Abstract
Long interspersed element 1 (L1) is the only currently active autonomous retroelement in the human genome. Along with the parasitic SVA and short interspersed element Alu, L1 is the source of DNA damage induced by retrotransposition: a copy-and-paste process that has the potential to disrupt gene function and cause human disease. The retrotransposition process is dependent upon the ORF2 protein (ORF2p). However, it is unknown whether most of the protein is important for retrotransposition. In particular, other than the Cys motif, the C terminus of the protein has not been intensely examined in the context of retrotransposition. Using evolutionary analysis and the Alu retrotransposition assay, we sought to identify additional amino acids in the C terminus important for retrotransposition. Here, we demonstrate that Gal4-tagged and untagged C-terminally truncated ORF2p fragments possess residual potential to drive Alu retrotransposition. Using sight-directed mutagenesis we identify that while the Y1180 amino acid is important for ORF2p- and L1-driven Alu retrotransposition, a mutation at this position improves L1 retrotransposition. Even though the mechanism of the contribution of Y1180 to Alu and L1 mobilization remains unknown, experimental evidence rules out its direct involvement in the ability of the ORF2p reverse transcriptase to generate complementary DNA. Additionally, our data support that ORF2p amino acids 1180 and 1250–1262 may be involved in the reported ORF1p-mediated increase in ORF2p-driven Alu retrotransposition.
Keywords: LINE-1, ORF2p, retrotransposition, Alu, retroelement
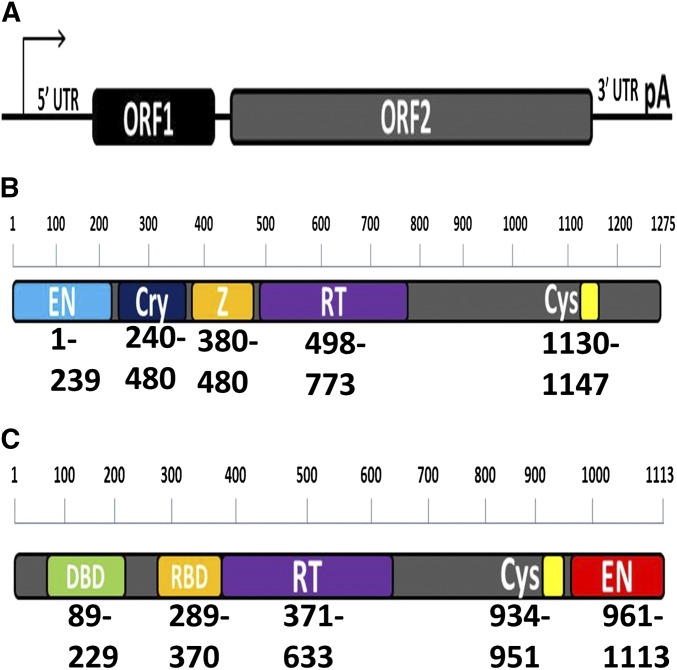
LONG interspersed element 1 (LINE-1 or L1) along with the short interspersed element (SINE) Alu represent the major, high-copy-number, currently active retrotransposons in the human genome. The less-abundant SVA is also active within the human genome (reviewed in Hancks and Kazazian 2016). L1 consists of a 5′ untranslated region (UTR) with a Pol II promoter, two open reading frames (ORFs), a 3′ UTR, and a poly(A) signal (Figure 1A). ORF1 codes for the ORF1 protein (ORF1p), which is the main structural protein of the L1 ribonucleoprotein particle (RNP), a necessary intermediate in the L1 replication cycle (Leibold et al. 1990; Kolosha and Martin 1997; Kolosha and Martin 2003; Martin et al. 2005; Alisch et al. 2006; Martin 2010; Callahan et al. 2012). ORF2 codes for the ORF2 protein (ORF2p), which contains the enzymatic machinery necessary for retrotransposition of both L1 and its parasite Alu (Mathias et al. 1991; Feng et al. 1996). While ORF1p is absolutely necessary for L1 retrotransposition (Moran et al. 1996), Alu is able to mobilize using only the enzymatically active ORF2p (Wallace et al. 2008a).
Figure 1.
Reported functional domains/regions in the L1 ORF2p and R2 ORFp. (A) Schematic representation of L1. L1 is comprised of a 5′ UTR containing internal Pol II promoter, ORF1, ORF2, a 3′ UTR, and a Poly(A) signal. ORF1 codes for the structural ORF1p, while ORF2 codes for the enzymatically active ORF2p. (B) Human ORF2p schematic. The human ORF2p has five functional regions, two of which are known to have enzymatic activity. The enzymatically active domains of the ORF2p are the EN domain (light blue) and RT domain (purple). The ORF2p also contains a Cysteine-rich domain (Cys, yellow) and Z domain (Z, orange). The Z domain contains a PCNA binding domain necessary for both L1 and Alu retrotransposition. The recently described Cryptic region (Cry, dark blue) contains amino acids important for retrotransposition. Amino acid boundaries of the domains are indicated below. Amino acid scale bar is displayed above the schematic. (C) Schematic of annotated functional domains/regions within the R2 ORFp. The R2 ORFp contains five annotated regions. These include two enzymatically active domains: an RT domain (purple) and an EN domain (red). The R2 ORFp also contains a DNA binding domain (DBD, green) and an RNA binding domain (RBD, orange), which has sequence homology to the ORF2p Z domain. The R2 ORFp also has a Cystein-rich motif (Cys, yellow), similar to the Cys domain of the L1 ORF2p.
The ORF2p contains five annotated regions. At the N terminus of the molecule is an APE-like endonuclease (EN) (Feng et al. 1996), which preferentially nicks AT-rich DNA sequences (Figure 1B). This is a necessary initiation step in both the L1 and Alu retrotransposition cycle phase known as target-primed reverse transcription (TPRT) (Luan et al. 1993). C-terminal to the EN domain is the Cryptic region, a recently characterized region of the molecule that was previously assumed to be linker sequence (Figure 1B) (Christian et al. 2016a,b). This region contains a conserved WD pair that is vital to the complementary DNA (cDNA) synthesis step of TPRT (Christian et al. 2016a). The Cry region may be involved in modulating the EN function (Christian et al. 2016b). It also contains a putative proliferating cell nuclear antigen (PCNA)-binding domain important for retrotransposition (Christian et al. 2016a). Following the Cryptic region is the Z domain, which although first identified through homology to regions in related proteins has now been shown to contain a PCNA-binding domain vital to retrotransposition of both L1 (Taylor et al. 2013) and Alu (Christian et al. 2016a) (Figure 1B). The reverse transcriptase (RT) domain (Mathias et al. 1991; Dombroski et al. 1994) is able to copy the L1 or Alu RNA into cDNA for integration into the new genomic location during TPRT (Christensen and Eickbush 2005) (Figure 1B). ORF2p also contains a C-terminal cysteine-rich domain (Cys), which has three conserved C residues and one conserved H residue (Figure 1B). These have been shown to be important for L1 retrotransposition (Moran et al. 1996). It was thought that the ORF2p molecule could only function as a full-length, contiguous molecule. Our recently published results have demonstrated that some ORF2p fragments can reconstitute Alu retrotransposition (Kines et al. 2014; Christian et al. 2016a). Here, we report additional data supporting that C-terminally truncated ORF2p’s have residual ability to drive Alu retrotransposition.
