Abstract
Bacterial small non-coding RNAs (sRNAs) are known as novel regulators involved in virulence, stress responsibility, and so on. Recently, a lot of new researches have highlighted the critical roles of sRNAs in fine-tune gene regulation in both prokaryotes and eukaryotes. Edwardsiella tarda (E. tarda) is a gram-negative, intracellular pathogen that causes edwardsiellosis in fish. Thus far, no sRNA has been reported in E. tarda. The present study represents the first attempt to identify sRNAs in E. tarda S08. Ten sRNAs were validated by RNA sequencing and quantitative PCR (qPCR). ET_sRNA_1 and ET_sRNA_2 were homolous to tmRNA and GcvB, respectively. However, the other candidate sRNAs have not been reported till now. The cellular abundance of 10 validated sRNA was detected by qPCR at different growth phases to monitor their biosynthesis. Nine candidate sRNAs were expressed in the late-stage of exponential growth and stationary stages of growth (36~60 h). And the expression of the nine sRNAs was growth phase-dependent. But ET_sRNA_10 was almost expressed all the time and reached the highest peak at 48 h. Their targets were predicted by TargetRNA2 and each sRNA target contains some genes that directly or indirectly relate to virulence. These results preliminary showed that sRNAs probably play a regulatory role of virulence in E. tarda.
Introduction
Edwardsiella tarda (E. tarda) is a common and important pathogen of freshwater and marine fish, which causes enormous economic losses to the world-wide aquaculture industry. The pathogenesis of E. tarda has been studied for a long time and the virulence factors include type III and type VI secretion systems (T3SS and T6SS) [1, 2], chondroitinase [3], nucleoid-associated protein [4], catalase [5], hemolysins [6, 7], flagella [8, 9], adhesion [10], sigma factors RpoN and RpoS [11] and quorum sensing [12, 13].
But the fundamental pathogenic mechanism of E. tarda still remains to be discovered. In recent years, some significant experimental and theoretical evidence suggested that small non-coding RNAs (sRNAs) could coordinate virulence gene regulations and pathogen survival during infecting the host [14–17]. At the same time, sRNAs are crucial players of regulatory cascades, coordinating the expression of virulence genes in response to environmental or other changes [16, 17]. They are able to adapt the expression of virulence genes to stress and metabolic requirements [17]. These sRNAs function either directly on virulence genes and/or on regulators of virulence genes [16].
While sRNAs have been well known for some time and some examples have been confirmed in Escherichia coli and other pathogenic bacteria [18–22], our knowledge of the networks involving sRNAs and controlling pathogenesis in E. tarda is still in its infancy. Here, we systematically identify sRNAs in E. tarda genome by RNA sequencing and bioinformatics prediction for the first time. Then, the cellular abundance of validated sRNA was detected by quantitative PCR (qPCR) at different growth phases to monitor their biosynthesis. In addition, the potential targets of sRNAs were also predicted by bioinformatics analysis. Our results will provide insight into the knowledge of virulence regulation of E. tarda and pave the way for eradicating edwardsiellosis.
Materials and methods
Ethics statement
E. tarda S08 (Accession no. KX279865) was isolated from diseased turbot. Disease outbreaks occurred on some marine turbot farms in Qingdao, China. The farm owners hoped us to determine the causative agents of these outbreaks and assess potential therapies for the treatment of these infections. So they provided a large number of diseased turbot to us for the study. This experiment as described was carried out in strict accordance with the approval of the Animal Care and Use Committee of the Institute of Oceanology, Chinese Academy of Sciences.
Bacterial strains and growth conditions
E. tarda S08 isolated from diseased turbot was used for most experiments. The strain was routinely cultured in Tryptic Soy Broth (TSB, Difco) or TSA medium supplemented with additional 1% NaCl at 28°C, 180 rpm. Colistin was added at a final concentration with 12.5 μg/mL when necessary. The growth in the TSB was determined by spectrophotometric values (OD540 nm) at the interval of 2 h. Then, the growth curve was plotted using optical density against time points (2 h, 4 h, ……, 72 h). While the cultures of series of time points at the interval of 6 h were collected for the next step experiments. All the samples were run in triplicate.
In silico prediction of sRNAs
The genome sequences of E. tarda S08 (data unpublished) and E. tarda EBI202 (Accession no. CP002154.1) were chosen for in silico prediction. The computational methods were applied for the prediction of sRNAs including sRNAscanner and sRNAPredict3. sRNAPredict3 identified sRNAs based on intergenic conservation and Rho-independent terminators in the closely related bacterial genomes. sRNAscanner computes the locations of the intergenic signals using the Positional Weight Matrix (PWM) strategy for the search of intergenic sRNAs. All the parameters were set as the default analytical criteria for the two methods.
sRNA extraction and RNA sequencing
E. tarda S08 was grown in TSB medium at 28°C and harvested with centrifugation (at 6, 000×g for 5 min) at the series of time points. Finally, all the samples from different time points were mixed together at equal volumes. The sRNAs were isolated from cell pellets with bacterial small RNA isolation kit (OMEGA, USA). All RNA was treated with RNase free DNase I and library was built for Illumina Hiseq 2000 platform with library constructions kit following the manufacturer’s protocol.
