Abstract
Histone post-translational modifications (PTMs) alter chromatin structure by promoting the interaction of chromatin-modifying complexes with nucleosomes. The majority of chromatin-modifying complexes contain multiple domains that preferentially interact with modified histones, leading to speculation that these domains function in concert to target nucleosomes with distinct combinations of histone PTMs. In Saccharomyces cerevisiae, the NuA3 histone acetyltransferase complex contains three domains, the PHD finger in Yng1, the PWWP domain in Pdp3, and the YEATS domain in Taf14; which in vitro bind to H3K4 methylation, H3K36 methylation, and acetylated and crotonylated H3K9, respectively. While the in vitro binding has been well characterized, the relative in vivo contributions of these histone PTMs in targeting NuA3 is unknown. Here, through genome-wide colocalization and by mutational interrogation, we demonstrate that the PHD finger of Yng1, and the PWWP domain of Pdp3 independently target NuA3 to H3K4 and H3K36 methylated chromatin, respectively. In contrast, we find no evidence to support the YEATS domain of Taf14 functioning in NuA3 recruitment. Collectively our results suggest that the presence of multiple histone PTM binding domains within NuA3, rather than restricting it to nucleosomes containing distinct combinations of histone PTMs, can serve to increase the range of nucleosomes bound by the complex. Interestingly, however, the simple presence of NuA3 is insufficient to ensure acetylation of the associated nucleosomes, suggesting a secondary level of acetylation regulation that does not involve control of HAT-nucleosome interactions.
Keywords: histone acetylation, histone methylation, NuA3, histone acetyltransferase, Saccharomyces cerevisiae
EUKARYOTIC DNA is packaged into a nucleoprotein structure known as chromatin, which consists of DNA, histones, and nonhistone proteins. Histones are extensively post-translationally modified, with specific modifications reflecting activities occurring on the underlying DNA. For example, genes transcribed by RNA polymerase II (RNAPII) have acetylated and H3K4 trimethylated histones (H3K4me3) at their 5′ ends and H3K36 trimethylated histones (H3K36me3) over the gene body (Liu et al. 2005; Pokholok et al. 2005). In contrast, histone H2A glutamine 105 methylation and H2A.X serine 139 phosphorylation are associated with RNA polymerase I transcription and DNA double-strand break repair, respectively (Rogakou et al. 1998; Tessarz et al. 2014). It is generally accepted that these modifications promote the biological processes to which they are associated.
Although some histone post-translational modifications (PTMs) can directly alter chromatin structure, most function as recognition sites for histone PTM binding domains (Yun et al. 2011). Histone PTM binding domains are found in complexes that facilitate transcription or alter chromatin structure, such as basal transcription factors, chromatin-remodeling complexes, and even enzymes that post-translationally modify histones. The majority of chromatin-modifying complexes have multiple histone PTM binding domains, the purpose of which has been the subject of much speculation. The generally weak affinity of these domains for their requisite histone PTMs has led to the hypothesis that multiple histone-binding interactions are required to stabilize the recruitment of complexes to chromatin (Yun et al. 2011). Indeed, it is suggested that multiple histone-binding domains function synergistically to translate a “code” of histone PTMs into a single biological outcome (Strahl and Allis 2000). Alternatively, multiple histone-binding domains could act independently to target a chromatin-binding complex to a range of genomic loci.
One complex containing multiple histone PTM binding domains is the NuA3 histone acetyltransferase (HAT) complex in Saccharomyces cerevisiae, which acetylates H3K14 and H3K23 (Howe et al. 2001). NuA3 contains six subunits: the catalytic subunit Sas3, Nto1, Eaf6, and three histone PTM-binding proteins: Yng1, Pdp3, and Taf14 (John et al. 2000; Howe et al. 2002; Taverna et al. 2006). Yng1 contains a PHD finger which binds to H3K4 mono-, di-, and trimethylation, with binding affinity increasing with the number of methyl groups (Martin et al. 2006; Taverna et al. 2006; Shi et al. 2007). Pdp3 contains a PWWP domain, which recognizes H3K36me3 (Gilbert et al. 2014). Taf14, through its YEATS domain, binds to acetylated (H3K9ac) and crotonylated (H3K9cr) histone H3K9 (Shanle et al. 2015; Andrews et al. 2016). While the in vitro binding of these proteins to histone PTMs has been well characterized (Supplemental Material, Table S1), the relative contributions these histone PTMs make to NuA3 targeting in vivo remains unknown.
In mammalian cells, two HAT complexes appear analogous to NuA3: the MOZ/MORF complex and the HBO1–BRPF1 complex. Both complexes contain bromodomain PHD finger protein 1 (BRPF1) or one of its paralogs, BRPF2 or 3 (Doyon et al. 2006; Lalonde et al. 2013). BRPF1/2/3 share sequence similarity with yNto1, but additionally contain carboxy-terminal, H3K36me3-specific PWWP domains (Vezzoli et al. 2010), and thus may serve the role of both yNto1 and yPdp3 in mammalian complexes. The MOZ/MORF complex also has an H3K4me2/3-specific PHD finger in its subunit, inhibitor of growth 5 (ING5), while the HBO1–BRPF1 uses ING4 or ING5 to bind to H3K4me2/3 (Doyon et al. 2006; Lalonde et al. 2013). Finally, although the MOZ/MORF and HBO1–BRPF1 complexes lack a Taf14 equivalent, a bromodomain within BRPF1/2/3 and a double PHD finger in MOZ/MORF show specificity for acetylated histones (Laue et al. 2008; Ali et al. 2012; Liu et al. 2012; Qiu et al. 2012). Thus, the histone PTM binding domains in NuA3 are conserved in analogous complexes in other organisms, although the relative contribution that these domains make in targeting NuA3 has yet to be determined.
In this study we show that NuA3 is primarily targeted to “midgene” regions via interactions with H3K36me3 and H3K4me1/2/3, while H3K9ac and H3K9cr are unlikely to play a role in recruitment. Simultaneous disruption of H3K4 and H3K36 methylation abolishes NuA3 recruitment to actively transcribed genes. In contrast, disruption of H3K4 or H3K36 methylation singularly results in partial loss of NuA3 recruitment, suggesting that these PTMs recruit NuA3 independently and arguing against a synergistic effect. Finally, we show that NuA3 occupancy does not dictate histone acetylation, indicating that controlled targeting is not the only mechanism for regulation of NuA3 function.