It is important to note that most of our knowledge of TPRT comes from in vitro studies of the related R2 retroelement protein (Xiong and Eickbush 1988; Luan et al. 1993; Yang et al. 1999; Christensen and Eickbush 2005; Christensen et al. 2005, 2006; Jamburuthugoda and Eickbush 2011, 2014), though there is some data concerning the TPRT process from the L1 ORF2p (Feng et al. 1996). The R2 protein is organizationally different from the L1 ORF2p. From the N to the C terminus, this molecule contains a DNA-binding domain that confers target site specificity (Xiong and Eickbush 1988; Luan et al. 1993), a Z domain equivalent that has RNA binding capabilities (Jamburuthugoda and Eickbush 2014), an RT domain (Xiong and Eickbush 1990), and a restriction EN-like EN (Figure 1C) (Burke et al. 1999). Even though the R2 and L1 molecules are structured differently, much of the biochemistry and mechanics of TPRT are assumed to be similar for both molecules. This assumption is supported by the unique hallmarks of TPRT seen in both retroelements’ replication cycles, as well as their phylogenetic proximity (Xiong and Eickbush 1990). Much more is known about the specific steps in R2 TPRT than L1 and Alu TPRT.
While there have been recent inroads into understanding the function of ORF2p amino acid sequence outside of the enzymatic domains (Taylor et al. 2013; Christian et al. 2016a), the contribution of much of the molecule to retrotransposition remains poorly understood. Specifically, the role of the area of the protein C-terminal to the RT domain (outside of the C and H residues in the Cys domain) in retrotransposition is in particular under studied. The paucity of data concerning this area of the protein is demonstrated by the fact that there have been only three articles that dealt with this portion of the protein (Moran et al. 1996; Wagstaff et al. 2011; Piskareva et al. 2013).
We have previously reported that truncated versions of the ORF2p or ORF2p with premature stop codons could retain some ability to elicit the DNA damage response and/or facilitate Alu retrotransposition (Kines et al. 2014; Christian et al. 2016a). While there has been some in vitro biochemical data implicating the amino acid sequence C-terminal to the RT domain in nucleic acid binding (Piskareva et al. 2013), its function in the retrotransposition process in the mammalian cellular environment remains unknown. We sought to investigate whether the unannotated portion of the C-terminal ORF2p sequence was important to retrotransposition, and if there were specific amino acids in this sequence important to retrotransposition. We approached the interrogation of the C-terminal end of the ORF2p using two methodologies. First, we generated truncated ORF2p expression fragments to establish en bloc areas of the ORF2p C terminus important to retrotransposition. In parallel, we used the evolutionarily related R2 C terminus as an anchor point for three relatively closely related ORF2p molecule C-terminal sequences (human, mouse, and rat) to identify potential individual amino acids that could be important to retrotransposition. We reasoned that these two approaches in parallel could aid in advancing our understanding of this under-studied area of the ORF2p. To investigate the importance of the C-terminal ORF2p amino acid sequence to retrotransposition, we used the Alu retrotransposition reporter assay in conjunction with truncated ORF2, and ORF2 with mutations in conserved areas of the extreme C terminus of the molecule. We chose to assess Alu mobilization because it relies on the function of the ORF2p and does not require ORF1p, simplifying the experimental design and interpretation of results. Our findings demonstrate that the unannotated C-terminal portion of the ORF2p is important to Alu retrotransposition, with the exception of the last 13 amino acids. Additionally, we identified a Y residue (Y1180) that is critical to Alu retrotransposition driven by either the ORF2p alone or the full-length L1. In contrast to Alu retrotransposition, a Y1180A mutation improved L1 mobilization in HeLa cells. The data presented here demonstrate that the Y1180 residue is not required for cDNA synthesis by the ORF2p molecule RT, but may be involved via an unknown mechanism in the coordinated function of ORF1p and ORF2p during retrotransposition.
Materials and Methods
Naming conventions
ORF2 fragments are named as described in Christian et al. (2016a). For C-terminally truncated fragments, the previously reported domains are used as the body of the name, followed by the number corresponding to the terminal amino acid as it would be in the full-length ORF2p, with the truncated ORF2p sequence denoted by a ∆. For example, the ORF2p fragment that contains the EN and RT domains that ends at ORF2p amino acid 773 is written as ENRT773∆. By convention, Cryptic and Z domains are not included in the EN- and RT-containing fragment names.
Cloning
Plasmids containing codon-optimized L1 sequence (L1PA1 Chang) reported in Wallace et al. (2008b) were used as templates to generate truncated ORF2 PCR fragments for subcloning into pcDNA 3.1/Hygro+ (Life Technologies) and pBind (Promega, Madison, WI). To generate untagged ORF2p fragment expression constructs, an NheI restriction site, Kozac-ATG, and TGA-HindIII restriction site were added 5′ and 3′ of the ORF2 DNA sequence of interest. PCR products were digested with NheI and HindIII and cloned into pcDNA3.1/Hygro+ (Life Technologies). To generate Gal4-tagged ORF2p fragment expression constructs, PCR primers were designed using the Flexi Vector Primer Design Tool (Promega). These PCR primers added a 5′ SgfI restriction site and a 3′ PmeI restriction site containing a valine (V) codon (GTT) and a stop codon (TAA) to the ORF2 sequence of interest. PCR products were digested with SgfI/PmeI blend (Promega; catalogue number R1852) and cloned into the pBind vector (Promega Checkmate Mammalian Two Hybrid System; catalog number C934A). Codon optimized C-terminal ORF2 sequences with mutations in the C-terminal domain were commercially synthesized (GenScript), PCR amplified, and subcloned into pcDNA 3.1/Hygro+ (Life Technologies) containing previously subcloned, codon-optimized, N-terminal ORF2 sequence; with the end result being ORF2p expression constructs with mutations in the C-terminal domain. To generate Gal4-tagged ORF2 Y1180A and ORF2 Y1189A expression constructs, the corresponding ORF2 sequences were subcloned into pBind (Promega) as described above for ORF2 C-terminal truncations.
Y1180A L1 and L1Neo mutants were generated using site-directed mutagenesis using the previously reported, codon-optimized L1 sequence (Wallace et al. 2008b) as previously described (Christian et al. 2016a).
Alu retrotransposition driven by truncated ORF2, C-terminally mutated ORF2, or full-length L1 and L1 retrotransposition
HeLa cells were maintained in minimum essential medium supplemented with 1% sodium pyruvate, L-glutamate, nonessential amino acid solution, and 10% fetal bovine serum as previously described (Belancio et al. 2006). A total of 500,000 cells were seeded 16–18 hr prior to transfection. Then, 0.8 μg of indicated ORF2 expression plasmid was cotransfected with 1 μg of the previously described Alu retrotransposition reporter construct (Dewannieux et al. 2003) using 6 μl PLUS reagent (Life Technologies) in 200 μl Dulbecco’s Modified Eagle Medium (DMEM) with 8 μl Lipofectamine reagent (Life Technologies) in 92 μl DMEM.
Alu retrotransposition driven by the wild-type or indicated mutant ORF2p was assessed by transfecting 500,000 HeLa cells seeded 16–18 hr prior to transfection with 0.4 μg of the AluNeo plasmid and 0.4 μg of either wild-type or specific mutant ORF2 expression plasmids using 4 μl of PLUS reagent and 8 μl of Lipofectamine.
Alu retrotransposition driven by the full-length L1 was assessed by transfecting 500,000 HeLa cells seeded 16–18 hr prior to transfection with 0.4 μg of the AluNeo plasmid and 0.8 μg of either wild-type or Y1180A L1 expression plasmids using 4 μl of PLUS reagent and 8 μl of Lipofectamine.
L1 retrotransposition was assessed by transfecting 500,000 HeLa cells seeded 16–18 hr prior to transfection with 0.2 μg of the wild-type or Y1180A L1Neo expression plasmids using 4 μl of PLUS reagent and 8 μl of Lipofectamine.