Promoter prediction and in silico validation of predicted sRNAs
The program BPROM was used to predict the promoters of the bacterial sRNAs. The promoter prediction was conducted to search 200 bp upstream of the sRNA start site. RNAfold program was used to carry out the secondary structure prediction based on the lowest folding energy. The sRNAs were blasted into Rfam database to assess the novelty.
Quantitative PCR assays
Total RNA was extracted using Trizol reagent (Life tech, USA) and then reverse transcribed using oligo dT and random mix primers (ToYoBo, Japan) according to the manufacturer’s protocol. Quantitative PCR was performed to validate the reliability of predicted sRNAs and check the expression abundance of the validated sRNAs at the different growth phages. The qPCR primer pairs for the 10 candidate sRNAs were designed using Primer Premier 6.0. 16 S ribosomal RNA gene was used as internal control for normalization of gene expression. Quantitative PCR was run on Bio-Rad CFX (USA) with initial denaturation of 3 min at 95°C and a subsequent run of 40 cycles each comprising 10 s at 95°C, 10 s at 62°C, and melt curve was performed to assess the primer specificity. The samples were run in triplicate. The 2-ΔΔCq method (relative quantization) was used in which Cq value (threshold cycle) was normalized to endogenous reference gene 16S (ΔCq = Cqtarget—Cqreference) [23]. Using student’s t test, data were considered statistically significant when p < 0.05.
Target prediction of validated sRNAs
Web-based program TargetRNA2 was used to predict the target genes for each validated sRNA. TargetRNA2 considers each mRNA in the replicon as a possible target of the sRNA. 80 bp before the start codon and 20 bp after the start codon were searched. After searching all mRNAs in the specified replicon for interactions with the sRNA, TargetRNA2 outputs a list of likely regulatory targets ranked by p-value.
Results
Bacterial growth condition of E. tarda S08
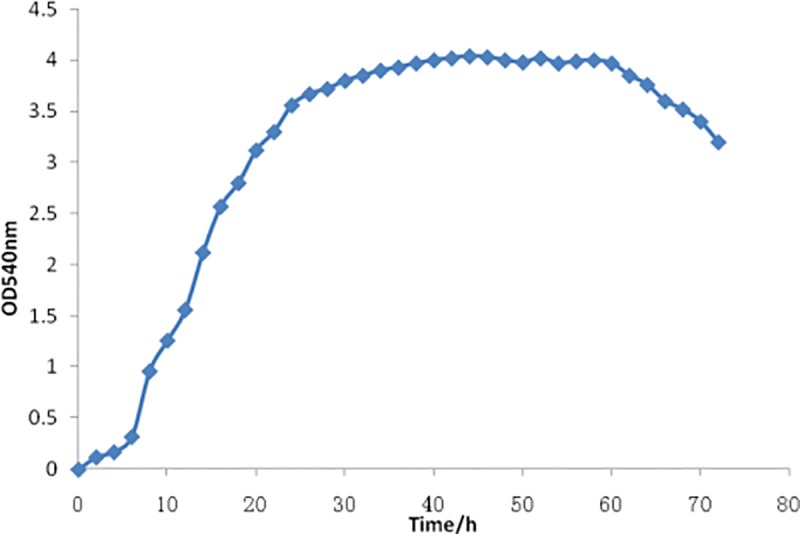
E. tarda S08 was cultured in TSB medium at 28°C, 180 rpm. The OD540nm value was monitored at the interval of 2 h and the growth curve was plotted (Fig 1). After 24 h, the strain was showed to grow into post-exponential phage and after 40 h into stationary phage. It entered into decline phase after 60 h.
Fig 1. Growth curve of E. tarda S08.
Bioinformatic prediction of sRNAs and RNA sequencing
Two computational methods were used to predict the sRNAs and the comparative results were provided as follows (Table 1). After aligned the results, a total of 10 sRNA candidates were predicted (>100 bp in length). Genomic location and the orientations of sRNAs were also analyzed. Table 2 categorized a detailed description of the candidate sRNAs.
Table 1. The statistic results of predicted sRNAs.
| Method | No. of prediction | Average length | Max length | Min length | CRISPR |
|---|---|---|---|---|---|
| sRNAPredict3 | 111 | 156 bp | 363 bp | 66 bp | 1 |
| sRNAscanner | 134 | 234 bp | 560 bp | 34 bp | - |
| RNA sequencing | 2668 | 83 bp | 150 bp | 50 bp | - |
Table 2. The feature description of 10 validated sRNAs.
