012). The naive LD-pruned GRM estimator shows slight improvement over the classic GRM estimator but loses to the LDAK on all occasions. The LD-weighted GRM estimator dominates both and in performance and loses to only on remote relatives. This reflects the amount of LD present in the selected marker panel in the combined population, and shows that appropriately adjusting for patterns of LD can significantly improve the accuracy of kinship estimation.
When inbreeding is present, performance of is the most affected among the Group-2 estimators. Glazner and Thompson (2015) noted that, in their example, this local IBD Day-Williams method failed to pick up short segments of complex (autozygous) IBD; the varying kinship levels across short distances seem to challenge this method (see File S1, section C). The performance of is also affected by inbreeding. Perhaps the higher IBD levels in inbred individuals conflict with the assumption of unrelatedness in the estimator derivation.
When the LD weights are combined with the local IBD methods, there is clear improvement in performance for the HMM method (compare to but less so for the Day-Williams local method (compare and As noted above, the HMM overestimates IBD in high-LD regions, so that the LD weights are beneficial.
Pedigree kinship is included in Table 1; it may be considered an estimator based only on the pedigree and not on genetic data. The MSE here represents the variation in realized kinship among pairs of individuals in the same pedigree relationship. The advantage of using genetic-data-based estimates of realized kinship instead of pedigree kinship is clear. Only on remote relatives (C2 and C3) do some of the marker-based estimates differ from the realized kinship by more than the pedigree values do. In fact, the results of Table 1 underplay the performance of the GRM estimators on remote relatives, since, for all the other estimators, estimates are constrained to be within the range. A comparison of constrained and unconstrained performance of the GRM estimators is shown in Table S6.
Discussion
We have shown that improved estimators of realized kinship can be obtained (1) by optimal weighting of markers, (2) by taking physical contiguity of genome into account, and (3) by weighting on the basis of LD. In practice, the choice of estimator largely depends on the availability of information. When one only has SNP genotypes and marker allele frequencies to work with, the two-step GRM estimator is both accurate and computationally efficient. If large genotyped samples from the relevant population are available, the LD-weighted GRM estimator is an attractive alternative. The LD weights can be computed very efficiently using existing optimization software, and the computation needs to be done only once for a population. If information on marker order or genetic positions is available, either the local Day-Williams estimator or the HMM estimator can offer a substantial increase in accuracy at the cost of longer computation time (see Methods of analysis). Once computed, these local IBD estimates across the genome can also be used in other analyses that use location-specific IBD: for example, in gene mapping.
In the derivation of optimal estimators, assumptions such as absence of inbreeding, linkage equilibrium, or unrelatedness were necessary to keep computations tractable. However, the proposed estimators applied well outside the initially assumed context. The two-step GRM estimator does not seem to be affected by the presence of inbreeding. It compares favorably to other estimators that use the same amount of information even when the assumption of linkage equilibrium is violated. The LD-weighted GRM estimator performed well on all relationship types considered, and is only slightly affected when related individuals happen to be inbred.
In our simulation study, relative pairs are generated as random draws from each relationship type. In practice, nonrandom sampling may create biases causing realized kinship to differ from the pedigree values. For instance, ascertainment by traits in human genetics and artificial selection in animal and plant genetics can result in sampled relatives being more (or less) genetically related than expected under the pedigree structure (Liu et al. 2003; Purcell et al. 2007). The comparison of pedigree vs. realized kinships in this article (Table 1) is thus an idealized best-case scenario.
Our simulation study used real haplotypes and a dense SNP marker panel. Thus LD is present in the simulated data. However, in assuming the variance form of Equation (6), the covariances due to LD are ignored (see Equation 5). To investigate the impact of LD on the relative performance of estimators, we compared the empirical and theoretical (no-LD) SDs for several estimators and relationship types. For any GRM estimator described in Linkage equilibrium and any relationship type (except the inbred cousins) listed in Table 1, the true variance is well approximated by empirical MSE. The theoretical (no-LD) variance (6) is computed using the simulation values of and and averaged across all 1000 pairs of that relationship type. Table S3 summarizes the results. We see that the factor by which the SD is underestimated by ignoring LD is generally smaller on remote relatives. More importantly, for a given relationship type, it is fairly consistent across estimators. This suggests that between GRM estimators that do not adjust for LD, relative efficiency computed under the assumption of linkage equilibrium can be a good approximation to the true relative values.
