Abstract
The protein phosphatase 2A (PP2A) is a conserved heterotrimeric enzyme that regulates several cellular processes including the DNA damage response and mitosis. Consistent with these functions, PP2A is mutated in many types of cancer and acts as a tumor suppressor. In mammalian cells, PP2A inhibition results in DNA double strand breaks (DSBs) and chromosome aberrations (CABs). However, the mechanisms through which PP2A prevents DNA damage are still unclear. Here, we focus on the role of the Drosophila twins (tws) gene in the maintenance of chromosome integrity; tws encodes the B regulatory subunit (B/B55) of PP2A. Mutations in tws cause high frequencies of CABs (0.5 CABs/cell) in Drosophila larval brain cells and lead to an abnormal persistence of γ-H2Av repair foci. However, mutations that disrupt the PP4 phosphatase activity impair foci dissolution but do not cause CABs, suggesting that a delayed foci regression is not clastogenic. We also show that Tws is required for activation of the G2/M DNA damage checkpoint while PP4 is required for checkpoint recovery, a result that points to a conserved function of these phosphatases from flies to humans. Mutations in the ATM-coding gene tefu are strictly epistatic to tws mutations for the CAB phenotype, suggesting that failure to dephosphorylate an ATM substrate(s) impairs DNA DSBs repair. In addition, mutations in the Ku70 gene, which do not cause CABs, completely suppress CAB formation in tws Ku70 double mutants. These results suggest the hypothesis that an improperly phosphorylated Ku70 protein can lead to DNA damage and CABs.
Keywords: Tws, PP2A-B55, chromosome aberrations, ATM, Ku70, Drosophila
COMPLEX biological processes require cellular activities to quickly switch from one state to another. One of the most important mechanisms that allows activation and silencing of these activities is reversible phosphorylation. Multiple kinases and phosphatases have been identified that work in concert to regulate fundamental processes such as the cell cycle and the DNA damage response (DDR) pathways (Freeman and Monteiro 2010). The protein phosphatase 2A (PP2A) is one of the major serine/threonine phosphatases; it regulates several cellular processes including DDR and mitosis. Consistent with these functions, accumulating evidence indicates that PP2A is mutated in many types of cancer and acts as a tumor suppressor (reviewed in Eichhorn et al. 2009; Khanna et al. 2013; Perrotti and Neviani 2013). PP2A is conserved from yeast to humans and is one of the most abundant enzymes, accounting for up to 1% of total cellular proteins in some tissues (Eichhorn et al. 2009; Shi 2009).
In mammals, PP2A is a heterotrimeric enzyme consisting of a core dimer, formed by a catalytic (C) and a structural (A) subunit, associated with a third regulatory B subunit, which governs subcellular localization and substrate specificity. PP2A A and PP2A C are encoded by two distinct genes, each of which produces two protein isoforms; the B subunits (B/B55, B′/B56, B″/PR72, and B‴/STRN) are encoded by four genes that also produce several isoforms. It has been estimated that the combinatorial association of the PP2A subunits can give rise to >90 different complexes, which are likely to mediate different physiological processes (Janssens and Goris 2001; Eichhorn et al. 2009). PP2A is a heterotrimeric complex also in Drosophila. The fly genome harbors two genes, PP2A-29B and microtubule star (mts), which encode the PP2A A and PP2A C subunits, respectively, and four genes that specify the B regulatory subunits: twins (tws; B/B55), widerborst (wdb; B′/B56 type 1), well rounded (wrd; B′/B56 type 2), and CG4733 (B″/PR72) (Mayer-Jaekel et al. 1992; Uemura et al. 1993; Snaith et al. 1996; Hannus et al. 2002; Viquez et al. 2006; see also FlyBase). The fly genome also contains the connector to kinase to AP-1 (cka) gene that encodes a protein partially homologous to B‴/STRN (Ribeiro et al. 2010; see also FlyBase).
Studies carried out in several metazoan organisms, including Drosophila and humans, have shown that PP2A plays an important role in the regulation of cell division. In some studies, the role of PP2A was addressed by inhibiting the enzyme activity with either okadaic acid or the small tumor antigen (ST) of Simian virus 40. Other studies exploited mutations or RNA interference (RNAi) to deplete specific PP2A subunits (see, for example, Tournebize et al. 1997; Chen et al. 2007; Kotadia et al. 2008; Foley et al. 2011). Work on the B55 subunit showed that the PP2A-B55 enzyme is responsible for dephosphorylation of many mitotic proteins, allowing mitotic exit and progression through interphase. The activity of this enzyme is high during interphase but is inhibited when cells enter mitosis to avoid premature reversion of cyclin-dependent kinase 1 (CDK1)-driven phosphorylation of mitotic proteins. PP2A-B55 is regulated by a conserved pathway involving the Greatwall (Gwl) kinase; in preparation of the M phase, CDK1 phosphorylates and activates Gwl, which in turn phosphorylates Endosulfine that binds and inactivates PP2A-B55 (Glover 2012; Hunt 2013; Lorca and Castro 2013; Williams et al. 2014).
Besides exerting a general control on mitotic progression and exit, PP2A has been implicated in specific aspects of mitosis. For example, PP2A inhibition with okadaic acid or ST, or disruption of its core subunits (A or C) affects centrosome behavior, spindle formation, and chromosome segregation in both Drosophila and vertebrates (Snaith et al. 1996; Tournebize et al. 1997; Kitajima et al. 2006; Tang et al. 2006; Chen et al. 2007; Kotadia et al. 2008). Defects in microtubule-kinetochore interaction and chromosome segregation have been also observed in HeLa and Drosophila cells depleted of the B56 regulatory subunits (Chen et al. 2007; Foley et al. 2011; Porter et al. 2013), while B55 depletion in Drosophila S2 cells counteracted Plk4 autophosphorylation leading to centrosome amplification (Brownlee et al. 2011).
In addition to its mitotic functions, PP2A plays important roles in nervous system maintenance and functioning, and in DNA repair (reviewed in Lambrecht et al. 2013). Studies in mammalian cells have shown that PP2A inhibitors or RNAi against the core subunits of the complex cause DNA double strand breaks (DSBs) and chromosome aberrations (CABs) (Chowdhury et al. 2005; Lankoff et al. 2006; Wang et al. 2009; Kalev et al. 2012). Several nonmutually exclusive hypotheses have been proposed to explain the role of PP2A in the repair of DSBs in mammalian cells. It has been suggested that loss of PP2A affects the dephosphorylation kinetics of γ-H2AX DNA repair foci associated with DSBs (Chowdhury et al. 2005). Immediately after their occurrence, DSBs recruit a series of DNA repair factors, starting with the Mre11-Rad50-Nbs (MRN) complex and the ATM kinase. ATM phosphorylates the histone variant H2AX (H2Av in Drosophila) at Ser139 to form γ-H2AX, which spreads in both directions from the DSB and helps recruiting additional DNA repair factors; leading to the formation of cytologically detectable foci, which disassemble after DNA repair, following γ-H2AX dephosphorylation (reviewed in Bekker-Jensen and Mailand 2010 and Polo and Jackson 2011). It has been shown that PP2A directly binds and dephosphorylates γ-H2AX at DNA repair foci and that in PP2A-deficient cells, γ-H2AX foci persist longer that in control cells, suggesting that foci persistence leads to incomplete DSB repair (Chowdhury et al. 2005). It has been also reported that PP2A-deficient cells display an increase in the level of ATM autophosphorylation/activation accompanied by upregulation of the ATM downstream kinase CHK2 and downregulation of the RAD51 and BRCA1 factors, which mediate the homologous recombination (HR) pathway of DSB repair (Kalev et al. 2012). Other studies suggested that PP2A dephosphorylates the Ku complex that mediates the nonhomologous end joining pathway (NHEJ), and that PP2A-deficient cells accumulate DSBs due to defects in this pathway (Wang et al. 2009). Finally, consistent with a general role of PP2A in DNA repair, several studies have shown that PP2A dephosphorylates not only ATM and CHK2 but also the ATR kinase and its downstream target CHK1, and it is required for the G2/M cell cycle arrest induced by DNA damage (Leung-Pineda et al. 2006; Yan et al. 2010).
Although most of the roles of PP2A are evolutionarily conserved, there are currently very few data suggesting an involvement of Drosophila PP2A in DNA repair. The only indication of a possible role of PP2A in the maintenance of chromosome integrity is provided by the observation that mutants in the B55-coding gene tws (also called aar) exhibit chromatin bridges at anaphase (Gomes et al. 1993; Mayer-Jaekel et al. 1993). Here we show that mutations in tws cause high frequencies of CABs and abnormal persistence of γ-H2Av DNA repair foci. We also show that mutations in the ATM-coding gene tefu and the ku70 gene are strictly epistatic to tws mutations for the CAB phenotype. Collectively, our genetic analyses suggest that failure of Tws-mediated dephosphorylation generates abnormally and/or untimely phosphorylated Ku70, which interferes with normal DNA repair leading to DNA damage and CABs.
Materials and Methods
Drosophila strains and crosses
tws430 was isolated by a cytological screen of larval brain squashes from a collection of 1680 EMS-induced late lethals generated in Charles Zuker’s laboratory (University of California, San Diego, CA). twsP and tws196 were obtained from M.L. Goldberg (Cornell University, Ithaca, NY), brca56E and brcaK0 from T. Schüpbach (Princeton University, Princeton, NJ), and ku70Ex8 from W. R. Engels (University of Wisconsin, Madison, WI); tefuatm6, mei-4129D, nbs1, His2av 810, and grapes1 mutations have been described previously (van Daal and Elgin 1992; Sullivan et al. 1993; Laurençon et al. 2003; Silva et al. 2004; Ciapponi et al. 2006). Df(3L)by62, Df(3L)ED54541, and Df(3L)BSC621, Pp4-19C, brca2KG03961, lig45, okr17-11, okrA19-10, mus301D4/SpnCD4, mei-9A1, rad511/SpnA1 were all obtained from the Bloomington Stock Center. Mutations and deficiencies on the third and second chromosomes were kept in stock over the TM6C Sb Tb and CyO-TbA (Lattao et al. 2011) balancers, respectively; homozygous and hemizygous mutant larvae were recognized for their non-Tubby phenotype. Mutations on the X chromosome were balanced over FM7-GFP and mutant larvae were recognized for their non-GFP phenotype. tws430 and brca2KG03961 double mutants were constructed by crossing tws430/TM6B; CyO-GFP/Sco females to MKRS/TM6B; brca2KG03961/CyO-GFP males; tws430/TM6B; brca2KG03961/CyO-GFP progeny were mated inter se to obtain a stable stock. To generate brca56E/brcaK0; tws430/tws430 and okr17-11/okrA19-10; tws430/tws430 double mutants, tws430/TM6B; brca56E/CyO-GFP and tws430/TM6B; okr17-11/CyO-GFP females were crossed to tws430/TM6B; brcaK0/CyO-GFP and tws430/TM6B; okrA19-10/CyO-GFP males, respectively. mei4129D tws430, lig45 tws430, and mei-9A1 tws430 double mutants were generated by crossing mei4129D/FM7-GFP; +/TM6B, lig45/FM7-GFP; +/TM6B and mei-9A1/FM7-GFP; +/TM6B females to w; tws430/TM6C males, respectively. The w/FM7-GFP; tws430/TM6B females resulting from this cross were then mated to mei4129D; tws430/TM6B, lig45; tws430/TM6B and mei-9A1; tws430/TM6B males to generate mei4129D/FM7-GFP; tws430/TM6B, lig45/FM7-GFP; tws430/TM6B and mei-9A1/FM7-GFP; tws430/TM6B females that were crossed to FM7-GFP; tws430/TM6B males to generate stable stocks. Double mutant larvae from these crosses were unambiguously identified on the basis of their non-GFP and/or non-Tubby phenotypes. tws430 tefuatm6, nbs1 tws430, tws430 H2Av810, tws430 rad511, tws430 ku70Ex8, and mus301D4 tws430 double mutants were generated by recombination and balanced over TM6B. To check for the presence of both mutations on the recombinant third chromosomes we performed complementation tests; the presence of both tws430 ku70Ex8 on the recombinant chromosome was also confirmed by PCR. Also, the ku70MT tws430 ku70Ex8 chromosome was constructed by recombination and the presence of ku70MT, tws430, and ku70Ex8 in this chromosome was confirmed by complementation and PCR analyses. We designate as ku70MT a ku70/irbp gene carrying the 2xTY1-sGFP-V5-Pre-TEV-BLRP-3xFLAG multi-tag at its C terminus. ku70MT is a component of the fosmid clone FlyFos 015211, which contains the nina, CG6723, Ranbp9, ku70/irbp, mgr, and mRpL40 genes and part of the pros gene. ku70/irbp was tagged in vitro, and the entire fosmid construct was then inserted into a fly stock carrying an attP landing site at 65B using a nanos-ΦC31 integrase source [Sarov et al. 2016; the ku70MT bearing line was obtained from the Vienna Drosophila Resource Center (VDRC) stock center]. Tip60 and Mrg15 RNAi flies were generated by crossing females bearing the RNAi construct (Tip60, 22233 from VDRC; Mrg15, 35241 from Bloomington Stock Center) to males carrying the 69B-Gal4 driver. To determine the effect of ku70 RNAi in a tws430 mutant background, we used the 110,409/kk VDRC construct (ku70-RNAi) mapping to the second chromosome; CyO-GFP/actin-GAL4, tws430/TM6B females were crossed to CyO-GFP/ku70-RNAi, tws430/TM6B males to obtain ku70-RNAi/actin-GAL4; tws430/tws430 larvae that were recognized for their non-GFP and non-Tubby phenotype. The Oregon R strain was used as wild-type control. All stocks were maintained, and crosses were made at 25° on standard Drosophila medium. The balancers and the genetic markers used in these crosses are described in detail in FlyBase (http://flybase.bio.indiana.edu/)
Chromosome cytology
Colchicine-treated larval brain metaphases for CAB scoring and noncolchicine-treated preparations for the analysis of anaphases and the mitotic index (MI) were obtained as described (Gatti and Goldberg 1991). The MI is the average number of mitotic figures per optic field; the frequency of anaphases is the ratio between the number of anaphases and the total number of mitotic figures observed (Gatti and Goldberg 1991). To perform the checkpoint assay, wild-type and mutant larvae were treated with 10 Gy of X rays. Brains from irradiated larvae were then dissected and fixed at the indicated times to determine the MI. The relative MIs were calculated normalizing the MIs of irradiated brains with respect to that of untreated brains. All fixed preparations were mounted in Vectashield H-1200 with DAPI (Vector Laboratories, Burlingame, CA) to stain the chromosomes.