| sRNA name | Start position | End position | SRNA length(bp) | Orientations | Up gene name | Down gene name |
|---|---|---|---|---|---|---|
| ET_sRNA_1 | 2877375 | 2877738 | 364 | + | small protein B, tmRNA-binding protein | putative integrase |
| ET_sRNA_2 | 797781 | 797578 | 204 | + | cysteine sulfinate desulfinase | DNA-binding transcriptional activator GcvA |
| ET_sRNA_3 | 2731396 | 2731835 | 440 | - | hypothetical protein | potassium-transporting ATPase subunit A |
| ET_sRNA_4 | 3081498 | 3081919 | 422 | + | lipid A biosynthesis palmitoleoyl acyltransferase | outer membrane lipoprotein |
| ET_sRNA_5 | 1285062 | 1285378 | 317 | + | putative DNA-binding transcriptional regulator | lysine transporter |
| ET_sRNA_6 | 1726729 | 1727058 | 330 | + | phage-related protein | hypothetical protein |
| ET_sRNA_7 | 1931296 | 1931693 | 398 | + | hypothetical protein | hypothetical protein |
| ET_sRNA_8 | 3480759 | 3481235 | 477 | - | 4-alpha-glucanotransferase | glucose-1-phosphate adenylyltransferase |
| ET_sRNA_9 | 284289 | 284627 | 339 | - | putative tartrate:succinate antiporter | hypothetical protein |
| ET_sRNA_10 | 1443879 | 1444365 | 487 | + | hypothetical protein | transcriptional activator |
| ET_sRNA_16s-internal | 3710745 | 3712281 | 376 | - | tRNA-Glu | putative GntR-famly transcriptional regulator |
Promoter and second structure analysis of candidate sRNAs
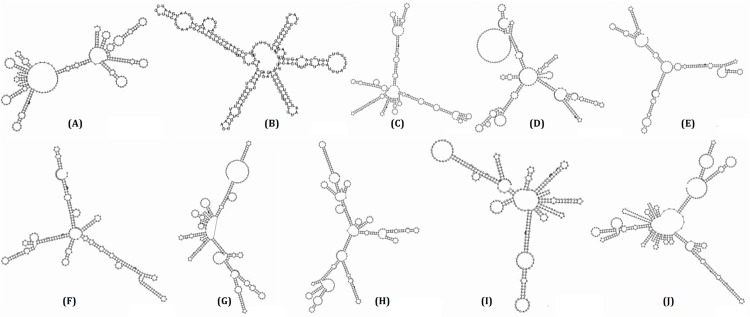
The web-based program, BPRORM, was implemented to perform the promoter analysis. By searching 200 bp upstream of the candidate sRNA start site for the -10 box and -35 box, the results showed that all 10 candidate sRNAs were successfully found the -10 and -35 promoter sites and corresponding TF binding sites. The average distance for the -10 box and -35 box were 53 and 76 bp upstream of the candidate sRNAs, respectively. Secondary structure analysis were carried out using RNAfold program and depicted in Fig 2. Next, the 10 candidate sRNAs were undergone to blast against Rfam database for the novelty. Two of 10 candidate sRNAs, named ET_sRNA_1 and ET_sRNA_2 (homologues to tmRNA and GcvB), showed the homology in Rfam. While the other candidate sRNAs were first found. The sequence of 10 sRNAs genes was analyzed for terminator prediction. Rho-independent terminators were predicted at the 3' end using ARNold (S1 File).
Fig 2. Second structure of ET_sRNA_1~ ET_sRNA_10.
(A) ET_sRNA_1 (B) ET_sRNA_2 (C) ET_sRNA_3 (D) ET_sRNA_4 (E) ET_sRNA_5 (F) ET_sRNA_6 (G) ET_sRNA_7 (H) ET_sRNA_8 (I) ET_sRNA_9 (J) ET_sRNA_10.
Experimental validation by qPCR assays under different growth phage
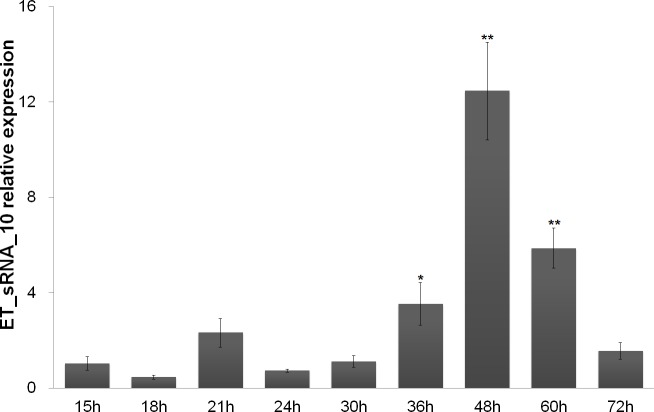
Further experimental validation was performed for the 10 candidate sRNAs. The qPCR primer sequences used for sRNA genes were listed in Table 3. The total RNA was extracted from different time points and reverse transcribed. The cDNAs were used as the templates for qPCR to assess the expression of candidate sRNAs. ET_sRNA_10 was almost expressed all the time and reached the highest peak at 48 h (Fig 3). However, the other nine sRNAs were expressed in the late-stage of exponential growth and stationary stages of growth (36~60 h). And their transcript level reached the highest point at the final phase of stationary growth (60 h) (S1–S9 Figs). This showed that the expression of the nine sRNAs was growth phase-dependent.