Materials and Methods
Yeast strains and plasmids
All strains used in this study were isogenic to S288C, and are listed in Table S2. All strains are available upon request. Yeast culture and genetic manipulations were performed using standard protocols. Genomic deletions were verified by PCR analysis and whole cell extracts were generated as previously described (Kushnirov 2000). The previously described kanMX-GAL1pr-flo8-HIS3 strains (Cheung et al. 2008) were generous gifts from Fred Winston.
Drug treatments
For H3K23ac chromatin immunoprecipitation (ChIP) with sequencing (ChIP-seq), bar1Δ cells were arrested in G1 by 2.5 hr treatment with 5 μM α-factor. Culture synchrony in G1 was confirmed by the appearance of “shmooing” cells, as seen under the microscope. Trichostatin A (TSA) was added for 15 min at 25 μM, from 5 mM DMSO stock.
ChIP-seq
The ChIP-seq protocol was based on that outlined in Maltby et al. (2012). Cell pellets were resuspended in lysis buffer (50 mM HEPES, pH 7.5, 140 mM NaCl, 1 mM EDTA, 2% Tritox X-100, 0.2% Na-deoxycholate, 1× Roche Protease Inhibitor Cocktail, 1 mM PMSF) and lysed by bead beating. Cell lysates were spun down at 15,000 × g for 30 min. The chromatin pellets were resuspended in NP-S buffer (0.5 mM spermidine, 1 mM β-ME, 0.075% NP-40, 50 mM NaCl, 10 mM Tris, pH 7.4, 5 mM MgCl2, 1 mM CaCl2) and digested with micrococcal nuclease (MNase) at 37° for 10 min to obtain predominantly mono-nucleosomal DNA. Reactions were stopped with the addition of EDTA to 10 mM and digested lysates were clarified by centrifugation at 9000 × g for 10 min. To extract insoluble chromatin, pellets were resuspended in 300 μl of lysis buffer with 0.2% SDS, and sonicated in a Diagenode Bioruptor at high output for four cycles of 30 sec. Extracts were then reclarified by centrifugation at 9000 × g for 10 min, and the supernatant pooled with the preexisting extract. The buffer composition of the lysate was adjusted to that of the original lysate, and 10% was set aside as input. The supernatant was precleared by incubation with magnetic Protein G Dynabeads (Life Technologies) for 1 hr at 4°. Precleared lysates were incubated with αHA (catalogue number 12013819001; Roche), αH3K4me3 (catalogue number ab1012; Abcam), αH3K23ac (catalogue number 39131; Active Motif), or αIgG (catalogue number PP64; Millipore, Bedford, MA) antibodies at 4° overnight. Immune complexes were precipitated by incubation with magnetic Protein G Dynabeads for 1 hr at 4°. Beads were washed twice with lysis buffer, high salt wash buffer (50 mM HEPES, pH 7.5, 640 mM NaCl, 1 mM EDTA, 2% Tritox X-100, 0.2% Na-deoxycholate), LiCl wash buffer (10 mM Tris-HCL, pH 8.0, 250 mM LiCl, 0.6% NP-40, 0.5% Na-deoxycholate, 1 mM EDTA), and once with TE. The immunoprecipitated DNA was subjected to Illumina HiSeq paired-end sequencing. Reads were aligned to the S. cerevisiae genome (Saccer3 genome assembly) using Burrows–Wheeler Aligner (BWA) (Li and Durbin 2010). Average gene profiles were generated using the sitepro tool in the Cis-regulatory Element Annotation System genome package (http://liulab.dfci.harvard.edu/CEAS/) and plotted using R. The H3K23ac signal represents the average of two independent replicates. For average plots showing the immunoprecipitation (IP) divided by input, a coverage value of one was added to both the IP and the input to avoid division by zero. Values past the polyadenylation sequence (Park et al. 2014) were excluded from the average calculation, and the fraction of genes included in the average calculation at any given distance from the transcription start site (TSS) was shown.
Published data sets
The data sets for the cooccurring nucleosomes and for H3K4me3 in the H3K36R mutant (Sadeh et al. 2016) were downloaded from the Sequence Read Archive (SRA) study SRP078243. Histone methylation and acetylation data sets from Weiner et al. (2015) were downloaded from SRA study SRP048526. H3K9ac data sets from Bonnet et al. (2014) were downloaded from SRA study SRP033513. The H3K4me3 data set from Maltby et al. (2012) was from our previous study. The fastq files were mapped to saccer3 using BWA (Li and Durbin 2010). The methylation data from Schulze et al. (2011) was downloaded from http://www.yeastgenome.org as mapped MAT scores. The Yng1 data from Taverna et al. (2006) was kindly provided by the authors as mapped intensity scores for IPs and inputs. The native elongating transcript sequencing (NET-seq) data from Churchman and Weissman (2011) was downloaded from SRA study SRP004431 and mapped to saccer3 using bowtie (Langmead et al. 2009). The processed data for Gcn5 from Xue-Franzén et al. (2013) was downloaded from the Gene Expression Omnibus (GEO) depository, series GSE36600. For average gene profiles, the +1 nucleosome was called using BEDTools (Quinlan and Hall 2010) as the closest consensus nucleosome position (Brogaard et al. 2012) to the TSS (Brogaard et al. 2012). Cell cycle-regulated genes (Eser et al. 2014) were excluded from analysis comparing G1-arrested and asynchronous data sets (File S1).
Nucleosome enrichments
For each data set the average coverage over genome-wide called nucleosome positions (Weiner et al. 2015) was calculated. When available, the IPs were normalized to input files. Spearman correlation matrix was calculated in R, considering all pairwise complete observations.
Gene peak enrichment
For each gene, 100-bp windows were constructed in 5-bp steps, from upstream edge of +1 nucleosome core particle (NCP) to the polyadenylation site to a maximum of 3000 bp. The average signal (IP coverage for data from Sadeh et al. 2016 and IP/input for all others) was calculated for each bin, and the center of the most enriched bin was defined as the peak enrichment.
Boxplots
Methylation enrichment was defined as having an IP/input of >1.5, and depletion corresponded to an IP/input of <0.75. Boxplots extend from the first to third quartiles, with whiskers extending to 1.5 times the interquartile range or to the extreme of the data. Notches extend ±1/58 IQR /sqrt(n) and give an approximation of the 95% confidence interval for the difference in two medians.