Tests for X-ray and hydroxyurea sensitivity
To determine the X-ray sensitivity of tws mutants, homozygous tws430 larvae and wild-type larvae were irradiated with 2.5 Gy. Then, 3 hr after irradiation (IR), larval brains were dissected in saline (NaCl 0.7%), incubated for 1 hr with colchicine (10−5 M), and then fixed. To determine the hydroxyurea (HU) sensitivity, brains from homozygous tws430 mutant larvae and wild-type larvae were incubated for 20 min in 1 mM HU dissolved in saline. They were then rinsed, incubated in saline for 3.5 hr, and fixed. Finally, 1 hr before fixation, colchicine was added to the saline to collect metaphases.
Immunostaining and γ-H2Av foci detection
For immunostaining, brains from third instar larvae were dissected and fixed as described in Bonaccorsi et al. (2000). To induce γ-H2Av and Tws foci, larvae were irradiated with 5 Gy of X rays; larval brains were then dissected and fixed at various postirradiation (PIR) times. Brain preparations were then rinsed several times in PBS 0.1% Triton (PBST), incubated overnight at 4° with primary antibodies diluted in PBST, rinsed in PBST, and then incubated for 1 hr at room temperature with the pertinent secondary antibodies. The primary antibodies were: rabbit anti-Histone H2AvD pS137 (1:100; Rockland code #600-401-914) and rat anti-Twins (1:50; a gift from T. Uemura, Kyoto University, Japan). Antibodies were detected with Alexa-Fluor-555-conjugated anti-rabbit (1:300 in PBSt; Molecular Probes, Eugene, OR) and FITC-conjugated anti-rat (1:20 in PBSt; Jackson Laboratories). Immunostained preparations were mounted in Vectashield H-1200 (Vector Laboratories) containing DAPI (4,6 diamidino-2-phenylindole). To quantify the foci, at least 400 cells were analyzed for each PIR fixation time. All cytological preparations were examined with a Carl Zeiss (Thornwood, NY) Axioplan fluorescence microscope, equipped with an HBO100W mercury lamp and a cooled charged-coupled device (CCD camera; Photometrics CoolSnap HQ).
Western blotting
Extracts for Western blotting were prepared by lysing samples of 20 brains in 150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 30 mM NaF, 25 mM b-glycerophosphate, 0.2 mM Na3VO4, Triton X-100 1%, and cOmplete Protease Inhibitor Cocktail (Roche). Extracts were immunoblotted according to Somma et al. (2002); blotted proteins were detected using rabbit anti-Histone H2AvD pS137 (1: 500 in TBST; Rockland code #600-401-914) and rat anti-Tws (a gift form T. Uemura; diluted 1:500 in TBST). To determine the kinetics of H2Av phosphorylation, larvae were irradiated with X rays (10 Gy) and 20 brain samples were collected at different PIR times. The loading control was always Giotto (Gio), a Drosophila Phosphatidylinositol transfer protein (Giansanti et al. 2006).
RNAi in Drosophila S2 cultured cells
RNAi treatments in S2 cells were performed as described in Somma et al. (2008). To produce double-stranded RNAs we used the following primers:
lig4 F: ATGTGACCACCA; lig4 R: ATGCCTTCGCGA.
mus-301 F: CAGTTGGACTGC; mus-301 R: CGATTCAGCTGC.
brca2 F: AACCGCATCAAC; brca2 R: AAGGCTTGGGAG.
Each of these primers contained the T7 polymerase binding sequence:
5′-TAATACGACTCACTATAGGGAGG-3′.
Nucleic acid extraction, PCR, and RT-PCR
Preparation of fly genomic DNA and RNA from S2 cells, PCR, RT-PCR, agarose gel electrophoresis, DNA sequencing, and sequence analysis were performed with standard procedures. RNA extraction was performed with the RNeasy Mini Kit (Qiagen, Hilden, Germany). For RT-PCR we used 20 ng of RNA to synthesize complementary DNAs (cDNAs) using the Superscript kit (Invitrogen, Carlsbad, CA). For cDNA amplification were used the following primers:
rp49 F: TACAGGCCCAAGATCGTGAA; rp49 R: ACGTTGTGCACCAGGAACTT.
brca2 F: CTGGACGACAAGGAGCAACC; brca2 R: TCAAGTCCAACAGACGTCGG.
lig4 F: ACGATCACGGCACCTTAACG; lig4 R: CTTGTCCTTGTACCACCCAC.
His2Av F: GCTGGCGGTAAAGCAGGCAA; His2Av R: AATGACGTTGCCCTTCCGCT.
mus-301 F: AAATGGTGGGACGAGCAGGT; mus-301 R: ACAGAATGCTGGCATCTGCC.
To test for the presence of kuMT and ku70Ex8, we used the following GFP and ku70 primers:
GFP F: AAGGGCGAGGAGCTGTTCA; GFP R: TTGTACAGCTCATCCATGCCCA.
Ku70 F: GACTCATCTTCGCCAACACCA; ku70 R: ATGATGCTGCTGGGCTTCAAG.
Data availability
All data needed to confirm the results presented in this study are included in the main article and in the supplemental material. Fly strains are available upon request.
Results
Mutations in tws induce CABs
In the course of a screen aimed at the isolation of new Drosophila mitotic mutants (see Materials and Methods), we identified a lethal mutation that causes frequent CABs in larval brain cells. Recombination and deletion mapping showed that this mutation is included in the 85F12–85F14 polytene chromosome interval and fails to complement mutations in the tws gene that maps to the same region (Supplemental Material, Figure S1A). We therefore named our mutation tws430 (430 is the number of the tws-bearing stock in the collection we screened). tws encodes eight transcripts that differ at the 5′ UTR; all these transcripts give rise to two polypeptides of 499 (a isoform) and 433 aa (b isoform) that differ only in 56 aa at the N terminus (Figure S1B). DNA sequencing showed that tws430 carries a G→A transition in a splice site at the 5′ of an intron shared by all transcripts (Figure S1B). Consequently, the first AG in the downstream exonic sequence is used as a splice site, leading to a stop codon that would result in truncated proteins of 142 and 86 amino acids (Figure S1C).
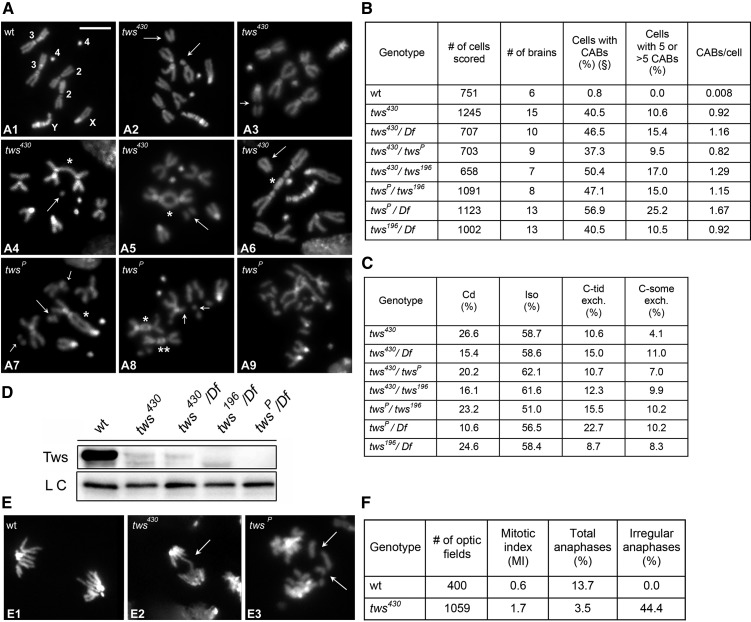
To define the frequency and pattern of CABs induced by mutations in tws we examined DAPI-stained brain preparations from third instar larvae of various genotypes: tws430/tws430, tws430/Df(3R)by62 (Df(3R)by62, henceforth designated as Df, is a deficiency that removes tws+), tws430/tws196, tws430/twsP, twsP/tws196, tws196/Df, and twsP/Df. The chromosomes carrying the tws196 and twsP also carry second site lethal mutations, which prevented the analysis of twsPand tws196 homozygotes. The brains of all mutant genotypes examined displayed very high frequencies of cells with CABs (ranging from 28 to 57%) compared to wild-type controls (0.8%). In all mutants, we observed metaphases displaying from one to five CABs and metaphases with more than five CABs often showing an extensive chromosome fragmentation (Figure 1, A and B). Since the latter cells did not permit a reliable evaluation of the type and number of CABs, in calculating the frequencies of CABs per cell we arbitrarily assumed that each of them contained only five CABs. Thus the CABs/cell frequencies reported in Figure 1B are an underestimation of the actual frequencies. An analysis of the CAB frequencies indicates that twsP is the strongest mutant allele (Figure 1B). Consistent with this result, Western blotting showed that the Tws protein is undetectable in twsP/Df mutant brains and strongly reduced with respect to control in brains from tws430/Df and tws196/Df mutants (Figure 1D).