Table 3. The qPCR primer sequences used for sRNA genes.
| sRNA name | Primers used for qPCR |
|---|---|
| ET_sRNA_1 | for: 5’actacgcactcgcagcttaataac |
| rev: 5’cggacagacacgccactaaca | |
| ET_sRNA_2 | for: 5’agacatggcggtggcgtaag |
| rev: 5’actaaatcactatggacagacagggta | |
| ET_sRNA_3 | for: 5’gcgatagaggacagcaacgataatg |
| rev: 5’aaccaacaggagtagcaccagtac | |
| ET_sRNA_4 | for: 5’ttacagcatgaagcatcggtcatagaa |
| rev: 5’gacggtgagtgagaggaagaggaa | |
| ET_sRNA_5 | for: 5’actcgctaataatccgccaaccatc |
| rev: 5’tttgtctgagccattagaaccctatcg | |
| ET_sRNA_6 | for: 5’cgacctcaagccgaacctcttc |
| rev: 5’atgttgccgctgccactacg | |
| ET_sRNA_7 | for: 5’accgctggagattccgctatgt |
| rev: 5’tgctacaactcactgccgtcac | |
| ET_sRNA_8 | for: 5’cgctacccgtttattccagcatcc |
| rev: 5’cgcctgtcatccgcaacaaca | |
| ET_sRNA_9 | for: 5’catcaggatggtggttctgagtca |
| rev: 5’cgccctctttaagtattcccattcaac | |
| ET_sRNA_10 | for: 5’cgctgatggatattccgccgatg |
| rev: 5’tggtgcttccctctgaacgatagtaa |
Fig 3. Quantitative PCR detection the transcript levels of ET_sRNA_10 under different growth phases.
Statistical significance (*p<0.05;**p<0.01) was obtained using ANOVA test.
Target prediction of validated sRNAs
Accurate prediction of sRNA targets plays an important role in studying sRNA function. The targets of 10 sRNAs were predicted by TargetRNA2 (S2 File). TargetRNA2 outputs a list of likely regulatory targets ranked by p-value (p≤0.05). A total of 385 potential targets were identified. We parsed the predicted mRNA targets based on their respective protein function (Table 4) [24]. Our result demonstrated that the majority of known targets for sRNAs were involved in metabolism (114), virulence (59), and transport (35). However, a large number of target genes were categorized as ‘other’ (49) and ‘hypothetical proteins’ (115), respectively (Table 4). Each sRNA targets contain a number of genes that directly or indirectly relate to virulence. The result preliminary shows that sRNAs probably play regulatory roles of virulence. Of course, the related work is being verified by experiments.
Table 4. sRNA target categorization.
| Target classification | Number of predicted targets by category |
|---|---|
| Cell division | 3 |
| Cell wall | 5 |
| Metabolism | 114 |
| Ribosomal protein | 3 |
| Virulence | 59 |
| Other | 49 |
| Transport | 35 |
| Hypothetical protein | 115 |
| T3SS | 1 |
| T6SS | 1 |
| Total | 385 |
Target genes are classified into ten categories based on either known or hypothetical function for E. tarda.
Discussion
E. tarda is associated with edwardsiellosis in cultured fish, resulting in heavy losses in aquaculture. The pathogenesis of E. tarda has been studied for a long time and some virulence factors have been identified. However, the fundamental pathogenic mechanism of E. tarda still remains to be discovered. More and more evidence shows that the use of sRNAs is among the strategies developed by bacteria to fine-tune gene expression. They are involved in many biological processes to regulate iron homeostasis [25–27], expression of outer membrane proteins [28, 29], quorum sensing [30, 31], and bacterial virulence [16, 17] through binding to their target mRNAs or proteins.
In this research, it is the first time to report the existence of small RNAs within the genome of E. tarda. In principle, four major computational methods were applied for the prediction of sRNA locations from bacterial genome sequences: (1) secondary structure and thermodynamic stability, (2) comparative genomics, (3) ‘Orphan’ transcriptional signals and (4) ab initio methods regardless of sequence or structure similarity [32]. Transcriptional signal-based sRNA prediction tools include sRNApredict [33], sRNAscanner [34], and sRNAfinder [35]. sRNAPredict depends on the promoter signals, transcription factor binding sites, rho-independent terminator signals predicted by TRANSTERMHP [36] and BLAST [37] outputs as predictive features of sRNAs. sRNApredict3 is recent version of the sRNApredict suite that is used in the efficient prediction of sRNAs, with a high level of specificity. Some researchers found that sRNAPredict provided the best performance by comprehensively considering multiple factors [38]. The main advantage with sRNAscanner is that it uses its own algorithm and the training PWM dataset to calculate the genomic locations of the promoter, transcription factor, and terminator signals. Moreover, the sensitivity and specificity profile of sRNAscanner was first evaluated through the Receiver Operator Characteristic (ROC) curves and confirmed its satisfactory performance [32]. In this research, we choose transcriptional signal-based sRNA prediction tools (sRNA predict3 and sRNA scanner) for in silico prediction.