For all experiments, cell-culture media was supplemented with 0.45 mg/ml G418 ∼24 hr post-transfection and colonies were stained after 2 weeks of G418 selection with crystal violet solution (0.2% crystal violet, 5% acetic acid, 2.5% isopropanol) and counted with Oxford Optronics ColCount. Statistical significance was assessed using Student’s t-test for paired samples (n = 3), with error bars denoting SD.
Immunoblot analysis: transfection, cell culture, and total protein harvest
ORF2:
A total of 2,000,000 cells were seeded 16–18 hr prior to transfection in T75 flasks. Then, 6 μg of appropriate ORF2 expression construct or appropriate empty vector (control) was transfected with 24 μl Lipofectamine reagent (Life Technologies) and 12 μl PLUS reagent (Life Technologies). Approximately 24 hr post-transfection, cells were washed once with phosphate buffered saline (PBS) and then harvested in 500 μl total lysis buffer (50 mM Tris, 150 mM NaCl, 10 mM EDTA, 0.5% SDS, 0.5% Triton X, pH 7.2) supplemented with 10 μl/ml of Halt Protease Inhibitor Cocktail, Phosphate Inhibitor Cocktail 2, and Phosphate Inhibitor Cocktail 3 (Sigma Chemical, St. Louis, MO). After one round of freeze (−80°)/thaw on ice, cells were sonicated three times with a Microson XL-2000 sonicator (Misonix) (10 sec sonication/10 sec rest on ice), and cell lysates were centrifuged at 4° at 14000 rpm for 15 min. Protein concentrations of cell lysates were determined using Bio-Rad (Hercules, CA) protein assay (Bradford method).
L1:
A total of 7 × 105 HeLa cells were seeded per T25 flask, and 16–18 hr after this they were transfected with 2 μg of a pCEP, L1PA1 wild-type, or Y1180A expression plasmids using 6 μl of PLUS reagent and 12 μl of Lipofectamine. After 3 hr the transfection reaction was replaced with serum containing HeLa media. The cells were harvested ∼24 hr later using total protein extraction protocol (Sokolowski et al. 2013).
Immunoblot analysis: Western blot analysis
ORF2:
Total cell lysate of mass 30 μg was heated at 85° for 5 min in Laemmli buffer without β-mercaptoethanol supplementation. Samples were fractionated on 3–8% Tris-Acetate gels (Life Technologies) in Figure 2B and Figure 5A or 4% Tris-Glycine gel (Life Technologies) in Figure 4A. Samples were then transferred to nitrocellulose membranes using the iBlot system (Life Technologies). Membranes were blocked in PBS-Tween (PBS, 0.1% Tween) with 5% blotting-grade blocker (Bio-Rad) and incubated with primary antibodies overnight at 4°. All Gal4-tagged constructs (Figure 2B and Figure 5A) detected using anti-Gal4 DNA binding domain antibodies (sc-577, 1:1000 dilution; Santa Cruz). Full-length ORF2p was detected using previously reported ORF2p antibodies (Kines et al. 2014, 2016). Membranes were washed three times in PBS-Tween for 5 min following overnight primary antibody incubation. Secondary antibody was applied for 1 hr at 25° (HRP-goat anti-rabbit or anti-mouse, 1:5000 in 3% blotting-grade blocker with PBS-Tween). Western blots were developed using the Immun-Star WesternC Kit (Bio-Rad). Images were captured using a Bio-Rad Gel Doc XR+ imager. GAPDH (sc-25778; Santa Cruz) detection was used as loading control (1:3000 in 3% blotting-grade blocker with PBS-Tween).
Figure 2.
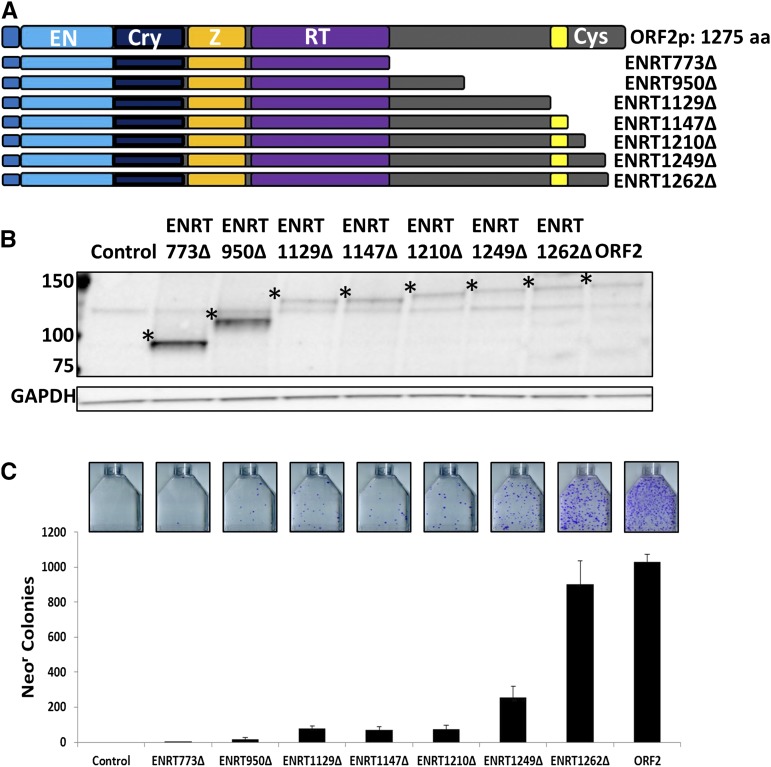
C-terminally truncated Gal4-tagged human ORF2p can drive variable levels of Alu retrotransposition. (A) Schematic of C-terminally truncated ORF2 constructs. Gal4 tag position indicated by blue-gray rectangle. (B) Western blot analysis of ORF2 fragments transiently transfected in HeLa cells. ORF2p fragments were detected using anti-Gal4 antibodies. Protein product of expected size is denoted by a * for each construct. Molecular weight markers are denoted at the left of the image. GAPDH was used as loading control. (C) Alu retrotransposition assay results. Neor colonies correspond to de novo retrotransposition events. Error bars denote SD of n = 3 experiments. Representative flasks for each ORF2 construct are shown above corresponding graph bars.
Figure 5.