Figure 1.
tws mutants exhibit frequent CABs. (A) Examples of CABs: (A1) Wild-type male metaphase. (A2) Isochromatid deletion (iso) of a major autosome (Au), arrows. (A3) Iso of the X chromosome, arrow. (A4) Asymmetric Au-Au chromatid exchange (*) with acentric fragment (AF; arrow). (A5) Au-Au dicentric chromosome (*) with the AF (arrow). (A6) Au-Au dicentric chromosome (*) with the AF (arrow). (A7) Au-XL dicentric chromosome (*) with AF (big arrow) and autosomal isochromatid deletion (small arrows). (A8) XR-Au (*) and Au-Au (**) dicentric chromosomes with the AFs; and chromatid deletion (Cd) of a major autosome (arrows). (A9) Metaphase with extensive chromosome fragmentation. Bar, 5 μm. (B) Frequencies and (C) types of CABs observed in tws mutants. (D) Western blotting showing strong reductions of the Tws protein in different tws mutants. (E) Examples of irregular anaphases: (E1) wild type, (E2) mutant anaphase with a chromatin bridge, (E3) mutant anaphase with lagging acentric fragments. (F) Mitotic parameters in wild-type and tws mutant brains. The MI is the average number of mitotic figures per optic field (see Materials and Methods). #, number; C-tid exch., chromatid exchange; C-some exch., chromosome exchange; LC, loading control; wt, wild type.
The types and frequencies of CABs observed in metaphases with one to five CABs are reported in Figure 1C. As shown in Figure 1, A and C, tws mutant cells displayed chromatid and isochromatid deletions and exchanges of chromatid and chromosome type; chromosome exchanges included mostly dicentric chromosomes with only a few translocations, as in Drosophila most exchanges involve the homologous chromosomes due to the somatic pairing, and chromosome type symmetrical exchanges between homologs are usually not detectable (Gatti et al. 1974). All dicentric chromosomes observed in tws mutants were accompanied by acentric fragments, indicating that they are not telomeric fusions (TFs) (Figure 1A). Thus PP2A-B55 downregulation results in chromosome-type CABs generated during G1 and chromatid-type CABs formed during S/G2. Notably, most aberrations and exchanges were “complete,” namely they contained all the elements that give rise to the CAB. For example, the large majority of isochromatid deletions consisted of both the centric and the acentric fragment. “Incomplete” isochromatid breaks consisting of either a centric fragment without the corresponding acentric element or of an acentric fragment associated with a normal chromosome complement were very rare. Incomplete isochromatid breaks are the expected outcome of the rupture of chromosome bridges during anaphase and are very frequent in Topoisomerase2 (Top2) mutants which exhibit anaphase bridges generated by failure to decatenate sister chromatids (see Mengoli et al. 2014 for a detailed analysis of the types of CABs generated by the rupture of anaphase bridges). Thus, we conclude that most CABs observed in tws mutants are not generated by breakage of anaphase bridges but by DNA lesions produced during the interphase that precedes the mitotic division examined.
We also asked whether tws mutations affect the cell cycle progression. Preparations from tws430 mutant brains not treated with colchicine and hypotonic solution showed an MI higher than controls (1.7 vs. 0.6) but a lower anaphase frequency (3.5 vs. 13.7% of control) (Figure 1, E and F). Consistent with previous work on aar/tws mutants (Gomes et al. 1993), 44% of the anaphases observed in tws mutant brains displayed chromatin bridges and lagging acentric chromosome fragments. These aberrant anaphases are probably originated by the dicentric chromosomes present in metaphase (Figure 1, A and C) and not by events directly occurring during anaphase, which would generate “incomplete aberrations” that are not found in brain preparations from tws mutants.
To obtain some insight into the mechanisms underlying CAB formation in tws mutants, we asked whether these mutants are sensitive to mutagenic agents such as X rays and HU. X rays directly induce DNA breaks; while HU inhibits the production of deoxyribonucleotides, causing replication fork collapse which ultimately results in DSBs. As shown in Table S1, tws mutants are sensitive to both agents. Treatments that produce ∼0.05 CABs/cell in wild-type controls resulted in ∼2.5-fold increases of the CAB frequency in tws mutants.
Collectively, these results indicate that the Tws/B55 PP2A subunit is required to prevent chromosome damage in Drosophila. Previous work showed that the human PP2A C catalytic subunit is also required to prevent CABs (Wang et al. 2009; Bouley et al. 2015). Altogether, these findings indicate that PP2A plays an evolutionarily conserved role in the maintenance of genome integrity.
PP2A is required for γ-H2Av dephosphorylation
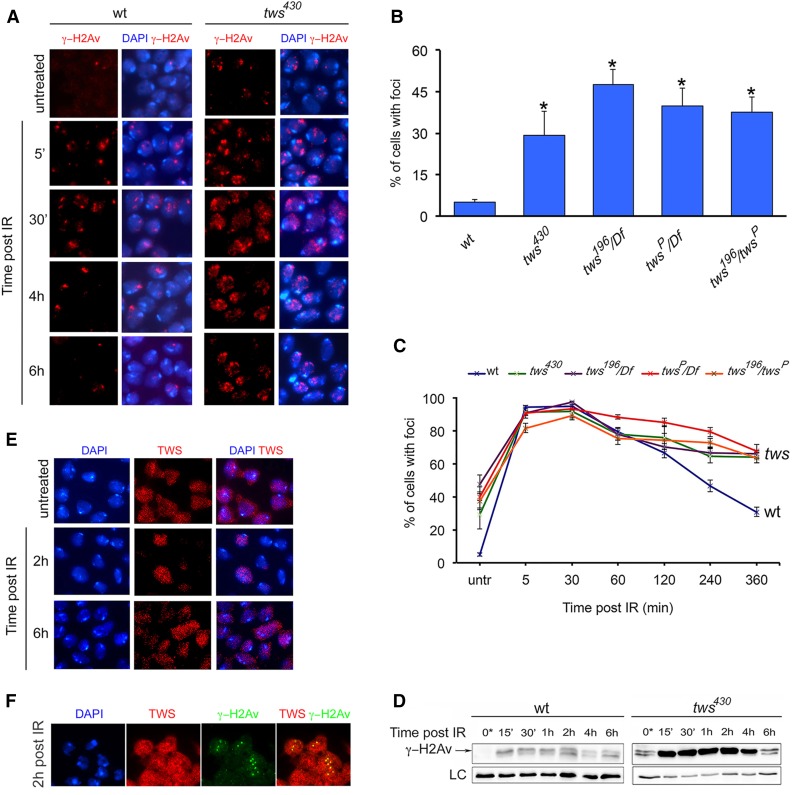
Because tws mutations exhibit CABs and previous work has shown that PP2A dephosphorylates γ-H2AX in mammalian cells (Chowdhury et al. 2005), we asked whether tws mutant cells exhibit γ-H2Av DNA repair foci. Brains were fixed and immunostained with the pS137 anti-phospho-histone antibody that specifically recognizes γ-H2Av (Madigan et al. 2002), and the frequency of cells with foci was quantified by microscope analysis. In brain cells from tws mutants, the average frequency of nuclei with γ-H2Av foci ranged from 30 to 45%, while only 5% of wild-type cells displayed γ-H2Av accumulations (Figure 2, A and B). Because γ-H2AX foci form around DSBs (Polo and Jackson 2011), these results indicate that tws downregulation results in DSBs.
Figure 2.
tws mutants exhibit delayed dissolution of γ-H2Av foci. (A) Examples and (B) frequencies of γ-H2Av foci observed in nuclei of wild-type (wt) and unirradiated tws mutant brains. Bars show the mean values of three independent experiments ± SEM; * significant in the Student’s t-test with P < 0.001. (A) Examples and (C) dissolution kinetics of X-ray-induced (5 Gy) γ-H2Av foci in wild-type and tws mutant brains; each PIR time represents the mean value from three independent experiments ± SEM; at least three brains per PIR time examined in each experiment. (D) Western blotting showing the persistence of γ-H2Av at 6 hr PIR in tws mutants; Giotto was used as a loading control (LC). (E) The Tws protein localizes in both the nucleus and the cytoplasm of untreated wild-type brain cells, it is enriched within the nuclei at 2 hr PIR, and diffuses back in the cytoplasm at 6 hr PIR. (F) Colocalization of Tws protein and γ-H2Av at 2 hr PIR.
We next asked whether tws mutations affect γ-H2Av dephosphorylation and increase the persistence of IR-induced γ-H2Av foci. We performed time course experiments to analyze the kinetics of X-ray-induced γ-H2Av foci in tws and wild-type brains. In both control and mutant brains, the frequency of nuclei with γ-H2Av-positive foci peaked at 5 min PIR and remained high at 30 min PIR to progressively decrease at 1 and 2 hr PIR. However, at 4 and 6 hr PIR, the frequencies of nuclei with foci remained high in tws mutants but dropped in wild-type controls; at 4 hr PIR, <50% wild-type nuclei were still displaying foci, whereas ∼80% of nuclei from tws mutants showed γ-H2Av foci; and at 6 hr PIR the frequencies of nuclei with foci were ∼30% in wild type and ∼70% in tws mutants (Figure 2, A and C). These findings are consistent with a Western blot analysis of H2Av phosphorylation. In blots from mock-treated brain extracts, the intensities of the γ-H2Av bands were high from 15 min to 2 hr PIR, but then decreased at 4 and 6 hr PIR. In blots from tws mutant brain extracts, the intensities of the γ-H2Av bands were much higher than those of controls and remained high until 4 hr PIR to decrease at 6 hr PIR (Figure 2D). Collectively, these results indicate that Drosophila PP2A has a role in γ-H2Av dephosphorylation and is required for the timely dissolution of γ-H2Av DNA repair foci. These findings are consistent with previous work showing that inhibition of PP2A in HeLa cells results in the persistence of Camptotecin-induced γ-H2AX foci (Chowdhury et al. 2005).
We also asked whether PP2A associates with γ-H2AX foci as occurs in mammalian cells (Chowdhury et al. 2005). We immunostained with an anti-Tws antibody wild-type cells before and after X-ray treatment. In unirradiated cells, the Tws protein was present in both the nucleus and the cytoplasm. However, at 2 hr PIR, Tws accumulated almost exclusively in the nucleus, where it self-aggregated forming cytologically detectable foci; at 6 hr PIR, Tws returned to the cytoplasm (Figure 2, E and F). We next analyzed brain cells immunostained with both anti-Tws and anti γ-H2Av antibodies. Although the number of γ-H2Av foci was lower than that of Tws foci, nearly all γ-H2Av foci colocalized with Tws aggregates (Figure 2F). These results provide further evidence for a role of Tws in γ-H2Av foci regulation.
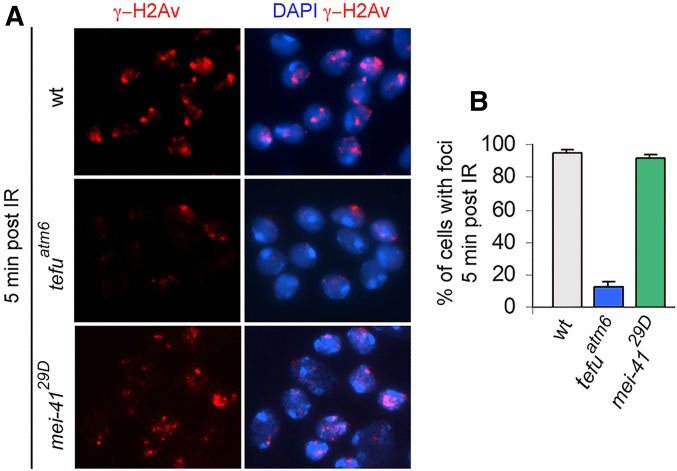
We finally asked whether the ATM and ATR kinases are required for γ-H2Av foci formation. Previous work on mammalian cells has shown that H2AX can be phosphorylated by these two kinases and the DNA-PK kinase (Burma et al. 2001; Ward and Chen 2001; Park et al. 2003; Chowdhury et al. 2008); the DNA-PK kinase does not have a Drosophila homolog (FlyBase). Wild-type and mutant brains were treated with X ray (5 Gy) and fixed 5 min PIR. Immunostaining with an anti γ-H2Av antibody revealed that tefuatm6 mutant cells display a significantly lower frequency of γ-H2Av foci compared to wild-type controls. In contrast, irradiated mei-41 mutants displayed the same frequency of cells with foci as nonmutant controls (Figure 3). These results indicate that formation of X-ray-induced γ-H2Av foci is mediated by the ATM kinase. They also suggest that ATR might not be involved in foci formation. However, we cannot completely exclude a minor role of ATR in γ-H2Av phosphorylation. mei-41 (ATR) and tefu (ATM) mutants are 10-fold and 3-fold more sensitive than wild type to X-ray-induced CABs, respectively (for mei-41 sensitivity see Gatti et al. 1980; to determine the tefu sensitivity we examined 300 metaphases from both wild-type and tefuatm6/tefuatm6 brains irradiated with 1 Gy of X rays). Thus, it is possible that the observed frequency of foci in irradiated mei-41 mutants is the combined outcome of an increased DNA damage and a low/moderate level of ATR-mediated H2Av phosphorylation.