Most of these tools are applied to locate the putative genomic sRNA locations followed by experimental validation of those transcripts. Then 10 sRNAs were validated by RNA sequencing and qPCR, of which 8 novel sRNAs were found. The other two sRNAs, ET_sRNA_1 and ET_sRNA_2, were homolous to tmRNA and GcvB, respectively. TmRNA (also known as 10Sa RNA or SsrA RNA) is a unique bi-functional RNA that acts as both a tRNA and an mRNA to enter stalled ribosomes and direct the addition of a peptide tag to the C terminus of nascent polypeptides. TmRNA is widely distributed among eubacteria and has also been found in some chloroplasts [39]. The sRNA GcvB was first described in E. coli as being transcribed from a promoter that is divergent from that encoding gcvA, which is a transcriptional regulator of the glycine-cleavage-system operon [40–43].
What's more, the cellular abundance of 10 validated sRNA was detected by qPCR at different growth phases to monitor their biosynthesis. ET_sRNA_10 was almost expressed all the time and reached the highest peak at 48 h, which indicated that ET_sRNA_10 was probably house-keeping sRNA. But the expression of the other nine sRNAs was growth phase-dependent and they were expressed in the late-stage of exponential growth and stationary stages of growth. It had been reported that the expression of some sRNAs in gram positive and negative pathogens was growth phase-dependent. The expression of 11 candidate sRNAs was characterized in Staphylococcus aureus strains under different experimental conditions, many of which accumulated in the late-exponential phase of growth [44]. The characteristics of 11 sRNAs were studied in Enterococcus faecalis V583, six of which were specifically expressed at exponential phase, two of which were observed at stationary phase, and three of which were detected during both phases [45]. The expression of twenty-four sRNAs was also phase- and media- dependent in Streptococcus pyogenes M49 [46]. In Clostridium difficile, the expression of six sRNAs was growth phase-dependent, three of which (RCd4, RCd5 and SQ1002) were induced at the onset of stationary phase, whereas three of which (RCd2, RCd6 and SQ1498) was high during exponential phase and decreased at the onset of stationary phase [47]. Among the twelve non-coding RNAs found in Listeria monocytogenes, two of these non-coding RNAs were expressed in a growth-dependent manner [48]. In Brucella melitensis, three validated sRNAs were significantly induced in the stationary phase [49]. In this research, nine sRNAs show growth phase-dependent expression profile. In addition, it has been reported that the expression of some virulence determinants and associated factors in E. tarda is also growth phase-dependent [50–52]. So, we speculate that some of growth phase-regulated E. tarda sRNAs may be involved in the control, as previously observed in some gram-positive and gram-negative bacteria [53–55].
Despite the abundance of sRNAs in all bacterial lineages, little is known about their function and mechanism of action within the bacterial genomes and only a few sRNAs have been assigned with functions till date [56]. Using TargetRNA2, we have predicted the target mRNAs of 10 sRNAs.
Functional categorization of the target genes regulated by sRNAs resulted in identification of genes involved in key pathways of cell division, cell wall, transport, virulence,type III secretion system, type VI secretion system, ribosomal protein, and metabolism. A majority of these pathways are critical for the growth and survival of E. tarda in the host cytoplasm. A significant number (29.87%) of predicted target genes were categorized as ‘hypothetical protein’, which is not surprising considering that nearly 30.89% of E. tarda EIB202 genes are still reported as hypothetical proteins.
Of course, the related work is being verified by experiments. The mutant strains E. tarda S08⊿SsrA, E. tarda S08⊿Gcv and E. tarda S08⊿ET_sRNA_10 have been constructed. The next step is going to verify in vivo regulation functions of sRNAs. Once the regulation function of virulence is further confirmed, the unique nature of sRNAs that can be exploited for the development of novel diagnostic tools and therapeutic interventions will maybe come true in the future [57].
Conclusion
This report presents the study of small non-coding RNAs on E. tarda for the first time. Ten sRNAs were validated by RNA sequencing and qPCR. ET_sRNA_1 and ET_sRNA_2 were homolous to tmRNA and GcvB, respectively. However, the other candidate sRNAs have not been reported till now. ET_sRNA_10 was almost expressed all the time and reached the highest peak at 48 h. However, the other nine sRNAs were expressed in the late-stage of exponential growth and stationary stages of growth (36~60 h), which showed that their expression was growth phase-dependent. And they probably played regulatory roles during the biological process. The targets of 10 sRNAs were also predicted by TargetRNA2. Each sRNA targets contain some genes that directly or indirectly relate to virulence. These results preliminary showed that sRNAs probably play a regulatory role of virulence in E. tarda. The related work is being verified by experiments.
Supporting information
The region in yellow and green shows start (5’) and stop (3’) codons respectively. 5’ and 3’ start and ending sites respectively are as predicted by SIPHT/ sRNAPredict3. The region in red shows Rho-independent terminators. The qPCR primer sites are shown in blue.