ChIP with quantitative PCR of Sas3
Cells were grown in 50 ml YPD to an OD600 of 0.8 and cross-linked with 1% formaldehyde for 30 min at room temperature. The reaction was stopped by the addition of 125 mM glycine, and incubated at room temperature for a further 15 min. Pellets were washed twice with cold PBS, resuspended in 600 μl of lysis buffer (50 mM HEPES, pH 7.5, 140 mM NaCl, 0.5 mM EDTA, 1% triton X-100, 0.1% sodium deoxycholate), and lysed mechanically by vortexing with glass beads for 25 min at 4°. The lysates were spun down by centrifugation at 15,000 × g for 30 min, the supernatant was discarded, and the pellet was washed and resuspended in 500 μl lysis buffer. The resuspended pellets were sonicated at high output for 30 sec on, with 30 sec break, for 30 cycles to obtain an average fragment length of 250 bp. A further 200 μl lysis buffer was added to each sample, and the lysates were clarified by centrifugation at 9000 × g for 10 min. Then, 10% of the lysate was reserved for input. Lysates were incubated with 1 μg anti-HA antibody (catalogue number 12CA5; Roche) at 4° overnight, followed by precipitation of immune complexes with 25 μl Protein G Dynabeads at 4° for 1 hr. Beads were washed twice with lysis buffer, high salt wash buffer (50 mM HEPES, pH 7.5, 640 mM NaCl, 1 mM EDTA, 2% Tritox X-100, 0.2% Na-deoxycholate), LiCl wash buffer (10 mM Tris-HCL, pH 8.0, 250 mM LiCl, 0.6% NP-40, 0.5% Na-deoxycholate, 1 mM EDTA), and once with TE. The DNA was eluted and processed as described previously (Maltby et al. 2012) and quantitative PCR (qPCR) was performed in technical triplicate using the primers listed in Table S3.
Data availability
The ChIP-seq data generated for this study have been deposited in the GEO database, GEO accession: GSE93059 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93059). See Table S2 for the list of strains and Table S3 for the list of primers used in this study.
Results
NuA3 is primarily bound to midgene regions of actively transcribed genes
The NuA3 histone acetyltransferase complex contains three histone PTM binding domains: the Yng1 PHD finger, the Pdp3 PWWP domain, and the Taf14 YEATS domain; which show specificity for H3K4me1/2/3, H3K36me3, and H3K9ac/H3K9cr, respectively (Taverna et al. 2006; Gilbert et al. 2014; Shanle et al. 2015; Andrews et al. 2016) (Table S1). To determine the relative contribution of each histone PTM in targeting the NuA3 complex, we reasoned that histone PTMs that promote the interaction of NuA3 with chromatin would colocalize with Sas3. To this end, we mapped NuA3-bound nucleosomes at high resolution in vivo using a MNase-based ChIP-seq approach, which has previously been used to map chromatin remodelers (Koerber et al. 2009; Floer et al. 2010; Yen et al. 2012; Ramachandran et al. 2015). We immunoprecipitated HA-tagged Sas3, in parallel with an untagged control, from cross-linked MNase-digested chromatin and performed paired-end sequencing. We could not detect any DNA in the untagged control mock IP, but nonetheless constructed a library and included the sample in the pool for sequencing. While sequencing of the Sas3 IP and inputs produced >8,000,000 DNA fragments, only 84,973 DNA fragments were recovered in the untagged control IP, confirming the specificity of our Sas3 ChIP-seq experiment.
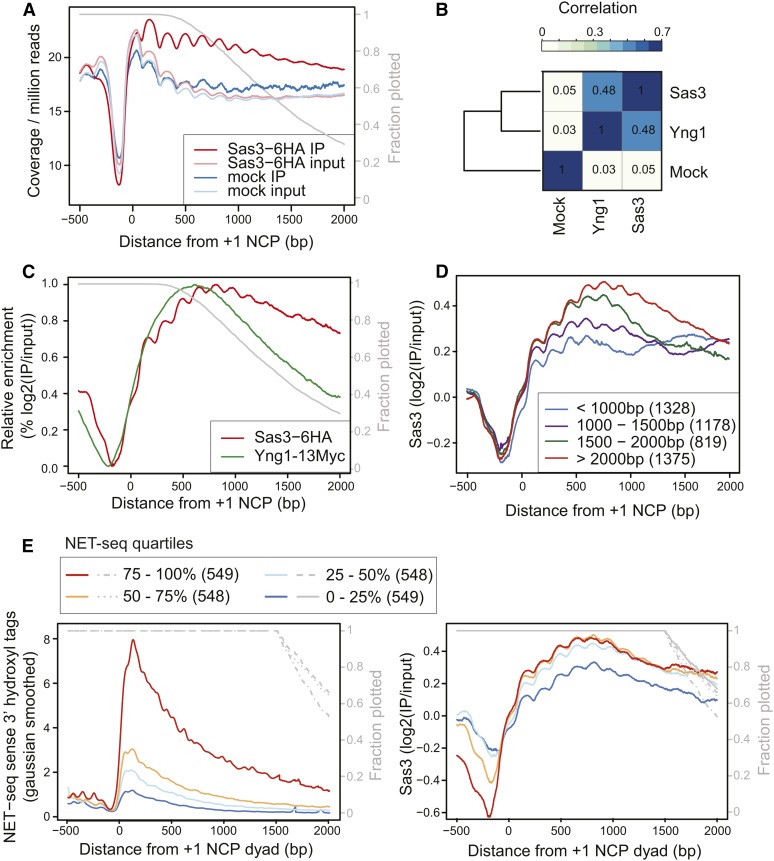
We mapped the sequenced DNA fragments to the S. cerevisiae genome, aligned genes by the +1 nucleosome, and calculated the average profile for each sample (Figure 1A). Relative to the input, Sas3 was enriched in the gene body but not at the +1 nucleosome, and this enrichment was specific to the Sas3 IP and was not observed for the untagged control. We next compared Sas3 occupancy to a previously reported sonication-based mapping of Yng1 (Taverna et al. 2006). Unsurprisingly, Yng1 and Sas3 levels correlated positively genome wide with a Spearman correlation coefficient of 0.48 (Figure 1B and Figure S1). Additionally, both of the Yng1 and Sas3 input-normalized distributions peaked in enrichment 600–800 bp downstream of the +1 nucleosome (Figure 1C). Thus our mapping of Sas3 agreed well with the previous mapping of Yng1, but the MNase-based approach increased the resolution of the assay, and we found that NuA3 was primarily recruited to midgene regions containing the +5 to +7 NCPs (Figure 1C). We also observed that Sas3 was enriched on long genes and modestly enriched on genes transcribed by RNAPII (Figure 1, D and E, and Figure S2), consistent with Sas3 recruitment to midgene regions of actively transcribed genes.