Figure 3.
Formation of Drosophila γ-H2Av foci is mediated by ATM. (A) Representative examples of nuclei from irradiated (5 Gy) wild-type, tefu (ATM), and mei-41 (ATR) mutant brains stained for γ-H2Av. (B) Frequencies of nuclei with γ-H2Av foci in irradiated wild-type, tefu, and mei-41 mutant brains. Bars show the mean values of three independent experiments ± SEM.
Relationships between foci dissolution and CAB formation in Drosophila
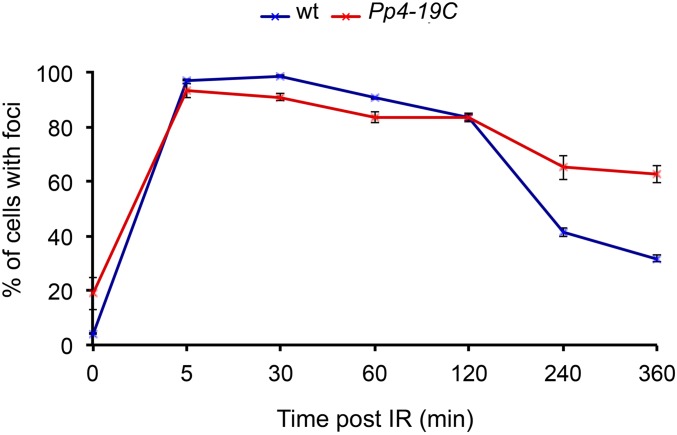
Previous work on mammalian cells has shown that both PP2A and PP4 can dephosphorylate γ-H2AX and promote foci dissolution; it has also been proposed that abnormal γ-H2AX foci persistence could result in DNA damage (Chowdhury et al. 2005, 2008). We thus asked whether Drosophila PP4 is also required for proper γ-H2Av foci behavior. We irradiated Pp4-19C larvae carrying a mutation in the catalytic subunit of PP4, and then followed the dynamics of γ-H2Av foci in a time course experiment. Both control and mutant brains displayed similar frequencies of nuclei with γ-H2Av foci until 2 hr PIR. However, at 4 and 6 hr PIR, mutant brains showed significantly higher frequencies of nuclei with foci than controls (∼65 vs. 40% at 4 hr, and ∼65 vs. 30% at 6 hr) (Figure 4). These findings indicate that Drosophila PP4 is required for γ-H2Av foci regression just like Tws (compare Figure 2C and Figure 4).
Figure 4.
PP4 is required for γ-H2Av foci regression in Drosophila. Each PIR time represents the mean value from three independent experiments ± SEM. Larvae were irradiated with X ray (5 Gy) and dissected brains were fixed at various PIR times; at least three brains per time examined in each experiment.
We then examined unirradiated Pp4-19C mutant brains for the presence of CABs and found a CAB frequency comparable to that of wild-type controls (0.006 CABs/cell, in 500 cells examined from 4 mutant brains). This finding suggests that a delay in γ-H2Av foci dissolution is not sufficient to induce CABs in Drosophila brain cells. An additional support for this conclusion comes from the analysis of mutants that disrupt the Tip60 pathway. In both Drosophila and humans, depletion of the TIP60 complex leads to foci persistence (Kusch et al. 2004; Jha et al. 2008). This complex mediates acetylation of γ-H2AX/H2Av, a post-translation modification that is thought to facilitate the access of phosphatases to the phospho-histone variant (Ikura et al. 2007). Specifically, an abnormal persistence of foci was observed in Drosophila cells depleted of the Tip60 and dMgr15 subunits of the complex (Kusch et al. 2004). We performed in vivo RNAi against Tip60 and Mrg15 using specific RNAi constructs and a 69B-GAL4 driver. RNAi flies died at the larval/pupal transition but their brains did not show an increase in CABs compared to wild-type controls (in Tip60 and Mrg15 RNAi brains, the frequencies of CABs were 0.002 and 0.003 per cell, respectively; 600 cells scored from at least three RNAi brains).
The tws and Pp4-29C genes control the G2/M checkpoint
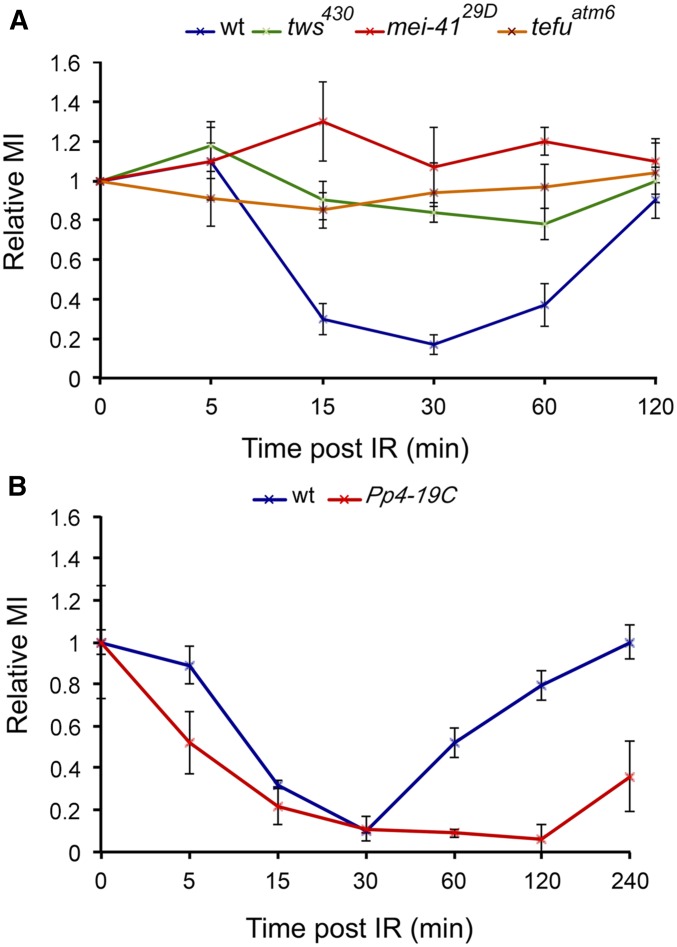
To further characterize the roles of Drosophila PP2A and PP4 in the DDR pathway, we asked whether they are involved in the regulation of the G2/M checkpoint like their mammalian counterparts (Nakada et al. 2008; Yan et al. 2010). The G2/M checkpoint is a complex signaling machinery that includes CDKs and opposing phosphatases; when this checkpoint detects lesions in the nuclear DNA, it arrests cell cycle progression to prevent cells from initiating mitosis with damaged DNA (Yasutis and Kozminski 2013). We performed a checkpoint assay, evaluating the MI in larval brains fixed at different times after X-ray exposure (10 Gy). In these experiments, we used the Oregon R wild-type strain as a negative control and the mei-41 (ATR) and tefu (ATM) mutant flies as positive controls, since they are known to have a defective G2/M checkpoint (Hari et al. 1995; Oikemus et al. 2004). As shown in Figure 5A, the MI of the Oregon R control brains dropped at 5–15 min PIR, remained low for ∼1 hr and came back to a normal value only at 2 hr PIR. In contrast, irradiated mei-41, tefu, and tws mutant brains did not show significant variations in the MI over time, indicating a defect in the G2/M checkpoint.
Figure 5.
PP2A-Tws and PP4 play different roles in the Drosophila G2/M DNA damage checkpoint. Each PIR time represents the mean value from three independent experiments ± SEM. Larvae were irradiated with X ray (10 Gy) and dissected brains were fixed at various PIR times; at least three brains per time examined in each experiment. The MI values reported in the graph are normalized with respect to the MI of untreated brains. Note that (A) tws is required for checkpoint signaling like tefu (ATM) and mei-41 (ATR), while (B) PP4 appears to be dispensable for signaling but necessary for checkpoint recovery. wt, wild type.
Differently from tws mutants, in Pp4-19C mutant brains, the MI dropped at 5–15 min PIR as occurs in controls, but then remained low for >3 hr, in contrast to wild-type brains where the cells began reentering mitosis at 30–60 min PIR (Figure 5B). These results indicate that PP2A and PP4 play different roles in the Drosophila G2/M checkpoint. PP2A appears to be required for initial checkpoint signaling like ATM and ATR. In contrast, PP4 is not involved in checkpoint initiation but it is required for termination of signaling and checkpoint recovery. A requirement of PP4 for checkpoint recovery has been previously demonstrated in budding yeast and human cells (Keogh et al. 2006; Nakada et al. 2008), pointing to a highly conserved function of this phosphatase in the regulation of the G2/M checkpoint.
Interactions between tws and the ATM kinase
We have shown that ATM mediates H2Av phosphorylation and that PP2A dephosphorylates this histone. In addition, previous work has shown that PP2A and ATM physically interact and that PP2A dephosphorylates ATM (Goodarzi et al. 2004; Kalev et al. 2012). We thus decided to use a genetic approach to investigate the relationships between PP2A and ATM in CAB formation. We performed an epistasis analysis by comparing the types and frequencies of chromosome abnormalities found in tefu (ATM) and tws single mutants with those observed in tws tefu double mutants. Consistent with our previous work (Ciapponi et al. 2006), single tefu null mutants (tefuatm6) showed 0.09 CABs/cell and an average of 0.5 TFs/cell (Table 1). Strikingly, tws430 tefuatm6 double mutants displayed CAB and TF frequencies very similar to those seen in the tefu single mutants, while the CAB frequency observed in tws430 single mutants was ∼10-fold higher than that seen in the double mutants (Table 1). Thus, mutations in tefu are perfectly epistatic to mutation in tws. These results indicate that ATM and PP2A function in the same pathway that leads to CABs in Drosophila tws mutants, and strongly suggest that CAB formation in these mutants depends on ATM activity. However, the role of ATM in CAB generation is unclear. In mammalian cells, quiescent ATM exists as a homodimer and is activated by autophosphorylation at serine 1981 (S1987 in mouse) and at least three additional sites. Abrogation of these modifications hampers the ATM activity in the DDR in human cells but not in mouse cells (reviewed in Shiloh and Ziv 2013). In addition, studies in human cells have shown that ATM phosphorylation at S1981 is dispensable for ATM recruitment at the DSBs but is required for ATM retention at DSBs (So et al. 2009). Based on these findings, we can envisage that dysregulation of ATM phosphorylation can interfere with the DNA repair processes, ultimately leading to DSBs and the ensuing CABs. Alternatively, it is possible that loss of B55/PP2A activity leads to an excessive and/or untimely phosphorylation of a specific ATM substrate(s), which would interfere with proper DNA repair, resulting in DNA damage. Should this hypothesis be correct, and should these “phospho-mutagens” exist, one would expect that loss-of-function mutations in the genes they specify would act as suppressors of the CAB phenotype elicited by tws mutations.