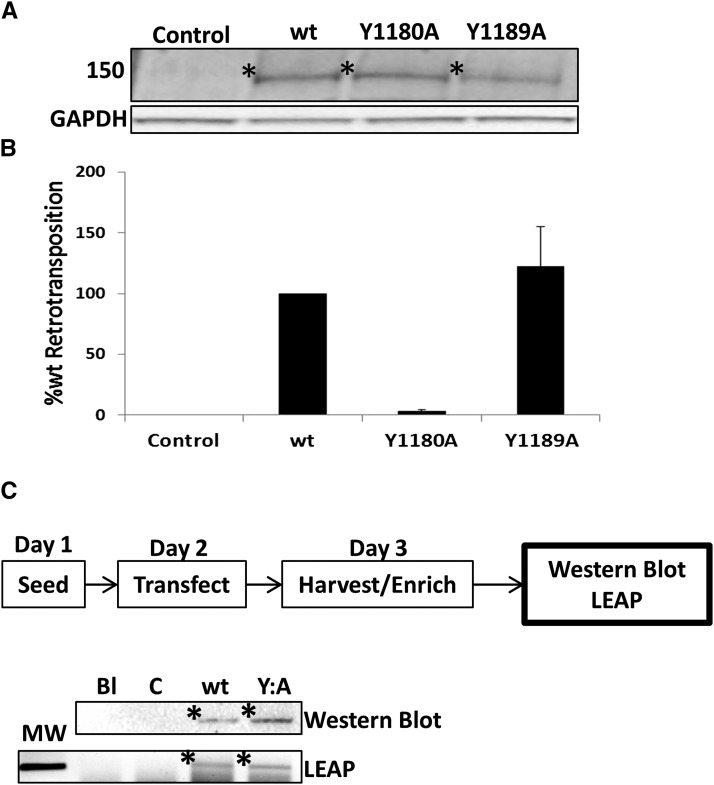
Amino acid Y1180 is not required for the RT-medicated cDNA synthesis by ORF2p. (A) Western blot analysis of the Gal4-tagged ORF2, ORF2 Y1180A, and ORF2 Y1189A transiently transfected in HeLa cells. ORF2p variants detected using anti-Gal4 antibodies. Protein products of expected size denoted by * for each ORF2 construct. Molecular weight markers denoted on the left. GAPDH used as loading control. (B) Alu retrotransposition assay results. Colony counts normalized to wild-type Gal4-tagged ORF2p-driven Alu retrotransposition levels. Neor colonies correspond to de novo retrotransposition events. Error bars denote SD determined using results from three independent experiments. Representative flasks are shown above corresponding graph bars. (C) LEAP analysis of wild-type and Y1180A Gal-4 tagged ORF2p. Displayed at the top is a schematic of the LEAP assay. Western blot analysis using anti-Gal4 antibodies of LEAP preparations shows that protein levels between the two ORF2p variants (wt and Y:A, *) are equivalent. Blank (Bl) is loaded with loading buffer only and control (C) is LEAP prepared on cells transfected with empty vector. LEAP analysis shows the presence of the PCR band of expected size in the ORF2p LEAP sample containing functional Y1180 (wt) and mutated Y1180 (Y:A) (*). MW, molecular weight marker; wt, wild type.
Figure 4.
Mutations in the C terminus of the untagged human ORF2p affect its ability to drive Alu retrotransposition. (A) Western blot analysis of steady-state levels of ORF2p containing C-terminal mutations following transient transfection of appropriate constructs into HeLa cells. ORF2p bands denoted by *. Control is cell lysate from HeLa cells transfected with empty vector. (B) Alu retrotransposition assay results, with colony counts normalized to the colony number obtained for Alu driven by wild-type (wt) ORF2p. Neor colonies correspond to de novo retrotransposition events. Error bars denote SD determined for the n = 3 experiments. Representative flasks are shown above corresponding graph bars.
ORF1:
A mass of 20 μg of total protein was combined with 10 μl of 2× Laemmli buffer and 1.6 μl 14.3 M β-mercaptoethanol, boiled for 5 min prior to fractionation on Bis-Tris 4–12% Midi Gels (Invitrogen, Carlsbad, CA) (Figure 6A, top), and transferred onto the nitrocellulose membranes (iBlot2 System; Invitrogen) after fractionation. Membranes containing fractionated protein samples were blocked for 1 hr in PBS-Tween containing 5% milk and incubated with primary anti-ORF1p antibody (TGNSKTQSASPPPK, dilution 1:5000) (Sokolowski et al. 2013) in 3% milk in PBS-Tween overnight at 4°. The detection of ORF1p was carried out using HRP-conjugated secondary antibodies (HRP-donkey anti-rabbit, sc-2317; Santa Cruz) at 1:5000 dilution in 3% milk in PBS-Tween for 1 hr at room temperature. Detection of GAPDH with Santa Cruz sc-25778 antibodies was used as a loading control. The Western blot ORF1p and GAPDH signals were quantitated using Bio-Rad Image Lab Software Version 4.1. The ORF1p signals were normalized to their respective GAPDH signals. The resulting ratio for the L1PA1 wild type was set at 100% to determine ORF1p levels generated by the Y1180 mutant.
Figure 6.
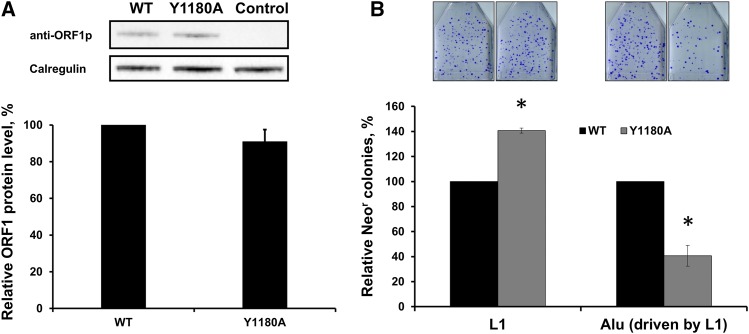
Y1180A mutation in the context of full-length L1. (A) Western blot analysis of the ORF1p expressed from wild-type and Y1180A mutated L1. (B) Relative retrotransposition of wild-type and Y1180A mutant L1 and Alu driven by wild-type (WT) and Y1180A mutant L1. * indicates statistical significance with P < 0.05 as determined by the t-test.
L1 element amplification protocol
Adapted from Kulpa and Moran (2006), Wagstaff et al. (2011), and Christian et al. (2016a). Briefly, HeLa cells were maintained as described above for Western blot analysis. Cells were seeded and transfected as described above with 6 μg of appropriate gal4-tagged ORF2 expression plasmids. Cells were washed as described above and harvested by scraping in 5 ml PBS. The mixture was pelleted at 6000 rpm for 5 min at 4° and supernatant removed. The resulting pellets were lysed in 500 μl of L1 element amplification protocol (LEAP) lysis buffer (1.5 mM KCl, 2.5 mM MgCl2, 5 mM Tris-Cl, 1% deoxycholate, 1% Triton X-100, EDTA, and RNAsin), incubated on ice for 5 min, and briefly centrifuged to remove debris. Supernatant was layered on top of an 8.5% sucrose cushion that was layered on top of a 17% sucrose cushion. Samples were centrifuged at 36,500 rpm for 2 hr at 4°. Supernatant was removed and pellet resuspended in 100 μl deionized water supplemented with Halt Protease Inhibitor and RNAsin. Protein concentration was assessed using the Bradford assay and samples brought to equal protein concentrations using glycerol. RT-PCR reaction was performed using 900 ng of sample as described in Kulpa and Moran (2006), Wagstaff et al. (2011), and Christian et al. (2016a).
Protein alignment
Amino acid sequence alignment was performed using MegAlign software (DNASTAR version 10.0.1). Sequences were aligned using the ClustalW method relative to the human ORF2p sequence. The amino acid sequence group match strength is represented by the histogram in Supplemental Material, Figure S4 (>50% sequences match). Sources of ORF2p and ORF2p-like molecule sequences with the amino acid numbers used for alignment and GenBank accession numbers are as follows: Mus musculus (AAA39398) amino acids 1156–1300, Rattus norvegicus (AAB41224) amino acids 1156–1300, and Bombyx mori (AAB59214) amino acids 934–1114. Aligned against human ORF2p amino acids 1130–1275.