Figure 1.
NuA3 is primarily bound to midgene regions of actively transcribed genes. (A) The average sequence coverage relative to 4701 +1 nucleosomes. (B) Genome-wide Spearman correlations (500-bp windows, 100-bp steps) for input-normalized Sas3 and Yng1 (Taverna et al. 2006). (C) Average enrichment for Yng1 and Sas3 relative to the +1 nucleosome. Signal represents the log2(IP/input) and plotted as relative signal within each sample. (D) Sas3 enrichment by gene length, with gene length defined as the distance from the +1 nucleosome to the polyadenylation site. (E) Sas3 enrichment by quartiles of sense NET-seq (Churchman and Weissman 2011) signal for genes longer than 1500 bp. Only genes longer than 1500 bp were plotted to avoid gene-length effects, but results are similar when all genes analyzed (Figure S2). Except for gene-length plot, all average plots only include data until the polyadenylation signal, and the gray line represents the fraction of genes still being plotted.
NuA3 is associated with H3K4me3, H3K4me2, H3K4me1, and H3K36me3 nucleosomes genome wide
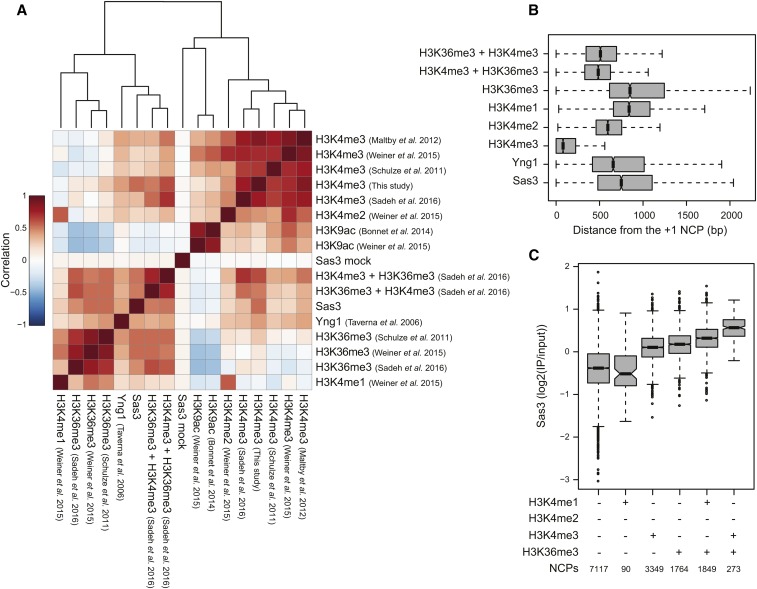
As domains within NuA3 have been reported to bind to H3K4me3, H3K4me2, H3K4me1, H3K36me3, H3K9ac, and H3K9cr in vitro (Table S1), we hypothesized that if these modifications recruit NuA3 to nucleosomes in vivo they will correlate positively with Sas3. To conduct this meta-analysis, we made use of published genome-wide studies of these histone PTMs to calculate nucleosome-based Spearman correlations with Sas3 and Yng1 (Figure 2A and Table S4) . Since crotonylation has not been mapped in S. cerevisiae, but colocalizes with acetylation in mammalian cells (Sabari et al. 2015), we used H3K9ac as a proxy for H3K9cr. To our surprise, H3K9ac did not correlate with Sas3 or Yng1, which suggests that despite Taf14 binding in vitro, it does not function in NuA3 recruitment in vivo. Conversely, H3K36me3, H3K4me3, H3K4me2, and H3K4me1 all correlated positively with Sas3 and Yng1, supporting their role in NuA3-nucleosome binding in vivo. Furthermore, cooccurring H3K4me3 and H3K36me3 nucleosomes, identified through sequential IPs (Sadeh et al. 2016), strongly correlated with Sas3 with Spearman correlation coefficients >0.5 (Figure 2A and Table S4). The dual H3K4me3 and H3K36me3 methylated nucleosomes also specifically grouped with Sas3 and Yng1 following hierarchical clustering, consistent with NuA3 binding to nucleosomes through the combined effects of H3K36me3 and H3K4 methylation.
Figure 2.
NuA3 is associated with H3K4me3, H3K4me2, H3K4me1, and H3K36me3 nucleosomes genome wide. (A) Spearman correlation matrix for Sas3, Yng1, and histone PTM enrichments at 66,360 genome-wide nucleosome positions. The rows and columns were sorted by hierarchical clustering, and the clustering is represented by dendrogram. (B) Gene peak enrichments relative to 4701 +1 nucleosome dyads for cooccurring H3K4me3 and H3K36me3 (Sadeh et al. 2016), H3K4me1/2/3 (Weiner et al. 2015), H3K36me3 (Weiner et al. 2015), Yng1 (Taverna et al. 2006), and Sas3. (C) Sas3 enrichment at nucleosome positioned enriched or depleted for H3K4me1/2/3 and H3K36me3 (Weiner et al. 2015).
Next, we compared the locations of peak enrichment for Sas3, Yng1, H3K36me3, and H3K4 methylation (Figure 2B). Sas3 and Yng1 had median peak enrichments at 750 and 657 bp downstream of the +1 dyad, respectively, which was in between the peak enrichments for H3K4me3 and H3K36me3. Notably, Yng1 and Sas3 peaked in enrichment slightly downstream of cooccurring H3K4me3 and H3K36me3; suggesting that H3K4me2 and H3K4me1 are also important for NuA3 targeting. Altogether the peak enrichments of Sas3 and Yng1 were consistent with NuA3 being primarily recruited to midgene regions through the combined effects of H3K36me3 and H3K4 mono-, di-, and trimethylation.