Table 1. Frequencies of CABs and TFs observed in larval brains of tws430 mutants and tws430-bearing double mutants.
| Genotype | No. of brains | No. of cells scored | Cells with CABsa (%) | Cells with ≥5 CABs (%) | CABs/cellb | TFs/cell |
|---|---|---|---|---|---|---|
| wtc | 5 | 460 | 0.4 | 0.0 | 0.004 | 0.0 |
| tws430c | 5 | 406 | 38.2 | 14.0 | 1.017 | 0.0 |
| tefuatm6c | 8 | 531 | 8.7d | 0.0 | 0.087 | 0.5 |
| tws430 tefuatm6c | 11 | 581 | 12.0d | 0.0 | 0.120 | 0.5 |
| nbs1c | 16 | 796 | 8.3 | 0.0 | 0.083 | 0.4 |
| nbs1 tws430e | 15 | 769 | 33.0 | 7.5 | 0.631 | 0.2 |
| ku70 Ex8c | 7 | 850 | 0.2 | 0.0 | 0.002 | 0.0 |
| tws430 ku70Ex8c | 15 | 1392 | 0.3 | 0.0 | 0.003 | 0.0 |
| ku70MT tws430 ku70Ex8c,f | 5 | 451 | 13.1 | 2.2 | 0.155 | 0.0 |
| ku70 RNAi tws430c,g | 5 | 513 | 2.7 | 0.0 | 0.027 | 0.0 |
| mei-9A1c | 10 | 950 | 4.0 | 0.0 | 0.040 | 0.0 |
| mei-9A1 tws430e | 12 | 423 | 24.3 | 1.7 | 0.391 | 0.0 |
| mei-4129Dc | 5 | 415 | 5.8 | 0.0 | 0.058 | 0.0 |
| mei-4129D tws430c | 8 | 632 | 47.6 | 10.4 | 0.950 | 0.0 |
| rad511c | 7 | 840 | 0.4 | 0.0 | 0.004 | 0.0 |
| tws430 rad511e | 16 | 1088 | 24.2 | 3.4 | 0.413 | 0.0 |
| grp1c | 10 | 650 | 0.0 | 0.0 | 0.000 | 0.0 |
| grp1 tws430e | 29 | 1137 | 32.9 | 7.4 | 0.664 | 0.0 |
Includes cells with more than five CABs.
The CABs/cell frequencies have been calculated assuming that the cells with more than five CABs contained only five CABs, and are therefore an underestimation of the actual frequencies.
Normal wandering third larvae with comparable body and brain sizes.
Not significantly different in the Chi square test.
Small larvae dying at the late second/early third instar.
tws430 ku70 Ex8/ku70MT tws430 ku70Ex8 heterozygous larvae.
ku70-RNAi construct/actin-GAL4; tws430/tws430.
Search for suppressors of the CAB phenotype of tws mutants
To identify the potential suppressors/modifiers of the tws-dependent CAB phenotype, we constructed a series of double mutants carrying both tws430 and a mutation in another gene involved in DNA repair. For this analysis, we selected genes whose protein products have two characteristics: they are involved in DNA repair and have phosphorylated mammalian orthologs. To assess the phosphorylation state of these gene products we exploited the extant literature and the PhosphoSitePlus database (Hornbeck et al. 2015). The list of the genes selected for double mutant analysis and their main features are reported in Table 2.
Table 2. Loss-of-function phenotypes of Drosophila and orthologous mouse and human genes required for DDR and DSBs repair.
| Drosophila gene name | Human gene name | Protein functiona | Loss-of-function phenotype in flies | Loss-of-function phenotype in mice//human diseases | Referencesb |
|---|---|---|---|---|---|
| HisH2Av | H2AFX | Nucleosome component; repair of DSBs | Lethal; CABs | Viable; growth retardation; male infertility; CABs//None | van Daal and Elgin 1992; Celeste et al. 2002; Vernì and Cenci 2015 |
| tefu | ATM | Serine/threonine kinase, DDR, DNA damage checkpoint | Lethal; CABs and TFs | Viable; CABs//ataxia telangiectasia; CABs | Oikemus et al. 2004; Silva et al. 2004; Song et al. 2004; Ciapponi et al. 2006; Shiloh and Ziv 2013 |
| mei-41 | ATR | Serine/threonine kinase, DDR, DNA replication; DNA damage checkpoint | Viable, female sterile; recombination defective; mutagen sensitive; CABs | Lethal; CABs//Seckel syndrome | Baker et al. 1976; Gatti 1979; Hari et al. 1995; Laurençon et al. 2003; Johnson-Schlitz et al. 2007; Wei and Rong 2007; Maréchal and Zou 2013 |
| grp | CHEK1 | Serine/threonine kinase, DDR, DNA damage checkpoint | Viable and fertile | Lethal//none | Fogarty et al. 1997; Liu et al. 2000; Takai et al. 2000 |
| nbs | NBN (NBS1) | Component of the rad50-Mre11-Nbs complex; DNA repair | Lethal; CABs and TFs | Lethal; CABs//Nijmegen breakage syndrome; CABs | Zhu et al. 2001; Bi et al. 2005; Ciapponi et al. 2006; Oikemus et al. 2006; Stracker and Petrini 2011 |
| ku70/irpb | XRCC6 (Ku70) | Forms a complex with Ku80; binds DNA ends; DNA helicase activity; NHEJ repair | Viable; telomere maintenance | Viable; telomere maintenance//none | Melnikova et al. 2005; Johnson-Schlitz et al. 2007; Fell and Schild-Poulter 2015 |
| lig4 | LIG4 | DNA ligase; NHEJ repair | Viable and fertile | Lethal//LIG4 syndrome | Gorski et al. 2003; Johnson-Schlitz et al. 2007; Wei and Rong 2007; Woodbine et al. 2014 |
| mei-9 | ERCC4 (XFP) | Forms a complex with ERCC1; endonuclease activity; DNA repair | Viable and fertile; mutagen sensitive; recombination defective; CABs | Lethal//Fanconi anemia, xeroderma pigmentosum, Cockayne syndrome, XFE progeroid syndrome | Baker et al. 1976; Gatti 1979; Sekelsky et al. 1995; Johnson-Schlitz et al. 2007; Wei and Rong 2007; Manandhar et al. 2015 |
| mus301/spnC | HELQ | DNA helicase; DNA repair | Viable, female sterile; mutagen sensitive; CABs (this report) | Viable, subfertile//none | Boyd et al. 1981; Johnson-Schlitz et al. 2007; Wei and Rong 2007; Adelman et al. 2013 |
| Blm/mus309 | BLM | RecQ-like DNA helicase; DNA replication; HR repair | Viable, male and female sterile; mutagen sensitive; CABs | Lethal//Bloom syndrome; CABs; high SCE rate | Boyd et al. 1981; Kusano et al. 2001; Johnson-Schlitz et al. 2007; Croteau et al. 2014; Cenci et al. 2015 |
| brca2 | BRCA2 | Binds single-stranded DNA; binds RAD51; HR repair | Viable, female sterile; HR repair; CABs (this report) | Lethal//breast-ovarian cancer susceptibility; Fanconi anemia D1 | Klovstad et al. 2008; Prakash et al. 2015 |
| spnA/rad51 | RAD51 | Binds DNA; binds BRCA2; HR repair | Viable, female sterile; HR repair | Lethal//breast cancer susceptibility | Tsuzuki et al. 1996; Staeva-Vieira et al. 2003; Johnson-Schlitz et al. 2007; Wei and Rong 2007 |
| okr/rad54 | RAD54L | Binds DNA; helicase activity; HR repair | Viable, female sterile; HR repair; CABs (this report) | Viable//tumor susceptibility | Kooistra et al. 1997; Ghabrial et al. 1998; Johnson-Schlitz et al. 2007; Wei and Rong 2007; Mazin et al. 2010 |
SCE, sister chromatid exchange.
All protein products of the human genes are phosphorylated (see text).
For Drosophila genes we reported the main references; for mammalian genes, when possible, we cited reviews.
We examined double mutants for tws and His2Av, nbs, or grapes (grp), whose protein products have mammalian orthologs that are phosphorylated by ATM (Shiloh and Ziv 2013). Nbs is a component of the MRN complex which binds to DSBs and participates in ATM activation, thus acting upstream of the major DNA repair pathways (Williams et al. 2010); grp is the Drosophila homolog of the Chk1 kinase which plays a major role in the DNA damage checkpoint (Fogarty et al. 1997; Patil et al. 2013). We also characterized the mei-41 tws double mutant; the Drosophila Mei-41/ATR kinase is not only involved in DNA damage signaling but has also a direct role in DSB repair (Oikemus et al. 2006; LaRocque et al. 2007; Benna et al. 2010).
Moreover, we analyzed the interactions between mutations in tws and mutations in genes known to be involved in either the NHEJ or the HR DSB repair pathway. These pathways are evolutionarily conserved from flies to humans, and several Drosophila genes functioning in these pathways have been characterized (Johnson-Schlitz et al. 2007; Wei and Rong 2007). In NHEJ, the DNA ends flanking the break are directly ligated, but this repair process often results in the insertion or loss of nucleotides at the site of ligation (Khanna and Jackson 2001; Mehta and Haber 2014). In Drosophila, NHEJ exploits the activities of at least two conserved genes: ligase4 (lig4) and ku70, which encodes a subunit of the Ku70/Ku80 heterodimer (Johnson-Schlitz et al. 2007; Wei and Rong 2007). HR is based on recombination with homologous genomic sequences and is not mutagenic, leading to an accurate repair of the DSB (Khanna and Jackson 2001; Mehta and Haber 2014). The factors that mediate HR in flies include Blm/Mus309 (a recQ-like protein homologous to the human BLM helicase responsible for the Bloom syndrome), the Drosophila homolog of the human tumor suppressor BRCA2, the conserved recombinases Okra (Okr, homologous to Rad54), and Spindle A (SpnA, homologous to Rad51) (Johnson-Schlitz et al. 2007; Wei and Rong 2007; Klovstad et al. 2008).
Finally, we examined double mutants in tws and either mei-9 or mus301/spnC, which encode an endonuclease orthologous to human XPF and a helicase homologous to human HELQ, respectively (Sekelsky et al. 1995; McCaffrey et al. 2006). mei-9 and mus301/spnC have been implicated in the SSA repair of DSBs, but there are conflicting results on their involvement in this process (Johnson-Schlitz et al. 2007; Wei and Rong 2007).
The tws430 His2av810, okr17-1/okrA19-10 tws430, lig45 tws430, brca2 KG03961 tws430, brca2ko/brca256E tws430, and mus301D4 tws430 double mutants displayed an early lethality (before the second instar), preventing cytological analysis of larval brain chromosomes. All other double mutants were lethal but reached a larval stage that allowed preparation of brain squashes for CAB scoring. Specifically, nbs1 tws430, mei-9A1 tws430, tws430 rad511, and grp1 tws430 died at the late second instar/early third instar larval stage, while, mei-4129D tws430, and tws430 ku70 Ex8 reached the mature third instar larval stage (wandering stage). We emphasize that larvae of mei-4129D tws430 and tws430 ku70Ex8 double mutants used for CAB analysis were comparable in body and brain size to larvae of mei-4129D, tws430, or ku70Ex8 single mutants. The frequency and types of CABs observed in single and double mutants are shown in Table 1 and Table S2. All mutants showed similar types and relative frequencies of chromatid- and chromosome-type CAB patterns (Table S2), suggesting that the CABs were generated throughout all phases of the cell cycle. However, the double mutants displayed different frequencies of CABs in relation to the tws single mutants. nbs1 tws430, mei-9A1 tws430, tws430 rad511, and grp1 tws430 double mutants displayed CAB frequencies significantly lower than the single tws mutants. mei-4129D tws430 double mutants showed a CAB frequency comparable to that seen in tws single mutants. Most strikingly, in tws430 ku70Ex8 double mutants CAB formation was completely suppressed and doubly mutant brains exhibited the same CAB frequency as wild-type controls. We note that the ku70Ex8 mutation is most likely a null allele, as it carries a very large deletion of the gene coding sequence (Johnson-Schlitz et al. 2007). However, ku70Ex8 homozygotes are viable, indicating that ku70 is not an essential gene in Drosophila.