Our study assumed population homogeneity so that allele frequencies and LD weights can be estimated once and applied to all pairs of relatives. This choice has the advantage of having a bigger pool of founder haplotypes available for sampling, and thus lower correlation among estimates from different simulation replicates. It also induces a more complex population structure in the simulated data, which is likely to influence performance of some estimators more than others. In limited additional experiments, we investigated performance of a subset of estimators on data simulated under either European ancestry or African ancestry, and compared results to those of Table 1 (see Table S7 and Table S8). Unsurprisingly, the GRM estimators generally benefit from the lesser structure in these more homogeneous pools of haplotypes. Relative performance among the GRM estimators that do not adjust for LD remains unchanged. The LD weighted GRM estimator loses to the two-step GRM estimator on close relatives, suggesting that the amount of LD adjustment (given the choice of marker panel and ancestry) is not enough to offset the effect of deviation from the assumption of unrelatedness. However, these results also suggest is robust to population substructure and admixture; compared to other GRM estimators, its performance is much less affected by the complex structure of the combined population (compare Table 1 with Table S7 and Table S8). The HMM estimator does better under African ancestry (than under combined ancestry), and worse under European ancestry. This is likely due to the higher LD in the European population; is sensitive to LD (Brown et al. 2012).
For convenience, we selected SNPs based on even genetic distance spacing and minor allele frequency (MAF). The relationship between genetic distance and LD is far from uniform, and our results on the impact of adjusting for LD show that it is a significant factor in our marker panel. The distribution of allele frequencies in our selected marker panel is quite similar to that in the commonly used OmniExpress24 genome-wide-association-study chip (see Figure S3). Compared to the OmniExpress24 autosomal markers, our selected marker panel has a slight underrepresentation for markers with MAF However, our panel was selected with a threshold of MAF whereas ∼ of the OmniExpress24 autosomal markers fall below this threshold. We found that naive pruning of markers by MAF generally reduces accuracy of the two-step GRM estimator but can sometimes improve accuracy of other Group-1 GRM estimators depending on relationship types. Even on MAF-pruned marker sets, always stands out among the Group-1 GRM estimators (results not shown). Overall, our selection of SNPs by genetic spacing and MAF create no strong biases in our estimator comparisons.
Even as SNP panels become denser, or sequence data become available, the issue of LD remains. Additional typed loci do not provide additional relatedness information without limit. Although methods that adjust for LD will gain from the use of additional markers, other methods may not benefit and can be adversely affected. In particular, non-LD-adjusted local methods such as the HMM estimator should be applied on an LD-pruned marker set to avoid overestimation of IBD. In human applications with nominally unrelated individuals, haplotypic similarities due to LD must be distinguished from cryptic relatedness. In animal and plant breeding applications, high levels of LD are a regular feature of artificially selected populations with a small effective founder population size. Therefore, methods for efficient kinship estimation in the presence of LD remain relevant even in the age of full genome sequencing.
Estimators developed in this article do not specifically deal with population substructure or admixture, but they can be generalized to do so. Thornton et al. (2012) and Conomos et al. (2016) proposed two different methods that estimate individual-specific allele frequencies which take population substructure and admixture into account. The estimated individual-specific allele frequencies were subsequently applied to the classic GRM estimator and the robust GRM estimator, respectively, for kinship estimation. The same logic may be applied to calculate LD weights that adjust for population substructure or admixture. These frequencies and LD weights can then be used with our proposed estimators. The merits of such an approach to address population substructure or admixture is a topic for future studies.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.197004/-/DC1.
Acknowledgments
We are grateful to Ellen Wijsman, Fiona Grimson, Aaron Baraff, Steven Lewis, and Alejandro Nato for valuable discussions. We also thank the two anonymous reviewers for their constructive comments. This research was supported in part by National Institutes of Health grants R37 GM-046255 and P01 GM-099568.
Appendix A: Reparameterization of the Global Day-Williams Estimator
The original form of the global Day-Williams estimator introduced in Day-Williams et al. (2011) is
where is the kinship between individual u and v, is the observed number of IBS matches between u and v, m is the total number of markers, is the reference allele frequency at marker i and is defined as
where and are the allelic types of the individual u at marker i, and and are the allelic types of the individual v at marker i.
Since we are working with genotypic data from pairs of individuals, indices of individuals within a pair are exchangeable, and so are indices of genes from the same individual at a given marker. Table A1 shows the correspondence between the Day-Williams and the GRM notations at each marker position. Note that and are equivalent for all possible genotype combinations. Therefore, we can rewrite as
which recovers the expression in Equation 4.