We next assessed Sas3 enrichment on nucleosomes containing H3K4me1/2/3 or H3K36me3 singularly or in combination (Figure 2C). In the absence of H3K36me3 or H3K4 methylation, Sas3 was depleted from nucleosomes. Likewise, nucleosomes solely enriched in H3K4me1 were also depleted in Sas3, albeit with the caveat that we only identified 90 such nucleosomes and these may represent noise within one of the data sets or be atypical cases in the genome. H3K4me2-enriched nucleosomes are in all but a handful of nucleosomes also enriched for H3K4me1 or H3K4me3, and we were unable to interrogate this modification in isolation. Nucleosomes enriched singularly with H3K36me3 or H3K4me3 were enriched for Sas3 to a similar extent, suggesting that each of these modifications recruit Sas3 to nucleosomes independently. Nucleosomes enriched with both H3K4me1 and H3K36me3 were enriched for Sas3 to a greater extent than H3K36me3 alone, suggesting that H3K4me1 does indeed function in Sas3 recruitment. Nucleosomes enriched for both H3K4me3 and H3K36me3 were enriched further still in Sas3 binding, consistent with NuA3 preferentially binding H3K4me3 over H3K4me1. We found similar results upon analysis of Yng1 (Figure S3). Altogether this analysis supports H3K36me3 and H3K4me1/2/3 functioning to recruit NuA3 to chromatin in vivo.
H3K4 and H3K36 methylation are necessary for Sas3 binding to active genes through Pdp3 and Yng1 respectively
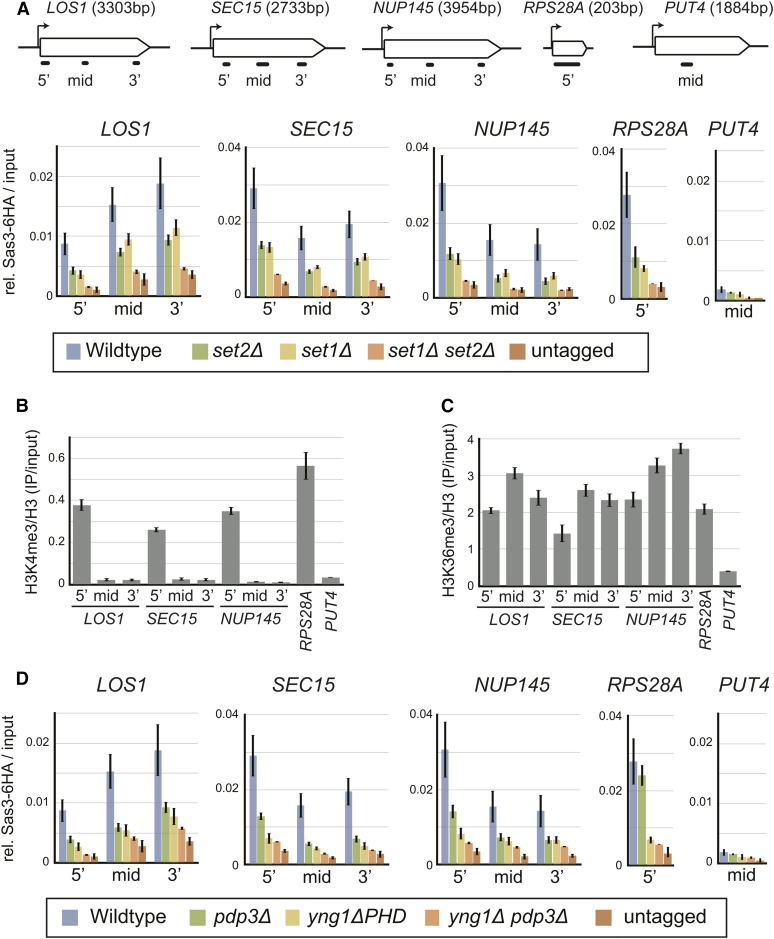
The genome-wide analysis suggested that H3K4 and H3K36 methylation are the major mechanisms for recruiting NuA3 to chromatin, and we next sought to test this directly. H3K4 and H3K36 methylation are entirely dependent on the histone methyltransferases Set1 and Set2, respectively, so we performed ChIP with qPCR (ChIP-qPCR) for Sas3 in strains lacking the methyltransferases singularly and in combination. Mutation of either SET1 or SET2 individually resulted in a partial loss of Sas3 at LOS1, SEC15, NUP145, and RPS28A (Figure 3A). However, Sas3 remained enriched over background at these four genes, and was also enriched relative to a repressed gene, PUT4. In contrast to the modest loss in the single mutants, the set1Δ set2Δ double mutant reduced Sas3 recruitment to background levels, demonstrating that Set1 and Set2 are necessary for Sas3 recruitment to chromatin. Furthermore, the Set1 dependence of Sas3 binding to the mid- and 3′ regions of LOS1, SEC15, and NUP145 occurred in regions depleted for H3K4me3 (Figure 3B) but enriched for H3K4me2 and H3K4me1 (Figure S4), which supports these modifications recruiting Sas3 in vivo. Set2-dependent binding of Sas3 occurred at regions containing H3K36me3 (Figure 3C), consistent with this PTM recruiting Sas3 in vivo. Additionally, from the single to double mutants, Sas3 displayed a stepwise reduction in binding, consistent with H3K36me3 and H3K4me1/2/3 recruiting Sas3 to chromatin in an independent and additive manner.
Figure 3.
H3K4 and H3K36 methylation are necessary for Sas3 binding to active genes through Pdp3 and Yng1, respectively. (A and D) Sas3 ChIP-qPCR in indicated strains at LOS1, SEC15, NUP145, RPS28A, and PUT4. Primer positions on genes are indicated in schematic. (B) H3K4me3 and (C) H3K36me3 ChIP-qPCR in wild-type cells. Values represent the mean of at least three independent replicates. Error bars represent the SEM.
To test if Sas3 recruitment to chromatin was dependent on the NuA3 domains reported to bind histone methylation, we performed ChIP-qPCR for Sas3 in yng1ΔPHD, pdp3Δ, and yng1ΔPHD pdp3Δ strains (Figure 3D). The pdp3Δ mutant had a similar reduction in Sas3 binding as the set2Δ mutant at all but RPS28A. However, disrupting H3K36 methylation at this locus caused a reduction in H3K4me3 (Figure S5), so the greater loss of Sas3 binding in the set2Δ compared to the pdp3Δ mutant was likely an indirect effect. Thus we demonstrate for the first time that Pdp3 is necessary for NuA3 recruitment to chromatin in vivo. The yng1ΔPHD mutation caused a reduction in Sas3 recruitment at 5′, mid-, and 3′ regions, consistent with Yng1 targeting NuA3 to H3K4me3, H3K4me2, and H3K4me1. The yng1ΔPHD pdp3Δ double mutant resulted in Sas3 recruitment comparable to the untagged control at all loci tested. Altogether, our ChIP-qPCR results support the hypothesis that Sas3 binds to actively transcribed chromatin due to binding of H3K4me1/2/3 and H3K36me3 by the Yng1 PHD finger and the Pdp3 PWWP domain, respectively.