(PDF)
(GZ)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Acknowledgments
The project was supported by The National Natural Science Foundation of China (41306165), The Natural Science Foundation of Jiangsu Province (BK20140442), A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, China Postdoctoral Foundation Funding (2011M501169) and the fifth-batch “521 High-Level Talent Education Project of Lianyungang”.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The project was supported by The National Natural Science Foundation of China (41306165), The Natural Science Foundation of Jiangsu Province (BK20140442), A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, China Postdoctoral Foundation Funding (2011M501169) and the fifth-batch “521 High-Level Talent Education Project of Lianyungang”.
References
- 1.Tan Y, Zheng J, Tung S, Rosenshine I, Leung K. Role of type III secretion in Edwardsiella tarda virulence. Microbiology. 2005; 151: 2301–2313. 10.1099/mic.0.28005-0 [DOI] [PubMed] [Google Scholar]
- 2.Zheng J, Leung K. Dissection of a type VI secretion system in Edwardsiella tarda. Molecular Microbiology. 2007; 66: 1192–1206. 10.1111/j.1365-2958.2007.05993.x [DOI] [PubMed] [Google Scholar]
- 3.Xiao J, Wang Q, Liu Q, Xu L, Wang X, Wu H, et al. Characterization of Edwardsiella tarda rpoS: effect on serum resistance, chondroitinase activity, biofilm formation, and autoinducer synthetases expression. Applied Microbiology and Biotechnology. 2009; 83(1): 151–160 10.1007/s00253-009-1924-9 [DOI] [PubMed] [Google Scholar]
- 4.Wang L, Xiao J, Cui S, Wang Q, Wu H, Liu Q, et al. HU-induced polymorphous filamentation in fish pathogen Edwardsiella tarda leading to reduced invasion and virulence in zebrafish. Veterinary Microbiology. 2014; 171: 165–174. 10.1016/j.vetmic.2014.03.030 [DOI] [PubMed] [Google Scholar]
- 5.Rao P, Yamada Y, Leung K. A major catalase (KatB) that is required for resistance to H2O2 and phagocyte-mediated killing in Edwardsiella tarda. Microbiology. 2003; 149: 2635–2644. 10.1099/mic.0.26478-0 [DOI] [PubMed] [Google Scholar]
- 6.Gao D, Cheng J, Zheng E, Li Y, Shao Z, Xu Z, et al. Eha, a transcriptional regulator of hemolytic activity of Edwardsiella tarda. FEMS Microbiology Letters. 2014; 353: 132–140. 10.1111/1574-6968.12420 [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Wang Q, Xiao J, Liu Q, Wu H, Zhang Y. Hemolysin EthA in Edwardsiella tarda is essential for fish invasion in vivo and in vitro and regulated by two-component system EsrA-EsrB and nucleoid protein HhaEt. Fish & Shellfish Immunology. 2010; 29: 1082–1091. [DOI] [PubMed] [Google Scholar]
- 8.Xie H, Lu J, Rolhion N, Holden D, Nie P, Zhou Y, et al. Edwardsiella tarda-induced cytotoxicity depends on its type three secretion system and flagellin. Infection and Immunity. 2014; 82: 3436–3445. 10.1128/IAI.01065-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu T, Su Y, Xu Y, He Y, Wang B, Dong X, et al. Mutations of flagellar genes fliC12, fliA and flhDC of Edwardsiella tarda attenuated bacterial motility, biofilm formation and virulence to fish. Journal of Applied Microbiology. 2014; 116: 236–244. 10.1111/jam.12357 [DOI] [PubMed] [Google Scholar]
- 10.Sakai T, Matsuyama T, Sano M, Iida T. Identification of novel putative virulence factors, adhesin AIDA and type VI secretion system, in atypical strains of fish pathogenic Edwardsiella tarda by genomic subtractive hybridization. Microbiology and Immunology. 2009; 53: 131–139. 10.1111/j.1348-0421.2009.00108.x [DOI] [PubMed] [Google Scholar]
- 11.Liu E, Ye J, Song SS, Wang K, Zhang Y, Zhang H. Impact of co-deficiency of RpoN and RpoS on stress tolerance, virulence and gene regulation in Edwardsiella tarda. Journal of Basic Microbiology. 2014; 54: 678–687. 10.1002/jobm.201300622 [DOI] [PubMed] [Google Scholar]
- 12.Morohoshi T, Inaba T, Kato N, Kanai K, Ikeda T. Identification of quorum-sensing signal molecules and the LuxRI homologs in fish pathogen Edwardsiella tarda. Journal of Bioscience and Bioengineering. 2004; 98: 274–281. 10.1016/S1389-1723(04)00281-6 [DOI] [PubMed] [Google Scholar]
- 13.Zhang M, Jiao X, Hu Y, Sun L. Attenuation of Edwardsiella tarda virulence by small peptides that interfere with LuxS/autoinducer type 2 quorum sensing. Applied and Environmental Microbiology. 2009; 75: 3882–3890. 10.1128/AEM.02690-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Current Opinion in Microbiology. 2010; 13: 24–33. 10.1016/j.mib.2010.01.001 [DOI] [PubMed] [Google Scholar]
- 15.Papenfort K, Vogel J. Regulatory RNA in bacterial pathogens. Cell Host & Microbe. 2010; 8: 116–127. [DOI] [PubMed] [Google Scholar]