Table A1. Correspondence between notations for all possible marker genotypes.
| Notation | Day-Williams | GRM | ||
|---|---|---|---|---|
| Genotype | ||||
| AA,AA | (1,1,1,1) | 1 | (2,2) | 1 |
| AA,AB | (1,1,1,2) | 0.5 | (2,1) | 0.5 |
| AA,BB | (1,1,2,2) | 0 | (2,0) | 0 |
| AB,AB | (1,2,1,2) | 0.5 | (1,1) | 0.5 |
| AB,BB | (1,2,2,2) | 0.5 | (1,0) | 0.5 |
| BB,BB | (2,2,2,2) | 1 | (0,0) | 1 |
Appendix B: Computations in Methods
General Case
Let and be the indicator functions that the two alleles of individual 1 at marker l are the reference allele, respectively, with l omitted from the notation. Define and similarly for individual 2. It is easy to see that and Recall that the basic assumptions are:
IBD genes are of the same allelic types, whereas non-IBD genes are of independent allelic types.
Either of the two genes from one individual is equal likely to be IBD to either of the two genes of the other individual at the same locus.
We have
and
Since
is unbiased for any and
Linkage Equilibrium
We now assume linkage equilibrium and absence of inbreeding. To derive an expression for defined in (5), note that
Calculation of the term involves probabilities of the underlying IBD states between the two genes of individual 1 and one gene of individual 2. Given the assumption of no inbreeding, there are only three possible IBD states with probabilities given in Table B1.
Table B1. IBD states and probabilities for the two genes of individual 1 and one random gene from individual 2.
| IBD state | (1,2,1) | (1,2,2) | (1,2,3) |
|---|---|---|---|
| Probability |
Following the above results, the closed-form expression of the variance of Equation 5 is derived in Box B1, assuming For general choices of is not symmetric about and is thus sensitive to the choice of reference allele. The part of that is responsible for this asymmetry is
Box 1.
Derivation of Vl(al) under the assumption of no inbreeding.
where can be a function of
Suppose, however, that is a weighted average of and 1: for Then
Then takes the value
which is symmetric about and it attains its minimum at conditional on and
Returning to the general form of and setting its derivative with respect to equal to 0, leads to
Thus the optimal is a weighted average of and 1, and is invariant to the choice of reference allele.
For fixed [and therefore fixed we can find the set of optimal weights by solving the following minimization problem:
With Lagrange multipliers λ,
The above expression holds true for all l, implying
The nonnegativity constraint is automatically satisfied as for all l.
Linkage Disequilibrium
We drop the assumptions of linkage equilibrium and absence of inbreeding, but assume instead that the two individuals are unrelated, so that are independent, for all l and m. Under the new assumptions, all of the results from General Case still hold, but most of the results from Linkage Equilibrium do not. Let the inbreeding coefficients of the two individuals be and respectively,
Analogous results hold for It can be verified (line 5 of Box B1) that
This time
since
Now
where
Setting
It then follows that
We assume that the matrix of squared LD correlations, is known. Correlation matrices are positive semidefinite. By the Schur product theorem, the Hadamard product of a correlation matrix and itself must also be positive semidefinite.