Sas3 occupancy does not dictate histone H3K23 acetylation
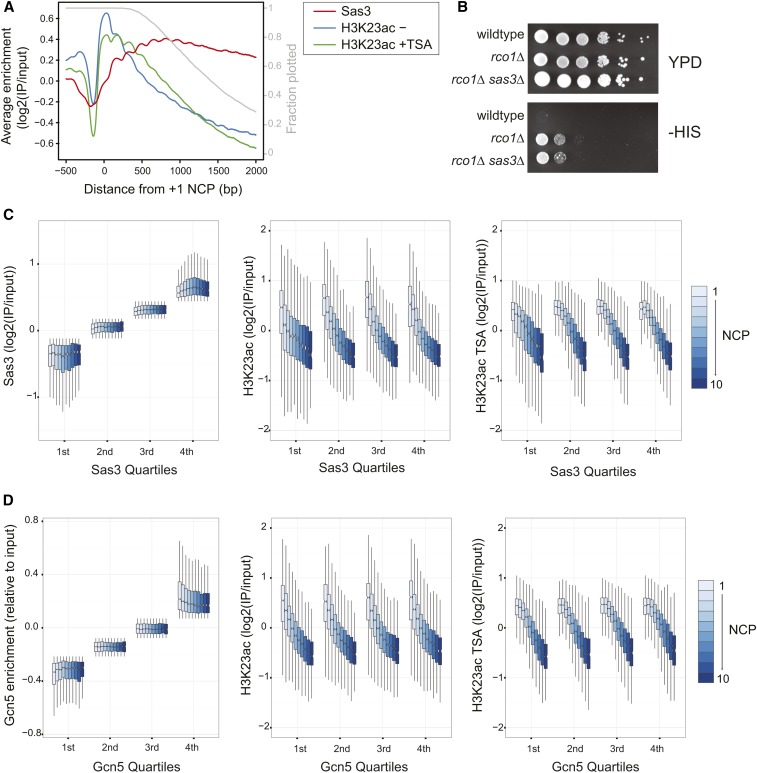
Histone H3K14 and H3K23 acetylation localize to the 5′ ends of genes, which is inconsistent with our demonstrated occupancy of Sas3 (Figure S6) (Weiner et al. 2015). One explanation for this discrepancy is the presence of the histone deacetylase complex (HDAC), Rpd3S, which deacetylates nucleosomes in the body of transcribed genes (Carrozza et al. 2005; Joshi and Struhl 2005; Keogh et al. 2005). To test whether HDAC inhibition could reveal acetylation in the bodies of genes, we performed ChIP-seq analysis for H3K23ac in cells treated with the HDAC inhibitor, TSA (Figure 4A). Following TSA treatment, H3K23ac spread further downstream of the TSS, but this PTM was still largely restricted to the 5′ ends of genes; contrary to NuA3 occupancy, which was found across the bodies of genes. Further, deletion of SAS3 failed to suppress cryptic transcription initiation of a reporter construct in a strain lacking the TSA-sensitive HDAC, RPD3S (Figure 4B) (Cheung et al. 2008), again suggesting that Sas3 was not responsible for acetylation at downstream targets.
Figure 4.
Sas3 occupancy does not dictate histone H3K23 acetylation. (A) The average enrichment relative to 4264 +1 nucleosomes for Sas3 and H3K23ac before and after 15 min incubation with 25 μM TSA. Cell cycle-regulated genes were excluded from this plot. Each gene is only included in the average calculation until its polyadenylation signal. The fractions of genes still contributing to the average profile are represented by the gray line. (B) Serial dilutions (10-fold ) of the indicated strains containing the kanMX-GAL1pr-flo8-HIS3 reporter were plated on rich media (YPD) and complete synthetic media lacking histidine, and incubated at 30° for 4 days. (C and D) HAT and H3K23ac before and after TSA enrichments by nucleosome position and by (C) Sas3 or (D) Gcn5 (Xue-Franzén et al. 2013) quartiles, represented as boxplots. Nucleosomes from cell cycle-regulated genes were excluded, leaving 33,942 nucleosomes from +1 to +10 positions relative to the TSS. Outliers were not plotted.
A disconnect between Sas3 occupancy and H3K23ac levels is surprising as it is generally thought that histone acetylation is regulated through control of HAT targeting. To further confirm these observations, we assessed the relationship between Sas3 and H3K23ac at the +1 to +10 nucleosomes, and while we did observe a modest association between Sas3 occupancy and H3K23ac (Figure S7A), the predominant determinant of H3K23ac was the nucleosome position relative to the TSS (Figure 4C). H3K23ac decreased into the gene body, and this was largely independent of Sas3 occupancy and was seen with or without TSA treatment. The H3 HAT, Gcn5, with Sas3, is necessary for global H3 acetylation (Howe et al. 2001) and so we asked if Gcn5 occupancy (Xue-Franzén et al. 2013) could explain the disparity between Sas3 occupancy and H3K23ac. Similar to Sas3, Gcn5 displayed a subtle association with H3K23ac (Figure S7B), but again this was a modest effect compared to gene position (Figure 4D). We observed similar effects when selecting for nucleosomes enriched for one or both of Sas3 and Gcn5 (Figure S8). Thus the lack of association of Sas3 with H3K23ac cannot be explained by Gcn5 occupancy, and the similar lack of association of Gcn5 with H3K23ac suggests that regulating HAT activity postrecruitment may be a general phenomenon. Collectively these results indicate that while histone methylation promotes the association of Sas3 with chromatin in gene bodies, this does not necessarily result in histone acetylation.
Discussion
In this study we investigated the relative contributions of histone PTMs in targeting NuA3 to chromatin. Using both genome-wide and locus-specific approaches we showed that H3K36me3 and H3K4me1/2/3 both independently and additively promoted the association of Sas3 with chromatin. We provide the first in vivo evidence for Pdp3 recruiting NuA3 to H3K36 trimethylated chromatin, and for Yng1 recruiting NuA3 to H3K4me2 and H3K4me1. The additive effects of H3K36me3 and H3K4me1/2/3 resulted in NuA3 being primarily recruited to the +5 to +7 nucleosomes, ∼700 bp into the gene. This is close to but slightly further into the gene body than where the mammalian homolog MOZ/MORF is predominantly found, ∼400 bp downstream of the TSS (Lalonde et al. 2013). This 5′ shift in MOZ/MORF localization could be due to the mammalian complex’s reduced affinity for H3K4me1 (Champagne et al. 2008) (Table S1) and H3K36me3 (Vezzoli et al. 2010; Wu et al. 2011) (Table S1), resulting in a much greater dependence on H3K4me2/3 for recruitment to chromatin. The differing methyl-histone-binding properties of NuA3 and MOZ/MORF coincide with differing genome-wide localization of H3K4me1 and H3K36me3. With longer mammalian genes, H3K36me3 can stretch >20 kb from the TSS, while H3K4me1 is associated with enhancers (Barski et al. 2007). Thus the reduction in binding affinity for H3K36me3 and H3K4me1 has maintained MOZ/MORF targeting close to, but downstream of, the TSS, which suggests a conserved function in this genomic region.