To confirm the epistatic relationship between mutations in ku70, which do not cause CABs, and mutations in tws, we first performed a rescue experiment. We generated a recombinant chromosome carrying ku70Ex8, tws430, and ku70+ insertion in region 65B; this insertion, designated as ku70MT, consists of a fosmid clone containing the ku70 gene fused with the 2xTY1-sGFP-V5-Pre-TEV-BLRP-3xFLAG multi-tag, as well as the nina, CG6723, Ranbp9, and mRpL40 genes (Sarov et al. 2016; see Materials and Methods). We then used this recombinant chromosome to generate ku70Ex8 tws430/ku70MT ku70Ex8 tws430 third instar larvae; these larvae displayed normal body and brain sizes and showed relatively high frequencies of CABs (0.16 per cell; Table 1) compared tws430 ku70Ex8 double mutants (0.003 per cell; Table 1). We next analyzed brain cells from tws430/tws430 larvae bearing a ku70 RNAi construct and an actin-GAL4 driver. These brains displayed a much lower frequency of CABs (0.027 per cell; Table1) than those of tws430 homozygotes (1.017 per cell; Table1). Collectively, these results confirm that mutations in ku70 are strictly epistatic to mutations in tws, indicating that the expression of a normal ku70 protein is essential for CAB formation in a tws mutant background.
Establishing epistasis relationships between the other couples of mutations (mei-4129D tws430, nbs1 tws430, mei-9A1 tws430, tws430 rad511, and grp1 tws430) is more difficult. Given that mei-41 has only a few CABs, it is not possible to determine whether the CAB frequency found in the mei-4129D tws430 double mutant is due to epistasis of tws over mei-41 or instead reflects an additive pattern of CABs. The relatively low CAB frequency observed in the double mutants bearing tws430 and either nbs1, grp1, mei-9A1, or rad511 would suggest epistasis of these mutations over tws430. However, it is likely that these doubly mutant larvae exhibit a relatively low CAB frequency because they die earlier (at the late second/early third instar stage) than tws430 homozygous larvae. It has been shown that mothers heterozygous for a mutation in a gene specifying an essential cell cycle function accumulate into the egg a substantial amount of wild-type product, which is progressively depleted during development but is usually sufficient to allow larvae to reach the third instar. As a result, young larvae contain a higher amount of wild-type product than old larvae and thus exhibit a milder phenotype (Gatti and Baker 1989; Gatti and Goldberg 1991). We verified this notion by Western blot analysis, showing that in third instar larvae the level of the Tws protein is substantially lower than in first or second instar larvae (Figure S2). Thus, the apparent epistasis of nbs, grp, mei-9 and rad51 over tws might simply reflect the fact that double mutants have a higher level of wild-type Tws protein than tws single mutants and are therefore less subject to DNA damage.
The reason why several double mutants die early during development is unclear. His2av810 and tws430 homozygotes both die as third instar larvae, and it is thus understandable that the double mutant dies earlier than either single mutant. However, okr17-1/okrA19-10 and brca2ko/brca256E heterozygous flies, and flies homozygous for lig45, brca2KG03961, or mus301D4 are viable, suggesting that these mutations interact synergistically with the tws430 mutant allele leading to early death. To ascertain whether the early lethal phase of these double mutants is due to an intolerable level of chromosome breakage, we exploited RNAi-mediated gene silencing in S2 cells. For each couple of mutations resulting in early lethality in flies, we performed single RNAi against each of the genes they identify and double RNAi against both genes. We included Blm in this analysis because the close map position of Blm and tws prevented the construction of a double mutant by recombination. In all cases, we checked the efficiency of RNAi and found that target messenger RNAs were not detectable by RT-PCR (Figure S3). We anticipate that S2 cells are not an ideal system for the analysis of CABs because they exhibit a relatively high frequency of cells (∼12%) with spontaneous CABs (Figure 6 and Figure S4). The main purpose of our RNAi experiments was to discover possible synergistic interactions between genes in CAB formation. S2 cells allow detection of such interactions, as they manage to undergo mitosis even in the presence of an extremely high frequency of CABs (Somma et al. 2008).
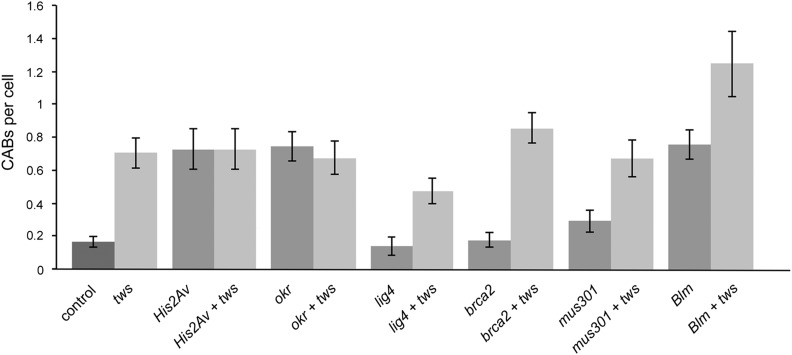
Figure 6.
Frequencies of CABs observed in S2 cells after RNAi against the indicated genes. At least 100 cells examined for each RNAi or double RNAi experiment; error bars indicate SEM. The CAB frequency observed in double Blm tws RNAi cells is significantly higher (P < 0.01) than that seen in tws RNAi cells. In the other double RNAi cells, the CAB frequencies are not significantly different from that detected after RNAi against tws only.
A comparison of the CAB frequencies induced by RNAi against His2Av, okr, lig4, brca2, mus301, or Blm with those elicited by double RNAi against tws and each of these genes did not reveal synergistic effects leading to extensive chromosome damage (Figure 6), suggesting that the embryonic lethality observed in the corresponding double mutants is not the consequence of an intolerable CAB level. RNAi against lig4, brca2, or mus301 did not cause a CAB increase compared to control, and double RNAi against tws and each of these genes resulted in a CAB frequency similar to that seen in tws RNAi cells (Figure 6); indicating that lig4, brca2, and mus301 are not involved in the pathway leading to CABs in Tws-depleted cells. RNAi against His2av or okr resulted in a substantial increase of CABs compared to control; His2av tws and okr tws double RNAi cells displayed CAB frequencies similar to those observed after RNAi against single genes, suggesting that His2av, okr, and tws function in the same CAB formation pathway. Finally, Blm tws double RNAi cells showed an additive CAB pattern with respect to Blm and tws RNAi cells (Figure 6), suggesting that Blm and tws function in parallel pathways leading to CABs.
Discussion
It is generally accepted that DNA DSBs are the lesions that lead to CAB formation and that the CAB frequency is a reliable measure of DNA damage (Obe et al. 2002; Durante et al. 2013). DSBs can be generated by the exposure to a variety of exogenous physical and chemical mutagens or by the action of endogenous agents such as viruses, reactive oxygen species, and mutations in genes required for proper DNA metabolism (Khanna and Jackson 2001; Mehta and Haber 2014). Many studies have established that CABs can promote carcinogenesis, and CABs are therefore considered as one of the main hallmarks of cancer (Kasparek and Humphrey 2011). However, despite their involvement in tumorigenesis, the mechanisms underlying CAB formation are still ill-defined. We believe that our results on Drosophila PP2A/B55 provide substantial insight into the role of phosphatases in the maintenance of genome integrity, suggesting possible mechanisms through which loss of phosphatase activity can lead to CABs.
Previous studies on Drosophila Tws focused on the role of this PP2A subunit in mitotic division (Gomes et al. 1993; Mayer-Jaekel et al. 1993; Chen et al. 2007; Brownlee et al. 2011; Wang et al. 2011; Kim et al. 2012). Here, we have shown that Tws is also required for the maintenance of chromosome integrity, and that tws mutants are threefold more sensitive than wild type to CAB induction by either X ray or HU. In addition, we have shown that Tws/PP2A is required for γ-H2Av dephosphorylation and DNA repair foci regression, and that it is involved in the DNA damage G2/M checkpoint. All these roles of PP2A have been previously described in mammalian cells (Chowdhury et al. 2005, 2008; Wang et al. 2009; Yan et al. 2010; Kalev et al. 2012) but have never been demonstrated in Drosophila. Thus, our results strongly suggest that Tws/PP2A serves a highly conserved function in the maintenance of genome integrity, and that Drosophila is an excellent model system for the analysis of this function.
Previous work in mammalian cells has shown that loss of PP2A leads to DSBs and CABs. However, there are at least three different hypotheses on the mechanisms leading to DNA damage in PP2A deficient cells. It has been proposed that PP2A depletion leads to an abnormal persistence of γ-H2AX foci impairing normal DSB repair (Chowdhury et al. 2005, 2008). It has been also suggested that PP2A downregulation inhibits the HR pathway by decreasing the expression of both BRCA1 and RAD51 (Kalev et al. 2012). Other studies have shown that the PP2A C catalytic subunit associates with Ku and promotes its dephosphorylation, and that PP2A C overexpression accelerates DSB repair in wild-type cells but not in Ku-depleted cells. These results have led to the hypothesis that PP2A-mediated Ku dephosphorylation activates Ku, promoting DSB repair and preventing CAB formation (Wang et al. 2009). However, it has been also reported that a specific phosphorylated form of Ku70 (pS27-S33 Ku70), which is overexpressed in aggressive forms of chronic lymphocytic leukemia, contributes to a faster, error-prone DNA repair process that results in a high CAB level. In addition, this study suggested that both DNA PK and ATM redundantly mediate S27 Ku70 phosphorylation (Bouley et al. 2015).
Our analysis of γ-H2Av foci regression in tws and Pp4-29C mutants argues against the possibility that a simple delay in foci dissolution can lead to CAB formation. We have also shown that rad51 null mutations do not cause CABs, ruling out the possibility that tws mutations lead to DNA damage through disruption of the rad51 function. Finally, our observation that null mutations in ku70 do not exhibit CABs exclude the possibility that tws mutations lead to CABs via inhibition of Ku activity. We found that mutations in both tefu (ATM) and ku70 are perfectly epistatic over tws mutations for the CAB phenotype. Specifically, we have shown that tws tefu double mutants exhibit the same chromosomal phenotype as tefu single mutants, namely a low CAB frequency (0.12 vs. 1.0 CABs/cell in tws mutants) and frequent TFs (0.5 per cell), which are absent in tws mutants. Most strikingly, we have shown that tws ku70 double mutants exhibit the same CAB frequency as ku70 single mutants, which is comparable to the wild-type frequency (∼0.005 per cell). One of the most straightforward explanations for these epistatic relationships is that Ku70 is a substrate of ATM and that, in the absence of the tws function, Ku70 is not properly dephosphorylated and as such interferes with the normal repair processes leading to DSBs and CABs.
A mutagenic activity of phosphorylated Drosophila Ku70 is consistent with the finding that a phosphorylated Ku70 form (pS27-S33 Ku70) leads to CAB formation in human cells by affecting the NHEJ DNA repair process (Bouley et al. 2015). However, we would like to point out that Drosophila Ku70 has only a limited homology with its human counterpart (only 21% identity) and that the “mutagenic” phosphorylation sites found in human Ku70 (S27 and S33) are not conserved in the fly protein. Thus, the mechanisms underlying the possible mutagenic activity of Drosophila phospho-Ku70 remain to be defined. At the moment, we can only speculate on these mechanisms, which could also involve Ku80, as in mammalian cells Ku70 and Ku80 form a binary complex and are mutually dependent for their stability (reviewed in Fell and Schild-Poulter 2015). The Ku complex binds the double-stranded DNA ends, forming a ring that encircles DNA and bridges the broken ends making them compatible. After end ligation, Ku keeps encircling repaired linear DNA and it is eventually removed. It is currently unclear whether Ku is removed through ubiquitin-mediated degradation or through direct DNA nicking which allows Ku to escape from the topological trap (Fell and Schild-Poulter 2015). In this scenario, it is not difficult to conceive that an abnormally persisting post-translational modification of Ku can affect its behavior and leads to DNA damage.