Data availability
The authors state that the information necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Gal4-tagged, C-terminally truncated ORF2p fragments support Alu retrotransposition
We have previously reported that ORF2 constructs with premature stop codons introduced downstream of the RT domain can support residual Alu retrotransposition (Kines et al. 2014). However, it remains unknown whether or not this residual Alu retrotransposition was from true truncated ORF2p fragments, or from full-length ORF2p generated by stop codon readthrough. We hypothesized that C-terminally truncated ORF2p could have the capacity to support limited Alu retrotransposition. To test this hypothesis, we generated ORF2 fragment expression constructs designed to express Gal4-tagged, C-terminally truncated ORF2p fragments (Figure 2A). The Gal4 tag was added because it does not significantly disrupt ORF2p function, but does allow detection of different ORF2p fragments with the same antibody (Christian et al. 2016a,b). Naming conventions for constructs include the core name of the fragment, which is comprised of the enzymatic domains contained in the fragment (EN and RT domains). The second part of the construct designation is derived from the number of the amino acid at which the corresponding protein fragment ends relative to full-length ORF2p, followed by a Δ. Break points were chosen to encompass only the EN and RT (ENRT) core domains (ENRT773Δ), to split the unannotated region of the ORF2p between the RT and Cys domains in half (ENRT950Δ), to contain the entire region between the RT and Cys domains while excluding the Cys domain (ENRT1129Δ), and to include all the annotated domains of the ORF2p (EN, Cryptic, Z, RT, and Cys), while excluding the unannotated extreme C-terminal region (ENRT1147Δ). Additional fragments were generated based on empirical data (ENRT1210Δ, ENRT1249Δ, and ENRT1262Δ).
HeLa cells were transiently transfected with the above-described expression plasmids, and total cellular lysate was subjected to Western blot analysis to determine if C-terminally truncated ORF2p fragments were expressed. All ENRT-containing fragments were detected at expected molecular weights using commercially available Gal4 antibodies (Santa Cruz) (Figure 2B). To test the retrotransposition potential of these ORF2 fragments, we used the previously reported AluNeo retrotransposition assay (Dewannieux et al. 2003). The Alu reporter construct uses an Alu element that contains a 3′-encoded Neomycin resistance (Neor) cassette that can generate Neo resistance only after successful L1-dependent reverse transcription and integration into the genome due to the presence of an artificial intron. HeLa cells were transiently transfected with the Alu reporter construct and an empty plasmid (control) or an indicated ORF2 fragment expression plasmid (Figure 2C). ENRT-containing constructs designed to express ORF2p fragments ending at amino acids 950, 1129, 1147, and 1210 were capable of supporting limited levels of Alu retrotransposition (Figure 2C: ENRT950Δ, ENRT1129Δ, ENRT1147Δ, ENRT1210Δ). Compared to these ENRT-containing ORF2 fragments, ENRT1249Δ was able to support a higher (albeit still significantly reduced) level of Alu retrotransposition. ENRT1262Δ was able to support robust Alu retrotransposition comparable to full-length ORF2p (Figure 2C).
ORF2p-driven Alu retrotransposition was previously reported to be increased by the coexpression of the L1 ORF1p (Wallace et al. 2008a). To test whether Alu retrotransposition driven by the C-terminally truncated ORF2p fragments is similarly affected by the ORF1p, the above-described AluNeo and constructs containing truncated ORF2 sequences were cotransfected with an ORF1 expression plasmid (Sokolowski et al. 2013). Interestingly, ENRT1262Δ was the only ORF2p fragment whose ability to drive Alu retrotransposition was increased in the presence of ORF1p (Figure S1 and File S1) in a manner similar to that of full-length ORF2p (Wallace et al. 2008a).
To rule out the possibility that the Gal4 tag may influence the ability of ENRT-containing fragments to support Alu retrotransposition, we generated ENRT1249Δ and ENRT1262Δ constructs without the Gal4 tag. These constructs were tested in the Alu retrotransposition assay. There was no statistically significant difference in the ability of the untagged ENRT1249Δ fragment to drive Alu retrotransposition when compared to the Gal4-tagged equivalent (Figure S2). The untagged ENRT1262Δ construct was able to drive Alu retrotransposition at a slightly greater efficiency (∼130%) than the Gal4-tagged equivalent (Figure S2). Indeed, no statistically significant difference was observed in the ability of the untagged ENRT1262Δ construct to drive Alu retrotransposition when compared to the full-length ORF2p (Figure S3). These data demonstrate that while the extreme C terminus of the ORF2p (amino acids 1263–1275) may be dispensable for Alu retrotransposition, the C terminus of the ORF2p may contain amino acids (outside of the previously studied Cys motif) important to retrotransposition.
The extreme C-terminal end of ORF2p contains evolutionarily conserved amino acids that share identity with amino acids in the EN domain of the related R2 element
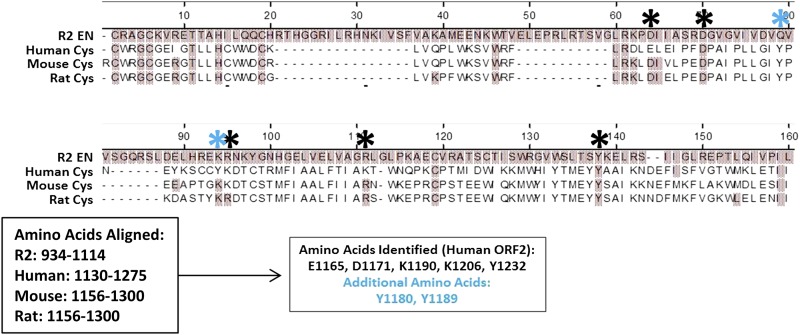
Many regions of the ORF2p have no known homology to other proteins in nature, making dissection of the importance of individual amino acids outside of the previously annotated ORF2p domains challenging. Previously we were able to identify and characterize an essential WD pair in the Cryptic region through a combination of functional genetic interrogation and evolutionary analysis of the protein sequence of ORF2p from taxonomically diverse species (Christian et al. 2016a). Similar direct comparison between the extreme C-terminal sequences of ORF2p is more challenging, as the extreme C terminus of the protein is more variable among many evolutionarily diverse species than the EN, Cryptic, or RT domains. However, the R2 ORFp and the L1 ORF2p share a common ancestor, with the R2-type retroelements having emerged first, and this comparison may be helpful in identifying residues important for L1 and/or Alu mobilization (Xiong and Eickbush 1990; Malik et al. 1999; Eickbush and Jamburuthugoda 2008). One of the primary differences between the R2-type and L1-type retroelements is the placement and type of EN employed. R2 elements have a C-terminal restriction EN-like EN (Yang et al. 1999), while L1 has an N-terminal APE-like EN (Feng et al. 1996). At some point, the APE-like EN was acquired and the C-terminal EN function was lost. However, there are some retroelements that possess both ENs (N- and C-terminal) (Kojima and Fujiwara 2005). Further, the CCHC motif is common to both L1 and R2 elements, supporting its conservation from R2-like retroelements into L1-like retroelements. We limited our analysis to the extreme C-terminal ends of ORF2p from three relatively closely related species (human, mouse, and rat) and aligned them to a distant, phylogenetically related anchor point of the EN domain of the R2 element (Figure 3 and Figure S4). We identified five amino acids that were conserved in the ORF2p (Figure 3, black asterisks: E1165, D1171, K1190, K1206, Y1232) and that could be considered analogous to catalytically essential amino acids in the R2 EN domain (Yang et al. 1999). We also included two additional Y residues (Figure 3, blue asterisks: Y1180, Y1189) to account for some ambiguity in the equivalency of Y1232 (human ORF2p) to the catalytically essential Y in the R2 EN domain.
Figure 3.
Amino Acid sequence alignment of C-termini of human, mouse, and rat L1 ORF2p with R2 ORFp from B. mori. The origin of aligned ORF2p/ORFp is indicated on the left. Amino acids identified as having strong identity to the R2 sequence are denoted by black * and named according to their position in the human ORF2p sequence (E1165, D1171, K1190, K1206, Y1232). Amino acids identified as additional possible equivalents to aligned Y (human Y1232) residue in the R2 ORFp denoted by blue * (Y1180, Y1189).