Unlike for histone methylation, we found no evidence to support a role for histone acetylation or crotonylation in NuA3 recruitment. While we did not test the effect of loss of acetylation or Taf14 on NuA3 binding, the complete loss of Sas3 recruitment in the absence of H3K4 and H3K36 methylation demonstrated that the YEATS domain was not sufficient for NuA3 recruitment. While it is possible that this domain has no function in NuA3, the retention of acetyl-binding domains in mammalian MOZ/MORF argues for a functional role. It is possible that binding to acetyl or crotonyl lysines of the histone tail may regulate NuA3’s activity, similarly to the role of histone methylation in stimulating the activity of the Rpd3S deacetylase complex (Govind et al. 2010). Alternatively, regulation could occur through binding to nonhistone substrates. Indeed, proteomic studies show that Nto1 and Taf14 both contain acetylated lysines (Henriksen et al. 2012), and so it is possible that the YEATS domain is binding to a modified lysine in the NuA3 complex. Such a role is seen for the Rsc4 bromodomain, which binds to an acetylated lysine in the complex to regulate its function (VanDemark et al. 2007; Choi et al. 2008).
Our results showed that Sas3 was localized across the body of transcribed genes, which is inconsistent with the predominantly TSS-proximal patterns of H3K14ac and K23ac (Weiner et al. 2015). This is unsurprising, however, as other studies have shown that HAT occupancy is a poor predictor of histone acetylation (Xue-Franzén et al. 2010; Rossetto et al. 2014). Instead, these results suggest that there is a level of regulation of histone acetylation that is independent of HAT recruitment. Molecular simulation studies predict histone tails to be tightly intertwined with nucleosomal DNA (Li and Kono 2016; Shaytan et al. 2016), and thus disruption of these interactions may be required for acetylation by available HATs. Interestingly, although RNAPII is also found across gene bodies, NET-seq and photoactivatable ribonucleoside-enhanced cross-linking and IP experiments show that RNAPII struggles to transcribe through the 5′ ends of genes (Churchman and Weissman 2011; Schaughency et al. 2014), where the majority of histone acetylation is found. Thus an attractive hypothesis is that histones are acetylated by available HATs in response to DNA unwrapping during slow RNAPII passage. Although other molecular mechanisms could explain our observations, our data underscores the fact that the presence of a histone PTM-binding domain within a chromatin-modifying complex does not ensure that the associated enzymatic activity will function on all nucleosomes with the requisite PTM.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.199422/-/DC1.
Acknowledgments
We acknowledge Sean Taverna for sharing chromatin immunoprecipitation with DNA microarray data. This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council (NSERC) and an operating grant from the Canadian Institutes of Health Research awarded to L.J.H. B.J.E.M. is a recipient of an NSERC Canada Graduate Scholarships award, while V.E.M. was supported by a University of British Columbia Four Year Fellowship.
Footnotes
Communicating editor: M. Hampsey
Literature Cited
- Ali M., Yan K., Lalonde M.-E., Degerny C., Rothbart S. B., et al. , 2012. Tandem PHD fingers of MORF/MOZ acetyltransferases display selectivity for acetylated histone H3 and are required for the association with chromatin. J. Mol. Biol. 424: 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews F. H., Shinsky S. A., Shanle E. K., Bridgers J. B., Gest A., et al. , 2016. The Taf14 YEATS domain is a reader of histone crotonylation. Nat. Chem. Biol. 12: 396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.-Y., Schones D. E., et al. , 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. [DOI] [PubMed] [Google Scholar]
- Bonnet J., Wang C.-Y., Baptista T., Vincent S. D., Hsiao W.-C., et al. , 2014. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 28: 1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogaard K., Xi L., Wang J.-P., Widom J., 2012. A map of nucleosome positions in yeast at base-pair resolution. Nature 486: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., et al. , 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592. [DOI] [PubMed] [Google Scholar]
- Champagne, K. S., N. Saksouk, P. V. Peña, K. Johnson, M. Ullah et al., 2008 The crystal structure of the ING5 PHD finger in complex with an H3K4me3 histone peptide. Proteins 72: 1371–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, V., G. Chua, N. N. Batada, C. R. Landry, S. W. Michnick et al., 2008 Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6: e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. K., D. E. Grimes, K. M. Rowe, and L. J. Howe, 2008 Acetylation of Rsc4p by Gcn5p is essential in the absence of histone H3 acetylation. Mol. Cell. Biol. 28: 6967–6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman, L. S., and J. S. Weissman, 2011 Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon, Y., C. Cayrou, M. Ullah, A.J. Landry, V. Côté et al., 2006 ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 21: 51–64. [DOI] [PubMed] [Google Scholar]
- Eser P., Demel C., Maier K. C., Schwalb B., Pirkl N., et al. , 2014. Periodic mRNA synthesis and degradation co-operate during cell cycle gene expression. Mol. Syst. Biol. 10: 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M., Wang X., Prabhu V., Berrozpe G., Narayan S., et al. , 2010. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141: 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert T. M., McDaniel S. L., Byrum S. D., Cades J. A., Dancy B. C. R., et al. , 2014. A PWWP domain-containing protein targets the NuA3 acetyltransferase complex via histone H3 lysine 36 trimethylation to coordinate transcriptional elongation at coding regions. Mol. Cell. Proteomics 13: 2883–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind C. K., Qiu H., Ginsburg D. S., Ruan C., Hofmeyer K., et al. , 2010. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell 39: 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen P., Wagner S. A., Weinert B. T., Sharma S., Bacinskaja G., et al. , 2012. Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol. Cell. Proteomics 11: 1510–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe L., Auston D., Grant P., John S., Cook R. G., et al. , 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 15: 3144–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe L., Kusch T., Muster N., Chaterji R., Yates J. R., et al. , 2002. Yng1p modulates the activity of Sas3p as a component of the yeast NuA3 histone acetyltransferase complex. Mol. Cell. Biol. 22: 5047–5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S., Howe L., Tafrov S. T., Grant P. A., Sternglanz R., et al. , 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 14: 1196–1208. [PMC free article] [PubMed] [Google Scholar]
- Joshi A. A., Struhl K., 2005. Eaf3 chromodomain interaction with methylated H3–K36 links histone deacetylation to Pol II elongation. Mol. Cell 20: 971–978. [DOI] [PubMed] [Google Scholar]