As noted in the text (Equation 13), we solve the following minimization problem for the LD weights,
When has a block-diagonal structure (e.g., each chromosome forms a block), we can rewrite the minimization problem in terms of subvectors and submatrices,
Since the form a partition of and the are independent submatrices of minimization can be implemented for each block independently. We can solve for in block i using the corresponding submatrix If for the block solutions, the final solution will be a concatenation of the block solutions rescaled by
Footnotes
Communicating editor: E. Eskin
Literature Cited
- 1000 Genomes Project Consortium , 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A. D., Weir B. S., 2007. A maximum-likelihood method for the estimation of pairwise relatedness in structured populations. Genetics 176: 421–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo R., 2008. Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci. 48: 1649–1664. [Google Scholar]
- Brown M. D., Glazner C. G., Zheng C., Thompson E. A., 2012. Inferring co-ancestry in population samples in the presence of linkage disequilibrium. Genetics 190: 1447–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Wijsman E. M., Weir B. S., 2009. Case-control association testing in the presence of unknown relationships. Genet. Epidemiol. 33: 668–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conomos M. P., Reiner A. P., Weir B. S., Thornton T. A., 2016. Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet. 98: 127–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J., de Los Campos G., Pérez P., Gianola D., Burgueño J., et al. , 2010. Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 186: 713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day-Williams A., Blangero J., Dyer T., Lange K., Sobel E., 2011. Linkage analysis without defined pedigrees. Genet. Epidemiol. 35: 360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Gianola D., Allison D. B., 2010. Predicting genetic predisposition in humans: the promise of whole-genome markers. Nat. Rev. Genet. 11: 880–886. [DOI] [PubMed] [Google Scholar]
- Gianola D., van Kaam J. B., 2008. Reproducing kernel Hilbert spaces regression methods for genomic assisted prediction of quantitative traits. Genetics 178: 2289–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazner C. G., Thompson E. A., 2015. Pedigree-free descent-based gene mapping from population samples. Hum. Hered. 80: 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard K. A., Yu C. E., Oshima J., Miki T., Nakura J., et al. , 1996. Toward localization of the Werner syndrome gene by linkage disequilibrium and ancestral haplotyping: lessons learned from analysis of 35 chromosome 8p11.1–21.1 markers. Am. J. Hum. Genet. 58: 1286–1302. [PMC free article] [PubMed] [Google Scholar]
- Hayes B. J., Visscher P. M., Goddard M. E., 2009. Increased accuracy of artificial selection by using the realized relationship matrix. Genet. Res. 91: 47–60. [DOI] [PubMed] [Google Scholar]
- Henderson C. R., 1976. A simple method for computing the inverse of a numerator relationship matrix used in prediction of breeding values. Biometrics 32: 69–83. [Google Scholar]
- Hill W. G., Weir B. S., 2011. Variation in actual relationship as a consequence of Mendelian sampling and linkage. Genet. Res. 93: 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S.-y., et al. , 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Goodman M., Muse S., Smith J. S., Buckler E., et al. , 2003. Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165: 2117–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin M., Dunning I., 2015. Computing in operations research using julia. INFORMS J. Comput. 27: 238–248. [Google Scholar]
- Matise T. C., Chen F., Chen W., De La Vega F. M., Hansen M., et al. , 2007. A second-generation combined linkage physical map of the human genome. Genome Res. 17: 1783–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan B. G., 2003. Maximum-likelihood estimation of relatedness. Genetics 163: 1153–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moltke I., Albrechtsen A., Hansen T., Nielsen F. C., Nielsen R., 2011. A method for detecting IBD regions simultaneously in multiple individuals: with applications to disease genetics. Genome Res. 21: 1168–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Price A. L., Reich D., 2006. Population structure and eigenanalysis. PLoS Genet. 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R. a., 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel E., Lange K., 1996. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am. J. Hum. Genet. 58: 1323–1337. [PMC free article] [PubMed] [Google Scholar]
- Speed D., Hemani G., Johnson M. R., Balding D. J., 2012. Improved heritability estimation from genome-wide SNPs. Am. J. Hum. Genet. 91: 1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sverdlov, S., 2014 Functional quantitative genetics and the missing heritability problem. Ph.D. Thesis, University of Washington, Seattle. [Google Scholar]
- Thompson E. A., 2000. Statistical Inference from Genetic Data on Pedigrees, Vol. 6 Institute of Mathematical Statistics, Beechwood, OH, and Alexandria, VA. [Google Scholar]
- Thompson, E. A., and S. Lewis, 2016 MORGAN, version 3.3.1. Monte Carlo genetic analysis package Available at: https://www.stat.washington.edu/thompson/Genepi/MORGAN/Morgan.shtml.
- Thornton T., Tang H., Hoffman T. J., Ochs-Balcom H. M., Caan B. J., et al. , 2012. Estimating kinship in admixed populations. Am. J. Hum. Genet. 91: 122–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M., 2007. Genomic measures of relationship and inbreeding. Interbull Bull 37: 33–36. [Google Scholar]
- VanRaden P. M., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423. [DOI] [PubMed] [Google Scholar]
- Visscher P. M., Medland S. E., Ferreira M. A. R., Morley K. I., Zhu G., et al. , 2006. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2: e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F., Lee S., Knowles M. R., Wright F. A., 2010. Quantification of population structure using correlated SNPs by shrinkage principal components. Hum. Hered. 70: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 1000 Genomes Project Phase 3 data are available at http://www.1000genomes.org/data. The Rutgers Map version 3a is available at http://compgen.rutgers.edu/download_maps.shtml. Information on the set of 169,751 SNP markers used in the simulation study is provided as File S2.