- 16.Romby P, Vandenesch F, Wagner E. The role of RNAs in the regulation of virulence-gene expression. Current Opinion in Microbiology. 2006; 9: 229–236. 10.1016/j.mib.2006.02.005 [DOI] [PubMed] [Google Scholar]
- 17.Toledo-Arana A, Repoila F, Cossart P. Small noncoding RNAs controlling pathogenesis. Current Opinion in Microbiology. 2007; 10: 182–188. 10.1016/j.mib.2007.03.004 [DOI] [PubMed] [Google Scholar]
- 18.Abu-Qatouseh L, Chinni S, Seggewiss J, Proctor R, Brosius J, Rozhdestvensky T, et al. Identification of differentially expressed small non-protein-coding RNAs in Staphylococcus aureus displaying both the normal and the small-colony variant phenotype. Journal of Molecular Medicine. 2010; 88: 565–575. 10.1007/s00109-010-0597-2 [DOI] [PubMed] [Google Scholar]
- 19.Acebo P, Martin-Galiano A, Navarro S, Zaballos A, Amblar M. Identification of 88 regulatory small RNAs in the TIGR4 strain of the human pathogen Streptococcus pneumoniae. RNA. 2012; 18: 530–546. 10.1261/rna.027359.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altuvia S. Identification of bacterial small non-coding RNAs: experimental approaches. Current Opinion in Microbiology. 2007; 10: 257–261. 10.1016/j.mib.2007.05.003 [DOI] [PubMed] [Google Scholar]
- 21.Pichon C, Felden B. Small RNA gene identification and mRNA target predictions in bacteria. Bioinformatics. 2008; 24: 2807–2813. 10.1093/bioinformatics/btn560 [DOI] [PubMed] [Google Scholar]
- 22.Raabe C, Hoe C, Randau G, Brosius J, Tang T, Rozhdestvensky T. The rocks and shallows of deep RNA sequencing: Examples in the Vibrio cholera RNome. RNA. 2011; 17: 1357–1366. 10.1261/rna.2682311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods. 2001; 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 24.Schroeder CL, Narra HP, Rojas M, Sahni A, Patel J, Khanipov K, et al. Bacterial small RNAs in the Genus Rickettsia. BMC Genomics. 2015;16:1075 10.1186/s12864-015-2293-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massé E, Vanderpool C, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. Journal of Bacteriology. 2005; 187: 6962–6971. 10.1128/JB.187.20.6962-6971.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massé E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Current Opinion in Microbiology. 2007; 10: 140–145. 10.1016/j.mib.2007.03.013 [DOI] [PubMed] [Google Scholar]
- 27.Vecerek B, Moll I, Bläsi U. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO Journal. 2007; 26: 965–975. 10.1038/sj.emboj.7601553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Molecular Microbiology. 2006; 59: 231–247. 10.1111/j.1365-2958.2005.04929.x [DOI] [PubMed] [Google Scholar]
- 29.Valentin-Hansen P, Johansen J, Rasmussen A. Small RNAs controlling outer membrane porins. Current Opinion in Microbiology. 2007; 10: 152–155. 10.1016/j.mib.2007.03.001 [DOI] [PubMed] [Google Scholar]
- 30.Lenz D, Miller M, Zhu J, Kulkarni R, Bassler B. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Molecular Microbiology. 2005; 58: 1186–1202. 10.1111/j.1365-2958.2005.04902.x [DOI] [PubMed] [Google Scholar]
- 31.Tu K, Bassler B. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi.Genes & Development. 2007; 21: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papenfort K, Vanderpool C. Target activation by regulatory RNAs in bacteria. FEMS Microbiology Reviews. 2015; 39(3): 362–378. 10.1093/femsre/fuv016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livny J, Fogel M, Davis B, Waldor M. sRNAPredict: an integrative computational approach to identify sRNAs in bacterial genomes. Nucleic Acids Research. 2005; 33: 4096–4105. 10.1093/nar/gki715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sridhar J, Sambaturu N, Sabarinathan R, Ou H, Deng Z, Sekar K, et al. sRNAscanner: A computational tool for intergenic small RNA detection in bacterial genomes. PLoS ONE. 2010; 5: e11970 10.1371/journal.pone.0011970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tjaden B. Prediction of small, noncoding RNAs in bacteria using heterogenous data. Journal of Mathematical Biology. 2008; 56: 183–200. 10.1007/s00285-007-0079-5 [DOI] [PubMed] [Google Scholar]
- 36.Kingsford C, Ayanbule K, Salzberg S. Rapid, accurate, computational discovery of Rho-independent transcriptional terminators illuminates their relationship to DNA uptake. Genome Biology. 2007; 8: R22 10.1186/gb-2007-8-2-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. Journal of Molecular Biology. 1990; 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 38.Li W, Ying X, Lu Q, Chen L. Predicting sRNAs and their targets in bacteria. Genomics Proteomics Bioinformatics. 2012; 10(5): 276–284. 10.1016/j.gpb.2012.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams K. The tmRNA website. Nucleic Acids Research. 2000; 28: 168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Busi F, Le Derout J, Cerciat M, Régnier P, Hajnsdorf E. Is the secondary putative RNAeRNA interaction site relevant to GcvB mediated regulation of oppA mRNA in Escherichia coli. Biochimie. 2010; 92: 1458–1461. 10.1016/j.biochi.2010.06.020 [DOI] [PubMed] [Google Scholar]