To the best of our knowledge, there are no examples in the literature of an improperly phosphorylated protein that acts as a mutagen. However, previous studies on DNA repair have provided examples in which the presence of an abnormal protein is more harmful than its absence. For example, the expression of catalytically inactive ATM in mice causes a stronger genomic instability and is more detrimental than ATM loss (Daniel et al. 2012; Yamamoto et al. 2012). Similarly, while loss of Topoisomerase II does not activate a G2/M checkpoint, catalytically inactive forms of this enzyme interrupt the DNA decatenation process and trigger a G2 arrest (Luo et al. 2009; Furniss et al. 2013).
In summary, our genetic analyses have shown that loss-of-function mutations in the tws gene lead to CABs in a Ku-dependent manner, raising the possibility that failure to dephosphorylate Ku leads to DNA damage. Identification of the putative mutagenic phospho-forms of Ku70, and possibly of Ku80, and definition of the mechanisms underlying their effects on DNA metabolism will be the goal for future studies.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.192781/-/DC1.
Acknowledgments
We thank T. Uemura for the anti-Tws antibody, and T. Schüpbach, W. Engels, and M. Goldberg for the brca2, ku70, and tws mutant alleles. C.M. was the recipient of a fellowship from Istituto-Pasteur Fondazione Cenci Bolognetti. This work was supported in part by a grant from Associazione Italiana per la Ricerca sul Cancro (IG 16020) to M.G.
Footnotes
Communicating editor: R. J. Duronio
Literature Cited
- Adelman C. A., Lolo R. L., Birkbak N. J., Murina O., Matsuzaki K., et al. , 2013. HELQ promotes RAD51 paralogue-dependent repair to avert germ cell loss and tumorigenesis. Nature 502: 381–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. S., Boyd J. B., Carpenter A. T., Green M. M., Nguyen T. D., et al. , 1976. Genetic controls of meiotic recombination and somatic DNA metabolism in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 73: 4140–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S., Mailand N., 2010. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst.) 9: 1219–1228. [DOI] [PubMed] [Google Scholar]
- Benna C., Bonaccorsi S., Wulbeck C., Helfrich-Forster C., Gatti M., et al. , 2010. Drosophila timeless2 is required for chromosome stability and circadian photoreception. Curr. Biol. 20: 346–352. [DOI] [PubMed] [Google Scholar]
- Bi X., Srikanta D., Fanti L., Pimpinelli S., Badugu R., et al. , 2005. Drosophila ATM and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proc. Natl. Acad. Sci. USA 102: 15167–15172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi S., Giansanti M. G., Gatti M., 2000. Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nat. Cell Biol. 2: 54–56. [DOI] [PubMed] [Google Scholar]
- Bouley J., Saad L., Grall R., Schellenbauer A., Biard D., et al. , 2015. A new phosphorylated form of Ku70 identified in resistant leukemic cells confers fast but unfaithful DNA repair in cancer cell lines. Oncotarget 6: 27980–28000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd J. B., Golino M. D., Shaw K. E., Osgood C. J., Green M. M., 1981. Third-chromosome mutagen-sensitive mutants of Drosophila melanogaster. Genetics 97: 607–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee C. W., Klebba J. E., Buster D. W., Rogers G. C., 2011. The Protein Phosphatase 2A regulatory subunit Twins stabilizes Plk4 to induce centriole amplification. J. Cell Biol. 195: 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burma S., Chen B. P., Murphy M., Kurimasa A., Chen D. J., 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276: 42462–42467. [DOI] [PubMed] [Google Scholar]
- Celeste A., Petersen S., Romanienko P. J., Fernandez-Capetillo O., Chen H. T., et al. , 2002. Genomic instability in mice lacking histone H2AX. Science 296: 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci G., Ciapponi L., Marzullo M., Raffa G. D., Morciano P., et al. , 2015. The analysis of pendolino (peo) mutants reveals differences in the fusigenic potential among Drosophila telomeres. PLoS Genet. 11: e1005260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Archambault V., Kar A., Lio P., D’Avino P. P., et al. , 2007. Multiple protein phosphatases are required for mitosis in Drosophila. Curr. Biol. 17: 293–303. [DOI] [PubMed] [Google Scholar]
- Chowdhury D., Keogh M. C., Ishii H., Peterson C. L., Buratowski S., et al. , 2005. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell 20: 801–809. [DOI] [PubMed] [Google Scholar]
- Chowdhury D., Xu X., Zhong X., Ahmed F., Zhong J., et al. , 2008. A PP4-phosphatase complex dephosphorylates gamma-H2AX generated during DNA replication. Mol. Cell 31: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciapponi L., Cenci G., Gatti M., 2006. The Drosophila Nbs protein functions in multiple pathways for the maintenance of genome stability. Genetics 173: 1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau D. L., Popuri V., Opresko P. L., Bohr V. A., 2014. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 83: 519–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J. A., Pellegrini M., Lee B. S., Guo Z., Filsuf D., et al. , 2012. Loss of ATM kinase activity leads to embryonic lethality in mice. J. Cell Biol. 198: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante M., Bedford J. S., Chen D. J., Conrad S., Cornforth M. N., et al. , 2013. From DNA damage to chromosome aberrations: joining the break. Mutat. Res. 756: 5–13. [DOI] [PubMed] [Google Scholar]
- Eichhorn P. J., Creyghton M. P., Bernards R., 2009. Protein phosphatase 2A regulatory subunits and cancer. Biochim. Biophys. Acta 1795: 1–15. [DOI] [PubMed] [Google Scholar]
- Fell V. L., Schild-Poulter C., 2015. The Ku heterodimer: function in DNA repair and beyond. Mutat. Res. Rev. Mutat. Res. 763: 15–29. [DOI] [PubMed] [Google Scholar]
- Fogarty P., Campbell S. D., Abu-Shumays R., Phalle B. S., Yu K. R., et al. , 1997. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr. Biol. 7: 418–426. [DOI] [PubMed] [Google Scholar]
- Foley E. A., Maldonado M., Kapoor T. M., 2011. Formation of stable attachments between kinetochores and microtubules depends on the B56–PP2A phosphatase. Nat. Cell Biol. 13: 1265–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman A. K., Monteiro A. N., 2010. Phosphatases in the cellular response to DNA damage. Cell Commun. Signal. 8: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furniss K. L., Tsai H. J., Byl J. A., Lane A. B., Vas A. C., et al. , 2013. Direct monitoring of the strand passage reaction of DNA topoisomerase II triggers checkpoint activation. PLoS Genet. 9: e1003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M., 1979. Genetic control of chromosome breakage and rejoining in Drosophila melanogaster: spontaneous chromosome aberrations in X-linked mutants defective in DNA metabolism. Proc. Natl. Acad. Sci. USA 76: 1377–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M., Baker B. S., 1989. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 3: 438–453. [DOI] [PubMed] [Google Scholar]
- Gatti M., Goldberg M. L., 1991. Mutations affecting cell division in Drosophila. Methods Cell Biol. 35: 543–586. [DOI] [PubMed] [Google Scholar]
- Gatti M., Tanzarella C., Olivieri G., 1974. Analysis of the chromosome aberrations induced by x-rays in somatic cells of Drosophila melanogaster. Genetics 77: 701–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M., Pimpinelli S., Baker B. S., 1980. Relationships among chromatid interchanges, sister chromatid exchanges, and meiotic recombination in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 77: 1575–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial A., Ray R. P., Schupbach T., 1998. okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev. 12: 2711–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti M. G., Bonaccorsi S., Kurek R., Farkas R. M., Dimitri P., et al. , 2006. The class I PITP giotto is required for Drosophila cytokinesis. Curr. Biol. 16: 195–201. [DOI] [PubMed] [Google Scholar]
- Glover D. M., 2012. The overlooked greatwall: a new perspective on mitotic control. Open Biol. 2: 120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes R., Karess R. E., Ohkura H., Glover D. M., Sunkel C. E., 1993. Abnormal anaphase resolution (aar): a locus required for progression through mitosis in Drosophila. J. Cell Sci. 104: 583–593. [DOI] [PubMed] [Google Scholar]
- Goodarzi A. A., Jonnalagadda J. C., Douglas P., Young D., Ye R., et al. , 2004. Autophosphorylation of ataxia-telangiectasia mutated is regulated by protein phosphatase 2A. EMBO J. 23: 4451–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski M. M., Eeken J. C., de Jong A. W., Klink I., Loos M., et al. , 2003. The Drosophila melanogaster DNA Ligase IV gene plays a crucial role in the repair of radiation-induced DNA double-strand breaks and acts synergistically with Rad54. Genetics 165: 1929–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannus M., Feiguin F., Heisenberg C. P., Eaton S., 2002. Planar cell polarization requires Widerborst, a B′ regulatory subunit of protein phosphatase 2A. Development 129: 3493–3503. [DOI] [PubMed] [Google Scholar]
- Hari K. L., Santerre A., Sekelsky J. J., McKim K. S., Boyd J. B., et al. , 1995. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell 82: 815–821. [DOI] [PubMed] [Google Scholar]
- Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., et al. , 2015. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43: D512–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T., 2013. On the regulation of protein phosphatase 2A and its role in controlling entry into and exit from mitosis. Adv. Biol. Regul. 53: 173–178. [DOI] [PubMed] [Google Scholar]
- Ikura T., Tashiro S., Kakino A., Shima H., Jacob N., et al. , 2007. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell. Biol. 27: 7028–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V., Goris J., 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353: 417–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S., Shibata E., Dutta A., 2008. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol. Cell. Biol. 28: 2690–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Schlitz D. M., Flores C., Engels W. R., 2007. Multiple-pathway analysis of double-strand break repair mutations in Drosophila. PLoS Genet. 3: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalev P., Simicek M., Vazquez I., Munck S., Chen L., et al. , 2012. Loss of PPP2R2A inhibits homologous recombination DNA repair and predicts tumor sensitivity to PARP inhibition. Cancer Res. 72: 6414–6424. [DOI] [PubMed] [Google Scholar]
- Kasparek T. R., Humphrey T. C., 2011. DNA double-strand break repair pathways, chromosomal rearrangements and cancer. Semin. Cell Dev. Biol. 22: 886–897. [DOI] [PubMed] [Google Scholar]
- Keogh M. C., Kim J. A., Downey M., Fillingham J., Chowdhury D., et al. , 2006. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature 439: 497–501. [DOI] [PubMed] [Google Scholar]
- Khanna K. K., Jackson S. P., 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27: 247–254. [DOI] [PubMed] [Google Scholar]
- Khanna A., Pimanda J. E., Westermarck J., 2013. Cancerous inhibitor of protein phosphatase 2A, an emerging human oncoprotein and a potential cancer therapy target. Cancer Res. 73: 6548–6553. [DOI] [PubMed] [Google Scholar]
- Kim M. Y., Bucciarelli E., Morton D. G., Williams B. C., Blake-Hodek K., et al. , 2012. Bypassing the Greatwall-Endosulfine pathway: plasticity of a pivotal cell-cycle regulatory module in Drosophila melanogaster and Caenorhabditis elegans. Genetics 191: 1181–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima T. S., Sakuno T., Ishiguro K., Iemura S., Natsume T., et al. , 2006. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441: 46–52. [DOI] [PubMed] [Google Scholar]
- Klovstad M., Abdu U., Schupbach T., 2008. Drosophila brca2 is required for mitotic and meiotic DNA repair and efficient activation of the meiotic recombination checkpoint. PLoS Genet. 4: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra R., Vreeken K., Zonneveld J. B., de Jong A., Eeken J. C., et al. , 1997. The Drosophila melanogaster RAD54 homolog, DmRAD54, is involved in the repair of radiation damage and recombination. Mol. Cell. Biol. 17: 6097–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotadia S., Kao L. R., Comerford S. A., Jones R. T., Hammer R. E., et al. , 2008. PP2A-dependent disruption of centrosome replication and cytoskeleton organization in Drosophila by SV40 small tumor antigen. Oncogene 27: 6334–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano K., Johnson-Schlitz D. M., Engels W. R., 2001. Sterility of Drosophila with mutations in the Bloom syndrome gene–complementation by Ku70. Science 291: 2600–2602. [DOI] [PubMed] [Google Scholar]