- Keogh M.-C., Kurdistani S. K., Morris S. A., Ahn S. H., Podolny V., et al. , 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605. [DOI] [PubMed] [Google Scholar]
- Koerber R. T., Rhee H. S., Jiang C., Pugh B. F., 2009. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol. Cell 35: 889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov V. V., 2000. Rapid and reliable protein extraction from yeast. Yeast 16: 857–860. [DOI] [PubMed] [Google Scholar]
- Lalonde M.-E., Avvakumov N., Glass K. C., Joncas F.-H., Saksouk N., et al. , 2013. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 27: 2009–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue K., Daujat S., Crump J. G., Plaster N., Roehl H. H., et al. , 2008. The multidomain protein Brpf1 binds histones and is required for Hox gene expression and segmental identity. Development 135: 1935–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Kono H., 2016. Distinct roles of histone H3 and H2A tails in nucleosome stability. Sci. Rep. 6: 31437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. L., Kaplan T., Kim M., Buratowski S., Schreiber S. L., et al. , 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3: e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Qin S., Zhang J., Ji P., Shi Y., et al. , 2012. Solution structure of an atypical PHD finger in BRPF2 and its interaction with DNA. J. Struct. Biol. 180: 165–173. [DOI] [PubMed] [Google Scholar]
- Maltby V. E., Martin B. J. E., Brind’Amour J., Chruscicki A. T., McBurney K. L., et al. , 2012. Histone H3K4 demethylation is negatively regulated by histone H3 acetylation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 109: 18505–18510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. G. E., Baetz K., Shi X., Walter K. L., MacDonald V. E., et al. , 2006. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol. Cell. Biol. 26: 7871–7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D., Morris A. R., Battenhouse A., Iyer V. R., 2014. Simultaneous mapping of transcript ends at single-nucleotide resolution and identification of widespread promoter-associated non-coding RNA governed by TATA elements. Nucleic Acids Res. 42: 3736–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok D. K., Harbison C. T., Levine S., Cole M., Hannett N. M., et al. , 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Liu L., Zhao C., Han C., Li F., et al. , 2012. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes Dev. 26: 1376–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S., Zentner G. E., Henikoff S., 2015. Asymmetric nucleosomes flank promoters in the budding yeast genome. Genome Res. 25: 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M., 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273: 5858–5868. [DOI] [PubMed] [Google Scholar]
- Rossetto D., Cramet M., Wang A. Y., Steunou A.L., Lacoste N., et al. , 2014. Eaf5/7/3 form a functionally independent NuA4 submodule linked to RNA polymerase II-coupled nucleosome recycling. EMBO J. 33: 1397–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari B. R., Tang Z., Huang H., Yong-Gonzalez V., Molina H., et al. , 2015. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 58: 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeh R., Launer-Wachs R., Wandel H., Rahat A., Friedman N., 2016. Elucidating combinatorial chromatin states at single-nucleosome resolution. Mol. Cell 63: 1080–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaughency P., Merran J., Corden J. L., 2014. Genome-wide mapping of yeast RNA polymerase II termination. PLoS Genet. 10: e1004632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze J. M., Hentrich T., Nakanishi S., Gupta A., Emberly E., et al. , 2011. Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev. 25: 2242–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanle E. K., Andrews F. H., Meriesh H., McDaniel S. L., Dronamraju R., et al. , 2015. Association of Taf14 with acetylated histone H3 directs gene transcription and the DNA damage response. Genes Dev. 29: 1795–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaytan A. K., Armeev G. A., Goncearenco A., Zhurkin V. B., Landsman D., et al. , 2016. Coupling between histone conformations and DNA geometry in nucleosomes on a microsecond timescale: atomistic insights into nucleosome functions. J. Mol. Biol. 428: 221–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Kachirskaia I., Walter K. L., Kuo J.-H. A., Lake A., et al. , 2007. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 282: 2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B. D., Allis C. D., 2000. The language of covalent histone modifications. Nature 403: 41–45. [DOI] [PubMed] [Google Scholar]
- Taverna S. D., Ilin S., Rogers R. S., Tanny J. C., Lavender H., et al. , 2006. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell 24: 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P., Santos-Rosa H., Robson S. C., Sylvestersen K. B., Nelson C. J., et al. , 2014. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 505: 564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDemark A. P., Kasten M. M., Ferris E., Heroux A., Hill C. P., et al. , 2007. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol. Cell 27: 817–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzoli A., Bonadies N., Allen M. D., Freund S. M. V., Santiveri C. M., et al. , 2010. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat. Struct. Mol. Biol. 17: 617–619. [DOI] [PubMed] [Google Scholar]
- Weiner A., Hsieh T.-H. S., Appleboim A., Chen H. V., Rahat A., et al. , 2015. High-resolution chromatin dynamics during a yeast stress response. Mol. Cell 58: 371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Zeng H., Lam R., Tempel W., Amaya M. F., et al. , 2011. Structural and histone binding ability characterizations of human PWWP domains. PLoS One 6: e18919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue-Franzén Y., Johnsson A., Brodin D., Henriksson J., Bürglin T. R., et al. , 2010. Genome-wide characterisation of the Gcn5 histone acetyltransferase in budding yeast during stress adaptation reveals evolutionarily conserved and diverged roles. BMC Genomics 11: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue-Franzén Y., Henriksson J., Bürglin T. R., Wright A. P. H., 2013. Distinct roles of the Gcn5 histone acetyltransferase revealed during transient stress-induced reprogramming of the genome. BMC Genomics 14: 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K., Vinayachandran V., Batta K., Koerber R. T., Pugh B. F., 2012. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell 149: 1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M., Wu J., Workman J. L., Li B., 2011. Readers of histone modifications. Cell Res. 21: 564–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChIP-seq data generated for this study have been deposited in the GEO database, GEO accession: GSE93059 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93059). See Table S2 for the list of strains and Table S3 for the list of primers used in this study.