The extreme C terminus of the human ORF2p contains a Y residue important for retrotransposition, but tolerates mutations in other conserved amino acids
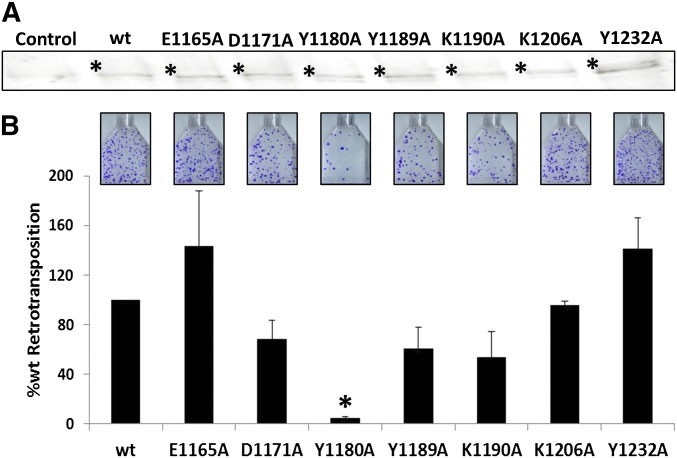
We wanted to test the importance of conserved amino acids identified in the extreme C terminus of the ORF2p to Alu retrotransposition (Figure 3 and Figure S4). To assess the impact of mutation of these amino acids on ORF2p expression, untagged ORF2 constructs with individual amino acids E1165, D1171, Y1180, Y1189 K1190, or Y1232 changed to A were transiently transfected into HeLa cells. Total cell lysate of these cells was subjected to Western blot analysis using ORF2p antibodies specific to amino acids 960–973 (Kines et al. 2014; Christian et al. 2016a; Kines et al. 2016). All constructs expressed their corresponding protein (Figure 4A). We tested the effect of these mutations on the ability of the ORF2p to drive Alu retrotransposition using the AluNeo retrotransposition assay. Only the Y1180A mutation significantly reduced Alu retrotransposition (Figure 4B). Our data demonstrate that while this Y is important for Alu retrotransposition, the ORF2p C terminus can tolerate mutations of other amino acids tested in this study.
Y1180 is important to Alu retrotransposition independent of RT-driven cDNA synthesis
We sought to address whether Y1180 was important to RT function. ORF2 containing the Y1180A or Y1189A mutations were cloned into expression plasmids that introduced a Gal4 N-terminal tag. The Gal4 tag was used in this context to take advantage of our laboratory’s previously optimized assay systems using Gal4-tagged ORF2p (Christian et al. 2016a), as well as to confirm our results using the untagged ORF2p (Figure 4) in an independent set of retrotransposition experiments. The Y1189A was chosen as a control because it is another Y residue that is in close proximity to Y1180, but it did not significantly affect Alu retrotransposition (Figure 4B). HeLa cells were transiently transfected with Gal4-tagged ORF2 constructs, and protein levels were analyzed for all constructs using anti-Gal4 antibodies (Santa Cruz). No difference in protein expression between the wild-type, Y1180A, and Y1189A ORF2p was observed (Figure 5A). To test the effect of the Y mutations in the Gal4-tagged ORF2 context on Alu retrotransposition, the Y1180A and Y1189A ORF2 constructs were transiently transfected into HeLa cells with the Alu retrotransposition reporter construct. The Y1180A mutation significantly reduced Alu retrotransposition supported by the Gal4-tagged ORF2p (Figure 5B) as it did in the untagged ORF2 (Figure 4B). Consistent with the untagged ORF2p results, the Y1189A mutation had no effect on Alu retrotransposition driven by the Gal4-tagged ORF2p (Figure 5B).
To test the effect of the Y1180 and Y1189 mutations on RT function, we used the previously described LEAP (Kulpa and Moran 2006; Wagstaff et al. 2011; Christian et al. 2016a). During LEAP, HeLa cells are transfected with ORF2p expression constructs. Transfected cells are then harvested and subjected to ultracentrifugation to purify cytoplasmid ribonucleoprotein particles (RNPs), consisting of at minimum the ORF2p and its parental messenger RNA (mRNA). These RNPs, which contain the ORF2p, can then be incubated with a specific primer for the 3′ end of the ORF2p mRNA that acts as a substrate for cDNA synthesis by the ORF2p RT. These cDNA products are then subjected to PCR to enable detection of the RT-generated cDNA. The Y1180A mutation had no effect on the ability of the ORF2p to create cDNA from its parent ORF2 mRNA (Figure 5C). These data demonstrate that though the Y1180 is important to retrotransposition, it is likely involved in retrotransposition steps other than cDNA synthesis.
Y1180 is important to Alu retrotransposition driven by full-length L1, but does not reduce L1 mobilization
We next tested the importance of the Y1180A mutation to both Alu retrotransposition driven by full-length L1 and L1 retrotransposition. The Y1180A mutation had no significant impact on the expression of the ORF1p from the full-length L1 relative to wild-type L1 control (Figure 6A). Similar to the Y1180A ORF2p, the Y1180A mutant L1 supported reduced levels of Alu retrotransposition (Figure 6B). The same mutation resulted in a 40% increase in L1 mobilization (Figure 6B). Although, as with Alu retrotransposition driven by the ORF2p, the Y1180A mutation significantly reduced Alu retrotransposition driven by full-length L1 (Figure 6B); the difference in the extent of this reduction (Figure 4, Figure 5, and Figure 6) prompted us to test the effect of ORF1p on the ability of this and other mutant ORF2p’s to drive Alu mobilization. Cotransfection of AluNeo and ORF2-containing plasmids with the ORF1p expression plasmid demonstrated that the reduced ability of the Y1180A mutant ORF2p to support Alu mobilization was improved by approximately five-fold by coexpression with ORF1p (Figure S5).
Discussion
While the enzymatic domains of the ORF2p have been fairly well characterized (Feng et al. 1996; Piskareva et al. 2003; Weichenrieder et al. 2004; Piskareva and Schmatchenko 2006; Monot et al. 2013) and the regions outside of the enzymatic domains have been shown to be important for retrotransposition (Moran et al. 1996; Taylor et al. 2013; Christian et al. 2016a), what most of the molecule does in the context of TPRT and the L1/Alu replication cycles is currently unknown. This study begins to elucidate the importance of the extreme C-terminal end of the ORF2p molecule outside of the Cys domain to retrotransposition, which has previously been shown to be required for retrotransposition (Moran et al. 1996).
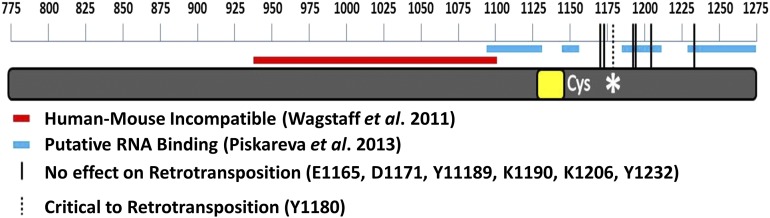
There is some biochemical data concerning the C terminus of the ORF2p indicating that, in general, the ORF2p sequence C-terminal to the RT domain can bind nucleic acids in vitro (Piskareva et al. 2013). Even though the study identified putative nucleic acid binding regions in the C-terminal sequence of the ORF2p (Figure 7), these regions were never shown to be involved in nucleic acid binding nor were they reported to be important for L1 or Alu retrotransposition in mammalian cells. Indeed, amino acids Y1189, K1190, K1206, and Y1232 were all predicted to be important for the putative nucleic acid binding motifs identified in Piskareva et al. (2013) (Figure 7), but individual mutations of these amino acids had no effect on Alu retrotransposition (Figure 4). These results suggest that, if this putative RNA binding region within the C-terminal portion of the ORF2p functions in this capacity in mammalian cells, there might be some degree of redundancy associated with RNA binding.