- Kusch T., Florens L., Macdonald W. H., Swanson S. K., Glaser R. L., et al. , 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306: 2084–2087. [DOI] [PubMed] [Google Scholar]
- Lambrecht C., Haesen D., Sents W., Ivanova E., Janssens V., 2013. Structure, regulation, and pharmacological modulation of PP2A phosphatases. Methods Mol. Biol. 1053: 283–305. [DOI] [PubMed] [Google Scholar]
- Lankoff A., Bialczyk J., Dziga D., Carmichael W. W., Gradzka I., et al. , 2006. The repair of gamma-radiation-induced DNA damage is inhibited by microcystin-LR, the PP1 and PP2A phosphatase inhibitor. Mutagenesis 21: 83–90. [DOI] [PubMed] [Google Scholar]
- LaRocque J. R., Jaklevic B., Su T. T., Sekelsky J., 2007. Drosophila ATR in double-strand break repair. Genetics 175: 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattao R., Bonaccorsi S., Guan X., Wasserman S. A., Gatti M., 2011. Tubby-tagged balancers for the Drosophila X and second chromosomes. Fly (Austin) 5: 369–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurençon A., Purdy A., Sekelsky J., Hawley R. S., Su T. T., 2003. Phenotypic analysis of separation-of-function alleles of MEI-41, Drosophila ATM/ATR. Genetics 164: 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung-Pineda V., Ryan C. E., Piwnica-Worms H., 2006. Phosphorylation of Chk1 by ATR is antagonized by a Chk1-regulated protein phosphatase 2A circuit. Mol. Cell. Biol. 26: 7529–7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Guntuku S., Cui X. S., Matsuoka S., Cortez D., et al. , 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 14: 1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Lorca T., Castro A., 2013. The Greatwall kinase: a new pathway in the control of the cell cycle. Oncogene 32: 537–543. [DOI] [PubMed] [Google Scholar]
- Luo K., Yuan J., Chen J., Lou Z., 2009. Topoisomerase IIalpha controls the decatenation checkpoint. Nat. Cell Biol. 11: 204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan J. P., Chotkowski H. L., Glaser R. L., 2002. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 30: 3698–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar M., Boulware K. S., Wood R. D., 2015. The ERCC1 and ERCC4 (XPF) genes and gene products. Gene 569: 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal A., Zou L., 2013. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 5: a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Jaekel R. E., Baumgartner S., Bilbe G., Ohkura H., Glover D. M., et al. , 1992. Molecular cloning and developmental expression of the catalytic and 65-kDa regulatory subunits of protein phosphatase 2A in Drosophila. Mol. Biol. Cell 3: 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Jaekel R. E., Ohkura H., Gomes R., Sunkel C. E., Baumgartner S., et al. , 1993. The 55 kd regulatory subunit of Drosophila protein phosphatase 2A is required for anaphase. Cell 72: 621–633. [DOI] [PubMed] [Google Scholar]
- Mazin A. V., Mazina O. M., Bugreev D. V., Rossi M. J., 2010. Rad54, the motor of homologous recombination. DNA Repair (Amst.) 9: 286–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey R., St Johnston D., Gonzalez-Reyes A., 2006. Drosophila mus301/spindle-C encodes a helicase with an essential role in double-strand DNA break repair and meiotic progression. Genetics 174: 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A., Haber J. E., 2014. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 6: a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova L., Biessmann H., Georgiev P., 2005. The Ku protein complex is involved in length regulation of Drosophila telomeres. Genetics 170: 221–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengoli V., Bucciarelli E., Lattao R., Piergentili R., Gatti M., et al. , 2014. The analysis of mutant alleles of different strength reveals multiple functions of topoisomerase 2 in regulation of Drosophila chromosome structure. PLoS Genet. 10: e1004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada S., Chen G. I., Gingras A. C., Durocher D., 2008. PP4 is a gamma H2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep. 9: 1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obe G., Pfeiffer P., Savage J. R., Johannes C., Goedecke W., et al. , 2002. Chromosomal aberrations: formation, identification and distribution. Mutat. Res. 504: 17–36. [DOI] [PubMed] [Google Scholar]
- Oikemus S. R., McGinnis N., Queiroz-Machado J., Tukachinsky H., Takada S., et al. , 2004. Drosophila atm/telomere fusion is required for telomeric localization of HP1 and telomere position effect. Genes Dev. 18: 1850–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikemus S. R., Queiroz-Machado J., Lai K., McGinnis N., Sunkel C., et al. , 2006. Epigenetic telomere protection by Drosophila DNA damage response pathways. PLoS Genet. 2: e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E. J., Chan D. W., Park J. H., Oettinger M. A., Kwon J., 2003. DNA-PK is activated by nucleosomes and phosphorylates H2AX within the nucleosomes in an acetylation-dependent manner. Nucleic Acids Res. 31: 6819–6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil M., Pabla N., Dong Z., 2013. Checkpoint kinase 1 in DNA damage response and cell cycle regulation. Cell. Mol. Life Sci. 70: 4009–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotti D., Neviani P., 2013. Protein phosphatase 2A: a target for anticancer therapy. Lancet Oncol. 14: e229–e238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S. E., Jackson S. P., 2011. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25: 409–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter I. M., Schleicher K., Porter M., Swedlow J. R., 2013. Bod1 regulates protein phosphatase 2A at mitotic kinetochores. Nat. Commun. 4: 2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R., Zhang Y., Feng W., Jasin M., 2015. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 7: a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro P. S., Josue F., Wepf A., Wehr M. C., Rinner O., et al. , 2010. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol. Cell 39: 521–534. [DOI] [PubMed] [Google Scholar]
- Sarov M., Barz C., Jambor H., Hein M. Y., Schmied C., et al. , 2016. A genome-wide resource for the analysis of protein localisation in Drosophila. eLife 5: e12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J. J., McKim K. S., Chin G. M., Hawley R. S., 1995. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics 141: 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., 2009. Serine/threonine phosphatases: mechanism through structure. Cell 139: 468–484. [DOI] [PubMed] [Google Scholar]
- Shiloh Y., Ziv Y., 2013. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14: 197–210. [PubMed] [Google Scholar]
- Silva E., Tiong S., Pedersen M., Homola E., Royou A., et al. , 2004. ATM is required for telomere maintenance and chromosome stability during Drosophila development. Curr. Biol. 14: 1341–1347. [DOI] [PubMed] [Google Scholar]
- Snaith H. A., Armstrong C. G., Guo Y., Kaiser K., Cohen P. T., 1996. Deficiency of protein phosphatase 2A uncouples the nuclear and centrosome cycles and prevents attachment of microtubules to the kinetochore in Drosophila microtubule star (mts) embryos. J. Cell Sci. 109: 3001–3012. [DOI] [PubMed] [Google Scholar]
- So S., Davis A. J., Chen D. J., 2009. Autophosphorylation at serine 1981 stabilizes ATM at DNA damage sites. J. Cell Biol. 187: 977–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somma M. P., Fasulo B., Cenci G., Cundari E., Gatti M., 2002. Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol. Biol. Cell 13: 2448–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somma M. P., Ceprani F., Bucciarelli E., Naim V., De Arcangelis V., et al. , 2008. Identification of Drosophila mitotic genes by combining co-expression analysis and RNA interference. PLoS Genet. 4: e1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. H., Mirey G., Betson M., Haber D. A., Settleman J., 2004. The Drosophila ATM ortholog, dATM, mediates the response to ionizing radiation and to spontaneous DNA damage during development. Curr. Biol. 14: 1354–1359. [DOI] [PubMed] [Google Scholar]
- Staeva-Vieira E., Yoo S., Lehmann R., 2003. An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J. 22: 5863–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker T. H., Petrini J. H., 2011. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 12: 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan W., Fogarty P., Theurkauf W., 1993. Mutations affecting the cytoskeletal organization of syncytial Drosophila embryos. Development 118: 1245–1254. [DOI] [PubMed] [Google Scholar]
- Takai H., Tominaga K., Motoyama N., Minamishima Y. A., Nagahama H., et al. , 2000. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(−/−) mice. Genes Dev. 14: 1439–1447. [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Shu H., Qi W., Mahmood N. A., Mumby M. C., et al. , 2006. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev. Cell 10: 575–585. [DOI] [PubMed] [Google Scholar]
- Tournebize R., Andersen S. S., Verde F., Doree M., Karsenti E., et al. , 1997. Distinct roles of PP1 and PP2A-like phosphatases in control of microtubule dynamics during mitosis. EMBO J. 16: 5537–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T., Fujii Y., Sakumi K., Tominaga Y., Nakao K., et al. , 1996. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA 93: 6236–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Shiomi K., Togashi S., Takeichi M., 1993. Mutation of twins encoding a regulator of protein phosphatase 2A leads to pattern duplication in Drosophila imaginal discs. Genes Dev. 7: 429–440. [DOI] [PubMed] [Google Scholar]
- van Daal A., Elgin S. C., 1992. A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol. Biol. Cell 3: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernì F., Cenci G., 2015. The Drosophila histone variant H2A.V works in concert with HP1 to promote kinetochore-driven microtubule formation. Cell Cycle 14: 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viquez N. M., Li C. R., Wairkar Y. P., DiAntonio A., 2006. The B′ protein phosphatase 2A regulatory subunit well-rounded regulates synaptic growth and cytoskeletal stability at the Drosophila neuromuscular junction. J. Neurosci. 26: 9293–9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Pinson X., Archambault V., 2011. PP2A-twins is antagonized by greatwall and collaborates with polo for cell cycle progression and centrosome attachment to nuclei in drosophila embryos. PLoS Genet. 7: e1002227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Gao F., Wang T., Flagg T., Deng X., 2009. A nonhomologous end-joining pathway is required for protein phosphatase 2A promotion of DNA double-strand break repair. Neoplasia 11: 1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I. M., Chen J., 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276: 47759–47762. [DOI] [PubMed] [Google Scholar]
- Wei D. S., Rong Y. S., 2007. A genetic screen for DNA double-strand break repair mutations in Drosophila. Genetics 177: 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B. C., Filter J. J., Blake-Hodek K. A., Wadzinski B. E., Fuda N. J., et al. , 2014. Greatwall-phosphorylated Endosulfine is both an inhibitor and a substrate of PP2A–B55 heterotrimers. eLife 3: e01695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. J., Lees-Miller S. P., Tainer J. A., 2010. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst.) 9: 1299–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbine L., Gennery A. R., Jeggo P. A., 2014. Reprint of “The clinical impact of deficiency in DNA non-homologous end-joining”. DNA Repair (Amst.) 17: 9–20. [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Wang Y., Jiang W., Liu X., Dubois R. L., et al. , 2012. Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice. J. Cell Biol. 198: 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Cao P. T., Greer P. M., Nagengast E. S., Kolb R. H., et al. , 2010. Protein phosphatase 2A has an essential role in the activation of gamma-irradiation-induced G2/M checkpoint response. Oncogene 29: 4317–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasutis K. M., Kozminski K. G., 2013. Cell cycle checkpoint regulators reach a zillion. Cell Cycle 12: 1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Petersen S., Tessarollo L., Nussenzweig A., 2001. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 11: 105–109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to confirm the results presented in this study are included in the main article and in the supplemental material. Fly strains are available upon request.