- 41.Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Research in Microbiology. 2009; 160: 278–287. 10.1016/j.resmic.2009.03.004 [DOI] [PubMed] [Google Scholar]
- 42.Pulvermacher S, Stauffer L, Stauffer G. Role of the sRNA GcvB in regulation of cycA in Escherichia coli. Microbiology. 2009; 155: 106–114. 10.1099/mic.0.023598-0 [DOI] [PubMed] [Google Scholar]
- 43.Stauffer LT, Stauffer GV. The Escherichia coli GcvB sRNA uses genetic redundancy to control cycA expression. ISRN Microbiology. 2012; 2012: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geissmann T, Chevalier C, Cros M, Boisset S, Fechter P, Noirot C, et al. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Research. 2009; 37: 7239–7257. 10.1093/nar/gkp668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shioya K, Michaux C, Kuenne C, Hain T, Verneuil N, Budin-Verneuil A, et al. Genome-wide identification of small RNAs in the opportunistic pathogen Enterococcus faecalis V583. PLoS ONE. 2011; 6: e23948 10.1371/journal.pone.0023948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patenge N, Billion A, Raasch P, Normann J, Wisniewska-Kucper A, Retey J, et al. Identification of novel growth phase- and media-dependent small non-coding RNAs in Streptococcus pyogenes M49 using intergenic tiling arrays. BMC Genomics. 2012; 13: 550 10.1186/1471-2164-13-550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soutourina O, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, et al. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genetics. 2013; 9: e1003493 10.1371/journal.pgen.1003493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Research. 2007; 35: 962–974. 10.1093/nar/gkl1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Ke Y, Xu J, Wang L, Wang T, Liang H, et al. Identification of a novel small non-coding RNA modulating the intracellular survival of Brucella melitensis. Frontiers in Microbiology. 2015; 6: 164 10.3389/fmicb.2015.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang M, Sun K, Sun L. Regulation of autoinducer 2 production and luxS expression in a pathogenic Edwardsiella tarda strain. Microbiology. 2008; 154: 2060–2069. 10.1099/mic.0.2008/017343-0 [DOI] [PubMed] [Google Scholar]
- 51.Sun Y, Zheng W, Hu Y, Sun B, Sun L. Edwardsiella tarda Eta1, an in vivo-induced antigen that is involved in host infection. Infection and Immunology. 2012; 80: 2948–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng W, Hu Y, Sun L. The two Dps of Edwardsiella tarda are involved in resistance against oxidative stress and host infection. Fish & Shellfish Immunology. 2011, 31: 985–992. [DOI] [PubMed] [Google Scholar]
- 53.Romby P, Charpentier E. An overview of RNAs with regulatory functions in Gram-positive bacteria. Cellular and Molecular Life Sciences. 2010; 67: 217–237. 10.1007/s00018-009-0162-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pichon C, du Merle L, Caliot M, Trieu-Cuot P, Le Bouguenec C. An in silico model for identification of small RNAs in whole bacterial genomes: characterization of antisense RNAs in pathogenic Escherichia coli and Streptococcus agalactiae strains. Nucleic Acids Research. 2012; 40: 2846–2861. 10.1093/nar/gkr1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sahr T, Rusniok C, Dervins-Ravault D, Sismeiro O, Coppee J, et al. Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biology. 2012; 9: 503–519. 10.4161/rna.20270 [DOI] [PubMed] [Google Scholar]
- 56.Warrier I, Hicks LD, Battisti JM, Raghavan R, Minnick MF.Identification of novel small RNAs and characterization of the 6S RNA of Coxiella burnetii.PLoS One. 2014; 9(6):e100147 10.1371/journal.pone.0100147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mraheil M, Billion A, Kuenne C, Pischimarov J, Kreikemeyer B, Engelmann S, et al. Comparative genome-wide analysis of small RNAs of major Gram-positive pathogens: from identification to application. Microbial Biotechnology. 2010; 3: 658–676. 10.1111/j.1751-7915.2010.00171.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The region in yellow and green shows start (5’) and stop (3’) codons respectively. 5’ and 3’ start and ending sites respectively are as predicted by SIPHT/ sRNAPredict3. The region in red shows Rho-independent terminators. The qPCR primer sites are shown in blue.
(PDF)
(GZ)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Statistical significance (*P≤0.05;**P≤0.01) was obtained using Anova test.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.