Figure 7.
Summary schematic of L1 ORF2p C-terminal region (aa 775–1275). Amino acid scale bar is displayed above. Region identified by Wagstaff et al. (2011) as nonmodular between human and mouse ORF2p is shown in red over ORF2p. Putative RNA binding regions identified by Piskareva et al. (2013) are shown in blue. Amino acids assessed in this manuscript whose mutation had no significant effect on Alu retrotransposition are shown by solid black vertical lines at their approximate location. Y1180, which is important for Alu retrotransposition, is denoted by a dashed black line and a *.
Most of the amino acids we identified for investigation in this study were conserved between the human, mouse, and rat ORF2p and the C-terminal EN domain of the related R2 element. However, without three-dimensional structures to superimpose the C-termini of the proteins from these species onto one another, there was some ambiguity as to the equivalence of several Y residues in the L1 ORF2p’s to the Y in the R2 EN domain which is necessary for EN activity. As such, we included other nearby Y residues in our analysis (Figure 3). Our data show that one of these Y residues, Y1180, is important to Alu retrotransposition (Figure 4 and Figure 5). It is possible that this Y is important for nucleic acid interactions in the ORF2p in a manner similar to the role that the putative equivalent Y residue plays in the related R2 molecule (Yang et al. 1999). However, further biochemical studies would be necessary to support or refute this possibility. Recently, it was discovered that phosphorylation is important to ORF1p function in retrotransposition (Cook et al. 2015). Y residues can be phosphorylated to regulate protein activity. Given that our data show that the effect of the Y1180A mutation has no effect on cDNA synthesis (Figure 5C), it is possible that this amino acid or its post-translational modification may be involved in integration steps downstream of cDNA synthesis. Alternatively, Y1180 may be required for coordination of ORF2p enzymatic and/or nucleic acid binding functions. Further studies, either through mass spectrographic analysis or introduction of phosphor-mimics to this amino acid position, will be needed to experimentally confirm these possibilities.
Strikingly, while the Y1180 amino acid appears to be important for Alu retrotransposition, it is not necessary for L1 retrotransposition (Figure 4 and Figure 6). This difference between the behaviors of these two retroelements has been observed before (Roy-Engel 2012). However, specific regions within the L1 proteins responsible for the effect have not been previously observed. Our data suggest that the observed discrepancy in Alu and L1 retrotransposition associated with the presence of the Y1180A mutation in the ORF2p may directly or indirectly involve ORF1p (Figure 6 and Figure S5). Thus, the identification of the Y1180 amino acid within the ORF2p is promising to be useful in investigating the ORF1/ORF2 interplay in retrotransposition and mechanistic differences between the Alu and L1 replication cycles.
While we did discover the importance of the Y1180 residue to Alu retrotransposition, it is worth noting that this region of the ORF2p can tolerate individual mutations at other conserved positions. This is similar to the recently described mutagenic tolerance of the ORF2p EN domain (Kines et al. 2016). However, there are several important differences between the two. The ORF2p EN domain has a catalytic activity, while the C terminus of ORF2p has no described catalytic function (despite the sequence similarity to the catalytically active R2 EN domain). Also, the ORF2p EN domain is fairly well conserved, while the protein sequence of the ORF2p extreme C-terminal region is more variable between species than the protein sequence for the EN domains. The ORF2p may be generally tolerant of mutagenesis across most of the molecule, excluding very essential components necessary for the L1 replication cycle. This characteristic would be shared by the distant relative of the ORF2p: the Pol proteins of retroviruses (Hattori et al. 1986; Xiong and Eickbush 1990; Eickbush and Malik 2002; Eickbush and Jamburuthugoda 2008). Alternatively, as mentioned above, all or some of these amino acids may play redundant roles in the ORF2p function. Therefore, although individual mutations at these positions do not significantly affect protein function in retrotransposition, a combination of mutations at assorted positions may impair ORF2p function.
Previously, it was thought that the ORF2p molecule could only function and have an impact on the host cell as a full-length, contiguous molecule. Our published findings support that some of the truncated ORF2p species may have a biological relevance. Namely, we have shown that EN-containing ORF2p fragments can be cytotoxic (Kines et al. 2014), ORF2p with stop codons introduced to generate EN- and RT-containing ORF2 fragments could drive residual Alu retrotransposition (Kines et al. 2014), and that untagged ORF2p fragments supplied separately from one another could also mobilize Alu with low efficiency (Christian et al. 2016a). Here, we show that the last 13 amino acids of the ORF2p are dispensable for robust Alu retrotransposition, while removal of the terminal 26 amino acids of the ORF2p resulted in a ∼70% reduction in Alu retrotransposition efficiency. Furthermore, we demonstrate that even though ENRT1129Δ lacks 146 C-terminal amino acids it can still support (albeit limited) Alu retrotransposition (Figure 2, Figure S1, and Figure S2). This result supports that the ORF2p sequence between the RT and Cys regions of the protein may have a specific function in retrotransposition. Further refinement of break points of these protein fragments, guided perhaps by evolutionary protein sequence analysis, could in the future define novel functional domains or regions of the C-terminal portion of the L1 ORF2p. Based on our data, one function of amino acids 1250–1262 may be to mediate the ORF1p-associated increase in Alu retrotransposition driven by ORF2p (Figure S1). This observation combined with the involvement of the position 1180 in ORF1-associated increase in Alu mobilization (Figure S5) further support that the C terminus of ORF2p may be physically or functionally involved with ORF1p during retrotransposition.
In summary and conclusion, our data demonstrate that C-terminally truncated ORF2p can drive Alu retrotransposition with various efficiencies (Figure 2 and Figure S1, and Figure S2). The efficiency of this retrotransposition depends on the degree of protein truncation, with the extreme C-terminal end of the protein (amino acids 1263–1275) apparently dispensable for ORF2p-driven Alu retrotransposition (Figure S2). We have also discovered that the Y1180 residue that is required for ORF2p-driven Alu retrotransposition (Figure 4 and Figure 5), is seemingly independent of having any measurable negative effect on cDNA synthesis (Figure 5C), but potentially involves ORF1p (Figure 6 and Figure S5). We also observed that this region of the ORF2p (amino acids 1165–1189) can tolerate individual mutations, despite containing regions essential for retrotransposition (Figure 3). Lastly, we identified a region in the ORF2p C terminus (amino acids 1250–1262) that may be responsible for the ability of ORF1p to increase Alu retrotransposition driven by ORF2p (Figure S1).
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.191403/-/DC1.
Acknowledgments
We would like to thank the members of the Consortium of Mobile Elements at Tulane for critical discussion. Funding for this work was provided by the Life Extension Foundation to V.P.B., a National Institutes of Health grant (P20 GM-103518) to V.P.B., and the Kay Yow Cancer Fund to V.P.B.
Author Contributions: C.M.C., M.S., and V.P.B. conceived and performed the experiments, performed data analysis, and wrote the manuscript. D.d.H. provided technical assistance with the L1 element amplification protocol. D.d.H. and K.J.K. cloned and initially characterized Gal4-tagged ORF2p and EN239Δ constructs.
Footnotes
Communicating editor: L. B. Jorde
Literature Cited
- Alisch R. S., Garcia-Perez J. L., Muotri A. R., Gage F. H., Moran J. V., 2006. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 20: 210–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio V. P., Hedges D. J., Deininger P., 2006. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 34: 1512–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke W. D., Malik H. S., Jones J. P., Eickbush T. H., 1999. The domain structure and retrotransposition mechanism of R2 elements are conserved throughout arthropods. Mol. Biol. Evol. 16: 502–511. [DOI] [PubMed] [Google Scholar]
- Callahan K. E., Hickman A. B., Jones C. E., Ghirlando R., Furano A. V., 2012. Polymerization and nucleic acid-binding properties of human L1 ORF1 protein. Nucleic Acids Res. 40: 813–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S. M., Eickbush T. H., 2005. R2 target-primed reverse transcription: ordered cleavage and polymerization steps by protein subunits asymmetrically bound to the target DNA. Mol. Cell. Biol. 25: 6617–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S. M., Bibillo A., Eickbush T. H., 2005. Role of the Bombyx mori R2 element N-terminal domain in the target-primed reverse transcription (TPRT) reaction. Nucleic Acids Res. 33: 6461–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S. M., Ye J., Eickbush T. H., 2006. RNA from the 5′ end of the R2 retrotransposon controls R2 protein binding to and cleavage of its DNA target site. Proc. Natl. Acad. Sci. USA 103: 17602–17607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian C. M., deHaro D., Kines K. J., Sokolowski M., Belancio V. P., 2016a Identification of L1 ORF2p sequence important to retrotransposition using Bipartile Alu retrotransposition (BAR). Nucleic Acids Res. 44: 4818–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian C. M., Kines K. J., Belancio V. P., 2016b The importance of L1 ORF2p cryptic sequence to ORF2p fragment-mediated cytotoxicity. Mob. Genet. Elements 6: e1198300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P. R., Jones C. E., Furano A. V., 2015. Phosphorylation of ORF1p is required for L1 retrotransposition. Proc. Natl. Acad. Sci. USA 112: 4298–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M., Esnault C., Heidmann T., 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35: 41–48. [DOI] [PubMed] [Google Scholar]
- Dombroski B. A., Feng Q., Mathias S. L., Sassaman D. M., Scott A. F., et al. , 1994. An in vivo assay for the reverse transcriptase of human retrotransposon L1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 4485–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush T. H., Jamburuthugoda V. K., 2008. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 134: 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush, T. H., and H. S. Malik, 2002 Origin and Evolution of Retrotransposons, pp. 1111–1144 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz. ASM Press, Washington, DC. [Google Scholar]
- Feng Q., Moran J. V., Kazazian H. H., Boeke J. D., 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87: 905–916. [DOI] [PubMed] [Google Scholar]
- Hancks D. C., Kazazian H. H., 2016. Roles for retrotransposon insertions in human disease. Mob. DNA 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Kuhara S., Takenaka O., Sakaki Y., 1986. L1 family of repetitive DNA sequences in primates may be derived from a sequence encoding a reverse transcriptase-related protein. Nature 321: 625–628. [DOI] [PubMed] [Google Scholar]
- Jamburuthugoda V. K., Eickbush T. H., 2011. The reverse transcriptase encoded by the non-LTR retrotransposon R2 is as error-prone as that encoded by HIV-1. J. Mol. Biol. 407: 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamburuthugoda V. K., Eickbush T. H., 2014. Identification of RNA binding motifs in the R2 retrotransposon-encoded reverse transcriptase. Nucleic Acids Res. 42: 8405–8415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kines K. J., Sokolowski M., deHaro D. L., Christian C. M., Belancio V. P., 2014. Potential for genomic instability associated with retrotranspositionally-incompetent L1 loci. Nucleic Acids Res. 42: 10488–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kines K. J., Sokolowski M., deHaro D. L., Christian C. M., Baddoo M., et al. , 2016. The endonuclease domain of the LINE-1 ORF2 protein can tolerate multiple mutations. Mob. DNA 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K. K., Fujiwara H., 2005. An extraordinary retrotransposon family encoding dual endonucleases. Genome Res. 15: 1106–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosha V. O., Martin S. L., 1997. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc. Natl. Acad. Sci. USA 94: 10155–10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosha V. O., Martin S. L., 2003. High-affinity, non-sequence-specific RNA binding by the open reading frame 1 (ORF1) protein from long interspersed nuclear element 1 (LINE-1). J. Biol. Chem. 278: 8112–8117. [DOI] [PubMed] [Google Scholar]
- Kulpa D. A., Moran J. V., 2006. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 13: 655–660. [DOI] [PubMed] [Google Scholar]
- Leibold D. M., Swergold G. D., Singer M. F., Thayer R. E., Dombroski B. A., et al. , 1990. Translation of LINE-1 DNA elements in vitro and in human cells. Proc. Natl. Acad. Sci. USA 87: 6990–6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan D. D., Korman M. H., Jakubczak J. L., Eickbush T. H., 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72: 595–605. [DOI] [PubMed] [Google Scholar]
- Malik H. S., Burke W. D., Eickbush T. H., 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16: 793–805. [DOI] [PubMed] [Google Scholar]
- Martin S. L., 2010. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA Biol. 7: 706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. L., Cruceanu M., Branciforte D., Wai-Lun Li P., Kwok S. C., et al. , 2005. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J. Mol. Biol. 348: 549–561. [DOI] [PubMed] [Google Scholar]
- Mathias S. L., Scott A. F., Kazazian H. H., Boeke J. D., Gabriel A., 1991. Reverse transcriptase encoded by a human transposable element. Science 254: 1808–1810. [DOI] [PubMed] [Google Scholar]
- Monot C., Kuciak M., Viollet S., Mir A. A., Gabus C., et al. , 2013. The specificity and flexibility of l1 reverse transcription priming at imperfect T-tracts. PLoS Genet. 9: e1003499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran J. V., Holmes S. E., Naas T. P., DeBerardinis R. J., Boeke J. D., et al. , 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87: 917–927. [DOI] [PubMed] [Google Scholar]
- Piskareva O., Schmatchenko V., 2006. DNA polymerization by the reverse transcriptase of the human L1 retrotransposon on its own template in vitro. FEBS Lett. 580: 661–668. [DOI] [PubMed] [Google Scholar]
- Piskareva O., Denmukhametova S., Schmatchenko V., 2003. Functional reverse transcriptase encoded by the human LINE-1 from baculovirus-infected insect cells. Protein Expr. Purif. 28: 125–130. [DOI] [PubMed] [Google Scholar]
- Piskareva O., Ernst C., Higgins N., Schmatchenko V., 2013. The carboxy-terminal segment of the human LINE-1 ORF2 protein is involved in RNA binding. FEBS Open Bio 3: 433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Engel A. M., 2012. LINEs, SINEs and other retroelements: do birds of a feather flock together? Front. Biosci. (Landmark Ed.) 17: 1345–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski M., Deharo D., Christian C. M., Kines K. J., Belancio V. P., 2013. Characterization of L1 ORF1p self-interaction and cellular localization using a mammalian two-hybrid system. PLoS One 8: e82021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. S., Lacava J., Mita P., Molloy K. R., Huang C. R., et al. , 2013. Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell 155: 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff B. J., Barnerssoi M., Roy-Engel A. M., 2011. Evolutionary conservation of the functional modularity of primate and murine LINE-1 elements. PLoS One 6: e19672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace N., Wagstaff B. J., Deininger P. L., Roy-Engel A. M., 2008a LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene 419: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace N. A., Belancio V. P., Deininger P. L., 2008b L1 mobile element expression causes multiple types of toxicity. Gene 419: 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenrieder O., Repanas K., Perrakis A., 2004. Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon. Structure 12: 975–986. [DOI] [PubMed] [Google Scholar]
- Xiong Y. E., Eickbush T. H., 1988. Functional expression of a sequence-specific endonuclease encoded by the retrotransposon R2Bm. Cell 55: 235–246. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H., 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9: 3353–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Malik H. S., Eickbush T. H., 1999. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl. Acad. Sci. USA 96: 7847–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that the information necessary for confirming the conclusions presented in the article are represented fully within the article.